Significance
Examination of mutants in linker domain of STAT3 suggests contacts with both the DNA binding and SH2 domains that may cause structural changes and affect pSTAT3-dependent transcription, opening the possibility of new targets for drug inhibition of pSTAT3.
Keywords: STAT3, linker domain, mutants
Abstract
Crystallography of the cores of phosphotyrosine-activated dimers of STAT1 (132–713) and STAT3 (127–722) bound to a similar double-stranded deoxyoligonucleotide established the domain structure of the STATs and the structural basis for activation through tyrosine phosphorylation and dimerization. We reported earlier that mutants in the linker domain of STAT1 that connect the DNA-binding domain and SH2 domain can prevent transcriptional activation. Because of the pervasive importance of persistently activated STAT3 in many human cancers and the difficulty of finding useful drug candidates aimed at disrupting the pY interchange in active STAT3 dimers, we have examined effects of an array of mutants in the STAT3 linker domain. We have found several STAT3 linker domain mutants to have profound effects of inhibiting STAT3 transcriptional activation. From these results, we propose (i) there is definite functional interaction of the linker both with the DNA binding domain and with the SH2 domain, and (ii) these putative contacts provide potential new targets for small molecule-induced pSTAT3 inhibition.
The activation/transcriptional action/inactivation cycle of the STAT transcription factors has been intensively studied (reviewed in refs. 1 and 2). Especially important conclusions relating to structure and function were based on mutagenesis studies and X-ray crystal structures that revealed five conserved regions of STATs 1, 3, and 5 cores (missing the first ∼130 and the last ∼30–50 aa of the full length 736- to 770-aa-long molecules) (3–5). From N to C termini, there are the N-terminal (crystal structure solved separately; refs. 6 and 7), coiled-coil, DNA binding, linker, and SH2 domains. In addition, there is an unstructured –COOH terminal region continuing after the SH2 domain through an unstructured region containing the activating phosphotyrosine and transcription activation domain and a conserved stretch of amino acids containing a serine that must be phosphorylated in the transcription of many genes (8).
The canonical pathway of STAT activation depends on a cytokine or a growth factor binding to its specific cell surface receptor leading to receptor tyrosine phosphorylation by an intrinsic or recruited tyrosine kinase, binding of a STAT through its SH2 domain, and subsequent tyrosine phosphorylation in the unstructured –COOH end of the STAT. Reciprocal binding of the STAT phosphotyrosine of one monomer to the SH2 domain of a partner forms the DNA-binding, transcriptionally active dimer that accumulates in the nucleus and functions after binding specific DNA-binding sites. Dephosphorylation in the nucleus and return to the cytoplasm completes the cycle.
Effective transcription complexes can include more than one DNA-binding transcription factor and almost always include coactivators that promote structural and biochemical changes in chromatin to drive transcription (9, 10). STAT proteins also engage other DNA-binding factors and coactivators (11). In fact, the original purified IFN-α–induced complex (12) consisted of STATs 1 and 2 plus a third 48-kDa protein similar to the myb protein (13). An interaction between an amino acid in the first helix in the STAT1 coiled-coil domain and p48 was found to be blocked by a mutation, K161A, that prevented in vivo transcription of IFNα target genes (14). The p48 protein is now recognized as a member of a large group of IRF (interferon regulated factors) proteins that are known to join STAT proteins in transcriptionally active DNA-binding complexes (reviewed in refs. 1 and 11). To date, no crystal structure of a STAT together with any of these cooperating proteins either on or off DNA has been reported.
Although it was discovered early that the STATs, as do most other TFs, interact with coactivators CBP/p300 transacetylase proteins (15, 16), functional interaction of STATs with other widely used coactivators such as the mediator has been fully documented in cells in only one case (17).
When the STAT1 gene was first cloned, two mRNAs were found, STAT1α and STAT1β produced through alternative choice of polyA sites requiring different splices. STAT1β is much less active in vivo than STAT1α (18), but in vitro, both proteins can drive transcription of a naked DNA template equally with the assistance of the mediator complex. STAT1α can also drive transcription of chromatin in vitro, presumably using the C-terminal transcription activation domain, the CBP/p300 binding site in its –COOH terminal region, but STAT1β lacking this site cannot drive chromatin transcription (19). At this point, the great majority of efforts to find inhibitors of STATs has been aimed at interrupting the phosphorylated STAT dimers or the inhibition of JAK kinases that do play a major but not exclusive role of tyrosine phosphorylation of the STATs (20, 21).
