Abstract
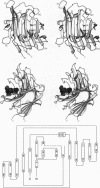
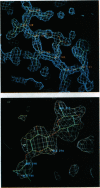
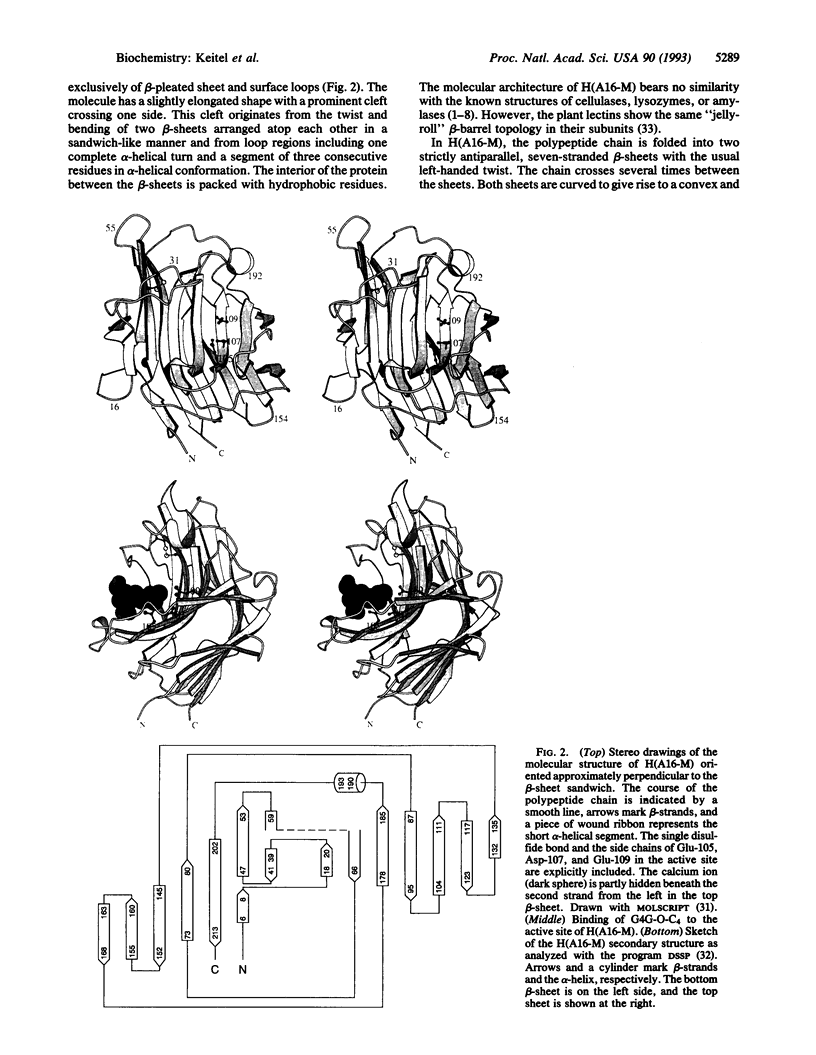
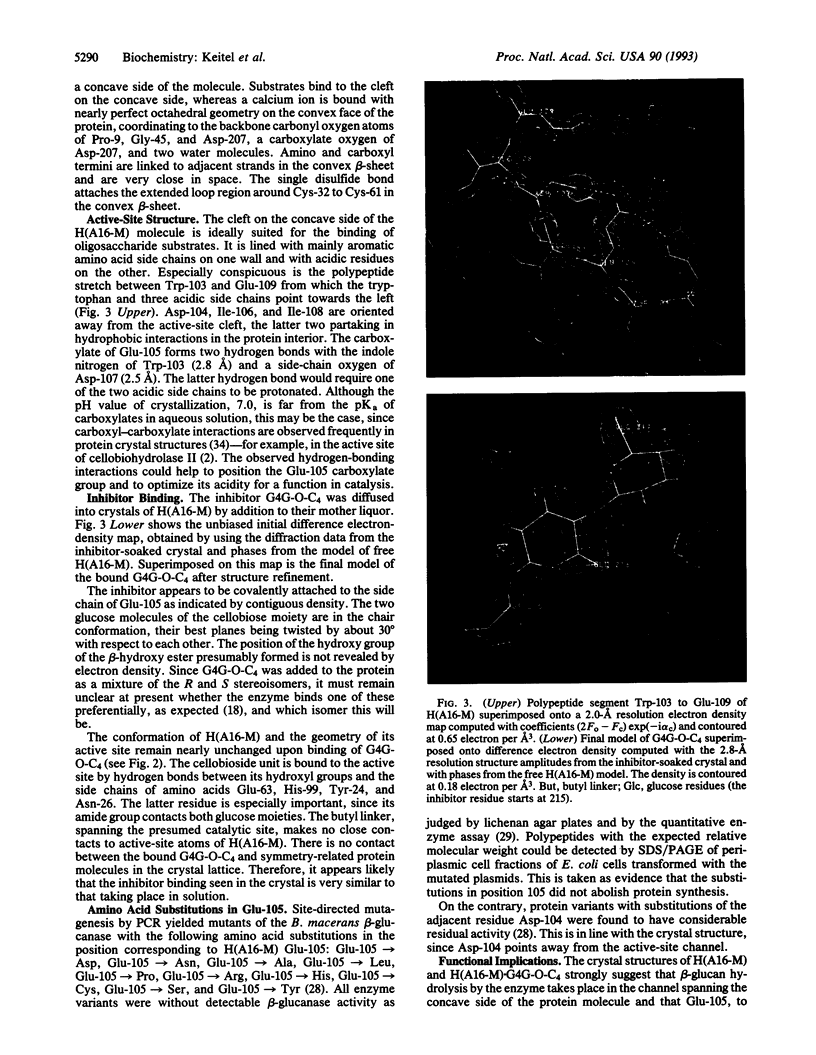
The three-dimensional structure of the hybrid Bacillus 1,3-1,4-beta-glucanase (beta-glucanase; 1,3-1,4-beta-D-glucan 4-glucanohydrolase, lichenase, EC 3.2.1.73) designated H(A16-M) was determined by x-ray crystallography at a resolution of 2.0 A and refined to an R value of 16.4% using stereochemical restraints. The protein molecule consists mainly of two seven-stranded antiparallel beta-pleated sheets arranged atop each other to form a compact, sandwich-like structure. A channel crossing one side of the protein molecule accommodates an inhibitor, 3,4-epoxybutyl beta-D-cellobioside, which binds covalently to the side chain of Glu-105, as seen in a crystal structure analysis at 2.8-A resolution of the protein-inhibitor complex (R = 16.8%). That Glu-105 may be indispensible for enzyme catalysis by H(A16-M) is suggested by site-directed mutagenesis of this residue, which inevitably leads to an inactive enzyme.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson M. A., Stone B. A. A new substrate for investigating the specificity of beta-glucan hydrolases. FEBS Lett. 1975 Apr 1;52(2):202–207. doi: 10.1016/0014-5793(75)80806-4. [DOI] [PubMed] [Google Scholar]
- Anderson W. F., Grütter M. G., Remington S. J., Weaver L. H., Matthews B. W. Crystallographic determination of the mode of binding of oligosaccharides to T4 bacteriophage lysozyme: implications for the mechanism of catalysis. J Mol Biol. 1981 Apr 25;147(4):523–543. doi: 10.1016/0022-2836(81)90398-3. [DOI] [PubMed] [Google Scholar]
- Artymiuk P. J., Blake C. C. Refinement of human lysozyme at 1.5 A resolution analysis of non-bonded and hydrogen-bond interactions. J Mol Biol. 1981 Nov 15;152(4):737–762. doi: 10.1016/0022-2836(81)90125-x. [DOI] [PubMed] [Google Scholar]
- Borriss R., Buettner K., Maentsaelae P. Structure of the beta-1,3-1,4-glucanase gene of Bacillus macerans: homologies to other beta-glucanases. Mol Gen Genet. 1990 Jul;222(2-3):278–283. doi: 10.1007/BF00633829. [DOI] [PubMed] [Google Scholar]
