Abstract
Parkinson’s disease is a common and usually sporadic neurodegenerative disorder. However, a subset of cases are inherited and, of these, mutations in the gene encoding Leucine-rich repeat kinase 2 (LRRK2) are the most frequent known genetic cause of disease. Here, we will discuss recent progress in understanding how LRRK2 mutations lead to disease and how this may have therapeutic implications. The effect of mutations on LRRK2 enzyme function provides-clues suggesting which functions of the protein are important to disease. Recent work has focused on the kinase and GTP binding domains of LRRK2 and it is assumed that these will be therapeutically important, although there is a substantial amount of work to be done to address this hypothesis.
Keywords: LRRK2, Parkinson’s disease, LRRK2 dysfunction, Kinase and GTPase toxicity, Enzymology, Substantia Nigra, Neurodegenerative diseases
Introduction
Neurodegenerative diseases represent a significant clinical problem, especially in older individuals as the population risk for disorders such as Alzheimer’s or Parkinson’s disease (PD) increases with age. Neurodegenerative diseases also have the insidious property of progressing with time, tending to involve increasing numbers of brain regions and therefore with more disabling and sometimes fatal symptoms. Therefore, both at the level of public health and for the individual person living with a neurodegenerative disorder, there is a genuine need for therapies that will intervene in the course of the disease.
Despite this, there are no clinically helpful tools to modify progression of these diseases. PD is a good example of this. At least part of the cause of the movement problems that a person with PD may suffer, including bradykinesia (slow movement), tremor, rigidity and postural instability is the loss of dopamine producing neurons in part of the midbrain, the substantia nigra pars compacta. Treatment with the dopamine precursor L-DOPA has been a generally effective strategy for these aspects of the disease, but this is a purely sympromatic approach. Progression is not halted, not all symptoms respond, and there are problems associated with prolonged L-DOPA use. Alternative drugs (eg dopamine agonists) are available and there are non-pharmacological approaches such as deep brain stimulation, but in most cases PD worsens over time. PD is well served by L-DOPA as at least there is reasonable symptomatic benefit, but for all of the major neurodegenerative conditions there is no cure.
These considerations lead to the question of why there are no anti-degenerative, or anti-progression, therapies for these conditions in current clinical use. There could be many reasons, and this article would probably stray into philosophy and politics to attempt to answer that question, but one reasonable argument is that we have simply not understood the etiology or root causes of neurodegeneration. As has been argued elsewhere (Ref. 1), because most cases of neurodegenerative disease are apparently sporadic, it has been hard to develop a way of even thinking about causality compared to, for example, infectious diseases. The imperfect argument is that understanding etiology should be a first priority and that developing therapeutics should incorporate our best understanding of causation as much as we can understand it.
Of course this is circular because if a disease is truly sporadic then etiology is going to involve guesswork. But, there is a subset of neurodegenerative conditions where etiology can be established with some certainty, the genetic forms. Even more intriguingly, there are inherited diseases that overlap clinically and pathologically with their sporadic counterparts suggesting a shared etiology. For PD, there are about half a dozen genes that appear to be causative for the condition and several chromosomal regions in the human genome that may yet yield genes (Ref. 2, 3). Furthermore, some of the genes that are found in familial PD also act as risk factors for the sporadic disease (Ref. 4, 5), strengthening the idea that etiology may be shared between the two forms of disease.
Summarizing the above points, it seems reasonable to examine genetic forms of PD for clues about the pathogenic process, recognizing that genes do not work alone and are influenced by aspects of their biological environment. From there, we can find out how the protein products of genes are changed by mutation and, using model systems try and understand how this might result in damage to neurons. Finally, and at the moment this last stage is hypothetical, we can then discuss possible therapeutic applications.
The aim of this review is to take one of the known genes for PD, Leucine-rich repeat kinase 2 (LRRK2), put it through its paces in the above scheme. Our focus here is on how a single gene product could lead to understanding about pathogenesis that might, in the future, have clinical implications. We will also try to identify some of the outstanding questions that should be answered before the full potential of the discovery of this gene can be realized.
The LRRK2 gene, protein and enzyme
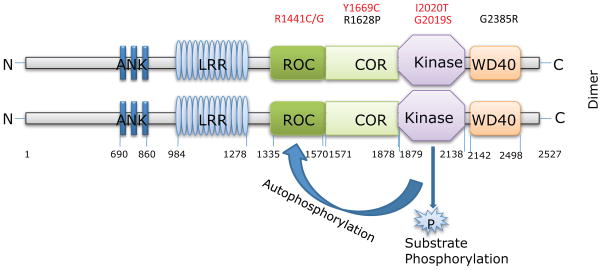
Before discussing mutations in LRRK2, we will first outline what is known about the wild type version. LRRK2 is a large gene on human chromosome 12 that has 51 exons and encodes a protein of 286kDa (shown diagramatically in Fig. 1). Prior to the cloning as a PD gene, LRRK2 and the homologous LRRK1 had been identified as a small family of protein kinases in the human genome (Ref. 6). LRRK1 and LRRK2 have a modular structure that includes a series of leucine-rich repeats towards their N-terminus, and so were named Leucine-Rich repeat kinases. The kinase domains of LRRK1/2 can be separated from most other kinases and are an offshoot of the tyrosine-like kinase family that includes other large modular kinases involved in diverse biological functions.
Figure 1.
Domains and mutations in LRRK2. The LRRK2 protein is shown diagrammatically with amino acid numbers below each domain. Abbreviataions are; ANK, ankyrin like repeats; LRR, leucine-rich repeats; ROC, Ras of complex proteins; COR, C-terminal of ROC. Above the dimer are positions of known pathogenic mutations (in red) and some possible risk variants (in black). The effects of kinase activity may be either phosphorylation of external substrates or autophosphorylation, as indicated. Whether the dimer is present in a head-to-head orientation as shown here is not known.
LRRK1/2 are also part of another group of proteins, the ROCO family. These are named because they have a tandem motif that includes a Ras-of complex proteins (ROC) followed by COR (C-terminal of ROC) domains (Ref. 7). The ROC domain is a GTPase, hydrolyzing GTP to GDP to act as a molecular motor of some kind. The COR domain is thought to regulate the activity of the ROC domain by bringing together two monomers, as demonstrated by the structure of a ROC-COR domain from a slime mold (Ref. 8).
