Abstract
Diatoms are major players in the global carbon cycle, and their metabolism is affected by ocean conditions. Understanding the impact of changing inorganic nutrients in the oceans on diatoms is crucial, given the changes in global carbon dioxide levels. Here, we present a genome-scale metabolic model (iMK1961) for Cylindrotheca closterium, an in silico resource to understand uncharacterized metabolic functions in this ubiquitous diatom. iMK1961 represents the largest diatom metabolic model to date, comprising 1961 open reading frames and 6718 reactions. With iMK1961, we identified the metabolic response signature to cope with drastic changes in growth conditions. Comparing model predictions with Tara Oceans transcriptomics data unraveled C. closterium’s metabolism in situ. Unexpectedly, the diatom only grows photoautotrophically in 21% of the sunlit ocean samples, while the majority of the samples indicate a mixotrophic (71%) or, in some cases, even a heterotrophic (8%) lifestyle in the light. Our findings highlight C. closterium’s metabolic flexibility and its potential role in global carbon cycling.
The predominant mixotrophic growth of diatoms in the marine environment can substantially affect the global carbon cycle.
INTRODUCTION
Diatoms are unicellular photosynthetic microscopic eukaryotes ubiquitous in freshwater and marine environments. These microalgae are responsible for around one-fifth of Earth’s carbon fixation (1) and account for about 25% of global oxygen release per year (2). Diatoms represent the most diverse group of phytoplankton, including nearly 200,000 different species (3). They have a distinct capability to generate porous silica cell walls surrounding the cells known as frustules (4). In addition, diatoms can accumulate high amounts of lipids for energy storage, making them potential candidates for generating commercially valuable products, such as biofuels and bioactives (5).
Marine plankton, including diatoms, uses a diverse array of nutritional strategies, encompassing photoautotrophy, heterotrophy, and mixotrophy. While some species exclusively harness CO2 in the presence of light (photoautotrophy), others sustain themselves solely on organic material (heterotrophy). However, recent literature indicates that many plankton members can adeptly engage in both phototrophic and heterotrophic modes concurrently (i.e., mixotrophy) (6). Mixotrophic capabilities provide a crucial advantage for marine plankton, enhancing their resilience amid fluctuating food webs in the ocean, thus highlighting the pivotal role of this trophic strategy (7). It was suggested that mixotrophy contributes to an ~35% increase of carbon flux to larger organisms, exerting a substantial influence on the global carbon cycle. Given the widespread distribution of diatoms in the oceans, unraveling their trophic strategies can deepen our understanding of their contribution to oceanic food webs and their broader impact on Earth’s biogeochemical processes.
Despite its low abundance (8), Cylindrotheca closterium, a meroplanktonic species, thrives in sunlit water columns across the globe as well as on benthic sediments (9). The ability of C. closterium for adapting to drastically varying environmental conditions and its ubiquitous distribution in the global oceans makes it a model diatom for filling knowledge gaps in diatom biology. Despite its importance for global nutrient cycling, the metabolic capabilities of C. closterium are not fully understood. Moreover, the metabolic strategies enabling this diatom to thrive in different but nutritionally distinct marine environments are currently unknown.
One of the vital computational tools for comprehending metabolic variations under different environmental conditions are genome-scale metabolic models (GEMs). A GEM contains a curated knowledge base, including detailed genome annotations, gene products, all known biochemical reactions in an organism, and physiological functions. GEMs of organisms from every domain of life, from microorganisms to humans, have been successfully applied to explore their metabolisms, to identify targets for metabolic engineering (10), to study variations in flux distribution through the metabolic network in different growth environments (11), to contextualize omics data, to elucidate strain-specific metabolic differences (12), and to study interactions in microbial communities (13–15). After our team reconstructed the first GEM of a diatom (Phaeodactylum tricornutum) in 2016 (16), models are now available for two other diatoms, Fragilariopsis cylindrus (17) and Thalassiosira pseudonana (18, 19).
Here, we reconstructed and manually curated a GEM of an environmental strain of C. closterium (iMK1961) to explore this diatom’s yet uncharacterized metabolic capabilities. Model-predicted fluxes were validated using experimental measurements under a broad range of growth conditions, including photoautotrophic, heterotrophic, and mixotrophic conditions. Photoautotrophic conditions were maintained by supplying light and CO2 to the cells, whereas under heterotrophic conditions, cells were cultivated in darkness without CO2 but with an organic carbon source. During mixotrophy, cells had access to light and CO2, as well as an organic carbon source simultaneously. To determine C. closterium’s role in different marine environments, we compared global ocean transcriptomics datasets (20–22) with the model predictions and revealed distinct metabolic responses of C. closterium to elevated nutrient concentrations in different parts of the ocean. Our framework demonstrated that in most ocean samples, C. closterium grows mixotrophically rather than photoautotrophically, shedding light on different strategies of this diatom to thrive under different nutritional conditions and providing insights in its ubiquitous abundance in the global oceans. Our findings also indicate that interactions between C. closterium and marine bacteria may contribute to its prevalent mixotrophic growth in open oceans.
RESULTS
Reconstruction of genome-scale metabolic network of C. closterium
While it is currently challenging to derive detailed knowledge about in situ metabolism from omics data alone, contextualization of omics data using GEMs has been an efficient tool to provide mechanistic understanding of microorganisms in pure cultures and simple communities (13, 15, 23). Here, we used a metabolic reconstruction pipeline (Fig. 1A) to reconstruct a draft metabolic network based on the annotated genome of C. closterium to shine light on its metabolism in situ. A functionally annotated genome was obtained from an environmental strain of C. closterium isolated from estuary samples collected at the Tijuana River Estuary (TJRE) outflow in San Diego County (CA, USA). The RAVEN and COBRApy/COBRA Toolboxes were used to reconstruct and refine the draft metabolic network (see Materials and Methods) (24–26). We identified reactions associated with metabolic genes in the genome of C. closterium based on homology between protein sequences of C. closterium and genomes of template organisms, namely P. tricornutum (16) and Chlamydomonas reinhardtii (27). Functionally annotated genes of C. closterium, which did not return any hit against template genes, were checked for assigned Enzyme Commission (EC) numbers. Next, we drew associated reactions from manually curated models in the BiGG database (28). This step expanded the reconstruction by 581 reactions, 684 metabolites, and 389 genes. These reactions were distributed among several subsystems, primarily including membrane lipid metabolism, purine metabolism, glycerophospholipid metabolism, cofactor, and prosthetic group biosynthesis, cell wall biosynthesis, nucleotide salvage pathway, nucleotides, fatty acid metabolism, oxidative phosphorylation, pyrimidine metabolism, alternate carbon metabolism, and pyruvate metabolism (table S12). Before incorporating these additional reactions into the reconstruction, each was manually checked and standardized using BiGG abbreviations (28, 29). All reactions in the reconstruction were localized in seven subcellular compartments (cytosol, chloroplast, the extracellular space, mitochondria, periplasm, peroxisome/glyoxysome, and thylakoid) (table S13). To predict the subcellular localization of proteins, we deployed a previously published pipeline (see Materials and Methods) (15).
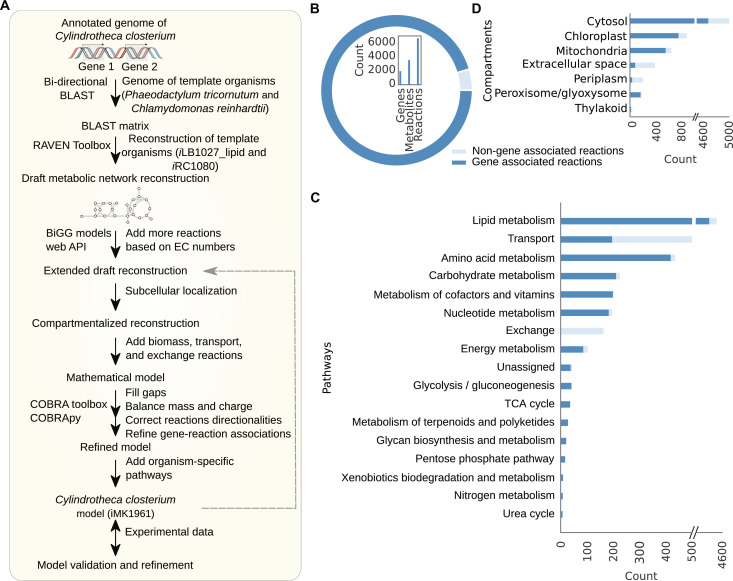
Fig. 1. Metabolic network reconstruction and features of iMK1961.
(A) Reconstruction pipeline of the GEM of C. clostridium (see Supplementary Text and Materials and Methods). (B) Features of iMK1961. The donut chart represents the proportions of the gene (cyan blue) and non-gene–associated reactions (light cyan blue) in iMK1961. The bar plot illustrates the number of genes, metabolites, and reactions in iMK1961. (C) Distribution of reactions among different functional categories. These categories are defined on the basis of KEGG pathways. Reaction distribution shows that lipid metabolism constitutes the largest portion of the total reactions in the network. (D) Reaction distribution per compartment. Among the seven compartments, the cytosol contains most of the reactions.
Information associated with each reaction, such as reaction directionality, charge and mass balances, and gene-protein-reactions (GPRs), was manually verified and corrected where needed using resources such as genome annotation, relevant literature, and reaction and protein databases (see Materials and Methods). The growth medium that was used to grow C. closterium in the laboratory was used to constrain the model during filling the gaps in the metabolic network of the draft reconstruction by adding a minimal set of reactions to enable the production of all biomass precursors and to connect the metabolites, which were causing dead ends in the network (tables S3 to S5). Further, gap-filling linked media nutrients (carbon and nitrogen) to the metabolic network using previously published data for C. closterium (see Materials and Methods). During this exercise, 65 additional reactions, mostly exchange and transport reactions, were added to the model (table S6).
Features of the GEM (iMK1961) for C. closterium
The model iMK1961 encompasses 1961 genes (representing around 10% of all functionally annotated C. closterium genes), 6718 reactions, and 3559 metabolites distributed across seven compartments. Features of iMK1961 are summarized in Fig. 1 (B to D) and tables S13 to S15. The model includes 6050 reactions (90% of total reactions) based on genome annotations, as well as 668 reactions (10% of total reactions) without genetic evidence (i.e., orphan reactions). However, these orphan reactions are essential for the biosynthesis of biomass precursors (Fig. 1B). All reactions in iMK1961 are categorized within 208 subsystems (table S13), and these subsystems are associated with 17 pathways (Fig. 1C). Among these pathways, lipid metabolism and amino acid metabolism include 75% of the total reactions, and the remaining 25% reactions are distributed among transport, carbohydrate metabolism, cofactors and vitamins metabolism, nucleotide metabolism, energy metabolism, exchange, glycolysis/gluconeogenesis, tricarboxylic acid (TCA) cycle, and metabolism of terpenoids and polyketides, glycan biosynthesis and metabolism, pentose phosphate pathway, xenobiotics biodegradation and metabolism, nitrogen metabolism, and the urea cycle (Fig. 1C). The largest number of reactions is localized in the cytosol (68%), followed by the chloroplast (12%), mitochondria (9%), extracellular space (5%), periplasm (3%), peroxisome/glyoxysome (2%), and thylakoid (1%) (Fig. 1D).
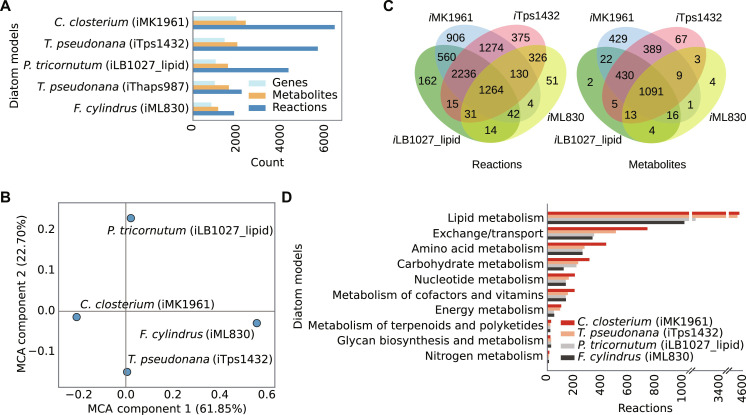
To date, four diatom GEMs have been published. Two GEMs (iTps1432 and iThaps987) (18, 19) have been developed for T. pseudonana, and two separate GEMs have been reconstructed for P. tricornutum (iLB1027) (16) and F. cylindrus (iML830) (fig. S1) (17). By comparing iMK1961 with other diatom GEMs based on the distribution of reactions among different pathways, iMK1961 comprises a notably higher number of reactions in lipid metabolism, transport, amino acid metabolism, carbohydrate metabolism, metabolism of cofactors and vitamins, nucleotide metabolism, and energy metabolism (Fig. 2). These additional reactions in iMK1961 aided the accurate prediction of phenotypic traits under a broad range of growth conditions (see section “Model validation”).
