Abstract
From an MCR fragment library, two novel chemical series have been developed as inhibitors of RET, which is a kinase involved in the pathology of medullary thyroid cancer (MTC). Structure activity relationship studies (SAR) identified two sub-micromolar tractable leads, 6g and 13g. 6g was confirmed to be a Type-II RET inhibitor. 13g and 6g inhibited RET in cells transformed by RET/C634. A RET DFG-out homology model was established and utilized to predict Type-II inhibitor binding modes.
Keywords: MCR, RET, Medicinal chemistry, Lead discovery, Targeted therapeutics, Kinase
1. Introduction
In 1985, the RET (Rearranged during transfection) gene was identified as a novel oncogene activated by DNA rearrangement. The isolated oncogene resulted from a recombination event between two unlinked human DNA segments, which occurs during the transfection process [1]. It was discovered that point mutations in RET lead to the development of medullary thyroid cancer (MTC) [2–9]. The identification of a small molecule inhibitor for the RET kinase represents an important strategy for the intervention of RET-driven cancers; however, the target has largely been neglected. Research manuscripts have been published on RET inhibition [10–16] and anticancer drugs vandetanib [17] and cabozantinib [18] were found to exhibit RET activity. However, both drugs were developed as vascular endothelial growth factor receptor 2 (VEGFR2) inhibitors and have VEGFR2 as the primary target. To improve efficacy and toxicity profiles, it is highly desirable to develop RET selective inhibitors for the treatment of MTC. We sought out to utilize MCR enabling chemistries to assist in the discovery of novel RET inhibitors.
Multicomponent reactions (MCRs) encompass a broad synthetic landscape where multiple inputs react to create a diversified final product [19,20]. The synthetic ease and facile analoging ability make MCR enabling chemistries an attractive tool for drug-discovery to rapidly identify novel, tractable leads. In the drug-discovery realm, MCR chemistry has been utilized to identify novel therapeutics for tuberculosis [21], viral infections [22], malaria [23], etc. Therefore, MCR enabling chemistries represent a highly validated means to generate biologically relevant chemotypes to treat human disease. Despite the large scope and application of kinase directed MCRs [24–27], no group has displayed the ability to generate RET kinase inhibitors utilizing MCR enabling chemistries. RET represents an unmet therapeutic target, and tailoring an MCR for lead development can help facilitate the discovery of novel inhibitor functionality.
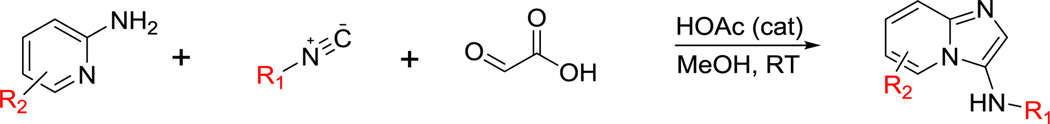
In order to utilize MCR enabling chemistries to target the RET kinase we have generated a kinase inhibitor library utilizing a modified MCR protocol. The MCR is based on the Groebke–Blackburn–Bienaymé reaction [28], but glyoxylic acid is employed in order to create monosubstitution on the 5 membered ring of imidazopyridine (Scheme 1) [29,30]. The modified MCR permits the rapid generation of diverse, drug-like scaffolds that contain kinase inhibitor functionality. By using this chemistry, we have synthesized more than 100 imidazopyridine analogues. The screening of these analogues, followed by non-MCR hit optimization, produced two novel chemical scaffolds that inhibited the RET kinase at sub-micromolar concentrations. We report herein the progress and development of RET kinase inhibitors derived from an imidazopyridine MCR chemical library.
Scheme 1.
Three component Groebke–Blackburn–Bienaymé reaction with glyoxylic acid.
2. Results and discussion
2.1. MCR hit generation
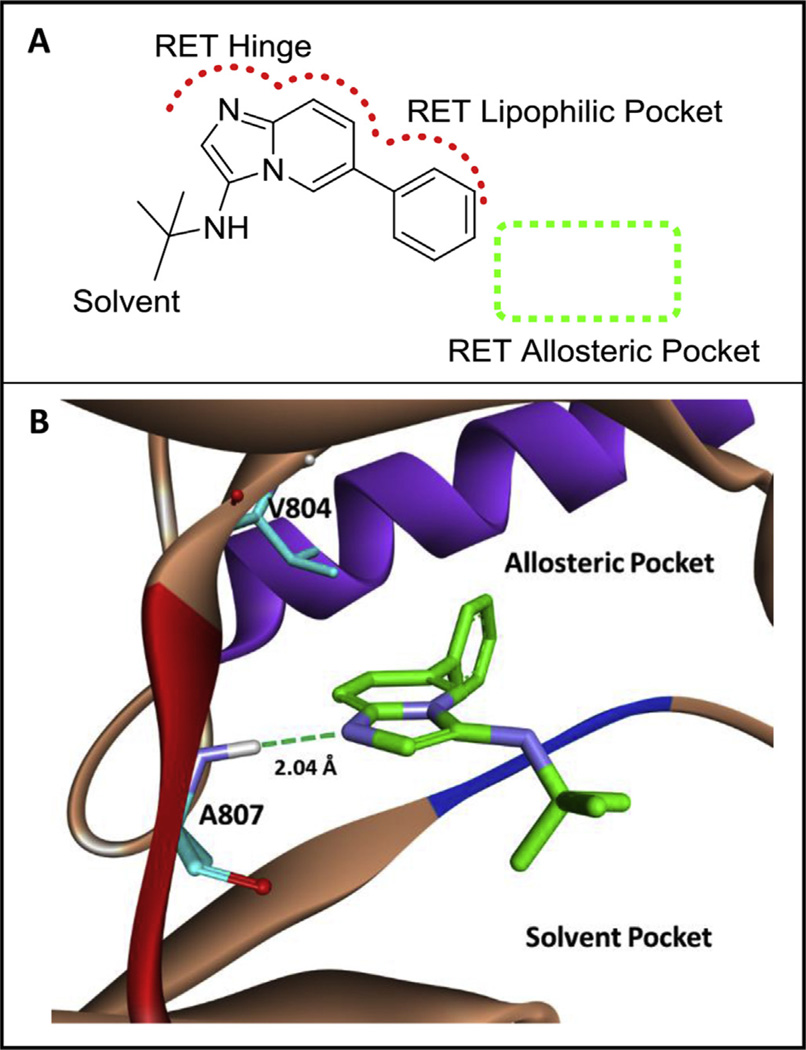
MCR kinase library screening was completed with a mid-throughput screening platform utilizing microfluidics technology established by Caliper® [31]. The assay contains a microfluidics chip that can separate phosphorylated peptide from unphosphorylated starting peptide. The starting peptide is FAM tagged, and can be tracked in each well using the EZ Reader® plate reader. With this screening technique, an MCR kinase library with 100 kinase fragments was screened and RET active 1 was identified (Fig. 1, A).
Fig. 1.
A) A novel RET hit (1, RET IC50 ~2.0 mM) from an MCR-based library. The scaffold is predicted to bind the kinase in an atypical, backward binding mode to that of ATP. B) Computational binding of 1 in the RET kinase. Compound 1 is predicted to bind the hinge of RET by hydrogen bonding to A807. The phenyl ring system of 1 accesses a lipophilic pocket close to the RET allosteric pocket. The RET hinge is shown in red, the DFG loop is shown in blue, and the c-Helix is shown in purple. Computational experiments were completed with AutoDock Vina [39]. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Based on the structure of compound 1, the warhead region (imidazopyridine) is predicted to bind to the RET hinge, while the phenyl ring system is predicted to engage a lipophilic pocket adjacent to the RET allosteric pocket. This is an atypical binding modality [32], permitting the fragment to bind the kinase ‘backward’ in comparison to the typical binding of ATP to the active site. Computational modeling showed the phenyl ring system was in close proximity to the allosteric pocket on RET and therefore adding allosteric functionality to compound 1 was hypothesized to increase potency (Fig. 1, B).
2.2. RET inhibitor optimization
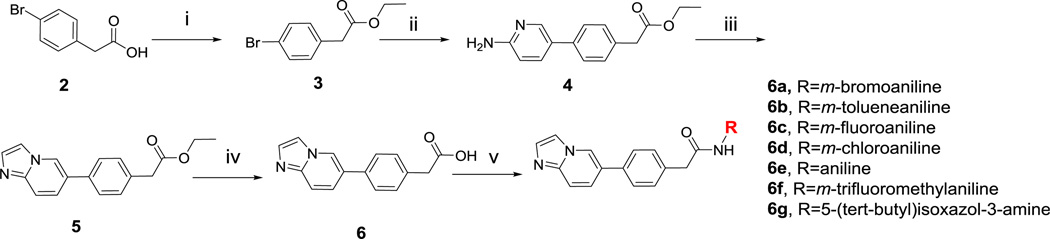
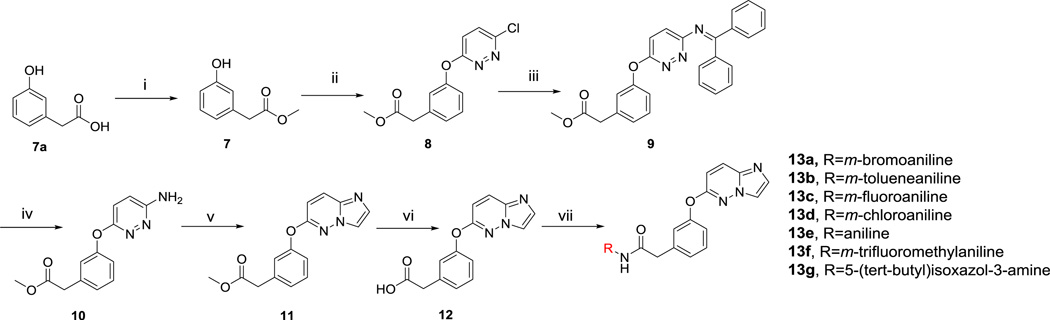
A synthetic protocol was developed to grow MCR active 1 into the RET allosteric pocket (Scheme 2). The solvent exposed tert-butyl moiety was replaced with a hydrogen to decrease lipophilicity at the solvent region. Using Fischer esterification, starting acid 2 was esterified to compound 3. Employing developed Suzuki coupling conditions, compound 4 was generated from a coupling reaction between an amino-pyridine boronic ester and compound 3. Compound 4 was cyclized to compound 5 using chloroacetaldehyde. After, compound 5 was saponified with base to produce 6 and subsequently coupled to various anilines using EDC to generate compounds 6a–6g.
