Abstract
Glioblastoma (GBM) is the most common and highly lethal primary malignant brain tumor in adults. There is a dire need for easily accessible, noninvasive biomarkers that can delineate underlying molecular activities and predict response to therapy. To this end, we sought to identify subtypes of GBM, differentiated solely by quantitative MR imaging features, that could be used for better management of GBM patients. Quantitative image features capturing the shape, texture, and edge sharpness of each lesion were extracted from MR images of 121 patients with de novo, solitary, unilateral GBM. Three distinct phenotypic “clusters” emerged in the development cohort using consensus clustering with 10,000 iterations on these image features. These three clusters—pre-multifocal, spherical, and rim-enhancing, names reflecting their image features—were validated in an independent cohort consisting of 144 multi-institution patients with similar tumor characteristics from The Cancer Genome Atlas (TCGA). Each cluster mapped to a unique set of molecular signaling pathways using pathway activity estimates derived from analysis of TCGA tumor copy number and gene expression data with the PARADIGM algorithm. Distinct pathways, such as c-Kit and FOXA, were enriched in each cluster, indicating differential molecular activities as determined by image features. Each cluster also demonstrated differential probabilities of survival, indicating prognostic importance. Our imaging method offers a noninvasive approach to stratify GBM patients and also provides unique sets of molecular signatures to inform targeted therapy and personalized treatment of GBM.
Introduction
Glioblastoma (GBM) is the most frequent and lethal primary malignant brain tumor in adults. Upon patient presentation with subacute and progressive neurologic signs and symptoms, gadolinium-enhanced cranial magnetic resonance imaging (MRI) is used as the main diagnostic modality for brain abnormalities (1). Characteristic hypointensity on T1-weighted images and heterogeneous enhancement following contrast infusion strongly suggest GBM. MR images demonstrate the extent and location of tumor involvement, which can determine the feasibility of, and approach used in surgical intervention. Although recent clinical trials are evaluating advanced MRI techniques to improve assessment of treatment response in GBM (2) or to evaluate changes in tumor blood flow following treatment (3) in known GBM cases, MR images are not currently being used to sub-classify GBM risk groups. Moreover, regardless of imaging findings, a tissue diagnosis is ultimately required for definitive histopathologic confirmation and to distinguish from other primary and metastatic brain tumors. Factors currently known to be associated with survival include age and Karnofsky performance status (KPS) (4), as well as O6-methylguanine–DNA methyltransferase (MGMT) promoter hypermethylation (5) and mutations in isocitrate dehydrogenase 1 (IDH1) or IDH2 (6, 7). Furthermore, gene expression-based molecular classification of GBM (8), epidermal growth factor receptor (EGFR) amplification (9) and CpG island methylator phenotype (CIMP) status (10) have emerged as potential, additional predictors of treatment response and outcome. While such genomic characterization that encompasses descriptions of gene expression profiles, underlying genomic abnormalities, and epigenetic modification has improved the clinical assessment of GBM (8, 10–12), there remains an unmet clinical need for easily accessible, surrogate biomarkers able to delineate accurately underlying molecular activities and predict response to therapy.
Tumor molecular heterogeneity poses a challenge to the accurate understanding of the underlying molecular activities in GBM (13, 14). Substantial intratumoral heterogeneity requires analysis of multiple regions of a tumor to capture its full clonal history. Recent advances in imaging analysis permit 3D quantitative characterization of the imaging phenotype of GBM tumors (15–18) that includes this heterogeneity. The emerging field of imaging genomics involves mapping image features to molecular data. In pioneering work, investigators have linked quantitative CT image features to gene expression data of non-small cell lung cancer to predict survival (19, 20). Similarly, a handful of groups have discovered associations between imaging and gene expression modules in GBM (15), and built models predicting survival by correlating qualitative imaging phenotypes with gene expression data alone (9) or with the addition of microRNA data (21).
In this study, we sought to establish image-based biomarkers of GBM subtypes, ultimately enabling imaging to substitute for intensive molecular analysis. Such an image-based approach would avoid the risks of biopsy and more comprehensively assess intratumoral heterogeneity. Here, we identify three GBM subtypes differentiated solely by quantitative MR imaging features and show that these subtypes have prognostic relevance and reflect distinct molecular pathways.
Results
Three MR imaging GBM subtypes exist
MR imaging data were obtained in 265 GBM patients, split into two different cohorts: the development cohort for subtype identification and a validation cohort. The development cohort consisted of 121 patients with solitary unilateral tumors evident on MRIs. The validation cohort, which was used to validate findings from the development cohort, was comprised of 144 subjects with solitary, unilateral brain lesions subjects from The Cancer Imaging Archive (TCIA). Table 1 summarizes the baseline characteristics of the two cohorts. The selection process did not materially alter the clinical characteristics (age, sex, KPS) of each cohort from those of the original cohorts (tables S1 and S2). For each subject in each cohort we applied a quantitative imaging pipeline, extracting gray-value histogram statistics, textures, sharpness of lesion boundaries, and metrics of compactness and roughness as described in Methods, and generated 388 image features representing both 2D and multi-slice 2D (aggregated 2D slices) characteristics of each lesion (table S3).
Table 1. Clinical and molecular characteristics of the development (Stanford) and validation (TCGA) cohorts.
Clinical data for the validation cohort were available on 114 subjects, and molecular data on 107 subjects (missing data are noted).
| Characteristic | Stanford Development Cohort (n=121) |
TCGA Validation Cohort (n=144) |
|
|---|---|---|---|
| Age, mean (SD) | 64.6 (14.1) | 58.5 (15.1) | |
| Sex, n male (%) | 69 (57) | 73 (64) | |
| KPS, n (%) | |||
| >70% | 65 (54) | 78 (68) | |
| 50–70% | 44 (37) | 19 (17) | |
| <50% | 11 (9) | 1 (1) | |
| Missing | 1 | 16 | |
| Mean ± SD | 71.7 ± 17 | 78.6 ± 12.3 | |
| Location, n (%) | |||
| Frontal | 42 (35) | ||
| Parietal | 23 (19) | ||
| Basal Ganglia | 3 (3) | ||
| Temporal | 49 (41) | ||
| Occipital | 4 (3) | ||
| EGFR amplification, n (%) | 21 (17) | 100 (69) | |
| Missing | 74 | 26 | |
| IDH1 mutation, n (%) | 4 (4) | ||
| Missing | 22 | ||
| MGMT hypermethylation, n (%) | 27 (22) | 25 (23) | |
| Missing | 73 | 60 | |
| Molecular subgroups, n (%) | |||
| Proneural | 32 (30) | ||
| Neural | 18 (17) | ||
| Mesenchymal | 31 (29) | ||
| Classical | 25 (23) | ||
| Missing | 1 | ||
Standard deviation is denoted as SD. Karnofsky Performance Status (KPS) is shown as three categories and a mean value. Tumor location, epidermal growth factor receptor (EGFR) amplification, isocitrate dehydrogenase 1 (IDH1) mutation, and O6-methylguanine–DNA methyltransferase (MGMT) promoter hypermethylation are tabulated as number (n) and percentage.
