Abstract
Background
Acquired resistance to BRAF inhibitors (BRAFi) is a near-universal phenomenon caused by numerous genetic and non-genetic alterations. In this study, we evaluated the spectrum, onset, pattern of progression, and subsequent clinical outcomes associated with specific mechanisms of resistance.
Methods
We compiled clinical and genetic data from 100 patients with 132 tissue samples obtained at progression on BRAFi therapy from 3 large, previously published studies of BRAFi resistance. These samples were subjected to whole exome sequencing and/or PCR-based genetic testing.
Results
Among 132 samples, putative resistance mechanisms were identified in 58%, including NRAS or KRAS mutations (20%), BRAF splice variants (16%), BRAFV600E/K amplifications (13%), MEK1/2 mutations (7%), and non-MAPK pathway alterations (11%). Marked heterogeneity was observed within tumors and patients; 18 of 19 patients (95%) with >1 progression biopsy had distinct/unknown drivers of resistance between samples. NRAS mutations were associated with vemurafenib use (p=0.045) and intracranial metastases (p=0.036), and MEK1/2 mutations correlated with hepatic progression (p=0.011). Progression-free survival and overall survival were similar across resistance mechanisms. The median survival after disease progression was 6.9 months, and responses to subsequent BRAF and MEK inhibition were uncommon (2 of 15; 13%). Post-progression outcomes did not correlate with specific acquired BRAFi resistance mechanisms.
Conclusions
This is the first study to systematically characterize the clinical implications of particular acquired BRAFi resistance mechanisms in patients with BRAF-mutant melanoma largest study to compile the landscape of resistance. Despite marked heterogeneity of resistance mechanisms within patients, NRAS mutations correlated with vemurafenib use and intracranial disease involvement.
Keywords: BRAF, vemurafenib, dabrafenib, resistance, acquired, MAPK, NRAS, amplification, splice, MEK1, MEK2, meta-analysis, spectrum
Introduction
Nearly half of melanomas harbor a valine to glutamine substitution in codon 600 of the serine-threonine kinase BRAF (1). These alterations confer constitutive activation of the mitogen activated-protein kinase (MAPK) pathway and thereby drive melanoma growth and progression (1, 2). Small molecule inhibitors of mutant BRAF (vemurafenib and dabrafenib) induce tumor regression and improve survival in melanoma patients compared to chemotherapy (3, 4).
The development of acquired resistance to BRAFi, however, is a significant obstacle to effective targeted therapy. To characterize and overcome acquired resistance, studies have identified numerous genetic and non-genetic drivers of resistance involving MAPK pathway reactivation and MAPK-redundant signaling. These include NRAS mutations (5), BRAFV600E/K amplification (6), alternate splicing of BRAF (7), MEK1/2 mutations (8), PI3K/AKT pathway dysregulation (9, 10), and overexpression of genes including COT, PDGFRβ, and others (5, 11–13). The translational value of these findings has been clearly demonstrated by successful co-targeting of BRAF and MEK, which has further improved clinical outcomes compared with single-agent BRAFi (14–17).
Efforts to systematically define the spectrum and frequency of these BRAFi-resistance mechanisms have been published recently (18–20). These studies established the high incidence of MAPK-reactivating alterations, but identified intra-patient and intra-tumoral heterogeneity of resistance, and a sizable subset of resistance unaccounted for by genomic profiling alone. Despite the value of these studies, each was limited by sample size to correlate specific mechanisms of resistance with corresponding clinical/biologic behavior. Moreover, the different studies reported discordant frequencies of particular resistance mechanisms (e.g. NRAS mutations). Thus, we combined published data from the three largest studies of acquired BRAFi-resistance in 100 melanoma patients to assess the landscape of resistance mechanisms and the corresponding clinical characteristics (18–20).
Methods
Patients and Study Design
Patients (n=100) and progression samples (n=132) were aggregated from previously-published studies conducted under IRB-approved protocols. These studies were led by University Hospital Essen (Essen, Germany) and the Broad Institute (Boston, MA, USA) (18), Melanoma Institute Australia (Sydney, NSW, Australia) (19), University of California, Los Angeles (USA) (20), and collaborators. All patients had advanced BRAFV600-mutant melanoma and received vemurafenib or dabrafenib as first-line MAPK-directed therapy through clinical trials or as commercially-available therapy. Nearly all patients experienced tumor regression and subsequent progressive disease, except seven with primary disease progression. Objective responses and disease progression were defined using Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1 (21). Baseline demographics, treatment duration, overall survival (OS), progression-free survival (PFS), prior therapies, and subsequent treatments were obtained by reviewing medical records and imaging results.
