Abstract
Objectives
We aim to develop non-invasive prenatal diagnosis (NIPD) for cystic fibrosis (CF) and determine costs and implications for implementation.
Methods
A next-generation sequencing assay was developed to detect ten common CF mutations for exclusion of the paternal mutation in maternal plasma. Using uptake data from a study exploring views on NIPD for CF, total test-related costs were estimated for the current care pathway and compared with those incorporating NIPD.
Results
The assay reliably predicted mutation status in all control and maternal plasma samples. Of carrier or affected adults with CF (n = 142) surveyed, only 43.5% reported willingness to have invasive testing for CF with 94.4% saying they would have NIPD. Using these potential uptake data, the incremental costs of NIPD over invasive testing per 100 pregnancies at risk of CF are £9025 for paternal mutation exclusion, and £26 510 for direct diagnosis.
Conclusions
We have developed NIPD for risk stratification in around a third of CF families. There are economic implications due to potential increased test demand to inform postnatal management rather than to inform decisions around termination of an affected pregnancy. © 2015 The Authors. Prenatal Diagnosis published by John Wiley & Sons, Ltd.
Introduction
Cystic fibrosis (CF) is a severe, autosomal recessive, multi-system condition, predominantly affecting the respiratory and digestive systems. CF is caused by mutations, of which there are more than 1900, in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. The prevalence of CF is 1 in 2500 to 1 in 3500 live births, and for people with Northern European ancestry, the carrier frequency is 1 in 25.1 Advances in multi-disciplinary care have improved long-term outcomes for people with CF. In the UK, median survival is now 43 years.2,3
Prenatal diagnosis of CF currently requires an invasive test to obtain fetal genetic material and so carries a small risk of miscarriage4 but is an option that is valued by carrier couples as it allows them to either plan and prepare for the birth of an affected child or make decisions about termination of pregnancy.5 Non-invasive prenatal diagnosis (NIPD) based on analysis of cell-free fetal DNA (cffDNA) in maternal plasma has been reported to exclude the paternal mutation in couples carrying different CF mutations.6–8 However, these reports described analyses developed for individual families, a labour-intensive approach for implementation into routine clinical practice. Here, we describe the development of a next-generation sequencing (NGS) assay designed to detect or exclude ten of the most common CF mutations, for use when each parent carries a different CFTR mutation and the paternal mutation is one of the ten included in this panel. To inform implementation strategies, we also report a cost analysis of NIPD for CF, which was informed by an exploration of stakeholder preferences through a questionnaire-based survey of potential service users and providers.9
Methods
Ethical approval
National Health Service Research Ethics Committee approval was obtained for the collection of maternal and paternal blood samples for NIPD test development (01/0095) and for the questionnaire study (10/H0714/3).
Development of the NIPD assay
Sample collection
Normal and heterozygous genomic DNA (gDNA) control samples with known CFTR mutations were ascertained from our Regional Genetics Laboratory records and used to assess test performance, before testing on maternal plasma samples collected, as part of a larger programme designed to develop standards for NIPD (RAPID RP-PG-0707-10107), from women undergoing invasive diagnostic prenatal testing because of a risk of CF. Outcome of invasive testing was known in all cases.
Sample processing and DNA extraction
Maternal plasma samples were separated from 20 mL of blood within 48 h of blood draw and processed for storage at −80 °C as previously described.10 The cell free DNA (cfDNA) was extracted from 4 mL of maternal plasma using the QIAsymphony into a final volume of 60 μL EB buffer (Qiagen). The gDNA was extracted from 2 mL of blood with the Quickgene-610L Nucleic Acid Isolation System (Fujifilm) into a final volume of 200 μL elution buffer.
Next-generation DNA sequencing
Ten CFTR mutations commonly seen in our regional genetics service, including those on the UK neonatal screening panel, were selected. A targeted amplicon approach was used. PCR primers were designed, using Primer 3 software, to amplify five amplicons of CFTR covering these ten mutations (Table1) and the ZFX/ZFY and HLA-B genes. Each amplicon was individually optimised. Mutation targets for each patient DNA sample were given a common molecular barcode and were amplified in individual single-plex PCRs. PCR was carried out using 10 μL of Phusion High-Fidelity PCR Master Mix (NEB), 500 nM of each primer in a final reaction volume of 20 μL. Cycling conditions were 98 °C for 1 min, followed by 42 cycles of 98 °C for 10 s, 64 °C for 30 s and 72 °C for 30 s, followed by 72 °C for 10 min. Amplicons were pooled and purified using a MinElute PCR Purification Kit (Qiagen), quantified using a Qubit dsDNA BR Assay Kit (Invitrogen) and amplicon quality assessed using a Bioanalyzer (Agilent). Purified PCR products were diluted to 2 nM in Elution Buffer (Qiagen), and samples were pooled to give a single 2 nM library. This pooled library was denatured using 2 M sodium hydroxide, diluted to 8 pM and mixed with an 8 pM PhiX control to give a 20% PhiX spike, providing sequence diversity. The library was sequenced with the Illumina MiSeq using a single-end 100-cycle protocol. After de-multiplexing, the number of reads containing the wild-type and mutant alleles was counted for each target site. The presence of fetal DNA was confirmed by the detection of paternal CFTR sequences, ZFY or paternal HLA-type sequences in the maternal plasma.
