Abstract
In metazoans, members of the insulin-like peptide (ILP) family play a role in multiple physiological functions in response to the nutritional status. ILPs have been identified and characterized in a wide variety of insect species. Insect ILPs that are mainly produced by several pairs of medial neurosecretory cells in the brain circulate in the hemolymph and act systemically on target tissues. Physiological and biochemical studies in Lepidoptera and genetic studies in the fruit fly have greatly expanded our knowledge of the physiological functions of ILPs. Here, we outline the recent progress of the structural classification of insect ILPs and overview recent studies that have elucidated the physiological functions of insect ILPs involved in nutrient-dependent growth during development.
Introduction
Nutrients are critical environmental signals influencing growth and development in animals. Although each cell in a multicellular organism responds directly to nutrition, the growth and development of the entire organism needs to be coordinated by adjusting growth between tissues and controlling the consumption of stored nutrients. The coordination of systemic organismal growth in response to the nutritional status is primarily mediated by the insulin-like peptide (ILP) family, which includes insulin and insulin-like growth factors (IGFs) in vertebrates, as well as multiple ILPs in invertebrates.
In vertebrates, insulin and IGFs regulate metabolism, growth and development in response to nutritional availability. Although insulin and IGFs have similar amino acid sequences, they have different physiological functions that are meditated by distinct receptor tyrosine kinases (RTKs), the insulin receptor and IGF-I receptor, respectively [1]. The major function of insulin is to control carbohydrate and lipid metabolism [2], whereas that of IGFs is to promote tissue and body growth during development [3]. Numerous studies have shown that the key regulator of the activities of insulin and IGFs is the nutritional status [4]. The production and secretion of insulin by pancreatic β -cells are tightly regulated by the nutrient status [5]. Nutritional availability also influences the production, serum concentration, and action of IGF-I in regulating appropriate tissue and body size [6]. Another class of ILP family peptides in vertebrates, relaxins and relaxin-like peptides, function through leucine-rich repeat-containing G protein-coupled receptors (GPCRs) and have multiple functions, especially associated with reproduction [7].
ILPs have been identified and characterized in a wide variety of invertebrate phyla and in arthropods, including insects [8]. In insects, ILPs are involved in multiple biological processes, including growth, metabolism, reproduction, immunity, behavior, stress resistance, diapause, and lifespan [8-15]. Recently, powerful genetic studies using the fruit fly Drosophila melanogaster have greatly enhanced our understanding of the conserved functions of ILPs, as well as their downstream signaling pathways called the insulin/IGF signaling (IIS) pathways [9-15]. In this review, we will first focus on the structural classification of ILPs in insects. We will then overview the recent progress in our understanding of the physiological functions of insect ILPs, especially as it relates to nutrient-dependent growth during development. Through this review, we aim to provide insights into the diverse yet conserved roles of insect ILPs in the coordination of systemic organismal growth, as well as tissue-specific growth, in response to the nutritional status during development.
Structural classification of insulin-like peptides in insects
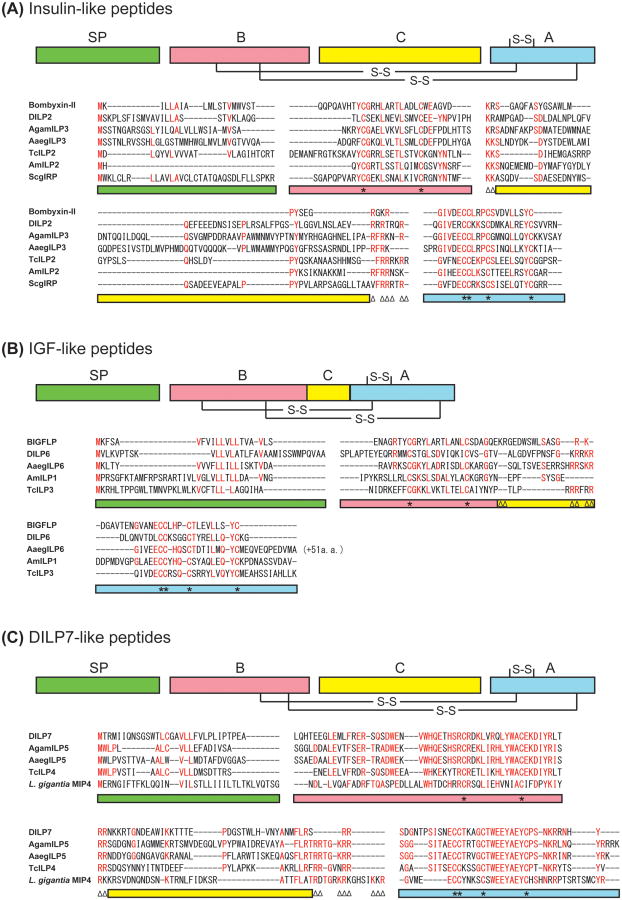
ILP family members have been identified in multiple insect species, with their numbers varying significantly between only one in some orthopteran species and more than 40 in the silkworm Bombyx mori [8, 16]. The amino acid sequences of insect ILPs are highly divergent between insect orders, except for some critical residues (such as cysteines) that are necessary for tertiary structure formation. However, they can be classified into at least three groups based mainly on the sequence features of their precursors: insulin-like peptides, IGF-like peptides, and DILP7-like peptides (Figure 1) [17, 18]. The first group, insulin-like peptides, shares the most common structural feature of the ILP family, and most insect ILPs are classified into this group (Figure 1A). The common feature of insulin-like peptides is a conserved domain organization of their precursors, consisting of a signal peptide, with a B-chain, C-peptide, and A-chain, similar to the vertebrate ILP family. After cleavage of the signal peptide, the C-peptide is most likely removed to generate a mature heterodimeric peptide consisting of the A- and B-chains, such as in vertebrate insulin or relaxins. The second group is the putative IGF-like peptides. The recently identified Bombyx IGF-like peptide (BIGFLP) retains the C-peptide, resulting in a single-chain polypeptide, which is similar to vertebrate IGFs [19]. One of the characteristic features of IGF-like peptides is that they have a relatively shortened C-peptide compared with other insect ILPs. The third group, DILP7-like peptides, is characterized by an unusually conserved sequence shared by several insects and even some molluscan species. Precursor polypeptides with long conserved sequences are found in Drosophila (DILP7), the African malaria mosquito Anopheles gambiae (AgamILP5), the yellow fever mosquito Aedes aegypti (AaegILP5), the red flour beetle Tribolium castaneum (TcILP4), and molluscs such as the owl limpet Lottia gigantea (MIP4) and the California sea hare Aplysia californica (MIP1) [20, 8]. To date, the conserved biological function of these unique ILPs has not yet been clarified. In Drosophila, DILP8 has recently been identified as a new ILP and has a unique effect on developmental timing and systemic body growth [21**, 22**, 23]. Compared with other Drosophila ILPs, DILP8 has atypical number of amino acid residues between two cysteine residues in the B chain, and between forth and fifth cysteine residues in the A chain. Although direct sequence comparisons show no clear DILP8 homologues in non-dipteran insects or nematoceran (mosquito) genomes [21**], it is possible that functional orthologs of DILP8 exist in other insects.