Melanoma Samples
All patients underwent biopsies of melanoma metastases that were present at baseline and enlarged on therapy, or arose during treatment. Most had matched pre-treatment biopsies and some had multiple biopsies obtained upon disease progression. Specimens were stored as FFPE or were snap-frozen per institutional standard operating procedures. Tumor DNA and RNA were extracted per institutional protocols.
Tumor Sequencing
Sequencing was performed by whole exome sequencing (WES in 56% of disease-progression samples or PCR-based sequencing in the remainder. Sequencing of pre-treatment samples corresponding to 107 of the progressing samples was performed (81%), including in all WES samples. Recurrent “hotspot” mutations in NRAS, MEK1, and MEK2 were assessed in all 132 progression samples. Quantitative genomic DNA PCR was performed to detect BRAFV600E/K amplifications in 120 samples (91%). Alternative splicing of BRAF was evaluated by Sanger detection of novel exon–exon boundaries in the cDNAs in 86 progression samples (65%). Recurrent AKT1 “hotspot” mutations were assessed in all samples, other mutations in the PI3K/AKT pathway were evaluated in the WES samples. WES data analysis has been previously described (18–20). Analyses performed in particular tumors are shown in Table S1.
Resistance Mechanisms
Mechanisms of acquired BRAFi resistance were limited to molecular alterations that were: 1) detected in the progression sample, 2) not present in the pre-treatment sample, or if baseline tissue was unavailable, prior establishment as a resistance mechanism had been performed, and 3) previously validated to confer BRAFi-resistance in vitro (Table S2). Mechanisms proposed in other publications as possible drivers of resistance without pre-clinical validation were not included.
Statistical Analysis
Associations between classes of resistance mechanisms and clinical variables were evaluated using multivariable logistic regression models. We classified resistance mechanisms as the following: 1) NRAS or KRAS mutations, 2) BRAF amplifications, 3) BRAF splice variants, 4) MEK1 or MEK2 mutations, and 5) non-MAPK alterations. The elastic net method was used for variable selection for building multivariable models. The elastic net is a generalization of the LASSO (least absolute shrinkage and selection operator), which provides variable selection in the p≫N case without being limited by sample size, and improves performance in the case of potentially correlated explanatory variables (22). We used the elastic net method for prescreening to discard those least contributing variables ignoring the covariance structure due to multiple samples in some patients (23). Following variable selection, generalized linear mixed-effects models (logit link or identity link depending on outcome variable type) were used for coefficient estimates to account for multiple biopsy specimens within patients.
PFS, OS, and survival after progression were calculated using the Kaplan-Meier method. Due to the co-occurrence of alterations among tumor specimens and within patients, each class of resistance mechanism was compared against all other patients using the logrank test. Cox mixed effects models were used to investigate baseline factors that influenced survival. All analyses were conducted using R version 3.1.1.
Results
Patients
We included 100 patients with 132 progression samples. Patients had a median age of 54 years; 70% had AJCC stage IV M1c melanoma, and 17% had brain metastases (Table 1). BRAFV600E was present in 87% of patients, and BRAFV600K/R in 13%.. Vemurafenib was the BRAFi received by 64% of patients and dabrafenib by 36%. The median PFS was 4.7 months (95% CI 3.8–5.6 months); median OS was 12.8 months (95% CI 10.7–15.0 months). The objective response rate was 72%.
Table 1.
