Abstract
We have previously demonstrated that the circadian clock protein period (Per)1 coordinately regulates multiple genes involved in Na+ reabsorption in renal collecting duct cells. Consistent with these results, Per1 knockout mice exhibit dramatically lower blood pressure than wild-type mice. The proximal tubule is responsible for a majority of Na+ reabsorption. Previous work has demonstrated that expression of Na+/H+ exchanger 3 (NHE3) oscillates with a circadian pattern and Na+-glucose cotransporter (SGLT)1 has been demonstrated to be a circadian target in the colon, but whether these target genes are regulated by Per1 has not been investigated in the kidney. The goal of the present study was to determine if Per1 regulates the expression of NHE3, SGLT1, and SGLT2 in the kidney. Pharmacological blockade of nuclear Per1 entry resulted in decreased mRNA expression of SGLT1 and NHE3 but not SGLT2 in the renal cortex of mice. Per1 small interfering RNA and pharmacological blockade of Per1 nuclear entry in human proximal tubule HK-2 cells yielded the same results. Examination of heterogeneous nuclear RNA suggested that the effects of Per1 on NHE3 and SGLT1 expression occurred at the level of transcription. Per1 and the circadian protein CLOCK were detected at promoters of NHE3 and SGLT1. Importantly, both membrane and intracellular protein levels of NHE3 and SGLT1 were decreased after blockade of nuclear Per1 entry. This effect was associated with reduced activity of Na+-K+-ATPase. These data demonstrate a role for Per1 in the transcriptional regulation of NHE3 and SGLT1 in the kidney.
Keywords: gene regulation, kidney, transcription, renal sodium reabsorption, Na+/H+ exchanger, Na+-glucose cotransporter 1, period 1
the circadian clock is an important regulator of multiple physiological functions including blood pressure (BP), the immune response, the sleep-wake cycle, metabolism, vascular function, and renal function (for reviews, see Refs. 36 and 37). Urinary Na+ excretion, renal blood flow, glomerular filtration rate, and plasma aldosterone levels have been shown to undergo rhythmic fluctuations (for a review, see Ref. 44). The circadian clock, at the molecular level, consists of four core proteins, which interact with one another to regulate expression of circadian target genes (13). These four core circadian proteins are CLOCK, Bmal1, period (Per; homologs 1–3), and cryptochrome (Cry; homologs 1–2). CLOCK and Bmal1 heterodimerize and interact with E box response elements to transcriptionally upregulate circadian target genes, including Per and Cry. Per and Cry interact and then repress the transcriptional activity of CLOCK and Bmal1 (1). The circadian clock also undergoes posttranslational modifications through the phosphorylation of Per proteins by circadian kinases, casein kinase 1 isoforms δ/ε (CK1δ/ε). Phosphorylation by CK1δ/ε allows Per1 nuclear entry (22, 33, 35).
We have previously demonstrated that Per1 positively regulates basal and aldosterone-mediated expression of the α-subunt of the renal epithelial Na+ channel (αENaC) (17, 18, 33, 35, 45). We also recently demonstrated that Per1 coordinately regulates the expression of multiple genes involved in the regulation of renal Na+ reabsorption in collecting duct cells (45). It was initially assumed that Per1 behaves primarily as a repressor of CLOCK and Bmal1 activity. However, we and others have shown that Per1 appears to activate gene expression in a manner that appears to be gene and tissue specific (9, 11, 17, 32, 33, 45). Concordantly, we proposed a putative mechanism for Per1 action involving repression of the circadian repressor Cry2 (33). These data predict that loss of Per1 should result in decreased renal Na+ reabsorption, with subsequent decreased plasma volume and decreased BP. Consequently, we have demonstrated that Per1 knockout (KO) mice have significantly lower BP compared with wild-type (WT) control mice (45) and increased urinary Na+ levels (18). Consistent with these results, we have recently shown that mice with reduced Per1 expression exhibit reduced plasma aldosterone levels and renal Na+ wasting (34).
Per1 appears to play an integral role in the regulation of αENaC and other genes that modulate renal Na+ reabsorption. We have also recently demonstrated that Per1 regulates the Na+-Cl− cotransporter and the with no lysine kinases in a model of the murine distal convoluted tubule (39). However, the role of Per1 in the proximal tubule has not been investigated. The proximal tubule is the major site of Na+ reabsorption in the nephron. The majority of Na+ is reabsorbed in this segment through the actions of Na+/H+ exchanger (NHE)3 (for a review, see Ref. 2). Na+-glucose cotransporter (SGLT)1 and SGLT2 also play an important role in the reabsorption of Na+ and glucose (for a review, see Ref. 48). Previous work has demonstrated that NHE3 mRNA oscillates with a circadian pattern in the mouse renal medulla (30, 40). Likewise, SGLT1 is regulated in a similar manner in the colon (6, 7). However, it is unknown if expression of NHE3, SGLT1, or SGLT2 are regulated by Per1.
The goal of the present study was to test whether Per1 contributes to the regulation of Na+ transporters in the proximal tubule via transcriptional modulation of NHE3 and SGLT1. Pharmacological blockade of nuclear Per1 entry in vivo led to decreased mRNA expression of NHE3 and SGLT1 in the renal cortex of mice. Using several additional, independent methods in the human proximal tubule cell line HK-2, we demonstrated a role for Per1 in the transcriptional regulation of NHE3 and SGLT1 but not SGLT2. Importantly, pharmacological blockade of Per1 nuclear entry resulted in decreased intracellular and membrane protein levels of NHE3 and SGLT1. Taken together, these data demonstrate a potential role for the circadian protein Per1 in the regulation of NHE3 and SGLT1 in human proximal tubule cells.
MATERIALS AND METHODS
Animals.
All animal use protocols were approved by the University of Florida and North Florida/South Georgia Veterans Administration Institutional Animal Care and Use Committees in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. WT and Per1 KO mice (129/sv) were originally provided by Dr. David Weaver (University of Massachusetts) (5). WT and Per1 heterozygous (Het) mice were bred in house by University of Florida Animal Care Services staff and maintained on a normal 12:12-h light-dark cycle. For in vivo PF670462 (CK1δ/ε inhibitor) experiments, weight-matched male WT 129/sv mice were given either vehicle (20% hydroxypropyl-β-cyclodextrin) or 30 mg/kg PF670462 subcutaneously every 12 h for 2.5 days starting at noon and euthanized at midnight 12 h after the last injection as previously described (34, 39). Mice were anesthetized by inhalant isoflourane, and tissues were collected and snap frozen in liquid nitrogen. Kidneys were later dissected, and the cortex was removed for RNA isolation or protein collection.
Cell culture.
HK-2 cells were kindly provided by Kirstan Meldrum (University of Florida, Gainesville, FL) (26, 27). HK-2 cells were maintained in DMEM-F-12, 10% FBS, and 1% penicillin-streptomycin at 37°C. For RNA silencing experiments, nontarget and Per1 small interfering (si)RNA (Dharmacon) were used according to the manufacturer's instructions. HK-2 cells were transfected using Dharmafect 1 (Dharmacon). CK1δ/ε inhibitor (PF670462) experiments were performed as described previously (33, 35). For the dose and time course, HK-2 cells were treated with either 0.1, 1, 10, or 100 μM PF670462 or vehicle (water) for 24, 48, or 72 h as previously described (35, 39). For nuclear entry experiments, HK-2 cells were treated with 10 μM PF670462 or vehicle for 24 h.
RNA isolation and quantitative real-time PCR.