Figure 1.
Predicted insulin-like, IGF-like and DILP7-like peptides in insects. (A) Amino acid sequences of the representatives of predicted insulin-like peptides from Bombyx (bombyxin-II), Drosophila (DILP2), Anopheles (AgamILP3), Aedes (AaegILP3), Apis (AmILP2), Tribolium (TcILP2), and Schistocerca (ScgIRP) are aligned. (B) Amino acid sequences of the representatives of predicted IGF-like peptides from B. mori (BIGFLP), Drosophila (DILP6), Aedes (AaegILP6), Apis (AmILP1), and Tribolium (TcILP3) are aligned. (C) Amino acid sequences of the representatives of predicted highly conserved ILP group (DILP7-like peptides) from Drosophila (DILP7), Anopheles (AgamILP5), Aedes (AaegILP5), Tribolium (TcILP4), and Lottia (molluscan insulin-related peptide 4, MIP4) are aligned. Highly conserved amino acid residues are shown in red. Color bars indicate the predicted domains in the precursor peptides: green, signal peptide; red, B-chain; yellow, C-peptide; blue, A-chain. Asterisks on the color bars below the alignment denote Cys residues, and paired triangles denote potential cleavage sites (dibasic amino acids).
Insect ILPs act on target tissues by activating an RTK named insulin-like receptor (InR), which shows high similarity with mammalian insulin and IGF type-I receptors. Although multiple ILPs exist in each insect genome, it typically encodes only one or two InRs [8]. However, it remains possible that some ILPs act through alternative receptors such as GPCRs. Recently, Veenstra hypothesized that the receptors for DILP7 and DILP8 in Drosophila are candidate relaxin receptors, LGR4 (CG34411) and LGR3 (CG31096), respectively [24]. If this is indeed the case, DILP7 and DILP8 can be classified as relaxin-like peptides, although they have no relaxin-specific GPCR-binding motif, RxxxRxxI/V, in the B-chain [25].
Nutrient-dependent secretion and the transcription of insulin-like peptides by medial neurosecretory cells in the brain
The principal ILP-producing cells that are tightly associated with nutrient-dependent systemic growth regulation are the several pairs of medial neurosecretory cells (mNSCs), also known as insulin-producing cells (IPCs), in the brain. In this section, we will focus on the regulatory mechanisms of the secretion and transcription of insect ILPs in brain mNSCs.
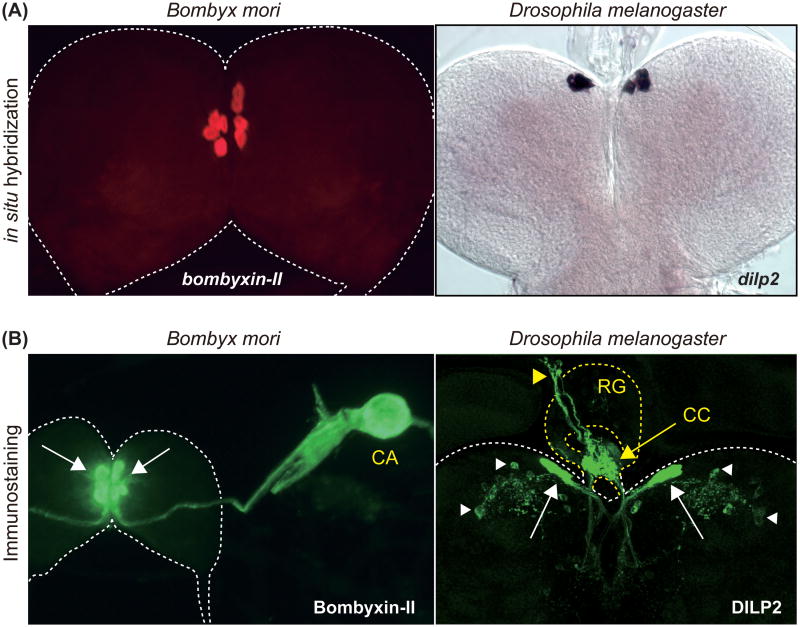
In Bombyx, bombyxins produced by mNSCs are axonally transported to and released from the neurohemal organ called the corpora allata (CA) [16] (Figure 2). In Drosophila, mNSCs extend processes to the dorsal vessel (insect heart) [26], allowing the direct release of DILPs into the circulating hemolymph. DILPs produced by brain mNSCs are also axonally transported to the corpora cardiaca (CC), a pair of neurohemal organs in the ring gland. A recent study shows that the mNSC-derived DILP2 is also released within the brain and received by other specific sets of neurons, including several neurosecretory cells [27]. As in Bombyx and Drosophila, ILPs are produced in brain mNSCs in other insect species, including Aedes [28, 29], the desert locust Schistocerca gregaria [30], and the migratory brown planthopper Nilaparvata lugens [31**], suggesting critical and evolutionary conserved functions of mNSC-derived ILPs in insects.
Figure 2.
Insulin-like peptides are mainly produced by brain mNSCs in insects. (A) Detection of bombyxin-II and dilp2 mRNA in the larval brain by in situ hybridization. bombyxin-II and dilp2 expression is observed in four and seven pairs of mNSCs in Bombyx and Drosophila, respectively. (B) Detection of Bombyxin-II and DILP2 localization in the larval brain by immunostaining. Bombyxin-II produced by mNSCs (white arrows) are axonally transported to the CA. DILP2 produced by mNSCs (white arrows) are axonally transported to the CC (yellow arrow) on the ring gland, and further transported to the dorsal vessel (yellow arrowhead). DILP2 signal can also be detected in specific sets of neurons within the brain (white arrowhead). CA, corpora allata; CC, corpora cardiaca; RG, ring gland.
