Abstract
Rationally designed combinations of targeted therapies for refractory cancers, such as activated B cell–like diffuse large B cell lymphoma (ABC DLBCL), are likely required to achieve potent, durable responses. Here, we used a pharmacoproteomics approach to map the interactome of a tumor-enriched isoform of HSP90 (teHSP90). Specifically, we chemically precipitated teHSP90-client complexes from DLBCL cell lines with the small molecule PU-H71 and found that components of the proximal B cell receptor (BCR) signalosome were enriched within teHSP90 complexes. Functional assays revealed that teHSP90 facilitates BCR signaling dynamics by enabling phosphorylation of key BCR signalosome components, including the kinases SYK and BTK. Consequently, treatment of BCR-dependent ABC DLBCL cells with PU-H71 attenuated BCR signaling, calcium flux, and NF-κB signaling, ultimately leading to growth arrest. Combined exposure of ABC DLBCL cell lines to PU-H71 and ibrutinib, a BCR pathway inhibitor, more potently suppressed BCR signaling than either drug alone. Correspondingly, PU-H71 combined with ibrutinib induced synergistic killing of lymphoma cell lines, primary human lymphoma specimens ex vivo, and lymphoma xenografts in vivo, without notable toxicity. Together, our results demonstrate that a pharmacoproteome-driven rational combination therapy has potential to provide more potent BCR-directed therapy for ABC DLCBL patients.
Introduction
Tumor growth and survival are maintained by molecular mechanisms linked either to somatic mutations or to normal proteins hijacked by tumors resulting in non-oncogene addiction. Targeting these proteins and pathways holds the promise to improve efficacy and decrease toxicity of anticancer treatments. However, even targeted therapies are unlikely to be curative as single agents, given the genetic complexity, clonal heterogeneity, feedback mechanisms, and propensity to develop resistance in most tumors. Tumor eradication will require combinations of agents that hit parallel or complementary pathways or enable more thorough suppression of complex signaling pathways. Design of rational combinations is challenging, given the massive numbers of drugs and the difficulty of defining the complement of proteins that drive tumor survival, independent of whether they are affected by somatic mutations.
These challenges are highly relevant to diffuse large B cell lymphoma (DLBCL), a heterogeneous group of tumors that are not curable with current therapies in 40% of patients (1, 2). One way to address the heterogeneity challenge is to identify and target mechanisms that drive lymphomagenesis in a broad cross section of patients, regardless of their complement of somatic mutations. In this regard, it is recognized that tumors acquire a biological dependence on stress proteins during malignant transformation. Recent data implicate high molecular weight stress protein complexes enriched in tumor cells as playing a key role in the survival of lymphoma cells (3–5). These complexes contain a modified form of the molecular chaperone protein HSP90. Tumor-enriched HSP90 (teHSP90) complexes preferentially associate with mutant and WT proteins required to maintain tumor survival, whereas the housekeeping form of HSP90 are not preferentially associated with tumor-associated proteins (5, 6). Recently, a small molecule called PU-H71 was developed that preferentially binds with high affinity to teHSP90 stress complexes (3, 4). PU-H71 disrupts the functionality of these teHSP90 complexes with little effect on housekeeping fractions of HSP90 (5). This endows PU-H71 with preferential binding to tumor cells versus normal cells and a greater therapeutic window than traditional nonselective HSP90 inhibitors. In particular, PU-H71 has potent activity against DLBCL cell lines, as well as primary human DLBCL cells grown ex vivo, without significant toxicity in mice (3). PU-H71 has moved into the clinical arena, where it is currently in phase I trials.
The high affinity of PU-H71 to teHSP90 stress complexes has been harnessed as a proteomics capture approach to identify the cellular complement of oncoproteins specifically contributing to the growth and survival of tumors (5). For example, PU-H71 pulldowns performed in leukemia cells driven by BCR-ABL capture BCR-ABL but not the normal counterpart ABL protein; both BCR-ABL and ABL are HSP90 clients, but only BCR-ABL is bound and stabilized by teHSP90 (5). Importantly, these and other initial studies suggested that teHSP90 complexes mediate functions distinct from the standard molecular chaperone housekeeping function of HSP90 (5).
Collectively, these data led us to hypothesize that the teHSP90 oncoproteome would point toward mechanisms for these complexes in DLBCL cells. We predicted that these mechanisms would lead to rational combinatorial therapies anchored by PU-H71 and geared toward more thorough suppression of key oncogenic pathways than can be achieved with single-agent therapy. Herein, using PU-H71 affinity capture proteomics in DLBCL cells, we identified the B cell receptor (BCR) pathway proteins as components of teHSP90 stress complexes. We demonstrate that multiple nodes of BCR signaling — including kinase phosphorylation, calcium signaling, and NF-κB activity — require teHSP90 function. Furthermore, we show that combinatorial targeting of teHSP90 and BCR signaling synergizes to inhibit the growth of activated B cell–like DLBCL (ABC DLBCL) cell lines, xenografts, and human patient samples.
Results
The teHSP90 oncoproteome in DLBCL cells.
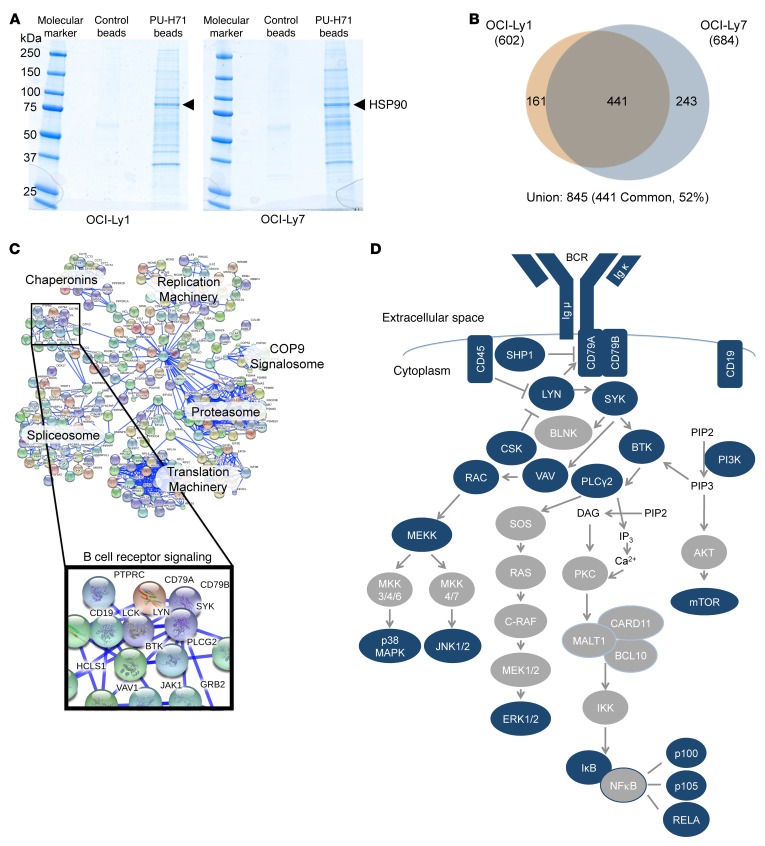
To interrogate the teHSP90 oncoproteome in DLBCL, we performed PU-H71 proteomics in DLBCL cells. We used agarose beads with either an inert chemical or beads that were covalently attached to PU-H71 to chemically precipitate teHSP90-client complexes in 2 DLBCL cell lines, OCI-Ly1 and OCI-Ly7. These complexes were resolved by polyacrylamide gel electrophoresis (Figure 1A) and analyzed by liquid chromatography followed by tandem mass spectrometry (LC-MS/MS). After subtracting common contaminants and the agarose proteome (7), we identified 602 and 684 teHSP90 client proteins in OCI-Ly1 and OCI-Ly7, respectively (Figure 1B and Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI80714DS1). Of the identified proteins, 52% were common to both cell lines.
Figure 1. teHSP90 pharmacoproteomics reveals multiple interactions with BCR pathway proteins.
(A and B) Lysates from OCI-Ly1 and OCI-Ly7 cells were subjected to chemical precipitation with PU-H71 or a control chemical followed by SDS PAGE and colloidal blue staining (A). The extracted proteins were examined by LC-MS/MS as shown in B. Venn diagram is used to illustrate the numbers of overlapping and unique proteins identified by LC-MS/MS in each cell line. (C) STRING representation of the union of proteins identified in OCI-Ly1 and OCI-Ly7 PU-H71 chemical precipitations at 0.99 confidence. Enriched protein networks are indicated by text, and the BCR pathway is magnified to reveal components captured by proteomics. (D) A representation of the BCR pathway is shown. Proteins with blue shading were identified as components of the teHSP90 interactome by LC-MS/MS.
