Abstract
The NBN gene, also known as NBS1, is located on the chromosome band 8q21.3, and encodes a 754-amino acid-long protein named nibrin. This protein is a member of the MRE1-RAD50-NBN nuclear complex, and is involved in numerous cell processes essential for maintaining genomic stability. Heterozygous variants in the NBN gene, including p.I171V, c.657del5 and p.R215W, have been described as risk factors for the development of several malignancies. However, there is no report regarding the association of these mutations with lung cancer thus far. Therefore, the present study aimed to evaluate whether there is an association between the heterozygous p.I171V, c.657del5 and p.R215W variants of the NBN gene and the risk of developing lung cancer. The frequency of these variants was estimated in a group of 453 adults diagnosed with non-small cell lung cancer (NSCLC) and in healthy controls (2,400 for p.I171V, 2,090 for c.657del5 and 498 for p.R215W). The p.I171V variant was assessed by restriction fragment length polymorphism analysis of polymerase chain reaction (PCR) products, using MunI (MfeI) restriction enzyme, whereas the c.657del5 and p.R215W variants were assessed by the PCR single-strand conformation polymorphism method. A significantly increased risk of developing lung cancer was observed for the p.I171V variant, which was present in 17 (3.75%) of the 453 cases of lung cancer and in 12 (0.5%) of the 2,400 healthy individuals (odds ratio, 7.759; P<0.0001). The results obtained indicated an association between the p.I171V mutation and the development of lung cancer. Therefore, this variant may be considered a risk factor for NSCLC. Prospective studies with larger groups of patients may reveal the potential impact of the p.I171V variant in the occurrence of lung cancer.
Keywords: NBN gene, nibrin, p.I171V, lung cancer risk
Introduction
The NBN gene, also known as NBS1, is located on the chromosome band 8q21.3, and encodes a 754-amino acids-long protein termed nibrin (1). This protein is a member of the MRE1-RAD50-NBN (MRN) nuclear complex, and is involved in numerous cell processes that are essential for maintaining genomic stability, including the detection and reparation of DNA double-strand breaks (DSBs), cell cycle checkpoint control, meiosis and induction of apoptosis (2–4). Therefore, any mutations disrupting its functionality may lead to genomic instability and promote tumorigenesis. Homozygous mutations in the NBN gene, resulting in the production of an abnormally short version of the nibrin protein, lead to Nijmegen breakage syndrome (NBS) and increased susceptibility to tumorigenesis (5–7). Heterozygous variants in this gene, including p.I171V (c.551A>G), c.657delACAAA and p.R215W (c.643C>T), have been described as risk factors for several malignancies, including leukemia, melanoma, breast, ovarian, prostate, larynx and colorectal cancer (8–12). To the best of our knowledge, there is no report regarding the association of the aforementioned mutations with lung cancer. Therefore, the present study aimed to evaluate whether there is an association between the heterozygous p.I171V, c.657del5 and p.R215W variants of the NBN gene and the risk of developing lung cancer.
Materials and methods
The frequency of constitutional mutations in the fifth and sixth exons of the NBN gene was estimated in a group of 453 adults (139 females and 314 males) diagnosed with lung cancer, and in healthy controls (2,400 for p.I171V, 2,090 for c.657del5 and 498 for p.R215W). The cohort of 453 cases of lung cancer were recruited in the Department of Thoracic Surgery of the University of Medical Sciences in Poznań (Poznań, Poland). All the subjects enrolled in the study were of Caucasian ancestry from the Wielkopolska region of Poland. The majority of the cases were males (69%), with a median age of 63.5 years. The diagnosis of non-small cell lung cancer (NSCLC) was confirmed by histopathological examination. Anonymous blood samples, collected on Guthrie cards (Whatman 903®; Whatman Inc., Dassel, Germany) drawn from the newborn screening program of the Wielkopolska region, were used as controls. The results for the control samples were obtained in previous studies (10,13,14). All the patients signed an informed consent form approved by the Ethics Committee of the University of Medical Sciences in Poznań (approval no. 802/10).