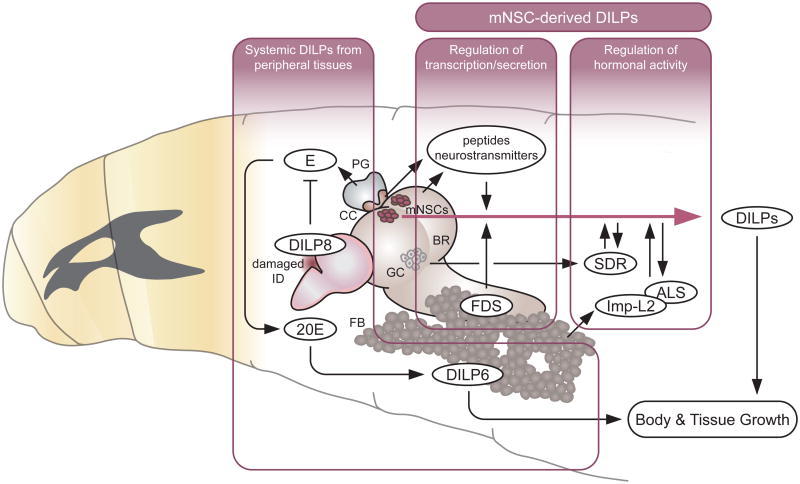
In Drosophila, as in mammals, the secretion and transcription of each DILP are regulated directly or indirectly by multiple cues, including by several hormones and neurotransmitters [32, 15] (Figure 3). The secretion of DILPs from mNSCs highly depends on the nutritional condition. During Drosophila larval development, the major nutrient for systemic growth and development is amino acids. The availability of amino acids is mainly sensed by the fat body [33], a functional equivalent of the vertebrate liver and adipocytes, which in turn remotely regulates DILP secretion from brain mNSCs through unknown humoral signals called fat body-derived signals (FDSs) [34]. In addition to the amino acid-inducible FDSs, a fat body-derived leptin-like protein called Unpaired 2 (Upd2) acts through GABAergic neurons to stimulate the secretion of DILPs in response to high-fat and high-sugar diets [35**]. Moreover, a fat body-derived small peptide called CCHamide-2 directly activates brain mNSCs to modulate DILP secretion primarily in response to glucose [36]. It has been suggested that the secretion of all DILPs is simultaneously induced by the depolarization of mNSCs [34]. However, a recent study demonstrated that the secretion of DILP3 is selectively stimulated by sugar during larval development [37*]. Although DILP2 and DILP5 secretion responds to amino acids [34], sugar stimulates the CC to release adipokinetic hormone (AKH), which acts directly on the mNSCs to promote the secretion of DILP3 [37*]. In addition to the hormones, dietary lipids derived from yeast can be a nutritional signal to regulate the release of DILP2 into hemolymph by modulating the activity of specific neurons in the brain [38].
Figure 3.
Systemic function of DILPs during larval development and its regulation by multiple factors (see text for details).
DILP, Drosophila insulin-like peptide; E, ecdysone; 20E, 20-hydroxyecdysone (active form of ecdysone); FDS, fat body-derived signal; SDR, secreted decoy of insulin receptor; Imp-L2, ecdysone-inducible gene L2; ALS, acid-labile subunit; mNSCs, median neurosecretory cells; BR, brain; GC, glial cells; PG, prothoracic gland; CC, corpora cardiaca; ID, imaginal discs; FB, fat body.
There are additional nutrient-dependent signals that affect mNSCs in adult Drosophila. In the adult fly, the CC also produce Limostatin (Lst), a peptide hormone that suppresses the secretion of DILPs from mNSCs [39]. Moreover, similar to the insulin release in mammals, a recent paper demonstrated that the secretion of DILPs is regulated by the direct sensing of glucose by GLUT1, the type-1 glucose transporter, which is expressed in mNSCs in the adult fly [40]. In Bombyx, glucose also stimulates the secretion of bombyxin into the hemolymph [41], suggesting the possibility that common ancestral mechanisms control the secretion of ILPs from insect mNSCs and mammalian, β -cells.
It has been suggested that the secretion and transcription of DILPs in mNSCs are regulated by different mechanisms [34, 40]. The transcription of each dilp gene in mNSCs is independently regulated [42], although a compensatory expression of dilp genes has also been demonstrated [43, 44*]. dilp2 expression in mNSCs is already detectable in the late embryonic stage, whereas the expression levels of dilp5 and -3 are upregulated in the 2nd and 3rd instar, respectively [42]. Importantly, in both larval and adult stages, dilp5 transcription is tightly regulated in a nutrient-dependent manner [42, 45, 34]. It has been shown that the transcription factors Eyeless (Ey) and Dachshund (Dac) synergistically and directly promote dilp5 expression in mNSCs [46, 47]. However, how these two transcription factors are involved in the nutrient-dependent expression of dilp5 remains unclear. Interestingly, Dach1/2 and Pax6, vertebrate homologs of the Drosophila Dac and Ey, have similar combinatorial effects on the activation of insulin expression in a mammalian β -cell-derived cell line [47].
Regulation of growth by the systemic and local action of insulin-like peptides during development
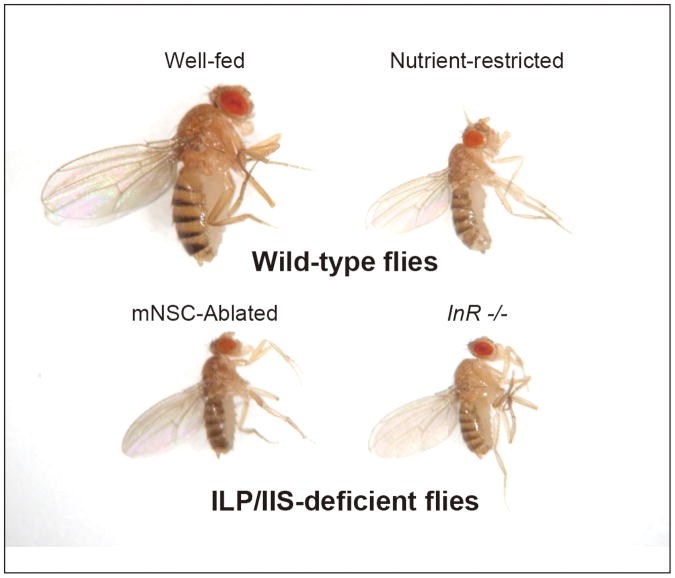
In insects, the larval feeding period is the specialized stage for systemic body growth. During this period, ILPs produced in mNSCs function as key signals that couple systemic and tissue-specific growth with nutritional availability. In Drosophila, for example, larval growth is severely retarded and the adult body size is significantly reduced, as if they are starved when mNSCs are genetically ablated [26], all mNSC-derived dilps are knocked out [44*], or InR is mutated (Figure 4). This phenotypic similarity clearly indicates that ILPs secreted from mNSCs are the major circulating hormones that activate InR to promote body growth during larval feeding period. In addition to the systemic growth-promoting effect, systemically-circulating DILPs also affect specific cell/tissue growth, such as tracheal branching [48] and stem/progenitor cell maintenance and proliferation in the lymph gland [49] during larval development. The requirement of systemic ILPs from mNSCs for germline stem cell maintenance and proliferation as well as ovarian development has also been reported in adult flies and mosquitoes [50, 29, 8].
Figure 4.
Nutrient-restricted or ILP/IIS-deficient flies show severe growth defect. Wild-type flies, wild-type adult female flies raised either on a nutrient-rich diet (Well-fed) or low-protein diet (Nutrient-restricted). ILP/IIS-deficient flies, a brain mNSC-ablated female fly (DILP-producing mNSCs in the brain were genetically ablated using a dilp2 promoter to express the pro-apoptotic gene, reaper) and an InR hypomorphic mutant female fly.