Based on consensus clustering of patients’ quantitative imaging features, we chose the solution with k=3 as optimal, using maximal consensus matrices, the consensus cumulative distribution function (CDF) curve, and the CDF progression graph when the Stanford and TCGA cohorts were used as development and validation cohorts, respectively (Fig. 1A–C), and vice versa (Fig. 1D–F). The three-cluster solution induced the largest k that induced the smallest incremental change in the AUC (Fig. 1, C and F) while still maximizing the consensus within clusters (Fig. 1, A and D) and minimizing the rate of ambiguity in cluster assignments across 10,000 iterations (Fig. 1, B and E). This resulted in 36 development cohort patients in Cluster 1 (30%), 51 patients in Cluster 2 (42%), and 34 patients in Cluster 3 (28%). When the development and validation cohorts were swapped, consensus clustering for k=3 yielded 25 patients in Cluster 1 (18%), 107 patients in Cluster 2 (74%), and 12 patients in Cluster 3 (8%).
Figure 1. Consensus matrix, cumulative distribution function curve, and delta curve for all clusters.
(A to C) The Stanford cohort as the development cohort and the TCGA cohort as the validation cohort. (D to F) The TCGA cohort as the development cohort and the Stanford cohort as the validation cohort. (A and D) Consensus matrices represented as heat maps for k=3 (Clusters 1, 2, and 3). Subjects are both rows and columns and consensus values range from 0 (never clustered together, white) to 1 (always clustered together, dark blue). The matrices are ordered by consensus-clustered groups, depicted as a dendrogram above the heatmap. (B and E) Cumulative distribution function (CDF) curve was one diagnostic tool used to select the optimal number of clusters in consensus clustering. The bottom left of the graph represents sample pairs rarely clustered together, whereas the upper right contains those almost always paired together. The middle segment represents sample pairs with ambiguous assignments across different clustering runs. The goal was to identify the lowest rate of ambiguous assignments (flat middle segment). (C and F) The delta curve depicts the CDF progression graph, plotting the relative change in area under the CDF curve, comparing k with k+1. The goal was to select the largest k that induced the smallest incremental change in the AUC. Data for the TCGA cohort as development, Stanford as validation, are in fig. S2.
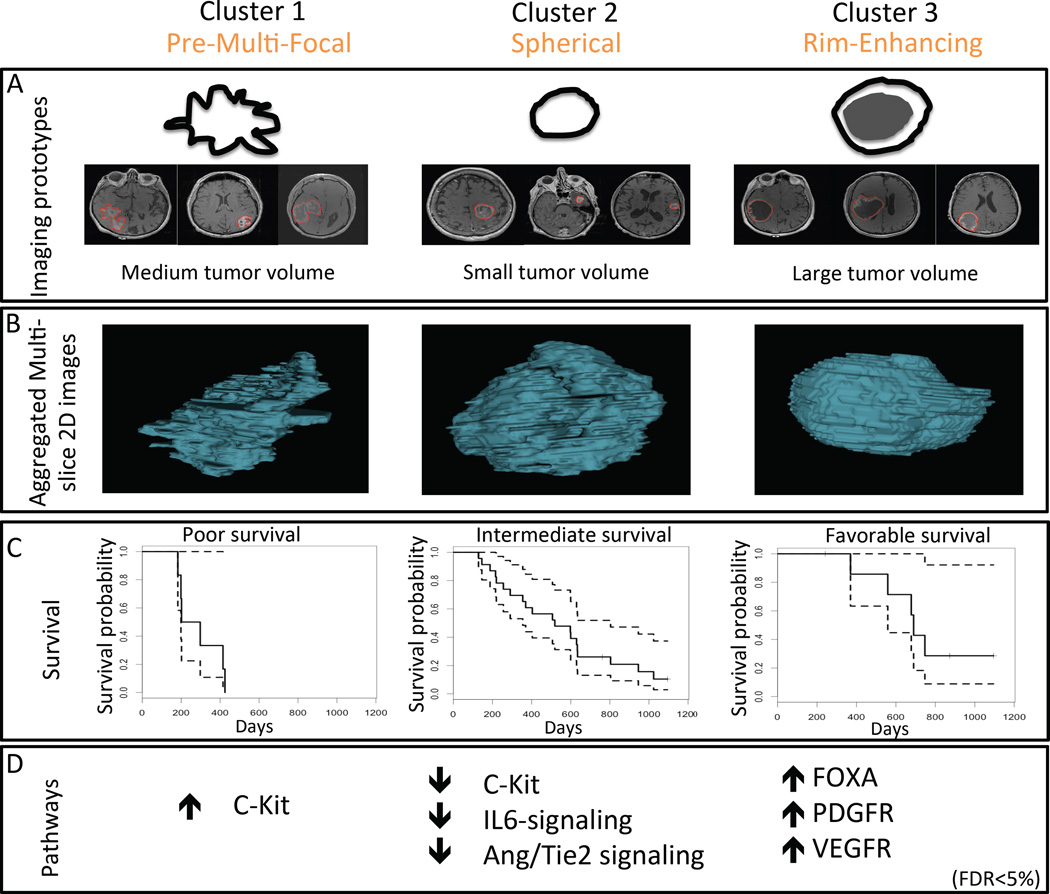
Next, we used Significance Analysis of Microarrays (SAM) (22) to identify the quantitative image features significantly associated with each subtype. In SAM, each feature represented a statistical descriptor of tumor pixel intensities and characteristics, and together they defined the multivariate image phenotypes (Fig. 2A, table S4). The top 24 two-dimensional (2D) and multi-slice 2D imaging features are in fig. S1. Cluster 1 was characterized by quantitative features that described the high irregularity of tumor shapes and many concavities along the tumor outlines. In addition, we observed that the excluded cases from the development cohort with multifocal lesions (n = 76) appeared to resemble features that defined Cluster 1. Therefore, we defined the phenotype of Cluster 1 as the “Pre-Multifocal GBM Cluster” (Fig. 2A). Cluster 2 was characterized by spherical tumors with regular edges that were well circumscribed. We defined the phenotype of Cluster 2 as the “Spherical GBM Cluster” (Fig. 2A). Cluster 3 was distinguished by prominence of central hypointensity encompassed by a hyperintense rim. We defined the phenotype of Cluster 3 as the “Rim-Enhancing GBM Cluster” (Fig. 2A). The aggregated multi-slice 2D renditions of the three clusters are shown in Fig. 2B.
Figure 2. GBM subtypes cluster by phenotypic MRI characteristics, correlate with survival, and associate with molecular pathways.