Fig. 2. Comparison of iMK1961 with previously published diatom GEMs.
(A) Comparative representation of genes, metabolites, and reactions in different diatom GEMs. C. closterium (iMK1961) and F. cylindrus GEM (iML830) include the highest and lowest number of reactions and metabolites. Apart from this plot, iThaps987 was not considered in this analysis because of its inconsistent reaction and metabolite identifiers. (B) The Multiple Correspondence Analysis (MCA) scatter plot shows dissimilarities between different diatom GEMs in terms of reactions and metabolites [n = 8803 (iMK1961), n = 7660 (iTps1432), n = 5909 (iLB1027_lipid), and n = 3005 (iML830)]. The scatter plot illustrates the first two components, which explained 61.85 and 22.70% of the variances. We compared the entire metabolic content (reactions and metabolites) of iMK1961 with previously published diatom GEMs using MCA (80). iMK1961 and iTps1432 (T. pseudonana) are placed closer to each other as they exclusively share 1274 reactions and 389 metabolites. F. cylindrus (iML830) is distinct from the other three GEMs because this model comprises the lowest number of reactions and metabolites. iMK1961 has 906 unique reactions and 429 unique metabolites, which are not present in any previous diatom models. These unique reactions are mostly distributed in pathways related to transport, lipid metabolism, amino acid metabolism, carbohydrate metabolism, cofactors and vitamins metabolism, nucleotide metabolism, energy metabolism, TCA cycle, and glycolysis/gluconeogenesis (fig. S2). (C) Venn diagrams represent the distinct and shared reactions and metabolites among different diatom GEMs. C. closterium GEM (iMK1961) includes the highest number of unique reactions and metabolites. It exclusively shared the maximum number of reactions with iTps1432 followed by iLB1027_lipid and iML830. (D) Comparison between diatom GEMs based on reaction distribution among different pathways. Reactions were defined in 10 functional categories based on KEGG metabolic pathways.
Model validation
To validate the prediction capability of iMK1961, we simulated in silico growth and phenotypes under several different growth conditions, including growth in the light and dark, different nutrients, and nutrient-limiting conditions. The model predictions were compared with experimental measurements collected from previous studies as well as those generated during this study (Fig. 3A and table S7). Growth of C. closterium under various trophic conditions was assessed experimentally on the basis of cell count or density or chlorophyll concentrations over time. We predicted the growth of C. closterium on the consumption of a broad range of nutrients as sole sources of carbon, nitrogen, phosphorus, and sulfur. This analysis suggested that the model can grow on 81 carbon, 43 nitrogen, 11 phosphorus, and 6 sulfur sources tested (table S7). Model predictions were compared with experimental biomass measurements based on 14 carbon and 15 nitrogen sources. In silico results have been confirmed experimentally with 100% accuracy (Fig. 3A). Under photoautotrophic (CO2 + light) conditions, our predictions on CO2 and 15 nitrogen source assimilation were found to be consistent with experimental measurements. Similarly, the model predicted growth on 13 carbon and 15 nitrogen sources under heterotrophic (dark + organic carbon source) and mixotrophic (CO2 + light + organic carbon source) conditions. These predictions were all confirmed experimentally (under at least one trophic condition).
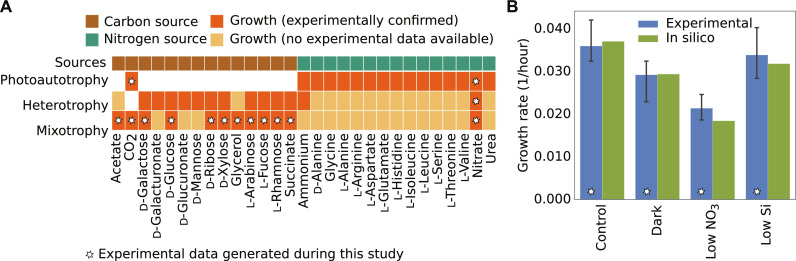
Fig. 3. Validation of iMK1961.
(A) Heatmap represents experimentally confirmed predictions of metabolic phenotypes on various nutrient conditions composed of different carbon, nitrogen sources in photoautotrophic and heterotrophic, and mixotrophic conditions. Nitrate was used as a nitrogen source to stimulate growth in all three trophic conditions on different carbon sources. Similarly, to stimulate growth on different nitrogen sources, CO2, succinate, and CO2 + succinate were used in photoautotrophic, heterotrophic, and mixotrophic conditions, respectively. White cells in the heatmap represent no growth data available based on the definition of photoautotrophy, heterotrophy, and mixotrophy. (B) Comparison between in silico and experimental growth rates. Experimental data were used to constrain the inputs of the model. The biomass-producing reaction was used as an objective function during each simulation.
Furthermore, we examined the accuracy of the model predictions by comparing in silico and experimental growth rates. As shown in Fig. 3B and table S8, GEM predictions agreed well with experimental results under one heterotrophic (dark) and three photoautotrophic growth conditions [control (light), low nitrogen, and low silicon] (see Materials and Methods and table S9). The experimental growth rates used in this analysis were exclusively determined as cell counts over time during.
Model predictions were rigorously validated through comparisons with experimental measurements gathered from previous studies as well as data generated during the course of this investigation. This comprehensive approach ensured the robustness and reliability of the model’s outputs, enhancing confidence in its predictive capabilities. By leveraging a combination of historical and newly acquired data, the study achieved a thorough evaluation of the model’s performance, affirming its utility for understanding and forecasting complex ecological dynamics.
Metabolic capabilities reveal flexibility in the C. closterium metabolism
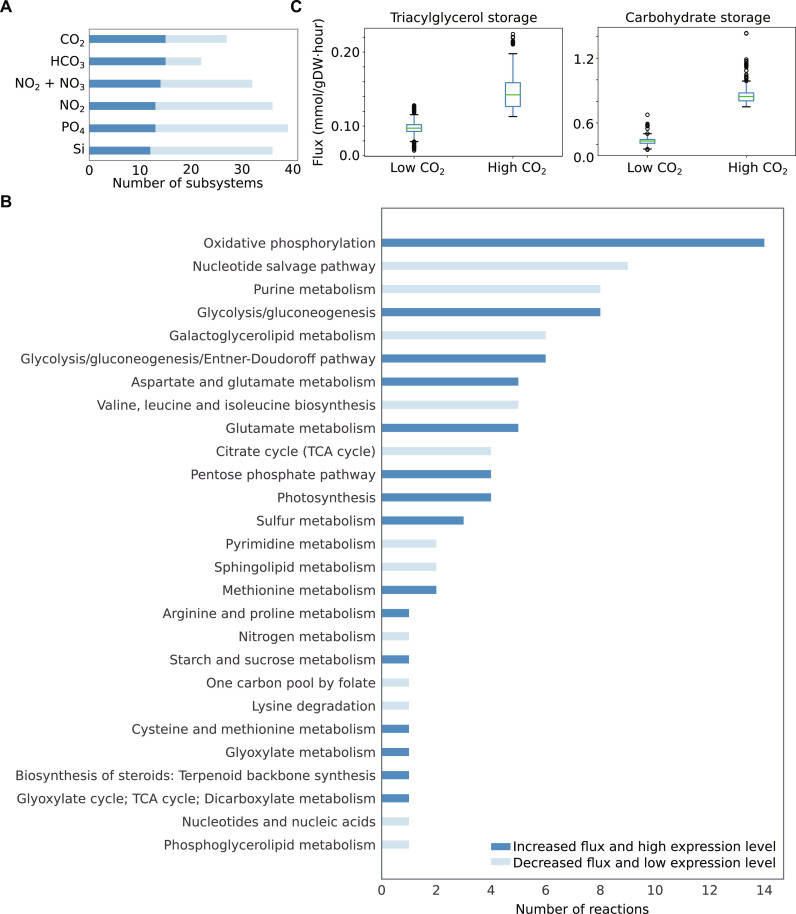
We deployed the model iMK1961 to gain insights into the ubiquitous abundance of C. closterium in different marine environments by studying its metabolic response to variation in nutrient concentrations. For this, we compared model predictions with transcriptomics data generated in the laboratory (30) and obtained from global ocean sampling (21). The optimized general parallel sampler (optGpSampler) (31) was used to predict all feasible fluxes through each reaction in the metabolic network under different nutrient conditions. We used publicly available microbiome and environmental data [Tara Oceans (21)] and selected six nutrient-related environmental factors (CO2, HCO3, NO2, NO2 + NO3, PO4, and Si) to simulate metabolic flux through each reaction under each nutrient condition. The samples in this dataset were divided into two categories based on low and high values of environmental factors, defined by comparing the measurement of each factor with its average value among different samples (table S10). Next, we constrained the model using these low and high values for each environmental factor (see Materials and Methods and table S11). This practice enabled us to mimic the nutritional conditions in the marine environment and simulate the response of C. closterium to varying nutrients. The subsystem-level differential flux distributions through the metabolic network were estimated under low and elevated concentrations of all six inorganic nutrients. The resulting flux distributions were used to classify the subsystems that carried increased and decreased fluxes on elevated nutrients compared to fluxes under the low concentration of nutrients (Fig. 4A and fig. S3). We also identified common and unique subsystems with differential fluxes by comparing flux distributions under different nutritional conditions. On average, we found 87 subsystems that carried differential fluxes under six nutritional conditions that represent about 42% of the total number of subsystems present in the model (table S16). On the basis of pairwise comparisons, two pairs (i.e., CO2 and HCO3 and NO2 + NO3 and NO2) shared more differential fluxes carrying subsystems compared to any other pairs (tables S17 and S18). Comparing flux distributions between all six conditions, only eight shared subsystems were found carrying differential fluxes, which indicated distinct metabolic responses of C. closterium to different growth environments (fig. S4).
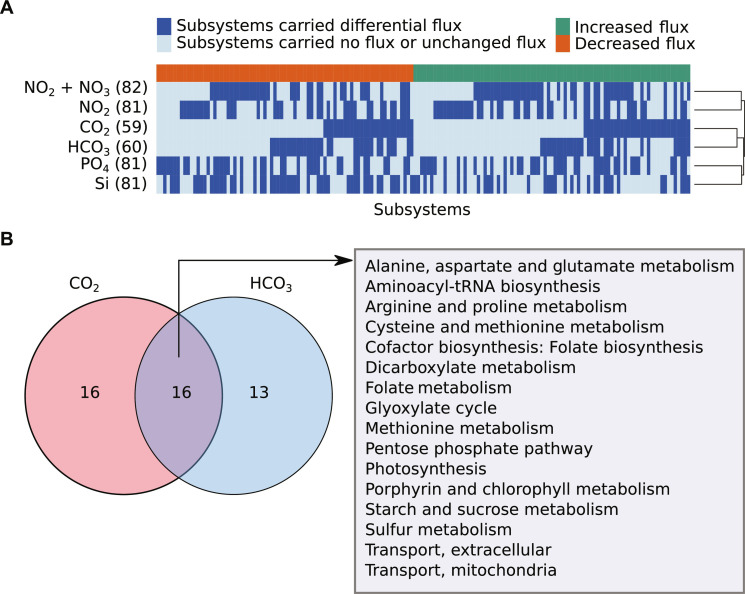
Fig. 4. Model predicted differential metabolic fluxes under elevated concentrations of nutrients.
(A) Heatmap represents differential flux-carrying subsystems under elevated CO2, HCO3, NO2, NO2 + NO3, PO4, and Si conditions. It shows subsystems separately with increased and decreased fluxes due to change in nutrient uptake fluxes from low to high levels. The numbers in parentheses denote the number of differential flux-carrying subsystems for each nutrient. The details of altered subsystems under varying growth conditions can be seen in table S19. We compared the metabolic flux distributions through the network when the model was simulated using the low and high concentrations of nutrients. Low and high uptake fluxes of nutrients to the model were used to mimic the low and high concentrations that were obtained from metadata of Tara Oceans global ocean microbiome data (table S11). The low and high concentrations of nutrients were defined by comparing with average concentration values for each nutrient (see Materials and Methods). (B) This Venn diagram represents shared and unique subsystems of C. closterium with increased flux distributions under the elevated concentration of CO2 and HCO3. The shared and unique subsystems in other pairs of conditions can be seen in figs. S5 to S7 and tables S17 and S18.