To identify the signaling networks most highly represented among components of teHSP90 complexes, we analyzed the union of interacting proteins using the Search Tool for the Retrieval of Interacting Genes/proteins (STRING) (8) with a CI of 0.99. Among the expected clusters of proteins linked to stress signaling complexes, we identified components of the proteasome and ribosomal proteins. However, teHSP90 complexes were also highly enriched for proteins that form part of the BCR signaling pathway (Figure 1C). An orthogonal pathway analysis approach using ingenuity pathway analysis (IPA) also identified the BCR pathway as highly represented in the teHs90 DLBCL interactome (P = 0.01) (Supplemental Table 2). Among components of the BCR pathway present within teHSP90 stress complexes were the BCR itself, as well as many canonical components, including CD79, SYK, BTK, SHP1, PLCγ2, CD45, LYN, VAV, RAC, PI3K, mTOR, CSK, MEKK, JNK1/2, and several NF-κB components (Figure 1D).
teHSP90 complexes maintain protein expression of core components of the BCR pathway in DLBCL cells.
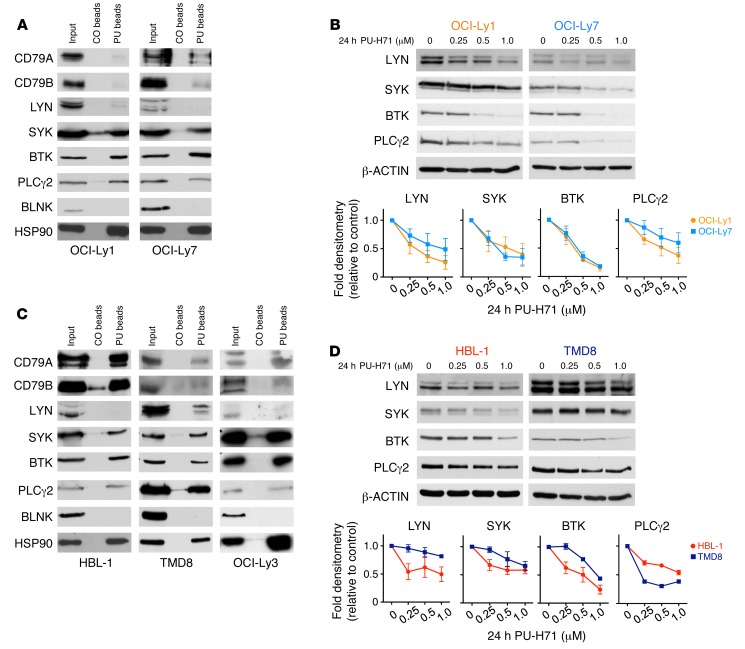
In order to validate teHSP90 pharmacoproteomics assays, we next performed PU-H71 chemical precipitations in OCI-Ly1 and OCI-Ly7 cells followed by immunoblot for many of the BCR pathway proteins identified by LC-MS/MS. All of these protein interactions were confirmed by chemical precipitation/immunoblot, demonstrating the robustness of the pharmacoproteomics assay. Additionally, B cell linker protein (BLNK), a scaffolding protein in the BCR pathway that was not present in the LC-MS/MS assays, failed to be captured as a component of teHSP90 complexes in these studies, further confirming the accuracy of the proteomic analysis (Figure 2A).
Figure 2. BCR pathway proteins interact with and require teHSP90 to maintain their expression.
(A and C) Lysates from OCI-Ly7 and OCI-Ly1 (A) or HBL-1, TMD8, and OCI-Ly3 (C) cells were subjected to chemical precipitation with PU-H71 (PU) or control chemical (CO) beads followed by immunoblotting for the indicated BCR pathway proteins. HSP90 is used as a positive control. (B and D) OCI-Ly1 and OCI-Ly7 (B) or HBL-1 and TMD8 (D) cells were exposed to vehicle or increasing doses of PU for 24 hours as indicated. Lysates were subjected to immunoblotting with the indicated antibodies. β-Actin was used as a loading control. The relative abundance of each protein relative to actin was quantified by densitometry for each dose and is shown in the line graphs as average ± SEM (n ≥ 3).
Classical HSP90-regulated client proteins depend on HSP90 function for their stability. To determine if the stability of the BCR proteins identified by LC-MS/MS was dependent on their association with teHSP90 complexes, we exposed OCI-Ly1 and OCI-Ly7 cells to increasing doses of PU-H71 for 24 hours and analyzed abundance of proximal enzymes of the BCR pathway by immunoblot. These proteins — LYN, SYK, BTK, and PLCγ2 — were depleted in a dose-dependent manner upon exposure to PU-H71, indicating their dependency on teHSP90 complexes to maintain their expression (Figure 2B).
The more aggressive, therapy-resistant ABC-like subtype of DLBCLs features chronic activation of BCR signaling, often due to the presence of somatic mutations of proteins in this pathway (9). As such, ABC DLBCLs are sensitive to targeted therapies that disrupt BCR signaling. We wondered whether these proteins were also associated with teHSP90 complexes in ABC DLBCL cell lines. If so, PU-H71 might serve as a candidate therapeutic agent for ABC DLBCLs. To test this, we next performed PU-H71 chemical precipitations followed by immunoblot for BCR pathway proteins in 3 ABC DLBCL cell lines, HBL-1, TMD8, and OCI-Ly3. We observed that the BCR proteins identified by LC-MS/MS in germinal center B cell–like DLBCL (GCB DLBCL) cell lines were also bound to teHSP90 complexes in these ABC DLBCL cells (Figure 2C). Moreover, SYK, BTK, and PLCγ2 proteins demonstrated dose-dependent expression reduction upon exposure to PU-H71 (Figure 2D). LYN was not present in teHSP90 complexes in HBL-1 cells. Accordingly, the effect of PU-H71 on LYN stability in these cells is minimal. PU-H71 also induced a minimal effect on LYN stability in TMD8 cells. Moreover, PU-H71 accelerated degradation of SYK and BTK in HBL-1 and OCI-Ly7 cells after protein translation blockade by cycloheximide, suggesting that the effect of PU-H71 is at least in part related to protein stability (Supplemental Figure 1A). In HBL-1 and TMD8, CD79B was more abundant in teHSP90 complexes than in the GCB DLBCL cell lines tested, which may be because it is mutated and constitutively active in these cell lines (9). Finally, we observed a time-dependent decrease in SYK and BTK protein abundance upon exposure to PU-H71 in OCI-Ly1, OCI-Ly7, HBL-1, and TMD8 cells (Supplemental Figure 1B).
We extended our studies to compare PU-H71 chemical precipitation in lysates of normal donor CD19+ cells isolated from peripheral blood to those performed in OCI-Ly1 cells. In CD19+ cells, PU-H71 chemical precipitation pulled down HSP90 and the client proteins SYK and BTK. However, these proteins were not enriched compared with input, whereas PU-H71 pulldowns in OCI-Ly1 cell lysates were enriched for HSP90 2-fold compared with input, and the clients SYK and BTK were enriched 2- and 3-fold, respectively (Supplemental Figure 2). Immunoprecipitations using an antibody to HSP90 in lysates of CD19+ cells and OCI-Ly1 cells effectively precipitated HSP90 but very little of the client proteins SYK or BTK compared with PU-H71 chemical precipitation (Supplemental Figure 2). These results are consistent with greater interactivity between teHSP90 and SYK or BTK in lymphoma cells compared with normal B cells.
PU-H71 disrupts activation of BCR signaling proteins.
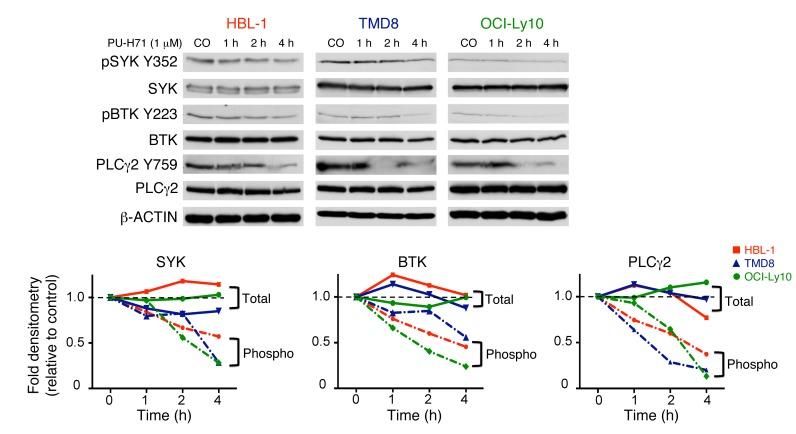
teHSP90 has been shown to mediate novel functions in protein physiology, independent of its effects on protein folding and stability (3, 5). For example, in B cell chronic lymphocytic leukemia (B-CLL), it maintains Lyn in an active conformation (10), whereas in chronic myelogenous leukemia (CML), it regulates the conformation of STAT5 to sustain elevated pSTAT5 species in the cell (5). Further, in DLBCL, teHSP90 was shown to maintain the BCL6 transcriptional repressor in a DNA-bound active conformation (3). We therefore hypothesized that teHSP90 is required for basal activity of the proximal enzymes of the BCR pathway independent of its function in maintaining protein expression. To test this notion, we treated 3 ABC DLBCL cell lines with PU-H71 at time points up to 4 hours, which is before reduction of protein abundance is observed. Immunoblots were performed for total and phosphorylated forms of SYK, BTK, and PLCγ2. Phospho-SYK, -BTK, and -PLCγ2 decreased in a time-dependent manner upon exposure to PU-H71, independent of total protein levels, in 3 ABC DLBCL cell lines (Figure 3).