Patient demographics
| Total Patients (n=100) | Number |
|---|---|
| Gender | |
| Male | 63 |
| Female | 37 |
| Age | 54 (median) |
| ECOG | |
| 0 | 51 |
| 1 | 42 |
| Stage | |
| M1a | 20 |
| M1b | 10 |
| M1c | 70 |
| LDH | |
| Elevated | 47 |
| Normal | 47 |
| Unknown | 6 |
| Disease sites (baseline) | |
| Brain | 17 |
| Lung | 52 |
| Liver | 52 |
| Skin/Subcutaneous | 77 |
| BRAF mutation | |
| V600E | 87 |
| V600K | 12 |
| V600R | 1 |
| BRAF inhibitor | |
| Vemurafenib | 64 |
| Dabrafenib | 36 |
| RECIST sum of disease in target lesions (mm) | 97 (median) |
Sequencing and identified mechanisms of resistance
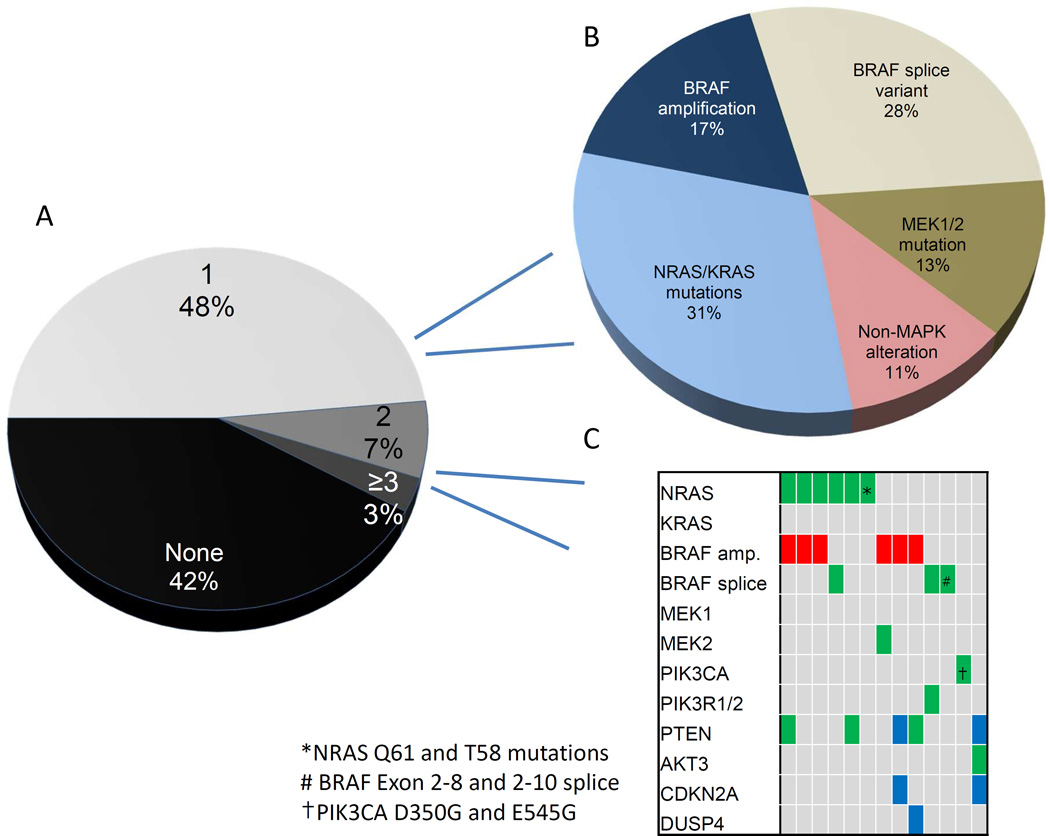
Validated mechanisms of resistance were identified in 77/132 progressing tumor samples (58%). Sixty-four samples (48%) had a single identified resistance mechanism, 9 (7%) had two identified alterations and 4 (3%) had 3 or more (Fig. 1A). Among all progression tumors, NRAS or KRAS mutations were identified in 26 samples (17% and 2%, respectively), BRAFV600E/K amplification in 17 (13%), BRAF splice variants in 21 (16%), MEK1/2 mutations in 9 (7%), and non-MAPK pathway alterations in 14 (11%) (Table 2). Of note, the BRAF splice variant was assessed in 65% of samples (n=86) and identified in 24% of these, suggesting that this may be the most common resistance mechanism. Non-MAPK pathway alterations largely occurred in the PI3K-AKT pathway but also included MITF amplification, and overexpression of PDGFR/IGF1R. CDKN2A deletion and DUSP4 loss occurred in 3 samples and were not included in further analyses given limited sample size. Several candidate “primary resistance” mechanisms were identified in pre-treatment samples from patients with primary disease progression or short duration of therapeutic benefit and are listed descriptively (Table S1, S3).
Figure 1.
(A) Number of identified resistance mechanisms per progression sample (B) Spectrum of resistance mechanisms in samples with only one identified alteration (C) Spectrum of resistance mechanisms in samples with >1 identified alteration co-occurring in the same sample
Legend: Green: mutation; Red: amplification; Blue: deletion; ^Distinct NRAS mutations identified in different samples
Table 2.
