Abstract
Sphere forming assays are routinely used for in vitro propagation and differentiation of stem cells. Because the stem cell clusters can become heterogeneous and polyclonal, they must first be dissociated into a single cell suspension for further clonal analysis or differentiation studies. The dissociated population is marred by the presence of doublets, triplets and semi-cleaved/intact clusters which makes identification and further analysis of differentiation pathways difficult. In this work, we use inertial microfluidics to separate the single cells and clusters in a population of chemically dissociated neurospheres. In contrast to previous microfluidic sorting technologies which operated at high flow rates, we implement the spiral microfluidic channel in a novel focusing regime that occurs at lower flow rates. In this regime, the curvature-induced Dean’s force focuses the smaller, single cells towards the inner wall and the larger clusters towards the center. We further demonstrate that sorting in this low flow rate (and hence low shear stress) regime yields a high percentage (> 90%) of viable cells and preserves multipotency by differentiating the sorted neural stem cell population into neurons and astrocytes. The modularity of the device allows easy integration with other lab-on-a-chip devices for upstream mechanical dissociation and downstream high-throughput clonal analysis, localized electroporation and sampling. Although demonstrated in the case of the neurosphere assay, the method is equally applicable to other sphere forming assays.
Introduction
Neural stem cells (NSCs) are self-renewing, multi-potent cells capable of differentiating into the major cell types (neurons and glia) of the nervous system. In vitro long term culture and propagation of NSCs is performed either as clusters1 or as an adherent monolayer of stem/progenitor cells.2 Such in vitro models are useful tools in identifying/measuring “stemness” of cells from different regions of the brain and in the development of cell-based therapies for neurological disorders like Alzheimer’s and Parkinson’s diseases. The neurosphere assay (NSA) has aggregates of free-floating cells – called neurospheres – that do not attach to the substrate and yield a heterogeneous cell population.3,4 The heterogeneity occurs, in part, because cells in the core are exposed to sub-optimal conditions and tend to differentiate, thus producing lineage-restricted progenitors in the assay.3 Precise and complete identification of the phenotypes expressed by the cell population is vital for NSCs to reach their full therapeutic potential. Also, the NSA has a population of polyclonal spheres, even at low plating densities, necessitating a rigorous clonal analysis with a single cell, per well, for stem cell identification.5 In order to induce differentiation or for clonal analysis, the neurospheres are first dissociated, either enzymatically or mechanically, neither of which produce a population of entirely single cells. The dissociated cell population invariably contains clusters, making identification (typically using immunostaining, although capacitance-based methods have been recently reported6) and clonal analysis difficult. Thus, it is desirable to have a screening step that separates the single cells from the clusters.
Several active and passive methods currently exist for sorting bioparticles. Separation methods involving membrane filtration7–9 can be expensive, in addition to having other issues like reduced cell viability and clogging. Fluorescence activated cell sorting (FACS) and magnetic activated cell sorting (MACS) need tagging with expensive antibodies. Other methods for sorting cells – such as dielectrophoresis,10,11 acoustophoresis12–14 and optical force switching15 - all involve active fields. They are usually limited by complex fabrication requirements and low throughput. Furthermore, the sorting efficiency decreases with increasing flow rate in active separation methods because the fields have less time to act on the flowing particles/cells. Passive sorting methods reported in the literature include pinched flow fractionation (PFF)16 and deterministic lateral displacement (DLD).17 Although DLD can achieve a separation resolution of 0.1 μm when sorting particles with a mean diameter of 1 μm, the separation resolution is lost when applied to bioparticles due to their elasticity.18
Inertial microfluidics has evolved as a passive, label-free, minimally invasive, high throughput method for sorting cells based on differences in size19–21 with multiple applications, reported in the literature, ranging from sorting of circulating tumor cells (CTCs),22 neuroblastoma cells23 to mesenchymal stem cells (MSCs).24 Inertial focusing was first reported by Segre and Silberberg in macroscale pipe flow.25 They observed focusing of mm sized particles in an annulus centered at a distance of ~ 0.6 times the radius of the channel’s cross section. The number of focusing positions reduces to four and two in micro-channels with square and rectangular cross sections, respectively.19 Addition of curvature26 or asymmetry27 along the length of the channel reduces the number of focusing positions to one. This is due to an additional drag force from a pair of counter-rotating Dean’s vortices28 in the channel’s cross-section. Typically, sorting is achieved either through differential inertial focusing (DIF)24,29,30 – where all particles equilibrate at different positions near the inner wall in a descending order of size – or through selective inertial focusing (SIF)22,31 – where only the particles/cells of interest are focused while the other (usually smaller) particles are entrained in the secondary vortices.