Scheme 2.
Synthesis of Compounds (6a–6g). i) EtOH, H2SO4,100 °C, 12 h. ii) Pd2(dba)3, P(Cy)3, Na2CO3, 4:1 DMF/H2O, MWI 130 °C, 30 min. iii) Chloroacetaldehyde, Na2CO3, EtOH, 100 °C, 48 h. iv) LiOH, 1:1 THF/H2O, 100 °C, 5 h. v) EDC, HOAt, DIPEA, DMF, rt.
As predicted, growing MCR active 1 into the allosteric region of RET drastically increased inhibitor potency as seen with compound 6g (RET IC50 = 0.21 ± 0.04 µM) (Table 1). Other compounds, 6e (RET IC50 = 18.8 ± 4.6 µM) and 6f (RET IC50 = 15.7 ± 2.9 µM), also displayed an increase in activity. The structure activity relationship (SAR) trend on RET is that bulky, lipophilic moieties at the meta position increase potency by engaging a conserved hydrophobic region. Compound 6f is >6 times more potent than 6b because the –CF3 moiety from compound 6f is more lipophilic and bulky than the –CH3 moiety on 6c. Interestingly, the SAR trend does not hold true for compound 6e, which contains a hydrogen atom at the meta position. Likely, the hydrophobicity of the unsubstituted aniline on 6e creates high-binding affinity at the hydrophobic region. From MCR active 1, a series of RET inhibitors were developed, which uncovered compound 6g, a RET lead inhibitor with sub-micromolar activity.
Table 1.
RET SAR for compounds 6a–g and 13a–g.a
| <!–Col Count:2– > Compound | RET IC50 (µM) |
|---|---|
| 6a | >100 |
| 6b | >100 |
| 6c | >100 |
| 6d | 67.8 ± 1.1 |
| 6e | 18.8 ± 4.6 |
| 6f | 15.7 ± 2.9 |
| 6g | 0.21 ± 0.04 |
| 13a | 22.3 ± 7.7 |
| 13b | 20.4 ± 5.6 |
| 13c | 19.3 ± 2.8 |
| 13d | 17.2 ± 3.1 |
| 13e | 14.3 ± 1.9 |
| 13f | 9.1 ± 2.6 |
| 13g | 0.75 ± 0.03 |
| Vandetanib | 0.102 ± 0.008 |
IC50 values represent the concentration required to inhibit enzyme activity by 50%. The screening was completed at 9 µM ATP concentration in duplicate.
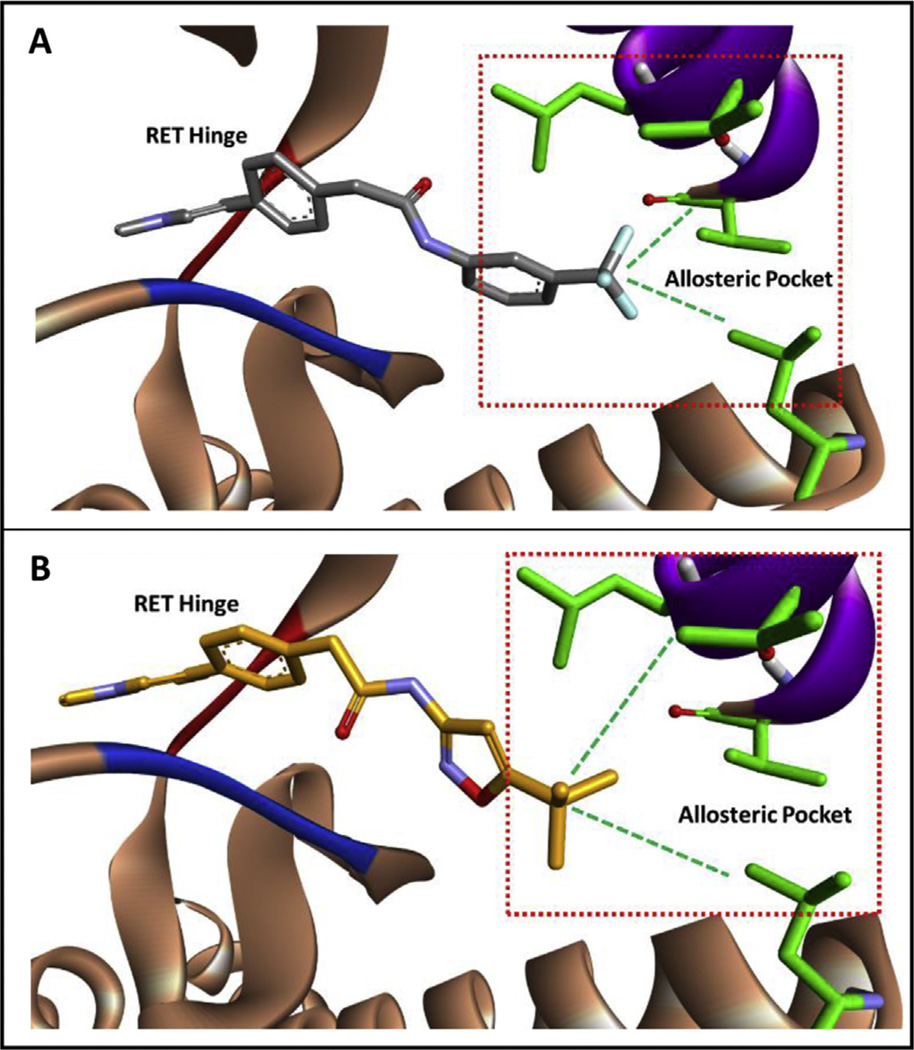
6g was strategically designed to engage the conserved hydrophobic region of RET (Fig. 2). Taking into account the increase in activity achieved with 6f, it was hypothesized that exchanging the –CF3 moiety with a tert-butyl group would have a favorable impact on activity (Fig. 2). Because of computational modeling results, 3-amino-5-tert-butylisoxazole was used as the amine input to generate compound 6g, which displayed ~75 times increase in activity when compared to 6f. It is hypothesized that the increase in activity is a direct result from better engagement in lipophilic contacts at the allosteric pocket (Fig. 2).
Fig. 2.
A) Computational modeling of 6f in the RET kinase. The –CF3 moiety engages hydrophobic regions at the back of the allosteric pocket. B) Computation modeling of 6g in the RET kinase. The tert-butyl group is more bulky than the –CF3 moiety and is predicted to better engage hydrophobic regions at the back of the RET allosteric pocket. The RET hinge is shown in red, the DFG loop is shown in blue, and the c-Helix is shown in purple. Computational experiments were completed with AutoDock Vina [39]. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
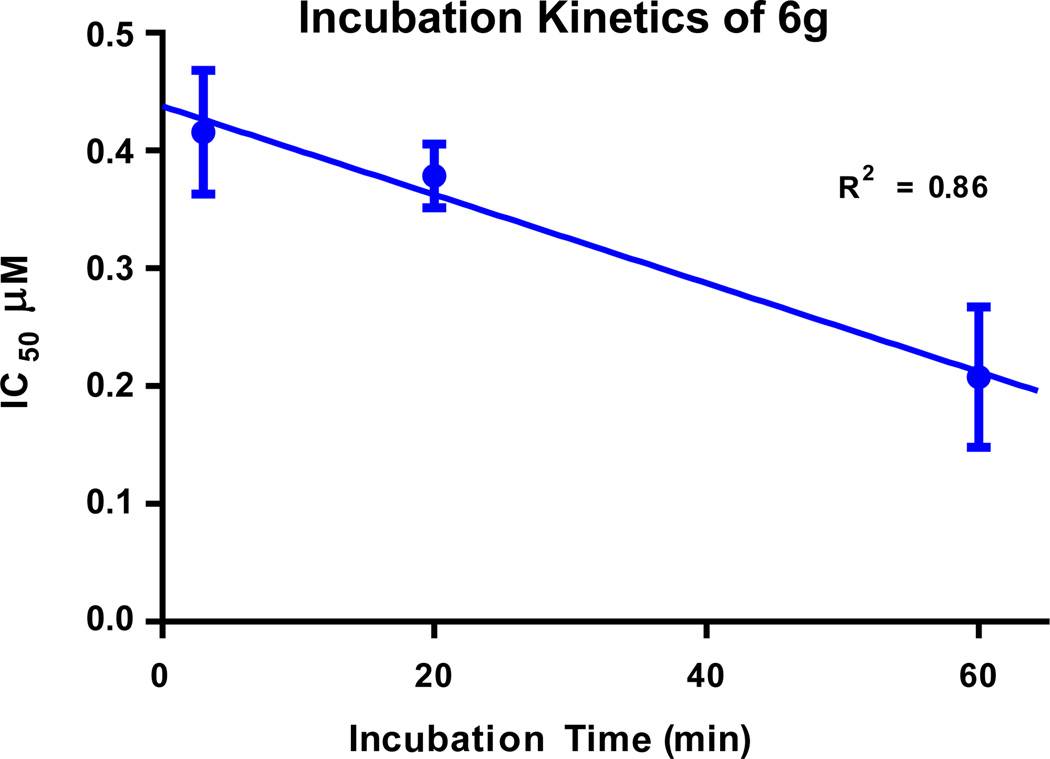
Compound 6g was subjected to an incubation assay to determine the kinetics of RET inhibition. In general, compounds that access the kinase allosteric pocket bind the DFG-out kinase conformation and incubation is typically required to achieve maximal inhibition [33]. With an increase in incubation time, 6g achieved greater RET inhibition (Fig. 3). A linear line was fit to the IC50 vs incubation time and the IC50 value for no incubation time (Y = 0) was determined to be 0.44 ± 0.03 µM. Therefore, through incubation, RET IC50 increases greater than 2 times and supports the hypothesis that 6g binds the DFG-out fold of RET. This suggests 6g is a Type-II kinase inhibitor that is not directly competitive with the binding of ATP.