One possible attribute of STATs has not been extensively studied during the STAT gene activation/inactivation cycle: how (or if) information is communicated in the course of STAT transcriptional activity by structural interactions among the three crucial domains, DNA binding, linker, and SH2 domains.
A window into such possible structural changes affecting function was opened with STAT1 when Yang et al. investigated mutations in highly conserved residues in the linker domain. One of these mutants (K544A/E545A) failed to stimulate transcription based on an accelerated off time from DNA of a normally tyrosine phosphorylated dimer (22, 23). Yang later showed that K544A was solely responsible for this effect (24). The question raised was if the domains do not interact structurally with each other how does a single amino acid change within the linker domain decrease DNA binding while leaving the protein intact and capable of normal phosphorylation and nuclear entry?
In addition, Bromberg et al. (25) had earlier found that mutants in the distal region of the SH2 domain of STAT3 (A661C/N663C) were weakly oncogenic and slightly more transcriptionally active than wild-type protein. Increased transcriptional activity of this mutant was proven by Li and Shaw to require Y701 phosphorylation (26). The basis for the small increased transcriptional activity and, presumably, the partial oncogenic capacity of STAT3C (as the mutant was named) was a more avid DNA binding of the STAT3C mutant leading to a slower off time from DNA than wild type and a concomitant slower dephosphorylation rate. Most recently human mutations, both positive- and negative-acting in STATs 1 and 3 (21, 27–31), have been reported especially in the SH2 domain, some of which have constitutive activity (31).
From these several results, there appears to be some communication between the DNA-binding domain and the linker domain and also between the linker domain and the SH2 domain that affects the STAT functions of DNA binding and transcriptional activation. Because of the great importance of frequent and persistent overactivity of STAT3 in a huge variety of human cancers (summarized in refs. 32 and 33), we have investigated and report now on the disruptive effects on transcription of additional mutations in the linker domain of STAT3.
Mutations in STAT3 Affecting Function: The K551 Residue
Crystal structures of STAT1 and STAT3 have been available since 1998 (3, 4). The STAT1 was human (3) and STAT3 was mouse (4), but their amino acid sequences are identical except for E at 706 in STAT3 (human) and D in STAT3 (mouse) plus an S insertion at 701. The structures showed a great similarity including in the linker domain (Fig. S1 A and C). The linker domains feature four major helical regions in both proteins (Fig. S1 B and D).
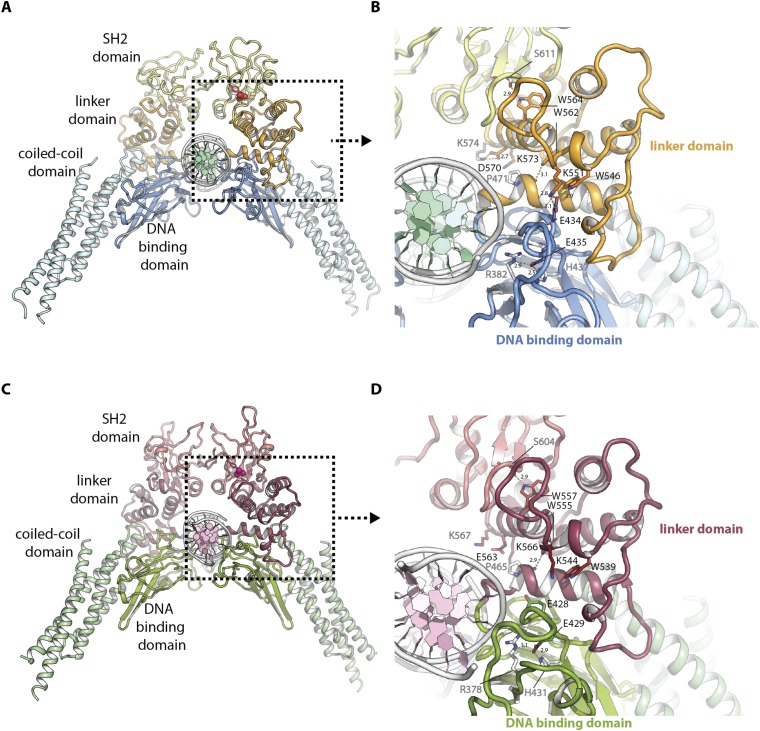
Fig. S1.