Three distinct image-based subtypes were derived from a development cohort (Stanford cohort) and validated in an independent validation cohort (TCGA cohort). (A) Imaging phenotypes are illustrated as simplified, representative pictograms for each cluster, although the multivariate combination of quantitative images features that characterize each cluster (table S4) cannot be fully visually exemplified. (B) Aggregate multi-slice 2D renditions of the three imaging subtypes (clusters). (C) Kaplan-Meier survival curves (solid lines) with 95% confidence intervals (dotted lines) derived from TCGA survival data are shown for each cluster in the TCGA cohort. Survival differences across the clusters: P = 0.004, Logrank test (n = 37 subjects across the clusters who underwent the same Stupp protocol treatment regimen). Cluster 1 was characterized by the least favorable survival (n=6), whereas Cluster 3 was marked by the most favorable survival (n=9); Cluster 2 was intermediate (n=22). (D) Molecular changes associated with each cluster. Arrows indicate up- or downregulation of sample pathways identified using PARADIGM. Table S6 provides a comprehensive list of significant regulatory pathways associated with each cluster at FDR < 5%.
In a separate analysis, we evaluated Fluid-Attenuated Inversion Recovery (FLAIR) data that we previously analyzed on 30 subjects (15). We determined that one quantitative FLAIR feature (“histogram-kurtosis”), which characterizes image homogeneity, was statistically significantly different among the three clusters (Kruskal-Wallis P = 0.01575).
GBM imaging subtypes confirmed in the heterogeneous validation cohort
We used the In-Group Proportions (IGP) statistic (23) to validate Clusters 1 to 3 in an independent, heterogeneous cohort. The IGP is a measure of cluster homogeneity and prediction accuracy and indicates whether a cluster in one dataset is similar to a cluster in another dataset; if the clusters are similar, the IGP approaches 100%. In IGP, the Pearson’s centered correlation coefficient between each datum and each centroid is calculated. The datum is then classified to the group whose centroid has the highest correlation with the datum. The IGP is defined to be the proportion of data in a group whose nearest neighbors (Pearson’s centered correlation) are also classified to the same group. All three clusters were statistically significant (P <0.001, <0.001, and <0.01 for Clusters 1, 2, and 3, respectively), denoting low probabilities that these clusters originated from a null distribution. The corresponding IGP values for Clusters 1 to 3 were 91%, 80%, and 80%, respectively. Only eight out of 144 subjects (six in Cluster 1 and two in Cluster 2) were assigned to a cluster in the validation cohort based on nearest neighbor assignment probabilities of less than 0.70.
Next, we used Prediction Analysis for Microarrays (PAM) (24) to define an image classifier on the three development cohort clusters and assign each sample in the validation cohort to one of the clusters. This assigned 41 validation subjects to the Pre-Multifocal Cluster 1 (21%), 63 to the Spherical Cluster 2 (64%) and 40 to the Rim-Enhancing Cluster 3 (15%). Repeating these procedures for k=3 by interchanging the development and validation cohorts led to poor consensus within clusters (Fig. 1D) and high rates of ambiguity of cluster assignments across the 10,000 iterations (Fig. 1D–E). Only Cluster 1 could be validated using IGP (P < 0.001, IGP 81%).
Secondary analysis including midline-crossing lesions does not change clusters
To address the impact of having excluded midline-crossing lesions, we performed a secondary analysis that included these lesions. The addition of midline-crossing lesions (n=19) did not alter the distribution of clusters. Cluster assignment remained grossly unchanged in 116 (97%) of the original 121 subjects in the development cohort (Table S5). Of the additional 19 subjects, three (16%) were assigned to Cluster 1, eight (42%) to Cluster 2, and eight (42%) to Cluster 3. The additional samples increased the total number of members in Cluster 1 to 39 (28% of 140), Cluster 2 to 59 (41%), and Cluster 3 to 43 (31%) from 30%, 42%, and 29%, respectively. Thus, midline-crossing lesions do not form their own cluster, but are subsumed under the existing three clusters.
GBM imaging subtypes are prognostic of survival, independent of established or putative clinical and molecular markers
We correlated the imaging-based GBM subtypes with overall survival of patients in the validation cohort treated on the Stupp protocol (25), which is standard first-line therapy consisting of concomitant radiotherapy with temozolomide, followed by adjuvant temozolomide alone. We observed significant differences in survival probabilities for the three subgroups in the TGCA cohort (Fig. 2C P = 0.004, Log rank test). Pre-Multifocal Cluster 1 had the poorest survival rate; Spherical Cluster 2 had an intermediate rate; and Rim-Enhancing Cluster 3 had the best survival rate.
We further examined the correlation of these image-based clusters with established or putative clinical and molecular risk factors of survival in the development cohort (Table 2A). There were no significant differences among the three clusters for age, sex, KPS, MGMT hypermethylation, or EGFR amplification. However, tumor volume was significantly different among the clusters; the smallest tumors were found in the Spherical Cluster 2, whereas the largest tumors were in the Rim-Enhancing Cluster 3 (P < 0.001, Kruskal-Wallis test) (Fig. 3). In spite of this association, tumor volume was not a sufficient independent predictor of the three clusters. The correlation coefficient between image-based subtypes and tumor volumes was 0.317 (95% CI 0.146–0.469). Moreover, in regression models, tumor volume was not a statistically significant predictor of image-based subtypes (p=0.923, p=0.0777, p=0.234 for Clusters 1, 2, and 3, respectively). Using tumor volume alone to predict image-based subtypes led to accuracy of 43.8%. We also found a nonsignificant correlation with tumor location (basal ganglia, frontal, occipital, parietal, or temporal) (P = 0.052, Fisher’s exact test).
Table 2. Clinical and molecular characteristics of the three imaging subtypes in both cohorts.
(A) The development (Stanford) cohort. (B) The validation (TCGA) cohort.