In holometabolous insects, many of the developing adult tissues, including the imaginal discs and primordia, undergo growth and differentiation during the wandering and pupal stages after larvae have stopped feeding [51]. During this period, the insect steroid hormone ecdysone plays critical roles in regulating adult tissue growth and differentiation. However, ILPs produced from the fat body also function, at least partially, as key systemic signals to regulate tissue and body growth. It has been shown in both Bombyx and Drosophila that the fat body predominantly produces ILPs called BIGFLP and DILP6, both of which are structurally classified as IGF-like peptides, during the post-feeding period in response to ecdysone [19, 52, 53] (Figure 3). dilp6 mutants show an approximately 10% reduction in the final adult body size and weight [52, 53, 44*], indicating that some additional systemic growth occurs during the post-feeding period by utilizing stored nutrients in the fat body that were accumulated during the larval feeding period. Therefore, even after larvae stop feeding, fat body-derived ILPs play critical roles in regulating growth in a systemic manner.
The sensitivity to ILPs or IIS differs among developmental stages and tissues, and these differences can control the relative sizes of the adult tissues [54, 55*]. In the butterfly Precis coenia and the tobacco hornworm Manduca sexta, the wing discs change their sensitivity to nutrition or IIS with developmental stages [56, 57]. These differences can be explained by the changes in ecdysone titer during the final larval stage, since ILPs and ecdysone act through separate but synergistic pathways to modulate the growth of their wing discs in vitro [58, 59]. In addition, different organs show different sensitivities to IIS during development. In Drosophila, the genital disc and the wing disc differ in their sensitivity to IIS [55*]. Similarly, in the rhinoceros beetle Trypoxylus dichotomus, knockdown of InR, specifically during the post-feeding period, caused a 16% reduction in horn length in the adult, but caused only a 2% reduction in wing length and no reduction in the genitalia [60**]. In Bombyx, BIGFLP promotes the growth of adult imaginal discs or primordia, but not of larval tissues, which are degenerated or reconstructed during metamorphosis in vitro [19]. In this way, change in the relative size of adult tissues is caused by developmental stage- and tissue-specific modifications in their response to ILPs or IIS.
Recently, DILP8 was identified as a humoral factor that is released from growth-retarded or damaged imaginal discs to inhibit ecdysone production and systemic body growth [21**, 22**]. DILP8 mutation abolishes the developmental delay caused by imaginal disc growth perturbation, while DILP8 overexpression delays the pupariation timing. These unique functions of DILP8 clearly distinguish this ILP from other ILPs and indicate that this peptide may activate a receptor other than InR, as discussed above.
Insect ILPs not only act systemically but also in a local manner [61, 62]. Recent studies have demonstrated that DILPs act locally within the central nervous system in a nutrient-dependent manner [63**, 64**, 65, 66*]. A subset of glial cells produce DILP6 during the larval feeding period, and its expression is inhibited by starvation. This nutrient-dependent expression and/or secretion of DILP6 activates the IIS in adjacent neural stem cells called neuroblasts, thereby leading to their exit from quiescence [63**, 64**, 66*]. Moreover, DILP6 has also been suggested to regulate the proliferation of perineural and cortex glia in the larval brain [65]. In the adult fly, midgut DILP3 functions as a local signal to regulate intestinal stem cell proliferation and growth in a nutrient-dependent manner [67]. In Bombyx, the ovariole sheath, which wraps around an array of follicles, produces BIGFLP during pupa-adult development [68] and may regulate early follicular growth in a paracrine manner.
Regulatory mechanisms of the activities of insulin-like peptides during development
In mammals, six classic IGF-binding proteins (IGFBPs) bind IGFs with high affinity and act as modulators of IGF activity. They act either to enhance or to dampen IIS by extending the half-life of IGFs, by changing their local and systemic availability, or by preventing them from binding to their receptor [69]. IGFBP3 can also interact with a third protein, called the acid-labile subunit (ALS), to form a trimeric complex in circulating blood. In addition, an IGFBP-related protein IGFBP7 (or IGFBP-rP), which shares approximately 30% similarity with IGFBP1 and IGFBP6 in its N-terminal domain, binds IGFs with a comparatively low affinity and also binds to the IGF-I receptor to act as a potent tumor suppressor in a wide variety of cancers [70, 71].
Insects also possess IGFBP-like proteins, including Neuroparsins [72] and Imp-L2 [73], which resemble mammalian IGFBP7. Although Neuroparsins and Imp-L2 show sequence homology to IGFBP7, this homology is restricted to different domains (Neuroparsins show similarity with the N-terminal domain, whereas Imp-L2 shows similarity with the C-terminal domain of IGFBP7). Therefore, Neuroparsins and Imp-L2 may have evolved from a common ancestral IGFBP7-like protein, although their functions are different. In Schistocerca, Neuroparsins directly bind to ILP (Scg-IRP) in vitro [30], suggesting that Neuroparsins act as potential modulators of ILP function. However, a recent study has shown that the mosquito neuroparsin-like factor called ovary ecdysteroidogenic hormone (OEH) promotes egg formation in parallel with ILP(s) by activating an RTK distinct from InR in Aedes [74*]. In Drosophila, Imp-L2 binds to circulating DILP2 and 5 and acts as a systemic inhibitor of IIS during development [75, 76] (Figure 3). Within the central nervous system, however, Imp-L2 functions as a positive regulator of DILP2-mediated IIS in some specific neurons [27]. Furthermore, two recent studies have demonstrated that Imp-L2 is secreted from tumors and creates insulin resistance in distant tissues, which drives a systemic wasting response in the adult fly [77, 78]. In parallel with mammals, the Drosophila homolog of ALS forms a trimeric complex with Imp-L2 and DILP2 in the circulating hemolymph, which inhibits IIS during larval development [79] (Figure 3).
In addition to IGFBP-like proteins, a secreted decoy of InR (SDR) that is structurally similar to the extracellular domain of InR has been identified in Drosophila [80*]. Like Imp-L2 and ALS, the secreted protein SDR can directly interact with several circulating DILPs to antagonize their activity during larval development (Figure 3). Phylogenetic analysis has shown that SDR is most closely related to InR among all Drosophila RTKs [80*], suggesting that SDR is duplicated from InR, but functions as a decoy receptor to negatively regulate IIS.
Although most insects including Drosophila only have a single InR gene, some insects have two InR genes. A recent study showed that two InR genes (InR1 and InR2) in the planthopper Nilaparvata have opposite functions [31**]. In the developing Nilaparvata wing, InR1 activates canonical IIS and leads to the development of long-winged adults. However, InR2 physically binds to InR1 and inhibits the function of InR1. This inhibition shuts down the IIS in the developing wing and leads to the development of short-winged adults. It is possible that a similar regulatory mechanism is conserved in insects with two InR genes, such as Tribolium (TcInR1 and -2) and the honey bee Apis mellifera (AmInR1 and -2) [8].
