Abstract
Saint Louis encephalitis virus (SLEV) is a member of the Japanese-encephalitis virus serocomplex of the genus Flavivirus. SLEV is broadly distributed in the Americas and the Caribbean Islands, where it is usually transmitted by mosquitoes of the genus Culex and primarily to birds and mammalian-hosts. Humans are occasionally infected by the virus and are dead-end hosts. SLEV causes encephalitis in temperate regions, while in tropical regions of the Americas, several human cases and a wide biological diversity of SLEV-strains have been reported. The phylogenetic analysis of the envelope (E) protein genes indicated eight-genotypes of SLEV with geographic overlap. The present paper describes the genotyping of two SLEV viruses detected in mosquito-pools collected in northern Colombia (department of Cordoba). We used reverse transcription-polymerase chain reaction to amplify a fragment of theE-gene to confirm the virus identity and completeE-gene sequencing for phylogenetic analysis and genotyping of the two-SLEV viruses found circulating in Córdoba. This is the first report of SLEV genotype IV in Colombia (Córdoba) in mosquitoes from a region of human inhabitation, implicating the risk of human disease due to SLEV infection. Physicians should consider SLEV as a possible aetiology for undiagnosed febrile and neurologic syndromes among their patients who report exposure to mosquito-bites.
Keywords: Saint Louis encephalitis virus, flavivirus, genotype, Colombia
Saint Louis encephalitis virus (SLEV) is a member of the Japanese encephalitis virus (JEV) serogroup of flaviviruses (family Flaviviridae) along with west Nile (WNV), Usutu and Murray Valley Encephalitis (MVEV) (Thiel et al. 2005). SLEV is generally transmitted by Culex mosquitoes (Culex pipiens, Culex tarsalis and Culex quinquefasciatus) and primarily to birds (Reisen 2003). However, this virus may utilise other mosquito vectors and vertebrate hosts in different geographical regions. For example, in Brazil, SLEV strains have been obtained from Culex declarator and Culex coronator (Hervé et al. 1986, Vasconcelos et al. 1991). There may be undiscovered vector species and alternative amplifier hosts besides birds.
SLEV is broadly distributed in the area from Canada to Argentina and is responsible for sporadic outbreaks in the United States of America (USA) (Reisen 2003), but it has a very low percentage of fatality (Tsai & Mitchell 1988). This epidemiological pattern is in contrast to the few human cases that have been reported in the tropical regions of the Americas (Pinheiro et al. 1981,Vasconcelos et al. 1998, Rocco et al. 2005), with only one exception of a SLEV outbreak in Córdoba, Argentina (Díaz et al. 2006, 2011). No neurological signs or symptoms have been reported in the tropics and most cases involve acute febrile illness or dengue-like disease symptoms (Mondini et al. 2007a, b).
In Colombia, SLEV activity was demonstrated serologically in humans living in Acandí (department of Chocó), a municipality located in Uraba’s Gulf, Caribbean region, and San Vicente del Chucurí (department of Santander, Middle Magdalena Valley) (Groot 1964). In the latter locality, one passeriform bird among the 140 tested using neutralisation tests was found to be SLEV antibody-positive (Groot 1964). Mattar et al. (2005) reported that sentinel equines were exposed to SLEV in 2004 in Córdoba and Sucre and both localities are located in the Caribbean region of Colombia. Subsequently, in a study with more extensive sampling of sentinel horses in five departments of the Caribbean region (departments of Córdoba, Sucre, Bolivar, Atlántico and Magdalena) from 2006-2007, SLEV-neutralising antibodies were found in 34.1% of 971 horses without neurologic signs or encephalitis (Mattar et al. 2011). Unfortunately, there is little information about mosquitoes regarding the genotypes of the viruses circulating in Colombia to help understand the absence of human cases or epidemic outbreaks.