Fig. 3.
Incubation assay to determine binding kinetics of compound 6g. IC50 without incubation: 0.44 ± 0.03 µM, IC50 after 3 min incubation: 0.42 ± 0.05 µM, IC50 after 20 min incubation: 0.38µ ± 0.03 µM, IC50 after 60 min incubation: 0.21 ± 0.06 µM.
Compounds based on the structure of 6g have very ridged, liner geometries and work was completed to identify a more flexible scaffold to define additional SARs on RET. An ether linker was hypothesized to be tolerated from a computational modeling study, and therefore a synthetic protocol was developed (Scheme 3).
Scheme 3.
Synthesis of Compounds (13a–g). i) EtOH, H2SO4,100 °C, 12 h. ii) NaH, DMF, 80 °C, 48 h. iii) Diphenylmethanimine, xantphos, Pd2(dba)3, K3PO4, dioxane, reflux, 12 h. iv) MeOH, 12 M HCl, 0 °C-RT, 12 h. v) Chloroacetaldehyde, Na2CO3, EtOH, 100 °C, 48 h. vi) LiOH, 1:1 THF/H2O, 100 °C, 5 h. vii) EDC, HOAt, DIPEA, DMF, RT.
Using Fischer esterification, compound 7 was generated from 7a. Compound 8 was synthetized employing a strong base to couple 7 with 3,6-dichloropyridazine. Buchwald conditions were utilized to convert compound 8 into compound 10, an amino-pyridazine. Cyclization of compound 10 with chloroacetaldehyde generated intermediate 11. Compound 11 was hydrolyzed with base to generate compound 12, which was subsequently coupled to various anilines using EDC to generate compounds 13a–g.
Through scaffold hopping from a novel MCR-based hit 1, compound 13g (RET IC50 =0.75 ± 0.03 µM) was identified, generating an additional RET lead inhibitor. The SAR on the scaffold based on compound 13g displayed a more direct trend than SAR from compound 6g (Table 1). On average, compounds generated from Scheme 3 displayed higher potency on RET, and bulkier groups at the meta position of the allosteric region produced compounds with higher potency. Similar to the case of compound 6g, compounds 13a–g engage a hydrophobic region in the allosteric pocket, which increases compound potency. Interestingly, despite having on average weaker RET inhibitors, Scheme 2 produced the most potent compound, 6g. Also, both 6g and 13g contain a t-butyl isoxazole structural moiety in the allosteric pocket and are at least 10-times more potent than any other compound with different substituents at the same region.
2.3. Cell-based studies
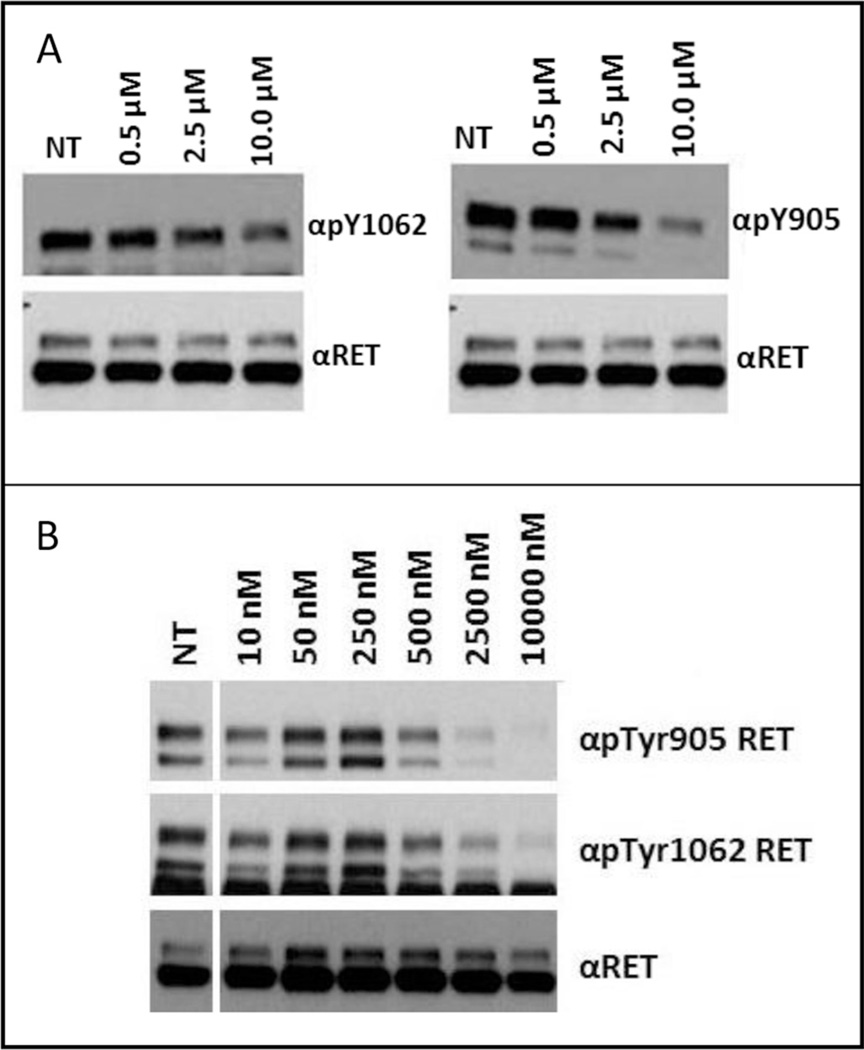
To further evaluate RET inhibition of 13g and 6g, the compounds were progressed into cell-based assays. The assay determined the amount of RET target inhibition by monitoring phosphorylation status of Y905 and Y1062 on the RET kinase domain (Fig. 4). Y1062 is responsible for activating PI3K/AKT and RAS/ERK pathways and is important to inhibit to block RET oncogene signaling. 13g was found active on RET in RAT1 cells transformed with a RET/C634R oncogene at an IC50 between 2.5 and 10 µM (Fig. 4, A). This value corresponds well to the determined biochemical IC50 of 0.75 ± 0.03 µM. 6g was found active on RET in RAT1 cells transformed with a RET/C634R oncogene at an IC50 between 0.25 and 0.50 µM (Fig. 4, B). Like 13g, the cell activity of 6g corresponds well to the determined biochemical IC50 of 0.21 ± 0.04 µM. Because 6g was shown to bind RET in the DFG-out fold (Fig. 3), there is not a large difference in biochemical vs cellular IC50s again suggesting the compound is not directly competitive with ATP. 6g represents a strong lead candidate that can be further developed into a RET advanced lead.
Fig. 4.
Serum-starved RAT1 RET/C634R cells were treated for 2 h with increasing concentrations of 13g (A), 6g (B) or left untreated (NT); cell lysates (50 µg) were immunoblotted with phospho-Y1062 (αpTyr1062) or-Y905 (αpTyr905) RET antibodies. Anti-RET (αRET) was used for normalization. 13g displayed an IC50 between 2.5 and 10.0 µM and 6g displayed an IC50 between 0.25 and 0.50 µM.
2.4. Homology model development
In order to better understand activity for compounds 6g and 13g a novel RET DFG-out homology model was generated utilizing Swiss-Model [34–36]. Crystal structure coordinates of a VEGFR-2 DFG-out structure was used as a template [37], and the RET amino acid sequence [38] was employed to build a RET DFG-out crystal structure. The resulting RET DFG-out model clearly displays the correct shift in the DFG-loop, which opens up the allosteric pocket of the kinase for inhibitors to access. There are two interchangeable folds of the RET kinase: DFG-in, the kinase is active and the allosteric pocket is closed; DFG-out, the kinase is not active and the allosteric pocket is open [33]. The developed DFG-out model served as a tool to evaluate binding modes of compounds 6g and 13g.