LRRK2 therefore is a single polypeptide with two enzyme activities, kinase and GTPase. The kinase activity of LRRK2 has been demonstrated by a number of different assays using recombinant protein in vitro (Ref. 9, 10, 11, 12, 13, 14). The GTPase activity is weak, and therefore more difficult to quantitate, but has measured reported in some studies (Ref. 15, 16, 17, 18) although negative results have also been reported (Ref. 19).
Kinases and GTPases both work to influence cellular signaling pathways, implying that LRRK2 is a signaling molecule of some kind. Furthermore, many kinases and GTPases work together and this may also be true for LRRK2 in the sense that the two domains communicate. There is some evidence that adding non-hydrolyzable GTP analogues, mimicking the GTP-bound state, increases kinase activity although this has been challenged with some negative reports (Ref. 20). In the other direction, the kinase domain of LRRK2 phosphorylates its ROC domain at several sites (Ref. 21, 22, 23, 24). One interpretation of this data is that the role of the kinase domain is to regulate GTP-dependent functions of LRRK2 (Ref. 4) as indicated in Figure 1. If correct, then this hypothesis suggests that kinase dead versions of LRRK2 will be active in terms of GTP-dependent function but would lack regulation that allows mutant effects to be expressed, which will be discussed below.
Because enzyme activities are measureable and because mutations are found in these domains (see below), these domains have been a major focus of attention. However, there are several other important regions of LRRK2 outside of the central LRR, ROC, COR and kinase domains. The N-terminus of LRRK2 is characterized by a series of repeats that have some homology to ARM/armadillo/HEAT motifs. These are characteristically longer in LRRK2 compared to LRRK1 and therefore may be an important distinguishing region between the two otherwise similar kinases. Towards the C-terminus is a putative WD40 repeat and a short C-terminal region; these again differ between LRRK1 and LRRK2. The significance of the LRRK2 repeats and WD40 domains are that, along with the LRR, they probably act as scaffolds for protein-protein interactions. Conceptually, these regions could be important in binding LRRK2 partners modified by the activity of kinase and GTPase regions. Although several LRRK2 interactors have been proposed, interestingly none yet have been shown to map to these regions.
One additional interesting aspect of the large LRRK2 protein is that it can assemble to form dimers and probably higher molecular complexes (Ref. 8, 9, 15, 22, 25, 26, 27, 28, 29, 30). Dimers are more active as a kinase than other forms (Ref. 9, 30) and there have been suggestions that dimerization is important for GTPase function (Ref. 8). Therefore, the transitions of LRRK2 between monomer, dimer and high molecular weight complex are likely to regulate the overall enzyme output.
LRRK2 is generally cytoplasmic, and never nuclear, at least when overexpressed in various cell lines and neurons (Ref. 10, 11, 12, 13, 14, 31, 32, 33). Part of the localization to the cytoplasm may be related to a proportion of the protein appears to be associated with cellular membranes including autophagic vesicles (Ref. 9, 14, 31, 32, 34, 35). Although the precise significance of these observations is not clear, the implication is that LRRK2 may transition between cytoplasmic and membrane-associated compartments as part of its putative signaling function.
LRRK2 mutations and molecular mechanisms
In 2002, Funayama et al reported a novel PARK locus linked to chromosome 12 (Ref. 36). The family, from the Sagamihara region of Japan, had clear evidence of autosomal dominant inheritance over several generations and relatively high, but incomplete, penetrance. Subsequently, several other families were linked to the same locus and two independent groups identified LRRK2 as the causal gene in 2004 (Ref. 37, 38). Importantly, the family that was used to generate the initial linkage was shown to have a mutation in LRRK2 (Ref. 39). Sequencing of additional cases throughout the world revealed a common and recurrent mutation in many small families (Ref. 40, 41, 42, 43, 44, 45, 46, 47, 48).
There are five unambiguously causal mutations in different domains of LRRK2. The original cloning papers identified R1441C and R1441G in the ROC domain as well as Y1699C in the COR domain. The very common mutation, G2019S, is found in the kinase domain and the original Japanese family has an I2020T mutation at the adjacent residue also in the kinase domain. The location of these mutations is shown in figure 1.
There are additional variants in the LRRK2 gene, which is large and polymorphic, but most of these are thought to be non-pathogenic. However, two variants are associated with altered risk of PD in Asian populations, R1628P in the COR domain and G2385R in the WD40 domain (Ref. 49). These variants are found in controls as well as cases, but at significantly different frequencies.
Given that most of the dominant mutations are in the enzymatic ROC-COR-kinase tridomain, a reasonable approach to understanding mutations is to examine if they affect enzyme activity. Of the known mutations, only G2019S has been consistently shown to increase kinase activity (Ref. reviewed in 50). Mutations in the ROC-COR domain cause a decrease in GTPase activity, where it can be measured (Ref. 16, 17, 20, 51).
A reasonable next question is whether these two observations are related to one another. It has been proposed that kinase activity is stimulated by GTP binding (Ref. 19, 52), in which case lower GTPase activity would lead to higher GTP-stimulated kinase activity. However, this is difficult to reconcile with the lack of effect of ROC-COR mutations on kinase activity and some studies have not seen an effect of addition of non-hydrolysable GTP analogues on kinase activity, as would be predicted (Ref. 20).
Another possibility is that instead of GTP stimulating kinase, the kinase domain is modulatory to GTP binding or hydrolysis. This is potentially supported by identification of sites in the ROC domain that are modified by autophosphorylation (Ref. 21, 22, 23). In this model, the kinase activity of LRRK2 would change GTPase activity such that the enzyme would be more likely to be in an active, GTP-bound state. Support for this comes from recent data showing that one of the autophosphorylation sites, T1410, subtly regulates GTP-binding in vitro (Ref. 24).