- Borriss R., Olsen O., Thomsen K. K., von Wettstein D. Hybrid bacillus endo-(1-3,1-4)-beta-glucanases: construction of recombinant genes and molecular properties of the gene products. Carlsberg Res Commun. 1989;54(2):41–54. doi: 10.1007/BF02907584. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Buisson G., Duée E., Haser R., Payan F. Three dimensional structure of porcine pancreatic alpha-amylase at 2.9 A resolution. Role of calcium in structure and activity. EMBO J. 1987 Dec 20;6(13):3909–3916. doi: 10.1002/j.1460-2075.1987.tb02731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einspahr H., Parks E. H., Suguna K., Subramanian E., Suddath F. L. The crystal structure of pea lectin at 3.0-A resolution. J Biol Chem. 1986 Dec 15;261(35):16518–16527. [PubMed] [Google Scholar]
- Fincher G. B., Lock P. A., Morgan M. M., Lingelbach K., Wettenhall R. E., Mercer J. F., Brandt A., Thomsen K. K. Primary structure of the (1-->3,1-->4)-beta-D-glucan 4-glucohydrolase from barley aleurone. Proc Natl Acad Sci U S A. 1986 Apr;83(7):2081–2085. doi: 10.1073/pnas.83.7.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosalbes M. J., Pérez-González J. A., González R., Navarro A. Two beta-glycanase genes are clustered in Bacillus polymyxa: molecular cloning, expression, and sequence analysis of genes encoding a xylanase and an endo-beta-(1,3)-(1,4)-glucanase. J Bacteriol. 1991 Dec;173(23):7705–7710. doi: 10.1128/jb.173.23.7705-7710.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson W. A. Stereochemically restrained refinement of macromolecular structures. Methods Enzymol. 1985;115:252–270. doi: 10.1016/0076-6879(85)15021-4. [DOI] [PubMed] [Google Scholar]
- Hofemeister J., Kurtz A., Borriss R., Knowles J. The beta-glucanase gene from Bacillus amyloliquefaciens shows extensive homology with that of Bacillus subtilis. Gene. 1986;49(2):177–187. doi: 10.1016/0378-1119(86)90278-7. [DOI] [PubMed] [Google Scholar]