Table 1.
Mutations in the cystic fibrosis transmembrane conductance regulator gene that have been included in the next-generation sequencing panel
| cDNA | Protein |
|---|---|
| c.489 + 1G > T | |
| c.1521_1523delCTT | p.(Phe508del) |
| c.1519_1521delATC | p.(Ile507del) |
| c.1624G > T | p.(Gly542*) |
| c.1646G > A | p.(Ser549Asn) |
| c.1647T > G | p.(Ser549Arg) |
| c.1652G > A | p.(Gly551Asp) |
| c.1657C > T | p.(Arg553*) |
| c.1679G > C | p.(Arg560Thr) |
| c.3846G > A | p.(Trp1282*) |
Estimation of assay applicability
The ten mutations included in the NGS assay have a combined allele frequency of 77%.11 We estimated how many couples would be eligible to use this assay using the reported allele frequency11 for each mutation on the panel to determine the probability that the father has the mutation and the mother does not have the mutation (Table2).
Table 2.
Probability that the father carries one of the ten common mutations and the mother carries a different mutation to the father
| CFTR mutation | Probability father has the mutation | Probability mother does not have the mutation | Probability father has the mutation and mother has a different mutation |
|---|---|---|---|
| c.489 + 1G > T | 0.009 | 0.991 | 0.008919 |
| p.Phe508del | 0.686 | 0.314 | 0.215404 |
| p.Ile507del | 0.003 | 0.997 | 0.002991 |
| p.Gly542* | 0.024 | 0.976 | 0.023424 |
| p.Ser549Asn | 0.001 | 0.999 | 0.000999 |
| p.Ser549Arga | — | — | — |
| p.Gly551Asp | 0.021 | 0.979 | 0.020559 |
| p.Arg553* | 0.009 | 0.991 | 0.008919 |
| p.Arg560Thr | 0.002 | 0.998 | 0.001996 |
| p.Trp1282* | 0.014 | 0.986 | 0.013804 |
| Total | 0.297015 |
Allele frequencies obtained from Bobadilla et al.11
CFTR, cystic fibrosis transmembrane conductance regulator.
Allele frequency not reported.
Health economics
To assess the economic consequences of implementing NIPD for prenatal diagnosis of CF, we estimated the total test-related costs of three different clinical pathways: the current invasive testing only pathway, NIPD for the paternal CF mutation and NIPD for direct diagnosis (Figure1). A questionnaire-based study of stakeholder views and preferences was undertaken to estimate the uptake of invasive testing and NIPD with detailed results published elsewhere.9 In brief, adult patients with CF and carriers of CF attending one children’s and one adult NHS regional specialist CF centre serving large geographical areas were invited to complete a questionnaire while waiting to see the clinician, or to complete it at home and return it via reply paid envelope. The questionnaire addressed views on key attributes of NIPD, assessed attitudes towards cffDNA testing for CF and asked whether they had had, or would have, an invasive test for CF, if they would have NIPD if available and what their reasons for testing are. The current price of prenatal molecular testing for CF (£370) and NIPD for paternal exclusion (£550) in our Regional Genetics laboratory were used. An estimated price of £750 for the NIPD test currently under development for direct diagnosis was used. Costs of sending in the NIPD sample (£5) and cost of performing the invasive test (£750), which includes chorionic villus sampling, counselling, quantitative fluorescent PCR (QF-PCR) and karyotyping, were obtained from our local fetal medicine unit. NHS reference costs were used to determine the phlebotomy costs, and the cost of feeding back the NIPD results was estimated from Unit Costs of Health and Social Care.13 The number of women undergoing each test and the total costs of the tests were calculated per 100 pregnancies at risk of CF. As the proportion of carrier parents eligible for NIPD for paternal exclusion is approximately 29.7%, in this pathway, we assumed that the remaining parents were offered invasive testing only. Lastly, we calculated the total number of procedure-related miscarriages for each pathway. Input parameters for the economic analysis are shown in Table3. In view of the uncertainty around the number of eligible parents for NIPD for parental exclusion, the uptake of NIPD and IPD and the costs of NIPD, we varied these parameters over a wide range in a sensitivity analysis on the incremental costs of each NIPD pathway compared with the current pathway.
Figure 1.
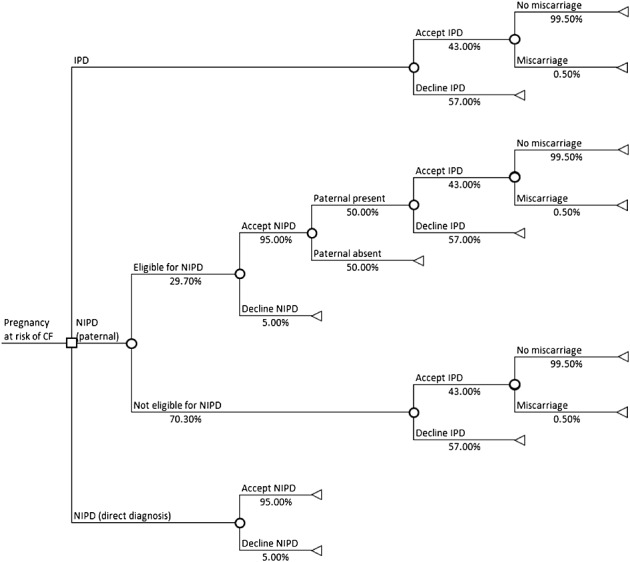
Decision tree depicting the clinical pathways of current invasive testing only, non-invasive prenatal diagnosis (NIPD) for the paternal cystic fibrosis (CF) mutation and NIPD for direct diagnosis
Table 3.