Figure 3. teHSP90 maintains activity of BCR proteins in ABC DLBCLs.
HBL-1, TMD8, and OCI-Ly10 cells were exposed to vehicle or PU-H71 (1 μM) for increasing times as indicated. Lysates were subjected to immunoblotting with the indicated antibodies. β-Actin was used as a loading control. The relative abundance of each phosphorylated protein relative to each total protein was quantified by densitometry for each timepoint and is shown on the line graphs. Dotted lines, phospho. The relative abundance of each total protein relative to actin was quantified by densitometry for each timepoint and is shown in the line graphs. Solid lines, total.
We wondered if PU-H71 is required for LYN activity. While several Src-family kinases are known to initiate BCR signaling, LYN mediates negative feedback on BCR signaling through phosphorylation of receptor immunoreceptor tyrosine–based inhibition motifs (ITIMs) and a negative regulatory site on SYK (11). It has been suggested that, in order to facilitate chronic active BCR signaling, CD79 mutations arose in ABC DLBCL to attenuate the negative feedback mechanism by LYN (9). We examined both activating (Y396) and inhibitory (Y507) phosphorylation sites on LYN and found low levels of pLYN Y396 but high levels in pLYN Y507 in ABC DLBCL cell lines, suggesting that LYN is in an “off” state in these cells, consistent with chronic active BCR signaling. Moreover, exposure to PU-H71 induced loss of pLYN Y507 (Supplemental Figure 3). This loss of LYN inhibition is consistent with the decreased activity of downstream kinases upon exposure to PU-H71 that we observed.
teHSP90 is required for BCR signalosome function.
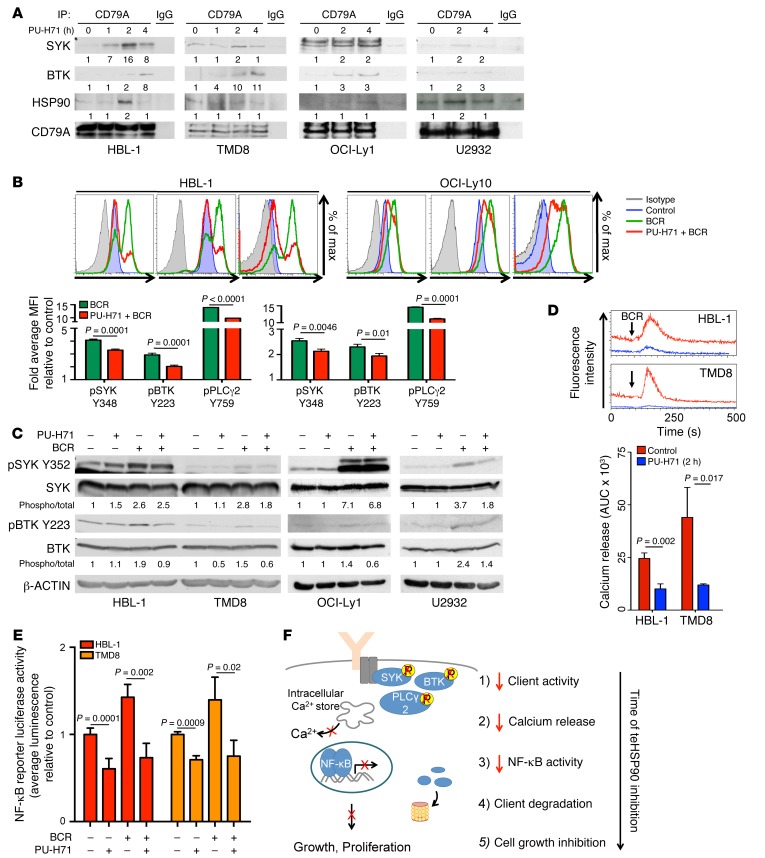
PU-H71 disrupts the dynamics of stress complexes by maintaining teHSP90 in the ATP-bound configuration attached to its partner proteins (5). Given the observed attenuation of SYK, BTK, and PLCγ2 phosphorylation upon PU-H71 exposure, we wondered whether teHSP90 was required for dynamic assembly of the BCR signalosome complex and whether PU-H71 would disrupt this effect. To address this question, we interrogated the BCR signalosome complex using coimmunoprecipitation assays in cells exposed to PU-H71 at the short timepoints where loss of SYK and BTK phosphorylation was observed independent of effects on total protein abundance. We observed a time-dependent increase in the amount of SYK and BTK coimmunoprecipitated with CD79A in cells exposed to PU-H71 (Figure 4A; inputs shown in Supplemental Figure 4B). The presence of HSP90 within the BCR signalosome was verified in these complexes by coimmunoprecipitation (Figure 4A; inputs shown in Supplemental Figure 4B). A similar effect was observed in OCI-Ly1 and U2932 cells, but SYK and BTK accumulated on CD79A to a lesser extent (Figure 4A; inputs shown in Supplemental Figure 4B). This PU-H71–induced accumulation of SYK and BTK on CD79A occurs at times when SYK and BTK exhibit reduced activity and when decreased association of the BCR with lipid rafts is observed, suggesting that this buildup of kinases on the BCR represents a loss of dynamic BCR complex function. Finally, using confocal microscopy and the Costes automated thresholding method (12), we observed a small but significant decrease in colocalization of the BCR with cholesterol-rich membrane (13–15) in cells treated with PU-H71 compared with vehicle (Supplemental Figure 4A). Because the BCR signalosome is most active in the context of membrane-lipid–rich regions (e.g., lipid rafts) following BCR ligation and aggregation (16), this decreased association is consistent with PU-H71–induced loss of SYK, BTK, and PLCγ2 activity. Collectively, these data suggest that teHSP90 contributes to BCR functionality by enabling dynamic interactions with its downstream mediators perhaps through localization of lipid-rich membrane structures.
Figure 4. Inhibition of teHSP90 induces broad attenuation of BCR signaling at multiple nodes.
(A) Lysates of HBL-1, TMD8, OCI-Ly1, and U2932 cells exposed to vehicle or PU-H71 (1 μM) were immunoprecipitated with antibodies to CD79A or IgG and immunoblotted with indicated antibodies. (B) HBL-1 and OCI-Ly10 cells treated with vehicle or PU-H71 (1 μM, 1 h) before BCR stimulation (IgM + IgG, 10 μg/ml, 15 min) were fixed, permeabilized, and stained with phospho-antibodies or isotype controls. Phospho-proteins were quantified using flow cytometry in 3 biological replicates using unpaired t test. (C) Lysates of HBL-1, TMD8, OCI-Ly1, and U2932 cells treated with vehicle or PU-H71 (1 μM, 1 h) before BCR stimulation (IgM + IgG, 10 g/ml, 15 min) were subjected to immunoblotting. (D) HBL-1 and TMD8 cells were treated with vehicle or PU-H71 (1 μM, 2 h) then incubated with a fluorescent calcium indicator (Fluo-4 AM, 2 μM, 30 min). Calcium release was measured over time by flow cytometry before and after BCR stimulation (IgM + IgG 10 μg/ml, 120 s). Three biological replicates; unpaired t test. (E) HBL-1 and TMD8 cells were transfected with an NF-κB luciferase reporter and treated with PU-H71 (1 μM) and/or BCR stimulation (IgM + IgG, 10 μg/ml, 16 hours). Luciferase activity was measured and is shown as average ± SEM relative to vehicle (n ≥ 3; unpaired t test). (F) Model of PU-H71 effects on BCR signaling at multiples nodes: BCR signalosome stability, kinase activity, calcium release, and NF-κB activity.
Signaling induced by BCR stimulation requires teHSP90 function.