If this scenario is correct, then the implication is that understanding GTP-dependent functions of LRRK2 is a critical step forward for the field. Many GTPases are active in the GTP-bound form, so one possibility is that GTP-bound LRRK2 is an effector of signaling, perhaps acting as a scaffold given that large portions of the protein have potential protein-protein interaction domains. Conceptually, a kinase hyperactive mutant would be more likely, and a kinase dead version less likely, to be switched into a higher affinity, GTP-dependent state. In other words, all mutants would be persistent GTP-dependent function and kinase dead mutations would counteract this effect. However, this remains speculative as there is little evidence that autophosphorylation occurs in vivo. This, along with some other outstanding questions, will be discussed below.
LRRK2 phenotypes in man and model organisms
LRRK2 mutations are usually associated with clinical parkinsonism, i.e. the typical movement problems of tremor, rigidity, bradykinesia and postural instability (Ref. 53). Where autopsies have been performed, prominent loss of melanized dopamine neurons in the substantia nigra pars compacta has been noted. As might be expected, movement problems in these patients respond to L-DOPA treatment.
The age at which symptoms are first noted in LRRK2 cases is variable but is generally when the patients are in their 50s and 60s (Ref. 49). This contrasts with genes for recessive parkinsonism, including PINK1 and parkin mutations, which tend to have onset nearer to the age of 30 (Ref. 54). Whether this distinction has a mechanistic basis is unclear at this time, but based on clinical symptoms and age at onset, LRRK2 mutation cases are therefore similar to sporadic PD and distinct from some other types of inherited parkinsonism. Also like sporadic PD, a subset of LRRK2 cases have clear evidence of involvement of other brain regions in the clinicopathological picture, including for example dementia (Ref. 53) or autonomic problems (Ref. 55).
The apparent homogeneity of clinical syndromes, with variation probably similar to that seen in sporadic PD, stands in contrast to the more variable pathology associated with LRRK2 mutations. This variability was emphasized in one of the original cloning papers (Ref. 38) and further variability has been discovered over time (Ref. 56). Most cases appear to have Lewy bodies, the characteristic α-synuclein deposition pathology seen in PD and related conditions, but an appreciable fraction have either no distinctive pathology or tau-positive lesions (Ref. reviewed in 57). The significance of this is unclear. It is possible that LRRK2 is informative for many pathological events, but equally it might be the case that pathological events are epiphenomena and LRRK2 does nothing to help resolve this problem.
There are some people who have LRRK2 mutations who never develop clinical parkinsonism in their lifetime. For example, there have been reports of relatively elderly persons with mutations who were found to be clinically normal even after extensive evaluation (Ref. 58). LRRK2 mutations therefore show age-dependent but incomplete penetrance. Overall penetrance has been estimated at 80% at the age of 70 years (Ref. 59), although lower estimates in some population based studies have also been reported (Ref. 60). A recent study in Basque families reported GTPase domain R1441G mutation penetrance of 83.4% by the age of 80 years (Ref. 61). The number of non-G2019S cases that have been examined is low and so firm conclusions are difficult to make, but it appears that with kinase domain and non-kinase domain mutations have similar but very variable ages at onset.
Increasing evidence suggests that Drosophila melanogaster (Ref. 62, 63, 64, 65, 66, 67, 68, 69, 70, 71) and Caenorhabditis elegans (Ref. 72, 73, 74, 75, 76) can be used as model organisms for investigating neurodegenerative diseases including PD. Here, we will only briefly review work in flies and worms and direct the interested reader to the original literature.
In Drosophila melanogaster, overexpression of mutant LRRK2 causes loss of dopaminergic neurons, retinal degeneration, motor impairment and shorter life whereas wild type protein produces less severe phenotypes(Ref. 62, 63, 68, 71). These animals can be used to identify pathways important in neurodgeneration and in various studies, a contribution of alterations in proteins translation (Ref. 63), microRNA synthesis (Ref. 62) and mitochondrial effectors (Ref. 69, 71) have each been suggested to modify LRRK2 related phenotypes. In C elegans models, overexpression of LRRK2 also causes phenotypic changes including axonal damage in neurons and these phenotypes also seem to involve altered mitochondrial function (Ref. 73).
One way to understand the normal function of LRRK2 is to knockout the gene. Knockout of the nearest homologous gene Drosophila produces variable effects with loss of dopamine cells reported in one study (Ref. 66) but this was not replicated in an independent laboratory (Ref. 77). Knockout of the C elegans homologue causes changes in axonal polarity (Ref. 74) and in neurite outgrowth in response to stress (Ref. 75). What complicates interpreting this data is that flies and worms have single LRRK homologues compared to the two distinct genes in vertebrates (Ref. 78) and at this time we cannot be sure if there are specific functions associated with LRRK2 in higher organisms.
Three independent mouse knockouts have been reported (Ref. 79, 80, 81). In all three published studies, the brain of the animals was reported to be grossly normal and there was no loss of dopamine neurons in the substantia nigra. Andres-Mateos et al further stressed LRRK2 KO mice by exposing them to the dopaminergic neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) and reported no differences between wild type and knockout animals. Therefore, LRRK2 appears to be dispensable for the survival of neurons under both basal and stressed conditions. This argues against a dominant negative effect of mutations, with the caveat that mice live a much shorter lifespan than humans, which may be an important difference for a disease with age-dependent penetrance.
In contrast to the lack of brain phenotypes, one of the reported knockout mice has significant pathology in the kidney (Ref. 81). Although the mechanistic details have not yet been worked out, several correlated events occur in these animals, namely changes in markers of autophagy, accumulation of α-synuclein, cell death and tissue damage. This report still needs to be confirmed by other groups, but it shows that LRRK2 has an important role in organs other than the brain. Whether there is similar kidney damage in human LRRK2 patients is not yet clear.
The other manipulation that has been performed by several groups is to express mutant forms of LRRK2, either under the endogenous promoter using a knockin strategy (Ref. 82) or using heterologous promoters (Ref. 80, 83, 84, 85, 86). Generally, phenotypes of these animals have been modest. No substantial loss of dopamine neurons in the substantia nigra was noted, although tau positive axonal pathology (Ref. 84, 85) and alterations in dopamine transmission (Ref. 82, 83, 84, 85) have been noted in different models, suggesting that these phenotypes may be early but consistent phenomena indicating some damage to the nigrostriatal system.