Input parameters for the economic analysis
| Parameter | Value | Source |
|---|---|---|
| Proportion of carrier parents eligible for paternal mutation NIPD | 29.7% | Table 1 – supplementary information |
| Uptake invasive testing | 43.0% | Questionnaire results |
| Uptake NIPD | 95.0% | Questionnaire results |
| Cost of invasive molecular testing for CF | £370 | Regional Genetics laboratory |
| Cost of counselling, invasive test and cytogenetics | £750 | Local fetal medicine unit |
| Cost of NIPD to exclude paternal CF mutation | £550 | Regional Genetics laboratory |
| Cost of NIPD to directly diagnose CF | £750 | Estimation from the Regional Genetics laboratory |
| Cost of phlebotomy | £4 | NHS reference costs15 |
| Cost of sending in NIPT sample | £5 | Local fetal medicine unit |
| Cost of feedback NIPT results | £27 | Unit Costs of Health and Social Care16 |
| Risk of procedure-related miscarriage with invasive testing | 0.5% | Tabor et al.4 |
Total costs of invasive testing were £370 + £750 = £1120. Total costs of NIPD were £550 + £4 + £5 + £27 = £586 for paternal exclusion and £750 + £4 + £5 + £27 = £786 for direct diagnosis. Costs of pretest genetic counselling and ultrasound for dating and exclusion of multiple pregnancies are not included as it applies equally to all scenarios.
NIPD, non-invasive prenatal diagnosis; CF, cystic fibrosis.
Results
NGS assay development
All eight mutations in the gDNA samples from CF carriers were reliably detected at an allele frequency of 50%. Three normal control samples gave either no or extremely low numbers of mutation reads (zero to 0.09% of wild-type reads) across all amplicons (Table4). Sequencing of three controls, using spiked mutant gDNA with three CFTR mutations with an expected mutant allele frequency of 10%, gave percentage mutant allele reads of 10.4%, 8.8% and 4.4% thus demonstrating detection of low level mutant allele targets (Table4).
Table 4.
The ten cystic fibrosis transmembrane conductance regulator mutations used on the next-generation sequencing panel showing the validation of the assay
| Amplicon | Target | Control samples | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| wt | wt | wt | c.489 + 1G > T | c.489 + 1G > Ta | p.Phe508del | p.Phe508del | p.Phe508dela | p.Ile507del | p.Gly542* | p.Gly542*a | p.Ser549Arg;p.Arg553* | p.Arg553* | p.Arg560Thr | p.Trp1282* | ||
| 1 | c.489 + 1G > T | |||||||||||||||
| wt | 63 507 | 80 847 | 87 904 | 3790 | 117 281 | - | - | - | - | - | - | - | - | - | - | |
| Mutation | 48 | 29 | 45 | 986 | 13 624 | - | - | - | - | - | - | - | - | - | - | |
| 2 | p.Phe508del | |||||||||||||||
| wt | 39 505 | 65 137 | 53 881 | - | - | 29 065 | 16 391 | 94 324 | - | - | - | - | - | - | - | |
| Mutation | 0 | 0 | 0 | - | - | 38 038 | 24 215 | 9114 | - | - | - | - | - | - | - | |
| 2 | p.Ile507del | |||||||||||||||
| wt | 39 505 | 65 137 | 53 881 | - | - | - | - | - | 102 390 | - | - | - | - | - | - | |
| Mutation | 6 | 13 | 0 | - | - | - | - | - | 73 954 | - | - | - | - | - | - | |
| 3 | p.Gly542* | |||||||||||||||
| wt | 35 070 | 76 550 | 61 765 | - | - | - | - | - | - | 11 479 | 67 557 | - | - | - | - | |
| Mutation | 8 | 15 | 5 | - | - | - | - | - | - | 9323 | 3093 | - | - | - | - | |
| 4 | p.Ser549Asn | |||||||||||||||
| wt | 43 652 | 63 265 | 62 241 | - | - | - | - | - | - | - | - | - | - | - | - | |
| Mutation | 9 | 9 | 6 | - | - | - | - | - | - | - | - | - | - | - | - | |
| 4 | p.Ser549Arg | |||||||||||||||
| wt | 43 652 | 63 265 | 62 241 | - | - | - | - | - | - | - | - | 6382 | - | - | - | |
| Mutation | 25 | 41 | 55 | - | - | - | - | - | - | - | - | 117 728 | - | - | - | |
| 4 | p.Gly551Asp | |||||||||||||||
| wt | 43 652 | 63 265 | 62 241 | - | - | - | - | - | - | - | - | - | - | - | - | |
| Mutation | 12 | 7 | 12 | - | - | - | - | - | - | - | - | - | - | - | - | |
| 4 | p.Arg553* | |||||||||||||||
| wt | 43 652 | 63 265 | 6 224 127 | - | - | - | - | - | - | - | - | 6382 | 7814 | - | - | |
| Mutation | 20 | 32 | - | - | - | - | - | - | - | - | 134 550 | 5825 | - | - | ||
| 4 | p.Arg560Thr | |||||||||||||||
| wt | 43 652 | 63 265 | 62 241 | - | - | - | - | - | - | - | - | - | - | 84 779 | - | |
| Mutation | 1 | 0 | 1 | - | - | - | - | - | - | - | - | - | - | 99 571 | - | |
| 5 | p.Trp1282* | |||||||||||||||
| wt | 68 398 | 55 077 | 72 883 | - | - | - | - | - | - | - | - | - | - | - | 12 500 | |
| Mutation | 9 | 13 | 12 | - | - | - | - | - | - | - | - | - | - | - | 11 719 | |
| Sex chr marker | ||||||||||||||||
| ZFX | 29 051 | 76 122 | 33 139 | - | 152 529 | 75 726 | 42 498 | 103 476 | 97 466 | - | - | 154 579 | - | 234 960 | 25 651 | |
| ZFY | 34 994 | 53 | 36 324 | - | 2 | 16 | 51 726 | 10 674 | 105 767 | - | - | 187 497 | - | 5 | 0 | |
- Mutation not tested.