2.5. RET computational modeling
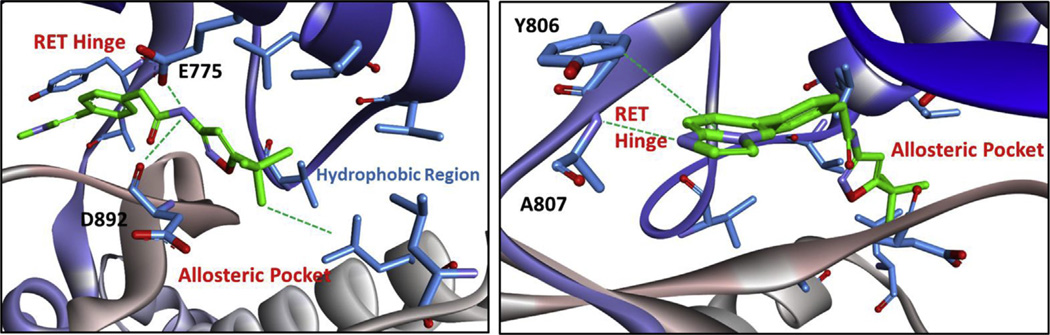
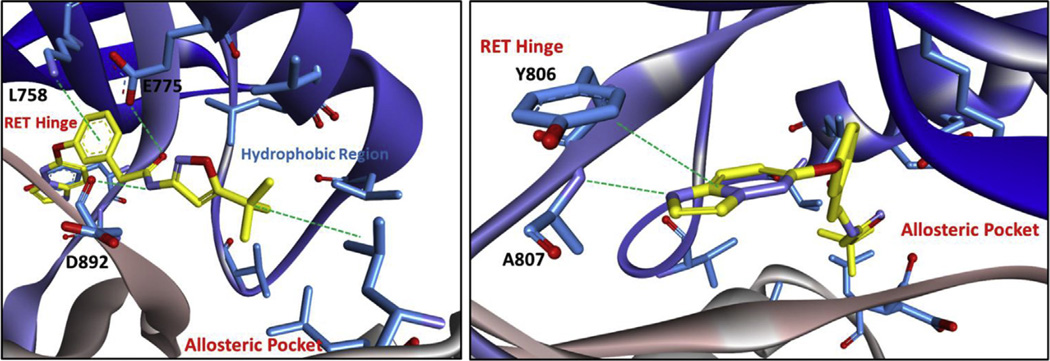
Compounds developed from the MCR-hit 1 were created to access the allosteric pocket of RET, which is only available in the DFG-out fold and a DFG-out homology model was developed and utilized to computationally model 6g and 13g (Figs. 5 and 6). Compound 6g is predicted to bind to the hinge of RET with imidazopyridine, making a hydrogen bond with A807 and pi–pi stacking with Y806. The amide of compound 6g sits on a ‘bridge’ held up by two hydrogen bonds with E775 and D892 (from the DFG motif), which allows the inhibitor to enter the allosteric pocket. This is a typical binding modality that is also observed with imatinib (PDB# 3K5V). Within the allosteric pocket, there is a hydrophobic region containing conserved valine, leucine, and isoleucine residues that the tert-butyl moiety engages (Fig. 5). Compound 13g is predicted to bind RET in a similar mode to compound 6g, where the imidazopyridine ring system makes a hydrogen bond with A807 and pi–pi stacks with Y806 at the RET hinge. At the ‘bridge’ region, the aryl-ring system from engages L758 through an ion-induced dipole interaction. Also at the ‘bridge’ region, E775 and D892 (from the DFG motif) hydrogen bond to 13g. This permits access into the allosteric pocket where the tert-butyl moiety can engage the hydrophobic region (Fig. 6). Because of similar biding-modes observed with compounds 6g and 13g, both compounds display somewhat similar potency. However, compound 6g was found ~3 times more potent than compound 13g in the RET biochemical assay. In cell-based assays, 6g was found ~10 times more potent than 13g.
Fig. 5.
Compound 6g computationally docked in the DFG-out RET homology model. RET: carbon = blue; oxygen = red; nitrogen = purple. Inhibitor: carbon = green; oxygen = red; nitrogen = purple. Computational experiments were completed with AutoDock Vina [39]. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
Compound 13g computationally docked in the DFG-out RET homology model. RET: carbon = blue; oxygen = red; nitrogen = purple. Inhibitor: carbon = yellow; oxygen = red; nitrogen = purple. Computational experiments were completed with AutoDock Vina [39]. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3. Conclusion
MCR fragment-based drug discovery has been utilized to uncover two novel kinase inhibitors (6g and 13g) from active fragment 1, and established ‘proof-of-concept’ for MCR-directed RET hit to lead generation. The binding geometries of both compounds are unique for the RET kinase and present as valid starting points for drug-discovery campaigns. A RET DFG-out homology model was generated and utilized to model both lead inhibitors. The tert-butyl moiety was found essential for efficient potency, and engages a hydrophobic region in the allosteric pocket of RET. An incubation based binding study was completed that identified 6g binds RET slowly, typical of inhibitors that bind the DFG-out form of the kinase. Further, 13g and 6g inhibited RET in RAT1 cells transformed by RET/C634R. Both compounds are being developed into advanced leads through optimization of potency and activity, and the work will be published in due course.
4. Experimental Section
4.1. General experimental
All solvents were reagent grade or HPLC grade and all starting materials were obtained from commercial sources and used without further purification. Purity of final compounds was assessed using a Shimadzu ultra-high throughput LC/MS system (SIL-20A, LC-20AD, LC-MS 2020, Phenomenex® Onyx Monolithic C-18 Column) at variable wavelengths of 254 nM and 214 nM (Shimadzu PDA Detector, SPD-MN20A) and was >95%, unless otherwise noted. The HPLC mobile phase consisted of a water–acetonitrile gradient buffered with 0.1% formic acid. 1H NMR spectra were recorded at 400 MHz and 13C spectra were recorded at 100 MHz, both completed on a Varian 400 MHz instrument (Model# 4001S41ASP). Compound activity and kinetics were determined with the EZ Reader II plate reader (PerkinElmer®, Walthman, USA). Vandetanib was obtained from LC Laboratories® (Woburn, USA, Lot# BTB-105). All compounds were purified using silicagel (0.035–0.070 mm, 60 Å) flash chromatography, unless otherwise noted. Microwave assisted reactions were completed in sealed vessels using a Biotage Initiator microwave synthesizer.
4.2. Synthesis and characterization
4.2.1. 5-Phenylpyridin-2-amine
5-Chloropyridin-2-amine (500 mg, 3.89 mmol), phenyl boronic acid (711 mg, 5.83 mmol), and Na2CO3 (11.67 mmol) were added to a mixture of 4:1 DMF/Water (10 mL). The resulting solution was stirred and degassed with argon for 5 min. P(Cy)3 (49.0 mg, 0.175 mmol) and Pd2(dba)3 (53.4 mg, 0.58 mmol) were then added and the reaction was heated under microwave irradiation for 30 min at 130 °C. The crude reaction was adsorbed onto silica and purified using flash chromatography utilizing a DCM/MeOH gradient to yield compound 5-phenylpyridin-2-amine (345 mg, 52.1%). 1H NMR(400 MHz, DMSO-d6) δ 8.23 (dd, J = 2.5, 0.6 Hz, 1H), 7.77 (dd, J = 8.0, 1.5 Hz, 1H), 7.69 (dd, J = 8.6, 2.6 Hz, 2H), 7.53 (d, J = 7.2 Hz, 2H), 7.38 (d, J = 7.2 Hz, 2H), 6.53 (d, J = 9.3 Hz, 1H), 6.10 (s, 2H). ESIMS m/z [M+H]+ 171.
4.2.2. N-(tert-butyl)-6-phenylimidazo[1,2-a]pyridin-3-amine (1)
Glyoxylic acid 50% solution in water (0.097 mL, 0.881 mmol), 5- phenylpyridin-2-amine (100 mg, 0.588 mmol), and acetic acid (5.08 µL, 0.088 mmol) were added to MeOH (1 mL) under stirring. After 15 min, 2-isocyano-2-methylpropane (0.070 mL, 0.617 mmol) was added to the reaction and was stirred overnight. After 12 h, the reaction was adsorbed onto silica and purified using flash chromatography using a DCM/MeOH gradient to yield compound 1 (90 mg, 57.7%). 1H NMR (400 MHz, DMSO-d6) δ 8.52 (dd, J = 1.8, 1.0 Hz, 1H), 7.99 (s, 1H), 7.75 (dd J = 8.0,1.5 Hz, 1H), 7.69 (dd J = 8.4, 1.2 Hz, 2H), 7.49 (t J = 8.0 Hz, 3H), 7.42 (dd J = 9.3,1.9 Hz, 1H), 7.15 (s, 1H), 4.61 (s, 1H), 1.17 (s, 9H). ESIMS m/z [M+H]+ 266. HPLC Purity >95% at 254 nM.
4.2.3. Ethyl 2-(4-bromophenyl)acetate (2)
2-(4-bromophenyl)acetic acid (15 g, 69.8 mmol) was added to ethanol (125 mL). Then, sulfuric acid (0.186 mL, 3.49 mmol) was added to the reaction and the reaction was heated to 80 °C for 12 h. The reaction was confirmed complete based on TLC. Solid NaHCO3 was added to the reaction and then the ethanol was evaporated. The product was extracted with ether and washed 2× with water and 1× with brine. The organic layer was collected, dried with MgSO4, and condensed to generate compound 2 as a clear oil (16.113 g, 95%). 1H NMR (400 MHz, CDCl3) δ 7.45 (d, J = 8.4 Hz, 2H), 7.16 (d, J = 8.4 Hz, 2H), 4.15 (q, J = 7.1 Hz, 2H), 3.56 (s, 2H), 1.25 (t, J = 7.1 Hz, 3H). ESIMS m/z [M+H]+ 243.
4.2.4. Ethyl 2-(4-(6-aminopyridin-3-yl)phenyl)acetate (3)
Compound 2 (1.270 g, 5.23 mmol), 5-(4,4,5,5-tetramethyl-1,3,2- dioxaborolan-2-yl)pyridin-2-amine (1 g, 4.54 mmol), and Na2CO3 (1.445 g, 13.63 mmol) were added to a 20 mL microwave vessel. The degassed 4:1 DMF/water v/v (10 mL) was then added to the reaction and the reaction was degassed with argon for 5 min. P(Cy)3 (0.057 g, 0.204 mmol) and Pd2(dba)3 (0.062 g, 0.068 mmol) were then added to the reaction vessel followed by degassing with argon for an additional 5 min. The reaction vessel was sealed and heated under microwave irradiation for 30 min at 130°C. TLC confirmed all of 5-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)pyridin-2-amine was consumed. The solvent from the reaction was evaporated and the reaction was adsorbed onto silica and purified with flash chromatography using a gradient from 100% DCM to 40% 4:1 DCM/ MeOH to yield compound 3 as yellow crystals (1.153 g, 99%). 1H NMR (400 MHz, DMSO-d6) δ 8.21 (d, J = 2.6 Hz, 1H), 7.66 (dd, J = 8.6, 2.6 Hz, 1H), 7.49 (d, J = 8.3 Hz, 2H), 7.26 (d, J = 8.3 Hz, 2H), 6.49 (d, J = 8.6 Hz, 1H), 6.02 (s, 2H), 4.06 (q,J = 7.1 Hz, 2H), 3.64 (s, 2H), 1.17 (t, J = 7.1 Hz, 3H). ESIMS m/z [M+H]+ 257.