Conclusion
Over the last decade, both genetic and biochemical analyses of the functions of insect ILPs have advanced our understanding of how animals coordinate their growth and metabolism, as well as how different cells/tissues communicate in response to nutrition. Although only a few model insects, such as Drosophila, have been used in such studies, evolutionarily and ecologically diversified insects can possess significant differences in the nutritional regulation of ILP functions. The recent application of powerful genetics such as RNAi, TALEN and CRISPR/Cas9 technologies in non-model insects should offer great opportunities for exploring new concepts and principles in the nutrition-dependent control of insect development in the future.
Highlights.
- Insulin-like peptides (ILPs) are encoded by multiple genes in insects.
- Insect ILPs are mainly produced by the brain medial neurosecretory cells (mNSCs).
- Transcription and secretion of ILPs are regulated by multiple nutritional signals.
- Insect ILPs regulate body and tissue growth by systemic as well as local actions.
- Activities of secreted ILPs are regulated by several binding proteins in hemolymph.
Acknowledgments
N. O. is supported by a JSPS Postdoctoral fellowship for Research Abroad and the Naito Foundation. N.Y. is supported by NIH grant R00 HD073239 from NICHD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nakae J, Kido Y, Accili D. Distinct and overlapping functions of insulin and IGF-I receptors. Endocr Rev. 2001;22:818–835. doi: 10.1210/edrv.22.6.0452. [DOI] [PubMed] [Google Scholar]
- 2.Saltiel AR, Kahn CR. Insulin signaling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 3.Stewart CE, Rotwein P. Growth, differentiation, and survival: multiple physiological functions for insulin-like growth factors. Physiol Rev. 1996;76:1005–26. doi: 10.1152/physrev.1996.76.4.1005. [DOI] [PubMed] [Google Scholar]
- 4.Taguchi A, White MF. Insulin-like signaling, nutrient homeostasis, and life-span. Annu Rev Physiol. 2008;70:191–212. doi: 10.1146/annurev.physiol.70.113006.100533. [DOI] [PubMed] [Google Scholar]
- 5.Prentki M, Matschinsky FM, Madiraju SR. Metabolic signaling in fuel-induced insulin secretion. Cell Metab. 2013;18:162–185. doi: 10.1016/j.cmet.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 6.Thissen JP, Ketelslegers JM, Underwood LE. Nutritional regulation of the insulin-like growth factors. Endocr Rev. 1994;15:80–101. doi: 10.1210/edrv-15-1-80. [DOI] [PubMed] [Google Scholar]
- 7.Bathgate RAD, Halls ML, van der Westhuizen ET, Callander GE, Kocan M, Summers RJ. Relaxin family peptides and their receptors. Physiol Rev. 2013;93:405–480. doi: 10.1152/physrev.00001.2012. [DOI] [PubMed] [Google Scholar]
- 8.Antonova Y, Arik AJ, Moore W, Riehle MR, Brown MR. Insulin-like peptides: structure, signaling, and function. In: Gilbert LI, editor. Insect Endocrinology. Elsevier/Academic Press; 2012. pp. 63–92. [Google Scholar]
- 9.Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- 10.Edgar BA. How flies get their size: genetics meets physiology. Nat Rev Genet. 2006;7:907–916. doi: 10.1038/nrg1989. [DOI] [PubMed] [Google Scholar]
- 11.Géminard C, Arquier N, Layalle S, Bourouis M, Slaidina M, Delanoue R, Bjordal M, Ohanna M, Ma M, Colombani J, Léopold P. Control of metabolism and growth through insulin-like peptides in Drosophila. Diabetes. 2006;552(Suppl. 2):S5–8. [Google Scholar]
- 12.Mirth CK, Riddiford LM. Size assessment and growth control: how adult size is determined in insects. Bioessays. 2007;29:344–355. doi: 10.1002/bies.20552. [DOI] [PubMed] [Google Scholar]
- 13.Broughton S, Partridge L. Insulin/IGF-like signalling, the central nervous system and aging. Biochem J. 2009;418:1–12. doi: 10.1042/BJ20082102. [DOI] [PubMed] [Google Scholar]
- 14.Teleman AA. Molecular mechanisms of metabolic regulation by insulin in Drosophila. Biochem J. 2010;425:13–26. doi: 10.1042/BJ20091181. [DOI] [PubMed] [Google Scholar]
- 15.Nãssel DR, Liu Y, Luo J. Insulin/IGF signaling and its regulation in Drosophila. Gen Comp Endocrinol. 2015 doi: 10.1016/j.ygcen.2014.11.021. In press. [DOI] [PubMed] [Google Scholar]
- 16.Mizoguchi A, Okamoto N. Insulin-like and IGF-like peptides in the silkmoth Bombyx mori: discovery, structure, secretion, and function. Front Physiol. 2013;4:217. doi: 10.3389/fphys.2013.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Predel R, Neupert S, Hauser F, Tanaka Y, Cazzamali G, Williamson M, Arakane Y, Verleyen P, Schoofs L, et al. Genomics, transcriptomics, and peptidomics of neuropeptides and protein hormones in the red flour beetle Tribolium castaneum. Genome Res. 2008;18:113–122. doi: 10.1101/gr.6714008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grönke S, Partridge L. The functions of insulin-like peptides in insects. In: Clemmons DR, Robinson ICAF, Christen Y, editors. IGFs: local repair and survival factors throughout life span. Heidelberg: Springer; 2010. pp. 105–124. [Google Scholar]
- 19.Okamoto N, Yamanaka N, Satake H, Saegusa H, Kataoka H, Mizoguchi A. An ecdysteroid-inducible insulin-like growth factor-like peptide regulates adult development of the silkmoth Bombyx mori. FEBS J. 2009;276:1221–1232. doi: 10.1111/j.1742-4658.2008.06859.x. [DOI] [PubMed] [Google Scholar]
- 20.Veenstra JA. Neurohormones and neuropeptides encoded by the genome of Lottia gigantea, with reference to other mollusks and insects. Gen Comp Endocrinol. 2010;167:86–103. doi: 10.1016/j.ygcen.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 21**.Garelli A, Gontijo AM, Miguela V, Caparros E, Dominguez M. Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science. 2012;336:579–582. doi: 10.1126/science.1216735. This paper (and an accompanying paper by Colombani et al. [22]) describes the discovery of a new ILP family member called DILP8 in Drosophila. By using microarray analysis, the authors found that dilp8 expression was increased in tumors in eye discs. They demonstrated that DILP8 is released from tumor or damaged imaginal discs to inhibit ecdysone production (Figure 3). dilp8 mutation abolishes the developmental delay caused by imaginal disc growth perturbation, while dilp8 overexpression delays the pupariation timing. A unique point demonstrated by this study is the function of DILP8 in regulation of individual developmental stability analyzed by left-right wing asymmetry. Readers are also directed to Colombani et al. [22]. [DOI] [PubMed] [Google Scholar]
- 22**.Colombani J, Andersen DS, Léopold P. Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science. 2012;336:582–585. doi: 10.1126/science.1216689. This paper (and an accompanying paper by Garelli et al. [21]) describes the discovery of new ILP family member called DILP8 in Drosophila. By using in vivo RNAi screening, the authors found that the knockdown of dilp8 rescued the developmental delay caused by wing disc growth perturbation. Both this study and Garelli et al. essentially come to the same conclusion that DILP8 is released from growth-retarded or damaged imaginal discs to induce developmental delay by inhibiting ecdysone production (Figure 3). This paper further shows that the possible function of DILP8 as a growth inhibitory endocrine signal that coordinates tissue growth rate. Readers are also directed to Garelli et al. [21]. [DOI] [PubMed] [Google Scholar]