| Characteristic | Cluster 1, Pre-Multifocal |
Cluster 2, Spherical |
Cluster 3, Rim- Enhancing |
P value | |
|---|---|---|---|---|---|
| (A) Development cohort | |||||
| Age, mean (SD) | 64.7 (17.7) | 66.0 (11.7) | 62.0 (13.0) | 0.324* | |
| Sex, n (%) | |||||
| Female | 14 | 24 | 14 | ||
| Male | 22 | 27 | 20 | 0.736# | |
| Mean KPS (SD) | 67.7 (19.2) | 73.7 (13.9) | 72.9 (18.1) | 0.439* | |
| Tumor volume, cm3 (SD) | 31.2 (30.1) | 22.6 (18.9) | 55.9 (32.7) | 4.49×10−7* | |
| MGMT hypermethylation, n | 8 | 12 | 7 | 0.747# | |
| Missing | 25 | 29 | 19 | ||
| EGFR amplification, n | 5 | 8 | 8 | 0.702# | |
| Missing | 24 | 33 | 17 | ||
| Location, n | 0.0532# | ||||
| Basal ganglia | 0 | 2 | 1 | ||
| Frontal | 9 | 20 | 13 | ||
| Occipital | 4 | 0 | 0 | ||
| Parietal | 7 | 6 | 10 | ||
| Temporal | 16 | 23 | 11 | ||
| (B) Validation cohort | |||||
| Age, mean (SD) | 58.6 (14.5) | 58.9 (16.5) | 57.6 (13.4) | 0.506* | |
| Sex, n (%) | |||||
| Female | 15 (37) | 15 (24) | 11 (28) | ||
| Male | 15 (37) | 37 (59) | 21 (52) | 0.171# | |
| Missing | 11 | 11 | 8 | ||
| Mean KPS ± SD | 78.3 ± 10.3 | 80 ± 12.6 | 76.6 ± 13.2 | 0.488* | |
| Missing | 7 | 6 | 3 | ||
| Molecular subgroups, n (%) | |||||
| Classical | 4 (10) | 13 (21) | 8 (20) | ||
| Mesenchymal | 7 (17) | 19 (30) | 5 (13) | ||
| Neural | 7 (17) | 3 (5) | 8 (20) | ||
| Proneural | 7 (17) | 12 (19) | 7 (18) | ||
| Missing | 16 | 16 | 12 | ||
| 0.088# | |||||
| CIMP, n (%) | 4 (10) | 1 (2) | 1 (3) | 0.121# | |
| Missing | 1 | 2 | 2 | ||
| IDH1 mutation, n (%) | 3 (10) | 1 (2) | 0 (0) | 0.216# | |
| Missing | 2 | 11 | 9 | ||
| MGMT hypermethylation, n (%) | 6 (21) | 13 (27) | 6 (21) | 0.496# | |
| Missing | 17 | 28 | 15 | ||
| EGFR amplification, n (%) | 27 (66) | 43 (68) | 30 (75) | ||
| Missing | 10 | 15 | 8 | ||
KPS: Karnofsky performance status; MGMT: O6-methylguanine–DNA methyltransferase promoter hypermethylation, EGFR: Epidermal growth factor receptor; IDH1 isocitrate dehydrogenase 1 mutation; and CIMP: CpG island methylator phenotype. For continuous values, data are means (SD).
P-values are derived from comparisons across the three clusters, using the Kruskal-Wallis test (*) for continuous variables or the Fisher’s exact test (#) for categorical variables. Where noted as missing, molecular features were not available for analysis.
Figure 3. Tumor volumes demonstrated by cluster for the development cohort (Stanford cohort).
The largest tumors were in Cluster 3, and the smallest in Cluster 2 (P < 0.001, Kruskal-Wallis test). An overlap in tumor volume is observed between Clusters 1 and 2.
We conducted a similar analysis for the validation cohort using molecular data from the TCGA (Table 2B). We observed no significant differences among the three clusters for age, sex, KPS, or other known or putative molecular factors that have been associated with survival, including MGMT hypermethylation, EGFR amplification, IDH mutation status, and CIMP.
Mapping canonical pathways to GBM subtypes
To estimate various signaling pathway activities as they pertain to the three Clusters, we used the Pathway Recognition Algorithm Using Data Integration on Genomic Models (PARADIGM) (26) to integrate gene expression and copy number variation (CNV) data of patients in the TCGA validation cohort. All three clusters were significantly associated with one or more regulatory pathways (Fig. 2D, Table 3, table S6, all false discovery rate (FDR) <5%). Notably, the Pre-Multifocal Cluster 1 was marked by only one association: upregulation of the c-Kit stem cell factor receptor pathway. The Spherical Cluster 2 was characterized by downregulation of 21 pathways, including c-Kit, VEGFR signaling, PDGFR-α signaling, FOXA transcriptional networks, and angiopoietin (Ang)/Tie2. Lastly, the Rim-Enhancing Cluster 3 was differentiated by upregulation of 31 pathways, including canonical WNT and PDGFR-β signaling and many of the pathways downregulated in Cluster 2, such as VEGFR signaling, FOXA, and Ang/Tie2 (Table 3, table S6).
Table 3. Selected pathways associated with each image-based cluster.
From the complete set of pathways significantly associated with each image-based cluster (table S6) are selected pathways that are either specifically differentially expressed among the three clusters or among the top ten for the cluster by fold change.
| Signaling pathway | Fold change |
q-value (%) |
|---|---|---|
| Cluster 1 | ||
| Signaling events mediated by stem cell factor receptor (c-Kit) | 1.033 | 0 |
| Cluster 2 | ||
| Signaling events mediated by c-Kit | 0.976 | 0 |
| Signaling events mediated by prolactin (PRL) | 0.997 | 4.5 |
| Platelet-derived growth factor receptor-alpha (PDGFR-α) signaling pathway | 0.997 | 0 |
| Forkhead box protein A2 (FOXA2) and FOXA3 transcription factor networks | 0.991 | 0 |
| Vascular endothelial growth factor receptor 1 (VEGFR1)–specific signals | 0.996 | 4.5 |
| Angiopoietin receptor Tie2-mediated signaling | 0.988 | 0 |
| Regulation of nuclear mothers against decapentaplegic homolog 2/3 (SMAD2/3) signaling | 0.995 | 4.5 |
| Signaling events mediated by protein-tyrosine phosphatase 1B (PTP1B) | 0.995 | 4.5 |
| Osteopontin-mediated events | 0.995 | 4.5 |
| Signaling events activated by hepatocyte growth factor receptor (c-Met) | 0.994 | 4.5 |
| Cluster 3 | ||
| FOXA2 and FOXA3 transcription factor networks | 1.013 | 0 |
| PDGFR-β signaling pathway | 1.013 | 0 |
| Canonical Wnt signaling pathway | 1.003 | 3.6197 |
| VEGFR1–specific signals | 1.004 | 0 |
| Angiopoietin receptor Tie2-mediated signaling | 1.009 | 3.6197 |
| Syndecan-1–mediated signaling events | 1.011 | 0 |
| IL6–mediated signaling events | 1.010 | 3.6197 |
| Osteopontin-mediated events | 1.010 | 0 |
| Fc-ε receptor I signaling in mast cells | 1.009 | 0 |
| Alpha-M beta-2 (αMβ2) Integrin signaling | 1.009 | 0 |
Discussion
We have identified three distinct clusters of unilateral, solitary GBM defined by quantitative MR image features. The clusters of GBM subtypes were discovered in one cohort and validated in a second cohort. In addition to distinguishing imaging phenotypes, or “clusters”—which we named Pre-Multifocal, Spherical, and Rim-Enhancing—the three clusters demonstrated significant differences in survival probabilities and in associations with canonical signaling pathways. Imaging-based markers of disease phenotype may therefore offer actionable knowledge for clinical decision-making and therapeutic targeting.
