Abstract
Signal transducer and activator of transcription 3 (STAT3) is constitutively activated in many cancers where it acts to promote tumor progression. A STAT3-specific transcription factor decoy has been developed to suppress STAT3 downstream signaling, but a delivery strategy is needed to improve clinical translation. Ultrasound-targeted microbubble destruction (UTMD) has been shown to enhance image-guided local delivery of molecular therapeutics to a target site. The objective of this study was to deliver STAT3 decoy to squamous cell carcinoma (SCC) tumors using UTMD to disrupt STAT3 signaling and inhibit tumor growth. Studies performed demonstrated that UTMD treatment with STAT3 decoy-loaded microbubbles inhibited STAT3 signaling in SCC cells in vitro. Studies performed in vivo demonstrated that UTMD treatment with STAT3 decoy-loaded microbubbles induced significant tumor growth inhibition (31-51% reduced tumor volume vs. controls, p < 0.05) in mice bearing SCC tumors. Furthermore, expression of STAT3 downstream target genes (Bcl-xL and cyclin D1) was significantly reduced (34-39%, p < 0.05) in tumors receiving UTMD treatment with STAT3 decoy-loaded microbubbles compared to controls. In addition, the quantity of radiolabeled STAT3 decoy detected in tumors eight hours after treatment was significantly higher with UTMD treatment compared to controls (70-150%, p < 0.05). This study demonstrates that UTMD can increase delivery of a transcription factor decoy to tumors in vivo and that the decoy can inhibit STAT3 signaling and tumor growth. These results suggest that UTMD treatment holds potential for clinical use to increase the concentration of a transcription factor signaling inhibitor in the tumor.
Keywords: Ultrasound Targeted Microbubble Destruction, STAT3 Decoy, STAT3 Signaling Knockdown, Tumor Growth Inhibition, Head and Neck Cancer
Introduction
Signal transducer and activator of transcription 3 (STAT3) is a transcription factor that is constitutively activated in a large number of cancers, including breast, head and neck, hepatocellular, lung, ovarian, pancreatic, prostate, and hematological malignancies 1-7. STAT3 is activated by phosphorylation in response to multiple stimuli, including growth factor receptors, cytokine receptors, and tyrosine kinases 8. Activated STAT3 protein dimerizes and translocates to the nucleus where it binds to promoter elements and upregulates expression of STAT3-responsive genes that promote cell proliferation, survival, migration/invasion, angiogenesis, and immune evasion 9-12. Blocking downstream STAT3 signaling has been shown to inhibit tumor progression, thus STAT3 inhibition represents an important therapeutic target and is an unmet clinical need 13-17. A decoy oligonucleotide has been developed that binds STAT3 protein in the cytoplasm or nucleus with high affinity, effectively competing for STAT3 binding to promoter elements in target genes and preventing STAT3 signaling. Studies have demonstrated that this STAT3 decoy is effective in suppressing growth of human head and neck squamous cell carcinoma (HNSCC) tumors in murine xenografts 18. Additionally, intratumoral injection of the STAT3 decoy has been shown to inhibit STAT3 signaling in HNSCC tumors in humans 18, 19. However, a clinically feasible intravenous delivery strategy is needed for translation of the STAT3 decoy therapy to the clinic.
Ultrasound-targeted microbubble destruction (UTMD) is an approach for image-guided local delivery of drugs or nucleic acids to a target site and holds potential for local delivery of STAT3 decoy to tumors. Microbubbles are encapsulated gas-filled microspheres (2-4 µm in diameter) currently approved for clinical use as ultrasound contrast agents for echocardiography. Recent efforts have focused on utilizing microbubbles for therapeutic applications 20-25. Microbubbles expand and contract in response to ultrasound pressure waves due to differences in compressibility between the internal gas core and the surrounding liquid. Above a critical ultrasound pressure threshold, the microbubble begins to expand more than it compresses, leading to non-linear volume oscillation and, if the acoustic pressure is high enough, microbubble collapse (i.e. “inertial cavitation”). Microbubble oscillation can generate microstreaming in the surrounding fluid and induce shear forces on nearby interfaces such as cell membranes 26, 27. Ultrasound stimulation of microbubble cavitation can also enhance vascular permeability. For example, Lin et al. found that ultrasound-stimulated microbubbles enhanced accumulation of 130-nm quantum dots in mouse colorectal adenocarcinomas for up to six hours 28. Kooiman et al. demonstrated that the permeability of a human umbilical vein endothelial layer increased for up to 12 hours following UTMD 29. Furthermore, microbubble cavitation can also induce transient pore formation in cell membranes (sonoporation). Yu, et al., used time-lapse confocal microscopy to visualize sonoporation 30, while Fan, et al., demonstrated uptake of propidium iodide by sonoporated cells 31. All of these effects can potentially act together to enhance delivery of molecular therapeutics to tumor cells upon insonification.
Our group has previously demonstrated UTMD-mediated delivery of nucleic acids to cancer cells in SCC tumors. We have reported UTMD-mediated delivery of a DNA plasmid encoding herpes simplex virus-1 thymidine kinase, a suicide gene, to inhibit the growth of SCC tumors 32. In addition, we have delivered siRNA against epithelial growth factor receptor (EGFR) to murine squamous cell carcinoma tumors using UTMD, resulting in reduced EGFR expression and significant tumor growth inhibition 33. However, to our knowledge UTMD has not been previously used to deliver a transcription factor decoy to tumors in vivo.
The objective of this study was to deliver a STAT3 decoy on cationic lipid-coated microbubbles to squamous cell carcinomas and inhibit STAT3 signaling in vitro and in vivo using UTMD treatment. The effect of STAT3 decoy MB + UTMD treatment on tumor growth and tumor accumulation of STAT3 decoy was also assessed.
Materials and Methods
Microbubble preparation
STAT3 decoy and mutant decoy double-stranded oligonucleotides were purchased from IDT technologies (Coralville, IA, USA). Oligonucleotides were self-annealed and ligated using T4 DNA ligase (New England Biolabs, Ipswich, MA, USA) to form cyclic decoy. The STAT3 decoy sequence and structure, illustrated in Fig. 1, contained two hexa-ethyleneglycol linkages (denoted spacer-18 in sequences below) to generate the completely circularized cyclic decoy in order to increase stability in circulation as previously described 19, 34. The complete sequence was 5´-GTAAATC(spacer-18)GATTTACGGGAAATG(spacer-18)CATTTCCC-3´ and the mutant decoy sequence, which differed by a single nucleotide base pair and served as a negative control, was 5´-TTAAATC(spacer-18)GATTTAAGGGAAATG(spacer-18)CATTTCCC-3´.