Two groups of models have shown more extensive neurodegenerative phenotypes. Using a transgenesis approach, Lin et al showed that while expressing mutant LRRK2 alone is not sufficient to trigger extensive neurodegeneration, co-expressing both mutant LRRK2 and mutant α-synuclein will cause neuronal loss in regions of the brain where the transgenes are active. Because Lin et al used a forebrain active CAMKII promoter, neuronal loss and reactive gliosis were seen in the striatum and cortex rather than the substantia nigra, so this is not a full model of Parkinson’s disease, but it does indicate that there are strategies that might result in mouse models that have useful phenotypes.
More recently, two groups have reported that transient overexpression of mutant LRRK2 using viral vectors will result in loss of dopamine neurons in the substantia nigra of mice (Ref. 87) or rats (Ref. 88). Importantly, wild type protein or kinase dead versions of LRRK2 had no effect, suggesting that simple overexpression of any similar large protein would not be sufficient to cause neurodegeneration. Therefore, this approach has the potential to provide a model that more fully replicates the phenotypes seen in human LRRK2 patients than are seen in conventional transgenic models.
Several groups have shown that LRRK2 mutations affect neurite branching in growing neurons. In primary neuronal cultures, overexpressing Y1699C or G2019S mutant LRRK2 overexpression cause significant reduction in neurite branching compare to control cells (Ref. 89, 90, 91). LRRK2 expression in flies also causes changes in neurites in vivo (Ref. 65). Wild type LRRK2 expressed at similar levels does not cause neurite shortening although knockout of LRRK2 causes increased neurite outgrowth (Ref. 89, 90, 91). It is therefore currently unclear if wild type human or mouse LRRK2 is important in these phenomena or whether there is a mutant specific effect.
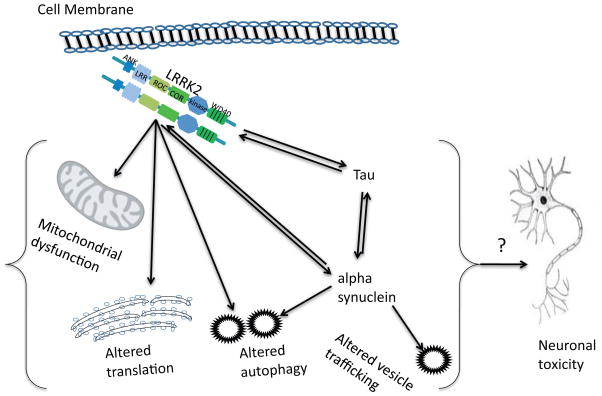
How mutant LRRK2 causes neuronal dysfunction and cell death is unclear, although several themes have arisen from the literature in recent years (Fig. 2). As discussed above, LRRK2 appears to have effects on vesicle function including potentially the autophagy-lysosome system and/or on synaptic vesicles. Evidence, particularly from Drosophila models, suggests an additional role in protein translation. Finally, there are poorly defined relationships to α-synuclein and tau that may be important in understanding LRRK2 in the context of other dominant genes associated with parkinsonism. It is not clear whether any or all of these effects are either necessary or sufficient for the detrimental effects of mutant LRRK2, but conceptually each might cause some neuronal dysfunction that collectively results in neuronal cell death (Fig. 2).
Figure 2.
Pathogenic pathways for LRRK2. LRRK2 has been suggested to directly or indirectly affect several cellular pathways, including mitochondria, protein translation and autophagy-lysosome function. Collectively, these may result in neurotoxicity. It is also likely important that there are genetic interactions between LRRK2 and tau and α-synuclein, which are other genes for autosomal dominant parkinsonism.
Clinical implications/applications
One of the major clinical implications of identifying LRRK2 as a relatively common gene for PD is that it highlights how powerful genetics is for understanding disease. In some populations, LRRK2 accounts for a significant proportion of all PD cases and so identification of this gene has implications for genetic testing and other clinical considerations (Ref. 92).
More broadly, evidence that LRRK2 dysfunction is related to α-synuclein (Ref. 80), the protein that defines the Lewy body pathology found in all cases of sporadic PD, suggests that inherited and sporadic PD may share pathogenic pathways. This supposition is potentially supported by the identification of common genetic variants around both α-synuclein and LRRK2 that are associated with altered risk of sporadic PD (Ref. 93, 94). If correct, then thinking about therapeutics, which is certainly the underlying motivation behind a great deal of work on neurodegenerative diseases, would not involve a separation between inherited and sporadic diseases. Rather, we could think about PD as a pathogenic entity and distinguish only between those cases with a strong genetic basis and those where genetics plays a more modest role.
At the current time, the idea that one may be able to develop treatments for sporadic PD based on the minority of inherited forms is speculative. But, it is also clear that developing tools for LRRK2 is one way to test this hypothesis. Specifically, it has been proposed that the detrimental effects of LRRK2 both in vitro (Ref. 12, 95) and in vivo (Ref. 87) rely in part on kinase activity. This leads to the suggestion that kinase inhibitors for LRRK2 may have therapeutic potential for people who have mutations in that gene (Ref. 96, 97, 98). Such tools would be invaluable and will be discussed in the next section of this review.
Research in progress and outstanding research questions
As discussed above, perhaps the key tool for understanding LRRK2 would be to have a highly specific, potent compound that inhibits kinase activity. Such a compound would need to be extremely well tolerated at inhibitory doses and would preferably be orally available and brain permeable. Although this is not yet available, the theoretical kinase inhibitor would allow for a test of the hypothesis that kinase activity is important for LRRK2 pathogenesis. If such compounds were available then they could be used to test the ancillary hypothesis that LRRK2 may be relevant for sporadic PD.