To further demonstrate that accumulation of SYK and BTK on the BCR is associated with a loss of BCR signalosome complex function, we pretreated cells with PU-H71 for 1 hour before stimulating the BCR with IgM + IgG and then measured the activity of SYK, BTK, and PLCγ2 by intracellular multicolor phosphoflow cytometry. In HBL-1 cells pretreated with PU-H71, we observed a 20%–30% decrease in SYK, BTK, and PLCγ2 activation following BCR stimulation (P ≤ 0.0001, Figure 4B). In OCI-Ly10 cells pretreated with PU-H71, we observed a 15%–25% reduction in antigen-induced kinase activation (P ≤ 0.01, Figure 4B). We used phosphoimmunoblot — an orthogonal but less quantifiable assay — to corroborate this finding in HBL-1, TMD8, OCI-Ly1, and U2932 cells. In these assays, we observed some variability between cell lines tested. PU-H71 inhibited BCR-stimulated activation of SYK by 36% and by 50% in TMD8 and U2932 cells, respectively, although there was minimal reduction in HBL-1 and OCI-Ly1. PU-H71 more powerfully inhibited BCR-stimulated activation of BTK in all 4 cell lines tested: 53% in HBL-1, 60% in TMD8, 45% in OCI-Ly1, and 42% in U2932 (Figure 4C). To further interrogate the reliance of BCR signaling on teHSP90 function, we used a fluorescent calcium indicator (Fluo-4 AM) to measure calcium mobilization, a direct output of proximal BCR signaling, in DLBCL cells treated with vehicle or PU-H71. These experiments revealed profound suppression of BCR-induced calcium flux in HBL-1 and TMD8 ABC DLBCL cells pretreated with PU-H71 (P = 0.017 and 0.002, respectively, Figure 4D). We observed similar but less potent attenuation of calcium flux in OCI-Ly1 and OCI-Ly7 cells (Supplemental Figure 4C). To determine whether impairment in BCR signaling was associated with downstream effects on NF-κB signaling, we measured basal and BCR-induced NF-κB activity in the presence and absence of PU-H71 in the Ramos-blue cell line engineered to express an NF-κB responsive reporter encoding alkaline phosphatase. In cells treated with PU-H71, we observed a significant decrease in NF-κB reporter activity (P = 0.02) and an even more profound suppression of NF-κB activity after BCR stimulation of these cells (P = 0.01, Supplemental Figure 4D). To interrogate the reliance of NF-κB signaling downstream of BCR signaling on HSP90 function in ABC DLBCL cells, we transfected HBL-1 and TMD8 cells with an NF-κB luciferase reporter and measured basal and BCR-induced NF-κB activity in the presence and absence of PU-H71. In HBL-1 and TMD8 cells treated with PU-H71, we observed a 40% and 30% decrease in NF-κB reporter activity (P = 0.0001, P = 0.0009, respectively) under basal conditions, with an even greater average inhibition of NF-κB activity following BCR stimulation (P = 0.002 HBL-1, P = 0.02 TMD8, Figure 4E). Collectively, these data reveal a critical role for teHSP90 stress complexes in BCR signaling. In the presence of PU-H71, the BCR exhibits deficient association with lipid rafts, while SYK and BTK accumulate on CD79A, the signaling moiety of the BCR. Due to these effects and possibly due to additional effects on downstream components, there is loss of signalosome kinase function, calcium flux, and NF-κB activity (Figure 4F).
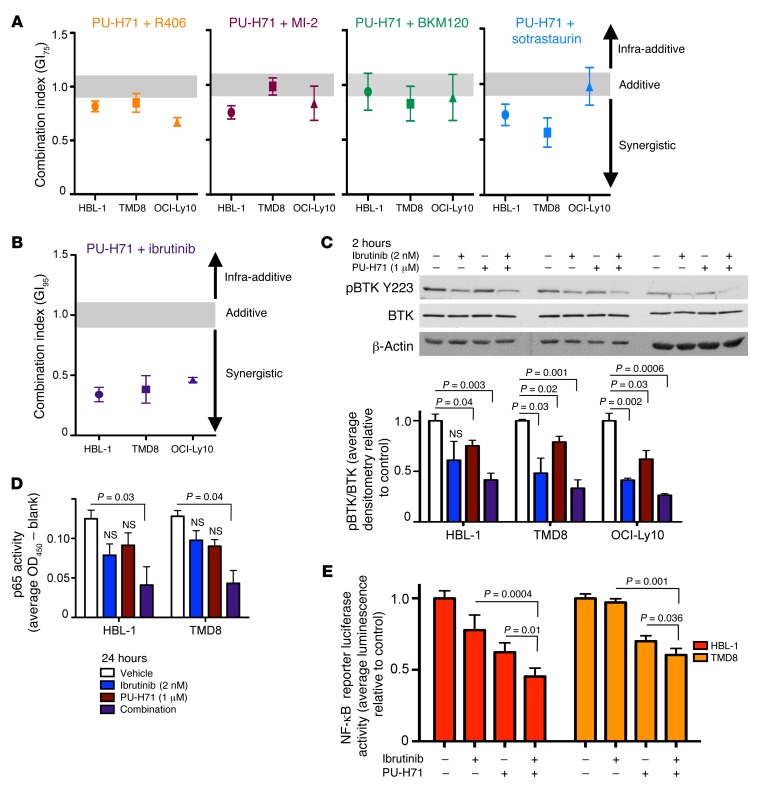
PU-H71 and inhibitors of BCR signaling synergize to inhibit growth of DLBCL.
For many therapeutic targets in cancer, it is imperative to achieve a high degree of suppression for maximal antitumor effect. Complex signaling networks like the BCR pathway tend to branch out, making their complete suppression a challenge. Moreover, complete suppression of the target of a specific inhibitor must be balanced against its off-target effects, which can become increasingly problematic with higher doses that may be needed for therapeutic effect. PU-H71 is different in its mechanism of action, which as shown above consists of broad attenuation of BCR signaling at multiple steps. Based on this notion, we hypothesized that combinatorial exposure of ABC DLBCL cells to the BCR attenuating activity of PU-H71, together with drugs that specifically target BCR components, would result in additive or synergistic cell growth inhibition. To test this idea, we treated 3 ABC DLBCL cell lines with PU-H71 in combination with inhibitors of BCR pathway proteins — including ibrutinib (BTK), R406 (SYK), BKM120 (PI3K), MI-2 (MALT1), and sotrastaurin (PKCβ) — and measured their effects on cell viability using a luminescent ATP-based assay. We calculated combination effect using the algorithm of Chou and Talalay (17). All of these combinations showed synergistic (combination index [CX] value below 0.9) or additive (CX value between 0.9 and 1.1) effect at concentrations sufficient to inhibit growth 75% (GI75, Figure 5A). The most potently synergistic combination was PU-H71 and ibrutinib, with marked synergy observed in all 3 cell lines (CX < 0.4, GI95, Figure 5B). The CARD11 mutant ABC DLBCL cell line OCI-Ly3 does not respond to ibrutinib. In viability assays with these cells, we observed that the combination of ibrutinib with PU-H71 did not inhibit cell growth more than PU-H71 alone (Supplemental Figure 5).
Figure 5. PU-H71 and ibrutinib synergize to inhibit ABC DLBCL growth through maximal inhibition of BCR signaling.
(A and B) HBL-1, TMD8, and OCI-Ly10 cells were treated with a dose curve of BCR pathway inhibitors indicated, PU-H71, or the combination. Cell viability was measured using a luminescent ATP method and normalized to vehicle-treated controls. Synergy was assessed using Compusyn. CX values < 0.9 are synergistic. Mean ± SEM; 3 biological replicates. (C) HBL-1, TMD8, and OCI-Ly10 cells were treated with vehicle, ibrutinib (2 nM), PU-H71 (1 μM), or the combination for 2 hours. Lysates were subjected to immunoblotting with the antibodies indicated. Relative protein abundance was quantified using densitometry. Data are presented as mean ± SEM; unpaired t test. (D) HBL-1 and TMD8 cells were treated with vehicle, ibrutinib (2 nM), PU-H71 (1 μM), or the combination for 24 hours. Nuclear extracts were probed for p65 DNA binding activity with TransAM ELISA. Data are presented as mean ± SEM (n = 3; unpaired t test). (E) HBL-1 and TMD8 cells were transfected with an NF-κB luciferase reporter and treated with PU-H71 (1 μM), ibrutinib (2 nM), or the combination for 16 hours. Luciferase activity was measured and is shown as average ± SEM relative to vehicle (n ≥ 3, unpaired t test).
We tested these drug combinations in 2 GCB DLBCL cell lines, OCI-Ly1 and OCI-Ly7. PU-H71 combined with R406 or BKM120 induced additive or weakly synergistic growth inhibition in these cells. Because these cells are resistant to ibrutinib and sotrastaurin, we could not calculate synergistic effect of these combinations. However, we observed that the combination treatment did not inhibit cell growth more than PU-H71 alone (Supplemental Figure 5).
