Abstract
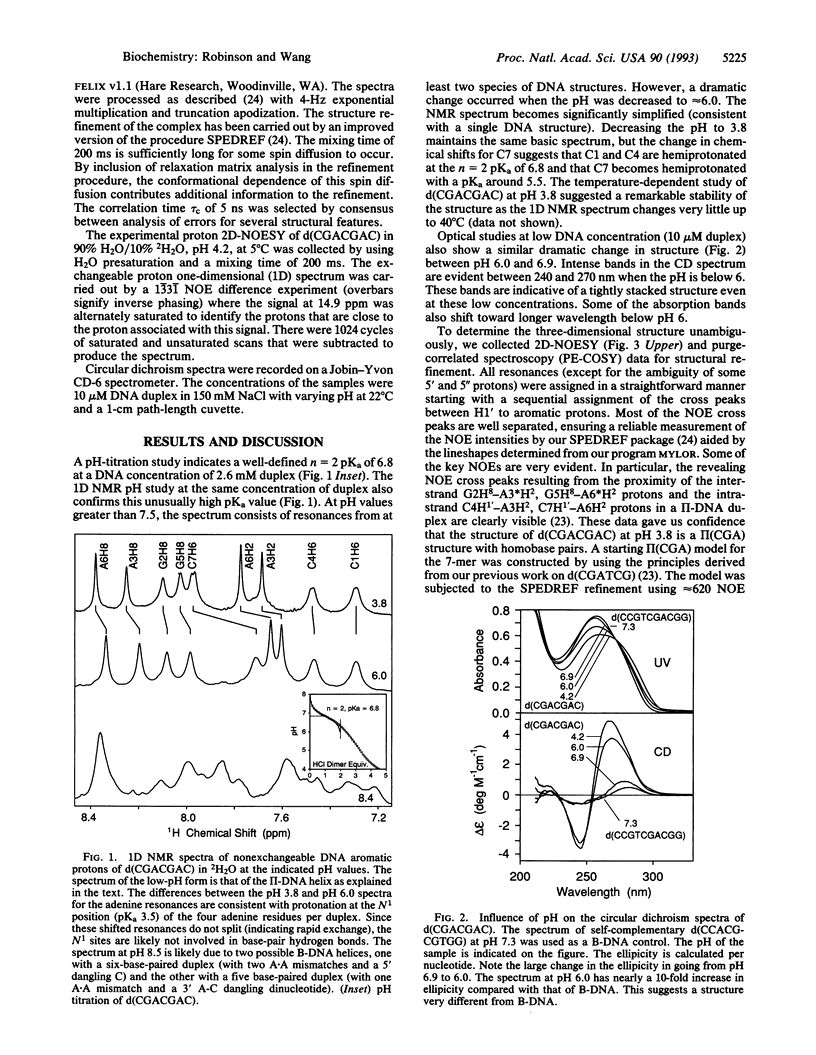
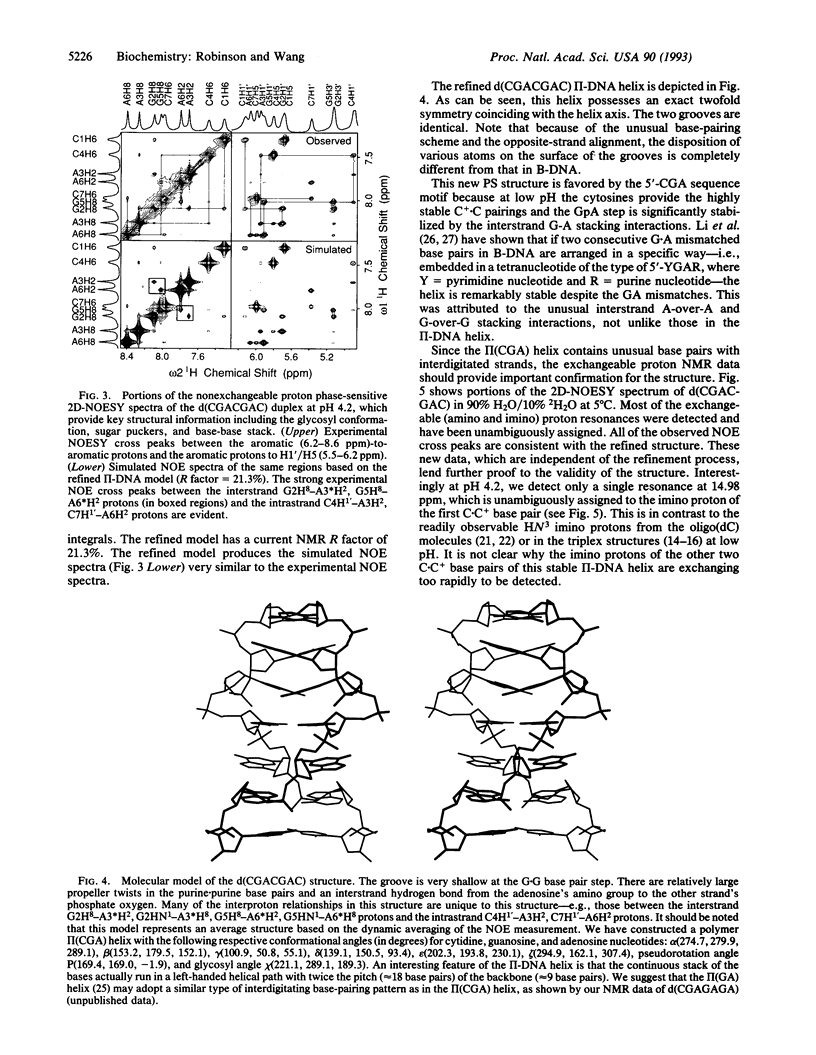
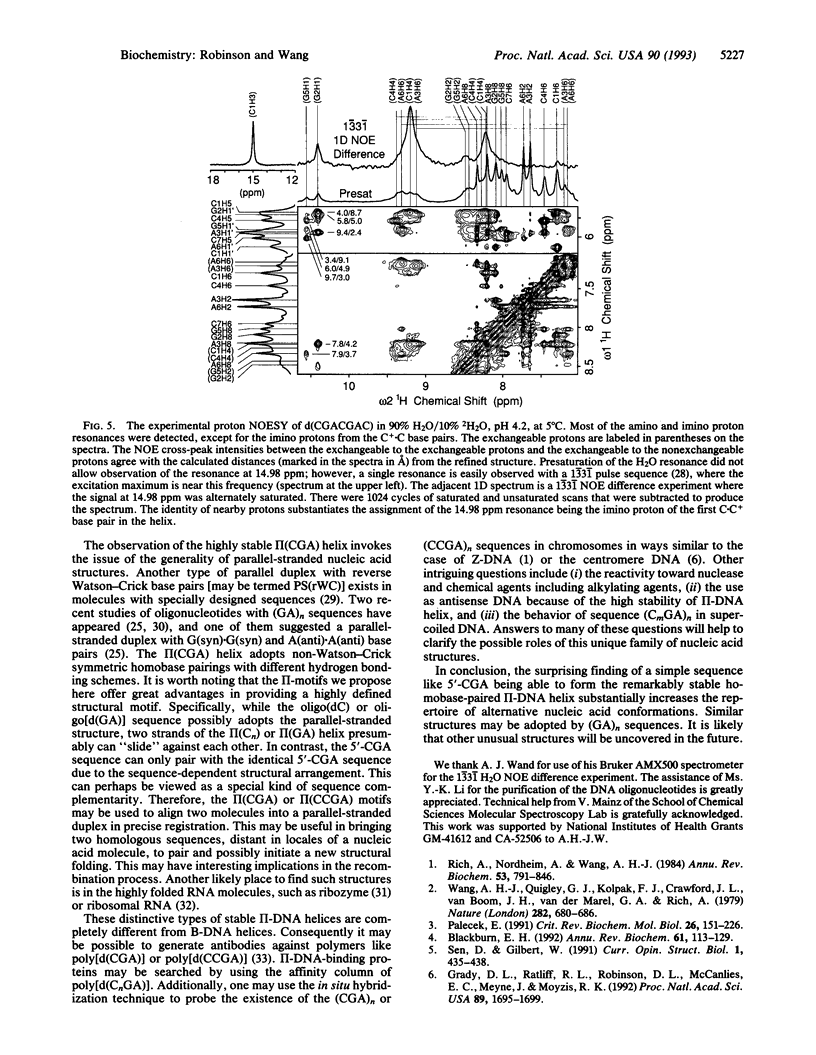
The structure of the non-self-complementary DNA heptamer d(CGACGAC) at low pH has been determined by the quantitative NMR refinement procedure designated SPEDREF (SPEctral-Driven REFinement). Acid-base titration of the molecule indicated a prominent n = 2 pKa near 6.8. In the pH range up to 6.0, the heptamer forms a remarkably stable double helix, which was conclusively shown to be an unusual homobase-paired parallel-stranded double helix (termed II-DNA). In this II-DNA helix, the 5'-CGA trinucleotide is the structural motif that accounts for the stability, with the C+-C hemiprotonated base pair (in which C+ is N3-protonated cytosine) providing for the alignment site and the unusual interstrand G-A base stack in the GpA step furnishing the additional stabilizing forces. The exchangeable proton data from two-dimensional nuclear Overhauser effect spectroscopy are in total agreement with the refined structure. We conclude that the 5'-CGA or other related sequences (e.g., 5'-CCGA) are powerful motifs in promoting the II-DNA or II-RNA conformations that may play certain biological functions.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banville D. L., Marzilli L. G., Wilson W. D. NMR investigation of DNA conformational changes on base protonation: use of Cu2+ and pyrazole as probes. Biochemistry. 1986 Nov 18;25(23):7393–7401. doi: 10.1021/bi00371a022. [DOI] [PubMed] [Google Scholar]
- Blackburn E. H. Telomerases. Annu Rev Biochem. 1992;61:113–129. doi: 10.1146/annurev.bi.61.070192.000553. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Bass B. L. Biological catalysis by RNA. Annu Rev Biochem. 1986;55:599–629. doi: 10.1146/annurev.bi.55.070186.003123. [DOI] [PubMed] [Google Scholar]