Since the first epidemic occurred in Saint Louis, Missouri (1933), samples of SLEV reflecting a wide geographic and temporal diversity of vertebrate hosts and mosquitoes have been collected (Baillie et al. 2008). These strains show varying levels of viraemia, neurovirulence and disease severity in mammalian and avian hosts (Monath et al. 1980). Phylogenetic analysis of complete envelope (E) gene sequences group SLEV strains into eight genotypes with geographic segregation (Kramer & Chandler 2001, Rodrigues et al. 2010) reflecting the genetic similarity between nearby strains, but showing intermixing of some viral strains between countries such as the USA, Argentina and Brazil (May et al. 2008, Auguste et al. 2009, Díaz et al. 2011). The classification of SLEV genotypes shows no correlation with phenotypic characteristics such as virulence (Kramer et al. 1997). Given that SLEV strains are known to differ in pathogenicity, understanding which strains are circulating in a particular geographic region may be important from an epidemiological perspective.
From 2011-2013, during a research program on the viral ecology of emerging arboviruses in a region with large populations of migratory birds in northern Córdoba, we found natural infections of mosquitoes carrying SLEV. The aim of this study was to amplify theE gene to characterise the genotype of SLEV circulating in northern Córdoba, including representative sequences of other genotypes previously published and to assess the relevance from an epidemiological context in the geographical region of study.
MATERIALS AND METHODS
Samples - During entomological surveillance for the detection of emerging and re-emerging arboviruses between 2011-2013 in San Bernardo del Viento (Córdoba, Colombia) (9º21’30.97”N 75º58’37.28”W) (Fig. 1), mosquitoes were collected using a manual aspirator from resting sites inside mangroves, mounted and identified by morphological keys and two legs were collected for DNA barcode methodology (Hebert et al. 2003, Kumar et al. 2007,Cywinska et al. 2010). Later, the mosquitoes were collected using CDC-light traps with dry ice as bait. All the insects sampled were separated into pools based on the morphological characteristics of vouchers previously collected, identified and characterised by DNA barcode. Some mosquito specimens were collected from each pool and their legs were removed. If a pool was found to be positive for arbovirus, the legs were used for DNA extraction and amplification of the cytochrome c oxidase I-DNA barcode as a method to corroborate their taxonomic identity (Hebert et al. 2003). All of the pools were triturated in minimum essential medium supplemented with 10% foetal bovine serum and 1% penicillin and centrifuged at 13,000 rpm for 30 min.
Fig. 1. : the study area showing San Bernardo del Viento (northern Córdoba), the geographic position of the locality where the mosquitoes were sampled (source: Virology Journal 2015 doi: 10.1186/s12985-015-0310-8).
Molecular characterisation - The supernatant from each pool was used for RNA extraction using the RNeasy kit (Qiagen, USA) and total RNA extracted from SLEV-positive pools, following the methodology described by Sánchez-Seco et al. (2005), was used for reverse transcription-polymerase chain reaction (RT-PCR) to amplify a region of theE gene using the following primers: SLE1497+(RRYATGGGYGAGTATGGRACAG)/SLE2517-(CTCCTCCACAYTTYARTTCACG). The RT-PCR products (cDNA) were used as the template for nested RT-PCR with the primers SLE2002+(TGGAYTGGACRCCGGTTGGAAG) and SLE2257-(CCAATRGATCCRAARTCCCACG) and the resulting 234-nucleotide (nt) fragment was sequenced (Ré et al. 2008). For genotyping the detected strains of SLEV, we amplified the complete E gene from the positive pool supernatants using primers and conditions described by Kramer and Chandler (2001).
Sequence analysis - Sequences were aligned with homologous sequences from other SLEV strains and other flaviviruses such as JEV, MVEV and WNV available in GenBank using CLUSTALW (Larkin et al. 2007). The Neighbour-joining (NJ) method in MEGA v.6.0 was used to construct phylogenetic trees using the Tamura-Nei model and a gamma distribution (Saitou & Nei 1987, Tamura et al. 2013). The jModelTest v.2.1 program (Darriba et al. 2012) was used to determine the best evolutionary model of nt substitution to estimate the phylogenetic tree according to the parameters to increase the likelihood without including many variables [Akaike information criterion (AIC)] (Posada & Buckley 2004). The genotypic lineages were reconstructed by the maximum likelihood (ML) method using PHYML v.3.0 (Guindon et al. 2010) and previously estimated parameters of nt substitution. Bootstrap replication (n = 1000) was applied to provide confidence for the clusters and phylogenetic groupings (Felsenstein 1985) of the NJ dendrogram and phylogenetic trees. DNAsp v.5.0 (Librado & Rozas 2009) was used to establish polymorphic sites between E sequences characterised in our study and reference sequences of the genotypes to which these sequences belonged and the genetic distances were estimated.