Figure 1.
Illustration of cyclic STAT3 decoy structure and sequence. Two hexa-ethyleneglycol linkages were added to generate the completely circularized cyclic decoy structure in order to increase stability in circulation. The mutant decoy contains a G to T substitution in the starred (*) position.
Nucleic acids were loaded onto cationic lipid-coated microbubbles via charge-charge interactions as follows: a lipid formulation of 1,2-distearoyl-sn-glycero-3-phosphocholine, 1,2-distearoyl-sn-glycero-3-ethylphosphocholine, 1,2-distearoyl-sn-glycero-3-phosphoglycerol (Avanti Lipids, Alabaster, AL, USA), and polyethylene glycol-40 stearate (Sigma-Aldrich, St. Louis, MO, USA) was dissolved in chloroform at a mole ratio of 100:43:1:4.5 and dried with argon gas. The dried lipid film was rehydrated to a concentration of 1.7 mg total lipid/mL in PBS containing 1 mM EDTA and sonicated with a Sonicator XL ultrasonic processor (Misonix, Farmingdale, NY, USA) at power setting 5 until lipids were dispersed. The lipid stock solution was diluted 1:4 in PBS/EDTA and added to a vial containing 10 µg of cyclic STAT3 decoy or mutant decoy. The vial was sealed and the headspace was filled with perfluorobutane gas prior to amalgamation using a dental amalgamator to form perfluorobutane gas-containing microbubbles. The microbubbles solution was washed with PBS until the subnatant was clear (2-4 washes at 53 relative centrifugal force), resulting in a microbubble concentration of 1.3×109/mL, a mean diameter of 2.2 ± 1.1 µm as measured with a Coulter Multisizer 3 (Beckman Coulter, Miami, FL, USA), and a zeta potential of 21.1 ± 0.7 mV as measured with a Zetasizer (Malvern Instruments, Malvern, United Kingdom).
Assessment of MB-associated STAT3 decoy stability against DNAse degradation
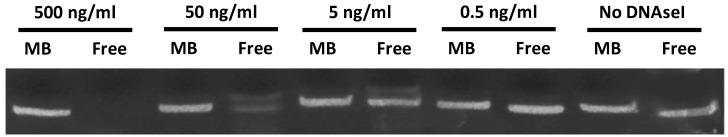
MB-associated cyclic STAT3 decoy and free decoy were incubated in DNAseI for 30 minutes at 37° C in digestion buffer (10 mM Tris, 2.5 mM MgCl2, 0.5 mM CaCl2) and the level of intact decoy was determined using 20% acrylamide-urea gel electrophoresis. MBs loaded with decoy DNA were washed 3 times to remove unbound DNA and exposed to a range of DNAseI concentrations from 0 to 500 ng/ml. As a control, 10 µg free decoy was also challenged with DNAseI at these same concentrations in digestion buffer.
UTMD-mediated STAT3 decoy delivery to SCC cells in vitro
To assess knockdown of STAT3 signaling in vitro, murine squamous cell carcinoma (SCC-VII) cells were transformed using a lentivirus containing a STAT3-responsive luciferase gene (Cignal Lenti Reporter Assays, Qiagen, Venlo, Netherlands) to produce SCC-STAT-luc cells. For in vitro UTMD studies, a 0.2 mL solution of PBS containing 2×108 microbubbles loaded with 2 µg of STAT3 decoy or mutant decoy was infused for five minutes into a 14 mL round-bottom polystyrene tube (BD, Franklin Lakes, NJ, USA) containing a 3 mL suspension of 1×106 SCC-STAT-luc cells while continuously mixing with a custom sample shaker. Ultrasound pulses were delivered for 7 minutes using an S3 transducer probe on a clinical diagnostic ultrasound system (Sonos 7500, Philips, Amsterdam, Netherlands). The sample was placed 5 mm from the transducer. The ultrasound system was operated in ultraharmonic mode (center frequency of 1.3 MHz), with the on-screen mechanical index set to 1.6. The system was time triggered, with 8 frames transmitted per US burst and a burst interval of 0.5 s. Immediately following UTMD, treatment cells were re-seeded in 60 mm2 Petri dishes in a cell incubator and harvested 8 hours later for assessment of luciferase expression. Cells were lysed using reporter lysis buffer and three freeze/thaw cycles. Luciferase activity was measured with a luciferase assay kit (Promega, Madison, WI, USA) and normalized to total protein as measured by Bradford assay (Thermo Fisher Scientific, Waltham, MA, USA). SCC-STAT-Luc cells were transfected with STAT3 decoy or mutant decoy using Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA) as a positive control. For cell viability measurements, trypan blue was added 5 minutes after UTMD and imaged microscopically to determine the percentage of non-viable cells.