- Høj P. B., Condron R., Traeger J. C., McAuliffe J. C., Stone B. A. Identification of glutamic acid 105 at the active site of Bacillus amyloliquefaciens 1,3-1,4-beta-D-glucan 4-glucanohydrolase using epoxide-based inhibitors. J Biol Chem. 1992 Dec 15;267(35):25059–25066. [PubMed] [Google Scholar]
- Høj P. B., Rodriguez E. B., Iser J. R., Stick R. V., Stone B. A. Active site-directed inhibition by optically pure epoxyalkyl cellobiosides reveals differences in active site geometry of two 1,3-1,4-beta-D-glucan 4-glucanohydrolases. The importance of epoxide stereochemistry for enzyme inactivation. J Biol Chem. 1991 Jun 25;266(18):11628–11631. [PubMed] [Google Scholar]
- Høj P. B., Rodriguez E. B., Stick R. V., Stone B. A. Differences in active site structure in a family of beta-glucan endohydrolases deduced from the kinetics of inactivation by epoxyalkyl beta-oligoglucosides. J Biol Chem. 1989 Mar 25;264(9):4939–4947. [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Keitel T., Granzin J., Simon O., Borriss R., Thomsen K. K., Wessner H., Höhne W., Heinemann U. Crystallization of the hybrid Bacillus (1-3,1-4)-beta-glucanase H(A16-M). J Mol Biol. 1991 Apr 20;218(4):703–704. doi: 10.1016/0022-2836(91)90259-9. [DOI] [PubMed] [Google Scholar]
- Kraulis J., Clore G. M., Nilges M., Jones T. A., Pettersson G., Knowles J., Gronenborn A. M. Determination of the three-dimensional solution structure of the C-terminal domain of cellobiohydrolase I from Trichoderma reesei. A study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing. Biochemistry. 1989 Sep 5;28(18):7241–7257. doi: 10.1021/bi00444a016. [DOI] [PubMed] [Google Scholar]
- Lloberas J., Perez-Pons J. A., Querol E. Molecular cloning, expression and nucleotide sequence of the endo-beta-1,3-1,4-D-glucanase gene from Bacillus licheniformis. Predictive structural analyses of the encoded polypeptide. Eur J Biochem. 1991 Apr 23;197(2):337–343. doi: 10.1111/j.1432-1033.1991.tb15916.x. [DOI] [PubMed] [Google Scholar]
- Matsuura Y., Kusunoki M., Harada W., Kakudo M. Structure and possible catalytic residues of Taka-amylase A. J Biochem. 1984 Mar;95(3):697–702. doi: 10.1093/oxfordjournals.jbchem.a134659. [DOI] [PubMed] [Google Scholar]