RESULTS
Of the 2,059 pools of 18 mosquitoes tested for flavivirus, just two pools were positive for SLEV by NS5/RT-PCR (Table I); all of the species were identified with morphological characters and DNA barcoding, using the legs of certain specimens chosen randomly, two amplicons obtained corresponded by DNA barcoding to species Mansonia titillans andCulex erraticus. The flavivirus amplicons were sequenced by the Sanger method (Macrogen, Korea) and later sequences were identified as SLEV using BLASTN. We confirmed this finding using nested RT-PCR and sequencing of a fragment of the E gene (Ré et al. 2008) (GenBank accessions KM103077-KM103078). DNA barcoding was accurate for the identified legs of specimens at the pool collected previously for RNA extraction and these sequences showed a high similarity percentage (> 99%) for species - Ma. titillans and Cx. erraticus, which were characterised by this methodology using voucher specimens collected in the same study zone.
TABLE I. Mosquito species, pools tested and positive pools for Flavivirus in San Bernardo del Viento (Córdoba, Colombia).
| Mosquito species | Pools tested (n) | Specimens tested (n) | Pools positive for SLEV (n) | Minimal infection rate |
|---|---|---|---|---|
| Aedes aegypti | 60 | 1,150 | - | - |
| Aedes scapularis | 30 | 512 | - | - |
| Anopheles albimanus | 10 | 35 | - | - |
| Anopheles aquasalis | 15 | 77 | - | - |
| Anopheles neomaculipalpus | 5 | 31 | - | - |
| Culex corniger | 28 | 477 | - | - |
| Culex coronator | 18 | 222 | - | - |
| Culex erraticus | 666 | 8,510 | 1 | 0,1175 |
| Culex group Salinarius | 23 | 312 | - | - |
| Culex quinquefasciatus | 67 | 1,322 | - | - |
| Culex section Spissipes | 19 | 234 | - | - |
| Deinocerites atlanticus | 529 | 5,511 | - | - |
| Deinocerites sp. 2 | 237 | 1,777 | - | - |
| Haemagogus section Splendes | 71 | 757 | - | - |
| Mansonia titillans | 185 | 1,074 | 1 | 0,9310 |
| Ochlerotatus taeniorhynchus | 47 | 102 | - | - |
| Psorophora confinnis | 41 | 622 | - | - |
| Sabethes cyaneus | 3 | 8 | - | - |
| Total | 2,059 | 22,733 | 2 | - |
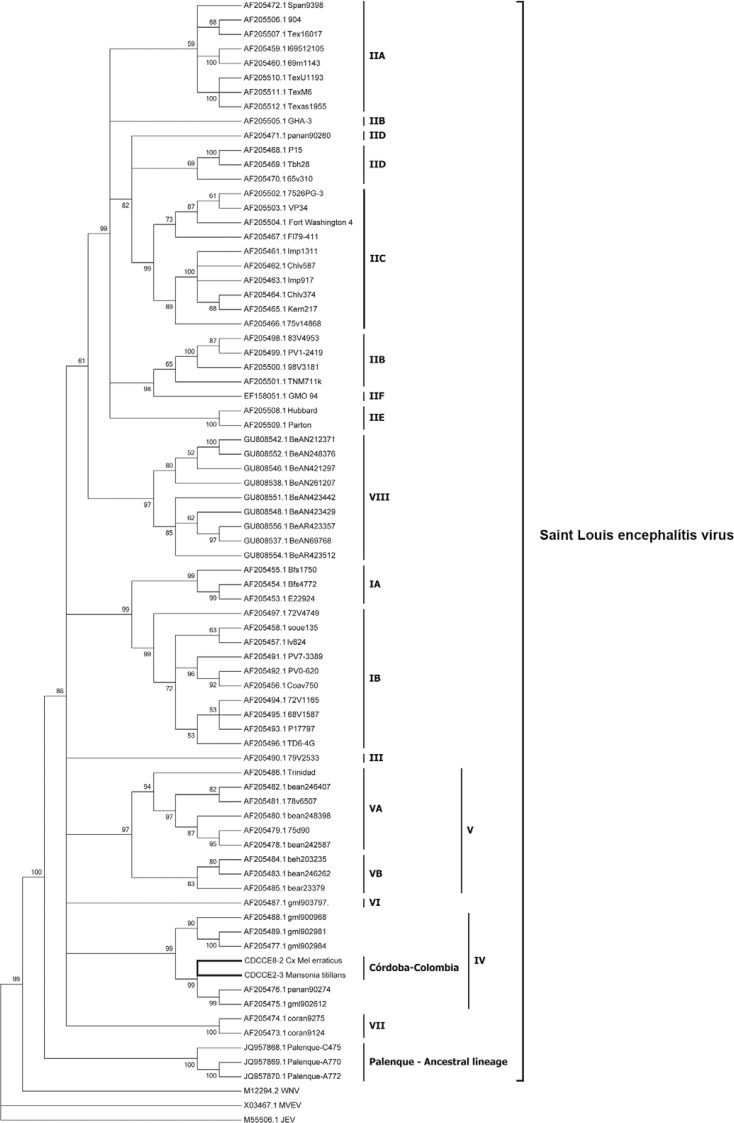
The E gene sequences obtained from the two samples were 1,617 nts in length (GenBank accessions KM103079-KM103080). A comparison with other representative genotypic lineages showed a high nt identity (88.2-97.7%). The strain with the most nt similarity with respect to CDCCE2-3 and CDCCE8-2 was the gml902612 strain, which was isolated from Panama in 1973 from a pool of Haemagogus equinus mosquitoes (Fig. 2).