As outlined in our hypothesis, we wondered if the combinatorial effect of PU-H71 with ibrutinib was linked to more powerful inhibition of BTK activity. To test this, we exposed 3 independent ABC DLBCL cell lines to PU-H71, ibrutinib, or the combination for 2 hours and measured BTK activity by phospho-immunoblot. The combination of PU-H71 and ibrutinib resulted in an additive inhibition of BTK activity (Figure 5C). To determine if the PU-H71–ibrutinib combination more potently suppresses BCR signaling downstream of BTK activity, we used p65 ELISA to measure NF-κB activity in nuclear lysates of ABC DLBCL cells treated with PU-H71, ibrutinib, or the combination for 24 hours. At the doses tested (average GI50 values), single drugs induced a modest reduction in p65 activity. In contrast, the PU-H71–ibrutinib combination significantly decreased p65 activity by 60% in HBL-1 and TMD8 cells in an additive manner (P = 0.03, P = 0.04, respectively, Figure 5D). To further interrogate the effect of the PU-H71–ibrutinib combination on NF-κB activity, we transfected HBL-1 and TMD8 cells with an NF-κB luciferase reporter construct and measured NF-κB activity in cells treated with PU-H71, ibrutinib, or the combination. We observed that the combination treatment decreased NF-κB activity more significantly than either drug alone (Figure 5E).
Ibrutinib enhances PU-H71 antilymphoma effect in vivo.
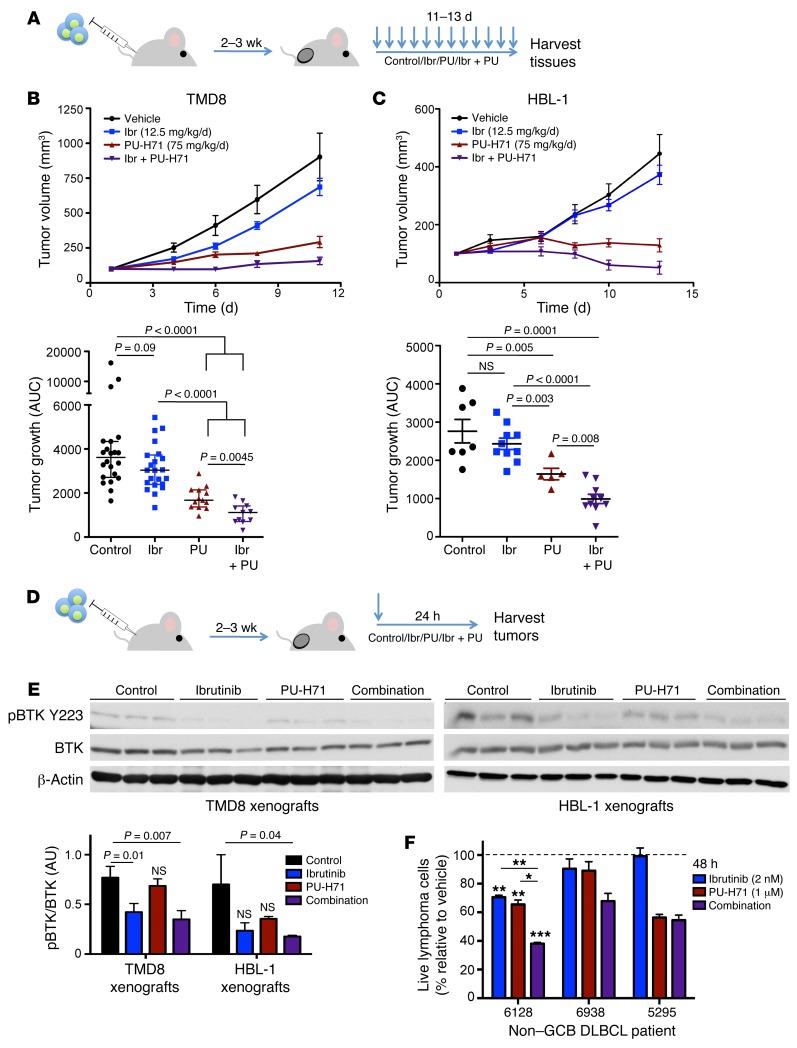
The PU-H71–ibrutinib drug combination is particularly suited for clinical translation because PU-H71 is in clinical trials and ibrutinib is an FDA-approved drug, prompting us to further investigate this therapeutic combination in ABC DLBCL in vivo. Importantly, we have shown that the concentrations of PU-H71 determined here to inhibit BCR signaling components (~0.5–1 μM) are readily achieved in the tumors of human patients using well tolerated doses in our phase I studies (18). We established HBL-1 and TMD8 xenografts in non-obese diabetic/severe combined immunodeficiency (NOD-SCID) mice. After tumors reached an average of 100 mm3 in volume, mice were randomized to receive vehicle, ibrutinib, PU-H71, or the combination. TMD8 and HBL-1 mice were treated daily, with 12.5 mg/kg ibrutinib (ad libitum), 75 mg/kg PU-H71 (i.p. injection), or the combination for 11–13 days (Figure 6A). As a single agent, ibrutinib inhibited the growth of both TMD8 and HBL-1 xenograft models, but this difference was not statistically significant as compared with vehicle. PU-H71 significantly reduced tumor growth in both TMD8 and HBL-1 xenografts (P < 0.0001, and P = 0.003, compared with vehicle). The PU-H71–ibrutinib combination suppressed TMD8 and HBL-1 xenograft growth significantly more than ibrutinib alone (P < 0.0001) or PU-H71 alone (P = 0.0045 and P = 0.008, respectively, Figure 6, B and C).
Figure 6. PU-H71 potentiates response to ibrutinib in ABC DLBCL in vivo.
(A) NOD-SCID mice were s.c. injected with 106 HBL-1 or TMD8 cells. When tumors were palpable, daily treatment began. (B and C) Tumor growth plots of TMD8 (B) and HBL-1 (C) xenografted mice treated with vehicle (saline), ibrutinib (Ibr; 12.5 mg/kg in chow, ad libidum), PU-H71 (PU; 75 mg/kg i.p. injection) or the combination (purple triangles). Right panels: Growth of each tumor was measured as area under the curve. Average tumor growth is represented on the y axis, which represents tumor volume (mm3)/time (days). Mean ± SEM, Mann-Whitney U test. (D) NOD-SCID mice were xenografted and treated as described above for 24 hours. (E) Lysates of tumors harvested from experiments in D were subjected to immunoblot with indicated antibodies. Relative abundance of phospho-BTK to total BTK was quantified using densitometry. Mean ± SD, unpaired t test. (F) Human non–GCB DLBCL patient samples cultured on an irradiated HK cell feeder layer were exposed to control, ibrutinib (2 nM every 24 hours), PU-H71 (1 μM), or the combination for 48 hours. Cell viability (represented as percentage of vehicle-treated cells) measured using flow cytometry is plotted on the y axis. Live cells are defined as CD20+/CD3– cells that are Annexin V/DAPI double negative. Sample 6128 was tested in 2 biological replicates, unpaired t test. Other samples were tested in 2 technical replicates. Data are presented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.0005.
In order to determine if the combination effect observed in vivo was linked to more potent inhibition of BTK, as we had observed in cell lines, we established new HBL-1 and TMD8 xenografts in NOD-SCID mice (n = 3/group) and administered a single dose of vehicle, ibrutinib, PU-H71, or the combination. Mice were sacrificed 24 hours after treatment, and tumors were harvested to assay for BTK activity by phospho-immunoblot (Figure 6D). In TMD8 xenografts, the single dose of PU-H71 had little effect on BTK phosphorylation. While ibrutinib reduced active BTK levels by 45% (P = 0.01), the PU-H71–ibrutinib combination significantly suppressed BTK phosphorylation by 55% (P = 0.007). In HBL-1 xenografts, we observed modest inhibition of BTK in both single-agent treatments, but these effects were not statistically significant. Once again, in animals treated with the PU-H71–ibrutinib combination, we observed a 75% inhibition of active BTK in their HBL-1 ABCL-DLBCL tumors (P = 0.04, Figure 6E). Taken together, the PU-H71–ibrutinib combination therapy results in enhanced antilymphoma effect in vivo, and pharmacodynamic studies indicate this was associated with more potent suppression of BTK activity.
To determine if the PU-H71–ibrutinib drug combination is toxic, we treated C57 Black mice (n = 3/group) for 12 days with vehicle, ibrutinib (12.5 mg/kg ad libitum), PU-H71 (75 mg/kg i.p. injection), or the combination. A second cohort of mice (n = 3/group) were maintained without drug exposure for an additional 14 days to serve as a washout comparison group for any observed toxicity in the treatment group (Supplemental Figure 6). Animals were weighed daily and sacrificed 24 hours after the last treatment, followed by a comprehensive histopathology and molecular analysis. There was no evidence of hematologic, renal, or hepatic toxicity, as determined by blood counts, blood chemistry, and liver-function testing (Supplemental Table 3). No microscopic evidence of toxicity was observed in the intestines, kidneys, spleens, livers, lungs, or hearts harvested from any of the mice (data not shown). The BM of control and ibrutinib-treated mice revealed no specific pathological findings, although there was evidence of hypocellularity in 1/3 PU-H71–only and 2/3 combination-treatment mice. BM in the PU-H71 and combination-treated mice in the washout group revealed normocellular BM, demonstrating that the observed toxicity is reversible when the drug is removed.