Mechanisms of resistance and method of testing
| Number | Percent | |
|---|---|---|
| Total progression samples | 132 | |
| Corresponding pre-treatment sample assessed | 109 | 83 |
| Analyses performed | ||
| PCR | 132 | 100 |
| WES | 74 | 56 |
| IHC | 10 | 8 |
| Identified Mechanism | 77 | 58 |
| Single Mechanism per sample | 64 | 48 |
| Two Mechanisms per sample | 9 | 7 |
| ≥3 Mechanisms per sample | 4 | 3 |
| Unknown | 55 | 42 |
| Specific mechanisms* | ||
| NRAS mutation | 23 | 17 (30) |
| KRAS mutation | 3 | 2 (4) |
| BRAF splice variants† | 21 | 16 (27) |
| BRAFV600E/K amplification‡ | 17 | 13 (22) |
| MEK1 mutation | 5 | 4 (6) |
| MEK2 mutation | 4 | 3 (5) |
| Other MAPK alterations# | 3 | 2 (4) |
| Non-MAPK alterations | 14 | 11 (18) |
Percentage of total samples; percentage of samples with identified alterations denoted in parentheses
BRAF splice variants were assessed in 65% of all samples
BRAF amplifications were assessed in 91% of all samples
Includes acquired DUSP4 loss and CDKN2A deletion.
Heterogeneity of BRAFi resistance mechanisms
We then investigated the intra-tumor and intra-patient heterogeneity of resistance. Although most progression samples had 0–1 detected alterations, 13 progression tumors, had >1 identified mechanism of resistance (10%) co-occurring in the same sample. We assessed whether specific resistance mechanisms occurred more frequently in isolation or arose concurrently with other alterations (Fig. 1B/C). NRAS mutations (20 of 26, 77%), BRAF splice variants (18 of 21, 86%), and MEK1/2 mutations (8 of 9, 89%) usually occurred in isolation. By contrast, BRAFV600E/K amplification (11 of 17, 65%) and non-MAPK alterations (7 of 14, 50%) more commonly co-occurred in tumors with other genetic changes (p=0.015). Among MAPK-alterations, NRAS, KRAS, MEK1, and MEK2 mutations arose in mutually exclusive fashion with each other, whereas BRAFV600E/K amplifications overlapped with NRAS mutations (n=3), non-MAPK alterations (n=4), and a MEK2 mutation. BRAF splice variants did not overlap with MEK1/2 mutations or BRAF amplifications. These results may suggest that certain mutations play complementary roles in driving resistance (e.g. BRAFV600E/K amplifications and non-MAPK alterations) whereas others have redundant signaling functions and are unlikely to co-occur.
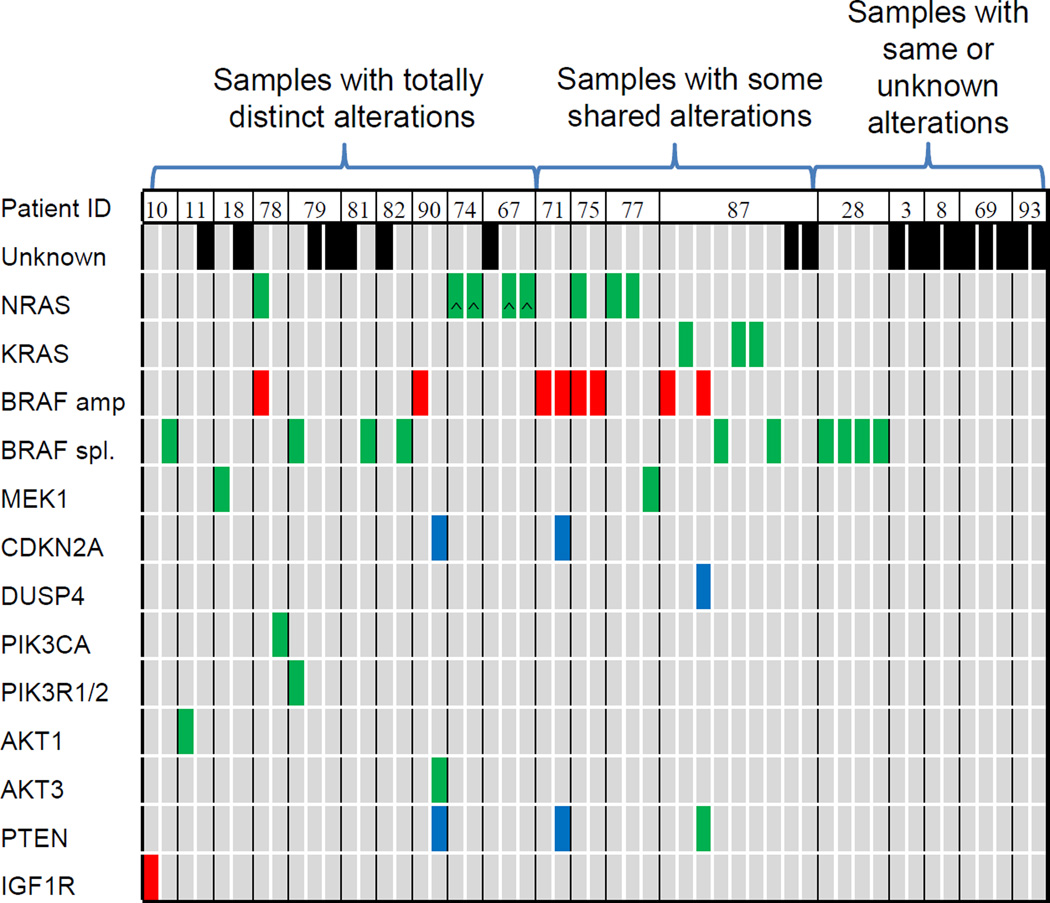
To investigate genetic heterogeneity within individual patients (as opposed to individual tumor samples), we evaluated 19 patients who underwent multiple biopsies during or after progression. Of these, 10 (53%) had completely distinct resistance mechanisms between separate biopsies, 4 (21%) had no identified alterations in any biopsy, and only 1 (5%) had the same identified mechanism in all progressing tumors (Fig. 2). Four patients (21%) were found to have some overlapping but not identical alterations, supporting the evolutionary development of resistance (20). Of these 19 patients, 12 had serial tissue acquisition at two distinct time points after progression. On average, earlier progression sample had fewer resistance mechanisms compared to the later sample (mean 0.42 vs. 0.83, p=0.054). These results may suggest that mutational complexity increases over time on BRAFi.