Fig. 2. : a phylogenetic tree estimated by the maximum likelihood analysis of 73 envelope sequences of Saint Louis encephalitis virus data under the GTR+I+G model of nucleotide substitution. Three outgroups were included in the analysis. The lineages are labelled according to the scheme of Kramer and Chandler (2001).
The consensus sequence obtained by aligning the E gene sequences was 1,498 nts in length and included representative viruses of the Japanese encephalitis serogroup that were used as outgroups. The best nt model substitution, according to the AIC, was GTR+I+G (-LnL = 11950.85, K = 140, AICc = 24208.46).
A phylogenetic tree of ML placed sequences CDCCE2-3 and CDCCE8-2 within SLEV lineage IV (Fig. 2) (bootstrap values > 90) and they were grouped with SLEV strains isolated in Panama fromHaemagogus lucifer,Hg. equinus and Mansonia dyari.
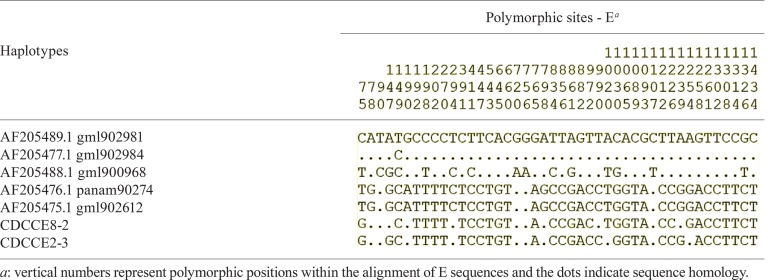
Forty-four polymorphic sites were identified in SLEV genotype IV: five were comprised of single nts, 37 were dimers and two were trimers. A substitution in position 75 was unique for CDCCE2-3 and CDCCE8-2 (Table II). Our sequences showed patterns of substitution similar to that observed in other sequences within close genetic distances and thereby supporting the strong relationships within this lineage (Table III).
TABLE II. Polymorphic sites between envelope (E) sequences belonging to genotype IV and then characterised from San Bernardo del Viento (Córdoba, Colombia).
TABLE III. Genetic distances estimated for sequences belonging to genotype IV and detected in San Bernardo del Viento (Córdoba, Colombia).
| Sequences - genotype IV | Genetic distances and SDa
|
||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| 1. AF205489.1_gml902981 | 0.001 | 0.002 | 0.005 | 0.005 | 0.004 | 0.004 | |
| 2. AF205477.1_gml902984 | 0.001 | 0.002 | 0.005 | 0.005 | 0.004 | 0.004 | |
| 3. AF205488.1_gml900968 | 0.010 | 0.010 | 0.004 | 0.004 | 0.004 | 0.004 | |
| 4. AF205476.1_panan90274 | 0.028 | 0.028 | 0.024 | 0.000 | 0.002 | 0.002 | |
| 5. AF205475.1_gml902612 | 0.028 | 0.028 | 0.024 | 0.000 | 0.000 | 0.002 | |
| 6. CDCCE8-2 | 0.023 | 0.022 | 0.023 | 0.005 | 0.005 | 0.002 | |
| 7. CDCCE2-3 | 0.024 | 0.023 | 0.023 | 0.005 | 0.005 | 0.003 | |
a: values in italics are the standard deviation (SD) and bold corresponds to the genetic distances estimated with the Tamura-Nei model.