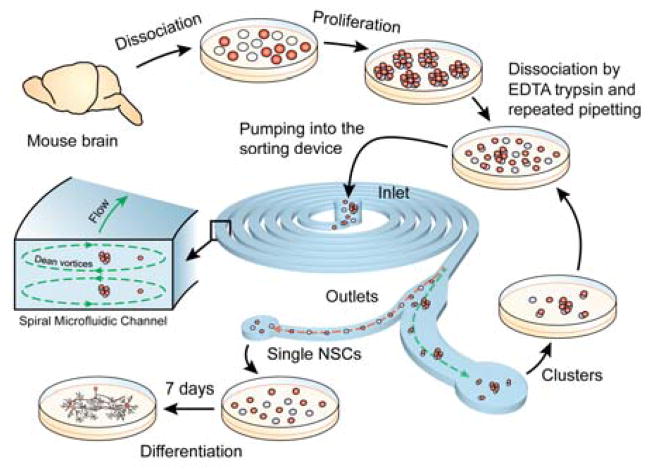
In this work, we use inertial microfluidics to isolate single cells from a mixed population of single cells and clusters produced from chemically dissociated neurospheres. Sorting single cells from the dissociated population presents two engineering challenges. There is a considerable disparity in the sizes of single cells (~ 8–14 μm) and clusters (~ 40–60 μm). Also, the dissociated population can have a distribution of sizes. NSCs, being primary cells, can be sensitive to shear stress. In the current work, we tune the Dean’s force to have opposite effects on single cells and clusters. Thus, single cells are focused near the inner wall while clusters are focused away from it in the middle of the microchannel (see Figure 1). We emphasize that the current methodology is also a case of DIF but is unique with respect to the use of Dean’s force. We show that the process is viable and preserves multipotency of the stem cells by differentiating the sorted population into neurons and astrocytes. While we demonstrate the current approach in the case of neurospheres, it is equally applicable to other sphere-forming cell populations32 where a dissociated single-cell population is needed.
Figure 1.
Overview of the experimental process. Neurospheres obtained from mice were first dissociated enzymatically followed by repeated pipetting. The dissociated cell sample was then pumped through the device. The sorted single cells were collected at the outlet and cultured in a low serum media to promote differentiation.
Materials and Methods
Device fabrication and assembly
The spiral microchannel was fabricated using soft lithographic procedures reported previously.33 Briefly, 130 μm thick SU-8 photo resist (2100, MicroChem) was patterned on a 4 inch Si wafer using photolithography (MA-6 mask aligner, 250 mJ/cm2 exposure energy). PDMS prepolymer (Slygard 184, Dow Corning) was mixed in 10:1 w/w ratio (base, curing agent) and then poured onto the SU-8 mold. After curing, the PDMS layer was peeled off the SU-8 mold. Holes for inlets and outlets were made using a biopsy punch. The PDMS layer was bonded to a glass slide using oxygen plasma treatment.
Simulations
Numerical simulations were performed using ANSYS Fluent (Pennsylvania, USA) to generate the secondary flow field in the microchannel. A 120° arc with a radius of 1.25 cm was used as the geometry for the simulation. Steady-state Navier-Stokes and the continuity equations were solved in the laminar flow regime. The fluid was assumed to be incompressible and material properties (density, viscosity) of cell media were approximated with that of water. A fully developed parabolic velocity profile corresponding to the desired flow rate was taken as the inlet boundary condition. Zero pressure was taken as the outlet boundary condition. No slip boundary condition was imposed on the walls.
Sorting experiments with microbeads
The 38 μm polyethylene microbead powder (Cospheric, SD~3 μm) was first dispersed in 0.1% (w/v) suspension of Tween 80 (Cospheric) surfactant for preventing particle aggregation. A 7.7 μm microbead suspension (Spherotech, SD~0.5 μm) provided at 1% (w/v) was diluted to 0.1% (w/v) using de-ionized water. Equal volumes of the two suspensions were mixed and then pumped through the device at different flow rates using a syringe pump (New Era Pump Systems). The focusing positions were observed near the outlets using a dark field microscope (Olympus-IX71).
Cell culture, enzymatic dissociation and differentiation into neurons and astrocytes
All procedures in this study involving animals were approved by the Northwestern University IUCUC prior to performance. Murine neurospheres were obtained by following previously reported protocols with minor changes.34 Briefly, E13 embryos from CD1 mice (Charles River, Wilmington, MA) were dissected and the ganglionic eminences isolated and grown in serum-free medium (SFM) with human recombinant epidermal growth factor (FGF, 20 ng/ml, Biosource) for 5–7 days until neurospheres developed. More than 91% of the isolated cells express nestin and Ki67 (see Figure S1 in SI). The neurospheres were then dissociated with 0.25% trypsin (Life Technologies) at 37 °C for 5 min, followed by incubation with soybean trypsin inhibitor (Life Technologies) at 37 °C for another 5 min. The vial was centrifuged for 5 min to remove the supernatant and then resuspended, by repeated pipetting (>100 times), in media containing SFM and FGF (1 ng/ml) to induce differentiation.
Poly-D-lysine (PDL) functionalization and sorting experiments with NSCs
For PDL functionalization, the 24-well plates (Falcon BD) were covered with at least 1 ml solution (per well) of PDL (20 μg/ml, Sigma) for 12 hours at room temperature and then washed thrice with PBS (Life Technologies). The eight outlets of the sorting device were each connected to different wells in the well plate. After neurosphere dissociation, the cell sample was pumped through the device using a syringe pump. The sorted cells were collected in the well plate and cultured in differentiation medium for 1–2 weeks before immunostaining. Initial experiments to observe focusing and sorting of NSCs using dark field microscopy were performed outside the bio-safety hood. Subsequent experiments were performed within the bio-safety hood to ensure sterility.