Figure 2.
Heterogeneity of resistance mechanisms within individual patients with multiple biopsies
Legend: Green: mutation; Red: amplification; Blue: deletion; Black: unknown
Clinical correlation with mechanisms of resistance
We then investigated whether baseline disease, treatment, and host factors were related to the development of particular resistance mechanisms. We developed multivariable logistic regression models assessing the impact of age, gender, LDH levels, sites of metastatic disease, drug (vemurafenib vs. dabrafenib), number of prior therapies, and burden of disease (RECIST sum of target lesion diameters). Several associations were identified; NRAS mutations occurred more often in patients who received vemurafenib (odds ratio 3.53, p=0.045), among patients who had received prior therapies (OR 2.57, p=0.003), and in those with brain metastases at baseline (OR 4.57, p=0.037). BRAFV600E/K amplifications also arose more frequently in pre-treated patients (OR 2.19, p=0.037). No clear associations were identified for other resistance mechanisms. The association between NRAS mutations and vemurafenib use is consistent with the higher incidence of cutaneous squamous cell carcinomas (cuSCCs) observed with vemurafenib compared with dabrafenib (3, 4, 24, 25). These cuSCCs usually harbor RAS mutations (26), suggesting that these otherwise similar agents may promote distinct resistant subclonal populations.
Next, we explored the timing and pattern of disease progression associated with particular resistance mechanisms. PFS did not significantly vary by group either in multivariable Cox regression models or in univariate analyses (Fig S1). Patients without known resistance mechanisms also had similar PFS. NRAS mutations were marginally associated with disease progression in the brain (OR 3.05, p=0.066) and negatively correlated with progression in the lungs (OR 0.25, p=0.036); MEK1/2 mutations were associated with hepatic progression (OR 7.61, p=0.011; 6 of 9 patients with MEK1/2 mutations had disease progression in the liver).
Effects of mechanisms of resistance on survival and response to subsequent therapies
We then assessed whether specific mechanisms of resistance influenced clinical outcomes following disease progression. Median survival following progression was 6.9 months (95% CI 5.3 – 8.5 months), and no statistically-significant differences were identified between patients harboring particular resistance mechanisms (Fig S2). OS from the start of treatment was also similar, regardless of the mechanism of resistance identified (Fig S3). Of interest, 6 patients experienced overall survival of > 1000 days (PFS range 300–672 days) and 5 had unknown drivers of resistance (19).