The current clinical standard for disease relapse surveillance is tracking changes clinically and with MR imaging (27–29). Initial treatment for newly diagnosed GBM is maximal surgical resection while preserving neurologic function. However, a major challenge in the management of GBM is the limited number of non-surgical therapeutic options, including targeted therapies against molecular derangements. Our results show the potential of imaging features to infer upregulated molecular activities for which targeted therapies exist. If the effectiveness of such therapies can be demonstrated, there may be a role of such agents as adjuvant therapy in the post-operative setting, primary treatment in surgically unresectable cases, and neo-adjuvant treatment for preoperative reduction of tumor volumes.
Based on these associations with canonical signaling pathways, the imaging subtypes have the potential to direct targeted therapy. Specifically, the Pre-Multifocal Cluster 1 was associated with upregulation of the c-Kit pathway, whereas the Rim-Enhancing Cluster 3 was characterized by upregulation of several pathways, including canonical Wnt, VEGFR, PDGFR, and angiopoietin/Tie2. This suggests the potential utility for patients in Cluster 1 of targeting c-Kit with tyrosine kinase inhibitors, such as imatinib and dasatinib. Similarly, while it has long been described that GBM overexpresses VEGF-A and are thus amenable to treatment with a VEGF inhibitor (30–37), there is strong rationale for the use of bevacizumab – a VEGF inhibitor – or anti-angiogenic multi-kinase inhibitors that target both VEGFR and PDGFR, such as sorafenib and sunitinib, in patients in Cluster 3. Clinical trials are currently investigating inhibitors of the Ang/Tie2 pathway, potentially expanding therapeutic options for members of Cluster 3 (38). These results provide intriguing data for retrospective validation and eventually for designing prospective clinical trials to confirm these hypotheses.
Conversely, absence of correlation suggests that the tumors in the cluster lack the potential therapeutic targets of the pathway. For instance, treatment options described for Clusters 1 and 3 will likely be ineffective in Cluster 2. Such knowledge makes clinical trials more efficient and spares patients the time and risk of undergoing trials of limited likelihood of success. Recently Chinot et al. reported the findings of a randomized, placebo-controlled, double-blind, phase 3 trial of bevacizumab as first-line therapy for patients with newly diagnosed glioblastoma in concert with standard chemoradiotherapy (39) and identified no overall survival difference in response to bevacizumab (40). As many new therapeutic approaches are often applied to all patients in a diagnostic category without individualization, or with suboptimal individualization, potential beneficial effects may be masked. We hypothesize that the Rim-Enhancing Cluster (Cluster 3), which upregulates the VEGFR signaling pathway, would be more likely to respond to bevacizumab.
The proposed imaging-based subtypes stratified survival in GBM patients treated on the Stupp protocol, which entails concurrent chemoradiotherapy, followed by adjuvant chemotherapy. Moreover, the imaging subtypes conferred survival differences, independent of established risk factors, thus offering additional independent prognostic information, similar to identifying significant factors while controlling for established risk factors. The quantitative MR image features used to define the three clusters thus constitute potential biomarkers for survival in GBM.
Our image-based clusters have the potential to be used for non-invasive treatment follow-up. The cluster phenotypes could be used as surrogate markers of underlying molecular activities for disease monitoring. In addition, the multivariate pathway activity profile can potentially be linked to a drug response profile. The association of distinct molecular activities with specific image-based GBM clusters suggests that image features alone may be used to track changes in disease activity, including providing insight into the natural progression of disease. The main technical challenge is integrating our multivariate image profile into the clinical workflow to assign patients to the three subtypes. Ideally, our methodology and image profile would be incorporated into the radiologists’ toolbox. This would require manual image segmentation, which is currently labor intensive.
Our study extended the earlier efforts in imaging genomics analysis of cancers. Previous studies have linked CT image features to gene expression data of lung cancer (19, 20) and MR image features with genomic data in GBM (15, 17, 21). Whereas previous work focused on genomic analysis before linking with an image phenotype, our study inverted this design by beginning with a characterization of imaging phenotypes of GBM tumors, followed by linking to signaling pathways. We believe that subtypes defined in this manner are more stable and less susceptible to sampling error resulting from assessing a limited portion of a molecularly heterogeneous tumor. Thus, our approach attempted to minimize the bias inherent in molecular sampling and maximize information captured from the entire 3D imaging phenotype of each tumor.
In this study we meticulously selected homogeneous study populations. We selected only those subjects with solitary, unilateral lesions and excluded patients with multifocal or midline-crossing lesions. We focused on this subset of patients with the greatest clinical need for imaging biomarkers that could differentiate prognostic groups and for whom personalization of treatment might be most beneficial. Moreover, the homogeneity of study subjects was designed to maximize signal detection.
However, we measured the impact of having excluded midline-crossing lesions by performing a secondary analysis that included these lesions. Subject cluster assignments remained largely unchanged, with a 97% retention of the original assignment (table S5), suggesting that quantitative image features for midline-crossing lesions do not differ materially enough to generate their own separate cluster. Conversely, the inclusion of multifocal lesions remains infeasible given that their analysis would necessitate biopsies at each tumor focus to overcome intertumoral molecular heterogeneity, and multifocal biopsies and molecular features are not routinely acquired.
Next, we used a heterogeneous validation cohort from TCGA that were acquired on multiple scanner models from three different manufacturers at four different institutions and successfully validated the imaging subtypes in a real-world setting. Swapping the development and validation cohorts did not result in the identification of robust cluster generation (Fig. S1), indicating that a homogeneous cohort is necessary for learning subtypes, but not for assigning subjects to imaging subtypes. Nevertheless, the successful validation in a more heterogeneous environment highlights the robustness of the model.
We also focused only on T1 MR imaging, the most widely used MR modality in patients diagnosed with GBM, to allow for broader applicability and use of the model in the clinical setting. We identified one quantitative FLAIR feature that characterizes image homogeneity as statistically significantly different among the three clusters. Larger studies will become possible as more FLAIR data become available in the future. Future work should include other MRI modalities to investigate if additional subtypes exist in the presence of multimodal imaging data.
In summary, the three GBM subtypes discovered here were distinguished by different expressions of molecular pathways and survival probabilities, which open possibilities for target identification and therapeutic development unique to each subtype. Quantitative imaging features may therefore serve as potential biomarkers for defining subtypes of GBM and potentially other cancers.
Materials and Methods
Study Design
We sought to identify novel subtypes of GBM, differentiated solely by quantitative MR imaging features, and discover signature canonical signaling pathways that are molecularly associated with each subtype. To do this, we separately analyzed two cohorts of subjects with GBM (Table 1). This study analyzed data that had already been collected and did not involve new patients randomized to different interventions or treatments. Thus there was no blinding. Also, we analyzed as many subjects as possible, applying stringent inclusion and exclusion criteria to homogenize our study cohorts as was most scientifically reasonable. Since this was not a prospective study, there were no a priori power calculations performed.