Immunostaining for neuronal (β-tubulin III) and astrocyte (GFAP) markers
The sorted, single cells in wells 1 and 2 of the well plate were fixed with ice cold methanol (−20 °C) for 10 min and washed thrice with PBS to remove any fixation media. The cells were first incubated, at room temperature, with a blocking buffer solution comprising PBS diluted with 1% BSA (Sigma) and 0.25% Triton X-100 (Sigma) for 1 hour. This was followed by incubation with primary antibodies specific to β-tubulin III and glial fibrillary acidic protein (GFAP) viz. monoclonal anti-β-tubulin III (Sigma) and polyclonal anti-GFAP (Dako) overnight at 4 °C. The cells were washed thrice with PBS and then incubated with secondary antibodies conjugated to Alexa Fluor 594 (Molecular Probes) for neurons and Alexa Fluor 488 (Invitrogen) for astrocytes in the dark at room temperature for 1 hour. The cells were gently washed with PBS, stained with Hoechst and washed again before fluorescent images were collected with an inverted microscope (Nikon Eclipse Ti-U) coupled to a CMOS camera (Andor Neo).
Results
Device design and modeling
Laminar flow at finite inertia (Reynolds number ~ 1–100) in a spiral microfluidic channel with a high aspect ratio can yield a single focusing position along the width of the channel for the particles/cells flowing through it. The focusing position is sensitive to both the diameter of the particle and the flow rate.35 Qualitatively, the equilibrium position can be explained as a balance of two forces: an inertial lift force (FL) and a curvature-induced Dean’s drag force (FD). A vector plot of the simulated velocity field is shown in Figure 2b. The curvature induces a pair of counter-rotating vortices which exert an additional drag force on the particles. Following the derivation in ref.27, the ratio between the two forces can be estimated as
Figure 2.
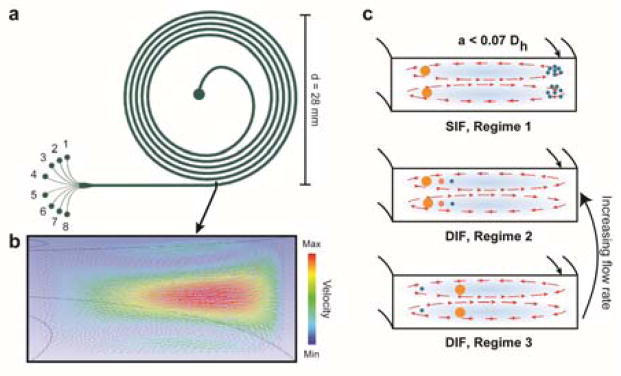
Schematics depicting the design and working principle of the sorting device a) Schematic of the device geometry, inlet and eight outlets. b) Numerical simulation showing the secondary flow field at the outlet. c) Schematic showing different focusing regimes in inertial microfluidics. In SIF, regime 1, only the particles of interest are focused while the others are entrained in the vortices. In DIF, regime 2, the particles are focused in descending order of size from the inner wall. In DIF, regime 3, the smaller particles are focused near the inner wall, while the larger ones are focused in the center. Regime 3 transitions to regime 2 with increasing flow rate.
where RC is the Reynold’s number of the flow, a is the radius of the particle, Dh is the hydraulic diameter of the channel, δ = Dh/2r is the curvature ratio and n is a fitting parameter. It should be noted that the estimate for FL is based on an asymptotic analysis by Asmolov36 and is valid for small, point-like particles. A different scaling for FL was reported by Di Carlo et al. based on direct numerical simulations.37 The force had a third-order power dependence (i.e. a3) on the particle diameter near the center and a sixth-order power dependence (i.e. a6) near the channel wall. In any case, the force scales exponentially with the m particle diameter i.e. am;m≥2 and does not affect the implications from this qualitative reasoning. The Dean’s drag force FD was estimated using an expression for the rotational flow velocity in curved pipes38–39 and by assuming that Stoke’s drag acts on the particle.
For a given particle size and flow rate, three different cases can operate :- 1) For the case where FL and FD are comparable, the Dean’s force pushes the particle and equilibrates it near the inner wall. 2) If FL≪FD, which is the case for particles satisfying a/Dh<0.07, the particle is entrained and circulated along the Dean’s vortices. 3) Lastly, if FL≫FD, which can occur for a particle with a large size, such as a neurosphere cluster, the curvature has a secondary effect and the particle is focused in the center (along the width) for a high aspect ratio channel. For a given particle size, case 3 occurs at a lower flow rate. As the flow rate is increased, the focusing position moves from the middle of the channel towards the inner wall. Thus, the three different cases can be realized for the same particle size by tuning the flow rate and the channel dimensions.