Finally, we assessed the influence of resistance mechanisms on response to subsequent therapy particularly focusing on MAPK-directed treatment. Fifteen patients received subsequent BRAF +MEK inhibition; two experienced partial responses (13%), and no mechanism of BRAFi resistance was identified in either patient. Stable disease was observed in an additional 7 patients (objective response + stable disease in 9 of 15 patients; 60%) and occurred in patients with various resistance mechanisms.. Patients with MAPK-only resistance drivers appeared to have more frequent disease stabilization (6 of 8; 75%) compared to patients with non-MAPK alterations (1 of 3, 33%). In addition, 21 patients also received ipilimumab following progression, and no patients responded (Table S3).
Discussion
In this study, we combined previously-published analyses of BRAFi resistance to form the largest cohort (to our knowledge) of 132 BRAFi progression melanoma tissue samples from 100 patients. This large population allowed us to correlate resistance mechanisms with clinical characteristics and outcomes. This study also provides a comprehensive landscape of the incidence and heterogeneity of genetic changes driving BRAFi-resistance.
Acquired resistance is a major hindrance to effective molecularly-targeted therapy. BRAFi in particular have been linked to a diverse array of genetic changes driving resistance with unclear therapeutic and prognostic implications. Assessing the resistance landscape in a large cohort with integrated clinical data has therefore been a critical need. We conclude that NRAS mutations, BRAFV600E/K amplifications, BRAF splice variants, and various non-MAPK alterations each occur in 11–24% of BRAFi-progression melanomas. We also observed marked tumor heterogeneity; although individual progression samples usually harbored only a single resistance mechanism, nearly all patients with multiple resistance biopsies had distinct or unknown drivers of resistance (95%). Such marked intra-patient genetic heterogeneity likely limited our ability to correlate individual genetic changes with clinical behavior.
Despite this, we identified clinically-relevant associations. First, resistance changes correlated with specific organ involvement in some cases (NRAS mutations and brain metastases; MEK1/2 mutations and hepatic progression). Second, NRAS mutations occurred more frequently in vemurafenib-treated patients, suggesting differences in subclonal activation between BRAFi. Third, the similar spectrum of resistance drivers between BRAFi monotherapy and those observed with BRAFi/MEKi therapy suggests that these clinical associations may be relevant for the now-preferred combination (19, 27–29). Finally, the intra-patient heterogeneity and lack of clear associations with post-progression outcomes implies that genetic profiling from a single post-progression biopsy to guide further therapy will be challenging.
Although this study was not designed to identify novel resistance mechanisms, several biologic insights were suggested. Some alterations, including MEK1P124 and PTEN mutations appear to mediate resistance in a complex, context-dependent fashion. For example, pre-existing MEK1P124 mutations diminished, but did not preclude clinical responses to BRAFi in this study and others (30). Conversely, in pre-clinical models, these mutations robustly mediate BRAFi-resistance (18). Similar dynamic changes should be considered in other drug-resistance states. In addition, patients with serial biopsies following progression had more alterations in the later post-progression sample compared to the earlier sample, potentially suggesting increasing mutational complexity arising on therapy. Finally, we observed that some resistance mechanisms tended to arise in isolation, whereas others tended to co-occur with other drivers, suggesting that particular alterations may differ in their ability to drive resistance in vivo.
Exploring the non-genetic and immune features of BRAFi-resistance will be a crucial next step. Despite comprehensive molecular characterization, >40% of tumor progression samples harbored no identified resistance drivers, including several patients with multiple progression samples analyzed by WES. Non-genetic (epigenetic) or transcriptome-based changes likely drive resistance in this substantial cohort. In addition, no patients responded to ipilimumab after BRAFi-failure, corroborating previous findings (31, 32). Thus, characterization of non-genetic drivers and the immune state of BRAFi-resistance is needed to complement important early findings (33–35).
Our study has several limitations. Sequencing was performed at three institutions with distinct protocols and analysis methods. Although most patients had baseline pre-treatment samples for comparison, nearly 20% did not. In addition, BRAF splice variants were assessed in 65% of progression samples. Finally, survival in this cohort was somewhat inferior to that observed in phase II/III BRAFi clinical trials, likely reflecting that progression must have occurred during the study timeframe.