UTMD-mediated STAT3 decoy delivery to murine squamous cell carcinoma in vivo
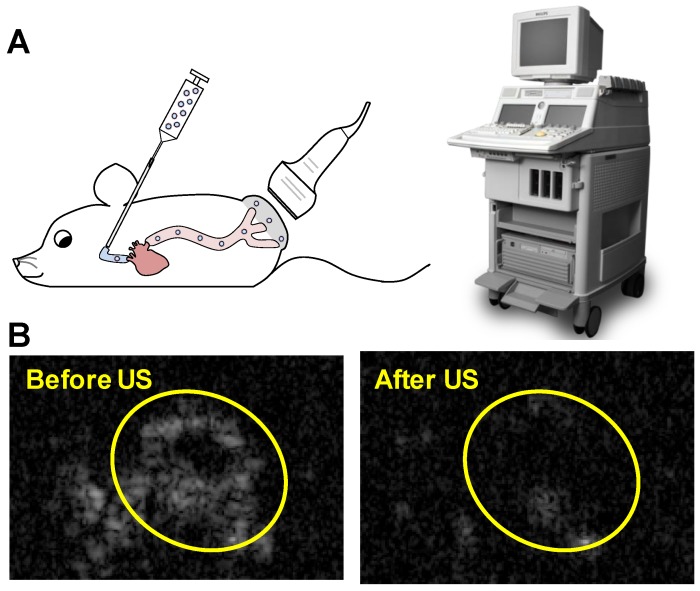
Animal procedures were approved by the University of Pittsburgh Institutional Animal Care and Use Committee. Squamous cell tumors were induced by subcutaneous injection of 1×106 cultured SCC-VII cells into immunocompetent C3H mice as previously described 32. The treatment protocol was as follows: mice were anesthetized using isoflurane and a catheter was placed in the internal jugular vein for MB infusion as previously described 33. A 0.5 mL volume of suspension in PBS containing 1×109 MBs prepared with 10 µg of STAT3 decoy or mutant decoy (maximum decoy loading) was infused for 20 minutes, with this duration chosen to avoid any possible hemodynamic effects associated with rapid volume loading. Non-ultrasound control groups received infusions of 0.5 mL PBS alone or PBS with 10 µg of free STAT3 decoy or microbubble-bound STAT3 decoy over 20 minutes. Ultrasound pulses were delivered for 25 minutes with the ultrasound system used for in vitro studies (S3 probe, Sonos 7500). MBs were detected in the tumor for up to five minutes following MB infusion, thus an additional five minutes of ultrasound was administered following MB infusion in order to insonify any residual MBs. The transducer probe was placed on the tumor as illustrated in Fig. 2A and the ultrasound system was operated in ultraharmonic mode (center frequency of 1.3 MHz) with an on-screen mechanical index of 1.6. The system was time triggered, with 4 frames per burst and a burst interval of 2 s to allow reperfusion of the tumor with microbubbles in between bursts. The treatment was monitored by imaging the tumor using a 15L8 transducer probe on a Sequoia 512 ultrasound imaging system (Siemens Ultrasound, Issaquah, WA, USA) operated in Contrast Pulse Sequencing (CPS) mode (mechanical index = 0.20 and frame rate of 5 Hz) to confirm MB destruction by therapy pulses and subsequent reperfusion of MBs in the tumor (representative images shown in Fig. 2B).
Figure 2.
(A) Experimental setup for in vivo studies: STAT3 decoy loaded MBs were infused intravenously into mice as the tumor was insonified with ultrasound pulses. (B) Representative Contrast Mode ultrasound images of tumor immediately before and after therapy ultrasound pulses were delivered, indicating that MBs perfuse tumor and are destroyed by the therapy ultrasound pulses.
For tumor growth inhibition studies (N=6-8 mice per group), treatment was initiated when tumor volumes were between 20-40 mm3 and animals received a total of three UTMD treatments at three-day intervals. Mice were euthanized 7 days after the last treatment, or if tumors grew to 1 cm3 in size or if the tumor ulcerated. For assessment of STAT3 downstream target gene knockdown (N=8 mice per group), UTMD treatment was performed when tumors reached 60-100 mm3, and mice were euthanized and tumors harvested 24 hours after treatment. To evaluate intratumoral delivery of STAT3 decoy (N=6-7 mice per group), UTMD delivery of radiolabeled STAT3 decoy was performed when tumors reached 100-200 mm3, and mice were euthanized and tumors were harvested and washed 8 hours after treatment.
Serial ultrasonic measurements of tumor volume
High resolution 3-dimensional ultrasound imaging was used to quantify tumor volume at 3-day intervals. Short axis cross-sectional images of the tumor were acquired at 0.197 mm step size using an automated scanning system with a 21 or 30 MHz transducer (Vevo 2100, VisualSonics, Toronto, Canada). Tumor outlines were manually drawn in representative images and volumes were computed from 3D reconstructions of the tumor outlines, as previously described 33. Doubling times (DT) were calculated as previously described 33 using DT = (ln 2)/k, where k was determined by fitting the tumor volume y as a function of time (t) from days 1 to 10 to the exponential function y(t) = X0·ekt, in which X0 represents the initial tumor volume.
STAT3 target gene expression
Western Blot analyses were conducted to detect expression of STAT3 target genes Bcl-xL and cyclin-D1, and the housekeeping gene β-tubulin 24 hours after UTMD. Tumors were homogenized in buffer containing 10 mM Tris, 5 mM EDTA, 50 mM NaCl, 30 mM Na4P2O7, 1 mM Na3VO4, 1% triton x-100, pH 7.6 supplemented with 1 Complete Mini Tablet (Roche Applied Science, Penzberg, Germany) per 10 ml. Protein lysates were then electrophoresed on a 10% precast gel (Bio-Rad, Hercules, CA) and transferred to PVDF membranes (Millipore, Billerica, MA) as per manufacturer's instructions. Membranes were blocked in 5% dry milk, and incubated with anti-Bcl-xL (1:1000 dilution), anti-cyclin-D1 (1:1000 dilution), or anti-β-tubulin (1:3000 dilution) in 1% dry milk. Membranes were then washed and incubated with goat anti-rabbit-HRP secondary antibody (1:5000 dilution), washed, and developed with ECL (Pierce, Thermo Fisher, Scientific, Rockford, IL, USA) as per manufacturer's instructions. Images were captured on x-ray film, and Bcl-xL and cyclin-D1 expression was quantified relative to β-tubulin as an internal control.
Labeling and detection of STAT3 decoy in tumors
STAT3 decoy was end labeled with [γ-32P] ATP using polynucleotide kinase (New England Biolabs, Ipswich, MA, USA) according to manufacturer's protocol. Labeled decoy was then self-annealed and ligated to generate 32P labeled cyclic STAT3 decoy, which was attached to microbubbles as described above. Eight hours after UTMD treatment, tumor samples were harvested and homogenized in RIPA buffer (25 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS), and radioactivity was quantified by scintillation counting. Decoy levels were expressed as percent of injected dose per gram of tissue.
Statistical methods
Statistical comparisons between two experimental groups were determined using a Student's t-test, with statistical significance defined as p < 0.05 (two-tailed). Statistical comparisons among the four experimental groups in vivo were performed using analysis of variance (ANOVA), with significance defined as p < 0.05. If ANOVA demonstrated a significant difference among the groups, post-hoc Tukey's test adjusting for multiple comparisons was performed to determine where the differences resided.