PU-H71 and ibrutinib combination yields enhanced killing of human non–GCB DLBCL patient samples ex vivo.
Whereas lymphoma cell lines have been used extensively for preclinical studies, it is important to underline that these cells may not be fully representative of primary human tumors. As part of an ongoing research protocol at Weill Cornell Medical College, we were able to obtain 3 viable primary non–GCB DLBCL specimens classified by Hans IHC criteria (19). These patient specimens where plated on a feeder layer of irradiated HK cells and treated with vehicle, 2 nM ibrutinib, 1 μM PU-H71, or the combination for 48 hours. Cell viability was measured by flow cytometry scoring CD20+ B cells for annexin V and DAPI to determine their viability. In patient sample 6128, the PU-H71–ibrutinib combination treatment produced significantly enhanced lymphoma-killing effect. In non–GCB DLBCL patient sample 6938, the PU-H71–ibrutinib combination resulted in even more potent killing relative to each drug alone, although statistics could not be generated since there were insufficient cells to perform multiple replicates (Figure 6F). Other samples tested were not sensitive to ibrutinib, and the combination treatment was not more effective than PU-H71 alone, similar to the CARD11 mutant ABC DLBCL cell line OCI-Ly3. The efficacy of this drug combination in ABC DLBCL in vitro, in vivo, and human patient samples ex vivo supports the rationale for translation of this combination therapy to human patients.
Discussion
In this work, we expand the scope of previously known HSP90 functions to include binding and facilitating signaling through the BCR signalosome in the clinically relevant context of ABC DLBCLs. This function is linked to a tumor-enriched class of HSP90 complexes that can be selectively targeted using the small molecule PU-H71. PU-H71 binds tightly to teHSP90 complexes in an ATP-competitive manner (5), causing teHSP90 to become locked in a partner protein–bound configuration (5). This highly stable binding enables enrichment of the teHSP90 proteome from cells using PU-H71 pulldowns (5). HSP90 immunoprecipitation brings down the same proteins but does not distinguish between those proteins contained in stress complexes versus those that are bound to housekeeping fractions of HSP90 (5). Hence, PU-H71 pharmacoproteomics can be used to specifically capture a unique functional class of HSP90 complex that exhibits greater association with biochemical effects specifically required by tumor cells to maintain their growth and survival (5, 20). Our current data confirm and underline that teHSP90 stress complexes carry out specific biochemical functions beyond the classical protein stability and folding functions attributed to HSP90.
Consistent with the locking-in effect of PU-H71, exposure of DLBCL cells to drug resulted in more stable association of CD79 with BTK and SYK, as well as disruption of the association of BCR complexes with lipid rafts. Collectively, these data are consistent with the notion that teHSP90 in some way is required to maintain the proper configuration of the BCR signalosome. BCR components must dynamically recycle their configuration during signaling, and teHSP90 may facilitate these dynamics. Clearly, in depth structural studies will be required in the future to understand these mechanisms. It has been noted, however, that CD79 mutations maintain surface BCR expression in ABC DLBCLs (9), consistent with their chronic active BCR status. Actual membrane-to-cytoplasm vesicle recycling of the BCR is only mildly perturbed by PU-H71 (data not shown). Hence, PU-H71 actions might be more intimately associated with perturbation of membrane complex protein interaction dynamics.
HSP90 isoforms are known to associate with membrane-bound proteins such as the BCR. For example, HSP90N has been shown to contain a putative myristylation sequence, which may explain its membrane localization (21). In T cells, HSP90 is required for the membrane association of the Src family kinase LCK (22). Activating mutations in LCK increase its dependence on HSP90 (23). CD79, the signaling moiety of the BCR, is often mutated in ABC DLBCLs, resulting in chronic activation of the pathway (9). Though the enzymes of BCR signaling — SYK, BTK, and PLCγ2 — do not harbor activating mutations, they form the BCR signalosome with CD79 and exhibit increased activity in the ABC DLBCLs. The biochemical dependence of the BCR signalosome on teHSP90 is consistent with the previously shown requirement of teHSP90 to maintain signaling pathways aberrantly activated in human leukemia and solid tumor cells (5). Notably, teHSP90 also plays a key role in maintaining viral proteins linked to virally induced lymphomagenesis (20). One minor point to note is that teHSP90 also associated with BCR components in GCB-type DLBCL cells. It has been previously shown that these cells exhibit tonic BCR activity and can be targeted by signaling inhibitors, such as SYK inhibitors (24). Hence, it is possible that teHSP90 is important both for tonic BCR activity in lymphoma cells and chronic active signaling that is more characteristic of ABC DLBCLs.
An important concept emerging from this work is the notion of teHSP90 as a general facilitator of BCR signaling. Indeed, our pharmacoproteomics assay indicated extensive interaction of teHSP90 with BCR signaling proteins at multiple levels and throughout multiple branches of the BCR signaling network. These findings correspond with the overall attenuation of BCR signaling, calcium flux, and NF-κB activity induced by PU-H71. The BCR pathway is well appreciated as a therapeutic target in ABC DLBCLs. Targeting with selective inhibitors against BTK, SYK, or MALT1 can suppress the growth of ABC DLBCL cells. While potentially effective, these treatments may not globally suppress all aspects of BCR signaling. Moreover, tumors can become resistant to drugs such as ibrutinib through acquisition of somatic mutations in BTK and PLCG2. Despite its rapid translation, a phase II clinical trial of ibrutinib monotherapy in ABC DLBCL patients revealed only a 41% overall response rate (25).
Based on these notions, we propose clinical translation of regimens that combine the BCR attenuator effects of PU-H71 discovered through our pharmacoproteomics approach together with the more punctual and incisive specific targeted approach represented by ibrutinib or similar agents. Indeed, the combination of PU-H71 was highly synergistic when combined with ibrutinib in cell lines, as well as in animal models and against primary human DLBCL cells. Synergy between these drugs was associated with more potent suppression of BTK and NF-κB activation. Naturally, PU-H71 suppression of other teHSP90 actions could also contribute to these synergistic effects, such as its effects on BCL6 and IKK (3, 26). If anything, this adds to the potential appeal of using PU-H71 to anchor combinatorial therapy for DLBCL. In further confirmation of these data, a recent high throughput combinatorial screening study identified HSP90 inhibitors as one of several classes of drugs that cooperate with ibrutinib to kill ABC DLBCLs (27). Finally, it is important to underline the lack of toxicity observed in animals exposed to the combination of PU-H71 and ibrutinib, which suggests this combination would be well tolerated in humans. PU-H71 is currently completing phase I testing in humans and will be available for phase II studies as a single agent or for rational combination therapy. We expect such studies to yield improved efficacy in humans compared with single BCR agent–targeted therapy.
Methods
Cell lines and reagents.
The DLBCL cell lines OCI-Ly1 and OCI-Ly7 were grown in Iscove’s Modified Dulbecco’s Medium (IMDM) containing 10% FBS and supplemented with penicillin/streptomycin. OCI-Ly10 was grown in IMDM with 20% FBS and penicillin/streptomycin. The DLBCL cell lines HBL-1, TMD8, and U2932 were grown in medium containing 90% RPMI and 10% FBS, supplemented with L-glutamine, HEPES, and penicillin/streptomycin. The cell line OCI-Ly3 was grown in medium containing 80% RPMI and 20% FBS, supplemented with L-glutamine, HEPES, and penicillin/streptomycin. CD19+ cells were purchased from AllCells. PU-H71 was synthesized as previously reported (5). Ibrutinib was provided by Pharmacyclics. R406, BKM120, and sotrastaurin were purchased from Selleck Chemicals. MI-2 was purchased from Albany Molecular Research Inc. Filipin and cycloheximide were purchased from Sigma-Aldrich. All human IgM and IgG were purchased from Jackson ImmunoResearch Laboratories Inc. A list of antibodies used is included in the Supplemental Material (Supplemental Table 4).
PU-H71 chemical precipitation.