Total RNA was isolated and DNase I-treated using Direct-Zol RNA Miniprep (Zymo Research) as per the manufacturer's instructions. DNase I-treated RNA (2 μg) samples were used as template for reverse transcription with a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The resulting cDNAs (20 ng) were then used as templates in quantitative real-time PCRs (Applied Biosystems) to evaluate changes in Per1, NHE3, SGLT1, SGLT2, and actin mRNA levels. Cycle threshold (Ct) values were normalized against β-actin, and relative quantification was performed using the ΔΔCt method (24). Fold change values were calculated as the change in mRNA expression levels relative to the control. Mouse and human TaqMan primer/probe sets were purchased from Applied Biosystems.
Membrane protein analysis, nuclear protein isolation, and Western blot analysis.
Membrane protein was isolated using the Cell Surface Protein Isolation Kit (Pierce) according to the manufacturer's instructions. HK-2 cells were grown to confluence and then treated with vehicle (water) or 10 μM PF670462 for 24 h. Nuclear extracts were isolated using the NE-PER Kit (Pierce) according to the manufacturer's instructions. For the Per1 nuclear entry experiments, protein concentrations were then quantified by BCA assay (Pierce). Western blots were performed as previously described (33–35). Proteins were separated on a 4–20% Tris·HCl Ready Gel (Bio-Rad) and transferred to a polyvinylidene difluoride membrane. The membrane was blocked with 2% nonfat dry milk in Tris-buffered saline plus 0.05% Rodeo Saddle Soap (TBS-S; USB) and incubated overnight at 4°C with anti-Per1 (1:500, Pierce), anti-NHE3 (Millipore), anti-SGLT1 (Millipore), or anti- β-actin (1:500, Santa Cruz Biotechnology) antibodies. β-Actin was used as a loading control. The membrane was washed with 2% nonfat dry milk in TBS-S for 15 min and then incubated with horseradish peroxidase-conjugated anti-rabbit secondary antibody and 2% nonfat dry milk in TBS-S for 1 h at 4°C. After incubation, the blot was washed with TBS-S for 15 min. Detection was performed using Novex ECL Chemiluminescent Substrate reagents (Invitrogen). Densitometry was performed using ImageJ (http://rsbweb.nih.gov/ij).
Heterogeneous nuclear RNA analysis.
Heterogeneous nuclear (hn)RNA analysis was performed as previously described (17, 18, 39). Primers were designed to amplify regions spanning the intron/exon boundary of NHE3, SGLT1, and GADPH genes. GAPDH was used as a cDNA loading control. Sequences and exon numbers are shown in Table 1. Total RNA was isolated from vehicle- and CK inhibitor-treated cells, treated with DNase I, and converted to cDNA as described above. PCRs were performed using 40 ng cDNA as a template. Reactions were heated to 95°C for 15 min to activate the Taq polymerase, and 35 amplification cycles were performed using the following parameters: 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min followed by a final 10-min extension at 72°C.
Table 1.
Sequences and exon numbers
| Gene | Spanning Exon | Forward Primer | Reverse Primer |
|---|---|---|---|
| NHE3 | 7 | 5′CTGTGACTCTGGGCCTAC-3′ | 5′ACTGCTCCGAGATGTTGG-3′ |
| SGLT1 | 3 | 5′GCTCCTTCTCCAATCCTC-3′ | 5′AGCAAAGAGGGAGGCTCC-3′ |
| GAPDH | 4 | 5′AAGAAATGTGCTTTGGGG-3′ | 5′GACTCCACGACGTACTCA-3′ |
NHE3, Na+/H+ exchanger 3; SGLT1, Na+-glucose tranporter 1.
Chromatin immunoprecipitation.
HK-2 cells were grown to ∼80% confluency and then treated with vehicle (water) or 10 μM PF670462 for 24 h. Chromatin immunoprecipitation (ChIP) was performed using the ChIP-IT Express Enzymatic Kit (Active Motif) according to the manufacturer's instructions and as previously described (38, 39). Chromatin concentrations were calculated, and equal amounts of vehicle-treated and 10 μM PF670462-treated chromatin were used per pulldown. Pulldowns were performed using 3 μg of either anti-Per1 (Pierce), anti-CLOCK (Pierce), anti-polymerase (Pol) II (Santa Cruz Biotechnology), or rabbit IgG (Bethyl). Complexes were incubated overnight at 4°C with end-over-end rotation. Immunoprecipitated DNA was amplified by End Point PCR. Band intensities were quantitated using densitometry, which was performed using ImageJ (http://rsbweb.nih.gov/ij). Bands were relativized to the relevant vehicle or PF670462-treated 10% input. Putative E boxes in NHE3 and SGLT1 promoters were analyzed using ALGGEN-PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3) (16, 28). For each gene, three primer sets were designed, as shown in Table 2.
Table 2.
Primer sets for each gene
| Forward Primer | Reverse Primer | |
|---|---|---|
| NHE3 | ||
| Primer set 1 | 5′GTCAGAAGACACATCCAT-3′ | 5′CTCATGCATACATCCC-3′ |
| Primer set 2 | 5′TGGAAACCTTGTGAGCGA-3′ | 5′ACCCCAGGCTCTGAGATG-3′ |
| Primer set 3 | 5′CAGGTTCCTGCTGAAGAC-3′ | 5′CGCAGCTCCTGGGATG-3′ |
| SGLT1 | ||
| Primer set 1 | 5′GACTGCAACCTCCACCT-3′ | 5′TTCTTAGTTTAGGCCGGG-3′ |
| Primer set 2 | 5′CGGCAAGAGAGAGTCCAA-3′ | 5′TTAGGGAGGGACTAGCTG-3′ |
| Primer set 3 | 5′CAAGTCCTTAGAAGGCAT-3′ | 5′AGGTAAGAGCTGTCCTGC-3′ |
Ouabain-sensitive 86Rb uptake.
Human kidney proximal tubule cells, HK-2 (American Type Culture Collection) or HKC11 (provided by Dr. Racusen, Johns Hopkins University), were treated with 10 μM PF670462 for 24 h in serum-free DMEM-F-12 containing 5 mM glucose. Ouabain-sensitive 86Rb uptake was measured as an index of Na+-K+-ATPase-mediated ion transport as previously described (20). Briefly, cells were treated with 5 μM monensin for 30 min to short-circuit Na+ channels so as to measure Na+-K+-ATPase-mediated ion transport at Vmax. A trace amount of 86Rb (≈1 μCi/ml 86RbCl) was added to the cells. Uptake was carried out for 10 min, after which cells were washed five to six times with ice-cold PBS. One-half of the cells were treated with 1 mM ouabain along with monensin for 30 min. Cells were lysed overnight in 0.5 N NaOH containing 0.1% Triton X-100 at 37°C. An aliquot (100 μl) of the lysate was used to measure radioactivity or to measure protein. The difference between 86Rb uptake measured in the presence of 1 mM ouabain and the absence of ouabain was used as a measure of Na+-K+-ATPase-mediated transport activity. Uptake data were calculated as nanomoles of 86Rb accumulated per milligram of protein per 10 min and expressed as percent differences from vehicle-treated cells.
Statistical analysis.
All data are presented as means ± SE with n = 3 or more. Statistical analyses were performed using Graphpad Prism (version 6). All graphs/plots were made with Graphpad Prism (version 6). An unpaired Student's t-test was used to compare differences between control and treated groups. P values of <0.05 were considered significant.
RESULTS
Pharmacological blockade of Per1 nuclear entry in vivo results in decreased mRNA expression of NHE3 and SGLT1 but not SGLT2.
