Abstract
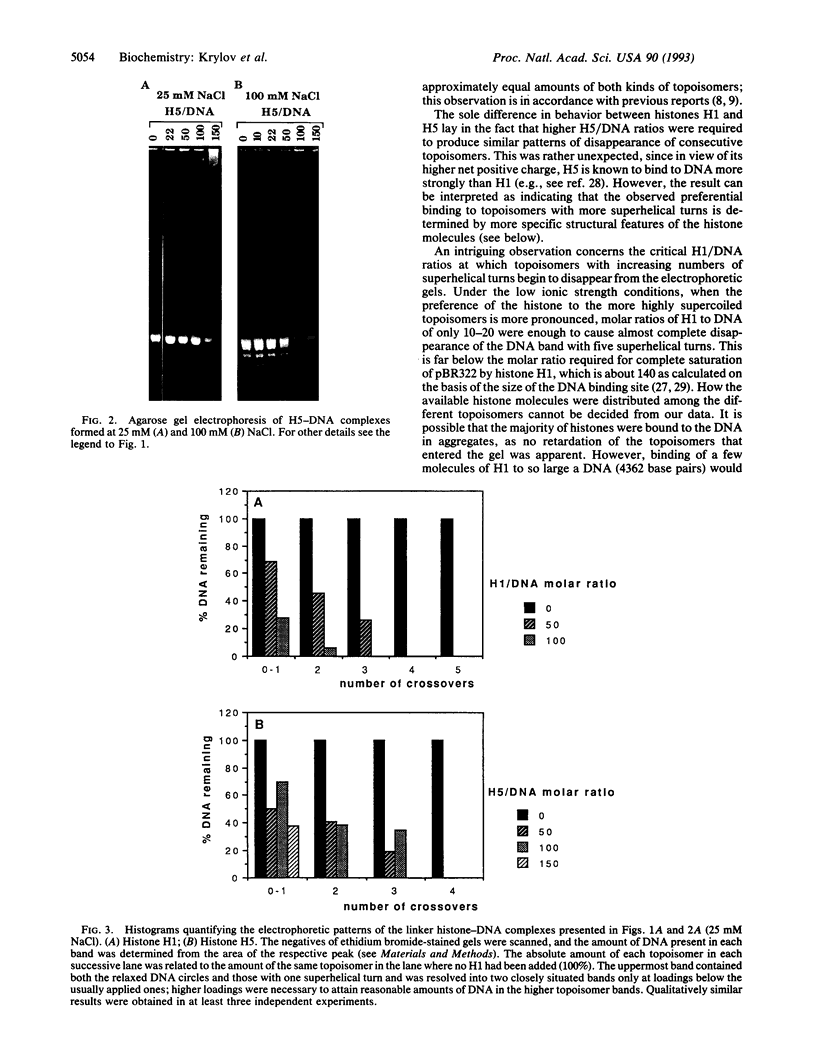
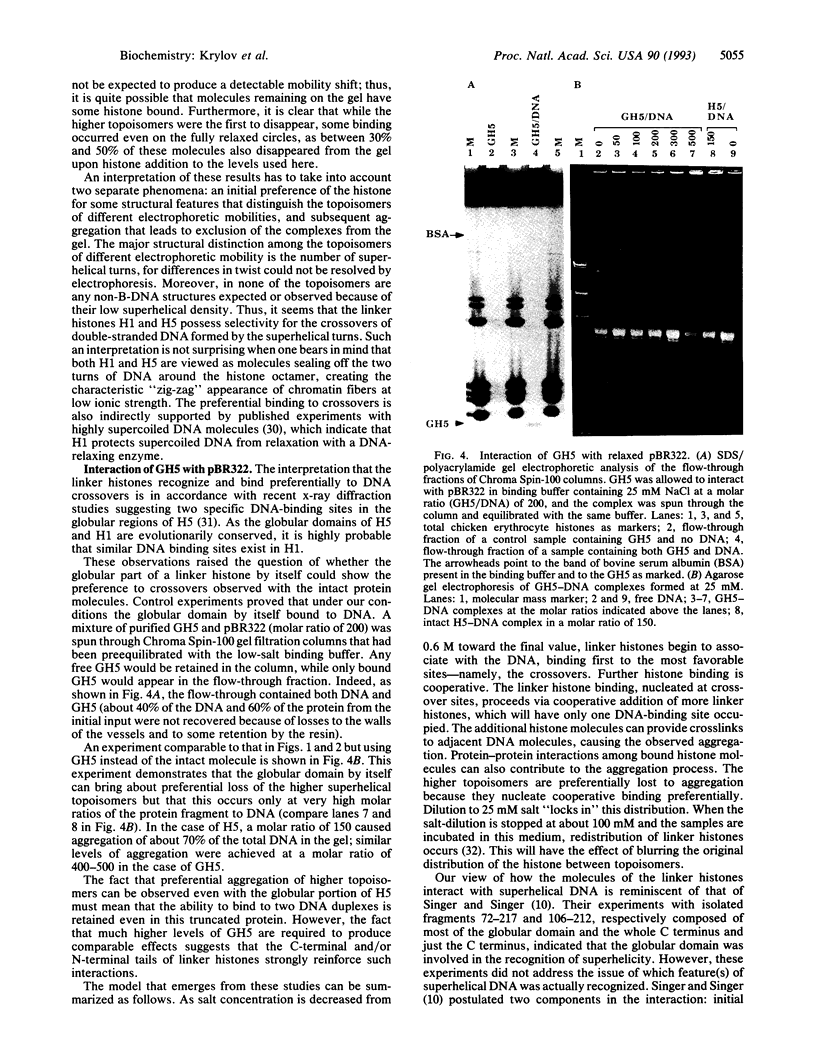
The interaction of the linker histones H1 and H5 from chicken erythrocyte chromatin with pBR322 was studied as a function of the number of superhelical turns in circular plasmid molecules. Supercoiled plasmid DNA was relaxed with topoisomerase I so that a population with a narrow distribution of topoisomers, containing from zero to five superhelical turns, was obtained. None of the topoisomers contained alternative non-B-DNA structures. Histone-DNA complexes formed at either 25 or 100 mM NaCl final concentration and at histone-DNA molar ratios ranging from 10 to 150 were analyzed by agarose gel electrophoresis. The patterns of disappearance of individual topoisomer bands from the gel were interpreted as an indication of preference of the linker histones for crossovers of double-helical DNA. This preference was observed at both salt concentrations, being more pronounced under conditions of low ionic strength. Isolated H5 globular domain also caused selective disappearance of topoisomers from the gel, but it did so only at very high peptide-DNA molar ratios. The observed preference of the linker histones for crossovers of double-helical DNA is viewed as a part of the mechanism involved in the sealing of the two turns of DNA around the histone octamer.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allan J., Hartman P. G., Crane-Robinson C., Aviles F. X. The structure of histone H1 and its location in chromatin. Nature. 1980 Dec 25;288(5792):675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- Banchev T., Srebreva L., Zlatanova J. Purification of histone H10 and its subfractions under non-denaturing conditions. Biochim Biophys Acta. 1991 Jan 23;1073(1):230–232. doi: 10.1016/0304-4165(91)90208-x. [DOI] [PubMed] [Google Scholar]
- Bina-Stein M., Singer M. F. The effect of H1 histone on the action of DNA-relaxing enzyme. Nucleic Acids Res. 1977 Jan;4(1):117–127. doi: 10.1093/nar/4.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina-Stein M., Vogel T., Singer D. S., Singer M. F. H5 Histone and DNA-relaxing enzyme of chicken erythrocytes. Interaction with superhelical DNA. J Biol Chem. 1976 Dec 10;251(23):7363–7366. [PubMed] [Google Scholar]