However important it is to have tools to inhibit kinase activity, it is not clear exactly where kinase activity fits into the overall picture of LRRK2 pathogenesis. One of the most critical basic science questions that must be answered is that of the authentic physiological substrate(s) of kinase activity, which has been debated for some time (Ref. 50). For a substrate to be considered authentic, then a specific phosphorylation site needs to be identified on the substrate that comes from a given kinase; that phosphorylation site should be responsive to inhibition of LRRK2 activity in vivo; and the reaction should also be able to be reconstituted in vitro using physiologically reasonable concentrations of substrate and enzyme. Is the autophosphorylation of the ROC domain really the key output of the protein, or is there a heterologous substrate that mediates damaging effects of LRRK2 mutations? And if the ROC-COR bidomain is critically important, as would be implied by the presence of pathological mutations, then what is its function?
Related to this, it is important to understand how other signaling pathways in the cell regulate LRRK2. Most signaling molecules receive input from initiating pathways as diverse as growth factors and oxidative stress, as well as signaling from other cellular pathways that is important in maintaining homeostasis. Understanding what regulates LRRK2 may give important hints as to normal function, as well as providing the practical benefit of increasing activity, making many of the currently used biochemical assays much more feasible.
Finally, how is LRRK2 related to α-synuclein? Because α-synuclein is such an important protein for sporadic PD as a major component of Lewy bodies it is important to understand whether LRRK2 pathogenesis requires the involvement of α-synuclein, thus linking two dominant genes with sporadic disease. This is likely to be a difficult problem, not least because not all cases with LRRK2 mutations have Lewy bodies. Clinical and pathological surveys of LRRK2 mutation carriers are needed to provide a stronger basis for understanding the pathology in this disease by cataloguing its variability.
Overall, therefore, LRRK2 has provided an important set of clues as to pathogenesis for an otherwise enigmatic disorder. Furthermore, there is at least one potential way forward to therapeutic avenues by targeting first the kinase activity of LRRK2. However, there are a number of important required next steps to move this idea forward into the clinic. Kinase inhibitor tools are required that have good specificity and potency for LRRK2 versus other kinases. These are starting to be reported in the literature (Ref. 99) and in conjunction with loss of function models such as knockout mice these will be useful in dissecting out the effects of normal LRRK2 function in the adult nervous system. We also need to understand how mutant LRRK2 diverges in function from the normal wild type protein as this might identify processes in cells on which mutant LRRK2 impinges that might provide intervention points for new therapeutic ideas.
Acknowledgments
Funding
This research was supported by the Intramural Research Program of the NIH, National Institute on Aging. The authors also thank the reviewers for their helpful and critical comments.
Footnotes
Websites
General information about PD research can be found at PDOnline: http://www.pdonlineresearch.org/
Up to date information on inherited forms of PD can be obtained from the OMIM (online inheritance in man) website. Additional data on the association of specific genetic variants with PD can be found at pdgene.
References
- 1.Cookson MR. The biochemistry of Parkinson’s disease. Annual Reviews of Biochemistry. 2005;74:29–52. doi: 10.1146/annurev.biochem.74.082803.133400. [DOI] [PubMed] [Google Scholar]
- 2.Cookson MR, Bandmann O. Parkinson’s disease: insights from pathways. Human Molecular Genetics. 2010;19:R21–7. doi: 10.1093/hmg/ddq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardy J. Genetic analysis of pathways to Parkinson disease. Neuron. 2010;68:201–6. doi: 10.1016/j.neuron.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cookson MR. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson’s disease. Nature Reviews Neuroscience. 2010;11:791–7. doi: 10.1038/nrn2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taymans JM, Cookson MR. Mechanisms in dominant parkinsonism: The toxic triangle of LRRK2, alpha-synuclein, and tau. Bioessays. 2010;32:227–35. doi: 10.1002/bies.200900163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manning G, et al. The protein kinase complement of the human genome. Science. 2002;298:1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 7.Marin I, van Egmond WN, van Haastert PJ. The Roco protein family: a functional perspective. FASEB Journal. 2008;22:3103–10. doi: 10.1096/fj.08-111310. [DOI] [PubMed] [Google Scholar]
- 8.Gotthardt K, et al. Structure of the Roc-COR domain tandem of C. tepidum, a prokaryotic homologue of the human LRRK2 Parkinson kinase. EMBO Journal. 2008;27:2239–49. doi: 10.1038/emboj.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berger Z, Smith KA, Lavoie MJ. Membrane localization of LRRK2 is associated with increased formation of the highly active LRRK2 dimer and changes in its phosphorylation. Biochemistry. 2010;49:5511–23. doi: 10.1021/bi100157u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dzamko N, et al. Inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser(910)/Ser(935), disruption of 14–3–3 binding and altered cytoplasmic localization. Biochemical Journal. 2010;430:405–13. doi: 10.1042/BJ20100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giasson BI, et al. Biochemical and pathological characterization of Lrrk2. Annals of Neurology. 2006;59:315–22. doi: 10.1002/ana.20791. [DOI] [PubMed] [Google Scholar]