Model-predicted flux distributions revealed that certain subsystems with increased fluxes were commonly found when the concentration of different carbon and nitrogen sources was elevated. For example, elevated levels of CO2 and HCO3 increased the activity of subsystems for photosynthesis, central carbon metabolism (including pentose phosphate pathway), and starch and sucrose metabolism (Fig. 4B). Similarly, elevated levels of nitrogen sources, such as NO2 + NO3 and NO2, reduced the activity of subsystems for nucleotide metabolism and lipid biosynthesis (including fatty acid biosynthesis and galactoglycerolipid metabolism) (fig. S7).
Comparison of environmental gene expression data with GEM predictions unravels C. closterium’s metabolic responses to distinct marine environments
Previously, it was observed that quantitative correlation between predicted metabolic fluxes and gene expression is key to unraveling variations in metabolism due to altered growth conditions (6). To gain knowledge about the functional role of C. closterium in various marine environments, we compared model-predicted reaction/subsystems fluxes (described in the previous section) with gene expression data for C. closterium obtained from global ocean sampling [Tara Oceans (21, 32)]. We found that 50 ± 7% of differential flux–carrying subsystems had associated genes that were differentially expressed (fig. S8). Similar to the approach used for analyzing differential fluxes, differentially expressed genes were determined by comparing read counts for individual genes between two groups of samples. These groups were divided on the basis of low and high values of six environmental factors: CO2, HCO3, NO2, NO2 + NO3, PO4, and Si (see Materials and Methods). The highest number (n = 39) of subsystems that had differentially expressed genes and differential flux reactions were found under elevated PO4 and the lowest number (n = 22) for elevated HCO3 levels. Agreement between differential flux and gene expression occurred in at least 12 highly expressed and 7 lowly expressed subsystems in one growth condition (Fig. 5A). For example, under elevated CO2 concentrations, reactions involved in photosynthesis and central carbon metabolism (involving glycolysis, Entner-Doudoroff pathway, TCA cycle, and the pentose phosphate pathway) and amino acid metabolism (involving arginine and proline metabolism, aspartate and glutamate metabolism, and cysteine and methionine metabolism) carried increased fluxes, which is consistent with environmental transcriptomic data, illustrating that high CO2 concentrations promotes up-regulation of photosynthesis and central carbon metabolism–related genes (Fig. 5B). To investigate how these up-regulated subsystems affect the entire carbon flow, we analyzed flux distributions. Our analysis postulated that elevated CO2 levels lead to an increase in growth rate and increased carbohydrate and lipid content. These predictions were in agreement with experimental observations performed under elevated CO2 concentrations (Fig. 5C) (33). Both model-predicted fluxes and expression data suggested that several reactions in glycolysis, such as fructose-bisphosphate aldolase, glyceraldehyde-3-phosphate dehydrogenase, pyruvate dehydrogenase, pyruvate kinase, and triose-phosphate isomerase were up-regulated because of elevated CO2 levels (table S20). These observations were corroborated by recent studies in other diatoms (33, 34).
Fig. 5. Intersection between differential model–predicted fluxes and gene expressions.
(A) Bar plot represents subsystems that carry differential (increased or decreased) fluxes and are associated with differentially expressed genes under elevated concentrations of nutrients, such as CO2, HCO3, NO2, NO2 + NO3, PO4, and Si. (B) As an example, under elevated CO2, 27 subsystems illustrated differential metabolic fluxes and differentially expressed genes. This bar plot represents the number of reactions involved in these subsystems. Affected subsystems under additional conditions are shown in figs. S9 to S13. (C) Model-predicted storage of TAG and carbohydrate in terms of accumulation fluxes of TAG and β-1,3 glucans (i.e., monomers of chrysolaminarin), respectively, under low and high CO2. Each box plot denotes all allowable possible fluxes that were estimated using a sampling method (31) while simulating the model (see Materials and Methods). Significance (P value <0.001) was computed using two-side Student’s t test.
Previously, it has been demonstrated that HCO3 transporters play an important role in maintaining uptake of HCO3 into cells under CO2-limiting conditions (35). As expected, we noticed that the subsystems involved in photosynthesis and central carbon metabolism involving TCA cycle, pentose phosphate pathway, and amino acid metabolisms such as alanine, aspartate and glutamate metabolism, arginine and proline metabolism, and cysteine and methionine metabolism, were all up-regulated under elevated HCO3 (fig. S9).
Under high nitrogen conditions (NO2 + NO3), the model predicted that subsystems associated with lipid biosynthesis [triacylglycerol (TAG) biosynthesis and fatty acid biosynthesis] carried decreased fluxes under elevated nitrogen conditions. The genes associated with these subsystems were transcriptionally down-regulated. This is consistent with previous studies, which demonstrated that low nitrogen stimulates lipid biosynthesis in various microalgae including P. tricornutum (36, 37), Scenedesmus acuminatus (38), T. pseudonana (39, 40), C. reinhardtii (41), Haematococcus pluvialis, and Nannochloropsis sp. (42). Our model indicates that under high nitrogen conditions, the diatom redirects more flux toward specific metabolic pathways, including carbon metabolism (glycolysis/gluconeogenesis, pentose phosphate pathway, fructose and mannose metabolism, and starch and sucrose metabolism), amino acid metabolism (valine, leucine, and isoleucine biosynthesis, glutathione metabolism, and arginine and proline metabolism), the urea cycle, and triacylglycerolipid degradation, compared to lipid biosynthesis.
There have been conflicting reports about the effect of phosphate on lipid (TAG) biosynthesis in green algae. For example, TAG biosynthesis and accumulation in C. reinhardtii have been reported for limiting phosphate (43) as well as for elevated phosphate concentrations (44). Here, we demonstrate on the basis of model-predicted fluxes that higher phosphate concentration is associated with decreased activity of TAG biosynthesis in C. closterium. Our data were corroborated by the low expression of genes involved in this subsystem from low-phosphate samples (fig. S12). Moreover, a higher activity of chlorophyll biosynthesis (photosynthesis) was noticed because of an increased concentration of phosphate. Similar metabolic behavior was observed in the algae Scenedesmus obliquus and Microcystis aeruginosa grown under high phosphate concentration (45). Furthermore, our analysis suggested that high Si levels affected lipid biosynthesis (including TAG and fatty acid biosynthesis) negatively (fig. S13). Previously, it was postulated that Si starvation promotes an increase in the accumulation of lipids in diatoms, such as P. tricornutum, Nitzschia sp., Thalassiosira weissflogii, and Cyclotella cryptica (46–48). Overall, the data highlight the ability of iMK1961 to accurately predict the metabolic responses of C. closterium to a variety of changing environmental conditions.
Widespread mixotrophic growth of C. closterium in the marine environment
Along with other protists, diatoms play a crucial role in the flow of carbon from surface water columns to marine sediments (1). Various diatoms have been identified to be exclusively photoautotrophs, while others can be solely heterotrophs (49); yet, other diatoms have been reported to have the ability to perform a combination of phototrophic and heterotrophic metabolism simultaneously, referred to as mixotrophy (CO2 + light + organic carbon source) (6, 50, 51). While mixotrophy has been demonstrated for only a few diatoms in the laboratory (52, 53), no studies currently exist to our knowledge that explore this distinct metabolism of diatoms in their natural environment. A mixotrophic lifestyle could provide clear advantage to planktonic microorganisms, but knowledge about its existence and abundance is currently lacking (54). Unraveling the various trophic modes of diatoms in the environment can contribute to our understanding of carbon flow in the oceans and provide insights into the role of diatom metabolism on global carbon cycling. We thus compared iMK1961 predictions and Tara Oceans global metatranscriptomics data to reveal the trophic strategies of C. closterium in marine environments (see Materials and Methods).
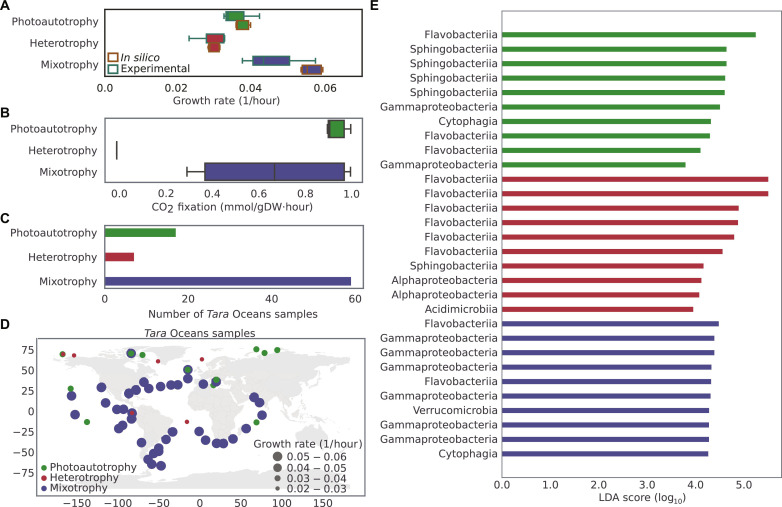
We simulated growth of C. closterium under photoautotrophic, heterotrophic, and mixotrophic conditions to investigate its metabolic versatility (table S9). Predicted flux distribution data were compared to examine the differences in growth and CO2 fixation under different nutrient conditions (Fig. 6, A and B) and identified the unique set of active reactions under each trophic condition (table S21 and figs. S14 to S16). Flux data suggested 34 and 47% higher growth in mixotrophy compared to photoautotrophy and heterotrophy, respectively [P value <0.01 (Wilcoxon rank sum/Mann-Whitney U test)]. Under mixotrophic growth CO2 fixation decreased by 25% compared to photoautotrophy as a fraction of C. closterium’s carbon requirement was fulfilled by assimilation of organic carbon. Further, transcription levels of genes associated with condition-specific active reactions were checked using metatranscriptomics data. The overlap between condition-specific active reactions and their associated expressed genes in a given sample was used to assign C. closterium’s metabolism in this sample (i.e., photoautotrophy, heterotrophy, or mixotrophy). Of a total of 146 Tara Oceans samples, 83 samples contained an overlap between nonzero fluxes of condition-specific reactions and expressions of associated genes. The results indicated that in the majority of samples (71%) C. closterium was exclusively growing mixotrophically (Fig. 6, C and D). Our results imply that mixotrophic growth of C. closterium is much more common than previously thought. In addition, 21% of samples indicated solely photoautotrophic growth, while only 8% revealed heterotrophic growth of C. closterium. We thus examined environmental factors associated with photoautotrophy, heterotrophy, and mixotrophy in C. closterium comparing metadata for ocean samples with varying compositions of nutrients and other environmental factors (e.g., temperature and depth). Samples associated with photoautotrophy and mixotrophy were not differentiated on the basis of measurements of any environmental factor. However, low concentrations of CO2/HCO3/PAR (photosynthetically active radiation) correlated with 50% of samples in which C. closterium grows heterotrophically (fig. S17). The lack of a clear correlation between physicochemical parameters and trophic mode could hint at the effect of microbe-microbe interactions on the metabolism of C. closterium. We have previously shown that environmental parameters but also single gene mutations alter the metabolism and interaction of phototrophic microbes with other organisms (15, 16). Moreover, earlier studies have identified certain bacterial taxa, such as Alpha-, Beta-, and Gammaproteobacteria, as well as Bacteroidetes, which potentially engage in interactions with diatoms (55). However, these interactions have been primarily observed with a limited number of genera, notably Roseobacter, Sulfitobacter, and Flavobacterium. We hypothesized that microbe-microbe interaction may be the driver behind C. closterium’s widespread mixotrophy and thus investigated whether trophic modes are associated with certain bacteria. We identified profoundly different populations of marine bacteria that were associated with C. closterium’s photoautotrophic, heterotrophic, or mixotrophic growth (Fig. 6E and table S26).
Fig. 6. Trophic modes of C. closterium in marine environments.
(A) Predicted and experimental growth rate of C. closterium under photoautotrophic, heterotrophic, and mixotrophic conditions. Extracellular succinate was used as an organic carbon source to stimulate growth under heterotrophic and mixotrophic conditions. Box plots represent all feasible biomass flux values that were determined by simulating iMK1961 using a uniform random sampling method (31) (see Materials and Methods) and experimentally measured growth rates. To simulate growth under trophic conditions, the model was constrained using experimentally measured uptake fluxes table S9. (B) CO2 fixation in C. closterium during photoautotrophy, heterotrophy, and mixotrophy. CO2 fixation was represented in terms of the uptake flux of CO2 while simulating the growth in different conditions. (C) The predicted flux distributions and metatranscriptomics data helped to identify trophic modes of C. closterium in the Tara Oceans samples. Bar plots represent the number of samples where solely photoautotrophic, heterotrophic, or mixotrophic growth of C. closterium was detected. Unique sets of active reactions, which overlapped with expressed genes in transcriptomics data for each trophic mode, were used to categorize samples in three different modes (table S21 and figs. S14 to S16). (D) The global distribution of various trophic modes of C. closterium was identified using iMK1961 and metatranscriptomics data from Tara Oceans samples. Dot size represents the predicted growth rate of C. closterium under the corresponding trophic condition. (E) To identify significantly differentially abundant marine prokaryotes between different trophic modes, linear discriminant analysis (LDA) effect size (79) was deployed on Tara Oceans metagenomic data. Bar plot represents 10 most differentially abundant prokaryotes in each trophic mode [P value <0.05 (Kruskal-Wallis test and pairwise Wilcoxon test); LDA score (log10) > 2.0] (see Materials and Methods). A complete list of differentially abundant prokaryotes can be seen in table S26.