- 23.Katsuyama T, Comoglio F, Seimiya M, Cabuy E, Paro R. During Drosophila disc regeneration, JAK/STAT coordinates cell proliferation with Dilp8-mediated developmental delay. Proc Natl Acad Sci USA. 2015;112:E2327–E2336. doi: 10.1073/pnas.1423074112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veenstra JA. The contribution of the genomes of a termite and a locust to our understanding of insect neuropeptides and neurohormones. Front Physiol. 2014;5:454. doi: 10.3389/fphys.2014.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bathgate RAD, Samuel CS, Burazin TCD, Layfield S, Claasz AA, Reytomas IGT, Dawson NF, Zhao C, Bond C, Summers RJ, et al. Human relaxin gene 3 (H3) and the equivalent mouse relaxin (M3) gene. J Biol Chem. 2002;277:1148–1157. doi: 10.1074/jbc.M107882200. [DOI] [PubMed] [Google Scholar]
- 26.Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- 27.Bader R, Sarraf-Zadeh L, Peters M, Moderau N, Stocker H, Köhler K, Pankratz MJ, Hafen E. The IGFBP7 homolog Imp-L2 promotes insulin signaling in distinct neurons of the Drosophila brain. J Cell Sci. 2013;126:2571–2576. doi: 10.1242/jcs.120261. [DOI] [PubMed] [Google Scholar]
- 28.Riehle MA, Fan Y, Cao C, Brown MR. Molecular characterization of insulin-like peptides in the yellow fever mosquito, Aedes aegypti: expression, cellular localization, and phylogeny. Peptide. 2006;27:2547–2560. doi: 10.1016/j.peptides.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 29.Brown MR, Clark KD, Gulia M, Zhao Z, Garczynski SF, Crim JW, Suderman RJ, Strand MR. An insulin-like peptide regulates egg maturation and metabolism in the mosquito Aedes aegypti. Proc Natl Acad Sci U S A. 2008;105:5716–5721. doi: 10.1073/pnas.0800478105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badisco L, Claeys I, Van Hiel M, Clynen E, Huybrechts J, Vandersmissen T, Van Soest S, Vanden Bosch L, Simonet G, Vanden Broeck J. Purification and characterization of an insulin-related peptide in the desert locust, Schistocerca gregaria: immunolocalization, cDNA cloning, transcript profiling and interaction with neuroparsin. J Mol Endocrinol. 2008;40:137–150. doi: 10.1677/JME-07-0161. [DOI] [PubMed] [Google Scholar]
- 31**.Xu HJ, Xue J, Lu B, Zhang XC, Zhuo JC, He SF, Ma XF, Jiang YQ, Fan HW, Xu JY, et al. Two insulin receptors determine alternative wing morphs in planthoppers. Nature. 2015;519:464–467. doi: 10.1038/nature14286. This elegant study shows the mechanisms of how the migratory brown planthoppers Nilaparvata can grow up to have either short or long wings, depending on environmental cues such as day length and temperature. Although most insect genomes only have a single InR gene, the authors found two InR genes, InR1 and InR2, in the brown planthopper. Surprisingly, the authors discovered that InR2 physically binds to InR1 and shuts down the insulin/IGF signaling in developing wings, leading to short-winged adults. How InR2 activity is regulated by environmental cues is an open question. [DOI] [PubMed] [Google Scholar]
- 32.Nässel DR, Kubrak OA, Liu Y, Luo J, Lushchak OV. Factors that regulate insulin producing cells and their output in Drosophila. Front Physiol. 2013;4:252. doi: 10.3389/fphys.2013.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Léopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114:739–749. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- 34.Géminard C, Rulifson EJ, Léopold P. Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. 2009;10:199–207. doi: 10.1016/j.cmet.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 35**.Rajan A, Perrimon N. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell. 2012;151:123–137. doi: 10.1016/j.cell.2012.08.019. This study is the first description of the mechanism of how a fat body-derived signal (FDS) regulates the secretion of DILPs from brain mNSCs. In Drosophila, it has been proposed that the fat body remotely regulates DILP secretion from brain mNSCs through unknown humoral signals [34]. The authors identified a leptin-like protein called Unpaired 2 (Upd2) as an FDS. Upd2 activates JAK/STAT signaling in GABAergic neurons that project onto mNSCs and modulates DILP secretion. Interestingly, Upd2 is produced by the fat body in response to high-fat and high-sugar diets but not to a protein-rich diet, suggesting that different nutrient-specific FDSs exist in Drosophila. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sano H, Nakamura A, Texada MJ, Truman JW, Ishimoto H, Kamikouchi A, Nibu Y, Kume K, Ida T, Kojima M. The nutrient-responsive hormone CCHamide-2 controls growth by regulating insulin-like peptides in the brain of Drosophila melanogaster. PLoS Genet. 2015;11:e1005209. doi: 10.1371/journal.pgen.1005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37*.Kim J, Neufeld TP. Dietary sugar promotes systemic TOR activation in Drosophila through AKH-dependent selective secretion of Dilp3. Nat Commun. 2015;6:4846. doi: 10.1038/ncomms7846. This paper shows the first evidence that the secretion of DILPs from brain mNSCs is selectively regulated by different nutritional cues. It has been suggested that the secretion of all DILPs is simultaneously induced by the depolarization of mNSCs [34]. Although the secretion of DILP2 and DILP5 responds to amino acids [34], the authors found that the secretion of DILP3 is selectively stimulated by sugar. Importantly, sugar stimulates the corpora cardiaca (CC) to release the adipokinetic hormone (AKH), which in turn acts on mNSCs to promote the secretion of DILP3. CC also produce Limostatin, a newly identified peptide hormone that suppresses DILP secretion from mNSCs in the adult fly [39]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brankatschk M, Dunst S, Nemetschke L, Eaton S. Delivery of circulating lipoproteins to specific neurons in the Drosophila brain regulates systemic insulin signaling. Elife. 2014;3:e02862. doi: 10.7554/eLife.02862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alfa RW, Park S, Skelly KR, Poffenberger G, Jain N, Gu X, Kockel L, Wang J, Liu Y, Powers AC, et al. Suppression of insulin production and secretion by a decretin hormone. Cell Metab. 2015;21:323–333. doi: 10.1016/j.cmet.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park S, Alfa RW, Topper SM, Kim GE, Kockel L, Kim SK. A genetic strategy to measure circulating Drosophila insulin reveals genes regulating insulin production and secretion. PLoS Genet. 2014;10:e1004555. doi: 10.1371/journal.pgen.1004555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masumura M, Satake SI, Saegusa H, Mizoguchi A. Glucose stimulates the release of bombyxin, an insulin-related peptide of the silkworm Bombyx mori. Gen Comp Endocrinol. 2000;118:393–399. doi: 10.1006/gcen.1999.7438. [DOI] [PubMed] [Google Scholar]