10% spiked genomic DNA sample.
In all four maternal plasma cell-free DNA samples, it was possible to accurately determine inheritance of the paternal mutant allele (Table5), and the results of NGS panel testing were concordant with mutation status determined by traditional testing in all cases. In families 2 and 3, very low (0.02%) or absence of paternal mutation counts in maternal plasma indicated transmission of the wild-type paternal allele. This is consistent with an unaffected fetus at 50% risk of having inherited the maternal mutation, which would be compatible with carrier status. In families 1 and 4, low but significant paternal mutation counts in the maternal plasma (15.9% and 5.9% respectively), which were not seen (family 1) or seen in very low level (0.02%) in the maternal gDNA (family 4), indicated that the fetus had inherited the paternal mutation and was at least a carrier of CF; further testing would be required to determine if the maternal mutation had been inherited.
Table 5.
Numbers of reads for the wild-type and mutant sequences with the next-generation sequencing assay for ten common cystic fibrosis transmembrane conductance regulator mutations in maternal plasma samples
| Family | Sample | Confirmed genotype | p.Phe508del | p.Gly551Asp | c.489 + 1G > T | p.(Arg553*) | Sex chromosome marker | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| wt | Mutant | wt | Mutant | wt | Mutant | wt | Mutant | ZFX | ZFY | |||
| 1 | Maternal gDNA | p.Gly551Asp/wt | 106 052 | 0 | 60 873 | 31 429 | - | - | - | - | 163 337 | 0 |
| 1 | Paternal gDNA (not available) | p.Phe508del/wt | - | - | - | - | - | - | - | - | - | - |
| 1 | Maternal plasma cfDNA | p.Phe508del/wt | 108 237 | 9364 | 84 808 | 62 779 | - | - | - | - | 120 054 | 11 060 |
| 2 | Maternal gDNA | p.Phe508del/wt | 58 949 | 88 064 | - | - | - | - | - | - | 137 041 | 11 |
| 2 | Paternal gDNA | c.489 + 1G > T | - | - | - | - | 131 471 | 68 657 | - | - | 114 436 | 145 574 |
| 2 | Maternal plasma cfDNA | p.Phe508del/wt | 54 003 | 68 117 | - | - | 108 880 | 21 | - | - | 26 298 | 0 |
| 3 | Maternal gDNA (not available) | p.(Arg553*)/wt | - | - | - | - | - | - | - | - | - | - |
| 3 | Paternal gDNA | p.Phe508del/wt | 12 813 | 21 653 | - | - | - | - | 23 012 | 335 | 17 693 | 23 163 |
| 3 | Maternal plasma cfDNA | p.(Arg553*)/wt | 45 550 | 0 | - | - | - | - | 13 427 | 13 878 | 34 688 | 2398 |
| 4 | Maternal gDNA | p.Phe508del/wt | 20 724 | 32 876 | 25 320 | 6 | - | - | - | - | 68 069 | 1 |
| 4 | Paternal gDNA (not available) | p.Gly551Asp/wt | - | - | - | - | - | - | - | - | - | - |
| 4 | Maternal plasma cfDNA | p.Phe508del/p.Gly551Asp | 28 076 | 26 776 | 22 616 | 1339 | - | - | - | - | 8700 | 29 |
- Mutation not tested.
gDNA, genomic DNA.
Applicability of the NGS assay using the ten mutations
An estimated 11.9% of carrier parents in the UK will be heterozygous for a mutation in this NGS panel. As the maternal mutation does not need to be on the panel, we estimate that 29.7% of CF carrier parents in the UK could be offered testing (Table2).