Gammaproteobacteria and Flavobacteria were more abundant in the top 10 differentially abundant bacteria associated with mixotrophy and heterotrophy compared to photoautotrophy (52). Gammaproteobacteria were previously reported to be associated with a mixotrophic lifestyle of diatoms (52). Flavobacteria have been previously isolated from hadal water and are known to contain large numbers of carbohydrate-active enzymes (56). Phycisphaerae, another group of bacteria with a profuse content of carbohydrate-active enzymes (57, 58), were present when C. closterium was growing mixotrophically (table S26). The association of C. closterium with Phycisphaerae and Flavobacteria of the NS5 and NS9 marine groups (59, 60) hints at a specific connection and could suggest a potential role of these bacteria in providing organic carbon for the diatom. In addition, C. closterium’s higher growth rates under mixotrophic conditions and the abundance of specific bacteria in these samples suggest that microbe-microbe interactions and, to a lesser extent, environmental factors could directly benefit the diatom and provide an ecological advantage.
DISCUSSION
How abundant mixotrophic growth is in the ocean and what role it has on global carbon cycling is currently not known. Unraveling the different trophic modes of phytoplankton in situ could provide an intimate understanding of global carbon cycling and insights into marine food webs (52). Exploring the trophic mode of ubiquitous diatoms, such as C. closterium, can contribute to estimate the magnitude of marine mixotrophy. While the trophic modes of a few diatoms were recently investigated under controlled laboratory settings (50), it is currently challenging to extrapolate these findings to actual activity in situ (6). Here, we introduce a method that deploys genome-scale metabolic modeling (61) and environmental metagenomics and metatranscriptomics to quantitatively unravel variations in the metabolism of the ubiquitous diatom C. closterium in situ. This approach successfully identifies whether inorganic carbon (photoautotrophic) or organic carbon (heterotrophic) is used for growth singly or concurrently (i.e., mixotrophy) and, furthermore, allows quantifying activities for each mode.
A GEM, iMK1961, of C. closterium has been reconstructed using the annotated genome of this diatom (Fig. 1; see Materials and Methods). iMK1961 represents the largest GEM for a diatom to date (Fig. 2 and fig. S18), containing 764 to 4553 more reactions compared to previous diatom GEMs (Fig. 2A) (16–19), accounting for C. closterium’s large genome among these diatoms (figs. S1 and S19). The rigorous reconstruction process allowed capturing a detailed metabolic network that can be used as a reference/template model for other model reconstructions of yet unexplored diatoms to investigate the metabolic diversity of diatoms (62).
Diatoms benefit from vast metabolic flexibility that allows them to grow in dramatically distinct environments, making them one of the most ecologically successful photosynthetic eukaryotes in the ocean (51). We predicted the growth of C. closterium under a broad range of growth environments including photoautotrophic, heterotrophic, and mixotrophic conditions using various carbon, nitrogen, phosphorus, and sulfur sources (Fig. 3A and table S7). High levels of agreement between model predictions and experimental measurements indicate that the model accurately represents the metabolic complexities of C. closterium and can elucidate survival strategies in different conditions (Fig. 3B and table S8). iMK1961 integrates multiple layers of biological data to understand the impact of perturbations on individual reactions and complex pathways and to determine consequences of different environmental conditions (Figs. 4 and 5).
By comparing Tara Oceans global ocean metatranscriptomics data (20, 21) with the model predictions, we revealed that mixotrophic growth in C. closterium is more prevalent in marine environments compared to photoautotrophic and heterotrophic growth (Fig. 6C). Considering the fact that C. closterium is widely distributed in global oceans (63), these results suggest that mixotrophy is widespread in the oceans (Fig. 6D). Although there is limited information available on the physiology of mixotrophs in the marine environment (6), mixotrophy has been postulated as advantageous for phytoplankton to survive in environments with changing nutrient availability (64). We correlated different trophic conditions with environmental parameters obtained by the Tara Oceans project (20, 21) to establish a link between physicochemical and biological factors and C. closterium‘s metabolism. However, the lack of correlation between physicochemical parameters and trophic modes suggests that microbe-microbe interactions may be the driver behind C. closterium’s mixotrophy. Certain bacteria, including species belonging to the classes Gammaproteobacteria and Phycisphaerae, were associated with mixotrophy, while Flavobacteria species were more abundant in samples of heterotrophic growth (Fig. 6E). This association hints at a potential role for certain bacteria in providing organic carbon for the diatom.
Heterotrophic growth has been reported not only for benthic [C. cryptica (65)] and meromictic diatoms (C. closterium) but also for diatoms that are commonly found in the open ocean, such as Nitzschia alba (66), suggesting that the ability to use organic molecules is not restricted to benthic diatoms. Since diatoms gain a clear growth advantage by using fixed carbon (Fig. 6), this lifestyle (mixotrophic as well as heterotrophic) could potentially be widespread among planktonic diatoms, contributing to their environmental success and their role as dominant members of global phytoplankton (3). While this work shines light on the role of C. closterium’s metabolism in the global ocean, more research is needed to explore how mixotrophy affect primary and secondary production in the oceans and what factors, such as other microorganisms, contribute and thus drive global carbon and nutrient cycles. Future investigations including a diverse set of ocean diatoms will help to delineate the effect of mixotrophy on diatom metabolism. In addition, these studies will reveal how various bacteria interact with diatoms, thus contributing to global carbon cycling. Collectively, our approach provides a framework to elucidate variations in trophic strategies of diatoms in the environment. In summary, we present a comprehensive metabolic model of the diatom C. closterium. Phenotypic and transcriptomic data–guided model predictions provided a framework to reveal the metabolic capabilities of C. closterium to survive in different growth conditions. This study demonstrated how the metabolic model and transcriptomics data can help us to understand the variations in metabolic activities of a diatom in elevated concentrations of nutrients in natural settings. The metabolic model presented in this study provides diverse and comprehensive coverage of a diatom’s metabolic network that can be seamlessly used as a template/reference model for generating metabolic reconstructions of additional diatoms. Thus, we expect this framework to be extended for the exploration of the metabolic capabilities of other diatoms. Along with fast-growing global ocean meta-omics data, we are confident that metabolic models will be a valuable tool to assist this type of holistic analysis. We envision this approach to aid big data analytics and to benefit the investigation of marine ecosystems and their response to changing environmental conditions (64).
MATERIALS AND METHODS
Functional genome annotation and subcellular localization of proteins
A functionally annotated genome of C. closterium TJRE Ct-21 was obtained from an environmental isolate. Briefly, Sediment was collected at the TJRE outflow in San Diego County (CA, USA) from 0- to 3-cm depth and stored at 4°C. In a 250-ml flask, 2 g of sediment was mixed with 50 ml of F/2 medium (supplemented with 880 μM NaNO3, 36 μM NaH2PO4, and 100 μM Na2SiO3) and incubated with 14:10 light:dark [150 microeinstein (μE) m−2 s−1] at 18°C for 3 days. Single cells of Cylindrotheca were individually picked using an inverted microscope (Olympus) coupled to a micromanipulator and transferred into 2 ml of F/2 medium for incubation in the same conditions.
High molecular weight DNA was extracted using cetyltrimethyl ammonium bromide (CTAB)-chloroform:isoamyl alcohol. A continuous long read (10- to 20-kb insert) genomic library for PacBio sequencing was constructed with the SMRTbell Templar prep kit (Pacific Biosciences) and sequenced on five SMRT cells on the PacBio Sequel platform at the University of Delaware Sequencing and Genotyping Center at the Delaware Biotechnology Institute. PacBio sequencing produced 1,242,306 reads that were cleaned, trimmed, and assembled using HGAP 4 Pipeline (PB SMRTportal ver 5.0.1.9585) using Falcon assembler with default options except: aggressive, expected genome size 150 Mb, seed coverage 30. The assembly generated 233 polished contigs, with a maximum contig length of 4.96 Mb, N50 contig length of 3.10 Mb, and a %GC of 46.82. Gene annotation was performed using MAKER (ver. 2.31.0) with three iterative gene model training rounds using Augustus (v. 3.2.3).
To predict the subcellular localization for proteins, we deployed a previously published pipeline (16). We used this pipeline with the recently updated version of SignalP 4.0 v-4.1 along with other tools such as HECTAR, Mitoprot v-1.101, predictNLS v-1.0.20, and TargetP v-1.1 (Supplementary Text). Protein sequences of C. closterium were used as input for this pipeline. The default parameters of all tools were used for processing protein sequences and assigning the compartments to each protein.
Draft reconstruction
To build a draft reconstruction, previously published metabolic networks of two photosynthetic organisms, namely P. tricornutum (iLB1027_lipid) (16) and C. reinhardtii (iRC1080) (27), were used as templates or reaction databases. Both template models were obtained from BiGG models (28). A draft reconstruction was built on the basis of homology between protein sequences of C. closterium and template organisms using best bidirectional BLAST hits (BBH) in the RAVEN Toolbox (24). On the basis of BBH, the reactions associated with homologous proteins were added to the reconstruction from template models. The draft reconstruction was analyzed using the COBRApy and COBRA Toolbox and curated manually to validate present information in the model and incorporate any missing information (25, 26). Apart from the C. closterium genome annotation and BiGG database (28), we also used more resources such as relevant literature (table S1), KEGG, modelSEED, MetaCyc, UniProt, BRENDA, IntEnz, TPDB, and TransportDB (Supplementary Text). MetaNetX was used to map reaction and metabolites identifiers from other databases to BiGG abbreviations (29). More reactions were added to the reconstruction from the BiGG database based on EC numbers associated with C. closterium proteins, which did not match any homologous protein in template genomes. All duplicate reactions and metabolites, which were included in the reconstruction because of different abbreviations for the same reactions, were removed at this stage.
Biomass composition
The biomass objective function (BOF) or biomass-producing reaction contains the biomass precursors that are produced within the metabolic network of the organism. Simulating a GEM using BOF allows us to predict the flux of biomass-producing reaction, which is considered as a proxy of growth rates on the given substrate consumption rates. The biomass-producing reaction is balanced for producing 1 g of biomass. This reaction in iMK1961 was reconstructed on the basis of the biomass precursors used in a previously published diatom model (16). In addition, we incorporated biosilica into the biomass-producing reaction of the model. The stoichiometric coefficients of this reaction were normalized using experimentally measured elemental ratios between carbon, nitrogen, and silicon (67). Furthermore, we used a previously published method that leveraged protein and DNA sequences from the C. closterium genome to further refine the representation of amino acids and nucleic acids within the biomass reaction (68). The biomass-producing reaction in iMK1961 contains metabolites that include amino acids, energy molecules, frustule components, lipids, nucleotides, and pigments (table S2). The model contains two biomass reactions: (i) representing growth in the presence of light (photoautotrophic or mixotrophic conditions) and (ii) a biomass reaction representing growth in the dark (heterotrophic conditions). Biomass reaction for dark conditions does not include the biosynthesis of pigments such as chlorophyll a, chlorophyll c1, and chlorophyll c2, TAGs, and chrysolaminarin, as previously reported (69).
Gap-filling
Because of incomplete genome annotations and different growth requirements, GEMs often contain gaps in the metabolic network. In the first step, the gaps between disconnected metabolites in the network were filled by a semiautomatic method by adding relevant reactions to the model from reaction databases such as KEGG. In the second step, we filled gaps in the model using a set of minimum reactions from the BiGG models (28) to enable the production of all biomass precursors using the media nutrients from laboratory experiments (tables S3 and S5). In the third step, on the basis of experimentally confirmed phenotypic capabilities of C. closterium, iMK1961 was gap-filled by adding missing pathways/reactions in the network to enable the related metabolic functions. In this step, mostly, these gap-filling reactions include transporters and exchange reactions (table S6). At the end of the gap-filling process, models contained some exogenous genes (genes from reference/template organisms’ model), which were either replaced by homologous genes in C. closterium genomes or removed from the model during the curation process. The reactions, which were added during gap-filling, were analyzed using flux balance analysis (FBA) and reactions, which were not active in any simulations and not associated with C. closterium genes, were removed from the model.