- 12.Greggio E, et al. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiology of Disease. 2006;23:329–41. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Nichols RJ, et al. 14–3–3 binding to LRRK2 is disrupted by multiple Parkinson’s disease-associated mutations and regulates cytoplasmic localization. Biochemical Journal. 2010;430:393–404. doi: 10.1042/BJ20100483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West AB, et al. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proceedings of the National Academy of Sciences USA. 2005;102:16842–7. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng J, et al. Structure of the ROC domain from the Parkinson’s disease-associated leucine-rich repeat kinase 2 reveals a dimeric GTPase. Proceedings of the National Academy of Sciences USA. 2008;105:1499–504. doi: 10.1073/pnas.0709098105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo L, et al. The Parkinson’s disease-associated protein, leucine-rich repeat kinase 2 (LRRK2), is an authentic GTPase that stimulates kinase activity. Experimental Cell Research. 2007;313:3658–70. doi: 10.1016/j.yexcr.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis PA, et al. The R1441C mutation of LRRK2 disrupts GTP hydrolysis. Biochemical and Biophysical Research Communications. 2007;357:668–71. doi: 10.1016/j.bbrc.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, et al. Leucine-rich repeat kinase 2 (LRRK2)/PARK8 possesses GTPase activity that is altered in familial Parkinson’s disease R1441C/G mutants. Journal of Neurochemistry. 2007;103:238–47. doi: 10.1111/j.1471-4159.2007.04743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito G, et al. GTP binding is essential to the protein kinase activity of LRRK2, a causative gene product for familial Parkinson’s disease. Biochemistry. 2007;46:1380–8. doi: 10.1021/bi061960m. [DOI] [PubMed] [Google Scholar]
- 20.Liu M, et al. Kinetic mechanistic studies of wild-type leucine-rich repeat kinase 2: characterization of the kinase and GTPase activities. Biochemistry. 2010;49:2008–17. doi: 10.1021/bi901851y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gloeckner CJ, et al. Phosphopeptide analysis reveals two discrete clusters of phosphorylation in the N-terminus and the Roc domain of the Parkinson-disease associated protein kinase LRRK2. Journal of Proteome Research. 2010;9:1738–45. doi: 10.1021/pr9008578. [DOI] [PubMed] [Google Scholar]
- 22.Greggio E, et al. The Parkinson’s disease kinase LRRK2 autophosphorylates its GTPase domain at multiple sites. Biochemical and Biophysical Research Communications. 2009;389:449–54. doi: 10.1016/j.bbrc.2009.08.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamikawaji S, Ito G, Iwatsubo T. Identification of the autophosphorylation sites of LRRK2. Biochemistry. 2009;48:10963–75. doi: 10.1021/bi9011379. [DOI] [PubMed] [Google Scholar]
- 24.Pungaliya PP, et al. Identification and characterization of a leucine-rich repeat kinase 2 (LRRK2) consensus phosphorylation motif. PLoS One. 2010;5:e13672. doi: 10.1371/journal.pone.0013672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greggio E, et al. The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. Journal of Biological Chemistry. 2008;283:16906–14. doi: 10.1074/jbc.M708718200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jorgensen ND, et al. The WD40 domain is required for LRRK2 neurotoxicity. PLoS One. 2009;4:e8463. doi: 10.1371/journal.pone.0008463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein CL, et al. Homo- and heterodimerization of ROCO kinases: LRRK2 kinase inhibition by the LRRK2 ROCO fragment. Journal of Neurochemistry. 2009;111:703–15. doi: 10.1111/j.1471-4159.2009.06358.x. [DOI] [PubMed] [Google Scholar]
- 28.Ko HS, et al. CHIP regulates leucine-rich repeat kinase-2 ubiquitination, degradation, and toxicity. Proceedings of the National Academy of Sciences USA. 2009;106:2897–902. doi: 10.1073/pnas.0810123106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu B, et al. Expression, purification and preliminary biochemical studies of the N-terminal domain of leucine-rich repeat kinase 2. Biochimica et Biophysica Acta. 2010 doi: 10.1016/j.bbapap.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 30.Sen S, Webber PJ, West AB. Dependence of leucine-rich repeat kinase 2 (LRRK2) kinase activity on dimerization. Journal of Biological Chemistry. 2009;284:36346–56. doi: 10.1074/jbc.M109.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alegre-Abarrategui J, et al. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Human Molecular Genetics. 2009;18:4022–34. doi: 10.1093/hmg/ddp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatano T, et al. Leucine-rich repeat kinase 2 associates with lipid rafts. Human Molecular Genetics. 2007;16:678–90. doi: 10.1093/hmg/ddm013. [DOI] [PubMed] [Google Scholar]
- 33.Miklossy J, et al. LRRK2 expression in normal and pathologic human brain and in human cell lines. Journal of Neuropathology and Experimental Neurology. 2006;65:953–63. doi: 10.1097/01.jnen.0000235121.98052.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biskup S, et al. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Annals of Neurology. 2006;60:557–69. doi: 10.1002/ana.21019. [DOI] [PubMed] [Google Scholar]
- 35.Higashi S, et al. Abnormal localization of leucine-rich repeat kinase 2 to the endosomal-lysosomal compartment in lewy body disease. Journal of Neuropathology and Experimental Neurology. 2009;68:994–1005. doi: 10.1097/NEN.0b013e3181b44ed8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Funayama M, et al. A new locus for Parkinson’s disease (PARK8) maps to chromosome 12p11.2–q13.1. Annals of Neurology. 2002;51:296–301. doi: 10.1002/ana.10113. [DOI] [PubMed] [Google Scholar]
- 37.Paisan-Ruiz C, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 38.Zimprich A, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–7. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 39.Funayama M, et al. An LRRK2 mutation as a cause for the parkinsonism in the original PARK8 family. Annals of Neurology. 2005;57:918–21. doi: 10.1002/ana.20484. [DOI] [PubMed] [Google Scholar]
- 40.Gilks WP, et al. A common LRRK2 mutation in idiopathic Parkinson’s disease. Lancet. 2005;365:415–6. doi: 10.1016/S0140-6736(05)17830-1. [DOI] [PubMed] [Google Scholar]
- 41.Goldwurm S, et al. The G6055A (G2019S) mutation in LRRK2 is frequent in both early and late onset Parkinson’s disease and originates from a common ancestor. Journal of Medical Genetics. 2005;42:e65. doi: 10.1136/jmg.2005.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Infante J, et al. LRRK2 G2019S is a common mutation in Spanish patients with late-onset Parkinson’s disease. Neuroscience Letters. 2006;395:224–6. doi: 10.1016/j.neulet.2005.10.083. [DOI] [PubMed] [Google Scholar]