In selected patient images, quantitative image features capturing the shape, texture, and edge sharpness of each lesion for each subject were extracted from T1 post-contrast MRI. We used Consensus Clustering to discover subtypes in the development cohort and performed 10,000 iterations to achieve high robustness and to minimize cluster-selection bias resulting from few iterations. Regions-of-interest (ROIs) on the MR imaging study that were too small (<10 pixels) to interpret were excluded. PAM and the IGP statistic were used to validate the reproducibility and robustness of our subtype classification in a second cohort of subjects (validation cohort). To map imaging subtypes to particular molecular signaling pathways, we performed SAM on pathway activity estimates derived from analysis of TCGA tumor CNV and gene expression data with the PARADIGM algorithm. We used SAM to correct for multiple-hypothesis testing. As described, we mitigated against biases in our analyses by the performance of numerous iterations of consensus clustering to ensure cluster stability, the use of an external validation cohort to validate our cluster findings, and correction for multiple hypothesis testing in SAM. Imaging subtypes were then correlated with survival and molecular characteristics, as well as traditional risk factors of GBM. For survival analysis we focused on TCGA cohort patients treated according to the Stupp protocol. Unlike the TCGA cohort, the Stanford cohort does not have a well characterized subgroup of patients who have undergone a homogeneous treatment plan (Stupp protocol), making the comparison of survival curves less interpretable. Because the Stanford cohort has undergone heterogeneous treatments due to diagnosis over a long period (2001–2010), meaningful survival curves could not be depicted.
Patient selection
The study was approved by Stanford’s Institutional Review Board. For the first cohort, we identified 364 single-institution subjects diagnosed with and treated for de novo GBM at the Stanford Medical Center between 2001 and 2010 and selected 121 that met the inclusion criteria below; this was defined as the development cohort. The second was a multi-institutional cohort from The Cancer Genome Atlas (TCGA) project, a federally funded cancer-specific repository of clinical, molecular, and imaging data, in which we selected 144 subjects out of 575 that met the inclusion criteria; this was defined as the validation cohort. Selection criteria for both cohorts included: i) diagnosis of de novo GBM, ii) minimum age of 18 years, iii) availability of preoperative MRI data, and iv) presence of solitary, unilateral tumors. For the development cohort alone, we further selected patients on the basis of similarity of their MR images with respect to machine model and slice thickness.
Quantitative imaging pipeline
MR images for the development cohort were obtained from the Stanford University Medical Center, whereas images for the validation cohort were obtained from TCIA, TCGA’s companion repository of imaging data for cases donated to the TCGA (www.cancerimagingarchive.net). Images from both cohorts underwent the same preprocessing procedures. Two blinded, board-certified neuroradiologists reached consensus regarding a three-dimensional tumor volume by delineating ROIs around areas of enhancement in each T1 post-contrast MR slice. We computed a set of quantitative image features, as previously reported in lung cancer and glioblastoma (15, 19), and applied the imaging feature extraction pipeline to the development and validation cohorts. Briefly, following linear normalization of image pixel intensities, our quantitative image feature pipeline extracted several features, including morphological characteristics, such as tumor shape, edge sharpness, and pixel intensity statistics on areas of enhancement from these ROIs. Two-dimensional and multi-slice 2D feature representation is described in Supplementary methods.
Subtype discovery
We used k-means consensus clustering for unsupervised class discovery on the development cohort to define imaging subtypes (41, 42). We performed 10,000 bootstraps with 80% item resampling on the quantitative image features and used the k-means clustering algorithm with the Euclidean distance metric to examine all resulting clusters from 2 to 10. We selected the number of clusters that yielded the most stable consensus matrices and the most unambiguous cluster assignments across permuted clustering runs. This established the optimal number of intrinsic unsupervised classes as defined by image features in the development cohort. We analyzed the discovered clusters using SAM, where its application on each discovered cluster identified specific image features defining each subtype. Significant associations were corrected for multiple testing using a FDR threshold of ≤15%.
Validation of clusters
To validate the reproducibility of the clusters derived from consensus clustering in the development cohort, we used the IGP analysis to demonstrate the existence of these clusters in the validation cohort, as described in Supplementary methods.
Mapping canonical pathways to imaging subtypes
We used the matched molecular data in the TCGA validation cohort to assign canonical signaling pathways to the discovered imaging subtypes, as described in Supplementary methods.
Evaluating imaging subtypes for traditional risk factors
We examined the subtypes for statistically significant differences in historically associated risk factors, as described in Supplementary methods.
Statistical analysis
In order to ensure cluster stability in our unsupervised analysis, we performed consensus clustering with 10,000 iterations and used CDF and CDF progression graphs to select the optimal number of clusters. Statistical analysis for determining the validity and reproducibility of the three clusters identified in the development cohort employed the IGP statistic in the validation cohort. When comparing across-cluster clinical and molecular feature differences in each cohort, we performed the Kruskal-Wallis test for continuous variables and the Fisher’s exact test for categorical variables (Tables 1 and 2). Parametric assumptions were not made for continuous variables. (The overall sample size and counts as low as 0 in some cells necessitated the use of the Fisher’s exact test rather than the Pearson chi-square test (43, 44)). For evaluation of statistically significant imaging and signaling pathway features associated with each cluster, we used SAM, which adjusts for multiple comparisons (tables S4 and S6). We reported results for FDR <15% for imaging features and FDR < 5% for signaling pathways. The Kaplan-Meier survival curves were compared using the logrank test.
Supplementary Material
Acknowledgments
We thank R. Tibshirani for reviewing the statistical methodologies used in our study.
Funding: Research reported in this publication was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Number R01EB020527, and by the National Cancer Institute under Award Number R01CA160251. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. H.I was supported by the Advanced Residency Training at Stanford (ARTS) Fellowship. S.D.C. gratefully acknowledges support for this study from Craig and Kimberly Darian and Carol Bade.
Footnotes
Supplementary Materials
Methods
Figure S1. Relative contributions of the top 24 imaging features in characterizing each of the clusters.
Table S1. Clinical characteristics of the Stanford cohort before and after selection of the development cohort.
Table S2. Clinical characteristics of the TCGA cohort before and after selection of the validation cohort.
Table S3. All 2D and multi-slice 2D quantitative MR image features used for analysis.
Table S4. Two-dimensional and multi-slice 2D quantitative MR image features significantly associated with each cluster.
Table S5. Cluster assignment by subjects in the development cohort with and without midline-crossing lesions.
Table S6. Regulatory signaling pathways significantly associated with each cluster.