Stakeholder views and preferences
One hundred and forty-two potential service users responded to the questionnaire (88.9%). For those affected with CF (n = 92) 45.7% were female, while for carriers of CF (n = 50), the majority were female (72%) and 91.7% had a child affected with CF. The group was well educated, 42.8% having college or other training and 21.7% a degree or equivalent.9 Of these 142 service users, 131 answered the questions regarding uptake of invasive or NIPD testing. More participants said they would decline (74/131) than would accept (57/131) invasive diagnostic testing for CF. The most common reasons for wanting prenatal diagnosis were ‘to prepare for the possible birth of a baby with CF’ (n = 33; 62.3%) and to ‘help make a decision about whether or not to continue the pregnancy’ (n = 17; 32.1%). Most participants (n = 130; 94.9%) said that they would choose NIPD for CF and 90% would be prepared to pay for it, with 49.2% prepared to pay up to £50, 39.0% prepared to pay £100–200 and 10.3% prepared to pay more than £200. One hundred and fourteen participants described potential benefits of NIPD. These fell into five main categories: no miscarriage risk (n = 90), simpler and less stressful test (n = 22), preparation for the birth of an affected child (n = 20), informing decisions around termination of pregnancy (n = 6), earlier testing (n = 6) and more people likely to accept a blood test (n = 9). The 115 responses to questions regarding concerns fell into three main categories: no concerns (n = 101), increase in terminations (n = 13) and increased pressure to terminate (n = 2). No significant differences were seen between responses of carriers and those affected with CF.
Health economics
Total costs were £586 for NIPD for paternal exclusion, £786 for direct diagnosis and £1120 for invasive testing. From the questionnaire responses, we estimated 43% of women at risk of having a baby with CF would have IPD at a total cost of £48 160 per 100 pregnancies at risk, and 28/100 would undergo NIPD for paternal exclusion if available with a total of 36/100 who would undergo invasive testing (six after NIPD, 30 without NIPD). The total cost for this pathway per 100 women was £57 185. We further estimated that 95 women would undergo NIPD for direct diagnosis of CF if available, and none would require invasive testing, costing in total £74 670. NIPD to exclude the paternal CF mutation would therefore increase the costs by £9025, while the incremental costs of NIPD to directly diagnose CF would be £26 510 if compared with the current pathway (Table6). The rate of miscarriages per 100 pregnancies was low, 0.22 in the current pathway, and implementation of paternal mutation NIPD could lower this to 0.18. No procedure-related miscarriages were expected for NIPD to directly diagnose CF. The sensitivity analysis (Table7) shows that the incremental costs of each NIPD pathway compared with the current pathway were higher than the base case if more parents were eligible for NIPD (for paternal exclusion pathway only) or if the uptake of NIPD or the costs of NIPD were higher. If the uptake of invasive testing was higher, the incremental costs of both NIPD pathways were lower than the base case value and vice versa. The NIPD pathways were more costly than the current pathway in each scenario.
Table 6.
Results of the economic analysis of prenatal diagnosis for cystic fibrosis per 100 pregnancies
| Pathway | Number of women undergoing NIPD | Total costs of NIPD | Number of women undergoing IPD | Total costs of IPD | Total costs (NIPD + IPD) | Number of procedure-related miscarriages |
|---|---|---|---|---|---|---|
| Current (invasive testing only) | 0.00 | £0 | 43.00 | £48 160 | £48 160 | 0.22 |
| NIPD (paternal exclusion) | 28.22 | £16 534 | 36.30 | £40 651 | £57 185 | 0.18 |
| Difference (paternal exclusion) compared with current | +28.22 | +£16 534 | −6.70 | −£7 509 | +£9 025 | −0.03 |
| NIPD (direct diagnosis) | 95.00 | £74 670 | 0.00 | £0 | £74 670 | 0.00 |
| Difference (direct diagnosis) compared with current | +95.00 | +£74 670 | −43.00 | −£48 160 | +£26 510 | −0.22 |
NIPD, non-invasive prenatal diagnosis; IPD, invasive prenatal diagnosis.
Table 7.
Sensitivity analysis describing the incremental costs of NIPD for paternal exclusion and NIPD for direct diagnosis compared with the current pathway (invasive testing only) at different levels of test uptake and costs
| Scenario | Incremental costs (paternal exclusion) | Incremental costs (direct diagnosis) |
|---|---|---|
| Base case scenario | £9025 | £26 510 |
| 20% eligible for paternal NIPD | £6077 | |
| 40% eligible for paternal NIPD | £12 154 | |
| 80% Uptake of NIPD | £5341 | £14 720 |
| 100% Uptake of NIPD | £10 252 | £30 440 |
| 30% Uptake of IPD | £11 295 | £41 070 |
| 60% Uptake of IPD | £6056 | £7470 |
| Paternal NIPD test costs £300 | £1971 | |
| Paternal NIPD test costs £800 | £16 078 | |
| Direct NIPD test costs £500 | £2760 | |
| Direct NIPD test costs £1000 | £50 260 |
NIPD, non-invasive prenatal diagnosis; IPD, invasive prenatal diagnosis.