4.2.5. Ethyl 2-(4-(imidazo[1,2-a]pyridin-6-yl)phenyl)acetate (4)
Compound 3 (1.165 g, 4.55 mmol) was added into a reaction vessel equipped with a stir bar. Ethanol (40 mL) and Na2CO3 (1.205 g, 11.36 mmol) were then added to the reaction vessel and the reaction was heated to 100°C in an oil bath. After 5 min, chloroacetaldehyde 50% solution in water (6.39 mL, 45.5 mmol) was added dropwise and the reaction was heated to 100°C for 48 h. TLC confirmed that all of starting material was consumed, the solvent was condensed and the reaction was adsorbed onto silica. The reaction was purified with flash chromatography using a gradient from 100% DCM to 40% 4:1 DCM/MeOH to yield compound 4 (1.210 g, 95%). 1H NMR (400 MHz, CDCl3) δ 8.30 (s, 1H), 7.69 (d, J = 9.3 Hz, 1H), 7.66 (d, J = 1.2 Hz, 1H), 7.63 (s, 1H), 7.52 (d, J = 8.2 Hz, 2H), 7.43 (dd, J = 9.3, 1.8 Hz, 1H), 7.40 (d, J = 8.2 Hz, 2H), 4.18 (q, J = 7.2 Hz, 2H), 3.67 (s, 2H), 1.29 (d, J = 7.2 Hz, 3H). ESIMS m/z [M+H]+ 281.
4.2.6. 2-(4-(Imidazo[1,2-a]pyridin-6-yl)phenyl)acetic acid (5)
Compound 4 (1.2 g, 4.28 mmol) was added to 1:1 THF/Water (40 mL) in a pressure reaction vessel. LiOH (1.798 g, 21.4 mmol) was then added and the reaction was heated to 100 °C for 5 h. TLC confirmed complete consumption of compound 4. All organic solvent was evaporated and the water solution was extracted with 5× DCM and all extracts were discarded. Then, the reaction was acidified with 3 M HCl to pH ~4. The acidified aqueous solution was extracted 10× with 4:1 DCM/IPA. All extracts were combined, dried, and condensed to yield compound 5 (455 mg, 42.1%). 1H NMR (400 MHz, DMSO-d6) δ 12.33 (s, 1H), 8.97 (s, 1H), 8.02 (s, 1H), 7.73–7.64 (m, 5H), 7.40 (d, J = 7.9 Hz, 2H), 3.65 (s, 1H). ESIMS m/z [M+H]+ 253.
4.2.7. N-(5-(tert-butyl)isoxazol-3-yl)-2-(4-(imidazo[1,2-a]pyridin-6-yl)phenyl)acetamide (6g)
Compound 5 (40 mg, 0.159 mmol), 5-(tert-butyl)isoxazol-3-amine (33.3 mg, 0.238 mmol), EDC (60.9 mg, 0.37 mmol), HOAt (21.56 mg, 0.159 mmol), and DIPEA (0.043 mL, 0.238 mmol) were all added to a reaction vessel. DMF (1 mL) was added and the reaction was stirred at room temperature overnight. After the reaction was complete, the organic layer was condensed and the crude reaction was transferred using DCM to a silica column and purified using DCM to 30% 4:1 DCM/MeOH. The desired compound 6g was isolated in moderate yield (20.4 mg, 34.4%).1H NMR (400 MHz, CDCl3) δ 9.83 (s, 1H), 8.29 (s, 1H), 7.70 (m, 2H), 7.64 (s, 1H), 7.53 (d, J = 7.6 Hz, 2H), 7.45 (d, J = 7.6 Hz, 2H), 7.40 (d, J = 9.4 Hz, 1H), 6.76 (s, 1H), 3.84 (s, 2H), 1.35 (s, 9H). 13C NMR (101 MHz, CDCl3) δ 181.78, 169.13, 157.97, 144.70, 136.53, 133.94, 133.72, 130.09, 127.44, 126.38, 125.10, 123.04, 117.76, 112.77, 93.42, 43.61, 33.05, 28.62. ESIMS m/z [M+H]+ 375. HPLC Purity 100%.
4.2.8. 2-(4-(Imidazo[1,2-a]pyridin-6-yl)phenyl)-N-(3-(trifluoromethyl)phenyl)acetamide (6f)
Compound 6f was synthesized according to the procedure outlined for 6g (17.5 mg, 27.9%). 1H NMR (400 MHz, DMSO-d6) δ 10.54 (s, 1H), 8.88 (s, 1H), 8.09 (s, 1H), 7.94 (s, 1H), 7.78 (d, J = 8.2 Hz, 1H), 7.62 (m, 4H), 7.56–7.48 (m, 2H), 7.44 (d, J = 7.9 Hz, 2H), 7.37 (d, J = 7.7 Hz, 1H), 3.72 (s, 2H).13C NMR (101 MHz, DMSO-d6) δ 170.06, 140.35, 130.44, 130.35, 126.91, 125.18, 124.79, 124.43, 123.05, 120.02 (q, J = 3.7 Hz), 117.33, 115.55 (q, J = 3.9 Hz), 114.07, 43.33. ESIMS m/z [M+H]+ 396. HPLC Purity 98%.
4.2.9. 2-(4-(Imidazo[1,2-a]pyridin-6-yl)phenyl)-N-phenylacetamide (6e)
Compound 6e was synthesized according to the procedure outlined for 6g (20.4 mg, 39.3%). 1H NMR (400 MHz, DMSO-d6) δ 10.18 (s, 1H), 8.88 (s, 1H), 7.95 (s, 1H), 7.64 (d, J = 8.2 Hz, 2H), 7.59 7.60–7.58 (m, 4H), 7.57–7.52 (m,1H), 7.44 (d, J = 8.2 Hz, 2H), 7.28 (t, J = 7.9 Hz, 2H), 7.02 (t, J = 7.4 Hz, 1H), 3.68 (s, 2H). 13C NMR (101 MHz, DMSO-d6) δ 169.41, 139.63, 135.97, 135.44, 134.10, 130.29, 129.16, 126.88, 125.23, 124.81, 124.41, 123.67, 119.55, 117.33, 114.07, 43.37. ESIMS m/z [M+H]+ 328. HPLC Purity 100%.
4.2.10. N-(3-chlorophenyl)-2-(4-(imidazo[1,2-a]pyridin-6-yl) phenyl)acetamide (6d)
Compound 6d was synthesized according to the procedure outlined for 6g (18.8 mg, 32.8%). 1H NMR (400 MHz, DMSO-d6) δ 10.38 (s, 1H), 8.88 (s, 1H), 7.95 (s, 1H), 7.81 (d, J = 1.7 Hz, 1H), 7.63 (m, 4H), 7.54 (d, J =9.4 Hz, 1H), 7.44 (m, 3H), 7.31 (t, J =8.1 Hz, 1H), 7.08 (dd, J =8.1, 2.1 Hz,1H), 3.69 (s, 2H).13C NMR (101 MHz, DMSO-d6) δ 169.84, 141.05, 135.60, 135.54, 134.11, 133.51, 130.89, 130.32, 126.90, 125.18, 124.79, 124.44, 123.40, 118.99, 117.91, 117.34, 114.05, 43.33. ESIMS m/z [M+H]+ 362. HPLC Purity 96%.
4.2.11. N-(3-fluorophenyl)-2-(4-(imidazo[1,2-a]pyridin-6-yl) phenyl)acetamide (6c)
Compound 6c was synthesized according to the procedure outlined for 6g (15.7 mg, 28.7%). 1H NMR (400 MHz, DMSO-d6) δ 10.41 (s, 1H), 8.88 (s, 1H), 7.94 (s, 1H), 7.64 (d, J = 8.2 Hz, 2H), 7.59 (m, 3H), 7.54 (dd, J = 9.4, 1.6 Hz, 1H), 7.43 (d, J = 8.2 Hz, 2H), 7.35–7.27 (m, 2H), 6.88–6.82 (m, 1H), 3.69 (s, 2H). 13C NMR (101 MHz, DMSO-d6) δ 169.82, 162.56 (d, J = 241.3 Hz), 141.33 (d, J = 11.1 Hz), 135.63, 135.53, 134.10, 130.82 (d, J = 9.6 Hz), 130.33, 126.90, 125.19, 124.81, 124.43, 117.33, 115.26 (d, J = 2.7 Hz), 114.08, 110.13 (d, J = 21.1 Hz), 106.30 (d, J = 26.3 Hz), 43.33. ESIMS m/z [M+H]+ 346. HPLC Purity 100%.
4.2.12. 2-(4-(Imidazo[1,2-a]pyridin-6-yl)phenyl)-N-(m-tolyl) acetamide (6b)
Compound 6b was synthesized according to the procedure outlined for 6g (19.2 mg, 35.5%). 1H NMR (400 MHz, DMSO-d6) δ 10.11 (s, 1H), 8.88 (s, 1H), 7.94 (s, 1H), 7.65–7.59 (m, 4H), 7.54 (dd, J = 9.4, 1.7 Hz, 1H), 7.45–7.41 (m, 3H), 7.38 (d, J = 7.8 Hz, 1H), 7.15 (t, J = 7.8 Hz, 1H), 6.83 (d, J = 7.8 Hz, 1H), 3.66 (s, 2H), 2.24 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 169.34, 139.56, 138.32, 136.02, 135.43, 134.10, 130.26, 129.00, 126.87, 125.22, 124.81, 124.41, 124.37, 120.09, 117.33, 116.74, 114.07, 43.40, 21.62. ESIMS m/z [M+H]+ 342. HPLC Purity 100%.
4.2.13. N-(3-bromophenyl)-2-(4-(imidazo[1,2-a]pyridin-6-yl) phenyl)acetamide (6a)
Compound 6a was synthesized according to the procedure outlined for 6g (17.1 mg, 26.5%). 1H NMR (400 MHz, DMSO-d6) δ 10.43 (s, 1H), 8.94 (s, 1H), 8.01 (s, 2H), 7.69 (m, 4H), 7.60 (d, J = 8.8 Hz, 1H), 7.55 (d, J = 7.9 Hz, 1H), 7.48 (d, J = 8.1 Hz, 2H), 7.31 (t, J = 7.9 Hz, 1H), 7.27 (d, J = 8.1 Hz, 1H), 3.75 (s, 2H). 13C NMR (101 MHz, DMSO-d6) δ 169.82, 141.19, 135.60, 135.53, 134.11, 131.20, 130.32, 126.90, 126.29, 125.18, 124.79, 124.44, 122.00, 121.85, 118.29, 117.34, 114.04, 43.33. ESIMS m/z [M+H]+ 406. HPLC Purity 100%.