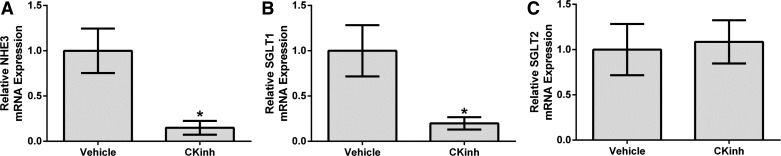
Per1 must be phosphorylated by CK1δ/ε to enter the nucleus (22). Our laboratory has previously shown that pharmacological inhibition of CK1δ/ε recapitulates the effects of Per1 knockdown, including decreased αENaC mRNA levels, protein levels, and ENaC activity (33, 35). To determine if Per1 regulates NHE3, SGLT1, and SGLT2 in vivo, WT mice were treated with vehicle or the CK1δ/ε inhibitor PF670462 as previously described (34). Kidneys were harvested, and the cortex was dissected. mRNA levels of NHE3, SGLT1, and SGLT2 were measured by quantitative real-time PCR. PF670462 treatment resulted in significantly decreased levels of NHE3 (Fig. 1A) and SGLT1 (Fig. 1B) compared with vehicle-treated mice. SGLT2 mRNA levels did not appear to be affected by PF670462 treatment (Fig. 1C). We have previously demonstrated that PF670462-treated mice had significantly reduced levels of nuclear Per1 in the kidney cortex (39).
Fig. 1.
Pharmacological blockade of period (Per)1 nuclear entry in vivo results in decreased Na+/H+ exchanger (NHE)3 and Na+-glucose transporter (SGLT)1 expression but not SGLT2 expression in the renal cortex. Weight-matched male wild-type (WT) 129/sv mice were injected subcutaneously with vehicle (20% hydroxypropyl-β-cyclodextrin) or 30 mg/kg PF670462 [casein kinase (CK)1 isoforms δ/ε (CKinh)]every 12 h for 2.5 days starting at noon and euthanized at midnight 12 h after the last injection, as previously described (34). Kidneys were harvested, the cortex was dissected, and mRNA expresssion of NHE3 (A), SGLT1 (B), or SGLT2 (C) was measured by quantitative real-time PCR. Values are means ± SE; n = 4. *P < 0.05 compared with WT mice.
Per1 siRNA-mediated knockdown or pharmacological inhibition of Per1 nuclear entry in vitro results in decreased mRNA expression of NHE3 and SGLT1 but not SGLT2.
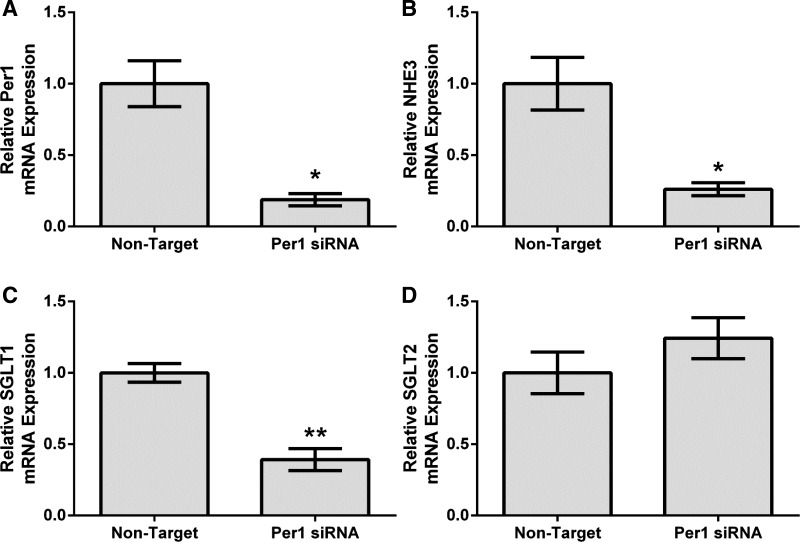
To further investigate our in vivo results, the human proximal tubule cell line HK-2 was used for subsequent experiments (19, 49). Per1 was knocked down using siRNA in HK-2 cells, and mRNA levels of NHE3, SGLT1, and SGLT2 were measured by quantitative real-time PCR. As expected, Per1 knockdown resulted in significantly decreased mRNA expression of Per1 (Fig. 2A), as we have previously demonstrated (17, 33, 45). Consistent with our in vivo results, Per1 knockdown resulted in significantly decreased expression of NHE3 (Fig. 2B) and SGLT1 (Fig. 2C) but not SGLT2 (Fig. 2D).
Fig. 2.
Per1 knockdown results in decreased NHE3 and SGLT1 expression but not SGLT2 expression in HK-2 cells in vitro. HK-2 cells were treated with nontarget (NT) or Per1 small interfering (si)RNA for 48 h. Quantitative real-time PCR was used to evaluate changes in Per1 (A), NHE3 (B), SGLT1 (C), or SGLT2 (D) mRNA expression in Per1 siRNA versus NT siRNA controls. Values are means ± SE; n = 3. *P < 0.05; **P < 0.01.
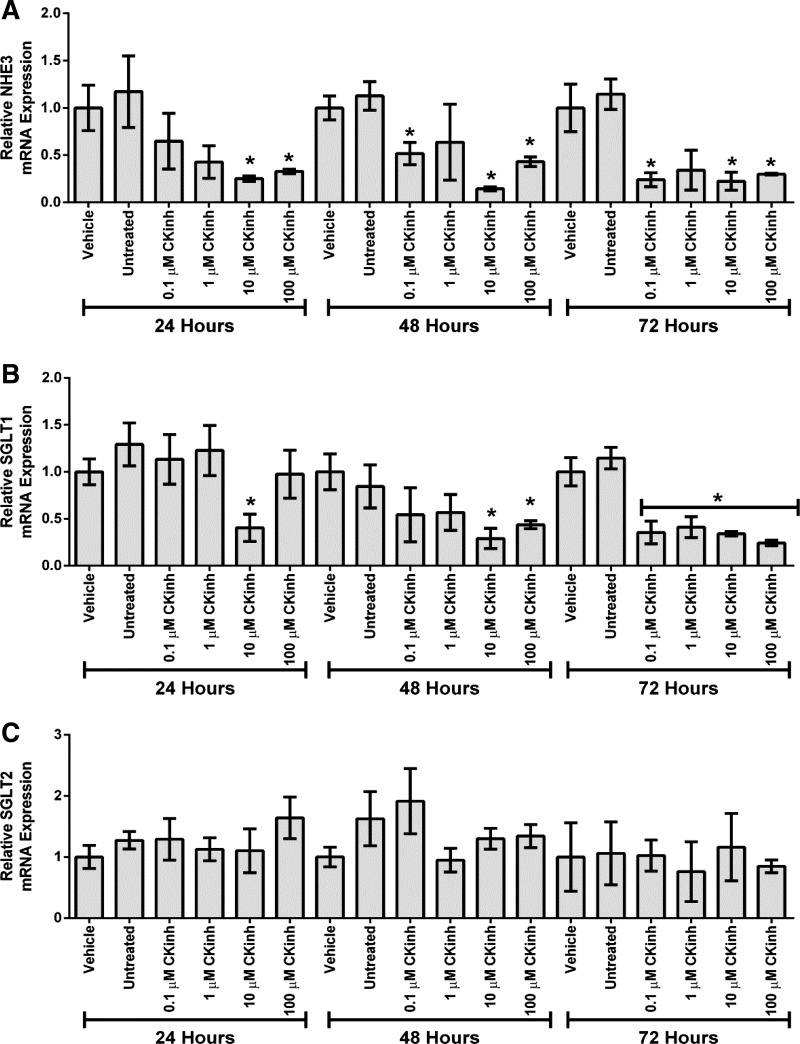
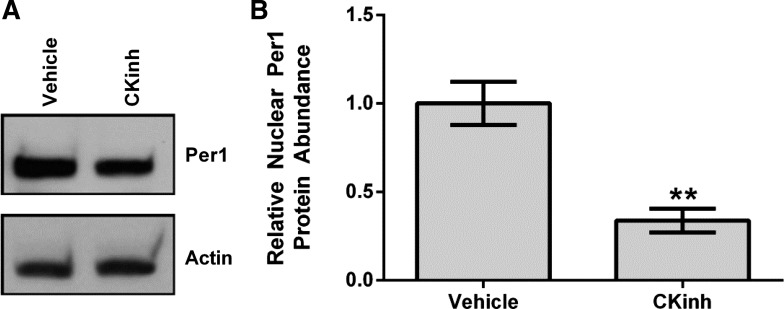
To further explore the potential role of Per1 in the regulation of NHE3 and SGLT1, HK-2 cells were treated with PF670462, and mRNA expression of NHE3, SGLT1, and SGLT2 was determined by quantitative real-time PCR. After 24 h, treatment with 10 μM PF670462 resulted in a significant reduction of NHE3 and SGLT1 mRNA (Fig. 3, A and B). SGLT2 expression was not affected at any time point or dose tested (Fig. 3C). To ensure that CK1δ/ε inhibition decreased Per1 nuclear entry, HK-2 cells were treated with vehicle or 10 μM PF670462 for 24 h, and nuclear extracts were collected. Western blot analysis was performed to assess nuclear Per1 levels. As expected, and as we have previously shown in other cell types (33, 35), nuclear Per1 levels were significantly decreased with CK1δ/ε inhibition (Fig. 4A; quantified in Fig. 4B).