- Dolinnaya N. G., Fresco J. R. Single-stranded nucleic acid helical secondary structure stabilized by ionic bonds: d(A(+)-G)10. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9242–9246. doi: 10.1073/pnas.89.19.9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards E. L., Patrick M. H., Ratliff R. L., Gray D. M. A.T and C.C+ base pairs can form simultaneously in a novel multistranded DNA complex. Biochemistry. 1990 Jan 23;29(3):828–836. doi: 10.1021/bi00455a033. [DOI] [PubMed] [Google Scholar]
- Grady D. L., Ratliff R. L., Robinson D. L., McCanlies E. C., Meyne J., Moyzis R. K. Highly conserved repetitive DNA sequences are present at human centromeres. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1695–1699. doi: 10.1073/pnas.89.5.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray D. M., Ratliff R. L., Vaughan M. R. Circular dichroism spectroscopy of DNA. Methods Enzymol. 1992;211:389–406. doi: 10.1016/0076-6879(92)11021-a. [DOI] [PubMed] [Google Scholar]
- Htun H., Dahlberg J. E. Single strands, triple strands, and kinks in H-DNA. Science. 1988 Sep 30;241(4874):1791–1796. doi: 10.1126/science.3175620. [DOI] [PubMed] [Google Scholar]
- INMAN R. B. TRANSITIONS OF DNA HOMOPOLYMERS. J Mol Biol. 1964 Sep;9:624–637. doi: 10.1016/s0022-2836(64)80171-6. [DOI] [PubMed] [Google Scholar]
- Johnston B. H. The S1-sensitive form of d(C-T)n.d(A-G)n: chemical evidence for a three-stranded structure in plasmids. Science. 1988 Sep 30;241(4874):1800–1804. doi: 10.1126/science.2845572. [DOI] [PubMed] [Google Scholar]
- Kallenbach N. R., Daniel W. E., Jr, Kaminker M. A. Nuclear magnetic resonance study of hydrogen-bonded ring protons in oligonucleotide helices involving classical and nonclassical base pairs. Biochemistry. 1976 Mar 23;15(6):1218–1224. doi: 10.1021/bi00651a007. [DOI] [PubMed] [Google Scholar]