4.2.14. Methyl 2-(3-hydroxyphenyl)acetate (7)
2-(3-hydroxyphenyl)acetic acid (15 g, 99 mmol) was added to methanol (197 mL). Sulfuric acid (3.94 mL, 73.9 mmol) was added and the reaction was heated reflux for three days. The reaction was cooled to room temperature and solid Na2CO3 was added until the pH was neutral. Solvent was evaporated and the reaction was redissolved in ether and washed 3× with water and 2× with brine. The organic layer was collected, dried, and condensed to generate compound 7 as a dark oil (14.764 g, 90%). 1H NMR (400 MHz, CDCl3) δ 7.17 (t, J = 7.8 Hz, 1H), 6.82 (d, J = 7.8 Hz, 1H), 6.77 (t, J = 1.8 Hz, 1H), 6.73 (dd, J = 8.1, 2.5 Hz, 1H), 5.56 (s, 1H), 3.70 (s, 3H), 3.58 (s, 2H). ESIMS m/z [M+H]+ 167.
4.2.15. Methyl 2-(3-((6-chloropyridazin-3-yl)oxy)phenyl)acetate (8)
Compound 7 (4.07 mL, 30.1 mmol) was added to DMF in a reaction vessel cooled to 0 °C. NaH 60% dispersion (1.565 g, 39.1 mmol) was added to the reaction and the reaction was stirred at 0°C for 45 min. 3,6-Dichloropyridazine (6.01 g, 39.1 mmol) was added and the reaction was raised to 80°C using an oil bath and was heated for 48 h. After the reaction was complete as judged by TLC, the solvent was evaporated and the reaction was adsorbed onto silica. The reaction was purified using flash chromatography on a silica column using a gradient from hexanes to EtOAc (50%). The desired product 8 was isolated in moderate yield (3.02 g, 36%). 1H NMR (400 MHz, CDCl3) δ 7.49 (d, J = 9.1 Hz, 1H), 7.38 (t, J = 7.9 Hz, 1H), 7.21–7.14 (m, 3H), 7.11 (ddd, J = 8.2, 2.4, 0.9 Hz, 1H), 3.70 (s, 3H), 3.65 (s, 2H). ESIMS m/z [M+H]+ 279.
4.2.16. Methyl 2-(3-((6-((diphenylmethylene)amino)pyridazin-3-yl)oxy)phenyl)acetate (9)
Compound 8 (2.768 g, 9.93 mmol), diphenylmethamine (1.980 g, 10.93 mmol), and potassium phosphate (10.53 g, 49.7 mmol) were added to dioxane (40 mL) and the reaction was degassed with argon for 10 min. Xantphos (0.287 g, 0.497 mmol) and Pd2(dba)3 (0.454 g, 0.497 mmol) were added to the reaction. The reaction was then sealed under positive argon pressure and heated under reflux for 12 h. After the overnight, the reaction was condensed and adsorbed onto silica. The reaction was purified with a silica column using flash chromatography with a hexane/EtOAc gradient to yield compound 9 (1.02 g, 24.5%). 1H NMR (400 MHz, CDCl3) δ 7.82 (d, J = 7.6 Hz, 2H), 7.52 (t, J = 7.3 Hz, 1H), 7.43 (t, J = 7.6 Hz, 2H), 7.31 (m, Hz, 4H), 7.19 (d, J = 6.5 Hz, 2H), 7.11–7.06 (m, 2H), 7.06–7.02 (m, 1H), 6.96 (d, J = 9.1 Hz, 1H), 6.88 (d, J = 9.1 Hz, 1H), 3.69 (s, 3H), 3.61 (s, 2H). ESIMS m/z [M+H]+ 424.
4.2.17. Methyl 2-(3-((6-aminopyridazin-3-yl)oxy)phenyl)acetate (10)
Compound 9 (1.02 g, 2.409 mmol) was dissolved in methanol (100 mL) and was cooled to 0 °C. 12 M HCl (2.007 mL, 24.09 mmol) was added to the reaction and stirred for 30 min at 0 °C. The reaction was removed from the ice bath and stirred at room temperature for 12 h. The reaction was purified with flash chromatography using a silica column and a gradient from DCM to 50% 4:1 DCM/MeOH. Compound 10 was isolated as a sticky-yellow oil (456 mg, 73%). 1H NMR (400 MHz, DMSO-d6) δ 7.30 (t, J = 7.8 Hz, 1H), 7.09 (d, J = 9.3 Hz, 1H), 7.02 (d, J = 7.8 Hz, 1H), 6.96–6.89 (m, 3H), 6.20 (s, 2H), 3.66 (s, 2H), 3.59 (s, 3H). ESIMS m/z [M+H]+ 260.
4.2.18. Methyl 2-(3-(imidazo[1,2-b]pyridazin-6-yloxy)phenyl) acetate (11)
Compound 11 was synthesized according to the procedure outlined for compound 4 (290 mg, 98%). 1H NMR (400 MHz, CDCl3) δ 7.95 (d, J = 9.7 Hz, 1H), 7.71 (s, 1H), 7.64 (s, 1H), 7.40 (t, J = 7.8 Hz, 1H), 7.19 (d, J = 7.8 Hz, 1H), 7.17–7.12 (m, 2H), 6.90 (d, J = 9.7 Hz, 1H), 3.71 (s, 3H), 3.67 (s, 2H). ESIMS m/z [M+H]+ 284. HPLC Purity 70%.
4.2.19. 2-(3-(Imidazo[1,2-b]pyridazin-6-yloxy)phenyl)acetic acid (12)
Compound 12 was synthesized according to the procedure outlined for 5(451.4 mg, 95%).1H NMR (400 MHz, DMSO-d6) δ 12.41 (s,1H), 8.14 (d, J = 9.6 Hz, 1H), 8.04 (s, 1H), 7.63 (s, 1H), 7.42–7.34 (m, 1H), 7.16–7.15 (m, 3H), 7.08 (d, J = 9.6 Hz,1H), 3.61 (s, 2H). ESIMS m/ z [M+H]+ 270.
4.2.20. N-(5-(tert-butyl)isoxazol-3-yl)-2-(3-(imidazo[1,2-b] pyridazin-6-yloxy)phenyl)acetamide (13g)
Compound 13g was synthesized according to the procedure outlined for 6g (14.7 mg, 33.7%).1H NMR (400 MHz, CDCl3) δ 9.70 (s, 1H), 7.93 (d, J = 9.6 Hz, 1H), 7.70 (s, 1H), 7.63 (s, 1H), 7.42 (t, J = 8.0 Hz), 7.30–7.24 (m, 2H), 7.16 (dd, J = 8.0, 2.2 Hz, 1H), 6.87 (d, J = 9.6 Hz, 1H), 6.72 (s, 1H), 3.81 (s, 2H), 1.31 (s, 9H). 13C NMR (101 MHz, CDCl3) δ 181.79, 168.69, 159.53, 157.90, 153.43, 135.79, 133.05, 130.26, 128.17, 126.50, 122.05, 120.16, 117.35, 111.47, 93.40, 43.83, 33.02, 28.57. ESIMS m/z [M+H]+ 392. HPLC Purity 100%.
4.2.21. 2-(3-(Imidazo[1,2-b]pyridazin-6-yloxy)phenyl)-N-(3-(trifluoromethyl)phenyl)acetamide (13f)
Compound 13f was synthesized according to the procedure outlined for 6g (17.2 mg, 37.4%).1H NMR (400 MHz, CDCl3) δ 8.28 (s, 1H), 7.87 (d, J = 9.6 Hz, 1H), 7.79 (s, 1H), 7.71 (d, J = 7.9 Hz, 1H), 7.63–7.61 (m, 2H), 7.44 (t, J = 7.9 Hz, 1H), 7.39 (t, J = 7.9 Hz, 1H), 7.33 (d, J = 7.6 Hz, 1H), 7.23 (d, J = 7.6 Hz, 1H), 7.21–7.14 (m, 2H), 6.86 (d, J = 9.6 Hz, 1H), 3.77 (s, 2H). 13C NMR (101 MHz, Chloroform-d) δ 168.91, 159.59, 153.48, 138.38, 137.36, 136.18, 133.07, 131.29 (q, J = 32.4 Hz), 130.40, 129.50, 128.09, 126.52, 122.87, 121.98, 120.94, 120.20, 117.31, 116.75–116.26 (m), 111.69, 44.23. ESIMS m/z [M+H]+ 413. HPLC Purity 100%.
4.2.22. 2-(3-(Imidazo[1,2-b]pyridazin-6-yloxy)phenyl)-N-phenylacetamide (13e)
Compound 13e was synthesized according to the procedure outlined for 6g (14.4 mg, 37.5%).1H NMR (400 MHz, CDCl3) δ 7.87 (d, J = 9.6 Hz, 2H), 7.61 (d, J = 6.0 Hz, 2H), 7.47 (d, J = 8.0 Hz, 2H), 7.41 (d, J = 8.0 Hz, 1H), 7.28–7.19 (m, 4H), 7.15 (d, J = 8.0 Hz, 1H), 7.08 (t, J = 7.4 Hz, 1H), 6.85 (d, J = 9.6 Hz, 1H), 3.74 (s, 2H). 13C NMR (101 MHz, CDCl3) δ 168.60, 159.59, 153.47, 137.76, 137.40, 136.60, 133.06, 130.33, 128.95, 128.12, 126.56, 124.49, 122.00, 120.04, 119.89, 117.24, 111.59, 44.32. ESIMS m/z [M+H]+ 345. HPLC Purity 100%.