Existing work on inertial microfluidic separation has mainly made use of the first two cases. One sorting approach is to focus larger particles/cells near the inner wall while the smaller ones are circulated along the vortices (SIF: regime 1, Figure 2c).22,31 This can be realized by ensuring flow conditions corresponding to case 1 for the larger particle and case 2 for the smaller ones. One caveat is that the number of spiral loops and the flow rate should be adjusted so that the smaller particles are diametrically opposite to the bigger ones near the outlet. For instance, Hou et al.22 were able to separate circulating tumor cells (CTCs, size: 10–20 μm) from red (8 μm) and white blood cells (7–12 μm) by making use of regime 1. The CTCs were focused near the inner wall where as the red blood cells and leukocytes were circulated along the vortices. Another separation methodology (henceforth referred to as regime 2, Figure 2c) focuses different particles near the inner wall by ensuring that FL and FD are comparable (case 1). Lee et al.29 used the connection between the cell size and cell cycle to fractionate human mesenchymal stem cells into different phases, making use of regime 2, where the particles/cells are differentially focused in a decreasing order of size near the inner wall.
In our design (regime 3, Figure 2c), the single cells are focused near the inner wall by realizing case 1, but the clusters are focused near the channel center by employing case 3. This occurs because the Dean’s force pushes the smaller particles towards the inner wall and the larger ones away from it. While the current method is also a case of DIF, it is novel in how the Dean’s force is used. There are two important reasons for using regime 3 in separating single NSCs from neurosphere clusters. The first is that, NSCs, being primary cells, can be extremely sensitive to shear stress. Regime 3 occurs at a lower flow rate and this translates to a smaller shear stress on the cells. As will be shown in the next subsection, an increase in the flow rate changes the focusing regime from 3 to 2. The second reason is that there could be a distribution of sizes for the neurosphere clusters. Because regime 2 focuses particles in a descending order of size, in the worst case scenario, the single cells would be pushed towards the outer wall which is not an equilibrium position.
It should be emphasized that the above qualitative analysis provides valuable insights into the focusing positions along the width of the cross section, but doesn’t indicate the number/location of focusing points along the depth. In addition, the force analysis assumes negligible disturbance to the primary flow field from the laden particles, which may not be true in the case of large neurosphere clusters. Indeed, design tools capable of predicting the experimental outcomes are greatly desired in the field of inertial microfluidics.21 For this work, the cross section of our device is 150 μm × 500 μm (height by width), the initial radius of the spiral is 1 cm and the device has a total of five turns. As we will demonstrate later, the chosen dimensions allow us to realize the three different focusing regimes in the same device, within flow rates of 1–3 ml/min. At the outlet, the channel is widened and further divided into 8 outlets to precisely monitor the focusing position at different flow rates.
Sorting microbeads as a model system
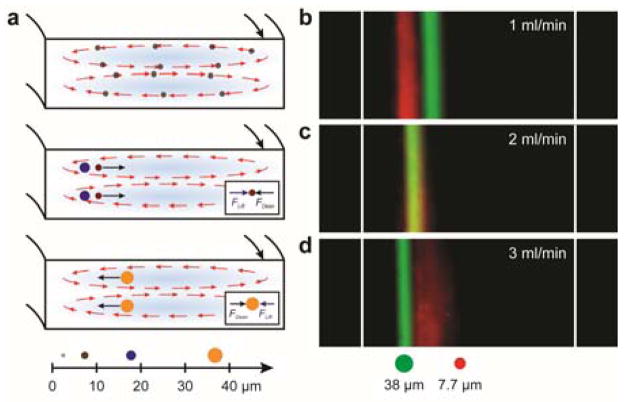
In our design, particles with diameter less than 7 μm will get entrained in the Dean’s vortices (Figure 3a, corresponds to case 2, discussed in the design section). Particles with intermediate sizes (10 – 25 μm) are focused near the inner wall at low flow rates (Figure 3a, case 1). Increasing the flow rate will cause outward movement of the particle’s focusing position. Particles with diameter 30 μm get focused in the middle of the channel (Figure 3a, case 3) at low flow rates. At higher flow rates, they move towards the inner wall (case 1). We demonstrate this focusing behavior using 7.7 μm (SD~0.5 μm) and 38 μm (SD~3 μm) microbeads as a model system. They also represent a single neural stem cell and a cluster, respectively. We monitored the focusing positions of the beads at different flow rates using dark-field microscopy. As shown in Figure 3b, at a flow rate of 1 ml/min, the 7.7 μm beads are focused towards the inner wall while the 38 μm beads are focused in the center of the microchannel (regime 3). As the flow rate is increased to 3 ml per min, there is a reversal in the focusing positions of the beads. The 38 μm beads are now focused near the inner wall. Also, the smaller beads become less tightly focused at a higher flow rate. Increasing the flow rate further will cause greater broadening and outward movement of the focused stream for the smaller beads whereas the larger beads will move closer to the inner wall.35 Thus, we employed a flow rate of 1 ml/min in all of our experiments with NSCs for two reasons: the smaller particles are more tightly focused at lower flow rates and since the shear stress increases with the flow rate, we sought to minimize it to avoid any deleterious effects on the viability and subsequent differentiation behavior of the sorted stem cells.
Figure 3.