DISCUSSION
Our analysis indicated that the two SLEV sequences characterised in San Bernardo del Viento from pools of Ma. titillans and Cx. erraticus belong to lineage IV of the classification proposed by Kramer and Chandler (2001). This is the first report of genotype IV in Colombia and outside of Panama.
Ma. titillans is a species characterised by eclectic feeding habits where domestic animals (pigs, horses and cows) and sylvatic fauna (mainly birds and rodents) have been identified as hosts by different methodologies (Lorosa et al. 2010, Navia-Gine et al. 2013). Cx. erraticus has a preference for a high diversity of birds, such as Passerinae and Ardeidae, followed by mammals with a high frequency of contact with humans, cows, foxes and also reptiles (Mendenhall et al. 2012). Unlike the preferences for birds by Cx. erraticus, for which there is strong evidence, Ma. titillans would be a candidate for bridge vector, bringing SLEV from sylvatic/rural to peridomestic environment where it could infect humans. Vector competence and vectorial capacity studies would be needed to fully assess the potential risk presented by SLEV-infected Ma. titillans.
The results allow us to postulate the existence of an enzootic cycle of SLEV lineage IV transmission without discarding the possible existence of other genotypes circulating in this zone. Genotypes VA/III/II are widely dispersed, probably by migratory birds, in Brazil, USA, Mexico and Argentina (Kramer & Chandler 2001, Auguste et al. 2009, Díaz et al. 2011). Interestingly, genotype IV has not been associated with geographic dispersion or equine/human cases of encephalitis and has not been detected in countries other than Panama and Colombia. This finding suggests that genotype IV, similar to other genotypes, has low virulence for equines and humans, which is consistent with previous observations of SLEV isolated from the Palenque region (Mexico) (Kopp et al. 2013) and could explain the absence of neurological disease in SLEV seropositive equines in the Córdoba (Mattar et al. 2005, 2011) with the exception of the recent isolation of SLEV-genotype VB from a neurologically ill horse in Brazil (Rosa et al. 2013).
SLEV does not exhibit a high level of genetic diversity and has a maximum of 10.1% nt sequence divergence and ≤ 5.5% intralineage divergence (May et al. 2008). Low divergence within genotype IV was also found in our sequences of CDCCE2-3/CDCCE8-2 and other members of genotype IV (rank 0.3-2.8%). These lower values may be associated with a lower selection pressure in genotypic lineages, different ecological dynamics of cycle transmission in diverse geographical locations and infectivity and low transmission rates in sylvatic and epidemic strains (Kramer & Chandler 2001,Ewald 2004, Ciota et al. 2007, May et al. 2008, Kopp et al. 2013). In our study, the SLEV-positive mosquito pools were collected from an undisturbed coastal mangrove ecosystem without fragmentation or deforestation. However, farms, livestock and human settlements border the mangroves. Changes in natural ecosystems may cause variation in the community structures of vertebrate hosts and vector mosquitoes, increasing human-mosquito-pathogen contact and resulting in occasional human cases with varied pathogenicity. In some situations, these changes may result in disease outbreaks, often referred to as “viral emergence” (Weaver 2005, Jones et al. 2008,Keesing et al. 2010, Weaver & Reisen 2010, Usme-Ciro et al. 2012).
The genetic characterisation of the E gene sequences of SLEV detected in mosquito pools provides an overview of the dispersion and evolution of SLEV in the Americas. This raises several questions about the ecology of SLEV that could identify driving factors in a future situation of viral emergence in human populations near conserved ecosystems with high significant diversity in reservoirs and mosquitoes. Additional research is needed on other aspects of the natural circulation of SLEV, such as temporal patterns, mammalian hosts, the relative roles of migratory vs. resident birds in the transmission cycle, mosquito vector species and blood preferences, with the ultimate goal of elucidating the viral traffic network and possible deviations that might result in the emergence of SLEV in northern Córdoba.