Results
Ultrasound and microbubble-mediated functional delivery of STAT3 decoy in vitro
In order to determine the ability of MBs to protect STAT3 decoy from degradation, and to confirm stable STAT3 decoy binding to MB, decoy bound to MBs was challenged with DNAse and degradation was assessed by acrylamide-urea gel electrophoresis. The amount of intact STAT3 decoy remaining after exposure of MB-associated decoy or free decoy to DNAseI is shown in Fig. 3. Microbubble association protected STAT3 decoy from DNAseI digestion at concentrations sufficient to completely degrade free decoy (Fig. 3).
Figure 3.
Acrylamide‐urea gel electrophoresis demonstrating resistance of circular decoy DNA bound to MBs against digestion by DNAseI. MBs loaded with decoy DNA were washed 3 times to remove unbound DNA and exposed to a range of DNAseI concentrations from 0 to 500 ng/ml (lanes 1, 3, 5, 7 and 9). As a control, free decoy was also challenged with DNAseI at these same concentrations (lanes 2, 4, 6, 8, and 10).
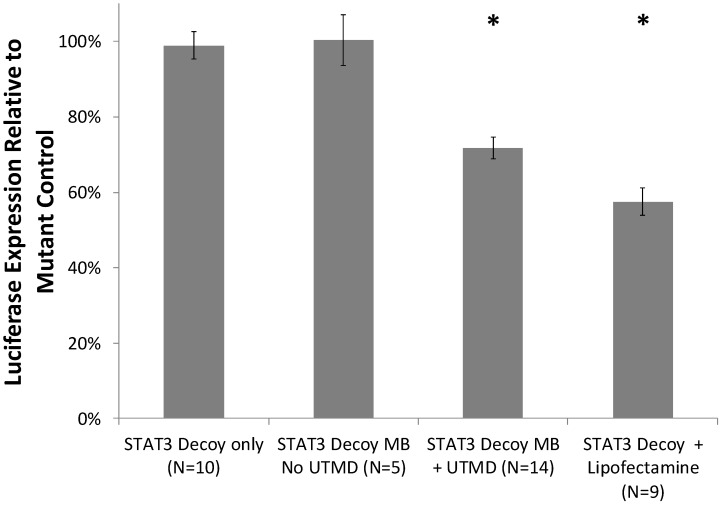
Cell viability with and without UTMD treatment was assessed with a trypan blue exclusion assay. STAT3 decoy MB + UTMD treatment resulted in 89% viability compared to untreated controls, indicating that toxicity was low but statistically higher than controls (p < 0.01). Knockdown of STAT3 signaling with and without UTMD treatment was assessed by measuring luciferase expression in SCC-STAT-luc cells, where luciferase expression is driven by a STAT3-responsive promoter. STAT3 decoy MB + UTMD treatment resulted in a 28 ± 11% reduction in luciferase activity (p < 0.01), while lipofectamine transfection with STAT3 decoy (positive control) knocked down luciferase activity by 42 ± 11% (p < 0.01, Fig. 4).
Figure 4.
Luciferase expression by cultured SCC-VII cells expressing luciferase driven by a STAT3-responsive promoter, after treatment with STAT3 decoy relative to expression after treatment with mutant decoy for each test condition. STAT3 decoy MB + UTMD treatment reduced luciferase expression by 28% relative to mutant decoy MB + UTMD treatment (p < 0.01), demonstrating inhibition of STAT3 signaling with STAT3 decoy MB + UTMD treatment in vitro.
Ultrasound and microbubble-mediated STAT3 target gene knockdown and tumor growth inhibition in vivo
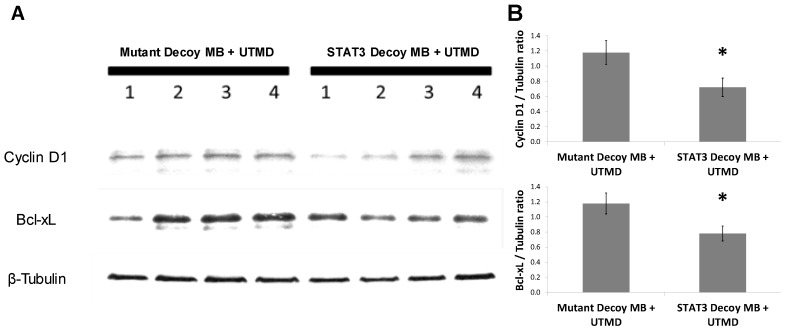
In order to determine the ability of UTMD to specifically silence STAT-responsive genes and inhibit tumor growth, STAT3 decoy was loaded onto MBs and intravenously injected into tumor-bearing mice concurrent with UTMD directed to the tumor (see Methods). Expression of STAT3 downstream target genes was reduced following UTMD treatment in vivo at the protein level. Western blot analysis 24 hours after treatment on tumors from 8 animals per group indicated that relative to mutant decoy MB + UTMD treatment, Bcl-xL and cyclin D1 expression were reduced by 34 ± 8% and 39 ±10%, respectively, in tumors after STAT3 decoy MB + UTMD treatment (Fig. 5).
Figure 5.
STAT3 decoy MB + UTMD treatment downregulates expression of Bcl-xL and cyclin D1 (STAT3 target genes) in tumors 24 hours after treatment. (A) Representative Western blot (N=4 per group). (B) Quantification of all Western blots (N=8 per group, Bcl-xL/Tubulin ratio: p=0.03 vs mutant decoy MB + UTMD treatment, cyclin D1/Tubulin ratio: p=0.04 vs mutant decoy MB + UTMD treatment).