- Carnevali F., Caserta M., Di Mauro E. Transitions in topological organization of supercoiled DNA domains as a potential regulatory mechanism. J Biol Chem. 1984 Oct 25;259(20):12633–12643. [PubMed] [Google Scholar]
- Caron F., Thomas J. O. Exchange of histone H1 between segments of chromatin. J Mol Biol. 1981 Mar 15;146(4):513–537. doi: 10.1016/0022-2836(81)90045-0. [DOI] [PubMed] [Google Scholar]
- Clark D. J., Thomas J. O. Differences in the binding of H1 variants to DNA. Cooperativity and linker-length related distribution. Eur J Biochem. 1988 Dec 1;178(1):225–233. doi: 10.1111/j.1432-1033.1988.tb14447.x. [DOI] [PubMed] [Google Scholar]
- Clark D. J., Thomas J. O. Salt-dependent co-operative interaction of histone H1 with linear DNA. J Mol Biol. 1986 Feb 20;187(4):569–580. doi: 10.1016/0022-2836(86)90335-9. [DOI] [PubMed] [Google Scholar]
- Garcia-Ramirez M., Leuba S. H., Ausio J. One-step fractionation method for isolating H1 histones from chromatin under nondenaturing conditions. Protein Expr Purif. 1990 Sep;1(1):40–44. doi: 10.1016/1046-5928(90)90043-x. [DOI] [PubMed] [Google Scholar]
- Higurashi M., Cole R. D. The combination of DNA methylation and H1 histone binding inhibits the action of a restriction nuclease on plasmid DNA. J Biol Chem. 1991 May 5;266(13):8619–8625. [PubMed] [Google Scholar]
- Iovcheva C., Dessev G. N. Interaction between lysine-rich histones and DNA. Mol Biol Rep. 1980 Mar 31;6(1):21–25. doi: 10.1007/BF00775749. [DOI] [PubMed] [Google Scholar]
- Kowalski D., Natale D. A., Eddy M. J. Stable DNA unwinding, not "breathing," accounts for single-strand-specific nuclease hypersensitivity of specific A+T-rich sequences. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9464–9468. doi: 10.1073/pnas.85.24.9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Miki B. L., Neelin J. M. The histones of rainbow trout erythrocytes include an erythrocyte-specific histone. Can J Biochem. 1975 Nov;53(11):1158–1169. doi: 10.1139/o75-161. [DOI] [PubMed] [Google Scholar]
- NEELIN J. M., CALLAHAN P. X., LAMB D. C., MURRAY K. THE HISTONES OF CHICKEN ERYTHROCYTE NUCLEI. Can J Biochem. 1964 Dec;42:1743–1752. doi: 10.1139/o64-185. [DOI] [PubMed] [Google Scholar]
- Palecek E. Local supercoil-stabilized DNA structures. Crit Rev Biochem Mol Biol. 1991;26(2):151–226. doi: 10.3109/10409239109081126. [DOI] [PubMed] [Google Scholar]