Sorting experiments on microbeads: a) Schematic showing the force balance for particles of different sizes in regime 3. (b–d) Dark field streaks (pseudo colored) showing focusing of 7.7 μm (red) and 38 μm (green) microbeads at different flow rates. Regime 3 operates at low flow rates (1 ml/min) while regime 2 becomes operative as the flow rate is increased (3 ml/min). The images were taken during the final turn of the spiral. For a flow rate of 1 ml/min, Reynold’s number ~ 130, and Dean’s number ~ 13
Sorting experiments on neural stem cells
a) Distribution of single cells and clusters in different channels at the outlet
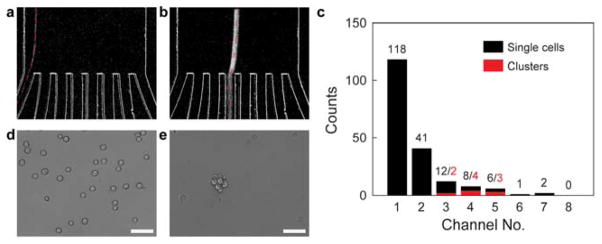
Prior to the sorting experiment, the neurospheres were enzymatically dissociated followed by repeated pipetting (> 100 repetitions). The dissociated cell sample was pumped at a flow rate of 1 ml/min through the device. The eight channels at the outlet were fed into eight different wells in a 6×4 well plate through plastic tubing. The wells were functionalized with PDL to promote attachment. In the ensuing discussion, the channels (and wells) are numbered starting from the inner wall i.e., channel 1 refers to the innermost channel and channel 8 refers to the outermost channel (see Figure 2a). The data were derived by processing time-lapse images using a custom MATLAB script. The dark field streaks were classified into either a single cell or a cluster based on a threshold for the full width at half maximum (FWHM) of the intensity. Figures 4a–b show dark field streaks corresponding to a single cell and a large cluster, respectively, from a sorting experiment. Single cells are focused into channels 1 and 2, while the clusters depending on their size, equilibrate near channels 3–5. The histogram in Figure 4c shows the number of single cells (black bars) and clusters (red bars) collected by different channels in a sorting experiment. Channels 1 and 2 isolate approximately 84% of the single cells present in the sample. Figures 4d–e show sample differential interference contrast images of the cells collected in different wells of the well plate. The images were taken an hour after the experiment was performed to allow the cells to adhere to the well plate. Well 1 contains single cells while clusters were collected in well 3. We observed occasional single cells in other wells also (data not shown) but could concentrate the majority of the single cell population (>80%) in wells 1, 2. The design of the device can be further modified by merging channels 1, 2 into a single channel and channels 3–8 into another. This will allow the neurospheres collected from channels 3–8 to be dissociated again and fed back into the device (see Figure 1). The presence of eight channels in the current design makes them a bit too narrow (~40 μm) and causes the bigger neurospheres (>50 μm) to undergo morphological changes as they pass through. Merging channels 3–8 into a single, wide channel will avoid this.
Figure 4.
Sorting experiments on NSCs: a) Dark field streak of a single NSC focused at outlet 1, highlighted by a red dashed line b) Dark field streak of a neurosphere focused into a middle outlet. c) Histogram of the single cells and clusters focused at different outlets. d) Single neural stem cells collected from outlet 1, cultured in a 6×4 well plate. e) Neurosphere collected from outlet 4 in the same experiment. All scale bars = 50 μm. The sorting experiment was performed at a flow rate of 1ml/min.
b) Effect of shear stress on viability
The shear stress τ experienced by the cells can be estimated as τ = 6μQ/wh2 where Q is the flow rate, μ is the dynamic viscosity of the fluid, w, h are the width and height of the channel. For Q=1 ml/min, τ≈9 Pa. To put this value in context, human mesenchymal stem cells (hMSCs) were previously sorted based on size.29 The device had similar dimensions (500 μm × 200 μm) but operated at a higher shear stress (Q = 2.5 ml/min). The sorting process was shown to be viable (~95%). We performed membrane integrity studies to estimate the number of viable cells after sorting. Because the dissociation process using trypsin and repeated pipetting could itself affect viability, we performed membrane integrity studies on both dissociated (control) and dissociated, sorted cells. We found that ~ 92% of the cells survive after just the dissociation step and (90±1)% after both dissociation and subsequent sorting (see Figure S2 in SI). Thus, when normalized with the control case, > 97% of the cell population remains viable after sorting. We also note that future work should further explore the effect of shear stress on DNA damage and mitochondrial function of the sorted cells.