The curation processes
GPR associations in the model were verified on the basis of gene annotations and protein sequences using BLAST. We assigned at least one subsystem to each reaction based on the KEGG pathway database. A modeling-specific subsystem, namely extracellular space having exchange reactions, was added to enable the uptake of medium metabolites and secretion of metabolic products. All reactions were balanced for mass and charge. Reaction reversibility was refined on the basis of published manually curated metabolic models (28). On the basis of quality control steps defined in Thiele et al. (61), we ensured that reconstruction was not able to generate ATP, NADPH, and NADH without the uptake of any substrates. Next, we refined exogenous genes present in GPRs by searching their homologs in C. closterium genomes using BLAST with a threshold of ⩽ 10−6 E value, ⩾ 40% identity, and ⩾ 80% alignment length. For transporter annotation and curation processes, mainly genome annotations (https://zenodo.org/doi/10.5281/zenodo.11053410 and https://github.com/manishsaini16/cylindrotheca-model/tree/main/Protein_sequences) and transport homology with a template diatom were used. Further, the annotations of transporters in the model were validated using BLAST against the Transporter Classification Database of each transporter. We further verified transporter information using other databases, such as UniProt, MetaCyc, and KEGG. Some intracellular transporters between different compartments were included into the model for resolving blocked pathways and to enable the synthesis of biomass precursors in known growth conditions. When experimental observations revealed C. closterium’s ability to catabolize certain substrates, we included the transporters for such substrates in the model, even in the absence of genetic evidence. While we curated transporters to the best of our knowledge, we are aware of existing challenges for annotating transporters and determining their specificity (70). Furthermore, we conducted a quality assessment using the Memote test suite (fig. S20) (71).
Model simulations
FBA was deployed to simulate GEMs by optimizing a predefined objective function in specified environmental constraints. The basic fundamentals of FBA have been defined previously (72). In the present work, a biomass-producing reaction was used as an objective function for simulating the model. All simulations were performed using the COBRA Toolbox and COBRApy with Gurobi Optimizer version 7.0.2 (Gurobi Optimization Inc., Houston, Texas) as a linear programming solver in MATLAB R2018a (The MathWorks Inc., Natick, MA) and Python 3.7.6, respectively. The uptake flux of media nutrients were constrained by experimentally determined measurements in FBA analysis.
The uptake rates of nutrients were calculated using the following expression, which used the slope of the consumption profile of experimentally measured nutrients during the exponential growth phase
where ΔC represents the change in nutrient concentration (millimole) over a specific time interval (Δt). Δt represents the time interval over which the nutrient concentration is measured. V represents the volume of the bacterial culture. The calculated uptake rates were then converted to fluxes (mmol/gDW·h) using the biomass dry weight (gDW) and used these fluxes as constraints in the model for condition-specific simulations.
Cell culture
In our study, control cultures were cultivated in F/2 nutrient media comprising 55 μM NaH2PO4, 100 μM Na2SiO3, and 880 μM NO3, supplemented with Aquil salts (73), without the addition of seawater. These cultures underwent incubation under a constant light intensity of 150 μE m−2 s−1 at 18°C, with filtered, aerated air throughout the experimental assays. Upon reaching mid-exponential phase (approximately 4 × 106 cells ml−1), cells were harvested via centrifugation (10 min at 3000g), followed by washing in the same media, a process repeated three times. The cells were subsequently resuspended in media tailored to each control and treatment and grown in triplicate cultures. The experimental conditions were as follows: Control: replete F/2 media (as described above) + light exposure (photoautotrophy); dark + succinate: replete F/2 media + dark conditions + succinate supplementation (heterotrophy); light + succinate: replete F/2 media + light + succinate supplementation (mixotrophy); low NO3: F/2 media with reduced nitrogen content (16.45 mg/liter NaNO3), representing 5% of the original nitrate concentration in F/2 media; low Si: F/2 media with reduced silicon content (10 μM Na2SiO3·9H2O), representing 10% of the original silica concentration in F/2 media. Dark conditions were maintained by covering the culture flasks with foil.
Sampling was conducted at five distinct time points (0, 24, 48, 72, and 96 hours), and subsamples were used for cell count and NO3 measurement. Cell counting was conducted using flow cytometry, while NO3 concentrations were determined using the Nitrite/Nitrate Assay Kit (Sigma-Aldrich) following the provided protocol. One-milliliter aliquot of culture sample was centrifuged at 5000g for 3 min and resuspended in 1-ml sterile 1× phosphate-buffered saline for counting. Ten microliters 10X SYBR green was added to the sample for detection by flow cytometry and incubated in the dark for 20 min at 37°C. Thirty microliters of AccuCount fluorescent particles (ACFP-70-10; Spherotech) was added to the samples for quantification. Samples were processed on an SH800 cell sorter (Sony Biotechnology) using a 130-μm chip with the threshold set on FL1 at 43% and gain settings as FSC = 3, BSC = 20.5%, and FL4 = 35%. C. closterium cells were gated from the background on an FL1-FL4 density plot. Final cell counts per microliter calculations were performed following the manufacturer’s instructions of the AccuCount counting beads.
Model validation
To ensure the accuracy of our model predictions across different growth conditions, we conducted tests on a wide range of carbon, nitrogen, phosphorus, and sulfur sources. To select the sources for this analysis, we considered the available carbon-, nitrogen-, phosphorus-, and sulfur-containing metabolites within the model. The transporters for these sources were determined either through genome annotations or by homology comparison with a template diatom (P. tricornutum). In cases where information regarding the transporter for a specific source was unavailable, we excluded that particular metabolite from the analysis. To validate our findings, we compared model-predicted growth and no-growth outcomes across various carbon and nitrogen sources with experimental data collected from both literature sources on C. closterium and our own experimental results (see table S7). When evaluating growth data sourced from existing literature, we adopted a criterion wherein growth of C. closterium on specific carbon or nitrogen substrates was deemed credible if the authors had quantified it in terms of either cell counts or chlorophyll measurements. For experimental data generated in the course of our study, growth was affirmed when there was a notable increase in cell counts exceeding fourfold within a span of 5 days. In addition, we used physiological data collected under four different conditions—control (light), dark, low nitrogen, and low silicon—to further verify our model’s predicted growth rates. For simulation with iMK1961, we broke down all components of the growth medium into individual metabolites (table S9) and used their uptake rates as input for our GEM. Nutrient uptake rates were then converted to mmol/gDW·h, which is the standard unit for all fluxes in our model. We set the biomass reaction as the objective function for our simulations.
Global oceans metatranscriptomics data analysis
Metatranscriptomic reads from the Tara Oceans expedition (ENA project PRJEB402), pico-, nano-, and microplankton size fractions were mapped onto the repeat-masked (RepeatMasker v4.0.7; www.repeatmasker.org) C. closterium TJRE Ct-21 genome assembly using the splicing-aware read mapper HISAT2 v2.1.0 (74), followed by SAMtools v1.8 sorting (75) and filtering by an in-house script to remove secondary or low-quality reads and reads consisting of more than ~70% nucleotide repeats using a higher-order Markov model entropy filter [adapted from Caballero et al. (76)]. Read counts to individual gene models were determined using BEDTools v2.30.0 (77), normalized to library size and gene length (rpkm) and averaged across all biosamples available for a given station and water layer.
Differentially expressed genes
Samples were divided into two categories based on the low and high values of environmental factors that included CO2, HCO3, NO2, NO2 + NO3, PO4, and Si (table S10). Low and high values of each environmental factor were defined by comparing with average values. The average count data were used to estimate the fold change and log2 transformation of fold changes between these two different groups of samples allowed to categorize the genes in up-regulated and down-regulated groups.
Sampling of feasible model-predicted fluxes and calculating differential fluxes
optGpSampler (31) on COBRApy platform was used to determine the distribution of all feasible fluxes of each reaction in the model under all environmental conditions. This method allowed us to uniformly sample the constrained solution space of the model to evaluate the full range of metabolic capabilities of genome-scale networks. The model was first constrained using low and high uptake fluxes (mmol/gDW·h) of each targeted nutrient. Six nutritional conditions, such as CO2, HCO3, NO2, NO2 + NO3, PO4, and Si, were tested. Except for one targeted nutrient, the constraints for uptake fluxes of all other nutrients were kept the same in both simulations using low and high uptake fluxes of targeted nutrients. Low uptakes of these nutrients were used from experimentally measured uptake rates of nutrients, and two times of these uptake rates were used to constrain the model with high uptake fluxes of the nutrients. The constraints of uptake fluxes in all growth conditions can be seen in table S11. All uniform random sampling was conducted with a step size of 100 for 5000 points. The fold changes and log2 transformation of fold changes between mean values of sampling fluxes of subsystems under low and elevated concentrations of targeted nutrient was determined. All negative values of log-transformed fold changes were considered as decreased fluxes, and all positive values were considered as increased fluxes. In addition, these data were filtered by using a minimal fold-change threshold of 0.2 between mean values of sampling fluxes of subsystems (78). Collectively, the fluxes that showed either an increase or decrease were categorized as differential fluxes. Differential fluxes can also be defined as
Fluxlow and Fluxhigh represent fluxes predicted using low and high uptake rates of the nutrients, respectively. Subsystems with a negative value of Fluxdiff were considered as down-regulated, indicating decreased fluxes, and subsystems with a positive flux value were regarded as up-regulated, indicating increased fluxes due to higher uptake of the nutrients.
Similarly, the sampling method was used to predict the metabolic fluxes for carbohydrate and TAG storage (Fig. 5C), and growth and CO2 fixation (Fig. 6, A and B) under different growth conditions such as low and high concentration of nutrients, and trophic modes like photoautotrophy, heterotrophy, and mixotrophy. Wilcoxon rank sum/Mann-Whitney U test was used to compare the reaction fluxes between different conditions. The differences with P values less than 0.01 (i.e., false discovery rate less than 1%) were considered as significant differences.
Analysis of trophic strategies of C. closterium in marine environments
During this analysis, iMK1961 was used to generate flux distributions under photoautotrophic (CO2 + light), heterotrophic (dark + organic carbon source), and mixotrophic (CO2 + light + organic carbon source) conditions. To simulate the growth of C. closterium under these three trophic conditions (table S9). For maintaining photoautotrophic conditions, the uptake flux of light and CO2 was provided to the model without any organic carbon in the media. Extracellular succinate was used as an organic carbon to maintain heterotrophic growth without light and CO2 in the media. For mixotrophic growth, the model was simulated using media including light, CO2, and succinate. Wilcoxon rank sum/Mann-Whitney U test was used to compare the growth rates between three trophic conditions. The differences with P values less than 0.01 (i.e., false discovery rate less than 1%) were considered as significant differences. The reaction fluxes under all three conditions were compared to identify the unique set of active reactions for each condition (tables S22 to S24). Three sets of condition-specific reactions were found under photoautotrophy, heterotrophy, and mixotrophy, respectively. The active reactions were defined as reactions that carry nonzero fluxes. Transcript counts of genes associated with these unique reactions were extracted from Tara Oceans metatranscriptomics data, and the average of transcript counts for each gene among all samples was compared with fluxes condition-specific reactions. This practice allowed us to filter the list of these reactions by screening out the reactions that carried nonzero flux, but their associated genes were not found expressed in global oceans (table S21 and figs. S14 to S16). After this filtering step, the remaining 22, 22, and 44 condition-specific reactions were used to identify photoautotrophy, heterotrophy, and mixotrophy of C. closterium, respectively, in each Tara Oceans sample. For example, if one or more photoautotrophy-specific reactions and their associated genes were found active in a sample, then this sample was defined under the category of photoautotrophy. The activity of a reaction and genes were defined by nonzero flux and expression values, respectively.
Statistical analysis of Tara oceans metagenomic data
Metagenomic data of Tara Oceans samples (20, 21) associated with photoautotrophy, heterotrophy, and mixotrophy of C. closterium (Fig. 6C) were analyzed using linear discriminant analysis (LDA) effect size (LEfSe) algorithm (79). The abundance data were obtained from Salazar et al. (20) and used as input for LEfSe. This analysis helped us to identify significantly differentially abundant marine prokaryotes between three trophic modes. The differences with P values less than 0.05 (i.e., false discovery rate less than 5%) using Kruskal-Wallis test and pairwise Wilcoxon test and logarithmic LDA score more than 2.0 were considered significant differences (Fig. 6E and table S26).
Acknowledgments
We extend our appreciation to A. Allen at Scripps Institution of Oceanography, the University of California San Diego, for invaluable review of this manuscript.