Author contributions: H.I. and O.G. conceived and designed the study; G.R.H., S.D.C., and D.L.R. created the Stanford GBM imaging database; A.S.A., L.A.M., J.J.L., T.L., E.M.W., A.H.F., S.R., S.E., T.D.A., and S.D.C. acquired imaging data and performed annotations and preprocessing; S.N. provided the software for generating quantitative image features; K.W.Y., S.D.C., and G.R.H. provided the single-institution patient cohort; H.I. and O.G. analyzed and interpreted the data; H.I. wrote the manuscript; O.G., G.R.H., L.A.M., S.N., K.W.Y., T.D.A., L.A.M., S.E., A.H.F., S.C., O.M., D.R., T.L. provided edits; all authors read and approved the manuscript.
Competing interests: O.G. and H.I. have filed a provisional patent on the method described in this manuscript. The authors declare that they have no competing interests.
Data and materials availability: TCGA molecular data are available at https://wiki.cancerimagingarchive.net/display/Public/TCGA-GBM. Corresponding imaging data are available at www.cancerimagingarchive.net.
References and Notes
- 1.Chang SM, Parney IF, Huang W, Anderson FA, Jr, Asher AL, Bernstein M, Lillehei KO, Brem H, Berger MS, Laws ER. Patterns of care for adults with newly diagnosed malignant glioma. Jama. 2005;293:557–564. doi: 10.1001/jama.293.5.557. [DOI] [PubMed] [Google Scholar]
- 2.Vanderbilt-Ingram Cancer Center. clinicaltrials.gov. Vanderbilt-Ingram Cancer Center; 2013. 3T MRI Biomarkers of Glioma Treatment Response. [Google Scholar]
- 3.Massachusetts General Hospital. clinicaltrials.gov. Massachusetts General Hospital; 2013. Bevacizumab with temozolomide PET and vascular MRI for GBM. [Google Scholar]
- 4.Laws ER, Parney IF, Huang W, Anderson F, Morris AM, Asher A, Lillehei KO, Bernstein M, Brem H, Sloan A, Berger MS, Chang S. Survival following surgery and prognostic factors for recently diagnosed malignant glioma: data from the Glioma Outcomes Project. Journal of Neurosurgery. 2003;99:467–473. doi: 10.3171/jns.2003.99.3.0467. [DOI] [PubMed] [Google Scholar]
- 5.Hegi ME, Liu L, Herman JG, Stupp R, Wick W, Weller M, Mehta MP, Gilbert MR. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26:4189–4199. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- 6.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. The New England journal of medicine. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanson M, Marie Y, Paris S, Idbaih A, Laffaire J, Ducray F, El Hallani S, Boisselier B, Mokhtari K, Hoang-Xuan K, Delattre JY. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 8.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O'Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haas-Kogan DA, Prados MD, Tihan T, Eberhard DA, Jelluma N, Arvold ND, Baumber R, Lamborn KR, Kapadia A, Malec M, Berger MS, Stokoe D. Epidermal Growth Factor Receptor, Protein Kinase B/Akt, and Glioma Response to Erlotinib. Journal of the National Cancer Institute. 2005;97:880–887. doi: 10.1093/jnci/dji161. [DOI] [PubMed] [Google Scholar]
- 10.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bredel M, Scholtens DM, Harsh GR, Bredel C, Chandler JP, Renfrow JJ, Yadav AK, Vogel H, Scheck AC, Tibshirani R, Sikic BI. A network model of a cooperative genetic landscape in brain tumors. Jama. 2009;302:261–275. doi: 10.1001/jama.2009.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tonjes M, Sill M, Bender S, Kool M, Zapatka M, Becker N, Zucknick M, Hielscher T, Liu XY, Fontebasso AM, Ryzhova M, Albrecht S, Jacob K, Wolter M, Ebinger M, Schuhmann MU, van Meter T, Fruhwald MC, Hauch H, Pekrun A, Radlwimmer B, Niehues T, von Komorowski G, Durken M, Kulozik AE, Madden J, Donson A, Foreman NK, Drissi R, Fouladi M, Scheurlen W, von Deimling A, Monoranu C, Roggendorf W, Herold-Mende C, Unterberg A, Kramm CM, Felsberg J, Hartmann C, Wiestler B, Wick W, Milde T, Witt O, Lindroth AM, Schwartzentruber J, Faury D, Fleming A, Zakrzewska M, Liberski PP, Zakrzewski K, Hauser P, Garami M, Klekner A, Bognar L, Morrissy S, Cavalli F, Taylor MD, van Sluis P, Koster J, Versteeg R, Volckmann R, Mikkelsen T, Aldape K, Reifenberger G, Collins VP, Majewski J, Korshunov A, Lichter P, Plass C, Jabado N, Pfister SM. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 13.Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavare S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, Suva ML, Regev A, Bernstein BE. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science (New York, N.Y.) 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gevaert O, Mitchell LA, Achrol AS, Xu J, Sebastian E, Steinberg GK, Cheshier SH, Napel S, Zaharchuk G, Plevritis SK. Glioblastoma Multiforme: Exploratory Radiogenomic Analysis by Using Quantitative Image Features. Radiology. 2014 doi: 10.1148/radiol.14131731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutman DA, Cooper LA, Hwang SN, Holder CA, Gao J, Aurora TD, Dunn WD, Jr, Scarpace L, Mikkelsen T, Jain R, Wintermark M, Jilwan M, Raghavan P, Huang E, Clifford RJ, Mongkolwat P, Kleper V, Freymann J, Kirby J, Zinn PO, Moreno CS, Jaffe C, Colen R, Rubin DL, Saltz J, Flanders A, Brat DJ. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology. 2013;267:560–569. doi: 10.1148/radiol.13120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diehn M, Nardini C, Wang DS, McGovern S, Jayaraman M, Liang Y, Aldape K, Cha S, Kuo MD. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proceedings of the National Academy of Sciences. 2008;105:5213–5218. doi: 10.1073/pnas.0801279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellingson BM, Lai A, Harris RJ, Selfridge JM, Yong WH, Das K, Pope WB, Nghiemphu PL, Vinters HV, Liau LM, Mischel PS, Cloughesy TF. Probabilistic radiographic atlas of glioblastoma phenotypes. AJNR. American journal of neuroradiology. 2013;34:533–540. doi: 10.3174/ajnr.A3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gevaert O, Xu J, Hoang CD, Leung AN, Xu Y, Quon A, Rubin DL, Napel S, Plevritis SK. Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data--methods and preliminary results. Radiology. 2012;264:387–396. doi: 10.1148/radiol.12111607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nair VS, Gevaert O, Davidzon G, Napel S, Graves EE, Hoang CD, Shrager JB, Quon A, Rubin DL, Plevritis SK. Prognostic PET 18F-FDG Uptake Imaging Features Are Associated with Major Oncogenomic Alterations in Patients with Resected Non–Small Cell Lung Cancer. Cancer Research. 2012;72:3725–3734. doi: 10.1158/0008-5472.CAN-11-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zinn PO, Majadan B, Sathyan P, Singh SK, Majumder S, Jolesz FA, Colen RR. Radiogenomic Mapping of Edema/Cellular Invasion MRI-Phenotypes in Glioblastoma Multiforme. PLoS ONE. 2011;6:e25451. doi: 10.1371/journal.pone.0025451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapp AV, Tibshirani R. Are clusters found in one dataset present in another dataset? Biostatistics (Oxford, England) 2007;8:9–31. doi: 10.1093/biostatistics/kxj029. [DOI] [PubMed] [Google Scholar]