c) Immunostaining to demonstrate preserved multipotency
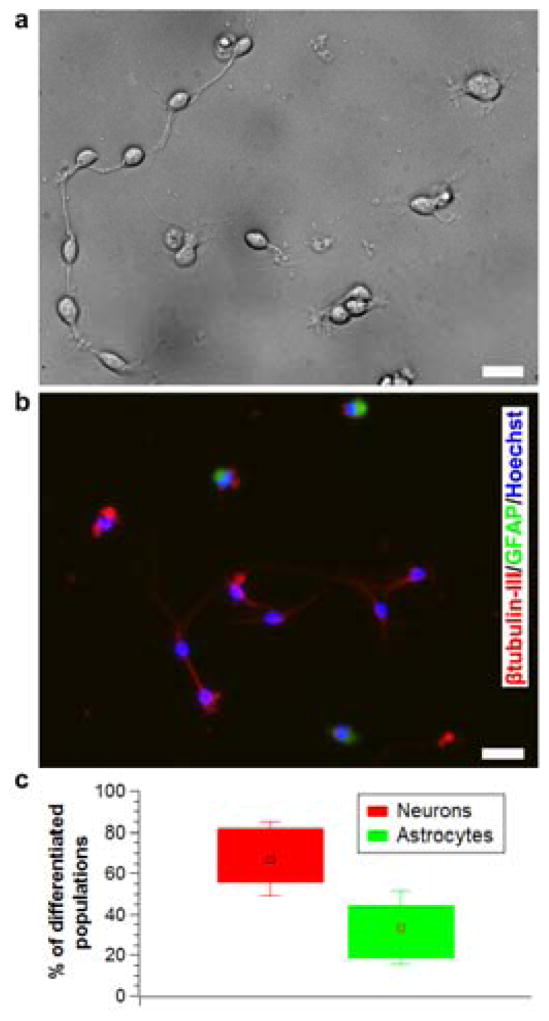
After sorting, the single cells were cultured (in the same well plate that was used to collect the sorted cells) in a low growth factor medium to promote differentiation. By the fifth day, the cells differentiated into a network of neurons and astrocytes showing morphological features such as axons, intermediate filaments and neuronal junctions (see Figure 5a). We confirmed that the differentiated cells contained both neurons and astrocytes by staining them for the neuron biomarker, β-tubulin III and astrocyte biomarker, GFAP (Figure 5b). This proved that the sorting process preserved multipotency of NSCs. All of the experiments were done on passage I NSCs and thus contained a higher relative percentage of neurons (~67%, see Figure 5c).
Figure 5.
Immunostaining of sorted single cells to analyze preserved multipotency. a) Differential interference contrast image showing differentiation of sorted NSCs (Day 5). b) Immunofluorescence image showing both neurons (red) and astrocytes (green) in the differentiated population. (Figures a, b correspond to different experiments) c) Relative percentage of neurons and astrocytes produced from different experiments. (Data reported as mean ± standard deviation, n = 12). All scale bars = 20 μm.
Conclusion
Stem cells, induced pluripotent stem cells (iPSCs) in particular, hold great promise in personalized medicine, drug screening and human disease modeling.40 Realization of these potential applications is currently hindered by our limited understanding of stem cell differentiation. Owing to the heterogeneity and polyclonality of the neurosphere, dissociation into a single-cell suspension is a critical step prior to rigorous clonal analysis or further study of differentiation pathways. Utilizing a novel focusing regime in inertial microfluidics, we have fabricated a sorting device and demonstrated separation of single stem cells with preserved multipotency. Because the device is modular, it can easily be integrated with other lab-on-a-chip devices. For instance, it can be integrated with a mechanical dissociation device41,42 upstream and a microfluidic device for high throughput clonal analysis43 or localized electroporation44 downstream, yielding a micro total analysis system (μTAS). In our experiments, chemical dissociation of the neurospheres was the most time consuming step. Microfluidic integration of mechanical and/or chemical dissociation functionality into the sorting device is a desirable feature, since it minimizes user intervention in the process. In addition to the NSA, in vitro assays with floating spheroid cell clusters are used in identifying and measuring stem-cell like behavior in cells from various organ systems – like the retina,45 pancreas,46 skin,47 etc. Sphere culture systems are also employed for in vitro culture of cancer stem cells or brain tumor cells.48 We believe the current method can be adapted to isolate singe cell populations in all these culture systems. Another limitation of the NSA is the low percentage of neurons produced with increasing passage numbers (less than 20%).3 Pure and enriched population of neurons may be necessary in cell therapy studies. Based solely on the size difference between neurons and glia (manifest in the forward scatter data), flow cytometry was previously used to isolate neurons with a purity of ~75%. 49 Exploiting this difference in the size of a neuron and an astrocyte, the current device can easily be extended to further sort neurons from the single cell population. A possible approach for achieving this would involve the following steps – In step 1, the single cells may be separated from the clusters using the method reported here. In step 2, the sorted single cells may be reflowed through the device, but at a higher flow rate (Q=3 ml/min) conforming to regime 2 (Figure 2). In this regime, the cells equilibrate in a descending order of size near the inner wall. Thus, the larger astrocytes will be collected in outlet 1 and the smaller neurons in outlet 2.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation under award IIP-1330151 and the National Institutes of Health under awards 1R41GM101833 and NS20778. S.S.P.N. is thankful to Dr. Changjin Huang for his help with immunostaining and membrane integrity tests.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
Author contributions
H.E., J.K and C.S conceived the project and supervised the experiments, analysis. F.Z designed and fabricated the sorting devices. T.M., S.S.P.N., W.K and J.G.V prepared NSCs. S.S.P.N and B.D performed the sorting experiments on microbeads and NSCs. B.D wrote the MATLAB code for analyzing the dark-field videos. S.S.P.N did the immunostaining. F.Z and S.S.P.N. carried out the numerical analysis. H.E., J.K., C.S., S.S.P.N., W.K., B.D., F.Z. and R.M. wrote the manuscript. All authors analyzed the data and discussed the results.