Discussion
Here, we report the successful development of NIPD for the detection or exclusion of a range of paternal CF mutations. This will reduce the need for invasive diagnostic testing, which will only be required if the paternal mutation is present, as if it is absent, the fetus cannot be affected with CF. This approach to NIPD for CF has been approved by the UK Genetic Testing Network, the regulatory body responsible for evaluating new genetic tests and recommending their use within the NHS.14
Our data clearly indicate the need to optimise PCR conditions for each mutation target and analyse maternal gDNA alongside the plasma sample to assess the level of background in any given sequencing run. In our spiking experiment, one sample showed a significant deviation from the expected level of 10%, and in others, the ratio of wild-type to mutant counts for some heterozygous control samples deviated from the expected level of 50%. It is unclear whether this is due to the quality of individual DNA samples or an intrinsic amplification bias of specific primer pairs. Furthermore, very low ‘background’ counts were sometimes observed for mutation targets not present in the DNA sample being tested (Table2, cases 2 and 4). It is likely that these are sequencing or PCR-related errors which have occurred during the early stages of amplification that are well documented in the literature.15 In cases where no paternal mutation is detected in maternal plasma, the presence of cffDNA should be confirmed by analysis of ZFX/ZFY and HLA-B sequences to control against false-negative results occurring because of very low levels of cffDNA in the maternal plasma.16
To date, the paternal exclusion assays for CF reported in the literature have been developed on a case-by-case basis, usually using PCR-based methodologies, which cannot be applied to multiple different mutations simultaneously in one assay.6–8 This approach cannot be readily implemented into a service laboratory where significant throughput of assays testing multiple cases or multiple mutations in a single run are required to optimise turnaround times and minimise costs. We have addressed this by developing an NGS assay that incorporates a panel of mutations, which also allows for expansion of the number of mutations tested for and therefore would be of use to more families. This assay can be used to simultaneously test different families for different mutations as well as testing other samples for different conditions. However, the high frequency of the CFTR mutation p.Phe508del means an estimated 47% of carrier parents will both carry this mutation and will not be eligible to utilise this test. NIPD for recessive disorders where the parental mutations are the same has been demonstrated using relative mutation dosage.10,17 These assays are reliant on precise single-molecule counting techniques and accurate assessment of the fetal fraction of cfDNA. The practical limitations of estimating fetal fraction, the number of repeat tests required and the need for a high proportion of cffDNA in the sample have hindered translation into clinical practice. Other approaches to NIPD for direct diagnoses of recessive conditions have been described, including the analysis of single-nucleotide polymorphisms by whole genome sequencing and relative haplotype dosage analysis18 and targeted NGS.19 However, costs and time requirements for testing with these methods currently inhibit widespread routine clinical implementation. An approach for offering direct diagnosis when parents carry the same mutation would be to sequence the CFTR mutations alongside several informative single-nucleotide polymorphisms around the CFTR locus; this could facilitate calculation of fetal fraction in the maternal plasma20 and relative mutation dosage could be used to determine inheritance.19 This approach is under development in our laboratory, and we have used the predicted costs of this assay in the economic analysis to estimate the cost of direct diagnosis of CF using NIPD.
Ideally, implementation of new technologies should be accompanied by stakeholder views and evaluation of costs. Our survey of potential service users showed that interest in NIPD was high, largely due to the improved safety, with over 90% of potential service users reporting they would have NIPD if it was available, compared with 43.5% who would currently consider invasive testing. However, while many stated results would guide decisions about termination, a large proportion would have the test to prepare for the birth of a baby affected with CF. This is compatible with other studies exploring the reproductive choices of couples at risk of CF.5,21–25 Consequently, it is likely that the high potential uptake of NIPD includes many couples who would currently decline invasive testing and who would want testing for information only, rather than to guide decisions around termination of pregnancy. These findings have clear consequences for the cost of prenatal testing for CF in the NHS as the likely increase in uptake means that the cost of a prenatal diagnostic service based on NIPD, whether for the exclusion of the paternal allele or for direct diagnosis, will be significantly higher than the current care pathway based on invasive testing, even if we take into account that hypothetical uptake of genetic tests can differ from actual uptake.26–28 When discussing prenatal testing for CF, professionals should be careful to include the fact that early postnatal diagnosis is now available as screening for CF now forms a routine part of neonatal screening in the UK and some other countries, as this may influence decisions of parents committed to continuing the pregnancy. The findings of this cost analysis are quite distinct to a study costing NIPD for fetal sex determination, which found that NIPD was cost neutral for the NHS as the cost of NIPD was offset by the reduction in invasive testing,29 the key difference being that current rates of invasive testing and termination for serious sex-linked disorders are high by comparison with those for CF.
The use of NIPD for CF for information only raises ethical questions around whether, in a state-funded health care system, directing resources to a test that would not change pregnancy management can be justified.30 In the UK, NIPD for fetal sex determination is used to direct invasive testing in pregnancies at risk of serious sex-linked conditions. However, this test has not been approved for use in women who are carriers of haemophilia, where termination of an affected pregnancy is increasingly rare and fetal gender could be determined by ultrasound at 20 weeks. Accordingly, some clinical services offer NIPD only when it is likely to change clinical management as the additional costs are not balanced by clinical benefits.31 The definition of clinical benefit is crucial in such policy decisions as there is evidence that there are psychological benefits to early testing and having information to plan and prepare.32,33 In addition, decisions may vary from the couple’s initial intention once they receive a result.