- Kremer E. J., Pritchard M., Lynch M., Yu S., Holman K., Baker E., Warren S. T., Schlessinger D., Sutherland G. R., Richards R. I. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991 Jun 21;252(5013):1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- Li Y., Zon G., Wilson W. D. NMR and molecular modeling evidence for a G.A mismatch base pair in a purine-rich DNA duplex. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):26–30. doi: 10.1073/pnas.88.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zon G., Wilson W. D. Thermodynamics of DNA duplexes with adjacent G.A mismatches. Biochemistry. 1991 Jul 30;30(30):7566–7572. doi: 10.1021/bi00244a028. [DOI] [PubMed] [Google Scholar]
- Luo J., Sarma M. H., Yuan R. D., Sarma R. H. NMR study of self-paired parallel duplex of d(AAAAACCCCC) in solution. FEBS Lett. 1992 Jul 20;306(2-3):223–228. doi: 10.1016/0014-5793(92)81005-7. [DOI] [PubMed] [Google Scholar]
- Miras-Portugal M. T., Delicado E. G., Casillas T., Sen R. P. Control of nucleoside transport in neural cells. Effect of protein kinase C activation. Adv Exp Med Biol. 1991;309A:435–438. doi: 10.1007/978-1-4899-2638-8_100. [DOI] [PubMed] [Google Scholar]
- Moser H. E., Dervan P. B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987 Oct 30;238(4827):645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Oberlé I., Rousseau F., Heitz D., Kretz C., Devys D., Hanauer A., Boué J., Bertheas M. F., Mandel J. L. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991 May 24;252(5009):1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- Palecek E. Local supercoil-stabilized DNA structures. Crit Rev Biochem Mol Biol. 1991;26(2):151–226. doi: 10.3109/10409239109081126. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Haniford D. B., Morgan A. R. A structural basis for S1 nuclease sensitivity of double-stranded DNA. Cell. 1985 Aug;42(1):271–280. doi: 10.1016/s0092-8674(85)80122-7. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I., Gao X., de los Santos C., Live D., Patel D. J. NMR structural studies of intramolecular (Y+)n.(R+)n(Y-)nDNA triplexes in solution: imino and amino proton and nitrogen markers of G.TA base triple formation. Biochemistry. 1991 Sep 17;30(37):9022–9030. doi: 10.1021/bi00101a016. [DOI] [PubMed] [Google Scholar]