Funding: We acknowledge the support of the Gordon and Betty Moore Foundation, grant number GBMF7000 (L.Z.A. and K.Z.), which made this work possible. Portions of computational analysis were performed on the BioMix cluster at the University of Delaware with support from Delaware INBRE [NIH-NIGMS P20 GM103446 (S.W.P.)] and the state of Delaware.
Author contributions: M.K. and K.Z. conceived and designed the study. M.K. and J.D.T.-B. developed the computational framework. M.K., J.D.T.-B., M.M.A.-B., A.P., M.N., C.Z., and F.K. curated the framework. M.K. implemented the framework and performed analyses. Z.F. analyzed global oceans metatranscriptomics data. C.L., M.K., S.M.S., A.V.L., and L.Z.A. designed and performed growth experiments. J.L.E., S.W.P., and L.Z.A. sequenced and annotated the genome. H.L. performed the HPLC analysis. M.K. and K.Z. discussed the results and wrote the manuscript with input from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The metatranscriptomic data used in this study were obtained from the Tara Oceans expedition (ENA project PRJEB402) (21, 32). Protein sequences of C. closterium used in GEM reconstruction are provided at: https://zenodo.org/doi/10.5281/zenodo.11053410 and https://github.com/manishsaini16/cylindrotheca-model/tree/main/Protein_sequences. The experimentally measured growth data used to constrain the model are provided in the Supplementary Materials. A genome-scale metabolic model of C. closterium (iMK1961) is provided at: https://zenodo.org/doi/10.5281/zenodo.11053410 and https://github.com/manishsaini16/cylindrotheca-model/tree/main/model in JSON and SBML format. Other custom codes or scripts for each analysis are available at: https://zenodo.org/doi/10.5281/zenodo.11053410 and https://github.com/manishsaini16/cylindrotheca-model, including source or input data for all simulations and information on the computational tools used in this study.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S20
Legends for tables S1 to S26
Legend for data S1
References
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S26
Data S1
REFERENCES AND NOTES
- 1.Y. Matsuda, P. G. Kroth, “Carbon fixation in diatoms” in The Structural Basis of Biological Energy Generation (Springer, 2014; https://link.springer.com/10.1007/978-94-017-8742-0_18), pp. 335–362. [Google Scholar]
- 2.Büchel C., How diatoms harvest light. Science 365, 447–448 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Guiry M. D., How many species of algae are there? J. Phycol. 48, 1057–1063 (2012). [DOI] [PubMed] [Google Scholar]
- 4.De Tommasi E., Gielis J., Rogato A., Diatom frustule morphogenesis and function: A multidisciplinary survey. Mar. Genomics 35, 1–18 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Wang S., Verma S. K., Hakeem Said I., Thomsen L., Ullrich M. S., Kuhnert N., Changes in the fucoxanthin production and protein profiles in Cylindrotheca closterium in response to blue light-emitting diode light. Microb. Cell Fact. 17, 110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen N. R., Mixotrophic plankton foraging behaviour linked to carbon export. Nat. Commun. 13, 1302 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ward B. A., Follows M. J., Marine mixotrophy increases trophic transfer efficiency, mean organism size, and vertical carbon flux. Proc. Natl. Acad. Sci. U.S.A. 113, 2958–2963 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malviya S., Scalco E., Audic S., Vincent F., Veluchamy A., Poulain J., Wincker P., Iudicone D., de Vargas C., Bittner L., Zingone A., Bowler C., Insights into global diatom distribution and diversity in the world’s ocean. Proc. Natl. Acad. Sci. U. S.A. 113, E1516–E1525 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H., Robinson J. L., Kocabas P., Gustafsson J., Anton M., Cholley P.-E., Huang S., Gobom J., Svensson T., Uhlen M., Zetterberg H., Nielsen J., Genome-scale metabolic network reconstruction of model animals as a platform for translational research. Proc. Natl. Acad. Sci. U.S.A. 118, e2102344118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J. E., Park S. J., Kim W. J., Kim H. J., Kim B. J., Lee H., Shin J., Lee S. Y., One-step fermentative production of aromatic polyesters from glucose by metabolically engineered Escherichia coli strains. Nat. Commun. 9, 79 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monk J. M., Lloyd C. J., Brunk E., Mih N., Sastry A., King Z., Takeuchi R., Nomura W., Zhang Z., Mori H., Feist A. M., Palsson B. O., iML1515, a knowledgebase that computes Escherichia coli traits. Nat. Biotechnol. 35, 904–908 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monk J. M., Charusanti P., Aziz R. K., Lerman J. A., Premyodhin N., Orth J. D., Feist A. M., Palsson B. Ø., Genome-scale metabolic reconstructions of multiple Escherichia coli strains highlight strain-specific adaptations to nutritional environments. Proc. Natl. Acad. Sci. U.S.A. 110, 20338–20343 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar M., Ji B., Zengler K., Nielsen J., Modelling approaches for studying the microbiome. Nat. Microbiol. 4, 1253–1267 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Kumar M., Ji B., Babaei P., Das P., Lappa D., Ramakrishnan G., Fox T. E., Haque R., Petri W. A., Bäckhed F., Nielsen J., Gut microbiota dysbiosis is associated with malnutrition and reduced plasma amino acid levels: Lessons from genome-scale metabolic modeling. Metab. Eng. 49, 128–142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuñiga C., Li T., Guarnieri M. T., Jenkins J. P., Li C.-T., Bingol K., Kim Y.-M., Betenbaugh M. J., Zengler K., Synthetic microbial communities of heterotrophs and phototrophs facilitate sustainable growth. Nat. Commun. 11, 3803 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levering J., Broddrick J., Dupont C. L., Peers G., Beeri K., Mayers J., Gallina A. A., Allen A. E., Palsson B. O., Zengler K., Genome-scale model reveals metabolic basis of biomass partitioning in a model diatom. PLOS ONE 11, e0155038 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lavoie M., Saint-Béat B., Strauss J., Guérin S., Allard A., Hardy S. V., Falciatore A., Lavaud J., Genome-scale metabolic reconstruction and in silico perturbation analysis of the polar diatom Fragilariopsis cylindrus predicts high metabolic robustness. Biology (Basel). 9, 30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Tol H. M., Armbrust E. V., Genome-scale metabolic model of the diatom Thalassiosira pseudonana highlights the importance of nitrogen and sulfur metabolism in redox balance. PLOS ONE 16, e0241960 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmad A., Tiwari A., Srivastava S., A genome-scale metabolic model of Thalassiosira pseudonana CCMP 1335 for a systems-level understanding of its metabolism and biotechnological potential. Microorganisms 8, 1396 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salazar G., Paoli L., Alberti A., Huerta-Cepas J., Ruscheweyh H.-J., Cuenca M., Field C. M., Coelho L. P., Cruaud C., Engelen S., Gregory A. C., Labadie K., Marec C., Pelletier E., Royo-Llonch M., Roux S., Sánchez P., Uehara H., Zayed A. A., Zeller G., Carmichael M., Dimier C., Ferland J., Kandels S., Picheral M., Pisarev S., Poulain J., Acinas S. G., Babin M., Bork P., Bowler C., de Vargas C., Guidi L., Hingamp P., Iudicone D., Karp-Boss L., Karsenti E., Ogata H., Pesant S., Speich S., Sullivan M. B., Wincker P., Sunagawa S., Acinas S. G., Babin M., Bork P., Boss E., Bowler C., Cochrane G., de Vargas C., Follows M., Gorsky G., Grimsley N., Guidi L., Hingamp P., Iudicone D., Jaillon O., Kandels-Lewis S., Karp-Boss L., Karsenti E., Not F., Ogata H., Pesant S., Poulton N., Raes J., Sardet C., Speich S., Stemmann L., Sullivan M. B., Sunagawa S., Wincker P., Gene expression changes and community turnover differentially shape the global ocean metatranscriptome. Cell 179, 1068–1083.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Vargas C., Audic S., Henry N., Decelle J., Mahé F., Logares R., Lara E., Berney C., Le Bescot N., Probert I., Carmichael M., Poulain J., Romac S., Colin S., Aury J.-M., Bittner L., Chaffron S., Dunthorn M., Engelen S., Flegontova O., Guidi L., Horák A., Jaillon O., Lima-Mendez G., Lukeš J., Malviya S., Morard R., Mulot M., Scalco E., Siano R., Vincent F., Zingone A., Dimier C., Picheral M., Searson S., Kandels-Lewis S., Acinas S. G., Bork P., Bowler C., Gorsky G., Grimsley N., Hingamp P., Iudicone D., Not F., Ogata H., Pesant S., Raes J., Sieracki M. E., Speich S., Stemmann L., Sunagawa S., Weissenbach J., Wincker P., Karsenti E., Boss E., Follows M., Karp-Boss L., Krzic U., Reynaud E. G., Sardet C., Sullivan M. B., Velayoudon D., Ocean plankton. Eukaryotic plankton diversity in the sunlit ocean. Science 348, 1261605 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Strzepek R. F., Nunn B. L., Bach L. T., Berges J. A., Young E. B., Boyd P. W., The ongoing need for rates: Can physiology and omics come together to co-design the measurements needed to understand complex ocean biogeochemistry? J. Plankton Res. 44, 485–495 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuñiga C., Li C.-T., Yu G., Al-Bassam M. M., Li T., Jiang L., Zaramela L. S., Guarnieri M., Betenbaugh M. J., Zengler K., Environmental stimuli drive a transition from cooperation to competition in synthetic phototrophic communities. Nat. Microbiol. 4, 2184–2191 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Wang H., Marcišauskas S., Sánchez B. J., Domenzain I., Hermansson D., Agren R., Nielsen J., Kerkhoven E. J., RAVEN 2.0: A versatile toolbox for metabolic network reconstruction and a case study on Streptomyces coelicolor. PLOS Comput. Biol. 14, e1006541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebrahim A., Lerman J. A., Palsson B. O., Hyduke D. R., COBRApy: COnstraints-based reconstruction and analysis for Python. BMC Syst. Biol. 7, 74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heirendt L., Arreckx S., Pfau T., Mendoza S. N., Richelle A., Heinken A., Haraldsdóttir H. S., Wachowiak J., Keating S. M., Vlasov V., Magnusdóttir S., Ng C. Y., Preciat G., Žagare A., Chan S. H. J., Aurich M. K., Clancy C. M., Modamio J., Sauls J. T., Noronha A., Bordbar A., Cousins B., El Assal D. C., Valcarcel L. V., Apaolaza I., Ghaderi S., Ahookhosh M., Ben Guebila M., Kostromins A., Sompairac N., Le H. M., Ma D., Sun Y., Wang L., Yurkovich J. T., Oliveira M. A. P., Vuong P. T., El Assal L. P., Kuperstein I., Zinovyev A., Hinton H. S., Bryant W. A., Aragón Artacho F. J., Planes F. J., Stalidzans E., Maass A., Vempala S., Hucka M., Saunders M. A., Maranas C. D., Lewis N. E., Sauter T., Palsson B. Ø., Thiele I., Fleming R. M. T., Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0. Nat. Protoc. 14, 639–702 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang R. L., Ghamsari L., Manichaikul A., Hom E. F. Y., Balaji S., Fu W., Shen Y., Hao T., Palsson B. Ø., Salehi-Ashtiani K., Papin J. A., Metabolic network reconstruction of Chlamydomonas offers insight into light-driven algal metabolism. Mol. Syst. Biol. 7, 518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King Z. A., Lu J., Dräger A., Miller P., Federowicz S., Lerman J. A., Ebrahim A., Palsson B. O., Lewis N. E., BiGG models: A platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 44, D515–D522 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moretti S., Tran V. D. T., Mehl F., Ibberson M., Pagni M., MetaNetX/MNXref: Unified namespace for metabolites and biochemical reactions in the context of metabolic models. Nucleic Acids Res. 49, D570–D574 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elagoz A. M., Ambrosino L., Lauritano C., De novo transcriptome of the diatom Cylindrotheca closterium identifies genes involved in the metabolism of anti-inflammatory compounds. Sci. Rep. 10, 4138 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Megchelenbrink W., Huynen M., Marchiori E., optGpSampler: An improved tool for uniformly sampling the solution-space of genome-scale metabolic networks. PLOS ONE 9, e86587 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sunagawa S., Acinas S. G., Bork P., Bowler C., Acinas S. G., Babin M., Bork P., Boss E., Bowler C., Cochrane G., de Vargas C., Follows M., Gorsky G., Grimsley N., Guidi L., Hingamp P., Iudicone D., Jaillon O., Kandels S., Karp-Boss L., Karsenti E., Lescot M., Not F., Ogata H., Pesant S., Poulton N., Raes J., Sardet C., Sieracki M., Speich S., Stemmann L., Sullivan M. B., Sunagawa S., Wincker P., Eveillard D., Gorsky G., Guidi L., Iudicone D., Karsenti E., Lombard F., Ogata H., Pesant S., Sullivan M. B., Wincker P., de Vargas C., Tara Oceans: Towards global ocean ecosystems biology. Nat. Rev. Microbiol. 18, 428–445 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Jensen E. L., Yangüez K., Carrière F., Gontero B., Storage compound accumulation in diatoms as response to elevated CO2 concentration. Biology 9, 5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu S., Gu W., Jia S., Wang L., Wang L., Liu X., Zhou L., Huang A., Wang G., Proteomic and biochemical responses to different concentrations of CO2 suggest the existence of multiple carbon metabolism strategies in Phaeodactylum tricornutum. Biotechnol. Biofuels 14, 235 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakajima K., Tanaka A., Matsuda Y., SLC4 family transporters in a marine diatom directly pump bicarbonate from seawater. Proc. Natl. Acad. Sci. U.S.A 110, 1767–1772 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yodsuwan N., Sawayama S., Sirisansaneeyakul S., Effect of nitrogen concentration on growth, lipid production and fatty acid profiles of the marine diatom Phaeodactylum tricornutum. Agric. Nat. Resour. 51, 190–197 (2017). [Google Scholar]