Functional architecture of mouse STAT3 and human STAT1 in complex with DNA. (A) Functional architecture of mouse STAT3 in complex with DNA (PDB ID code 1bg1) with coiled-coil domain (light blue), DNA-binding domain (blue), linker domain (orange), SH2 domain (yellow), and DNA (white and green). Phosphotyrosine is highlighted in red. (B) Detailed view of A showing the interface between mouse STAT3 and DNA. Side chains of residues mutated in this study are displayed as colored sticks with black type, residues interacting with them are displayed as white sticks with gray type. Hydrogen bonds are highlighted by yellow dashes and distances labeled in angstroms. (C) Functional architecture of human STAT1 in complex with DNA (PDB ID code 1bf5) with coiled-coil domain (light green), DNA binding domain (green), linker domain (mauve), SH2 domain (light mauve), and DNA (white and light pink). Phosphotyrosine is highlighted in pink. (D) Detailed view of C showing the interface between human STAT1 and DNA. Side chains of residues equivalent to those mutated in mouse STAT3 in this study are displayed as colored sticks with black type, residues interacting with them are displayed as white sticks with gray type. Hydrogen bonds are highlighted by yellow dashes and distances labeled in angstroms.
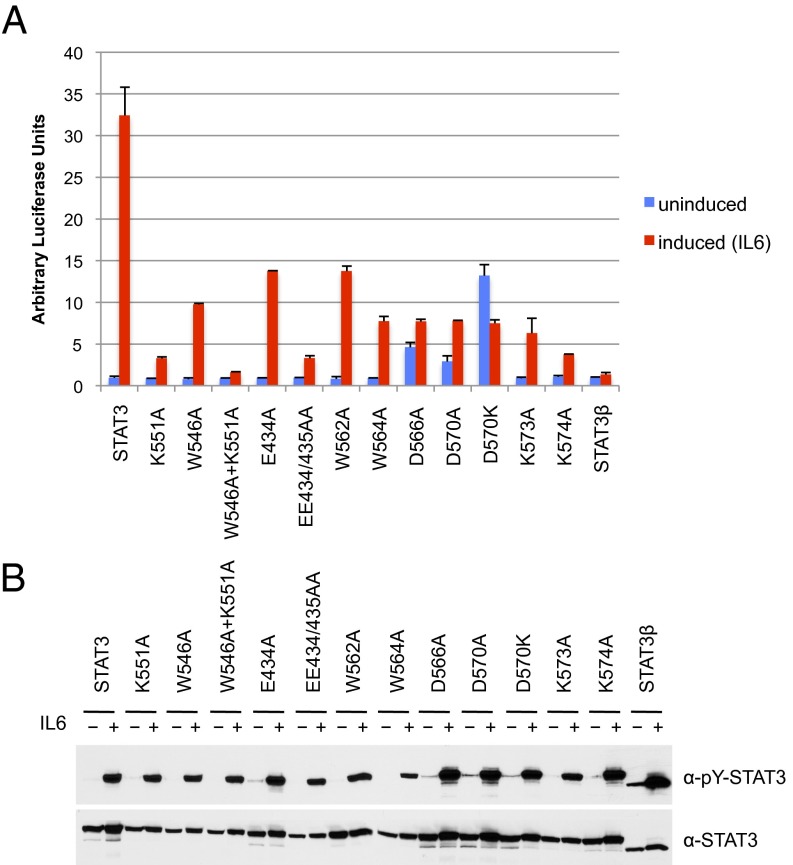
To begin a search for STAT3 linker domain mutants that might affect function, we naturally started with K551, which is the cognate residue to K544 identified earlier in STAT1 (22–24) that was required for effective strong DNA binding and transcriptional activation. We mutated K551 in STAT3 to alanine and transfected a transcriptional reporter construct into a STAT3-negative cell line (A4; provided by G. Stark, Cleveland Clinic, Cleveland) to test for IL-6 transcriptional induction. After IL-6 induction for 4 h, the wild-type protein gave a robust (∼35-fold) transcriptional increase (Fig. 1A). As a control, STAT3β, known to be ineffective in in vivo transcriptional activation (18), gave less than a twofold increase above background. The STAT3 mutant K551A gave a very weak transcriptional response (less than 10% of wild type in many different experiments). Thus, residue K551 in STAT3 clearly has a key functional role (Fig. 2).