DNA was extracted from blood using the columns provided in the QIAamp DNA Blood Mini Kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's instructions, and from Guthrie cards, as described previously (13,15). The mutation in the fifth exon of the NBN gene (p.I171V) was assessed by restriction fragment length polymorphism (RFLP) analysis of polymerase chain reaction (PCR) products, using MunI (MfeI) restriction enzyme (Fermentas, Thermo Fisher Scientific, Inc., Pittsburgh, PA, USA). PCR reactions were performed in a total volume of 25 µl containing 2.5 µl of 10X PCR buffer with 15 mM MgCl2 (Sigma-Aldrich, St. Louis, MO, USA), 6 pM of each primer, 2 mM of each deoxynucleotide triphospahate (dNTP; Sigma-Aldrich, Steinheim, Germany), 1.5 units of Taq DNA polymerase (Sigma-Aldrich, St. Louis, MO, USA) and 1.5 µl of DNA template. Amplification conditions involved initial denaturation for 5 min at 95°C, followed by 35 cycles of 25 sec at 95°C, 35 sec at 54°C and 55 sec at 72°C, with a 10-min final extension at 72°C. Next, 10 µl of the PCR products were digested for 3 h at 37°C, and analyzed by electrophoresis on a 2% agarose gel, in the presence of ethidium bromide. The endonuclease MunI recognizes and cleaves the sequence CA*ATTG, which occurs only once in each of the analyzed fragments. The mutations in the sixth exon of the NBN gene (c.657del5 and p.R215W) were assessed by the PCR single-strand conformation polymorphism (SSCP) method. PCR reactions were performed in a total volume of 25 µl containing 2.5 µl of 10X PCR buffer with 15 mM MgCl2, 6 pM of each primer, 2 mM of each dNTP, 1.5 units of Taq DNA polymerase and 1.5 µl of DNA template. Amplification conditions involved initial denaturation for 3 min at 95°C, followed by 5 cycles of 25 sec at 95°C, 25 sec at 50°C and 55 sec at 72°C; then 30 cycles of 25 sec at 94°C, 35 sec at 48°C, 45 sec at 72°C, with a 6-min final extension at 72°C. Next, 4 µl of the PCR products were mixed with 9 µl of the loading buffer, and denatured for 5 min at 95°C, cooled and separated on 7% non-denaturing polyacrylamide gel for 40 h 4°C. Those samples exhibiting positive results were subsequently sequenced using the Sanger method. The conditions of PCR before sequencing were the same as those before RFLP analysis for p.I171V and SSCP analysis for c.657-661del and p.R214W. PCR products were cleaned up using NucleoSpin® Gel and PCR Clean-up kit (Macherey-Nagel, Inc., Düren, Germany) according to the manufacturer's protocol. The sequencing reaction protocol included 45 cycles of 10 sec at 94°C, 5 sec at 52°C and 3 min at 60°C. The sequences of all the primers used for PCR analysis in the present study are provided in Table I.
Table I.
Characteristics of the primers used for PCR.
| Variant | Exon | Primer sequence | Product size (bp) | Tb (°C) |
|---|---|---|---|---|
| p.I171V | F: 5′-TTATGGATGTAAACAGCCTC-3′ | |||
| c.511A>G | 5 | R: 5′-TACCGAACTATAACACAGCA-3′ | 328 | 54 |
| c.657-661del | F: 5′-CAGATAGTCACTCGGTTTACAA-3′ | |||
| c.657_651delACAAA | 6 | R: 5′-TTCTTTAGGAAAATTTAGCT-3′ | 228 | 50 |
| p.R215W | F: 5′-CAGATAGTCACTCGGTTTACAA-3′ | |||
| c.643C>T | 6 | R: 5′-ACAACTACTGATAAGAGTTA-3′ | 273 | 50 |
PCR, polymerase chain reaction; F, forward; R, reverse; bp, base pairs; Tb, binding temperature.
Statistical analysis
The allele frequencies among the groups were compared using the χ2 and Fisher's exact tests, using GraphPad software version 5.03 (GraphPad Software Inc., La Jolla, CA, USA). The odds ratios (ORs) for the relative risk conferred by a particular variant were estimated with 95% confidence intervals (CI). P<0.05 was considered to indicate a statistically significant difference.
Results
The p.I171V variant in the fifth exon and the c.657del5 and p.R215W variants in the sixth exon of the NBN gene were analyzed in adult patients diagnosed with lung cancer (N=453) and in healthy controls (N=2,400 for p.I171V, N=2,090 for c.657del5 and N=498 for p.R215W).