Fig. 3.
Pharmacological blockade of Per1 nuclear entry results in decreased NHE3 and SGLT1 expression but not SGLT2 expression in vitro. A and B: HK-2 cells were treated with the CK1δ/ε inhibitor PF670462 at either 0.1, 1, 10, or 100 μM for 24, 48, or 72 h. Quantitative real-time PCR was used to evaluate changes in NHE3 (A), SGLT1 (B), or SGLT2 (C) expression in PF670462-treated cells versus vehicle (water)-treated cells. Values are means ± SE; n = 3. *P < 0.05.
Fig. 4.
Pharmacological blockade of Per1 nuclear entry results in decreased nuclear Per1 in vitro. A: nuclear extracts were collected from HK-2 cells treated with 10 μM of the CK1δ/ε inhibitor PF670462 or water for 24 h. Western blot analysis was performed using anti-Per1 (nuclear Per1: ∼50 kDa) or anti-β-actin (∼42 kDa) antibodies as loading controls. Data are representative of three independent experiments. B: densitometry analysis was used to quantitate the level of Per1 in A. Values are means ± SE; n = 3. **P < 0.01.
Per1 siRNA-mediated knockdown or pharmacological inhibition of Per1 nuclear entry results in decreased transcription of NHE3 and SGLT1.
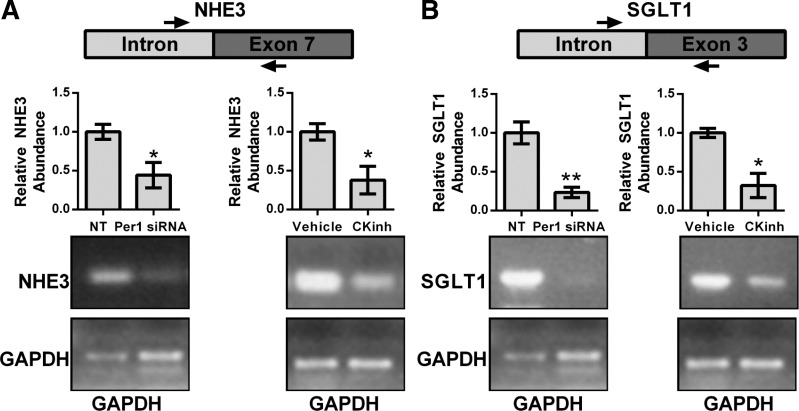
Measurement of short-lived hnRNA is a measure of transcriptional activity (10, 23). To assess if the effect of CK1δ/ε inhibition or Per1 knockdown on NHE3 and SGLT1 was transcriptional, hnRNA levels were assessed by PCR amplification of intron-exon junctions using cDNA templates from HK-2 cells treated with either PF670462 for 24 h or Per1 siRNA for 48 h. Per1 siRNA-mediated knockdown or blockade of Per1 nuclear entry led to significantly decreased hnRNA expression of both NHE3 (Fig. 5A) and SGLT1 (Fig. 5B).
Fig. 5.
Pharmacological blockade of Per1 nuclear entry results in decreased transcription of NHE3 and SGLT1. Primers were designed to amplify regions of heterozygous nuclear (hn)RNA spanning the intron/exon boundary of the NHE3 (A) and SGLT1 (B). GAPDH was used as a cDNA loading control. Arrows are representative of primer location. Bands were quantitated using ImageJ densitometry (http://rsbweb.nih.gov/ij). Signal strength was normalized to the relevant vehicle or CKinh-treated GAPDH control. Values are means ± SE; n = 3. *P < 0.05. **P < 0.01.
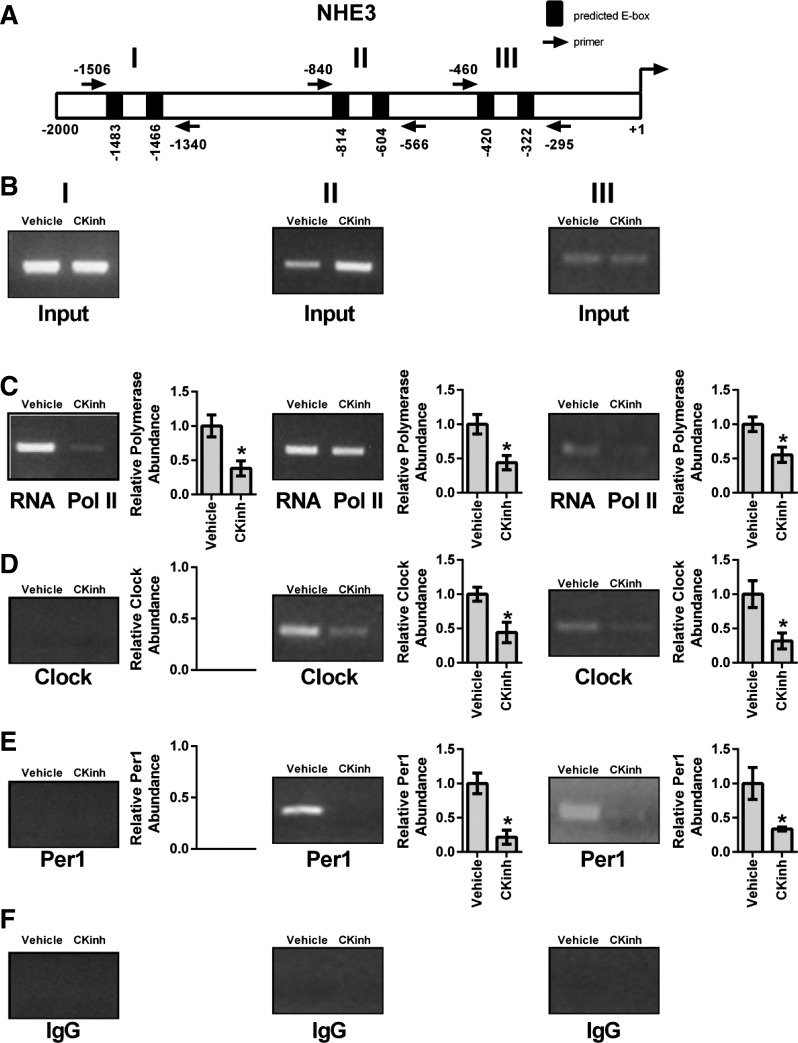
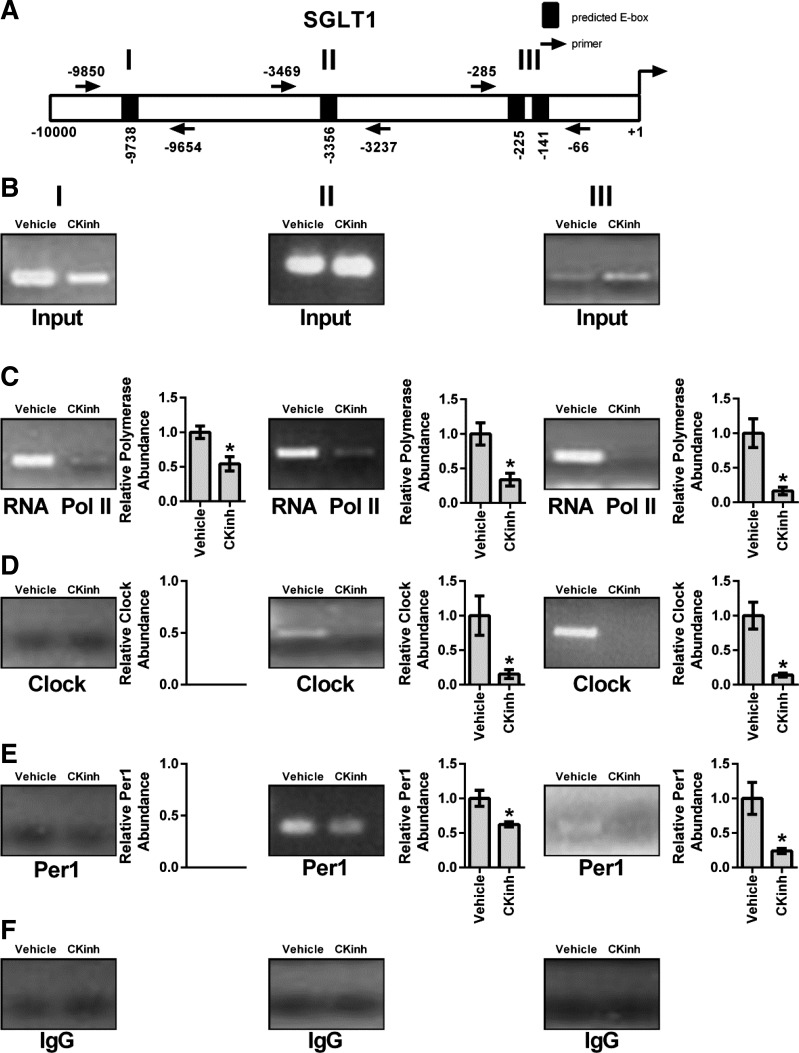
Pharmacological inhibition of Per1 nuclear entry results in decreased interactions of Per1 and CLOCK with promoters of NHE3 and SGLT1.