- 24.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, Wesseling P, Villa S, Eisenhauer E, Gorlia T, Weller M, Lacombe D, Cairncross JG, Mirimanoff RO. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. The Lancet. Oncology. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 26.Vaske CJ, Benz SC, Sanborn JZ, Earl D, Szeto C, Zhu J, Haussler D, Stuart JM. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26:i237–i245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macdonald DR, Cascino TL, Schold SC, Jr, Cairncross JG. Response criteria for phase II studies of supratentorial malignant glioma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1990;8:1277–1280. doi: 10.1200/JCO.1990.8.7.1277. [DOI] [PubMed] [Google Scholar]
- 28.Wen PY, Macdonald DR, Reardon DA, Cloughesy TF, Sorensen AG, Galanis E, Degroot J, Wick W, Gilbert MR, Lassman AB, Tsien C, Mikkelsen T, Wong ET, Chamberlain MC, Stupp R, Lamborn KR, Vogelbaum MA, van den Bent MJ, Chang SM. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:1963–1972. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 29.Nabors LB, Ammirati M, Bierman PJ, Brem H, Butowski N, Chamberlain MC, DeAngelis LM, Fenstermaker RA, Friedman A, Gilbert MR, Hesser D, Holdhoff M, Junck L, Lawson R, Loeffler JS, Maor MH, Moots PL, Morrison T, Mrugala MM, Newton HB, Portnow J, Raizer JJ, Recht L, Shrieve DC, Sills AK, Jr, Tran D, Tran N, Vrionis FD, Wen PY, McMillian N, Ho M. Central nervous system cancers. Journal of the National Comprehensive Cancer Network : JNCCN. 2013;11:1114–1151. doi: 10.6004/jnccn.2013.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plate KH, Breier G, Weich HA, Risau W. Vascular endothelial growth factor is a potential tumour angiogenesis factor in human gliomas in vivo. Nature. 1992;359:845–848. doi: 10.1038/359845a0. [DOI] [PubMed] [Google Scholar]
- 31.Hicklin DJ, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23:1011–1027. doi: 10.1200/JCO.2005.06.081. [DOI] [PubMed] [Google Scholar]
- 32.Berkman RA, Merrill MJ, Reinhold WC, Monacci WT, Saxena A, Clark WC, Robertson JT, Ali IU, Oldfield EH. Expression of the vascular permeability factor/vascular endothelial growth factor gene in central nervous system neoplasms. The Journal of clinical investigation. 1993;91:153–159. doi: 10.1172/JCI116165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chinot OL, de La Motte Rouge T, Moore N, Zeaiter A, Das A, Phillips H, Modrusan Z, Cloughesy T. AVAglio: Phase 3 trial of bevacizumab plus temozolomide and radiotherapy in newly diagnosed glioblastoma multiforme. Advances in therapy. 2011;28:334–340. doi: 10.1007/s12325-011-0007-3. [DOI] [PubMed] [Google Scholar]
- 34.Vredenburgh JJ, Desjardins A, Herndon JE, 2nd, Marcello J, Reardon DA, Quinn JA, Rich JN, Sathornsumetee S, Gururangan S, Sampson J, Wagner M, Bailey L, Bigner DD, Friedman AH, Friedman HS. Bevacizumab plus irinotecan in recurrent glioblastoma multiforme. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2007;25:4722–4729. doi: 10.1200/JCO.2007.12.2440. [DOI] [PubMed] [Google Scholar]
- 35.Kreisl TN, Kim L, Moore K, Duic P, Royce C, Stroud I, Garren N, Mackey M, Butman JA, Camphausen K, Park J, Albert PS, Fine HA. Phase II trial of single-agent bevacizumab followed by bevacizumab plus irinotecan at tumor progression in recurrent glioblastoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:740–745. doi: 10.1200/JCO.2008.16.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai A, Tran A, Nghiemphu PL, Pope WB, Solis OE, Selch M, Filka E, Yong WH, Mischel PS, Liau LM, Phuphanich S, Black K, Peak S, Green RM, Spier CE, Kolevska T, Polikoff J, Fehrenbacher L, Elashoff R, Cloughesy T. Phase II study of bevacizumab plus temozolomide during and after radiation therapy for patients with newly diagnosed glioblastoma multiforme. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:142–148. doi: 10.1200/JCO.2010.30.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narayana A, Gruber D, Kunnakkat S, Golfinos JG, Parker E, Raza S, Zagzag D, Eagan P, Gruber ML. A clinical trial of bevacizumab, temozolomide, and radiation for newly diagnosed glioblastoma. J Neurosurg. 2012;116:341–345. doi: 10.3171/2011.9.JNS11656. [DOI] [PubMed] [Google Scholar]
- 38.Cascone T, Heymach JV. Targeting the angiopoietin/Tie2 pathway: cutting tumor vessels with a double-edged sword? Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:441–444. doi: 10.1200/JCO.2011.38.7621. [DOI] [PubMed] [Google Scholar]
- 39.Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R, Carpentier AF, Hoang-Xuan K, Kavan P, Cernea D, Brandes AA, Hilton M, Abrey L, Cloughesy T. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. The New England journal of medicine. 2014;370:709–722. doi: 10.1056/NEJMoa1308345. [DOI] [PubMed] [Google Scholar]
- 40.Colman H, Zhang L, Sulman EP, McDonald JM, Shooshtari NL, Rivera A, Popoff S, Nutt CL, Louis DN, Cairncross JG, Gilbert MR, Phillips HS, Mehta MP, Chakravarti A, Pelloski CE, Bhat K, Feuerstein BG, Jenkins RB, Aldape K. A multigene predictor of outcome in glioblastoma. Neuro-oncology. 2010;12:49–57. doi: 10.1093/neuonc/nop007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monti S, Tamayo P, Mesirov JP, Golub T. Consensus Clustering: A Resampling-Based Method for Class Discovery and Visualization of Gene Expression Microarray Data. Machine Learning. 2003;52:91–118. [Google Scholar]
- 43.Agresti A. Categorical Data Analysis. 3rd Edition. John Wiley & Sons; 2013. [Google Scholar]
- 44.McDonald JH. Handbook of Biological Statistics. 3rd ed. Baltimore, Maryland: Sparky House Publishing; 2014. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.