- Singer D. S., Singer M. F. Studies on the interaction of H1 histone with superhelical DNA: characterization of the recognition and binding regions of H1 histones. Nucleic Acids Res. 1976 Oct;3(10):2531–2547. doi: 10.1093/nar/3.10.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Rees C., Finch J. T. Cooperative binding of the globular domains of histones H1 and H5 to DNA. Nucleic Acids Res. 1992 Jan 25;20(2):187–194. doi: 10.1093/nar/20.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasmel H. Influence of supercoiling on DNA structure: laser Raman spectroscopy of the plasmid pBR322. Biopolymers. 1985 Jun;24(6):1001–1008. doi: 10.1002/bip.360240607. [DOI] [PubMed] [Google Scholar]
- Vogel T., Singer M. F. Interaction of f1 histone with superhelical DNA. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2597–2600. doi: 10.1073/pnas.72.7.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel T., Singer M. F. The effect of superhelicity on the interaction of histone f1 with closed circular duplex DNA. J Biol Chem. 1976 Apr 25;251(8):2334–2338. [PubMed] [Google Scholar]
- Weintraub H. Assembly and propagation of repressed and depressed chromosomal states. Cell. 1985 Oct;42(3):705–711. doi: 10.1016/0092-8674(85)90267-3. [DOI] [PubMed] [Google Scholar]
- Yaneva J., Zlatanova J. Histone H1 interacts specifically with certain regions of the mouse alpha-globin gene. DNA Cell Biol. 1992 Mar;11(2):91–99. doi: 10.1089/dna.1992.11.91. [DOI] [PubMed] [Google Scholar]
- Yaneva J., Zlatanova J., Paneva E., Srebreva L., Tsanev R. Interaction of histones H1 and H1(0) with superhelical and linear DNA. FEBS Lett. 1990 Apr 24;263(2):225–228. doi: 10.1016/0014-5793(90)81379-3. [DOI] [PubMed] [Google Scholar]
- Zlatanova J. Histone H1 and the regulation of transcription of eukaryotic genes. Trends Biochem Sci. 1990 Jul;15(7):273–276. doi: 10.1016/0968-0004(90)90053-e. [DOI] [PubMed] [Google Scholar]
- Zlatanova J., Van Holde K. Histone H1 and transcription: still an enigma? J Cell Sci. 1992 Dec;103(Pt 4):889–895. doi: 10.1242/jcs.103.4.889. [DOI] [PubMed] [Google Scholar]
- Zlatanova J., Yaneva J. Histone H1-DNA interactions and their relation to chromatin structure and function. DNA Cell Biol. 1991 May;10(4):239–248. doi: 10.1089/dna.1991.10.239. [DOI] [PubMed] [Google Scholar]