- 42.Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr Biol. 2002;12:1293–1300. doi: 10.1016/s0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- 43.Broughton S, Alic N, Slack C, Bass T, Ikeya T, Vinti G, Tommasi AM, Driege Y, Hafen E, Partridge L. Reduction of DILP2 in Drosophila triages a metabolic phenotype from lifespan revealing redundancy and compensation among DILPs. PLoS ONE. 2008;3:e3721. doi: 10.1371/journal.pone.0003721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44*.Grönke S, Clarke DF, Broughton S, Andrews TD, Partridge L. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 2010;6:e1000857. doi: 10.1371/journal.pgen.1000857. The authors generated mutants for all dilp genes (except dilp8, which was not identified at that time), and systematically and extensively analyzed their phenotypes including body growth, metabolism, stress resistance, lifespan and fecundity in Drosophila. This study finds synergy, redundancy, compensation and functional differentiation between each DILP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Min KJ, Yamamoto R, Buch S, Pankratz M, Tatar M. Drosophila lifespan control by dietary restriction independent of insulin-like signaling. Aging Cell. 2008;7:199–206. doi: 10.1111/j.1474-9726.2008.00373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clements J, Hens K, Francis C, Schellens A, Callaerts P. Conserved role for the Drosophila Pax6 homolog Eyeless in differentiation and function of insulin-producing neurons. Proc Natl Acad Sci U S A. 2008;105:16183–16188. doi: 10.1073/pnas.0708330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okamoto N, Nishimori Y, Nishimura T. Conserved role for the Dachshund protein with Drosophila Pax6 homolog Eyeless in insulin expression. Proc Natl Acad Sci USA. 2012;109:2406–2411. doi: 10.1073/pnas.1116050109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Linneweber G, Jacobson J, Busch KE, Hudry B, Christov CP, Dormann D, Yuan M, Otani T, Knust E, de Bono M, et al. Neuronal control of metabolism through nutrient-dependent modulation of tracheal branching. Cell. 2014;156:69–83. doi: 10.1016/j.cell.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shim J, Mukherjee T, Banerjee U. Direct sensing of systemic and nutritional signals by haematopoietic progenitors in Drosophila. Nat Cell Biol. 2012;14:394–400. doi: 10.1038/ncb2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ables ET, Laws KM, Drummond-Barbosa D. Control of adult stem cells in vivo by a dynamic physiological environment: diet-dependent systemic factors in Drosophila and beyond. WIREs Dev Biol. 2012;1:657–674. doi: 10.1002/wdev.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nijhout HF, Riddiford LM, Mirth C, Shingleton AW, Suzuki Y, Callier V. The developmental control of size in insects. WIREs Dev Biol. 2013;3:113–134. doi: 10.1002/wdev.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okamoto N, Yamanaka N, Yagi Y, Nishida Y, Kataoka H, O'Connor MB, Mizoguchi A. A fat body-derived IGF-like peptide regulates postfeeding growth in Drosophila. Dev Cell. 2009;17:885–891. doi: 10.1016/j.devcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slaidina M, Delanoue R, Grönke S, Partridge L, Léopold P. A Drosophila insulinlike peptide promotes growth during nonfeeding states. Dev Cell. 2009;17:874–884. doi: 10.1016/j.devcel.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shingleton AW, Estep CM, Driscoll MV, Dworkin I. Many ways to be small: different environmental regulators of size generate distinct scaling relationships in Drosophila melanogaster. Proc R Soc B. 2009;276:2625–2633. doi: 10.1098/rspb.2008.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55*.Tang HY, Smith-Caldas MSB, Driscoll MV, Salhadar S, Shingleton AW. FOXO regulates organ-specific phenotypic plasticity in Drosophila. PLoS Genet. 2011;7:e1002373. doi: 10.1371/journal.pgen.1002373. The authors previously showed that genitalia resist the reduction in size that is caused by the loss of nutrition and insulin/IGF signaling components in Drosophila [54]. This study shows that the expression level of the transcription factor FoxO is important to maintain tissue-specific nutritional plasticity and insulin sensitivity. This work suggests that the extent of phenotypic plasticity is caused by changes in the expression of genes that couple nutritional status to tissue growth. Readers are also directed to Emlen et al. [60]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miner AL, Rosenberg AJ, Nijhout HF. Control of growth and differentiation of the wing imaginal disk of Precis coenia (Lepidoptera: Nymphalidae) J Insect Physiol. 2000;46:251–258. doi: 10.1016/s0022-1910(99)00177-8. [DOI] [PubMed] [Google Scholar]
- 57.Tobler A, Nijhout HF. A Switch in the Control of Growth of the Wing Imaginal Disks of Manduca sexta. PLoS ONE. 2010;5:e10723. doi: 10.1371/journal.pone.0010723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nijhout HF, Grunert LW. Bombyxin is a growth factor for wing imaginal disks in Lepidoptera. Proc Natl Acad Sci USA. 2002;99:15446–15450. doi: 10.1073/pnas.242548399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nijhout HF, Smith WA, Schachar I, Subramanian S, Tobler A, Grunert LW. The control of growth and differentiation of the wing imaginal disks of Manduca sexta. Dev Biol. 2007;302:569–576. doi: 10.1016/j.ydbio.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 60**.Emlen DJ, Warren IA, Johns A, Dworkin I, Lavine LC. A mechanism of extreme growth and reliable signaling in sexually selected ornaments and weapons. Science. 2012;337:860–864. doi: 10.1126/science.1224286. This study clearly shows how the sensitivity of insulin/IGF signaling can affect the nutrient-dependent tissue growth in the male rhinoceros beetle Trypoxylus dichotomus. In rhinoceros beetles, growth of the horn, a sexually selected weapon, is more sensitive to larval nutrition than other tissues such as wings and genitalia. The authors found that knockdown of InR shows little or moderate effects on genital and wing size, whereas it shows more significant effects on horn size. The authors also make an interesting argument on why and how trait exaggeration has evolved in the context of sexual selection. [DOI] [PubMed] [Google Scholar]
- 61.Shim J, Gururaja-Rao S, Banerjee U. Nutritional regulation of stem and progenitor cells in Drosophila. Development. 2013;140:4647–4656. doi: 10.1242/dev.079087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu J, Spéder P, Brand AH. Control of brain development and homeostasis by local and systemic insulin signaling. Diabetes Obes Metab. 2014;16:16–20. doi: 10.1111/dom.12337. [DOI] [PubMed] [Google Scholar]