As described above, regulation of target genes by circadian clock proteins is mediated through interaction of these proteins with E box response elements in the promoters of target genes. Therefore, to assess if Per1 and CLOCK interact with endogenous promoters of NHE3 and SGLT1, ChIP was performed in HK-2 cells treated with either vehicle or 10 μM PF670462 for 24 h. Three primer sets per gene were designed to amplify short regions containing putative E box sites upstream of the transcription start site (Figs. 6A and 7A). Loading was demonstrated by performing PCR on 10% input (Figs. 6B and 7B). IgG was used as a negative control (Panel F Figs. 6 and 7). CK1δ/ε inhibition led to significantly decreased binding of RNA Pol II at all three sites in the NHE3 promoter, indicative of decreased transcription, further corroborating our mRNA and hnRNA data (Fig. 6C). CK1δ/ε inhibition led to significant decreases of CLOCK and Per1 with the second and third promoter regions, but neither CLOCK nor Per1 appeared to interact with the promoter region amplified by the first primer set (Fig. 6, D and E). On the SGLT1 promoter, CK1δ/ε inhibition led to significantly decreased RNA Pol II binding at all three promoter regions (Fig. 7C). As with NHE3, CK1δ/ε inhibition led to significant decreases of CLOCK and Per1 interactions with the second and third promoter regions, but there was no apparent interaction with the promoter region amplified by the first primer set (Fig. 7, D and E).
Fig. 6.
Pharmacological blockade of Per1 nuclear entry leads to decreased occupancy of CLOCK and Per1 on the NHE3 promoter in HK-2 cells. Chromatin immunoprecipitation experiments were performed using HK-2 cells treated with either vehicle (water) or 10 μM PF670462 for 24 h. End-point PCR was performed using primer sets flanking three putative E boxes in the NHE3 promoter (A). B: input experiments. Chromatin immunoprecipitations were performed using anti-polymerase (Pol) II (Santa Cruz Biotechnology; C), anti-CLOCK (Pierce; D), anti-Per1 (Pierce; E), or rabbit IgG (Bethyl, negative control; F) antibodies. Bands were quantitated using densitometry, which was performed using ImageJ (http://rsbweb.nih.gov/ij). Signal strength was normalized to the relevant vehicle- or CKinh-treated input control. Values are means ± SE; n = 3. *P < 0.05.
Fig. 7.
Pharmacological blockade of Per1 nuclear entry leads to decreased occupancy of CLOCK and Per1 on the SGLT1 promoter in HK-2 cells. Chromatin immunoprecipitation experiments were performed using HK-2 cells treated with either vehicle (water) or 10 μM PF670462 for 24 h. End-point PCR was performed using primer sets flanking three putative E boxes in the SGLT1 promoter (A). B: input experiments. Chromatin immuprecipitations were performed using anti-Pol II (Santa Cruz Biotechnology; C), anti-CLOCK (Pierce; D), anti-Per1 (Pierce; E), or rabbit IgG (Bethyl, negative control; F) antibodies. Bands were quantitated using densitometry, which was performed using ImageJ (http://rsbweb.nih.gov/ij). Signal strength was normalized to the relevant vehicle- or CKinh-treated input control. Values are means ± SE; n = 3. *P < 0.05.
Pharmacological blockade of Per1 nuclear entry results in decreased membrane and intracellular protein expression of NHE3 and SGLT1.