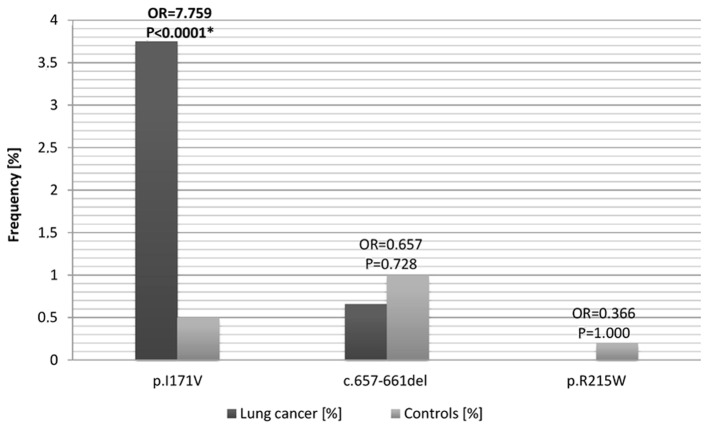
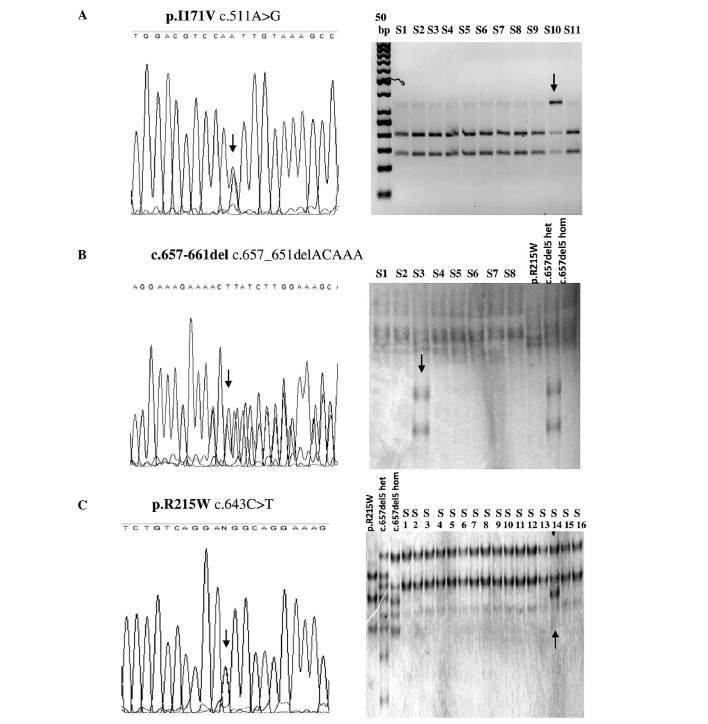
The p.I171V (c.551A>G) variant was identified in 17 of the 453 cases of lung cancer and in 12 of the 2,400 healthy individuals (Table II and Fig. 1). These results indicate a significantly higher incidence of the p.I171V variant in patients with lung cancer, compared to healthy controls (OR=7.759, P<0.0001). Of the 17 patients with cancer exhibiting this variant, 2 were females (2/139) and 15 were males (15/314). The heterozygous c.657del5 mutation was identified in 3 of the 453 patients with lung cancer (OR=0.657, P=0.728) and in 5 of the 2,090 controls (Table II and Fig. 1). The p.R215W (c.634C>T) mutation was identified in 1 case from the control group, and was absent in the group of patients with lung cancer (Table II). The results of the RFLP, SSCP and sequencing analyses conducted on the relevant samples are presented in Fig. 2.
Table II.
Frequency of the NBN variants and results of the logistic regression analysis performed in patients diagnosed with lung cancer versus healthy controls.
| Variant | Patients with lung cancer (%) | Healthy controls (%) | OR (95% CI) | P-value |
|---|---|---|---|---|
| p.I171V | 17/453 (3.75) | 12/2,400 (0.50) | 7.759 (3.679–16.360) | <0.0001a |
| c.657-661del | 3/453 (0.66) | 21/2,090 (1.00) | 0.657 (0.195–2.212) | 0.728 |
| p.R215W | 0/453 (0.00) | 1/498 (0.20) | 0.366 (0.015–9.006) | 1.000 |
NBN, nibrin; OR, odds ratio; CI, confidence interval.
Statistically significant difference.
Figure 1.