- 43.Kachergus J, et al. Identification of a novel LRRK2 mutation linked to autosomal dominant parkinsonism: evidence of a common founder across European populations. American Journal of Human Genetics. 2005;76:672–80. doi: 10.1086/429256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lesage S, et al. LRRK2 G2019S as a cause of Parkinson’s disease in North African Arabs. New England Journal of Medicine. 2006;354:422–3. doi: 10.1056/NEJMc055540. [DOI] [PubMed] [Google Scholar]
- 45.Lesage S, et al. G2019S LRRK2 mutation in French and North African families with Parkinson’s disease. Annals of Neurology. 2005;58:784–7. doi: 10.1002/ana.20636. [DOI] [PubMed] [Google Scholar]
- 46.Lesage S, et al. LRRK2 haplotype analyses in European and North African families with Parkinson disease: a common founder for the G2019S mutation dating from the 13th century. American Journal of Human Genetics. 2005;77:330–2. doi: 10.1086/432422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ozelius LJ, et al. LRRK2 G2019S as a cause of Parkinson’s disease in Ashkenazi Jews. New England Journal of Medicine. 2006;354:424–5. doi: 10.1056/NEJMc055509. [DOI] [PubMed] [Google Scholar]
- 48.Rajput A, et al. Parkinsonism, Lrrk2 G2019S, and tau neuropathology. Neurology. 2006;67:1506–8. doi: 10.1212/01.wnl.0000240220.33950.0c. [DOI] [PubMed] [Google Scholar]
- 49.Kumari U, Tan EK. LRRK2 in Parkinson’s disease: genetic and clinical studies from patients. FEBS Journal. 2009;276:6455–63. doi: 10.1111/j.1742-4658.2009.07344.x. [DOI] [PubMed] [Google Scholar]
- 50.Greggio E, Cookson MR. Leucine-rich repeat kinase 2 mutations and Parkinson’s disease: three questions. ASN Neuro. 2009;1 doi: 10.1042/AN20090007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daniels V, et al. Insight into the mode of action of the LRRK2 Y1699C pathogenic mutant. Journal of Neurochemistry. 2011;116:304–15. doi: 10.1111/j.1471-4159.2010.07105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.West AB, et al. Parkinson’s disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Human Molecular Genetics. 2007;16:223–32. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- 53.Haugarvoll K, Wszolek ZK. Clinical features of LRRK2 parkinsonism. Parkinsonism and Related Disorderss. 2009;15(Suppl 3):S205–8. doi: 10.1016/S1353-8020(09)70815-6. [DOI] [PubMed] [Google Scholar]
- 54.Alcalay RN, et al. Frequency of known mutations in early-onset Parkinson disease: implication for genetic counseling: the consortium on risk for early onset Parkinson disease study. Arch Neurol. 2010;67:1116–22. doi: 10.1001/archneurol.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goldstein DS, et al. Neurocirculatory and nigrostriatal abnormalities in Parkinson disease from LRRK2 mutation. Neurology. 2007;69:1580–4. doi: 10.1212/01.wnl.0000268696.57912.64. [DOI] [PubMed] [Google Scholar]
- 56.Wider C, Dickson DW, Wszolek ZK. Leucine-rich repeat kinase 2 gene-associated disease: redefining genotype-phenotype correlation. Neurodegenerative Diseases. 2010;7:175–9. doi: 10.1159/000289232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cookson MR, Hardy J, Lewis PA. Genetic neuropathology of Parkinson’s disease. International Journal Clinical Experimetnal Pathology. 2008;1:217–31. [PMC free article] [PubMed] [Google Scholar]
- 58.Kay DM, et al. Escaping Parkinson’s disease: a neurologically healthy octogenarian with the LRRK2 G2019S mutation. Movement Disorders. 2005;20:1077–8. doi: 10.1002/mds.20618. [DOI] [PubMed] [Google Scholar]
- 59.Dachsel JC, Farrer MJ. LRRK2 and Parkinson disease. Archives of Neurology. 2010;67:542–7. doi: 10.1001/archneurol.2010.79. [DOI] [PubMed] [Google Scholar]
- 60.Clark LN, et al. Frequency of LRRK2 mutations in early- and late-onset Parkinson disease. Neurology. 2006;67:1786–91. doi: 10.1212/01.wnl.0000244345.49809.36. [DOI] [PubMed] [Google Scholar]
- 61.Ruiz-Martinez J, et al. Penetrance in Parkinson’s disease related to the LRRK2 R1441G mutation in the Basque country (Spain) Mov Disord. 2010;25:2340–5. doi: 10.1002/mds.23278. [DOI] [PubMed] [Google Scholar]
- 62.Gehrke S, et al. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637–41. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Imai Y, et al. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO Journal. 2008;27:2432–43. doi: 10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kanao T, et al. Activation of FoxO by LRRK2 induces expression of proapoptotic proteins and alters survival of postmitotic dopaminergic neuron in Drosophila. Human Molecular Genetics. 2010;19:3747–58. doi: 10.1093/hmg/ddq289. [DOI] [PubMed] [Google Scholar]
- 65.Lee S, et al. LRRK2 kinase regulates synaptic morphology through distinct substrates at the presynaptic and postsynaptic compartments of the Drosophila neuromuscular junction. Journal of Neuroscience. 2010;30:16959–69. doi: 10.1523/JNEUROSCI.1807-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee SB, et al. Loss of LRRK2/PARK8 induces degeneration of dopaminergic neurons in Drosophila. Biochemical and Biophysical Research Communications. 2007;358:534–9. doi: 10.1016/j.bbrc.2007.04.156. [DOI] [PubMed] [Google Scholar]
- 67.Lin CH, et al. LRRK2 G2019S mutation induces dendrite degeneration through mislocalization and phosphorylation of tau by recruiting autoactivated GSK3ss. Journal of Neuroscience. 2010;30:13138–49. doi: 10.1523/JNEUROSCI.1737-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu Z, et al. A Drosophila model for LRRK2-linked parkinsonism. Proceedings of the National Academy of Sciences USA. 2008;105:2693–8. doi: 10.1073/pnas.0708452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ng CH, et al. Parkin protects against LRRK2 G2019S mutant-induced dopaminergic neurodegeneration in Drosophila. Journal of Neuroscience. 2009;29:11257–62. doi: 10.1523/JNEUROSCI.2375-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tain LS, et al. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nature Neuroscience. 2009;12:1129–35. doi: 10.1038/nn.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Venderova K, et al. Leucine-Rich Repeat Kinase 2 interacts with Parkin, DJ-1 and PINK-1 in a Drosophila melanogaster model of Parkinson’s disease. Human Molecular Genetics. 2009;18:4390–404. doi: 10.1093/hmg/ddp394. [DOI] [PubMed] [Google Scholar]
- 72.Hsu CH, et al. MKK6 binds and regulates expression of Parkinson’s disease-related protein LRRK2. Journal of Neurochemistry. 2010;112:1593–604. doi: 10.1111/j.1471-4159.2010.06568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saha S, et al. LRRK2 modulates vulnerability to mitochondrial dysfunction in Caenorhabditis elegans. Journal of Neuroscience. 2009;29:9210–8. doi: 10.1523/JNEUROSCI.2281-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sakaguchi-Nakashima A, et al. LRK-1, a C. elegans PARK8-related kinase, regulates axonal-dendritic polarity of SV proteins. Current Biology. 2007;17:592–8. doi: 10.1016/j.cub.2007.01.074. [DOI] [PubMed] [Google Scholar]