References
- 1.Reynolds BA, Weiss S. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 2.Sun Y, Pollard S, Conti L, Toselli M, Biella G, Parkin G, Willatt L, Falk A, Cattaneo E, Smith A. Mol Cell Neurosci. 2008;38:245–258. doi: 10.1016/j.mcn.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 3.Conti L, Cattaneo E. Nat Rev Neurosci. 2010;11:176–187. doi: 10.1038/nrn2761. [DOI] [PubMed] [Google Scholar]
- 4.Campos LS. Journal of Neuroscience Research. 2004;78:761–769. doi: 10.1002/jnr.20333. [DOI] [PubMed] [Google Scholar]
- 5.Singec I, Knoth R, Meyer RP, Maciaczyk J, Volk B, Nikkhah G, Frotscher M, Snyder EY. Journal. 2006;3:801–806. doi: 10.1038/nmeth926. [DOI] [PubMed] [Google Scholar]
- 6.Lee R, Kim IS, Han N, Yun S, Park KI, Yoo KH. Sci Rep-Uk. 2014;4 doi: 10.1038/srep06319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schirhagl R, Fuereder I, Hall EW, Medeiros BC, Zare RN. Lab on a Chip. 2011;11:3130–3135. doi: 10.1039/c1lc20353c. [DOI] [PubMed] [Google Scholar]
- 8.Wei HB, Chueh BH, Wu HL, Hall EW, Li CW, Schirhagl R, Lin JM, Zare RN. Lab on a Chip. 2011;11:238–245. doi: 10.1039/c0lc00121j. [DOI] [PubMed] [Google Scholar]
- 9.Zheng S, Lin H, Liu JQ, Balic M, Datar R, Cote RJ, Tai YC. J Chromatogr A. 2007;1162:154–161. doi: 10.1016/j.chroma.2007.05.064. [DOI] [PubMed] [Google Scholar]
- 10.Kim U, Shu CW, Dane KY, Daugherty PS, Wang JYJ, Soh HT. P Natl Acad Sci USA. 2007;104:20708–20712. doi: 10.1073/pnas.0708760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durr M, Kentsch J, Muller T, Schnelle T, Stelzle M. Electrophoresis. 2003;24:722–731. doi: 10.1002/elps.200390087. [DOI] [PubMed] [Google Scholar]
- 12.Thevoz P, Adams JD, Shea H, Bruus H, Soh HT. Analytical Chemistry. 2010;82:3094–3098. doi: 10.1021/ac100357u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding XY, Peng ZL, Lin SCS, Geri M, Li SX, Li P, Chen YC, Dao M, Suresh S, Huang TJ. P Natl Acad Sci USA. 2014;111:12992–12997. doi: 10.1073/pnas.1413325111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nilsson A, Petersson F, Jonsson H, Laurell T. Lab on a Chip. 2004;4:131–135. doi: 10.1039/b313493h. [DOI] [PubMed] [Google Scholar]
- 15.Wang MM, Tu E, Raymond DE, Yang JM, Zhang HC, Hagen N, Dees B, Mercer EM, Forster AH, Kariv I, Marchand PJ, Butler WF. Nature Biotechnology. 2005;23:83–87. doi: 10.1038/nbt1050. [DOI] [PubMed] [Google Scholar]
- 16.Yamada M, Nakashima M, Seki M. Analytical Chemistry. 2004;76:5465–5471. doi: 10.1021/ac049863r. [DOI] [PubMed] [Google Scholar]
- 17.Huang LR, Cox EC, Austin RH, Sturm JC. Science. 2004;304:987–990. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- 18.Davis JA, Inglis DW, Morton KJ, Lawrence DA, Huang LR, Chou SY, Sturm JC, Austin RH. P Natl Acad Sci USA. 2006;103:14779–14784. doi: 10.1073/pnas.0605967103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Carlo D. Lab Chip. 2009;9:3038–3046. doi: 10.1039/b912547g. [DOI] [PubMed] [Google Scholar]
- 20.Amini H, Lee W, Di Carlo D. Lab Chip. 2014;14:2739–2761. doi: 10.1039/c4lc00128a. [DOI] [PubMed] [Google Scholar]
- 21.Martel JM, Toner M. Annu Rev Biomed Eng. 2014;16:371–396. doi: 10.1146/annurev-bioeng-121813-120704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou HW, Warkiani ME, Khoo BL, Li ZR, Soo RA, Tan DS, Lim WT, Han J, Bhagat AA, Lim CT. Sci Rep. 2013;3:1259. doi: 10.1038/srep01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuntaegowdanahalli SS, Bhagat AA, Kumar G, Papautsky I. Lab Chip. 2009;9:2973–2980. doi: 10.1039/b908271a. [DOI] [PubMed] [Google Scholar]
- 24.Lee WC, Shi H, Poon Z, Nyan LM, Kaushik T, Shivashankar GV, Chan JK, Lim CT, Han J, Van Vliet KJ. Proc Natl Acad Sci U S A. 2014;111:E4409–4418. doi: 10.1073/pnas.1402306111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segre G, Silberberg A. Nature. 1961;189:209. [Google Scholar]