Allele frequency and results of logistic regression analysis corresponding to the p.I171V, c.657del5 and p.R215W variants of the NBN gene in the group of patients with lung cancer (N=453) vs. controls (p.I171V, N=2,400; c.6578del5, N=2,090; p.R215W, N=498). P<0.05 was considered to indicate a statistically significant difference. OR, odds ratio; NBN, nibrin; *P<0.0001 vs control.
Figure 2.
(A) Sequencing results and RFLP analysis (using MunI restriction enzyme) corresponding to the p. I171V variant. Sequencing and SSCP analysis of the variants (B) c.657del5 and (C) p.R215W. The variants are indicated by an arrow. RFLP, restriction fragment length polymorphism; SSCP, single-strand conformation polymorphism; S, sample; p.R215W, reference sequence for heterozygous p.R215W substitution; c.657del5 het, reference sequence for heterozygous c.657del5 mutation; c.657del5 hom, reference sequence for homozygous c.657del5 mutation.
Discussion
Previous studies have reported an association between heterozygous variants in the NBN gene and susceptibility to certain malignancies (16), including acute lymphoblastic leukemia (ALL), melanoma, breast, ovarian, prostate, colorectal and larynx cancer (8–12). The present study aimed to investigate whether variants in the NBN gene are associated with elevated risk of lung cancer incidence in adults.
The p.I171V variant in the fifth exon and the c.657del5 and p.R215W variants in the sixth exon of the NBN gene were analyzed in 453 adult patients diagnosed with lung cancer, and in healthy controls (2,400, 2,090 and 498 for p.I171V, c.657del5 and p.R215W, respectively).
A significantly increased risk of developing lung cancer was observed for the p.I171V (c.551A>G) variant, which was present in 17 of the 453 cases of lung cancer and in 12 of the 2,400 healthy individuals. These results indicate a higher incidence of the p.I171V variant in patients with lung cancer (OR=7.759, P<0.0001), and suggest that p.I171V may be a risk allele for lung cancer. Of the 17 patients with cancer who exhibited this variant, 2 were females (2/139) and 15 were males (15/314). The pathogenicity of the p.I171V variant is presumably associated with its location in the highly conserved breast cancer 1 (BRCA1) carboxy-terminal (BRCT) domain of the NBN gene. This BRCT domain, together with the forkhead-associated domain, participates in the translocation of the MRN complex to the sites of DSBs (17). Cerosaletti and Concannon (18) demonstrated that mutations in these domains result in disorders in the formation of nuclear clusters and phosphorylation of nibrin following irradiation. The occurrence of the p.I171V variant in the NBN gene has been previously linked with elevated risk of several types of cancer. A significantly higher frequency of the p.I171V variant was observed in ALL, breast, larynx and colorectal cancer, and in multiple primary tumors of the head and neck (9,13,15). However, such an association was not observed for solid malignancies in children (11). By contrast, Ciara et al (19) reported an association between this variant and childhood medulloblastoma. Nonetheless, the in vitro studies conducted on cells derived from heterozygous carriers of the p.I171V variant have demonstrated that, per se, this variant does not play a crucial role in tumorigenesis (20).
In the present study, the occurrence of the c.657del5 mutation in the sixth exon of the NBN gene was also analyzed. The heterozygous variant of this mutation was identified in 3 of 453 patients with lung cancer and in 5 of 2,090 controls. The occurrence of the c.657del5 variant, known as the Slavic mutation, results in the production of two truncated fragments of the nibrin protein (20). Thus, homozygous c.657del5 has a deleterious impact on nibrin function and leads to NBS, while the heterozygous variant has been associated with susceptibility to tumorigenesis (21). A higher frequency of occurrence of the heterozygous c.657del5 mutation has been observed in patients with melanoma and non-Hodgkin lymphoma (22). However, other studies have reported that c.657del5 did not occur more frequently in breast (9,22), colorectal (22) and larynx cancer (13), which suggests that this variant is not a risk factor for these malignancies. Similarly, in the present study, no association between the occurrence of the c.657del5 mutation and the risk of developing lung cancer was observed.
The p.R215W (c.634C>T) mutation of the NBN gene was identified in 1 case from the control group, and it was absent in the group of patients with lung cancer. This mutation was described for the first time in patients diagnosed with ALL (23). Previous studies conducted on a Caucasian population demonstrated that p.R215W carriers have an increased risk of non-Hodgkin lymphoma, colorectal, breast and prostate cancer (22,24,25). However, the results of the present study did not indicate that carriers of the p.R215W mutation have an elevated risk of developing lung cancer.