Notably, most respondents raised no concerns about NIPD for CF, with a very few worried about the potential for NIPD to increase the number of terminations for CF. These findings are in line with those from qualitative interviews with carriers of other single-gene disorders.32
Study limitations
The numbers of clinical cases tested are small, and further cases are needed for ongoing validation. The general applicability of the questionnaire study findings may be limited as the study was conducted in only two regional centres and some groups of potential service users are not well represented, including carrier fathers and carriers without children or whose child with CF has died. Furthermore, responses may vary from those made in real life.
Conclusions
We have successfully developed an NGS assay to allow NIPD to be used for risk stratification in a significant proportion of CF families. Consideration of stakeholders’ views and cost-effectiveness alongside test development indicates that introduction of NIPD for CF would be welcomed and uptake is likely to be high. However, as many would use NIPD for CF to inform postnatal management rather than decisions around termination of an affected pregnancy, there are potential economic implications, and further work is required to resolve ethical issues that might arise. These findings may have implications for NIPD for other conditions and highlight the need for prospective consideration of the ethical and economic issues that may arise as more tests are developed.
Acknowledgments
We thank the parents of children with CF, adults with CF who completed a questionnaire or donated a blood sample for the study and the health professionals who helped with recruitment.
What’s Already Known About this Topic?
Non-invasive prenatal diagnosis of some single-gene disorders, including CF, is possible by analysis of cell-free fetal DNA in maternal plasma.
What Does this Study Add?
Use of a next-generation sequencing panel to detect ten common cystic fibrosis (CF) mutations could provide a flexible approach to non-invasive prenatal diagnosis in around 30% of parents who are carriers of CF mutations.
Potential service users would welcome the introduction of non-invasive prenatal diagnosis for CF.
As uptake is likely to be high, increased costs for prenatal testing must be considered by policy makers.
References
- Southern KW, Munck A, Pollitt R, et al. A survey of newborn screening for cystic fibrosis in Europe. J Cyst Fibros. 2007;6:57–65. doi: 10.1016/j.jcf.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Rowe SM, Miller S, Sorscher EJ. Cystic fibrosis. N Engl J Med. 2005;352:1992–2001. doi: 10.1056/NEJMra043184. [DOI] [PubMed] [Google Scholar]
- UK CF Registry. Annual Data Report 2012. Bromley: UK Cystic Fibrosis Trust; 2013. [Google Scholar]
- Tabor A, Alfirevic Z. Update on procedure-related risks for prenatal diagnosis techniques. Fetal Diagn Ther. 2010;27:1–7. doi: 10.1159/000271995. [DOI] [PubMed] [Google Scholar]
- Henneman L, Bramsen I, Van Os TA, et al. Attitudes towards reproductive issues and carrier testing among adult patients and parents of children with cystic fibrosis (CF) Prenat Diagn. 2001;21:1–9. doi: 10.1002/1097-0223(200101)21:1<1::aid-pd967>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Gonzalez MC, Garcia-Hoyos M, Trujillo MJ, et al. Prenatal detection of a cystic fibrosis mutation in fetal DNA from maternal plasma. Prenat Diagn. 2002;22:946–8. doi: 10.1002/pd.439. [DOI] [PubMed] [Google Scholar]
- Bustamante-Aragones A, Gallego-Merlo J, Trujillo-Tiebas MJ, et al. New strategy for the prenatal detection/exclusion of paternal cystic fibrosis mutations in maternal plasma. J Cyst Fibros. 2008;7:505–10. doi: 10.1016/j.jcf.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Nasis O, Thompson S, Hong T, et al. Improvement in sensitivity of allele-specific PCR facilitates reliable noninvasive prenatal detection of cystic fibrosis. Clin Chem. 2004;50:694–701. doi: 10.1373/clinchem.2003.025981. [DOI] [PubMed] [Google Scholar]
- Hill M, Suri R, Nash E, et al. Preferences for prenatal tests for cystic fibrosis: a discrete choice experiment to compare the views of adult patients, carriers of cystic fibrosis and health professionals. J Clin Med. 2014;3:176–90. doi: 10.3390/jcm3010176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett AN, McDonnell TC, Chan KC, et al. Digital PCR analysis of maternal plasma for non-invasive detection of sickle cell anemia. Clin Chem. 2012;58:1026–32. doi: 10.1373/clinchem.2011.178939. [DOI] [PubMed] [Google Scholar]
- Bobadilla JL, Macek M, Jr, Fine JP, et al. Cystic fibrosis: a worldwide analysis of CFTR mutations – correlation with incidence data and application to screening. Hum Mutat. 2002;19:575–606. doi: 10.1002/humu.10041. [DOI] [PubMed] [Google Scholar]
- Department of Health. NHS reference costs 2012–2013. Available from https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/261154/nhs_reference_costs_2012-13_acc.pdf Accessed October 2014.
- Curtis L. Unit Costs of Health and Social Care 2013. PSSRU Available from http://wwwpssruacuk/project-pages/unit-costs/ Accessed October 2014.
- UK Genetic Testing Network. Supporting genetic testing in the NHS. Third biennial report of the UKGTN. 2012. http://ukgtn.nhs.uk/fileadmin/_migrated/tt_news/news_files/UKGTN_3rd_Biennial_Report_2012_01.pdf Accessed October 2014.