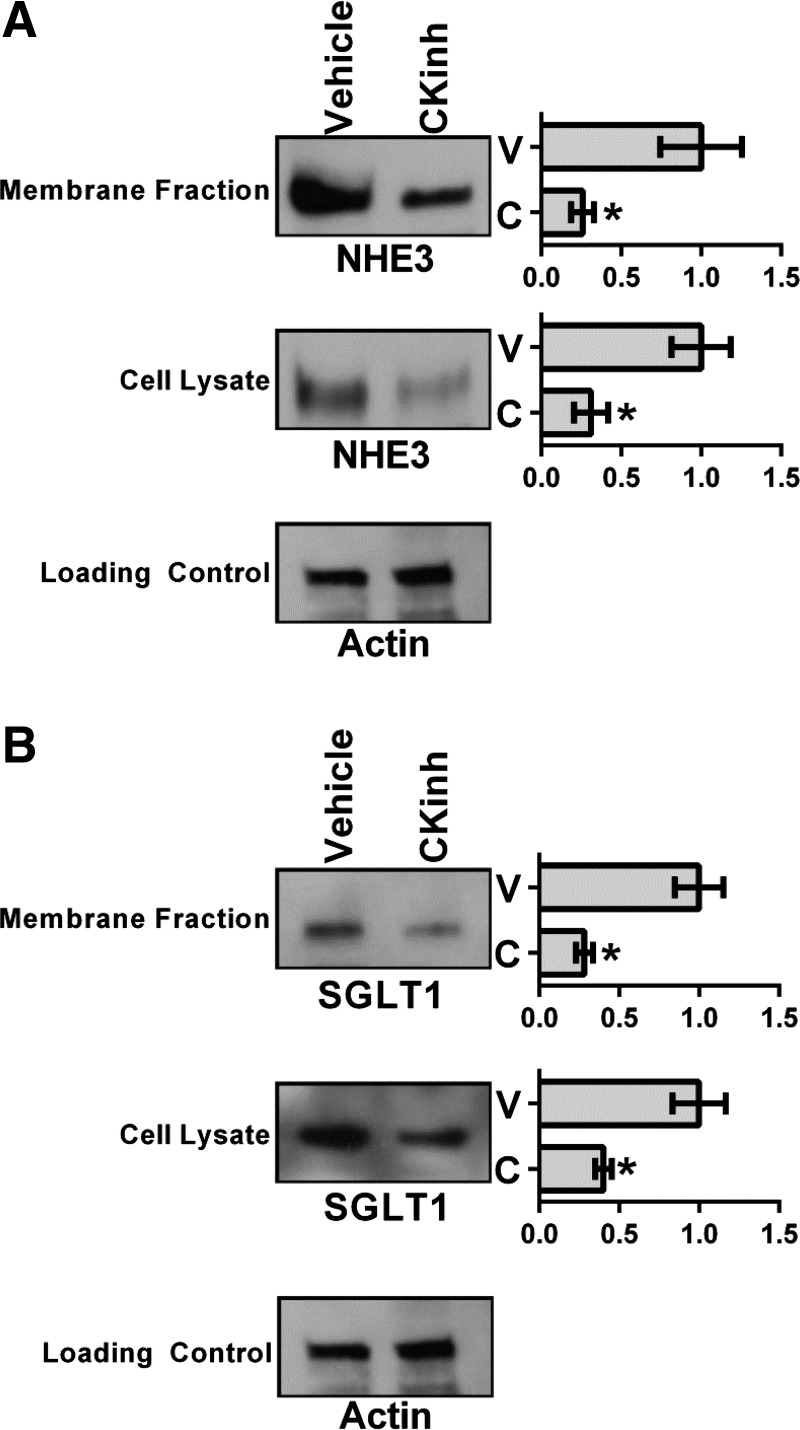
To test whether the effects of CK1δ/ε inhibition on NHE3 and SGLT1 expression extended to the level of protein, plasma membrane and intracellular fractions were isolated from HK-2 cells treated with vehicle or 10 μM PF670462 for 24 h. Western blot analysis was performed to determine changes in NHE3 and SGLT1 protein levels from intracellular and membrane fractions. CK1δ/ε inhibition led to significant decreases of both NHE3 and SGLT1 in both membrane and intracellular fractions (Fig. 8, A and B), consistent with the hypothesis that inhibition of Per1 nuclear entry leads to decreased transcription of NHE3 and SGLT1 (18, 33, 34, 39).
Fig. 8.
Pharmacological blockade of Per1 nuclear entry results in decreased membrane and intracellular protein expression of NHE3 and SGLT1 in HK-2 cells. Membrane and intracellular extracts were collected from HK-2 cells treated with 10 μM of the CK1δ/ε inhibitor PF670462 or water for 24 h. Western blot analysis was performed using anti-NHE3 (∼85 kDa; A), anti-SGLT1 (∼75 kDa; B) or anti-β-actin (∼42 kDa) antibodies as a loading control. Data are representative of three independent experiments. Densitometry analysis was used to quantitate the levels of NHE3 and SGLT1 in A and B. Values are means ± SE; n = 3. *P < 0.05.
Human proximal tubule cell line Na+-K+-ATPase activity decreases with pharmacological inhibition of CK1δ/ε.
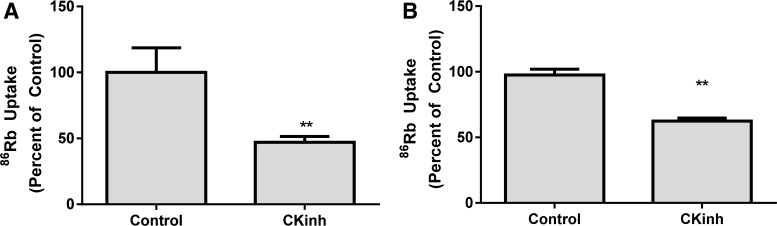
To test the effect of CK1δ/ε inhibition on ion transport, Na+-K+-ATPase activity was measured as ouabain-sensitive 86Rb uptake in two cell lines representative of human proximal tubule cells [HK-2 and HKC11 (41)]. Ouabain-sensitive Na+-K+-ATPase activity was significantly decreased in both cell lines after treatment with 10 μM PF670462 (Fig. 9). Although a direct effect of CK1δ/ε on the Na+ pump cannot be ruled out, decreased Na+-K+-ATPase activity indirectly provides support for the hypothesis that pharmacological blockade of CK1δ/ε results in decreased NHE3 and SGLT1 activity.
Fig. 9.
Na+-K+-ATPase activity is reduced in HK-2 and HKC11 cells after pharmacological blockade of CK1δ/ε. HK-2 (A) and HKC11 (B) cells both showed significantly reduced Na+-K+-ATPase activity after treatment with the CK1δ/ε inhibitor. Cells were treated with 10 μM PF670462 for 24 h in serum-free DMEM-F-12 containing 5 mM glucose. Cells were then treated with 5 μM monensin to short circuit Na+ channels and thus measure Na+-K+-ATPase transport at Vmax. Half of the cells were treated with 1 uM ouabain for 30 min followed by the addition of 86RbCl for 10 min to determine Na+-K+-ATPase-specific activity. Cells were then lysed, and protein was measured to normalize uptake to total protein. Values are calculated as nmoles of 86Rb accumulated per milligram of protein per 10 min and expressed as percent activity relative to vehicle-treated control cells. Values are means ± SE; n = 6 in A and 12 in B. **P < 0.01.
DISCUSSION
The present study demonstrated a role for Per1 in the transcriptional regulation of NHE3 and SGLT1 in proximal tubule cells. Pharmacological blockade of Per1 nuclear entry in vivo and in vitro resulted in decreased mRNA expression of NHE3 and SGLT1. ChIP and hnRNA assays performed in HK-2 cells consistently demonstrated that the effect of Per1 on NHE3 and SGLT1 appears to involve a transcriptional mechanism. Importantly, we demonstrated that the regulation of NHE3 and SGLT1 by Per1 extends to the protein level. Pharmacological blockade of nuclear Per1 entry resulted in decreased protein expression of NHE3 and SGLT1 at the membrane and in the cytosol. Consistent with decreased expression of NHE3 and SGLT1 at the apical membrane, which would predict decreased apical Na+ reabsorption, pharmacological inhibition of CK1δ/ε was associated with decreased Na+-K+-ATPase activity in two different proximal tubule cell lines, HK-2 and HKC11. Taken together, these data suggest a role for Per1 in the positive regulation of genes that function in Na+ reabsorption in proximal tubule cells; this model is entirely consistent with our previously proposed model for Per1 action on ENaC in cortical collecting duct cells (45) and on the Na+-Cl − cotransporter in distal convoluted cells (39).
Previous work has demonstrated that NHE3 is a potential circadian target in the mouse renal medulla (30, 40), whereas other studies have demonstrated that SGLT1 is similarly regulated in the colon (6, 7). However, it was unknown if NHE3 and SGLT1 were regulated by Per1 in the kidney. For the first time, we have demonstrated that both NHE3 and SGLT1 are Per1 targets in a cell model of the human proximal tubule. Along with our previous data on Per1 regulation of transporters in the cortical collecting duct (17, 18, 33, 35, 38, 45) and in the distal convoluted tubule (39), these data predict that loss of Per1 should result in decreased renal Na+ reabsorption, leading to decreased plasma volume and decreased BP. Consistent with these predictions, we have recently shown that mice with reduced levels of Per1 expression exhibit renal Na+ wasting (34) and that Per1 KO mice display lower BP compared with WT control mice (45).