Fig. 1.
Point mutations in the STAT3 linker domain inhibit transcriptional activity in response to IL-6. (A) STAT3-null A4 cells were transfected with a α2-macroglobulin reporter luciferase plasmid and the indicated expression constructs for wild-type or mutant STAT3 proteins. Cells were serum starved for 18 h, left untreated (blue bars) or treated (red bars) with IL-6 (80 ng/mL) and soluble IL-6 receptor (100 ng/mL) for 4 h before analysis of luciferase expression. Values were normalized against renilla luciferase activity of the cotransfected internal control plasmid. Data derived from one of three independent experiments with duplicate points (±SD). (B) Western blot analysis of Y705 phosphorylation (Top) and total STAT3 (Bottom) in cell lysates from one experiment subjected to reporter assay analysis shown in A.
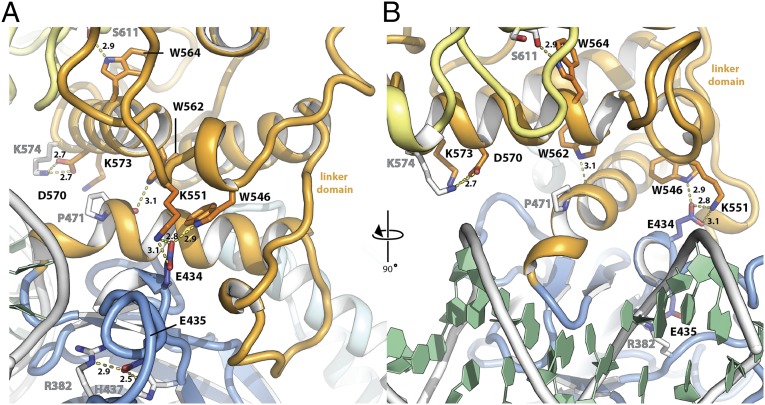
Fig. 2.
Location of mutated residues in the STAT3 linker domain structure. (A) Side view of mouse Stat3 in complex with DNA (PDB ID code 1bg1). DNA binding domain (blue), linker domain (orange), SH2 domain (yellow), and DNA (white and green) are highlighted. Side chains of residues mutated in this study are displayed as colored sticks with black type, residues interacting with them are displayed as white sticks with gray type. Hydrogen bonds are highlighted by yellow dashes and distances labeled in angstroms. (B) Front view of mouse Stat3 in complex with DNA, rotated 90° with respect to A.
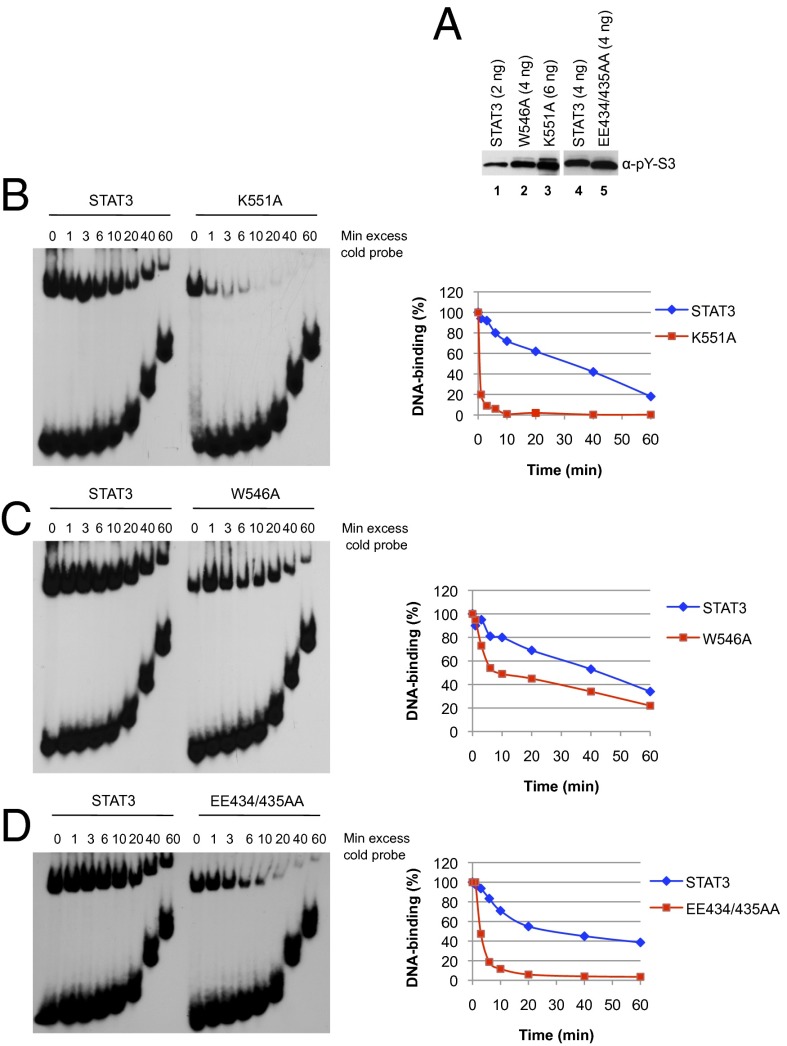
We tested the STAT3 K551A mutant for its DNA off rate by binding to a 32P-labeled M67 SIE nucleotide probe for 15 min followed by addition of unlabeled oligonucleotide and sampled at increasing amounts of time to check for DNA binding. Just as was the case for STAT1 mutated in residue K544, the off rate for STAT3 K551A was much faster than the wild-type protein whether tested as tyrosine phosphorylated pure protein or in a nuclear extract from transfected IL-6–treated cells (Fig. 3B and data not shown). After the unlabeled oligonucleotide chase, there was a 50–60% loss of bound wild-type STAT3 only after 40 min, but a virtual absence of bound STAT3 K551A in 1 min (Fig. 3B). It is further noteworthy that to obtain a DNA bound signal at the start of the off-time assay required three times as much pSTAT3 K551A as wild-type pSTAT3 (Fig. 3A).
Fig. 3.