- Nakamura K, Oshima T, Morimoto T, et al. Sequence-specific error profile of Illumina sequencers. Nucleic Acids Res. 2011;39:e90. doi: 10.1093/nar/gkr344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun FM, Chiu RW, Allen Chan KC, et al. Microfluidics digital PCR reveals a higher than expected fraction of fetal DNA in maternal plasma. Clin Chem. 2008;54:1664–72. doi: 10.1373/clinchem.2008.111385. [DOI] [PubMed] [Google Scholar]
- Lun FM, Tsui NB, Chan KC, et al. Non-invasive prenatal diagnosis of monogenic diseases by digital size selection and relative mutation dosage on DNA in maternal plasma. Proc Natl Acad Sci U S A. 2008;105:19920–5. doi: 10.1073/pnas.0810373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y, Chan K, Sun H, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2:61ra91. doi: 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- Lam KW, Jiang P, Liao GJ, et al. Noninvasive prenatal diagnosis of monogenic diseases by targeted massively parallel sequencing of maternal plasma: application to beta-thalassemia. Clin Chem. 2012;58:1467–75. doi: 10.1373/clinchem.2012.189589. [DOI] [PubMed] [Google Scholar]
- New MI, Tong YK, Yuen T, et al. Non-invasive prenatal diagnosis of congenital adrenal hyperplasia using cell-free fetal DNA in maternal plasma. J Clin Endocrinol Metab. 2014;99:E1022–30. doi: 10.1210/jc.2014-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denayer L, Welkenhuysen M, Evers-Kiebooms G, et al. Risk perception after CF carrier testing and impact of the test result on reproductive decision making. Am J Med Genet. 1997;69:422–8. doi: 10.1002/(sici)1096-8628(19970414)69:4<422::aid-ajmg17>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Sawyer SM, Cerritelli B, Carter LS, et al. Changing their minds with time: a comparison of hypothetical and actual reproductive behaviors in parents of children with cystic fibrosis. Pediatrics. 2006;118:e649–56. doi: 10.1542/peds.2005-2551. [DOI] [PubMed] [Google Scholar]
- Dudding T, Wilcken B, Burgess B, et al. Reproductive decisions after neonatal screening identifies cystic fibrosis. Arch Dis Child Fetal Neonatal Ed. 2000;82:F124–7. doi: 10.1136/fn.82.2.F124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotet V, de Braekeleer M, Roussey M, et al. Neonatal screening for cystic fibrosis in Brittany, France: assessment of 10 years’ experience and impact on prenatal diagnosis. Lancet. 2000;356:789–94. doi: 10.1016/S0140-6736(00)02652-0. [DOI] [PubMed] [Google Scholar]
- Myring J, Beckett W, Jassi R, et al. Shock, adjust, decide: reproductive decision making in cystic fibrosis (CF) carrier couples – a qualitative study. J Genet Couns. 2011;20:404–17. doi: 10.1007/s10897-011-9363-z. [DOI] [PubMed] [Google Scholar]
- Sanderson SC, O’Neill SC, Bastian LA, et al. What can interest tell us about uptake of genetic testing? Intention and behavior amongst smokers related to patients with lung cancer. Public Health Genomics. 2010;13:116–24. doi: 10.1159/000226595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman C, Croyle RT, Tercyak KP, et al. Genetic testing: psychological aspects and implications. J Consult Clin Psychol. 2002;70:784–97. doi: 10.1037//0022-006x.70.3.784. [DOI] [PubMed] [Google Scholar]
- Ropka ME, Wenzel J, Phillips EK, et al. Uptake rates for breast cancer genetic testing: a systematic review. Cancer Epidemiol Biomarkers Prev. 2006;15:840–55. doi: 10.1158/1055-9965.EPI-05-0002. [DOI] [PubMed] [Google Scholar]
- Hill M, Taffinder S, Chitty LS, et al. Incremental cost of non-invasive prenatal diagnosis versus invasive prenatal diagnosis of fetal sex in England. Prenat Diagn. 2011;31:267–73. doi: 10.1002/pd.2680. [DOI] [PubMed] [Google Scholar]
- Deans Z, Hill M, Chitty LS, et al. Non-invasive prenatal testing for single gene disorders: exploring the ethics. Eur J Hum Genet. 2012;21:713–8. doi: 10.1038/ejhg.2012.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill M, Compton C, Lewis C, et al. Determination of foetal sex in pregnancies at risk of haemophilia: a qualitative study exploring the clinical practices and attitudes of health professionals in the United Kingdom. Haemophilia. 2011;18:575–83. doi: 10.1111/j.1365-2516.2011.02653.x. [DOI] [PubMed] [Google Scholar]
- Hill M, Compton C, Karunaratna M, et al. 2012;23:1012–21. doi: 10.1007/s10897-014-9725-4. Client views and attitudes to non-invasive prenatal diagnosis for sickle cell disease, thalassaemia and cystic fibrosis. J Genet Couns 2014. [DOI] [PubMed] [Google Scholar]
- Lewis C, Hill M, Skirton H, et al. Non-invasive prenatal diagnosis for fetal sex determination: benefits and disadvantages from the service users’ perspective. Eur J Hum Genet. 2012;20:1127–33. doi: 10.1038/ejhg.2012.50. [DOI] [PMC free article] [PubMed] [Google Scholar]