Acquired resistance to targeted therapy remains a major unanswered challenge of cancer therapeutics. This study provides a comprehensive characterization of the genetic landscape of acquired BRAFi-resistance in advanced melanoma. Furthermore, it provides insight into the clinical implications of these alterations and suggests further investigation into immune and epigenetic changes accompanying resistance.
Supplementary Material
Highlights.
We examined clinical implications of BRAFi-resistance mechanisms in 100 patients
Mitogen-activated protein kinase pathway mechanisms predominated and overlapped
Acquired NRAS mutations were associated with vemurafenib use and brain metastases
MEK1/2 mutations correlated with hepatic disease progression
Post-progression outcomes were similar across resistance mechanisms
Acknowledgements
Funding was provided by K12 CA0906525 (DJ), P01 CA168585, the Dr. Robert Vigen Memorial Fund, the Ressler Family Foundation (ZE, WH, XK, AR, RSL), the Melanoma Research Alliance, Stand Up To Cancer, SWOG/Hope Foundation, and the Cancer Research Institute (to RSL).
Conflicts of Interest: DJ: Advisory board: Genoptix. AMM: honoraria and travel support; GSK and BMS. LZ: Honoraria: Bristol Myers Squibb, Roche, GlaxoSmithKline, Merck Serono, MSD Sharp & Dohme, Novartis; Advisory board: Bristol Meyers Squibb, Roche, MSD Sharp & Dohme; Travel support: Bristol Meyers Squibb, Roche, Teva. SMG: research funding: University of Zürich; travel support: MSD, Roche, BMS; Advisory board: MSD. CL: Honoria: Bristol Myers Squib, Roche, MSD Sharp & Dohme; Advisory board: Bristol Meyers Squibb, Roche, MSD Sharp & Dohme, Amgen, Novartis; Travel support: Bristol Meyers Squibb, Roche. CB: Honoraria: BMS, GSK, MSD, Novartis, Roche; Advisory board: Amgen, AstraZeneca, BMS, GSK, MSD, Novartis, Roche; Travel support: BMS, MSD, Roche; RG: Grants: Novartis, Pfizer, Roche, Johnson&Johnson. Honoraria: Roche, BMS, GSK, Novartis, MerckSerono, MSD, Almirall, Amgen, Galderma, Janssen, Boehringer Ingelheim, Pfizer, LEO. HG: Consulting : BMS, Roch, GSK, Amgen, MSD, Novartis. Travel support: BMS, Roche. JAS: Consulting : Merck, Research funding: BMS, GSK. AR: Consulting Merck, Amgen, Novartis, GSK, Genentech. Stock: Kite Pharma, Compugen. Research Funding : Merck. RL: Consulting: GSK, Research funding : GSK. GVL: Consulting: GSK, BMS, Novartis, Roche/Genentech, Amgen, Merck, Provectus. Travel support: Roche/Genentech. DS : Honoraria/Consulting/Travel: GSK, BMS, MSD, Amgen, Novartis, Roche.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 3.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 5.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi H, Hong A, Kong X, Koya RC, Song C, Moriceau G, et al. A Novel AKT1 Mutant Amplifies an Adaptive Melanoma Response to BRAF Inhibition. Cancer Discov. 2014;4:69–79. doi: 10.1158/2159-8290.CD-13-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paraiso KH, Xiang Y, Rebecca VW, Abel EV, Chen YA, Munko AC, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res. 2011;71:2750–2760. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haq R, Yokoyama S, Hawryluk EB, Jonsson GB, Frederick DT, McHenry K, et al. BCL2A1 is a lineage-specific antiapoptotic melanoma oncogene that confers resistance to BRAF inhibition. Proc Natl Acad Sci U S A. 2013;110:4321–4326. doi: 10.1073/pnas.1205575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marusiak AA, Edwards ZC, Hugo W, Trotter EW, Girotti MR, Stephenson NL, et al. Mixed lineage kinases activate MEK independently of RAF to mediate resistance to RAF inhibitors. Nat Commun. 2014;5:3901. doi: 10.1038/ncomms4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK Inhibition in Melanoma with BRAF V600 Mutations. N Engl J Med. 2012 doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ribas A, Gonzalez R, Pavlick A, Hamid O, Gajewski TF, Daud A, et al. Combination of vemurafenib and cobimetinib in patients with advanced BRAF(V600)-mutated melanoma: a phase 1b study. Lancet Oncol. 2014;15:954–965. doi: 10.1016/S1470-2045(14)70301-8. [DOI] [PubMed] [Google Scholar]