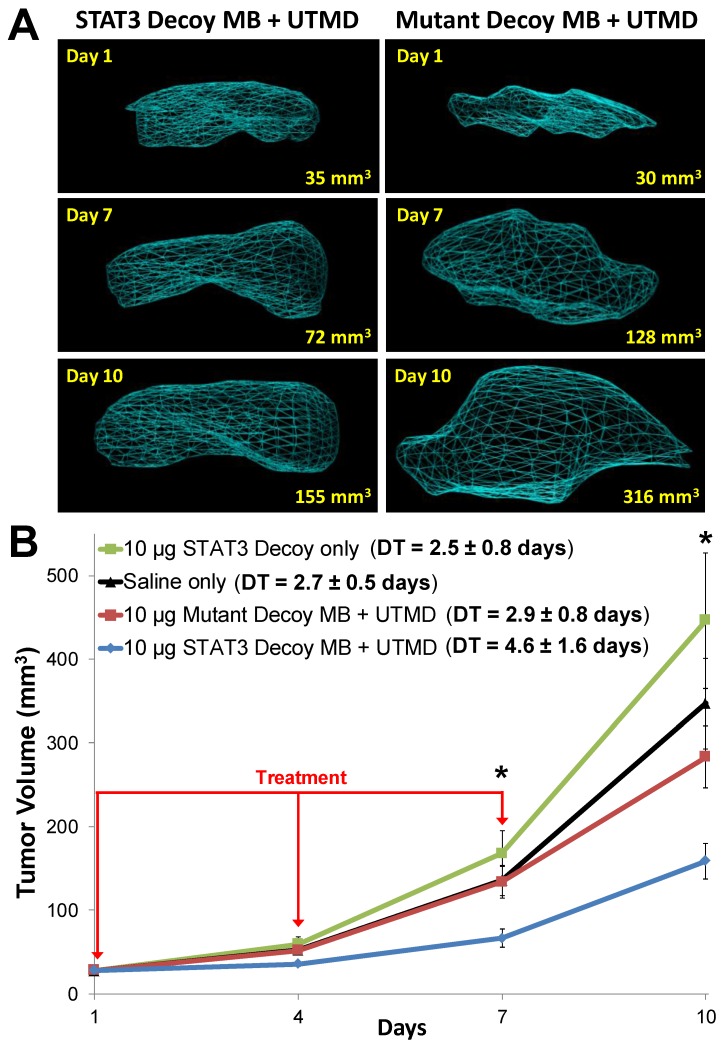
Representative 3D reconstructions of tumor volumes indicate that tumor growth was inhibited after STAT3 decoy MB + UTMD treatment compared to mutant decoy MB + UTMD treatment (Fig. 6A). As shown on the tumor growth curves summarizing all the tumors (Fig. 6B), tumor doubling time was significantly prolonged after STAT3 decoy MB + UTMD treatment (4.6 ± 1.6 days) compared to control groups receiving STAT3 decoy only (2.5 ± 0.8 days), saline only (2.7 ± 0.5 days), or mutant decoy MB + UTMD treatment (2.9 ± 0.8 days). The difference in tumor doubling time between STAT3 decoy MB + UTMD treatment and each control group was statistically significant (ANOVA p < 0.01, post-hoc Tukey's test p < 0.02 for STAT3 decoy MB + UTMD vs. each control group). There were no statistically significant differences in tumor doubling times between control groups (post-hoc Tukey's test p > 0.9 between each control group). Tumor volumes in mice receiving STAT3 decoy MB + UTMD treatment were 31-51% smaller than mutant decoy MB + UTMD treatment at 4, 7, and 10 days after the first treatment.
Figure 6.
(A) Representative 3D reconstructions of tumor volumes acquired using a high resolution ultrasound imaging system, indicating that tumor growth was blunted after treatment with STAT3 decoy MB + UTMD compared to mutant decoy MB + UTMD. (B) Tumor volume plotted as a function of time for each treatment group. Tumor growth was significantly inhibited after STAT3 decoy MB + UTMD treatment compared to control groups (ANOVA p = 0.003, N=6-8 per group). DT represents tumor doubling time. Asterisks (*) indicate ANOVA p < 0.05.
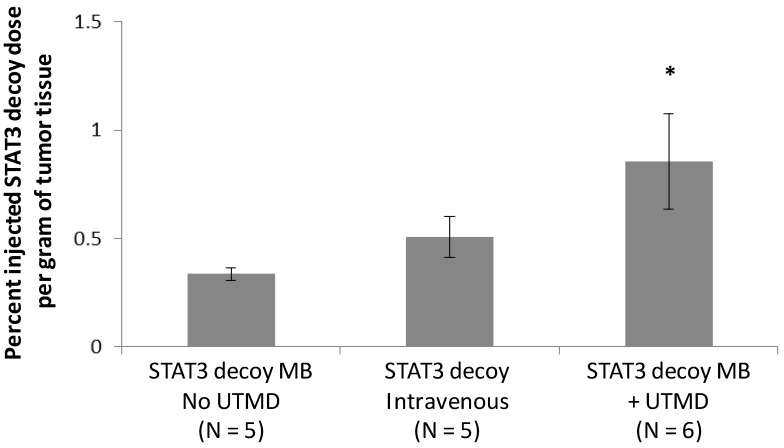
Radiolabeled STAT3 decoy was used to quantify delivery of decoy to the tumor. Despite the fact that the same total amount of STAT3 decoy (10 µg free decoy or loaded on MB) was administered to all mice, STAT3 decoy MB + UTMD treatment nearly doubled the intratumoral radiolabeled decoy concentration eight hours after treatment compared to infusion of radiolabeled STAT3 decoy alone or STAT3 decoy MBs without UTMD (p < 0.05, Fig. 7).
Figure 7.
Amount of radiolabeled STAT3 decoy in tumors treated with UTMD after decoy + MB infusion compared to infusion of STAT3 decoy MB without ultrasound or STAT3 decoy only.
Discussion
The main finding of this study is that in the presence of ultrasound directed at a murine squamous cell carcinoma, MBs carrying STAT3 decoy significantly inhibited tumor growth compared to control treatment groups. There were no statistically significant differences in tumor growth among the control groups that received saline infusion only, STAT3 decoy infusion only, or mutant decoy MB + UTMD. Tumor suppression was associated with decreased STAT3 target gene expression compared to that seen when mutant decoy was delivered on MBs in the presence of identical ultrasound conditions. Importantly, we found preferential intratumoral accumulation of STAT3 decoy compared to that seen when an equivalent amount of unbound decoy was intravenously delivered, or was bound to MBs but not insonified. To our knowledge, this is the first demonstration of successful targeted in vivo STAT3 decoy delivery to tumors. Further, while many have posited preferential nucleic acid delivery achieved via UTMD, this is the first study to quantify nucleic acid delivery via UTMD.
We first performed in vitro studies to determine the efficacy of our delivery platform. As we previously demonstrated with other nucleic acid payloads 32, 33, attachment of the STAT3 decoy to the MB conferred protection against digestion by nuclease attack, likely through the tight association of decoy to the lipid shell. The STAT3 decoy must avoid degradation in circulation in order to accumulate in the tumor and achieve therapeutic effect. To this end, a circular decoy has been previously developed which reduces nuclease degradation. However, attachment of the STAT3 decoy to microbubbles provided further protection against nuclease attack and may reduce the required dose for therapeutic effect while enhancing concentration in the tumor(s). Protection against nuclease degradation is an important attribute of our microbubble vector system, as it allows delivery of nucleic acids through a simple peripheral intravenous injection, thus precluding the need for more invasive intratumoral injection or intra-arterial injection typically required to minimize nucleic acid degradation during blood circulation 19.