4.2.23. N-(3-bromophenyl)-2-(3-(imidazo[1,2-b]pyridazin-6-yloxy)phenyl)acetamide (13d)
Compound 13d was synthesized according to the procedure outlined for 6g (19.4 mg, 41.1%).1H NMR (400 MHz, CDCl3) δ 8.03 (s, 1H), 7.88 (d, J = 9.6 Hz, 1H), 7.72 (s, 1H), 7.62 (s, 2H), 7.45–7.40 (m, 2H), 7.24–7.05 (m, 4H), 6.86 (d, J = 9.6 Hz,1H), 3.74 (s, 2H).13C NMR (101 MHz, CDCl3) δ 168.71, 159.59, 153.49, 139.07, 137.39, 136.24, 133.07, 130.54, 130.24, 128.14, 128.07, 127.41, 126.52, 122.86, 121.98, 120.18, 118.28, 117.28, 111.64, 44.46. ESIMS m/z [M+H]+ 423. HPLC Purity 100%.
4.2.24. N-(3-chlorophenyl)-2-(3-(imidazo[1,2-b]pyridazin-6-yloxy) phenyl)acetamide (13c)
Compound 13c was synthesized according to the procedure outlined in for 6g (16 mg, 37.9%). 1H NMR (400 MHz, CDCl3) δ 8.00 (s, 1H), 7.88 (d, J = 9.6 Hz, 1H), 7.62 (s, 2H), 7.58 (t, J = 1.8 Hz, 1H), 7.43 (t, J = 7.8 Hz, 1H), 7.34 (d, J = 8.0 Hz, 1H), 7.24–7.13 (m, 4H), 7.06 (d, J = 8.0 Hz, 2H), 6.86 (d, J = 9.6 Hz, 1H), 3.74 (s, 2H). 13C NMR (101 MHz, CDCl3) δ 168.70, 159.59, 153.50, 138.93, 137.39, 136.25, 134.58, 133.07, 130.55, 129.95, 128.11, 126.52, 124.48, 121.98, 120.19, 119.89, 117.78, 117.26, 111.63, 44.47. ESIMS m/z [M+H]+ 379. HPLC Purity 100%.
4.2.25. 2-(3-(Imidazo[1,2-b]pyridazin-6-yloxy)phenyl)-N-(m-tolyl) acetamide (13b)
Compound 13b was synthesized according to the procedure outlined for 6g(20.4 mg, 51.1%). 1H NMR(400 MHz, CDCl3) δ 7.88 (d, J = 9.6 Hz, 1H), 7.82 (s, 1H), 7.61 (d, J = 5.5 Hz, 2H), 7.42 (t, J = 7.8 Hz, 1H), 7.33 (s, 1H), 7.25–7.19 (m, 3H), 7.17–7.13 (m, 2H), 6.90 (d, J = 7.5 Hz, 1H), 6.85 (d, J = 9.6 Hz, 1H), 3.73 (s, 2H), 2.28 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 168.57, 159.60, 153.46, 138.89, 137.67, 137.40, 136.65, 133.06, 130.45, 128.76, 128.12, 126.56, 125.30, 122.01, 120.53, 120.02, 117.23, 116.96, 111.58, 44.54, 21.43. ESIMS m/z [M+H]+ 359. HPLC Purity 100%.
4.2.26. N-(3-fluorophenyl)-2-(3-(imidazo[1,2-b]pyridazin-6-yloxy) phenyl)acetamide (13a)
Compound 13a was synthesized according to the procedure outlined for 6g (21.0 mg, 52.0%). 1H NMR (400 MHz, CDCl3) δ 8.19 (s, 1H), 7.88 (d, J = 9.6 Hz, 1H), 7.66–7.60 (m, 2H), 7.48–7.40 (m, 2H), 7.24–7.11 (m, 5H), 6.86 (d, J = 9.6 Hz, 1H), 6.78 (t, J = 8.3 Hz, 1H), 3.74 (s, 2H). 13C NMR (101 MHz, Chloroform-d) δ 168.75, 162.89 (d, J = 244.9 Hz), 159.59, 153.46, 139.40 (d, J = 10.8 Hz), 137.39, 136.34, 133.04 (d, J = 9.7 Hz), 130.35, 130.00 (d, J = 8.6 Hz), 128.12, 126.52, 121.95, 120.11, 117.30, 114.99 (d, J = 2.8 Hz), 111.26, 111.20, 107.28 (dd, J = 26.7, 7.4 Hz), 44.27. ESIMS m/z [M+H]+ 363. HPLC Purity 100%.
4.3. RET biochemical inhibition assay
Kinase activity was measured in a microfluidic assay that monitors the separation of a phosphorylated product from substrate. The assay was run using a 12-sipper chip on a Caliper EZ Reader II (PerkinElmer®, Walthman, USA) with separation buffer (100 mM HEPES, 10 mM EDTA, 0.015% Brij-35, 0.1% CR-3 [PerkinElmer®, Walthman, USA]). In 96-well polypropylene plates (Greiner, Frickenhausen, Germany) compound stocks (20 mM in DMSO) were diluted into kinase buffer (50 mM HEPES, 0.075% Brij-35, 0.1% Tween 20,2 mM DTT, 10 mM MgCl2, and 0.02% NaN3) in 12-point ½log dilutions (2 mM–6.32 nM). After, 1 µL was transferred into a 384-well polypropylene assay plate (Greiner, Frickenhausen, Germany). The RET enzyme (Invitrogen™, Grand Island, USA) was diluted in kinase buffer to a concentration of 2 nM and 5 µL of the enzyme mixture was transferred to the assay plate. The inhibitors/ RET enzyme were incubated for 60 min with minor shaking. A substrate mix was prepared containing ATP (Ambresco®, Solon, USA) and 5FAM tagged RET peptide (peptide #22, 5’ FAM-EPLYWSFPA, PerkinElmer®, Walthman, USA) dissolved in kinase buffer, and 5 µL of the substrate mix was added to the assay plate. Running concentrations were as follows: ATP (9 µM), peptide (1.5 µM), compound 12-point ½log dilutions (0.2 mM–0.632 nM). For positive control, no inhibitor was added. For negative control, no enzyme was added. For running control, vandetanib (LC Laboratories®, Lot# BTB-105) was utilized. The plate was run until 10–20% conversion based on the positive control wells. The following separation conditions were utilized: upstream voltage −500 V; downstream voltage, −1900 V; chip pressure −0.8. Percent inhibition was measured for each well comparing starting peptide to phosphorylated product peaks relative to the baseline. Dose response curves, spanning the IC50 dose, were generated in GraphPad Prisim 6 and fit to an exponential one-phase decay line and IC50 values were obtained from the half-life value of the curve. IC50 values were generated in duplicate and error was calculated from the standard deviation between values.
4.4. RET kinetic assay
The assay was completed identically to the RET Biochemical Inhibition Assay, but compounds were incubated with RET for 60, 20, and 3 min before addition of the substrate mix. The IC50 verses incubation time graph was fit to a liner equation (y = mx + b) in GraphPad Prisim 6 and extrapolated to determine 0 min incubation.
4.5. RET homology model development
A VEGFR-2 DFG-out crystal structure [36] and the amino acid sequence of RET [37] was obtained. Using SwissModel Automatic Modelling Mode (swissmodel.expassy.org) [33–35], the RET amino acid sequence was employed to build a RET DFG-out homology model using the VEGFR2 DFG-out structure as a template. The resulting RET DFG-out homology model displayed the predicated shift in the DFG-out motif and harbored a newly accessible allosteric pocket.
4.6. RET computational modeling
Computational modeling studies were completed using Auto-Dock Vina [38], AutoDock Tools, and Discovery Studio 3.5. Using AutoDock Tools, the RET homology model was prepared as follows: 1) All hydrogens were added as ‘Polar Only’ 2) A grid box for the ATP binding site was created (center x = −25.881, center y = 9.55, center z = −10.927/size x = 16, size y = 44, size z = 18). Compounds to be computationally modeled were assigned appropriate rotatable bonds using AutoDock Tools. To computational model the compounds, AutoDock Vina [38] was employed. After the modeling study, the results were visualized and analyzed with Discovery Studio 3.5.
4.7. Cell cultures
RAT1 fibroblasts expressing RET/C634R point mutant cells have been described previously and were grown in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA) [40].
4.8. Protein studies
Immunoblotting experiments were performed according to standard procedures. Cells were harvested in lysis buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM EGTA, 1.5 mM MgCl2, 10 mM NaF, 10 mM sodium pyrophosphate, 1 mM Na3VO4, 10 µg of aprotinin/ml, 10 µg of leupeptin/ml) and clarified by centrifugation at 10,000× g. Protein concentration was estimated with a modified Bradford assay (Bio-Rad Laboratories, Berkeley, CA, USA). Antigens were revealed by an enhanced chemiluminescence detection kit (ECL, Amersham Pharmacia Biotech). Anti-RET is a polyclonal antibody raised against the tyrosine kinase protein fragment of human RET. Anti-pTyr905 and anti-pTyr1062 are phospho-specific affinity-purified polyclonal antibodies that recognize RET proteins phosphorylated at Y905 and Y1062, respectively. [17]
Supplementary Material
Acknowledgments
This work was supported by a training grant from The National Institutes of Health (3T32GM008804-10S1), University of Arizona startup funding, ACS-IRG grant 4288802, The Caldwell Health Sciences Research Fellowship, and the Associazione Italiana Ricerca sul Cancro AIRC.
Abbreviations
- RET
rearranged during transfection
- MTC
medullary thyroid cancer
- VEGFR-2
vascular endothelial growth factor receptor 2
- MCR
multicomponent reaction
- SAR
structure activity relationship
- DFG
tyrosine-phenylalanine-glycine
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- DTT
dithiothreitol
- ATP
adenosine triphosphate
- DMF
dimethyl formamide
- DMSO
dimethyl sulfoxide
- FAM
carboxyfluorescein
- DCM
dichloromethane
- MeOH
methanol
- TLC
thin layer chromatography
- NMR
nuclear magnetic resonance
- HPLC
high-performance liquid chromatography
- THF
tetrahydrofuran
- IPA
isopropyl alcohol
- EDC
1-ethyl-3-(3-dimethylaminopropyl)carbodiimide
- HAOt
1-hydroxy-7-azabenzotriazole
- DIPEA
N,N-diisopropylethylamine
- EtOAc
ethyl acetate
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ejmech.2014.09.023.