- 16.Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, et al. Combined BRAF and MEK Inhibition versus BRAF Inhibition Alone in Melanoma. N Engl J Med. 2014 doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- 17.Larkin J, Ascierto PA, Dreno B, Atkinson V, Liszkay G, Maio M, et al. Combined Vemurafenib and Cobimetinib in BRAF-Mutated Melanoma. N Engl J Med. 2014;371:1867–1876. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 18.Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, et al. The Genetic Landscape of Clinical Resistance to RAF Inhibition in Metastatic Melanoma. Cancer Discov. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rizos H, Menzies AM, Pupo GM, Carlino MS, Fung C, Hyman J, et al. BRAF Inhibitor Resistance Mechanisms in Metastatic Melanoma: Spectrum and Clinical Impact. Clin Cancer Res. 2014;20:1965–1977. doi: 10.1158/1078-0432.CCR-13-3122. [DOI] [PubMed] [Google Scholar]
- 20.Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, et al. Acquired Resistance and Clonal Evolution in Melanoma during BRAF Inhibitor Therapy. Cancer Discov. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 22.Zou H, Hastie T. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society. 2005;67:301–320. [Google Scholar]
- 23.Fernandez G. Statistical Data Mining Using SAS Applications. 2 ed. CRC Press; 2010. [Google Scholar]
- 24.Menzies AM, Kefford RF, Long GV. Paradoxical oncogenesis: are all BRAF inhibitors equal? Pigment Cell Melanoma Res. 2013;26:611–615. doi: 10.1111/pcmr.12132. [DOI] [PubMed] [Google Scholar]
- 25.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su F, Viros A, Milagre C, Trunzer K, Bollag G, Spleiss O, et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med. 2012;366:207–215. doi: 10.1056/NEJMoa1105358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villanueva J, Infante JR, Krepler C, Reyes-Uribe P, Samanta M, Chen HY, et al. Concurrent MEK2 mutation and BRAF amplification confer resistance to BRAF and MEK inhibitors in melanoma. Cell Rep. 2013;4:1090–1099. doi: 10.1016/j.celrep.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagle N, Van Allen EM, Treacy DJ, Frederick DT, Cooper ZA, Taylor-Weiner A, et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 2014;4:61–68. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moriceau G, Hugo W, Hong A, Shi H, Kong X, Yu CC, et al. Tunable-Combinatorial Mechanisms of Acquired Resistance Limit the Efficacy of BRAF/MEK Cotargeting but Result in Melanoma Drug Addiction. Cancer Cell. 2015;27:240–256. doi: 10.1016/j.ccell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlino MS, Fung C, Shahheydari H, Todd JR, Boyd SC, Irvine M, et al. Preexisting MEK1P124 mutations diminish response to BRAF inhibitors in metastatic melanoma patients. Clin Cancer Res. 2015;21:98–105. doi: 10.1158/1078-0432.CCR-14-0759. [DOI] [PubMed] [Google Scholar]
- 31.Ascierto PA, Simeone E, Giannarelli D, Grimaldi AM, Romano A, Mozzillo N. Sequencing of BRAF inhibitors and ipilimumab in patients with metastatic melanoma: a possible algorithm for clinical use. J Transl Med. 2012;10:107. doi: 10.1186/1479-5876-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ackerman A, McDermott DF, Lawrence DP, et al. Outcomes of patients with malignant melanoma treated with immunotherapy prior to or after vemurafenib. J Clin Oncol. 2012;30:8569. [Google Scholar]
- 33.Kakavand H, Wilmott JS, Menzies AM, Vilain R, Haydu LE, Yearley JH, et al. PD-L1 expression and tumor-infiltrating lymphocytes define different subsets of MAPK inhibitor treated melanoma patients. Clin Cancer Res. 2015 doi: 10.1158/1078-0432.CCR-14-2023. [DOI] [PubMed] [Google Scholar]
- 34.Tompers Frederick D, Piris A, Cogdill AP, Cooper ZA, Lezcano C, Ferrone CR, et al. BRAF inhibition is associated with enhanced melanoma antigen expression and a more favorable tumor microenvironment in patients with metastatic melanoma. Clin Cancer Res. 2013 doi: 10.1158/1078-0432.CCR-12-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu-Lieskovan S, Robert L, Homet Moreno B, Ribas A. Combining Targeted Therapy With Immunotherapy in BRAF-Mutant Melanoma: Promise and Challenges. J Clin Oncol. 2014 doi: 10.1200/JCO.2013.52.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.