PU-H71 and control (HSP90 inactive chemical [2-methoxyethlamine]) beads were synthesized as previously reported (5). Before use, PU-H71 or control beads were washed 3 times in Felts buffer (20 mM HEPES, 50 mM KCl, 5 mM MgCl2, 0.01% [w/v] NP-40, freshly prepared 20 mM Na2MoO4, and protease inhibitors; Roche Diagnostics). For cell lysis, 40 million (immunoblot) or 100 million (proteomics) cells were incubated in Felts buffer on ice for 30 minutes, then vortexed for 15 seconds and centrifuged at 14,000 rpm for 15 minutes at 4°C. Supernatants were precleared by incubating with 100 μl of washed control beads for 1 hour rocking at 4°C. For proteomics, lysates were precleared for 8 hours and again with fresh beads overnight. Following preclearing, half of each sample was incubated with 100 μl of either control or PU-H71 beads at 4°C overnight. Samples with PU-H71 beads were incubated in amber eppendorf tubes. Following incubation, bead conjugates were washed 3 times with Felts buffer and resolved by PAGE, followed by standard immunoblotting procedure. For proteomics, gels were stained with colloidal blue, cut into bands under sterile conditions, frozen on dry ice, and analyzed by MS.
MS.
In-gel trypsin-mediated digestion was carried out using standard procedures. Tryptic peptides were resolved on a nano-capillary reverse phase column and directly infused into a linear ion-trap mass spectrometer (LTQ Orbitrap XL, Thermo Electron Corp.). The mass spectrometer was set to collect one survey scan (MS1), followed by MS/MS spectra on the 9 most intense ions observed in an MS1 scan. Proteins were identified by searching the tandem mass spectra against human protein database using the X!Tandem/TransProteomic Pipeline software suite. All proteins identified with a probability of ≥0.9 were retained for further analysis.
Cell lysis for immunoblot.
Cells were treated at a concentration of 2 × 106/ml, harvested, washed with PBS, and resuspended in lysis buffer (10 mM HEPES-KOH pH 7.9, 1.5 mM MgCl2, 10 mM KCl, freshly prepared 1.5 mM Na2MoO4, 1% Triton-X, protease and phosphatase inhibitors, and 0.5 mM PMSF).
IP.
Cells were plated at a concentration of 2 × 106/ml and treated with vehicle or PU-H71. Cells were harvested, washed with PBS, and lysed in IP Buffer (25 mM HEPES pH 7.5, 150 mM NaCl, 0.2% NP-40, 10% glycerol, 1 mM DTT, 20 mM Na2MoO4, and protease inhibitors) for 30 minutes on ice, and then centrifuged at high speed for 15 minutes at 4°C to remove insoluble proteins. Protein concentration of supernatants was measured using the DC Protein Assay Kit (Bio-Rad). Input (10 μg) was saved at –20°C. For each IP, 1 mg of protein was precleared for 1 hour at 4°C with Protein G Sepharose beads (Sigma-Aldrich) washed in IP buffer. Precleared lysates were precipitated with 10 μg of either αCD79A (clone HM47, Santa Cruz Biotechnologies Inc.) or mouse IgG (catalog ab14813, Abcam) tumbling overnight at 4°C. The next day, 20 μl of washed, blocked Protein G Sepharose beads were added to immunoprecipitates and tumbled for 1 hour at 4°C. Beads were then washed 3 times in IP buffer, resuspended in 2× SDS loading buffer, and boiled for 5 minutes. SDS loading buffer (5×) was added to inputs and boiled for 5 minutes. Inputs and immunoprecipitates were resolved by standard immunoblot procedure. For HSP90 IPs, 10 μg of antibody (clone H9010, StressMarq Biosciences Inc.) was used to precipitate cell lysates. For assays with CD19+ cells, 275 μg of lysate was used per IP or CP, and 25 μg was used as input.
BCR-lipid raft colocalization.
Cells were treated with vehicle or PU-H71 for 4 hours. After washing with PBS, cells were fixed with 4% paraformaldehyde/0.1% glutaraldehyde/1× PBS for 20 minutes at room temperature. Cells were stained with 10 μg/ml F(ab′)2 fragment goat anti-human IgM (catalog 109-006-129, Jackson ImmunoResearch Laboratories Inc.) in 0.1% bovine serum albumin and 0.02% sodium azide in PBS for 20 minutes at room temperature. After washing with PBS, cells were stained with Alexa Fluor 647 donkey anti-goat IgG (catalog A21447, Invitrogen), washed with PBS, and incubated with 50 μg/ml filipin (catalog 70440, Cayman Chemical) in PBS for 1 hour at room temperature. Cells were then affixed to slides by cytocentrifugation at 1,300 rpm for 5 minutes and mounted with ProLong Gold antifade reagent (Invitrogen). Cells were imaged at ×60 using the Fluoview FV10i confocal microscope (Olympus) and analyzed for colocalization using the JACoP plugin (12) with ImageJ (NIH).
Multicolor intracellular phosphoflow cytometry.
Cells were treated with vehicle or 1 μM PU-H71 for 1 hour, then with 10 μg/ml IgG + IgM (catalog 309-005-107, Jackson Immunoresearch Laboratories Inc.) for 15 minutes. Cells were fixed for 15 minutes at 37°C by directly adding BD Phosflow Fix Buffer I (BD Biosciences). Cells were harvested, centrifuged at 1,500 rpm for 5 minutes, and permeabilized in BD Phosflow Perm Buffer III (BD Biosciences) for 30 minutes on ice. After the addition of wash/stain buffer (1% FBS, 0.09% NaN3 in 1× PBS), cells were centrifuged at 1,500 rpm for 5 minutes at 4°C. Each sample was divided for staining in equal amounts of either fluorescent-conjugated specific-phospho antibodies or appropriate isotype controls in wash/stain buffer on ice. Antibodies are listed in supplementary material. Data was acquired on MacsQuant Flow Cytometer (Miltenyi Biotec) and analyzed using FlowJo software (Tree Star Inc.).
Calcium release.
Cells were treated with vehicle or 1 μM PU-H71 for 2 hours, then incubated with 2 μM Fluo-4 AM (catalog F14217, Invitrogen) for 30 minutes at 37°C. Fluorescent indicator was washed out of cells with fresh media, and cells were recovered for 30 minutes at 37°C. Cells were collected in PBS without calcium, and fluorescence was measured on an LSR-II flow cytometer (BD Biosciences) for 2 minutes; then, IgG + IgM (10 μg/ml) (catalog 309-005-107, Jackson Immunoresearch Laboratories Inc.) was added and fluorescence measurement continued for at least 5 more minutes. Calcium release was quantified using FlowJo software as area under the curve for the first 4 minutes.
NF-κB activity.
In HBL-1 and TMD8 cells, 2 μg of (NF-κB)5-Luc2CP-pGL4 plasmid per 107 cells was transfected using nucleofection (Amaxa Biosystems, Lonza). Eight hours after transfection, cells were plated at a density of 2 × 106/ml and treated overnight as indicated. Lysates were submitted to luciferase assay following manufacturer’s protocol (Promega). Ramos-Blue cells were grown and assayed according to manufacturer’s instructions (InvivoGen). For TransAM p65 ELISA, nuclear extracts were prepared and p65 ELISA was performed according to manufacturer’s instructions (Active Motif).
Cell viability assays and synergy experiments.
DLBCL cell lines were plated in 384-well plates at concentrations sufficient to keep untreated cells in exponential growth during the time of drug exposure. Cells were treated with 6 doses of each drug or combination in triplicate. For combination treatments, cells were exposed to a dose curve of each drug or their combination in constant ratio. Cell viability was determined by either a fluorometric resazurin reduction method (CellTiter-Blue, Promega) or an ATP luminescent method (CellTiter-Glo, Promega). Fluoresence (560excitation/590emission) or luminescence was measured with the Synergy4 microplate reader (BioTek). Cell viability of drug-treated cells was normalized to vehicle-treated controls. Compusyn software (Biosoft) was used to plot dose-effect curves and calculate CX values.
Xenografts.
Eight-week-old NOD-SCID mice were injected with 106 cells in 1:1 PBS/matrigel. Tumors grew until palpable (2–3 weeks) and then treatment began. Mice were injected i.p. with vehicle or 75 mg/kg PU-H71 daily. Ibrutinib was administered via chow at a dose of 12.5 mg/kg/day ad libitum. Control and PU-H71–treated mice were given vehicle chow. Tumors were measured with digital calipers, and animals were weighed every other day. Animals were sacrificed after treatment, and tumors were weighed and harvested for molecular investigation (IHC, Western blot).
Toxicity.
Eight-week-old C57 Black 6 mice were treated with vehicle, ibrutinib (12.5 mg/kg/day ad libitum), PU-H71 (75 mg/kg/day i.p. injection), or PU-H71–ibrutinib combination for 12 days. Animals were weighed every other day. After 12 days, 3 animals per group were sacrificed, and organs (intestine, spleen, kidney, liver, lung, heart, and BM) were harvested. Tissues were fixed in 4% formalin, washed in 70% ethanol, and sent for sectioning and staining with H&E. Stained tissues were microscopically analyzed by specialized pathologists. Blood was also collected at the time of sacrifice for complete blood count test and chemical analysis.