- Rich A., Nordheim A., Wang A. H. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- Rippe K., Fritsch V., Westhof E., Jovin T. M. Alternating d(G-A) sequences form a parallel-stranded DNA homoduplex. EMBO J. 1992 Oct;11(10):3777–3786. doi: 10.1002/j.1460-2075.1992.tb05463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe K., Jovin T. M. Parallel-stranded duplex DNA. Methods Enzymol. 1992;211:199–220. doi: 10.1016/0076-6879(92)11013-9. [DOI] [PubMed] [Google Scholar]
- Robinson H., Wang A. H. A simple spectral-driven procedure for the refinement of DNA structures by NMR spectroscopy. Biochemistry. 1992 Apr 7;31(13):3524–3533. doi: 10.1021/bi00128a029. [DOI] [PubMed] [Google Scholar]
- Robinson H., van der Marel G. A., van Boom J. H., Wang A. H. Unusual DNA conformation at low pH revealed by NMR: parallel-stranded DNA duplex with homo base pairs. Biochemistry. 1992 Nov 3;31(43):10510–10517. doi: 10.1021/bi00158a014. [DOI] [PubMed] [Google Scholar]
- Sklenár V., Feigon J. Formation of a stable triplex from a single DNA strand. Nature. 1990 Jun 28;345(6278):836–838. doi: 10.1038/345836a0. [DOI] [PubMed] [Google Scholar]
- Stollar B. D. The origin and pathogenic role of anti-DNA autoantibodies. Curr Opin Immunol. 1989;2(4):607–612. doi: 10.1016/0952-7915(90)90019-d. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Gutell R., Gupta R., Noller H. F. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983 Dec;47(4):621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos C., Rosen M., Patel D. NMR studies of DNA (R+)n.(Y-)n.(Y+)n triple helices in solution: imino and amino proton markers of T.A.T and C.G.C+ base-triple formation. Biochemistry. 1989 Sep 5;28(18):7282–7289. doi: 10.1021/bi00444a021. [DOI] [PubMed] [Google Scholar]