References
- 1.Takahashi M, Ritz J, Cooper GM. Cell. 1985;42:581–588. doi: 10.1016/0092-8674(85)90115-1. [DOI] [PubMed] [Google Scholar]
- 2.Mulligan LM, Kwok JB, Healey CS, Elsdon MJ, Eng C, Gardner E, Love DR, Mole SE, Moore JK, Papi L, Ponder MA, Telenius H, Tunnacliffe A, Ponder BJ. Nature. 1993;363:458–460. doi: 10.1038/363458a0. [DOI] [PubMed] [Google Scholar]
- 3.Mulligan LM, Eng C, Healey CS, Clayton D, Kwok JB, Gardner E, Ponder MA, Frilling A, Jackson CE, Lehnert H, Neumann HPH, Thibodeau SN. Nat. Genet. 1994;6:70–74. doi: 10.1038/ng0194-70. [DOI] [PubMed] [Google Scholar]
- 4.Eng C, Smith DP, Mulligan LM, Nagai MA, Hea-ley CS, Ponder MA, Gardner E, Scheumann GF, Jackson CE, Tunnacliffe A, Ponder BA. J. Hum. Mol. Genet. 1994;3:237–241. doi: 10.1093/hmg/3.2.237. [DOI] [PubMed] [Google Scholar]
- 5.Hofstra RM, Landsvater RM, Ceccherini I, Stulp RP, Stelwagen T, Luo Y, Pasini B, Höppener JW, van Amstel HK, Romeo G, Lips CJM, Buys CHCM. Nature. 1994;367:375–376. doi: 10.1038/367375a0. [DOI] [PubMed] [Google Scholar]
- 6.Santoro M, Carlomagno F, Romano A, Bottaro DP, Dathan NA, Grieco M, Fusco A, Vecchio G, Matoskova B, Kraus MH, Di Fiore PP. Science. 1995;267:381–383. doi: 10.1126/science.7824936. [DOI] [PubMed] [Google Scholar]
- 7.Michiels FM, Chappuis S, Caillou B, Pasini A, Tal-bot M, Monier R, Lenoir GM, Feunteun J, Billaud M. Proc. Natl. Acad. Sci. 1997;94:3330–3335. doi: 10.1073/pnas.94.7.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acton DS, Velthuyzen D, Lips CJ, Höppener JW. Oncogene. 2000;19:3121–3125. doi: 10.1038/sj.onc.1203648. [DOI] [PubMed] [Google Scholar]
- 9.Drosten M, Hilken G, Böckmann M, Rödicker F, Mise N, Cranston AN, Dahmen U, Ponder BA, Pützer BM. J. Natl. Cancer Inst. 2004;96:1231–1239. doi: 10.1093/jnci/djh226. [DOI] [PubMed] [Google Scholar]
- 10.Rizzi E, Cassinelli G, Dallavalle S, Lanzi C, Cinci-nelli R, Nannei R, Cuccuru G, Zunino F. Bioorg. Med. Chem. Lett. 2007;17:3962–3968. doi: 10.1016/j.bmcl.2007.04.091. [DOI] [PubMed] [Google Scholar]
- 11.Robinett RG, Freemerman AJ, Skinn MA, Shew-chuk L, Lackey K. Bioorg. Med. Chem. Lett. 2007;17:5886–5893. doi: 10.1016/j.bmcl.2007.07.104. [DOI] [PubMed] [Google Scholar]
- 12.Cincinelli R, Cassinelli G, Dallavalle S, Lanzi C, Merlini L, Botta M, Tuccinardi T, Martinelli A, Penco S, Zunino F. J. Med. Chem. 2008;51:7777–7787. doi: 10.1021/jm8007823. [DOI] [PubMed] [Google Scholar]
- 13.Mologni L, Rostagno R, Brussolo S, Knowles PP, Kjaer S, Murray-Rust J, Rosso E, Zambon A, Scapozza L, McDonald NQ, Lucchini V, Gambacorti-Passerini C. Bioorg. Med. Chem. 2010;18:1482–1496. doi: 10.1016/j.bmc.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 14.Brandt W, Mologni L, Preu L, Lemcke T, Gambacorti-Passerini C, Kunick C. Eur. J. Med. Chem. 2010;45:2919–2927. doi: 10.1016/j.ejmech.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 15.Diner P, Alao JP, Soderlund J, Sunnerhagen P, Grotli M. J. Med. Chem. 2012;55:4872–4876. doi: 10.1021/jm3003944. [DOI] [PubMed] [Google Scholar]
- 16.Sartini S, Coviello V, Bruno A, La Pietra V, Marinelli L, Simorini F, Taliani S, Salerno S, Marini AM, Fioravanti A, Orlandi P, Antonelli A, Da Settimo F, Novellino E, Bocci G, La Motta C. J. Med. Chem. 2014;57:1125–1135. doi: 10.1021/jm401358b. [DOI] [PubMed] [Google Scholar]
- 17.Carlomagno F, Vitagliano D, Guida T, Ciardiello F, Tortora G, Vecchio G, Ryan AJ, Fontanini G, Fusco A, Santoro M. Cancer Res. 2002;62:7284–7290. [PubMed] [Google Scholar]
- 18.Yakes FM, Chen J, Tan J, Yamaguchi K, Shi Y, Yu P, Qian F, Chu F, Bentzien F, Cancilla B, Orf J, You A, Laird AD, Engst S, Lee L, Lesch J, Chou YC, Joly AH. Mol. Cancer Ther. 2011;10:2298–2308. doi: 10.1158/1535-7163.MCT-11-0264. [DOI] [PubMed] [Google Scholar]
- 19.Armstrong RW, Combs AP, Tempest PA, Brown SD, Keating TA. Acc. Chem. Res. 1996;29:123–131. [Google Scholar]
- 20.Dömling A, Ugi I. Angew. Chem. Int. Ed. 2000;39:3168–3210. doi: 10.1002/1521-3773(20000915)39:18<3168::aid-anie3168>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 21.Dömling A, Achatz S, Beck B. Bioorg. Med. Chem. Lett. 2007;17:5483–5486. doi: 10.1016/j.bmcl.2007.04.066. [DOI] [PubMed] [Google Scholar]
- 22.Fan X, Zhang X, Bories C, Loiseau PM, Torrence PF. Bioorg. Chem. 2007;35:121–136. doi: 10.1016/j.bioorg.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 23.Musonda CC, Taylor D, Lehman J, Gut J, Rosenthal PJ, Chibale K. Bioorg. Med. Chem. Lett. 2004;14:3901–3905. doi: 10.1016/j.bmcl.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 24.Serry AM, Luik S, Laufer S, Abadi AH. J. Med. Chem. 2008;51:7777–7787. [Google Scholar]
- 25.Akritopoulou-Zanze I, Wakefield BD, Gasiecki A, Kalvin D, Johnson EF, Kovar P, Djuric SW. Bioorg. Med. Chem. Lett. 2011;21:1480–1483. doi: 10.1016/j.bmcl.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Rao VK, Chhikara BS, Tiwari R, Shirazi AN, Parang K, Kumar A. Bioorg. Med. Chem. Lett. 2012;22:410–414. doi: 10.1016/j.bmcl.2011.10.124. [DOI] [PubMed] [Google Scholar]
- 27.Szymanski W, Zwolinska M, Klossowski S, Młynar-czuk-Biały I, Biały L, Issat T, Malejczyk J, Ostaszewski R. Bioorg. Med. Chem. 2014;22:1773–1781. doi: 10.1016/j.bmc.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Groebke K, Weber L, Mehlin F. Synlett. 1998:661–663. [Google Scholar]
- 29.Sharma A, Li HY. Synlett. 2011:1407–1412. [Google Scholar]
- 30.Nichol GS, Sharma A, Li HY. Acta Crystallogr. 2011;E67:1224. [Google Scholar]
- 31. http://www.caliperls.com/products/labchip-systems/labchip-ez-reader.htm.
- 32.Wu Z, Fraley ME, Bilodeau MT, Kaufman ML, Tasber ES, Balitza AE, Hartman GD, Coll KE, Rickert K, Shipman J, Shi B, Sepp-Lerenzino L, Thomas KA. Bioorg. Med. Chem. Lett. 2004;14:909–912. doi: 10.1016/j.bmcl.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 33.Pargellis C, Tong L, Churchill L, Cirillo PF, Gilmore T, Graham AG, Grob PM, Hickey ER, Moss N, Pav S, Regan J. Nat. Struct. Biol. 2002;9:268–272. doi: 10.1038/nsb770. [DOI] [PubMed] [Google Scholar]
- 34.Arnold K, Bordoli L, Kopp J, Schwede T. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 35.Schwede T, Kopp J, Guex N, Peitsch MC. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guex N, Peitsch MC. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 37.Hasegawa M, Nishigaki N, Washio Y, Kano K, Harris PA, Sato H, Mori I, West RI, Shibahara M, Toyoda H, Wang L, Nolte RT, Veal JM, Cheung M. J. Med. Chem. 2007;50:4453–4470. doi: 10.1021/jm0611051. [DOI] [PubMed] [Google Scholar]
- 38.Knowles PP, Murray-Rust J, Kjer S, Scott RP, Hanrahan S, Santoro M, Ibanez CF, McDonald NQ. J. Biol. Chem. 2006;281:33577–33587. doi: 10.1074/jbc.M605604200. [DOI] [PubMed] [Google Scholar]
- 39.Trott O, Olson AJ. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carlomagno F, Guida T, Anaganti S, Vecchio G, Fusco A, Ryan AJ, Billaud M, Santoro M. Oncogene. 2004;23:6056–6063. doi: 10.1038/sj.onc.1207810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.