Increasing evidence supports an important role for the circadian clock in the regulation of BP control and renal Na+ handling (for reviews, see Refs. 8, 36, 37, and 44). Per2 KO mice display decreased 24-h diastolic BP, increased heart rate, and reduced diurnal dipping (47). Bmal1 KO mice have reduced BP in the active phase, leading to a blunting of circadian BP variation (12). CLOCK KO mice are hypotensive and display dysregulated urinary Na+ excretion with mild diabetes insipidus (29, 50). Cry1/2 KO mice display salt-sensitive hypertension partially due to increased aldosterone production and serum aldosterone levels (14). This was potentially due to increased expression of 3-β-dehydrogenase isomerase, an enzyme in the aldosterone synthesis pathway. We have shown that Per1 KO mice have significantly lower BP than WT control mice (45) and excrete more Na+ in their urine (18, 34). Mice with reduced levels of Per1 also have decreased plasma levels of aldosterone and decreased renal Na+ reabsorption (34). While a multitude of factors contribute to the BP phenotypes of these animals [including, but not limited to, the heart (for a review, see Ref. 15), sympathetic activity (43, 46), the vasculature (4), and/or nitric oxide (3, 21)], accumulating data from multiple laboratories suggest that salt handling by the kidney is likely to play an important role.
Consistent with our findings, a recent human study (25) demonstrated that Per1 was overexpressed in the renal medulla of human hypertensive patients compared with normotensive control subjects. Given the magnitude of BP and renal Na+ phenotypes in Per1 KO or Per1 heterozygous mice, it is perhaps not surprising that multiple Na+ transporter genes along the nephron are regulated by Per1. Taken together, the data presented here demonstrate a novel role for Per1 in the transcriptional regulation of NHE3 and SGLT1 in the kidney in vivo and in proximal tubule cells in vitro. Determining the functional effects of this regulation will require further investigation. Reduced NHE3 expression may lead to a reduced glomerular filtration rate (42), which was not tested in the present study. On the other hand, KO of NHE3 in a rodent model was associated with reduced BP without alterations in the autoregulation of the glomerular filtration rate (31). Per1 appears to regulate gene expression in a manner consistent with driving Na+ reabsorption in cortical collecting duct cells (17, 18, 33, 35, 38, 45) and distal convoluted tubule cells (39). Now, for the first time, we have demonstrated a role for Per1 in the regulation of Na+ transporter gene expression in proximal tubule cells.
GRANTS
This work was supported by National Institutes of Health (NIH) Grants DK-085193 and DK-098460 and the ASN Foundation for Kidney Research (to M. L. Gumz), American Heart Association Predoctoral fellowship 13PRE16910096 (to J. Richards) and University of Florida Hypertension Center NIH Training Grant 2-T32-HL-083810 (to K. Solocinski).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: K.S., J.R., and M.L.G. conception and design of research; K.S., J.R., S.C.A., K.-Y.C., S.J.K., and M.L.G. performed experiments; K.S., J.R., K.-Y.C., and M.L.G. analyzed data; K.S., J.R., S.J.K., and M.L.G. interpreted results of experiments; K.S. and J.R. prepared figures; K.S. and J.R. drafted manuscript; K.S., J.R., S.C.A., S.J.K., and M.L.G. edited and revised manuscript; K.S., J.R., S.J.K., and M.L.G. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors kindly thank Kirstan Meldrum for providing the HK-2 cells.
Present address of J. Richards: National Cancer Institute, Bethesda, MD.
REFERENCES
- 1.Albrecht U, Eichele G. The mammalian circadian clock. Curr Opin Genet Dev 13: 271–277, 2003. [DOI] [PubMed] [Google Scholar]
- 2.Alexander RT, Dimke H, Cordat E. Proximal tubular NHEs: sodium, protons and calcium? Am J Physiol Renal Physiol 305: F229–F236, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anea CB, Cheng B, Sharma S, Kumar S, Caldwell RW, Yao L, Ali MI, Merloiu AM, Stepp DW, Black SM, Fulton DJ, Rudic RD. Increased superoxide and endothelial NO synthase uncoupling in blood vessels of Bmal1-knockout mice. Circ Res 111: 1157–1165, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anea CB, Zhang M, Stepp DW, Simkins GB, Reed G, Fulton DJ, Rudic RD. Vascular disease in mice with a dysfunctional circadian clock. Circulation 119: 1510–1517, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30: 525–536, 2001. [DOI] [PubMed] [Google Scholar]
- 6.Balakrishnan A, Stearns AT, Ashley SW, Rhoads DB, Tavakkolizadeh A. PER1 modulates SGLT1 transcription in vitro independent of E-box status. Dig Dis Sci 57: 1525–1536, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhutta HY, Deelman TE, Ashley SW, Rhoads DB, Tavakkoli A. Disrupted circadian rhythmicity of the intestinal glucose transporter SGLT1 in Zucker diabetic fatty rats. Dig Dis Sci 58: 1537–1545, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonny O, Firsov D. Circadian regulation of renal function and potential role in hypertension. Curr Opin Nephrol Hypertens 22: 439–444, 2013. [DOI] [PubMed] [Google Scholar]
- 9.Bose S, Boockfor FR. Episodes of prolactin gene expression in GH3 cells are dependent on selective promoter binding of multiple circadian elements. Endocrinology 151: 2287–2296, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H, Kilberg MS. Alignment of the transcription start site coincides with increased transcriptional activity from the human asparagine synthetase gene following amino acid deprivation of HepG2 cells. J Nutr 136: 2463–2467, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen R, Schirmer A, Lee Y, Lee H, Kumar V, Yoo SH, Takahashi JS, Lee C. Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol Cell 36: 417–430, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curtis AM, Cheng Y, Kapoor S, Reilly D, Price TS, Fitzgerald GA. Circadian variation of blood pressure and the vascular response to asynchronous stress. Proc Natl Acad Sci USA 104: 3450–3455, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol 72: 517–549, 2010. [DOI] [PubMed] [Google Scholar]
- 14.Doi M, Takahashi Y, Komatsu R, Yamazaki F, Yamada H, Haraguchi S, Emoto N, Okuno Y, Tsujimoto G, Kanematsu A, Ogawa O, Todo T, Tsutsui K, van der Horst GT, Okamura H. Salt-sensitive hypertension in circadian clock-deficient Cry-null mice involves dysregulated adrenal Hsd3b6. Nat Med 16: 67–74, 2010. [DOI] [PubMed] [Google Scholar]
- 15.Durgan DJ, Young ME. The cardiomyocyte circadian clock: emerging roles in health and disease. Circ Res 106: 647–658, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM, Messeguer X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res 31: 3651–3653, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gumz ML, Cheng KY, Lynch IJ, Stow LR, Greenlee MM, Cain BD, Wingo CS. Regulation of αENaC expression by the circadian clock protein period 1 in mpkCCD(c14) cells. Biochim Biophys Acta 1799: 622–629, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gumz ML, Stow LR, Lynch IJ, Greenlee MM, Rudin A, Cain BD, Weaver DR, Wingo CS. The circadian clock protein period 1 regulates expression of the renal epithelial sodium channel in mice. J Clin Invest 119: 2423–2434, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holthouser KA, Mandal A, Merchant ML, Schelling JR, Delamere NA, Valdes RR Jr, Tyagi SC, Lederer ED, Khundmiri SJ. Ouabain stimulates Na-K-ATPase through a sodium/hydrogen exchanger-1 (NHE-1)-dependent mechanism in human kidney proximal tubule cells. Am J Physiol Renal Physiol 299: F77–F90, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khundmiri SJ, Dean WL, McLeish KR, Lederer ED. Parathyroid hormone-mediated regulation of Na+-K+-ATPase requires ERK-dependent translocation of protein kinase Cα. J Biol Chem 280: 8705–8713, 2005. [DOI] [PubMed] [Google Scholar]