In summary, in the present study, a significantly higher frequency of the p.I171V variant in the fifth exon of the NBN gene was observed in patients diagnosed with NSCLC. The results obtained indicate an association between the occurrence of the p.I171V mutation and the development of lung cancer. Therefore, this variant may be considered as a risk factor for lung cancer. Prospective studies on a larger group of patients may reveal the potential impact of the p.I171V variant on the development of lung cancer.
Acknowledgements
The present study was partially supported by the National Science Centre grant no. 2011/01/D/NZ5/02841.
Abbreviations
- ALL
acute lymphoblastic leukemia
- BRCT
BRCA1 carboxy-terminal
- CI
confidence intervals
- DSBs
DNA double-strand breaks
- FHA
forkhead-associated domain
- NBS
Nijmegen breakage syndrome
- NSCLC
non-small cell lung cancer
- OR
odds ratio
- RFLP
restriction fragment length polymorphism
- SSCP
single-strand conformation polymorphism
References
- 1.Weizmann Institute of Science: GeneCards: NBN gene. Rehovot, Israel: Weizmann Institute of Science; 2014. [Google Scholar]
- 2.Assenmacher N, Hopfner KP. MRE11/RAD50/NBS1: Complex activities. Chromosoma. 2004;113:157–166. doi: 10.1007/s00412-004-0306-4. [DOI] [PubMed] [Google Scholar]
- 3.D'Amours D, Jackson SP. The Mre11 complex: At the crossroads of dna repair and checkpoint signalling. Nat Rev Mol Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 4.Tauchi H, Matsuura S, Kobayashi J, Sakamoto S, Komatsu K. Nijmegen breakage syndrome gene, NBS1, and molecular links to factors for genome stability. Oncogene. 2002;21:8967–8980. doi: 10.1038/sj.onc.1206136. [DOI] [PubMed] [Google Scholar]
- 5.Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, III, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: Linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/S0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 6.Matsuura S, Tauchi H, Nakamura A, Kondo N, Sakamoto S, Endo S, Smeets D, Solder B, Belohradsky BH, Der Kaloustian VM, et al. Positional cloning of the gene for Nijmegen breakage syndrome. Nat Genet. 1998;19:179–181. doi: 10.1038/549. [DOI] [PubMed] [Google Scholar]
- 7.Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanová E, Cooper PR, Nowak NJ, et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/S0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 8.Mosor M, Ziółkowska I, Pernak-Schwarz M, Januszkiewicz-Lewandowska D, Nowak J. Association of the heterozygous germline I171V mutation of the NBS1 gene with childhood acute lymphoblastic leukemia. Leukemia. 2006;20:1454–1456. doi: 10.1038/sj.leu.2404285. [DOI] [PubMed] [Google Scholar]
- 9.Roznowski K, Januszkiewicz-Lewandowska D, Mosor M, Pernak M, Litwiniuk M, Nowak J. I171V germline mutation in the NBS1 gene significantly increases risk of breast cancer. Breast Cancer Res Treat. 2008;110:343–348. doi: 10.1007/s10549-007-9734-1. [DOI] [PubMed] [Google Scholar]
- 10.Ziólkowska I, Mosor M, Wierzbicka M, Rydzanicz M, Pernak-Schwarz M, Nowak J. Increased risk of larynx cancer in heterozygous carriers of the I171V mutation of the NBS1 gene. Cancer Sci. 2007;98:1701–1705. doi: 10.1111/j.1349-7006.2007.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nowak J, Mosor M, Nowicka K, Rembowska J, Januszkiewicz D. Is the NBN gene mutation I171V a potential risk factor for malignant solid tumors in children? J Pediatr Hematol Oncol. 2011;33:e248–e249. doi: 10.1097/MPH.0b013e3181faf886. [DOI] [PubMed] [Google Scholar]