- 37.Yang Z.-K., Ma Y.-H., Zheng J.-W., Yang W.-D., Liu J.-S., Li H.-Y., Proteomics to reveal metabolic network shifts towards lipid accumulation following nitrogen deprivation in the diatom Phaeodactylum tricornutum. J. Appl. Phycol. 26, 73–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y., Wu H., Sun M., Peng Q., Li A., Photosynthetic physiological performance and proteomic profiling of the oleaginous algae Scenedesmus acuminatus reveal the mechanism of lipid accumulation under low and high nitrogen supplies. Photosynth. Res. 138, 73–102 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Hockin N. L., Mock T., Mulholland F., Kopriva S., Malin G., The response of diatom central carbon metabolism to nitrogen starvation is different from that of green algae and higher plants. Plant Physiol. 158, 299–312 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jian J., Zeng D., Wei W., Lin H., Li P., Liu W., The combination of RNA and protein profiling reveals the response to nitrogen depletion in Thalassiosira pseudonana. Sci. Rep. 7, 8989 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Msanne J., Xu D., Konda A. R., Casas-Mollano J. A., Awada T., Cahoon E. B., Cerutti H., Metabolic and gene expression changes triggered by nitrogen deprivation in the photoautotrophically grown microalgae Chlamydomonas reinhardtii and Coccomyxa sp. C-169. Phytochemistry 75, 50–59 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Recht L., Zarka A., Boussiba S., Patterns of carbohydrate and fatty acid changes under nitrogen starvation in the microalgae Haematococcus pluvialis and Nannochloropsis sp. Appl. Microbiol. Biotechnol. 94, 1495–1503 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Dean A. P., Sigee D. C., Estrada B., Pittman J. K., Using FTIR spectroscopy for rapid determination of lipid accumulation in response to nitrogen limitation in freshwater microalgae. Bioresour. Technol. 101, 4499–4507 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Qari H. A., Oves M., Fatty acid synthesis by Chlamydomonas reinhardtii in phosphorus limitation. J. Bioenerg. Biomembr. 52, 27–38 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Chen M., Li J., Dai X., Sun Y., Chen F., Effect of phosphorus and temperature on chlorophyll a contents and cell sizes of Scenedesmus obliquus and Microcystis aeruginosa. Limnology 12, 187–192 (2011). [Google Scholar]
- 46.Zhou B., Ma J., Chen F., Zou Y., Wei Y., Zhong H., Pan K., Mechanisms underlying silicon-dependent metal tolerance in the marine diatom Phaeodactylum tricornutum. Environ. Pollut. 262, 114331 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Cheng J., Feng J., Sun J., Huang Y., Zhou J., Cen K., Enhancing the lipid content of the diatom Nitzschia sp. by 60 Co-γ irradiation mutation and high-salinity domestication. Energy 78, 9–15 (2014). [Google Scholar]
- 48.d’Ippolito G., Sardo A., Paris D., Vella F. M., Adelfi M. G., Botte P., Gallo C., Fontana A., Potential of lipid metabolism in marine diatoms for biofuel production. Biotechnol. Biofuels 8, 28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Armstrong E., Rogerson A., Leftley J. W., The abundance of heterotrophic protists associated with intertidal seaweeds. Estuar. Coast. Shelf Sci. 50, 415–424 (2000). [Google Scholar]
- 50.Villanova V., Spetea C., Mixotrophy in diatoms: Molecular mechanism and industrial potential. Physiol. Plant. 173, 603–611 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Bailleul B., Berne N., Murik O., Petroutsos D., Prihoda J., Tanaka A., Villanova V., Bligny R., Flori S., Falconet D., Krieger-Liszkay A., Santabarbara S., Rappaport F., Joliot P., Tirichine L., Falkowski P. G., Cardol P., Bowler C., Finazzi G., Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524, 366–369 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Stoecker D. K., Hansen P. J., Caron D. A., Mitra A., Mixotrophy in the marine plankton. Ann. Rev. Mar. Sci. 9, 311–335 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Meyer N., Rydzyk A., Pohnert G., Pronounced uptake and metabolism of organic substrates by diatoms revealed by pulse-labeling metabolomics. Front. Mar. Sci. 9, 821167 (2022). [Google Scholar]
- 54.Caron D. A., Mixotrophy stirs up our understanding of marine food webs. Proc. Natl. Acad. Sci. U.S.A. 113, 2806–2808 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Amin S. A., Parker M. S., Armbrust E. V., Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 76, 667–684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu J., Xue C.-X., Sun H., Zheng Y., Meng Z., Zhang X.-H., Carbohydrate catabolic capability of a Flavobacteriia bacterium isolated from hadal water. Syst. Appl. Microbiol. 42, 263–274 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Kallscheuer N., Wiegand S., Kohn T., Boedeker C., Jeske O., Rast P., Müller R.-W., Brümmer F., Heuer A., Jetten M. S. M., Rohde M., Jogler M., Jogler C., Cultivation-independent analysis of the bacterial community associated with the calcareous sponge Clathrina clathrus and isolation of Poriferisphaera corsica Gen. Nov., Sp. Nov., belonging to the barely studied class Phycisphaerae in the Phylum Planctomycetes. Front. Microbiol. 11, 602250 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fukunaga Y., Kurahashi M., Sakiyama Y., Ohuchi M., Yokota A., Harayama S., Phycisphaera mikurensis gen. nov., sp. nov., isolated from a marine alga, and proposal of Phycisphaeraceae fam. nov., Phycisphaerales ord. nov. and Phycisphaerae classis nov. in the phylum Planctomycetes. J. Gen. Appl. Microbiol. 55, 267–275 (2009). [DOI] [PubMed] [Google Scholar]
- 59.Priest T., Heins A., Harder J., Amann R., Fuchs B. M., Niche partitioning of the ubiquitous and ecologically relevant NS5 marine group. ISME J. 16, 1570–1582 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buchan A., LeCleir G. R., Gulvik C. A., González J. M., Master recyclers: Features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Thiele I., Palsson B. Ø., A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 5, 93–121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.M. Kumar, C. Zuniga, J. D. Tibocha-Bonilla, S. R. Smith, J. Coker, A. E. Allen, K. Zengler, “Constraint-based modeling of diatoms metabolism and quantitative biology approaches” in The Molecular Life of Diatoms (Springer International Publishing, 2022; https://link.springer.com/10.1007/978-3-030-92499-7_26), pp. 775–808. [Google Scholar]
- 63.Stock W., Vanelslander B., Rüdiger F., Sabbe K., Vyverman W., Karsten U., Thermal niche differentiation in the benthic diatom Cylindrotheca closterium (Bacillariophyceae) complex. Front. Microbiol. 10, 1395 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hutchins D. A., Fu F., Microorganisms and ocean global change. Nat. Microbiol. 2, 17058 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Pahl S. L., Lewis D. M., Chen F., King K. D., Growth dynamics and the proximate biochemical composition and fatty acid profile of the heterotrophically grown diatom Cyclotella cryptica. J. Appl. Phycol. 22, 165–171 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Armstrong E., Rogerson A., Leftley J. W., Utilisation of seaweed carbon by three surface-associated heterotrophic protists, Stereomyxa ramosa, Nitzschia alba and Labyrinthula sp. Aquat. Microb. Ecol. 21, 49–57 (2000). [Google Scholar]
- 67.Brzezinski M. A., The Si:C:N ratio of marine diatoms: Interspecific variability and the effect of some environmental variables. J. Phycol. 21, 347–357 (2004). [Google Scholar]
- 68.Lachance J.-C., Lloyd C. J., Monk J. M., Yang L., Sastry A. V., Seif Y., Palsson B. O., Rodrigue S., Feist A. M., King Z. A., Jacques P.-É., BOFdat: Generating biomass objective functions for genome-scale metabolic models from experimental data. PLOS Comput. Biol. 15, e1006971 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jallet D., Caballero M. A., Gallina A. A., Youngblood M., Peers G., Photosynthetic physiology and biomass partitioning in the model diatom Phaeodactylum tricornutum grown in a sinusoidal light regime. Algal Res. 18, 51–60 (2016). [Google Scholar]
- 70.Cunha E., Lagoa D., Faria J. P., Liu F., Henry C. S., Dias O., TranSyT, an innovative framework for identifying transport systems. Bioinformatics 39, btad466 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lieven C., Beber M. E., Olivier B. G., Bergmann F. T., Ataman M., Babaei P., Bartell J. A., Blank L. M., Chauhan S., Correia K., Diener C., Dräger A., Ebert B. E., Edirisinghe J. N., Faria J. P., Feist A. M., Fengos G., Fleming R. M. T., García-Jiménez B., Hatzimanikatis V., van Helvoirt W., Henry C. S., Hermjakob H., Herrgård M. J., Kaafarani A., Kim H. U., King Z., Klamt S., Klipp E., Koehorst J. J., König M., Lakshmanan M., Lee D.-Y., Lee S. Y., Lee S., Lewis N. E., Liu F., Ma H., Machado D., Mahadevan R., Maia P., Mardinoglu A., Medlock G. L., Monk J. M., Nielsen J., Nielsen L. K., Nogales J., Nookaew I., Palsson B. O., Papin J. A., Patil K. R., Poolman M., Price N. D., Resendis-Antonio O., Richelle A., Rocha I., Sánchez B. J., Schaap P. J., Sheriff R. S. M., Shoaie S., Sonnenschein N., Teusink B., Vilaça P., Vik J. O., Wodke J. A. H., Xavier J. C., Yuan Q., Zakhartsev M., Zhang C., MEMOTE for standardized genome-scale metabolic model testing. Nat. Biotechnol. 38, 272–276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Orth J. D., Thiele I., Palsson B. Ø., What is flux balance analysis? Nat. Biotechnol. 28, 245–248 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morel F. M. M., Rueter J. G., Anderson D. M., Guillard R. R. L., AQUIL: A chemically defined phytoplankton culture medium for trace metal studies. J. Phycol. 15, 135–141 (1979). [Google Scholar]
- 74.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Caballero J., Smit A. F. A., Hood L., Glusman G., Realistic artificial DNA sequences as negative controls for computational genomics. Nucleic Acids Res. 42, e99–e99 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.B. G. Galuzzi, L. Milazzo, C. Damiani, Best Practices in Flux Sampling of Constrained-Based Models (Springer, 2023; 10.1007/978-3-031-25891-6_18), pp. 234–248. [DOI]
- 79.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., Huttenhower C., Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Monk J., Nogales J., Palsson B. O., Optimizing genome-scale network reconstructions. Nat. Biotechnol. 32, 447–452 (2014). [DOI] [PubMed] [Google Scholar]