We also evaluated the potential for cytoxic bioeffects ensuing from ultrasound-induced MB cavitation 35, independent of STAT3 decoy effects. With the ultrasound parameters that were used, there was a slight loss of viability of the SCC-STAT-luc cells, which was less than that seen when we initially tested more aggressive acoustic regimes (data not shown). Although enhanced cytotoxicity from the delivery platform itself may not necessarily be an undesirable “side effect” when used to treat cancer in vivo, we specifically employed an acoustic regime that was not associated with excessive cytotoxicity for two reasons: First, we were interested in determining anti-tumor efficacy resulting exclusively from enhanced delivery of the STAT3 decoy. Second, to the extent that non-tumor tissue may unavoidably fall within the ultrasound beam, we deemed it critical that the acoustic parameters themselves be non-toxic to normal tissue for clinical translational purposes. Having judged our in vitro viability data to be sufficiently supportive of these two objectives, we then proved that STAT3 decoy MB + our acoustic regime suppressed STAT3 signaling in SCC-STAT-luc cells.
Our subsequent in vivo studies of murine SCC demonstrate that STAT3 decoy MB + our acoustic regime (1) inhibited STAT3 target gene expression; (2) inhibited tumor growth; and (3) enhanced STAT3 decoy accumulation in tumor tissue. Specifically, STAT3 target genes Bcl-xL and cyclin D1 expression were reduced by about 1/3 upon treatment (when compared to mutant decoy MB + UTMD) and it took about twice as long for tumors to double (when compared to i.v. STAT3 decoy, mutant decoy MB + UTMD, or saline placebo). These observations were paralleled by the finding that compared to IV delivery of STAT3 decoy, delivery using UTMD resulted in nearly twice as much tumor accumulation of the decoy.
STAT3 downstream genes Bcl-xL and cyclin D1 promote cancer cell survival and proliferation through inhibition of apoptosis and cell cycle progression, respectively, and are frequently overexpressed in human squamous cell carcinomas 36-38. Delivery of a Bcl-xL antisense oligonucleotide to carboplatin-resistant squamous cell carcinomas resulted in inhibition of cell growth with carboplatin treatment 39. Inhibition of cyclin D1 with shRNA reduced the cisplatin dose required to induce squamous cell carcinoma death 40. Other STAT3 target genes also promote cancer progression, including matrix metalloproteinases, IFN-γ-inducible protein-10, and HIF-1α 8. STAT3 signaling is thus a central signaling molecule that controls the expression of dozens of genes implicated in cancer progression, and is an important therapeutic target. No drugs currently exist that target STAT3 signaling, making STAT3 decoy an attractive anti-cancer therapeutic approach that can address an unmet clinical need.
Unfortunately, a lack of strategies for effective delivery of oligonucleotides has limited clinical translation. Our previous studies demonstrated the efficacy of STAT3 decoy in HNSCC when delivered via intratumoral injections, resulting in suppression of tumor growth in HNSCC xenografts accompanied by inhibition of STAT3 signaling in the tumors 18, 19. However, repeat intratumoral injection is not clinically feasible. The STAT3 decoy has also been delivered with daily intravenous injections in preclinical studies but the precise concentration of the decoy delivered to the tumor is unknown with systemic administration 19, 34. In the present study, significant tumor growth inhibition was achieved after delivering only 10 µg of STAT3 decoy with UTMD treatment, a 10-25 fold reduction compared to prior studies using different delivery strategies. This suggests that overall, ultrasound and MB treatment - perhaps through MB protection of the decoy from nuclease degradation and ultrasound-mediated augmentation of intratumoral accumulation as shown in our study -- locally enhances functional delivery of STAT3 decoy and thus reduces the required dose for efficacy.
We previously demonstrated that UTMD-mediated delivery of a siRNA against epidermal growth factor receptor (EGFR) caused significant tumor growth inhibition in this same murine SCC model compared to delivery of a control siRNA. Similarly, this study demonstrates that UTMD-mediated delivery of STAT3 decoy also inhibits tumor growth. However, an important difference between the two studies is that the STAT3 decoy acts on preformed STAT3 protein whereas siRNA functions through RNA interference; therefore STAT3 signaling is likely to be suppressed more efficiently with the decoy approach. In addition, whereas drugs targeting upstream growth factor receptors such as EGFR are already clinically available 41, the current study offers, for the first time, a clinically feasible mechanism to specifically target STAT3 signaling.
Several limitations to our study should be noted. The MB dose used in this study (1×109 MBs in 20 g mice) is higher than the weight-adjusted MB doses used clinically for diagnostic echocardiography applications (typically ~8×109 MBs for Definity in an averaged size 70 kg male patient). It was assumed in this proof-of-concept study that a higher dose would confer greatest efficacy, hence the selection of the relatively high dose of maximally decoy-loaded microbubbles. Future studies will focus on quantifying dose-response effects, and in particular, whether lower microbubble doses will produce similar tumor inhibitory effects. Although UTMD treatment with STAT3 decoy-loaded MBs caused significant tumor growth inhibition compared to delivery of a mutant decoy control, complete growth inhibition was not achieved in this fast growing murine SCC-VII cell line. When used clinically, treatment efficacy may be further improved by modification of treatment interval or MB dose. Further, the efficacy of UTMD treatment with STAT3 decoy may be further improved when delivered in combination with chemo- and/or radiotherapy. The targeting advantage achieved by ultrasound is potentially disadvantageous when treating more disseminated disease. Our UTMD platform is ideally suited for areas were tumors have been identified and are amenable to ultrasound delivery. Widely disseminated metastatic disease will likely require additional systemic therapy, but would not preclude using UTMD with STAT3 decoy to locally increase decoy concentrations in the sites of major tumor burden. Finally, the efficacy of UTMD treatment with STAT3 decoy in human tumors is not known. Future studies will need to evaluate the efficacy of UTMD-mediated STAT3 decoy delivery to xenografted human head and neck cancers in nude mice.