- Murphy N., McConnell D. J., Cantwell B. A. The DNA sequence of the gene and genetic control sites for the excreted B. subtilis enzyme beta-glucanase. Nucleic Acids Res. 1984 Jul 11;12(13):5355–5367. doi: 10.1093/nar/12.13.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen O., Borriss R., Simon O., Thomsen K. K. Hybrid Bacillus (1-3,1-4)-beta-glucanases: engineering thermostable enzymes by construction of hybrid genes. Mol Gen Genet. 1991 Feb;225(2):177–185. doi: 10.1007/BF00269845. [DOI] [PubMed] [Google Scholar]
- Planas A., Juncosa M., Lloberas J., Querol E. Essential catalytic role of Glu134 in endo-beta-1,3-1,4-D-glucan 4-glucanohydrolase from B. licheniformis as determined by site-directed mutagenesis. FEBS Lett. 1992 Aug 17;308(2):141–145. doi: 10.1016/0014-5793(92)81262-k. [DOI] [PubMed] [Google Scholar]
- Rouvinen J., Bergfors T., Teeri T., Knowles J. K., Jones T. A. Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science. 1990 Jul 27;249(4967):380–386. doi: 10.1126/science.2377893. [DOI] [PubMed] [Google Scholar]
- Sawyer L., James M. N. Carboxyl-carboxylate interactions in proteins. Nature. 1982 Jan 7;295(5844):79–80. doi: 10.1038/295079a0. [DOI] [PubMed] [Google Scholar]
- Schimming S., Schwarz W. H., Staudenbauer W. L. Structure of the Clostridium thermocellum gene licB and the encoded beta-1,3-1,4-glucanase. A catalytic region homologous to Bacillus lichenases joined to the reiterated domain of clostridial cellulases. Eur J Biochem. 1992 Feb 15;204(1):13–19. doi: 10.1111/j.1432-1033.1992.tb16600.x. [DOI] [PubMed] [Google Scholar]
- Teather R. M., Erfle J. D. DNA sequence of a Fibrobacter succinogenes mixed-linkage beta-glucanase (1,3-1,4-beta-D-glucan 4-glucanohydrolase) gene. J Bacteriol. 1990 Jul;172(7):3837–3841. doi: 10.1128/jb.172.7.3837-3841.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. C. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]
- Zhang H., Scholl R., Browse J., Somerville C. Double stranded DNA sequencing as a choice for DNA sequencing. Nucleic Acids Res. 1988 Feb 11;16(3):1220–1220. doi: 10.1093/nar/16.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]