- 26.Yoon DH, Ha JB, Bahk YK, Arakawa T, Shoji S, Go JS. Lab on a Chip. 2009;9:87–90. doi: 10.1039/b809123d. [DOI] [PubMed] [Google Scholar]
- 27.Di Carlo D, Irimia D, Tompkins RG, Toner M. Proc Natl Acad Sci U S A. 2007;104:18892–18897. doi: 10.1073/pnas.0704958104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger SA, Talbot L, Yao LS. Annu Rev Fluid Mech. 1983;15:461–512. [Google Scholar]
- 29.Lee WC, Bhagat AA, Huang S, Van Vliet KJ, Han J, Lim CT. Lab Chip. 2011;11:1359–1367. doi: 10.1039/c0lc00579g. [DOI] [PubMed] [Google Scholar]
- 30.Russom A, Gupta AK, Nagrath S, Di Carlo D, Edd JF, Toner M. New J Phys. 2009;11:75025. doi: 10.1088/1367-2630/11/7/075025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhagat AA, Kuntaegowdanahalli SS, Papautsky I. Lab Chip. 2008;8:1906–1914. doi: 10.1039/b807107a. [DOI] [PubMed] [Google Scholar]
- 32.Pastrana E, Silva-Vargas V, Doetsch F. Cell Stem Cell. 2011;8:486–498. doi: 10.1016/j.stem.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia YN, Whitesides GM. Annu Rev Mater Sci. 1998;28:153–184. [Google Scholar]
- 34.Bonaguidi MA, McGuire T, Hu M, Kan LX, Samanta J, Kessler JA. Development. 2005;132:5503–5514. doi: 10.1242/dev.02166. [DOI] [PubMed] [Google Scholar]
- 35.Guan GF, Wu LD, Bhagat AAS, Li ZR, Chen PCY, Chao SZ, Ong CJ, Han JY. Sci Rep-Uk. 2013;3 doi: 10.1038/srep01475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asmolov ES. J Fluid Mech. 1999;381:63–87. [Google Scholar]
- 37.Di Carlo D, Edd JF, Humphry KJ, Stone HA, Toner M. Physical Review Letters. 2009;102 doi: 10.1103/PhysRevLett.102.094503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ookawara S, Higashi R, Street D, Ogawa K. Chem Eng J. 2004;101:171–178. [Google Scholar]
- 39.Ookawara S, Street D, Ogawa K. Chem Eng Sci. 2006;61:3714–3724. [Google Scholar]
- 40.Nishikawa S, Goldstein RA, Nierras CR. Nat Rev Mol Cell Biol. 2008;9:725–729. doi: 10.1038/nrm2466. [DOI] [PubMed] [Google Scholar]
- 41.Lin CH, Lee DC, Chang HC, Chiu IM, Hsu CH. Anal Chem. 2013;85:11920–11928. doi: 10.1021/ac402724b. [DOI] [PubMed] [Google Scholar]
- 42.Wallman L, Akesson E, Ceric D, Andersson PH, Day K, Hovatta O, Falci S, Laurell T, Sundstrom E. Lab Chip. 2011;11:3241–3248. doi: 10.1039/c1lc20316a. [DOI] [PubMed] [Google Scholar]
- 43.Roccio M, Gobaa S, Lutolf MP. Integr Biol (Camb) 2012;4:391–400. doi: 10.1039/c2ib00070a. [DOI] [PubMed] [Google Scholar]
- 44.Kang W, Giraldo-Vela JP, Nathamgari SS, McGuire T, McNaughton RL, Kessler JA, Espinosa HD. Lab Chip. 2014;14:4486–4495. doi: 10.1039/c4lc00721b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tropepe V, Coles BL, Chiasson BJ, Horsford DJ, Elia AJ, McInnes RR, van der Kooy D. Science. 2000;287:2032–2036. doi: 10.1126/science.287.5460.2032. [DOI] [PubMed] [Google Scholar]
- 46.Seaberg RM, Smukler SR, Kieffer TJ, Enikolopov G, Asghar Z, Wheeler MB, Korbutt G, van der Kooy D. Nat Biotechnol. 2004;22:1115–1124. doi: 10.1038/nbt1004. [DOI] [PubMed] [Google Scholar]
- 47.Toma JG, Akhavan M, Fernandes KJ, Barnabe-Heider F, Sadikot A, Kaplan DR, Miller FD. Nat Cell Biol. 2001;3:778–784. doi: 10.1038/ncb0901-778. [DOI] [PubMed] [Google Scholar]
- 48.Piccirillo SG, Vescovi AL. Expert Opin Biol Ther. 2007;7:1129–1135. doi: 10.1517/14712598.7.8.1129. [DOI] [PubMed] [Google Scholar]
- 49.Azari H, Osborne GW, Yasuda T, Golmohammadi MG, Rahman M, Deleyrolle LP, Esfandiari E, Adams DJ, Scheffler B, Steindler DA, Reynolds BA. PLoS One. 2011;6:e20941. doi: 10.1371/journal.pone.0020941. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.