- 75.Samann J, et al. Caenorhabditits elegans LRK-1 and PINK-1 act antagonistically in stress response and neurite outgrowth. Journal of Biological Chemistry. 2009;284:16482–91. doi: 10.1074/jbc.M808255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yao C, et al. LRRK2-mediated neurodegeneration and dysfunction of dopaminergic neurons in a Caenorhabditis elegans model of Parkinson’s disease. Neurobiology of Disease. 2010;40:73–81. doi: 10.1016/j.nbd.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang D, et al. Dispensable role of Drosophila ortholog of LRRK2 kinase activity in survival of dopaminergic neurons. Molecular Neurodegeneration. 2008;3:3. doi: 10.1186/1750-1326-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marin I. Ancient origin of the Parkinson disease gene LRRK2. J Mol Evol. 2008;67:41–50. doi: 10.1007/s00239-008-9122-4. [DOI] [PubMed] [Google Scholar]
- 79.Andres-Mateos E, et al. Unexpected lack of hypersensitivity in LRRK2 knock-out mice to MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) Journal of Neuroscience. 2009;29:15846–50. doi: 10.1523/JNEUROSCI.4357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin X, et al. Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson’s-disease-related mutant alpha-synuclein. Neuron. 2009;64:807–27. doi: 10.1016/j.neuron.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tong Y, et al. Loss of leucine-rich repeat kinase 2 causes impairment of protein degradation pathways, accumulation of alpha-synuclein, and apoptotic cell death in aged mice. Proceedings of the National Academy of Sciences USA. 2010;107:9879–84. doi: 10.1073/pnas.1004676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tong Y, et al. R1441C mutation in LRRK2 impairs dopaminergic neurotransmission in mice. Proceedings of the National Academy of Sciences USA. 2009;106:14622–7. doi: 10.1073/pnas.0906334106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li X, et al. Enhanced striatal dopamine transmission and motor performance with LRRK2 overexpression in mice is eliminated by familial Parkinson’s disease mutation G2019S. Journal of Neuroscience. 2010;30:1788–97. doi: 10.1523/JNEUROSCI.5604-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li Y, et al. Mutant LRRK2(R1441G) BAC transgenic mice recapitulate cardinal features of Parkinson’s disease. Nature Neuroscience. 2009;12:826–8. doi: 10.1038/nn.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Melrose HL, et al. Impaired dopaminergic neurotransmission and microtubule-associated protein tau alterations in human LRRK2 transgenic mice. Neurobiology of Disease. 2010;40:503–17. doi: 10.1016/j.nbd.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Winner B, et al. Adult neurogenesis and neurite outgrowth are impaired in LRRK2 G2019S mice. Neurobiology of Disease. 2011;41:706–16. doi: 10.1016/j.nbd.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee BD, et al. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson’s disease. Nature Medicine. 2010;16:998–1000. doi: 10.1038/nm.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dusonchet J, et al. A Rat Model of Progressive Nigral Neurodegeneration Induced by the Parkinson’s Disease-Associated G2019S Mutation in LRRK2. Journal of Neuroscience. 2011;31:907–12. doi: 10.1523/JNEUROSCI.5092-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dachsel JC, et al. A comparative study of Lrrk2 function in primary neuronal cultures. Parkinsonism Relat Disord. 2010;16:650–5. doi: 10.1016/j.parkreldis.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.MacLeod D, et al. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–93. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 91.Parisiadou L, et al. Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. J Neurosci. 2009;29:13971–80. doi: 10.1523/JNEUROSCI.3799-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nuytemans K, et al. Genetic etiology of Parkinson disease associated with mutations in the SNCA, PARK2, PINK1, PARK7, and LRRK2 genes: a mutation update. Human Mutation. 2010;31:763–80. doi: 10.1002/humu.21277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Satake W, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nature Genetics. 2009;41:1303–7. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 94.Simon-Sanchez J, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nature Genetics. 2009;41:1308–12. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smith WW, et al. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nature Neuroscience. 2006;9:1231–3. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- 96.Liu M, et al. Development of a mechanism-based high-throughput screen assay for leucine-rich repeat kinase 2--discovery of LRRK2 inhibitors. Analytical Biochemistry. 2010;404:186–92. doi: 10.1016/j.ab.2010.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nichols RJ, et al. Substrate specificity and inhibitors of LRRK2, a protein kinase mutated in Parkinson’s disease. Biochemical Journal. 2009;424:47–60. doi: 10.1042/BJ20091035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reichling LJ, Riddle SM. Leucine-rich repeat kinase 2 mutants I2020T and G2019S exhibit altered kinase inhibitor sensitivity. Biochemical and Biophysical Research Communications. 2009;384:255–8. doi: 10.1016/j.bbrc.2009.04.098. [DOI] [PubMed] [Google Scholar]
- 99.Deng X, et al. Characterization of a selective inhibitor of the Parkinson’s disease kinase LRRK2. Nat Chem Biol. 2011;7:203–5. doi: 10.1038/nchembio.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
Further reading and resources
- Cookson MR. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson’s disease. Nature Reviews Neuroscience. 2010;11:791–7. doi: 10.1038/nrn2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greggio E, Cookson MR. Leucine-rich repeat kinase 2 mutations and Parkinson’s disease: three questions. ASN Neuro. 2009;1 doi: 10.1042/AN20090007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- These two previous reviews explore the biochemistry of LRRK2 in more depth and reference additional articles for the interested reader.