ACKNOWLEDGEMENTS
To Diego Leon Arias-Builes, for the technical assistance in entomology collections, to Juan David Suaza-Vasco, for the separation and identification of mosquito pools, to Dr Luis Adrián Díaz, for the recommendations of molecular protocols for SLEV, and to Drs Scott Weaver and Robert Tesh, for the recommendations, suggestions and ideas.
Footnotes
Financial support: COLCIENCIAS (111599326198) (Convocatory 528 - Scholarships for PhD students to RL-H)
REFERENCES
- Auguste AJ, Pybus OG, Carrington CV. Evolution and dispersal of St. Louis encephalitis virus in the Americas. Infect Genet Evol. 2009;9:709–715. doi: 10.1016/j.meegid.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Baillie GJ, Kolokotronis SO, Waltari E, Maffei JG, Kramer LD, Perkins SL. Phylogenetic and evolutionary analyses of St. Louis encephalitis virus genomes. Mol Phylogenet Evol. 2008;47:717–728. doi: 10.1016/j.ympev.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Ciota A, Lovelace A, Ngo K, Le A, Maffei JG, Franke M, Payne A, Jones S, Kauffman E, Kramer L. Cell-specific adaptation of two flaviviruses following serial passage in mosquito cell culture. Virology. 2007;357:165–174. doi: 10.1016/j.virol.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cywinska A, Hannan M, Kevan P, Roughley R, Iranpour M, Hunter F. Evaluation of DNA barcoding and identification of new haplomorphos in Canadian derflies and horseflies. Med Vet Entomol. 2010;24:382–410. doi: 10.1111/j.1365-2915.2010.00896.x. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. 772Nat Methods. 2012;9 doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz LA, Nemeth NM, Bowen RA, Almiron WR, Contigiani MS. Comparison of Argentinean Saint Louis encephalitis virus non-epidemic and epidemic strain infections in an avian model. PLoS Negl Trop Dis. 2011;5: doi: 10.1371/journal.pntd.0001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz LA, Ré V, Almirón WR, Farías A, Vázquez A, Sanchez-Seco MP, Aguilar J, Spinsanti L, Konigheim B, Visintin A, Garciá J, Morales MA, Tenorio A, Contigiani M. Genotype III Saint Louis encephalitis virus outbreak, Argentina, 2005. Emerg Infect Dis. 2006;12:1752–1754. doi: 10.3201/eid1211.060486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald PW. Evolution of virulence. Infect Dis Clin North Am. 2004;18:1–15. doi: 10.1016/S0891-5520(03)00099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Groot H. Estudios sobre virus transmitidos por artrópodos en Colombia. Rev Acad Colomb Cien Exac Nat. 1964;12:197–217. [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–310. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hebert PD, Cywinska A, Ball SL, de Waard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé JP, Dégallier N, Travassos da Rosa APA, Pinheiro FP, Sá GC., Filho . Aspectos ecológicos das arboviroses. In Instituto Evandro Chagas: 50 anos de contribuição às ciências biológicas e à medicina tropical. Vol. I. Fundação Serviços de Saúde Pública; Belém: 1986. pp. 409–437. [Google Scholar]
- Jones K, Patel N, Levy M, Storeygard A, Balk D, Gittleman J, Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–994. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keesing F, Belden L, Daszak P, Dobson A, Harvell D, Holt R, Hudson P, Jolles A, Jones K, Mitchell C, Myers S, Bogich T, Ostfeld R. Impacts of biodiversity on the emergence and transmission of infectious disease. Nature. 2010;468:647–652. doi: 10.1038/nature09575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp A, Gillespie TR, Hobelsberger D, Estrada A, Harper JM, Miller R, Eckerle I, Muller M, Podsiadlowski L, Leendertz F, Drosten C, Junglen S. Provenance and geographic spread of St. Louis encephalitis virus. mBio 4 2013: doi: 10.1128/mBio.00322-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer LD, Chandler LJ. Phylogenetic analysis of the envelope gene of St. Louis encephalitis virus. Arch Virol. 2001;146:2341–2355. doi: 10.1007/s007050170007. [DOI] [PubMed] [Google Scholar]