In conclusion, we have demonstrated that UTMD can be utilized to intravenously deliver a transcription factor decoy -- STAT3 decoy -- and inhibit tumor growth in vivo. Our data suggest that UTMD treatment locally enhances the inhibition of STAT3 signaling for therapy at relatively low doses, precludes the need for intratumoral injection to achieve effective tumor decoy concentrations, and thus may facilitate clinical translation of transcription factor decoy strategies for treating cancers characterized by hyperactivation of STAT3, including head and neck cancer.
Acknowledgments
This study was supported by funding from the National Institutes of Health (R01EB016516 (FSV), R21CA167373 (FSV), P50 CA097190 (JG) and S10RR027383).
Author Contributions
Conception and design: AC, CM, FSV, JG, JK.
Development of Methodology: AC, BH, CM, FSV, JK, LL, MS, XC.
Acquisition of data: AC, BH, JK, LL.
Analysis and interpretation of results: AC, CM, FSV, JG, JK.
Writing, review, and/or revision of the manuscript: AC, FSV, JG, JK, XC.
References
- 1.Frank DA. STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 2007;251:199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 2.Grandis JR, Drenning SD, Zeng Q, Watkins SC, Melhem MF, Endo S, Johnson DE, Huang L, He Y, Kim JD. Constitutive activation of Stat3 signaling abrogates apoptosis in squamous cell carcinogenesis in vivo. Proc Natl Acad Sci U S A. 2000;97:4227–32. doi: 10.1073/pnas.97.8.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X, Sun Y, Pireddu R, Yang H, Urlam MK, Lawrence HR, Guida WC, Lawrence NJ, Sebti SM. A novel inhibitor of STAT3 homodimerization selectively suppresses STAT3 activity and malignant transformation. Cancer Res. 2013;73:1922–33. doi: 10.1158/0008-5472.CAN-12-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song L, Turkson J, Karras JG, Jove R, Haura EB. Activation of Stat3 by receptor tyrosine kinases and cytokines regulates survival in human non-small cell carcinoma cells. Oncogene. 2003;22:4150–65. doi: 10.1038/sj.onc.1206479. [DOI] [PubMed] [Google Scholar]
- 5.Liu P, Kimmoun E, Legrand A, Sauvanet A, Degott C, Lardeux B, Bernuau D. Activation of NF-kappa B, AP-1 and STAT transcription factors is a frequent and early event in human hepatocellular carcinomas. J Hepatol. 2002;37:63–71. doi: 10.1016/s0168-8278(02)00064-8. [DOI] [PubMed] [Google Scholar]
- 6.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–88. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 7.Lian JP, Word B, Taylor S, Hammons GJ, Lyn-Cook BD. Modulation of the constitutive activated STAT3 transcription factor in pancreatic cancer prevention: effects of indole-3-carbinol (I3C) and genistein. Anticancer Res. 2004;24:133–7. [PubMed] [Google Scholar]
- 8.Huang S. Regulation of metastases by signal transducer and activator of transcription 3 signaling pathway: clinical implications. Clin Cancer Res. 2007;13:1362–6. doi: 10.1158/1078-0432.CCR-06-2313. [DOI] [PubMed] [Google Scholar]
- 9.Kim D, Lee IH, Kim S, Choi M, Kim H, Ahn S, Saw PE, Jeon H, Lee Y, Jon S. A specific STAT3-binding peptide exerts antiproliferative effects and antitumor activity by inhibiting STAT3 phosphorylation and signaling. Cancer Res. 2014;74:2144–51. doi: 10.1158/0008-5472.CAN-13-2187. [DOI] [PubMed] [Google Scholar]
- 10.Masuda M, Suzui M, Yasumatu R, Nakashima T, Kuratomi Y, Azuma K, Tomita K, Komiyama S, Weinstein IB. Constitutive activation of signal transducers and activators of transcription 3 correlates with cyclin D1 overexpression and may provide a novel prognostic marker in head and neck squamous cell carcinoma. Cancer Res. 2002;62:3351–5. [PubMed] [Google Scholar]
- 11.Abdulghani J, Gu L, Dagvadorj A, Lutz J, Leiby B, Bonuccelli G, Lisanti MP, Zellweger T, Alanen K, Mirtti T, Visakorpi T, Bubendorf L, Nevalainen MT. Stat3 promotes metastatic progression of prostate cancer. Am J Pathol. 2008;172:1717–28. doi: 10.2353/ajpath.2008.071054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei D, Le X, Zheng L, Wang L, Frey JA, Gao AC, Peng Z, Huang S, Xiong HQ, Abbruzzese JL, Xie K. Stat3 activation regulates the expression of vascular endothelial growth factor and human pancreatic cancer angiogenesis and metastasis. Oncogene. 2003;22:319–29. doi: 10.1038/sj.onc.1206122. [DOI] [PubMed] [Google Scholar]
- 13.Sen M, Grandis JR. Nucleic acid-based approaches to STAT inhibition. Jakstat. 2012;1:285–91. doi: 10.4161/jkst.22312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang T, Niu G, Kortylewski M, Burdelya L, Shain K, Zhang S, Bhattacharya R, Gabrilovich D, Heller R, Coppola D, Dalton W, Jove R, Pardoll D, Yu H. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med. 2004;10:48–54. doi: 10.1038/nm976. [DOI] [PubMed] [Google Scholar]
- 15.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–46. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 16.Burke WM, Jin X, Lin HJ, Huang M, Liu R, Reynolds RK, Lin J. Inhibition of constitutively active Stat3 suppresses growth of human ovarian and breast cancer cells. Oncogene. 2001;20:7925–34. doi: 10.1038/sj.onc.1204990. [DOI] [PubMed] [Google Scholar]