- 81.Morris E. P., Kromkamp J. C., Influence of temperature on the relationship between oxygen- and fluorescence-based estimates of photosynthetic parameters in a marine benthic diatom (Cylindrotheca closterium). Eur. J. Phycol. 38, 133–142 (2003). [Google Scholar]
- 82.Alcoverro T., Conte E., Mazzella L., Production of mucilage by the adriatic epipelic diatom Cylindrotheca closterium (Bacillariophyceae) under nutrient limitation. J. Phycol. 36, 1087–1095 (2000). [Google Scholar]
- 83.Rijstenbil J. W., Effects of UVB radiation and salt stress on growth, pigments and antioxidative defence of the marine diatom Cylindrotheca closterium. Mar. Ecol. Prog. Ser. 254, 37–48 (2003). [Google Scholar]
- 84.Rijstenbil J. W., UV- and salinity-induced oxidative effects in the marine diatom Cylindrotheca closterium during simulated emersion. Mar. Biol. 147, 1063–1073 (2005). [Google Scholar]
- 85.Staats N., Stal L. J., Mur L. R., Exopolysaccharide production by the epipelic diatom Cylindrotheca closterium: Effects of nutrient conditions. J. Exp. Mar. Bio. Ecol. 249, 13–27 (2000). [DOI] [PubMed] [Google Scholar]
- 86.Wang S., Sirbu D., Thomsen L., Kuhnert N., Ullrich M. S., Thomsen C., Comparative lipidomic studies of Scenedesmus sp. (Chlorophyceae) and Cylindrotheca closterium (Bacillariophyceae) reveal their differences in lipid production under nitrogen starvation. J. Phycol. 55, 1246–1257 (2019). [DOI] [PubMed] [Google Scholar]
- 87.Araújo C. V. M., Diz F. R., Lubián L. M., Blasco J., Moreno-Garrido I., Sensitivity of Cylindrotheca closterium to copper: Influence of three test endpoints and two test methods. Sci. Total Environ. 408, 3696–3703 (2010). [DOI] [PubMed] [Google Scholar]
- 88.Almeyda M. D., Scodelaro Bilbao P. G., Popovich C. A., Constenla D., Leonardi P. I., Enhancement of polyunsaturated fatty acid production under low-temperature stress in Cylindrotheca closterium. J. Appl. Phycol. 32, 989–1001 (2020). [Google Scholar]
- 89.Van Bergeijk S. A., Van der Zee C., Stal L. J., Uptake and excretion of dimethylsulphoniopropionate is driven by salinity changes in the marine benthic diatom Cylindrotheca closterium. Eur. J. Phycol. 38, 341–349 (2003). [Google Scholar]
- 90.Erdogan A., Demirel Z., Dalay M. C., Eroglu A. E., Fucoxanthin content of Cylindrotheca closterium and its oxidative stress mediated enhancement. Turkish J. Fish. Aquat. Sci. 16, 499–506 (2016). [Google Scholar]
- 91.Orcutt D. M., Patterson G. W., Effect of light intensity upon lipid composition of Nitzschia closterium (Cylindrotheca fusiformis). Lipids 9, 1000–1003 (1974). [DOI] [PubMed] [Google Scholar]
- 92.Affan A., Heo S., Jeon Y., Lee J., Optimal growth conditions and antioxidative activities of Cylindrotheca closterium (Bacillariophyceae). J. Phycol. 45, 1405–1415 (2009). [DOI] [PubMed] [Google Scholar]
- 93.Brembu T., Mühlroth A., Alipanah L., Bones A. M., The effects of phosphorus limitation on carbon metabolism in diatoms. Philos. Trans. R. Soc. B Biol. Sci. 372, 20160406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ruocco N., Nuzzo G., D’Ippolito G., Manzo E., Sardo A., Ianora A., Romano G., Iuliano A., Zupo V., Costantini M., Fontana A., Lipoxygenase pathways in diatoms: Occurrence and correlation with grazer toxicity in four benthic species. Mar. Drugs 18, 66 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ryabushko V. I., Zheleznova S. N., Nekhoroshev M. V., Effect of nitrogen on fucoxanthin accumulation in the diatom Cylindrotheca closterium (Ehrenb.) Reimann et Lewin. Int. J. Algae 19, 79–84 (2017). [Google Scholar]
- 96.Han T., Song P., Shi R., Qi Z., Li J., Huang H., Optimal nutrient availability could alleviate diatom Cylindrotheca closterium fouling during seedling cultivation of Sargassum hemiphyllum. Aquaculture 552, 738020 (2022). [Google Scholar]
- 97.Becker A. E., Copplestone D., Cadmium uptake from sediment by Cylindrotheca closterium and the effect of diatom presence on partitioning of cadmium between sediment and water: A laboratory study. Limnol. Oceanogr. 64, 2550–2568 (2019). [Google Scholar]
- 98.Humphrey G., Subba Rao D., Photosynthetic rate of the marine diatom Cylindrotheca closterium. Mar. Freshw. Res. 18, 123 (1967). [Google Scholar]
- 99.Zhang F., Chi J., Influences of nutritional conditions on degradation of dibutyl phthalate in coastal sediments with Cylindrotheca closterium. Mar. Pollut. Bull. 153, 111021 (2020). [DOI] [PubMed] [Google Scholar]
- 100.Roncarati F., Rijstenbil J. W., Pistocchi R., Photosynthetic performance, oxidative damage and antioxidants in Cylindrotheca closterium in response to high irradiance, UVB radiation and salinity. Mar. Biol. 153, 965–973 (2008). [Google Scholar]
- 101.Wolfstein K., de Brouwer J., Stal L., Biochemical partitioning of photosynthetically fixed carbon by benthic diatoms during short-term incubations at different irradiances. Mar. Ecol. Prog. Ser. 245, 21–31 (2002). [Google Scholar]
- 102.Wolfstein K., Stal L., Production of extracellular polymeric substances (EPS) by benthic diatoms: Effect of irradiance and temperature. Mar. Ecol. Prog. Ser. 236, 13–22 (2002). [Google Scholar]
- 103.Suman K., Kiran T., Devi U. K., Sarma N. S., Culture medium optimization and lipid profiling of Cylindrotheca, a lipid- and polyunsaturated fatty acid-rich pennate diatom and potential source of eicosapentaenoic acid. Bot. Mar. 55, 289–299 (2012). [Google Scholar]
- 104.Underwood G. J. C., Boulcott M., Raines C. A., Waldron K., Environmental effects on exopolymer production by marine benthic diatoms: Dynamics, changes in composition, and pathways of production. J. Phycol. 40, 293–304 (2004). [Google Scholar]
- 105.Admiraal W., Laane R., Peletier H., Participation of diatoms in the amino acid cycle of coastal waters; uptake and excretion in cultures. Mar. Ecol. Prog. Ser. 15, 303–306 (1984). [Google Scholar]
- 106.De Brouwer J. F. C., Stal L. J., Daily fluctuations of exopolymers in cultures of the benthic diatoms Cylindrotheca closterium and Nitzschia sp. (Bacillariophyceae). J. Phycol. 38, 464–472 (2002). [Google Scholar]
- 107.Zheleznova S. N., Production characteristics of the diatom Cylindrotheca closterium (Ehrenb.) Reimann et Lewin grown in an intensive culture at various nitrogen sources in the medium. Mar. Biol. J. 4, 33–44 (2019). [Google Scholar]
- 108.Nilsson C., Sundback K., Sundback K., Amino acid uptake in natural microphytobenthic assemblages studied by microautoradiography. Hydrobiologia 332, 119–129 (1996). [Google Scholar]
- 109.Staats N., De Winder B., Mur L. R., Stal L. J., Isolation and characterization of extracellular polysaccharides from the epipelic diatoms Cylindrotheca closterium and Navicula salinarum. Eur. J. Phycol. 34, 161–169 (1999). [Google Scholar]
- 110.Urbani R., Magaletti E., Sist P., Cicero A. M., Extracellular carbohydrates released by the marine diatoms Cylindrotheca closterium, Thalassiosira pseudonana and Skeletonema costatum: Effect of P-depletion and growth status. Sci. Total Environ. 353, 300–306 (2005). [DOI] [PubMed] [Google Scholar]
- 111.Wolf Y. I., Koonin E. V., A tight link between orthologs and bidirectional best hits in bacterial and archaeal genomes. Genome Biol. Evol. 4, 1286–1294 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kanehisa M., Sato Y., Kawashima M., KEGG mapping tools for uncovering hidden features in biological data. Protein Sci. 31, 47–53 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Seaver S. M. D., Liu F., Zhang Q., Jeffryes J., Faria J. P., Edirisinghe J. N., Mundy M., Chia N., Noor E., Beber M. E., Best A. A., DeJongh M., Kimbrel J. A., D’haeseleer P., McCorkle S. R., Bolton J. R., Pearson E., Canon S., Wood-Charlson E. M., Cottingham R. W., Arkin A. P., Henry C. S., The ModelSEED Biochemistry Database for the integration of metabolic annotations and the reconstruction, comparison and analysis of metabolic models for plants, fungi and microbes. Nucleic Acids Res. 49, D575–D588 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Caspi R., Billington R., Keseler I. M., Kothari A., Krummenacker M., Midford P. E., Ong W. K., Paley S., Subhraveti P., Karp P. D., The MetaCyc database of metabolic pathways and enzymes - A 2019 update. Nucleic Acids Res. 48, D445–D453 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bateman A., Martin M.-J., Orchard S., Magrane M., Agivetova R., Ahmad S., Alpi E., Bowler-Barnett E. H., Britto R., Bursteinas B., Bye-A-Jee H., Coetzee R., Cukura A., Da Silva A., Denny P., Dogan T., Ebenezer T., Fan J., Castro L. G., Garmiri P., Georghiou G., Gonzales L., Hatton-Ellis E., Hussein A., Ignatchenko A., Insana G., Ishtiaq R., Jokinen P., Joshi V., Jyothi D., Lock A., Lopez R., Luciani A., Luo J., Lussi Y., MacDougall A., Madeira F., Mahmoudy M., Menchi M., Mishra A., Moulang K., Nightingale A., Oliveira C. S., Pundir S., Qi G., Raj S., Rice D., Lopez M. R., Saidi R., Sampson J., Sawford T., Speretta E., Turner E., Tyagi N., Vasudev P., Volynkin V., Warner K., Watkins X., Zaru R., Zellner H., Bridge A., Poux S., Redaschi N., Aimo L., Argoud-Puy G., Auchincloss A., Axelsen K., Bansal P., Baratin D., Blatter M.-C., Bolleman J., Boutet E., Breuza L., Casals-Casas C., de Castro E., Echioukh K. C., Coudert E., Cuche B., Doche M., Dornevil D., Estreicher A., Famiglietti M. L., Feuermann M., Gasteiger E., Gehant S., Gerritsen V., Gos A., Gruaz-Gumowski N., Hinz U., Hulo C., Hyka-Nouspikel N., Jungo F., Keller G., Kerhornou A., Lara V., Le Mercier P., Lieberherr D., Lombardot T., Martin X., Masson P., Morgat A., Neto T. B., Paesano S., Pedruzzi I., Pilbout S., Pourcel L., Pozzato M., Pruess M., Rivoire C., Sigrist C., Sonesson K., Stutz A., Sundaram S., Tognolli M., Verbregue L., Wu C. H., Arighi C. N., Arminski L., Chen C., Chen Y., Garavelli J. S., Huang H., Laiho K., McGarvey P., Natale D. A., Ross K., Vinayaka C. R., Wang Q., Wang Y., Yeh L.-S., Zhang J., Ruch P., Teodoro D., UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chang A., Jeske L., Ulbrich S., Hofmann J., Koblitz J., Schomburg I., Neumann-Schaal M., Jahn D., Schomburg D., BRENDA, the ELIXIR core data resource in 2021: New developments and updates. Nucleic Acids Res. 49, D498–D508 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fleischmann A., IntEnz, the integrated relational enzyme database. Nucleic Acids Res. 32, 434D–4437D (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Saier M. H., Reddy V. S., Moreno-Hagelsieb G., Hendargo K. J., Zhang Y., Iddamsetty V., Lam K. J. K., Tian N., Russum S., Wang J., Medrano-Soto A., The Transporter Classification Database (TCDB): 2021 update. Nucleic Acids Res. 49, D461–D467 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Elbourne L. D. H., Tetu S. G., Hassan K. A., Paulsen I. T., TransportDB 2.0: A database for exploring membrane transporters in sequenced genomes from all domains of life. Nucleic Acids Res. 45, D320–D324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Petersen T. N., Brunak S., von Heijne G., Nielsen H., SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 (2011). [DOI] [PubMed] [Google Scholar]
- 121.Gschloessl B., Guermeur Y., Cock J. M., HECTAR: A method to predict subcellular targeting in heterokonts. BMC Bioinformatics 9, 393 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Claros M. G., MitoProt, a Macintosh application for studying mitochondrial proteins. Bioinformatics 11, 441–447 (1995). [DOI] [PubMed] [Google Scholar]
- 123.Cokol M., Nair R., Rost B., Finding nuclear localization signals. EMBO Rep. 1, 411–415 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Emanuelsson O., Brunak S., von Heijne G., Nielsen H., Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971 (2007). [DOI] [PubMed] [Google Scholar]
- 125.Grant B., Madgwick J., Dal P. G., Growth of Cylindrotheca closterium var. californica (Mereschk.) Reimann & Lewin on nitrate, ammonia, and urea. Mar. Freshw. Res. 18, 129 (1967). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S20
Legends for tables S1 to S26
Legend for data S1
References
Tables S1 to S26
Data S1