- 63**.Chell JM, Brand AH. Nutrition-responsive glia control exit of neural stem cells from quiescence. Cell. 2011;143:1161–1173. doi: 10.1016/j.cell.2010.12.007. This study is the first intensive description of the mechanisms of how neuroblast reactivation is triggered by nutritional signals. The authors found that DILP2 and DILP6 are expressed in the nutrient-dependent manner by glial cells overlying the quiescent neuroblasts. In addition, they found that the insulin/IGF signaling pathway in neuroblasts trigger them to exit quiescence. See also Sousa-Nunes et al. [64]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64**.Sousa-Nunes R, Yee LL, Gould AP. Fat cells reactivate quiescent neuroblasts via TOR and glial insulin relays in Drosophila. Nature. 2011;471:508–512. doi: 10.1038/nature09867. This study clearly shows the local function of DILPs within the central nervous system. The authors found that nutrient-dependent production of DILPs in glial cells regulates neuroblast reactivation but not systemic larval growth. This study also demonstrates the existence of amino acid-inducible fat body-derived signals (FDSs) required for neuroblast reactivation. See also Chell and Brand [63]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Avet-Rochex A, Kaul AK, Gatt AP, McNeill H, Bateman JM. Concerted control of gliogenesis by InR/TOR and FGF signalling in the Drosophila post-embryonic brain. Development. 2012;139:2763–2772. doi: 10.1242/dev.074179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66*.Spéder P, Brand AH. Gap junction proteins in the blood-brain barrier control nutrient-dependent reactivation of Drosophila neural stem cells. Dev Cell. 2014;30:1–13. doi: 10.1016/j.devcel.2014.05.021. In addition to the previous study by Chell and Brand [63], this work shows the mechanisms of how neuroblast reactivation is triggered by glia-derived DILP6. Although the full implications of this study were not included in our review, the authors provide interesting data regarding the functions of gap junctions in the surface glial cells. They show that two Drosophila innexins, Ogre and Inx2, are required for calcium oscillations in surface glial cells to regulate the transcriptional status as well as secretory abilities of DILP6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O'Brien LE, Soliman SS, Li X, Bilder D. Altered modes of stem cell division drive adaptive intestinal growth. Cell. 2011;147:603–614. doi: 10.1016/j.cell.2011.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Okamoto N, Yamanaka N, Endo Y, Kataoka H, Mizoguchi A. Spatiotemporal patterns of IGF-like peptide expression in the silkmoth Bombyx mori predict its pleiotropic actions. Gen Comp Endocrinol. 2011;173:171–182. doi: 10.1016/j.ygcen.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 69.Hwa V, Oh Y, Rosenfeld RG. The insulin-like growth factor-binding protein (IGFBP) superfamily. Endocr Rev. 1999;20:761–787. doi: 10.1210/edrv.20.6.0382. [DOI] [PubMed] [Google Scholar]
- 70.Burger AM, Leyland-Jones B, Banerjee K, Spyropoulos DD, Seth AK. Essential roles of IGFBP-3 and IGFBP-rP1 in breast cancer. Eur J Cancer. 2005;41:1515–1527. doi: 10.1016/j.ejca.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 71.Evdokimova V, Tognon CE, Benatar T, Yang W, Krutikov K, Pollak M, Sorensen PH, Seth A. IGFBP7 binds to the IGF-1 receptor and blocks its activation by insulin-like growth factors. Sci Signal. 2012:5–ra92. doi: 10.1126/scisignal.2003184. [DOI] [PubMed] [Google Scholar]
- 72.Badisco L, Claeys I, Van Loy T, Van Hiel M, Franssens V, Simonet G, Vanden Broeck J. Neuroparsins, a family of conserved arthropod neuropeptides. Gen Comp Endocrinol. 2007;153:64–71. doi: 10.1016/j.ygcen.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 73.Sloth Andersen A, Hertz Hansen P, Schaffer L, Kristensen C. A new secreted insect protein belonging to the immunoglobulin superfamily binds insulin and related peptides and inhibits their activities. J Biol Chem. 2000;275:16948–16953. doi: 10.1074/jbc.M001578200. [DOI] [PubMed] [Google Scholar]
- 74*.Vogal KJ, Brown MR, Strand MR. Ovary ecdysteroidogenic hormone requires a receptor tyrosine kinase to activate egg formation in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2015;112:5057–5062. doi: 10.1073/pnas.1501814112. This paper describes the discovery of the receptor of a neuroparsin-like factor called ovary ecdysteroidogenic hormone (OEH) in the yellow fever mosquito Aedes aegypti. Because of its partial sequence similarity with vertebrate IGF-binding proteins, neuroparsins were initially suggested to function as ILP binding proteins, which could affect ILP binding to InR. However, the authors found that OEH activates ovaries to produce ecdysteroid independently of InR by binding to another receptor tyrosine kinase (RTK). Surprisingly, this OEH receptor can be found in mosquito genomes as well as the genome of Drosophila mojavensis, but not in that of Drosophila melanogaster. This is consistent with the previous knowledge that D. mojavensis and most other Drosophila species have a neuroparsin gene in their genomes, but melanogaster subgroup species do not. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Honegger B, Galic M, Köhler K, Wittwer F, Brogiolo W, Hafen E, Stocker H. Imp-L2, a putative homolog of vertebrate IGF-binding protein 7, counteracts insulin signaling in Drosophila and is essential for starvation resistance. J Biol. 2008;7:10. doi: 10.1186/jbiol72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alic N, Hoddinott MP, Vinti G, Partridge L. Lifespan extension by increased expression of the Drosophila homologue of the IGFBP7 tumour suppressor. Aging Cell. 2011;10:137–147. doi: 10.1111/j.1474-9726.2010.00653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kwon Y, Song W, Droujinine IA, Hu Y, Asara JM, Perrimon N. Systemic organ wasting induced by localized expression of the secreted insulin/IGF antagonist ImpL2. Dev Cell. 2015;33:36–46. doi: 10.1016/j.devcel.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Figueroa-Clarevega A, Bilder D. Malignant Drosophila tumors interrupt insulin signaling to induce cachexia-like wasting. Dev Cell. 2015;33:47–55. doi: 10.1016/j.devcel.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arquier N, Géminard C, Bourouis M, Jarretou G, Honegger B, Paix A, Léopold P. Drosophila ALS regulates growth and metabolism through functional interaction with insulin-like peptides. Cell Metab. 2008;7:333–338. doi: 10.1016/j.cmet.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 80*.Okamoto N, Nakamori R, Murai T, Yamauchi Y, Masuda A, Nishimura T. A secreted decoy of InR antagonizes insulin/IGF signaling to restrict body growth in Drosophila. Genes Dev. 2013;27:87–97. doi: 10.1101/gad.204479.112. The authors identified a novel ILP-binding protein named secreted decoy of InR (SDR) that acts as a negative regulator of insulin/IGF signaling in Drosophila. Interestingly, SDR encodes a protein that shows high similarities with the extracellular domain of InR. It will be interesting to analyze the functional relationship and differences between SDR and the other ILP-binding protein complex Imp-L2/ALS [75, 76, 79]. [DOI] [PMC free article] [PubMed] [Google Scholar]