- 17.Yue P, Turkson J. Targeting STAT3 in cancer: how successful are we? Expert Opin Investig Drugs. 2009;18:45–56. doi: 10.1517/13543780802565791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sen M, Joyce S, Panahandeh M, Li C, Thomas SM, Maxwell J, Wang L, Gooding WE, Johnson DE, Grandis JR. Targeting Stat3 abrogates EGFR inhibitor resistance in cancer. Clin Cancer Res. 2012;18:4986–96. doi: 10.1158/1078-0432.CCR-12-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sen M, Thomas SM, Kim S, Yeh JI, Ferris RL, Johnson JT, Duvvuri U, Lee J, Sahu N, Joyce S, Freilino ML, Shi H, Li C, Ly D, Rapireddy S, Etter JP, Li PK, Wang L, Chiosea S, Seethala RR, Gooding WE, Chen X, Kaminski N, Pandit K, Johnson DE, Grandis JR. First-in-human trial of a STAT3 decoy oligonucleotide in head and neck tumors: implications for cancer therapy. Cancer Discov. 2012;2:694–705. doi: 10.1158/2159-8290.CD-12-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blomley MJ, Cooke JC, Unger EC, Monaghan MJ, Cosgrove DO. Microbubble contrast agents: a new era in ultrasound. Bmj. 2001;322:1222–5. doi: 10.1136/bmj.322.7296.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiessling F, Fokong S, Koczera P, Lederle W, Lammers T. Ultrasound microbubbles for molecular diagnosis, therapy, and theranostics. J Nucl Med. 2012;53:345–8. doi: 10.2967/jnumed.111.099754. [DOI] [PubMed] [Google Scholar]
- 22.McDannold N, Arvanitis CD, Vykhodtseva N, Livingstone MS. Temporary disruption of the blood-brain barrier by use of ultrasound and microbubbles: safety and efficacy evaluation in rhesus macaques. Cancer Res. 2012;72:3652–63. doi: 10.1158/0008-5472.CAN-12-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watson KD, Lai CY, Qin S, Kruse DE, Lin YC, Seo JW, Cardiff RD, Mahakian LM, Beegle J, Ingham ES, Curry FR, Reed RK, Ferrara KW. Ultrasound increases nanoparticle delivery by reducing intratumoral pressure and increasing transport in epithelial and epithelial-mesenchymal transition tumors. Cancer Res. 2012;72:1485–93. doi: 10.1158/0008-5472.CAN-11-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alkins R, Burgess A, Ganguly M, Francia G, Kerbel R, Wels WS, Hynynen K. Focused ultrasound delivers targeted immune cells to metastatic brain tumors. Cancer Res. 2013;73:1892–9. doi: 10.1158/0008-5472.CAN-12-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sirsi SR, Borden MA. Advances in Ultrasound Mediated Gene Therapy Using Microbubble Contrast Agents. Theranostics. 2012;2:1208–22. doi: 10.7150/thno.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marmottant P, Hilgenfeldt S. Controlled vesicle deformation and lysis by single oscillating bubbles. Nature. 2003;423:153–6. doi: 10.1038/nature01613. [DOI] [PubMed] [Google Scholar]
- 27.Collis J, Manasseh R, Liovic P, Tho P, Ooi A, Petkovic-Duran K, Zhu Y. Cavitation microstreaming and stress fields created by microbubbles. Ultrasonics. 2010;50:273–9. doi: 10.1016/j.ultras.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Lin CY, Huang YL, Li JR, Chang FH, Lin WL. Effects of focused ultrasound and microbubbles on the vascular permeability of nanoparticles delivered into mouse tumors. Ultrasound Med Biol. 2010;36:1460–9. doi: 10.1016/j.ultrasmedbio.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Kooiman K, van der Steen AF, de Jong N. Role of intracellular calcium and reactive oxygen species in microbubble-mediated alterations of endothelial layer permeability. IEEE Trans Ultrason Ferroelectr Freq Control. 2013;60:1811–5. doi: 10.1109/TUFFC.2013.2767. [DOI] [PubMed] [Google Scholar]
- 30.Hu Y, Wan JM, Yu AC. Membrane perforation and recovery dynamics in microbubble-mediated sonoporation. Ultrasound Med Biol. 2013;39:2393–405. doi: 10.1016/j.ultrasmedbio.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Fan Z, Liu H, Mayer M, Deng CX. Spatiotemporally controlled single cell sonoporation. Proc Natl Acad Sci U S A. 2012;109:16486–91. doi: 10.1073/pnas.1208198109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carson AR, McTiernan CF, Lavery L, Hodnick A, Grata M, Leng X, Wang J, Chen X, Modzelewski RA, Villanueva FS. Gene therapy of carcinoma using ultrasound-targeted microbubble destruction. Ultrasound Med Biol. 2011;37:393–402. doi: 10.1016/j.ultrasmedbio.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carson AR, McTiernan CF, Lavery L, Grata M, Leng X, Wang J, Chen X, Villanueva FS. Ultrasound-targeted microbubble destruction to deliver siRNA cancer therapy. Cancer Res. 2012;72:6191–9. doi: 10.1158/0008-5472.CAN-11-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sen M, Paul K, Freilino ML, Li H, Li C, Johnson DE, Wang L, Eiseman J, Grandis JR. Systemic administration of a cyclic signal transducer and activator of transcription 3 (STAT3) decoy oligonucleotide inhibits tumor growth without inducing toxicological effects. Mol Med. 2014;20:46–56. doi: 10.2119/molmed.2013.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newman CM, Bettinger T. Gene therapy progress and prospects: ultrasound for gene transfer. Gene Ther. 2007;14:465–75. doi: 10.1038/sj.gt.3302925. [DOI] [PubMed] [Google Scholar]
- 36.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558–72. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 37.Pena JC, Thompson CB, Recant W, Vokes EE, Rudin CM. Bcl-xL and Bcl-2 expression in squamous cell carcinoma of the head and neck. Cancer. 1999;85:164–70. doi: 10.1002/(sici)1097-0142(19990101)85:1<164::aid-cncr23>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 38.Stacey DW. Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr Opin Cell Biol. 2003;15:158–63. doi: 10.1016/s0955-0674(03)00008-5. [DOI] [PubMed] [Google Scholar]
- 39.Itoh M, Noutomi T, Chiba H, Mizuguchi J. BcI-xL antisense treatment sensitizes Bcl-xL-overexpressing squamous cell carcinoma cells to carboplatin. Oral Oncol. 2002;38:752–6. doi: 10.1016/s1368-8375(02)00047-7. [DOI] [PubMed] [Google Scholar]
- 40.Kothari V, Mulherkar R. Inhibition of cyclin D1 by shRNA is associated with enhanced sensitivity to conventional therapies for head and neck squamous cell carcinoma. Anticancer Res. 2012;32:121–8. [PubMed] [Google Scholar]
- 41.Hansen AR, Siu LL. Epidermal growth factor receptor targeting in head and neck cancer: have we been just skimming the surface? J Clin Oncol. 2013;31:1381–3. doi: 10.1200/JCO.2012.47.9220. [DOI] [PubMed] [Google Scholar]