- Kramer LD, Presser SB, Hardy JL, Jackson AO. Genotypic and phenotypic variation of selected Saint Louis encephalitis viral strains isolated in California. Am J Trop Med Hyg. 1997;57:222–229. doi: 10.4269/ajtmh.1997.57.222. [DOI] [PubMed] [Google Scholar]
- Kumar NP, Rajavel AR, Natarajan R, Jambulingam P. DNA barcodes can distinguish species of Indian mosquitoes (Diptera: Culicidae) J Med Entomol. 2007;44:1–7. doi: 10.1603/0022-2585(2007)44[1:dbcdso]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. CLUSTALW and CLUSTALX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- López RH, Soto SU, Gallego-Gómez JC. Evolutionary relationships of west Nile virus detected in mosquitoes from a migratory bird zone of Colombian Caribbean. 10.1186/s12985-015-0310-8.Virology Journal. 20152015 doi: 10.1186/s12985-015-0310-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorosa E, Faria M, de Oliveira L, Alencar J, Marcondes C. Blood meal identification of selected mosquitoes in Rio de Janeiro, Brazil. J Am Mosq Control Assoc. 2010;26:18–23. doi: 10.2987/09-5914.1. [DOI] [PubMed] [Google Scholar]
- Mattar S, Edwards E, Laguado J, González M, Alvarez J, Komar N. West Nile virus infection in Colombian horses. Emerg Infect Dis. 2005;11:1497–1498. doi: 10.3201/eid1109.050426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattar S, Komar N, Young G, Alvarez J, Gonzalez M. Seroconversion for West Nile and St. Louis encephalitis viruses among sentinel horses in Colombia. Mem Inst Oswaldo Cruz. 2011;106:976–979. doi: 10.1590/s0074-02762011000800012. [DOI] [PubMed] [Google Scholar]
- May FJ, Li L, Zhang S, Guzman H, Beasley DW, Tesh RB, Higgs S, Raj P, Bueno R, Randle Y, Chandler L, Barret A. Genetic variation of St. Louis encephalitis virus. J Gen Virol. 2008;89:1901–1910. doi: 10.1099/vir.0.2008/000190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall I, Tello S, Neira L, Castillo L, Ocampo C, Wesson D. Host preference of the arbovirus vector Culex erraticus (Diptera: Culicidae) at Sonso Lake, Cauca Valley department, Colombia. J Med Entomol. 2012;49:1092–1102. doi: 10.1603/me11260. [DOI] [PubMed] [Google Scholar]
- Monath TP, Cropp CB, Bowen GS, Kemp GE, Mitchell CJ, Gardner JJ. Variation in virulence for mice and rhesus monkeys among St. Louis encephalitis virus strains of different origin. Am J Trop Med Hyg. 1980;29:948–962. doi: 10.4269/ajtmh.1980.29.948. [DOI] [PubMed] [Google Scholar]
- Mondini A, Bronzoni R, Cardeal I, Santos T, Lázaro E, Nunes S, Silva G, Madrid M, Rahal P, Figueiredo L. Simultaneous infection by DENV-3 and SLEV in Brazil. J Clin Virol. 2007a;40:84–86. doi: 10.1016/j.jcv.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Mondini A, Cardeal IL, Lázaro E, Nunes SH, Moreira CC, Rahal P, Maia IL, Franco C, Góngora DV, Góngora-Rubio F, Cabrera EM, Figueiredo LT, da Fonseca FG, Bronzoni RV, Chiaravalloti F, Neto, Nogueira ML. Saint Louis encephalitis virus, Brazil. Emerg Infect Dis. 2007b;13:176–178. doi: 10.3201/eid1301.060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navia-Gine W, Loaiza J, Miller W. Mosquito-host interactions during and after an outbreak of equine viral encephalitis in eastern Panama. PLoS ONE. 2013;8: doi: 10.1371/journal.pone.0081788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro FP, LeDuc JW, Travassos da Rosa APA, Leite OF. Isolation from St. Louis encephalitis virus from a patient in Belém, Brazil. Am J Trop Med Hyg. 1981;30:145–148. doi: 10.4269/ajtmh.1981.30.145. [DOI] [PubMed] [Google Scholar]