Primary cell treatment.
We obtained deidentified human tissues in accordance with and approval from the Institutional Review Board of the New York Presbyterian Hospital, New York, New York. Samples were classified as non–GCB DLBCL using Hans classification (19) (IHC for BCL6, CD10, and MUM1, with 30% cutoff for positivity). Single-cell suspensions from lymph node biopsies were thawed and resuspended in Advanced RPMI supplemented with 20% human serum, GlutaMAX (2×), glycine (5 mM), and penicillin/streptomycin. Cell number and viability was determined by counting with trypan blue. Irradiated HK cells (2,000 rad) in DMEM supplemented with 10% FBS and penicillin/streptomycin were adhered to tissue culture plates at 37°C. Media was aspirated, and patient samples were plated on the HK feeder layer. Samples were exposed to different drugs or vehicle for 48 hours and stained with FITC-conjugated anti-CD20, PE-Cy7–conjugated anti-CD3, and APC-conjugated anti–annexin V anti-human antibodies (BD Biosciences). DAPI was used for the exclusion of dead cells. Data was acquired on MacsQuant Flow Cytometer and analyzed using FlowJo software package. CD20+/CD3– cells that were Annexin V/DAPI double-negative were considered live.
Statistics.
Two-tailed unpaired t test was used unless otherwise stated. Specifically, 2-tailed Mann-Whitney U test was used for xenograft growth experiment analysis. All statistical analyses were carried out using Prism software (GraphPad Software).
Study approval.
Deidentified patient tissues were obtained in accordance with and approval from the Institutional Review Board (IRB) of the Weill Cornell Medical College. The Research Animal Resource Center of the Weill Cornell Medical College approved all mouse procedures.
Author Contributions
RLG, SNY, GC, LC, and AM designed experiments. RG and SNY conducted experiments. RG, SNY, JG, GC, LC and AM analyzed data. TT and GC provided PU-H71 and PU-H71 beads. BC provided ibrutinib and ibrutinib chow. WT and JPL provided human patient samples. KE-J acquired and analyzed MS data. RS analyzed histopathological samples. The manuscript was written by RG, LC, GC and AM and edited by SNY, TT, BC, WT, JPL, KEJ, RS and JG.
Supplementary Material
Acknowledgments
We would like to thank the members of the Melnick lab for their support and constructive discussions, and Cihangir Duy and Tharu Fernando for providing technical assistance. R.L. Goldstein is funded by NRSA 5 F31 CA174239-02. A. Melnick and G. Chiosis are funded by R01 CA155226. G. Chiosis is funded by U01 AG032969, R01 CA172546, and R21 CA158609. L. Cerchietti is supported by the Malvin Peace Sevin Research Scholar Award and the Irma T. Hirschl Career Scientist Award, and is the Raymond and Beverly Sackler Scholar and Scholar of the American Society of Hematology. A. Melnick is a Burroughs Wellcome Clinical Translational Scientist.
Footnotes
Conflict of interest: Memorial Sloan-Kettering Cancer Center holds the intellectual rights to PU-H71. Samus Therapeutics, of which Gabriela Chiosis has partial ownership, has licensed PU-H71.
Reference information:J Clin Invest. 2015;125(12):4559–4571. doi:10.1172/JCI80714.
References
- 1.Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100(17):9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coiffier B, et al. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(4):235–242. doi: 10.1056/NEJMoa011795. [DOI] [PubMed] [Google Scholar]
- 3.Cerchietti LC, et al. A purine scaffold Hsp90 inhibitor destabilizes BCL-6 and has specific antitumor activity in BCL-6-dependent B cell lymphomas. Nat Med. 2009;15(12):1369–1376. doi: 10.1038/nm.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caldas-Lopes E, et al. Hsp90 inhibitor PU-H71, a multimodal inhibitor of malignancy, induces complete responses in triple-negative breast cancer models. Proc Natl Acad Sci U S A. 2009;106(20):8368–8373. doi: 10.1073/pnas.0903392106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moulick K, et al. Affinity-based proteomics reveal cancer-specific networks coordinated by Hsp90. Nat Chem Biol. 2011;7(11):818–826. doi: 10.1038/nchembio.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mollapour M, et al. Asymmetric Hsp90 N domain SUMOylation recruits Aha1 and ATP-competitive inhibitors. Mol Cell. 2014;53(2):317–329. doi: 10.1016/j.molcel.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moorhead GB, et al. Displacement affinity chromatography of protein phosphatase one (PP1) complexes. BMC Biochem. 2008;9: doi: 10.1186/1471-2091-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen LJ, et al. STRING 8 — a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database issue):D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis RE, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trentin L, et al. Geldanamycin-induced Lyn dissociation from aberrant Hsp90-stabilized cytosolic complex is an early event in apoptotic mechanisms in B-chronic lymphocytic leukemia. Blood. 2008;112(12):4665–4674. doi: 10.1182/blood-2008-02-139139. [DOI] [PubMed] [Google Scholar]
- 11.Gauld SB, Cambier JC. Src-family kinases in B-cell development and signaling. Oncogene. 2004;23(48):8001–8006. doi: 10.1038/sj.onc.1208075. [DOI] [PubMed] [Google Scholar]
- 12.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224(pt 3):213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 13.Chen L, et al. SYK inhibition modulates distinct PI3K/AKT- dependent survival pathways and cholesterol biosynthesis in diffuse large B cell lymphomas. Cancer Cell. 2013;23(6):826–838. doi: 10.1016/j.ccr.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gimpl G, Gehrig-Burger K. Cholesterol reporter molecules. Biosci Rep. 2007;27(6):335–358. doi: 10.1007/s10540-007-9060-1. [DOI] [PubMed] [Google Scholar]
- 15.Karnell FG, Brezski RJ, King LB, Silverman MA, Monroe JG. Membrane cholesterol content accounts for developmental differences in surface B cell receptor compartmentalization and signaling. J Biol Chem. 2005;280(27):25621–25628. doi: 10.1074/jbc.M503162200. [DOI] [PubMed] [Google Scholar]
- 16.Cheng PC, Brown BK, Song W, Pierce SK. Translocation of the B cell antigen receptor into lipid rafts reveals a novel step in signaling. J Immunol. 2001;166(6):3693–3701. doi: 10.4049/jimmunol.166.6.3693. [DOI] [PubMed] [Google Scholar]
- 17.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 18.Gerecitano JM, et al. Using 124-I-PU-H71 PET imaging to predict intratumoral concentration in patients on a phase I trial of PU-H71. J Clin Oncol. 2013;31(suppl): [Google Scholar]
- 19.Hans CP, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 20.Nayar U, et al. Targeting the Hsp90-associated viral oncoproteome in gammaherpesvirus-associated malignancies. Blood. 2013;122(16):2837–2847. doi: 10.1182/blood-2013-01-479972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grammatikakis N, et al. The role of Hsp90N, a new member of the Hsp90 family, in signal transduction and neoplastic transformation. J Biol Chem. 2002;277(10):8312–8320. doi: 10.1074/jbc.M109200200. [DOI] [PubMed] [Google Scholar]
- 22.Bijlmakers MJ, Marsh M. Hsp90 is essential for the synthesis and subsequent membrane association, but not the maintenance, of the Src-kinase p56(lck) Mol Biol Cell. 2000;11(5):1585–1595. doi: 10.1091/mbc.11.5.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giannini A, Bijlmakers MJ. Regulation of the Src family kinase Lck by Hsp90 and ubiquitination. Mol Cell Biol. 2004;24(13):5667–5676. doi: 10.1128/MCB.24.13.5667-5676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L, et al. SYK-dependent tonic B-cell receptor signaling is a rational treatment target in diffuse large B-cell lymphoma. Blood. 2008;111(4):2230–2237. doi: 10.1182/blood-2007-07-100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wilson WH, et al. The Bruton’s Tyrosine Kinase (BTK) inhibitor, Ibrutinib (PCI-32765), has preferential activity in the ABC subtype of relapsed/refractory de novo diffuse large B-cell lymphoma (DLBCL): interim results of a multicenter, open-label, phase 2 study. Presented at: 54th Annual ASH Annual Meeting and Exposition; December 8–11, 2012; Atlanta, Georgia, USA. [Google Scholar]
- 26.Qu Z, Wang S, Teng R, Yi X. PU-H71 effectively induces degradation of IκB kinase β in the presence of TNF-α. Mol Cell Biochem. 2014;386(1–2):135–142. doi: 10.1007/s11010-013-1852-y. [DOI] [PubMed] [Google Scholar]
- 27.Mathews Griner LA, et al. High-throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B-cell-like diffuse large B-cell lymphoma cells. Proc Natl Acad Sci U S A. 2014;111(6):2349–2354. doi: 10.1073/pnas.1311846111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.