- 12.Cybulski C, Górski B, Debniak T, Gliniewicz B, Mierzejewski M, Masojć B, Jakubowska A, Matyjasik J, Złowocka E, Sikorski A, et al. NBS1 is a prostate cancer susceptibility gene. Cancer Res. 2004;64:1215–1219. doi: 10.1158/0008-5472.CAN-03-2502. [DOI] [PubMed] [Google Scholar]
- 13.Mosor M, Ziółkowska-Suchanek I, Nowicka K, Dzikiewicz-Krawczyk A, Januszkiewicz-Lewandowska D, Nowak J. Germline variants in MRE11/RAD50/NBN complex genes in childhood leukemia. BMC Cancer. 2013;13:457–464. doi: 10.1186/1471-2407-13-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ziółkowska I, Mosor M, Nowak J. Regional distribution of heterozygous 657del5 mutation carriers of the NBS1 gene in Wielkopolska province (Poland) J Appl Genet. 2006;47:269–272. doi: 10.1007/BF03194635. [DOI] [PubMed] [Google Scholar]
- 15.Nowak J, Mosor M, Ziółkowska I, Wierzbicka M, Pernak-Schwarz M, Przyborska M, Roznowski K, Pławski A, Słomski R, Januszkiewicz D. Heterozygous carriers of the I171V mutation of the NBS1 gene have a significantly increased risk of solid malignant tumours. Eur J Cancer. 2008;44:627–630. doi: 10.1016/j.ejca.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Gao P, Ma N, Li M, Tian Q-B, Liu DW. Functional variants in NBS1 and cancer risk: Evidence from a meta-analysis of 60 publications with 11 individual studies. Mutagenesis. 2013;28:683–697. doi: 10.1093/mutage/get048. [DOI] [PubMed] [Google Scholar]
- 17.Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 18.Cerosaletti KM, Concannon P. Nibrin forkhead-associated domain and breast cancer C-terminal domain are both required for nuclear focus formation and phosphorylation. J Biol Chem. 2003;278:21944–21951. doi: 10.1074/jbc.M211689200. [DOI] [PubMed] [Google Scholar]
- 19.Ciara E, Piekutowska-Abramczuk D, Popowska E, Grajkowska W, Barszcz S, Perek D, Dembowska-Bagińska B, Perek-Polnik M, Kowalewska E, Czajńska A, et al. Heterozygous germ-line mutations in the NBN gene predispose to medulloblastoma in pediatric patients. Acta Neuropathol. 2010;119:325–334. doi: 10.1007/s00401-009-0608-y. [DOI] [PubMed] [Google Scholar]
- 20.Dzikiewicz-Krawczyk A, Mosor M, Januszkiewicz D, Nowak J. Impact of heterozygous c.657-661del, p.I171V and p.R215W mutations in NBN on nibrin functions. Mutagenesis. 2012;27:337–343. doi: 10.1093/mutage/ger084. [DOI] [PubMed] [Google Scholar]
- 21.Seemanová E, Jarolim P, Seeman P, Varon R, Digweed M, Swift M, Sperling K. Cancer risk of heterozygotes with the NBN founder mutation. J Natl Cancer Inst. 2007;99:1875–1880. doi: 10.1093/jnci/djm251. [DOI] [PubMed] [Google Scholar]
- 22.Steffen J, Varon R, Mosor M, Maneva G, Maurer M, Stumm M, Nowakowska D, Rubach M, Kosakowska E, Ruka W, et al. Increased cancer risk of heterozygotes with NBS1 germline mutations in Poland. Int J Cancer. 2004;111:67–71. doi: 10.1002/ijc.20239. [DOI] [PubMed] [Google Scholar]
- 23.Seemanová E, Sperling K, Neitzel H, Varon R, Hadac J, Butova O, Schröck E, Seeman P, Digweed M. Nijmegen breakage syndrome (NBS) with neurological abnormalities and without chromosomal instability. J Med Genet. 2006;43:218–224. doi: 10.1136/jmg.2005.035287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varon R, Reis A, Henze G, von Einsiedel HG, Sperling K, Seeger K. Mutations in the Nijmegen Breakage Syndrome gene (NBS1) in childhood acute lymphoblastic leukemia (ALL) Cancer Res. 2001;61:3570–3572. [PubMed] [Google Scholar]
- 25.Hebbring SJ, Fredriksson H, White KA, Maier C, Ewing C, McDonnell SK, Jacobsen SJ, Cerhan J, Schaid DJ, Ikonen T, et al. Role of the Nijmegen breakage syndrome gene in familial and sporadic prostate cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:935–938. doi: 10.1158/1055-9965.EPI-05-0910. [DOI] [PubMed] [Google Scholar]