- 21.Kunieda T, Minamino T, Miura K, Katsuno T, Tateno K, Miyauchi H, Kaneko S, Bradfield CA, FitzGerald GA, Komuro I. Reduced nitric oxide causes age-associated impairment of circadian rhythmicity. Circ Res 102: 607–614, 2008. [DOI] [PubMed] [Google Scholar]
- 22.Lee HM, Chen R, Kim H, Etchegaray JP, Weaver DR, Lee C. The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc Natl Acad Sci USA 108: 16451–16456, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipson KE, Baserga R. Transcriptional activity of the human thymidine kinase gene determined by a method using the polymerase chain reaction and an intron-specific probe. Proc Natl Acad Sci USA 86: 9774–9777, 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 25: 402–408, 2001. [DOI] [PubMed] [Google Scholar]
- 25.Marques FZ, Campain AE, Tomaszewski M, Zukowska-Szczechowska E, Yang YH, Charchar FJ, Morris BJ. Gene expression profiling reveals renin mRNA overexpression in human hypertensive kidneys and a role for microRNAs. Hypertension 58: 1093–1098, 2011. [DOI] [PubMed] [Google Scholar]
- 26.Matsui F, Rhee A, Hile KL, Zhang H, Meldrum KK. IL-18 induces profibrotic renal tubular cell injury via STAT3 activation. Am J Physiol Renal Physiol 305: F1014–F1021, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meldrum KK, Zhang H, Hile KL, Moldower LL, Dong Z, Meldrum DR. Profibrotic effect of interleukin-18 in HK-2 cells is dependent on stimulation of the Toll-like receptor 4 (TLR4) promoter and increased TLR4 expression. J Biol Chem 287: 40391–40399, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18: 333–334, 2002. [DOI] [PubMed] [Google Scholar]
- 29.Nikolaeva S, Pradervand S, Centeno G, Zavadova V, Tokonami N, Maillard M, Bonny O, Firsov D. The circadian clock modulates renal sodium handling. J Am Soc Nephrol 23: 1019–1026, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishinaga H, Komatsu R, Doi M, Fustin JM, Yamada H, Okura R, Yamaguchi Y, Matsuo M, Emoto N, Okamura H. Circadian expression of the Na+/H+ exchanger NHE3 in the mouse renal medulla. Biomed Res 30: 87–93, 2009. [DOI] [PubMed] [Google Scholar]
- 31.Noonan WT, Woo AL, Nieman ML, Prasad V, Schultheis PJ, Shull GE, Lorenz JN. Blood pressure maintenance in NHE3-deficient mice with transgenic expression of NHE3 in small intestine. Am J Physiol Regul Integr Comp Physiol 288: R685–R691, 2005. [DOI] [PubMed] [Google Scholar]
- 32.Oster H, Baeriswyl S, Van Der Horst GT, Albrecht U. Loss of circadian rhythmicity in aging mPer1−/−mCry2−/− mutant mice. Genes Dev 17: 1366–1379, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richards J, All SC, Skopis G, Cheng KY, Compton B, Srialluri N, Stow LR, Jeffers LA, Gumz ML. Opposing actions of Per1 and Cry2 in the regulation of Per1 target gene expression in the liver and kidney. Am J Physiol Regul Integr Comp Physiol 305: R735–R747, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richards J, Cheng KY, All SC, Skopis G, Jeffers LA, Lynch IJ, Wingo CS, Gumz ML. A role for the circadian clock protein per1 in the regulation of aldosterone levels and renal sodium retention. Am J Physiol Renal Physiol 305: F1697–F1704, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richards J, Greenlee MM, Jeffers LA, Cheng KY, Guo L, Eaton DC, Gumz ML. Inhibition of αENaC expression and ENaC activity following blockade of the circadian clock-regulatory kinases CK1δ/ε. Am J Physiol Renal Physiol 303: F918–F927, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richards J, Gumz ML. Advances in understanding the peripheral circadian clocks. FASEB J 26: 3602–3613, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richards J, Gumz ML. Mechanism of the circadian clock in physiology. Am J Physiol Regul Integr Comp Physiol 304: R1053–R1064, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richards J, Jeffers LA, All SC, Cheng KY, Gumz ML. Role of Per1 and the mineralocorticoid receptor in the coordinate regulation of αENaC in renal cortical collecting duct cells. Front Physiol 4: 253, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richards J, Ko B, All S, Cheng KY, Hoover RS, Gumz ML. A role for the circadian clock protein Per1 in the regulation of the NaCl co-transporter (NCC) and the with-no-lysine kinase (WNK) cascade in mouse distal convoluted tubule cells. J Biol Chem 289: 11791–1806, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saifur Rohman M, Emoto N, Nonaka H, Okura R, Nishimura M, Yagita K, van der Horst GT, Matsuo M, Okamura H, Yokoyama M. Circadian clock genes directly regulate expression of the Na+/H+ exchanger NHE3 in the kidney. Kidney Int 67: 1410–1419, 2005. [DOI] [PubMed] [Google Scholar]
- 41.Salyer SA, Parks J, Barati MT, Lederer ED, Clark BJ, Klein JD, Khundmiri SJ. Aldosterone regulates Na+, K+ ATPase activity in human renal proximal tubule cells through mineralocorticoid receptor. Biochim Biophys Acta 1833: 2143–2152, 2013. [DOI] [PubMed] [Google Scholar]
- 42.Schnermann J. Sodium transport deficiency and sodium balance in gene-targeted mice. Acta Physiol Scand 173: 59–66, 2001. [DOI] [PubMed] [Google Scholar]
- 43.Sheward WJ, Naylor E, Knowles-Barley S, Armstrong JD, Brooker GA, Seckl JR, Turek FW, Holmes MC, Zee PC, Harmar AJ. Circadian control of mouse heart rate and blood pressure by the suprachiasmatic nuclei: behavioral effects are more significant than direct outputs. PLos One 5: e9783, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stow LR, Gumz ML. The circadian clock in the kidney. J Am Soc Nephrol 22: 598–604, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stow LR, Richards J, Cheng KY, Lynch IJ, Jeffers LA, Greenlee MM, Cain BD, Wingo CS, Gumz ML. The circadian protein period 1 contributes to blood pressure control and coordinately regulates renal sodium transport genes. Hypertension 59: 1151–1156, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki J, Ogawa M, Tamura N, Maejima Y, Takayama K, Maemura K, Honda K, Hirata Y, Nagai R, Isobe M. A critical role of sympathetic nerve regulation for the treatment of impaired daily rhythm in hypertensive Dahl rats. Hypertens Res 33: 1060–1065, 2010. [DOI] [PubMed] [Google Scholar]
- 47.Vukolic A, Antic V, Van Vliet BN, Yang Z, Albrecht U, Montani JP. Role of mutation of the circadian clock gene Per2 in cardiovascular circadian rhythms. Am J Physiol Regul Integr Comp Physiol 298: R627–R634, 2010. [DOI] [PubMed] [Google Scholar]
- 48.Wallner EI, Wada J, Tramonti G, Lin S, Kanwar YS. Status of glucose transporters in the mammalian kidney and renal development. Ren Fail 23: 301–310, 2001. [DOI] [PubMed] [Google Scholar]
- 49.Yan Y, Haller S, Shapiro A, Malhotra N, Tian J, Xie Z, Malhotra D, Shapiro JI, Liu J. Ouabain-stimulated trafficking regulation of the Na/K-ATPase and NHE3 in renal proximal tubule cells. Mol Cell Biochem 367: 175–183, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zuber AM, Centeno G, Pradervand S, Nikolaeva S, Maquelin L, Cardinaux L, Bonny O, Firsov D. Molecular clock is involved in predictive circadian adjustment of renal function. Proc Natl Acad Sci USA 106: 16523–16528, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]