- Posada D, Buckley T. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Ré V, Spinsanti L, Farías A, Díaz A, Vázquez A, Aguilar J, Tenorio A, Contigiani M. Reliable detection of St. Louis encephalitis virus by RT-nested PCR. Enferm Infecc Microbiol Clin. 2008;26:10–15. doi: 10.1157/13114389. [DOI] [PubMed] [Google Scholar]
- Reisen WK. Epidemiology of St. Louis encephalitis virus. Adv Virus Res. 2003;61:139–183. doi: 10.1016/s0065-3527(03)61004-3. [DOI] [PubMed] [Google Scholar]
- Rocco IM, Santos CLS, Bisordi I, Petrella S, Pereira LE, Souza RP, Coinbra LM, Bessa TA, Oshiro FM, Lima L, Cerroni M, Marti A, Barbosa V, Gizelda K, Suzuki A. St. Louis encephalitis: first virus isolation from a human in Sao Paulo state, Brazil. Rev Inst Med Trop Sao Paulo. 2005;47:281–285. doi: 10.1590/s0036-46652005000500008. [DOI] [PubMed] [Google Scholar]
- Rodrigues SG, Nunes MR, Casseb SM, Prazeres AS, Rodrigues DS, Silva MO, Cruz AC, Tavares JC, Neto, Vasconcelos PF. Molecular epidemiology of Saint Louis encephalitis virus in the Brazilian Amazon: genetic divergence and dispersal. J Gen Virol. 2010;91:2420–2427. doi: 10.1099/vir.0.019117-0. [DOI] [PubMed] [Google Scholar]
- Rosa R, Azevedo E, Elias R, Simas T, Furtini R, Quaresma M, Teixeira M, Alves T, Lima R. Isolation of Saint Louis encephalitis virus from a horse with neurological disease in Brazil. PLoS Negl Trop Dis. 2013;7: doi: 10.1371/journal.pntd.0002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sánchez-Seco MP, Rosario D, Domingo C, Hernández L, Valdes K, Guzmán MG, Tenorio A. Generic RT-nested-PCR for detection of flaviviruses using degenerated primers and internal control followed by sequencing for specific identification. J Virol Meth. 2005;126:101–109. doi: 10.1016/j.jviromet.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel HJ, Collett MS, Gould EA, Heinz FX, Meyers G, Purcell RH, Rice CM, Houghton M. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA.eds. Virus taxonomy. Eighth report of the International Committee for the Taxonomy of Viruses. Academic Press; San Diego: 2005. Flaviviridae; pp. 981–998. [Google Scholar]
- Tsai TF, Mitchell CJ. Monath TP.ed. The arboviruses: epidemiology and ecology. CRC Press; Boca Raton: 1988. St. Louis encephalitis; pp. 431–458. [Google Scholar]
- Usme-Ciro J, Hoyos-López R, Gallego-Gómez JC. Lunet N.ed. Epidemiology - Current perspectives on research and practice. InTech; Rijeka: 2012. Viral evolutionary ecology: conceptual basis of a new scientific approach for understanding viral emergence; pp. 119–127. [Google Scholar]
- Vasconcelos PF, da Rosa JF, da Rosa AP, Dégallier N, Pinheiro FP, Sá GC., Filho Epidemiology of encephalitis caused by arbovirus in the Brazilian Amazonia. Rev Inst Med Trop Sao Paulo. 1991;33:465–476. [PubMed] [Google Scholar]
- Vasconcelos PFC, Travassos da Rosa APA, Pinheiro FP, Shope RE, Travassos da Rosa JF S, Rodrigues SG, Degallier N, Travassos da Rosa ES. APA. Vasconcelos PFC, JFS . An overview of arbovirology in Brazil and neighbouring countries. Instituto Evandro Chagas; Belém: 1998. Arboviruses pathogenic for man in Brazil; pp. 72–99. [Google Scholar]
- Weaver S. Host range, amplification and arboviral disease emergence. Arch Virol. 2005;(Suppl.):33–44. doi: 10.1007/3-211-29981-5_4. [DOI] [PubMed] [Google Scholar]
- Weaver S, Reisen W. Present and future arboviral threats. Antiviral Res. 2010;85:328–345. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]