Mutations in the STAT3 linker domain impair binding of tyrosine phosphorylated STAT3 to DNA. (A) A4 cells were transfected with expression constructs for internally Flag-tagged wild-type and mutant STAT3 proteins carrying a Flag epitope between the first two helices of the coiled-coil domain between amino acid residues 197 and 198. Following serum starvation for 18 h and IL-6 stimulation for 30 min, whole-cell extracts were prepared and subjected to affinity purification on M2-agarose. Bound Flag-tagged STAT3 proteins were eluted with Flag peptide, and purified proteins were analyzed by Western blotting with P-Y705 STAT3 antibody. Protein amounts shown correspond to the DNA-binding signals detected by EMSA in B–D. B and C used WT STAT3 shown in lane 1 of A, D used a different WT STAT3 preparation shown in lane 4 of A. (B–D) Analysis of DNA binding stability by EMSA. Purified wild-type P-STAT3 in comparison with P-K551A (B), P-W546A (C), and P-EE434/435AA (D) was incubated for 15 min with 32P-labeled m67SIE probe. Excess of unlabeled competitor was then added at time 0, and STAT3–DNA complexes were loaded onto the running gels at the indicated times. Signals were quantified by using ImageJ software. Each graph represents the average of two to three independent experiments.
STAT3 Mutagenesis in Other Sites of the Linker Domain
The Yang et al. experiments (22, 23) had explored other STAT1 mutants of highly conserved residues in the linker domain of STAT1. We proceeded to check the effects of mutations in some of these same and additional sites in the STAT3 linker domain. There are 105 residues in the linker, 62 of which are identical to those in STAT1. The four longer helical elements of the linker domain include 53 residues, 45 of which are the same in STAT1 and STAT3. There is an ∼10-aa loop between the last two helices, which is also highly conserved and includes the K551(STAT3) and K544 (STAT1) residues discussed above.
In the STAT1 studies, a number of conserved tryptophans were mutated because of the suggestion that this amino acid plays an outsized role in stabilizing protein structure (34). The STAT1 residues W539, W555, and W557 are cognate in STAT3 with W546, W562, and W564. The first of these residues is in the highly conserved loop just upstream of the final long helix in each of the two STATs, which, as noted, contains the K551 (STAT3) and K544 (STAT1). The other tryptophan residues are in the last helix of the linker domain (Fig. 2).
The STAT1 W539A greatly reduced transcriptional activation (22), and the mutation in the cognate W546A in STAT3 produced a threefold reduction (Fig. 1A). Also, the off time of the STAT3 W546A was approximately twofold faster than wild-type protein (Fig. 3C). Inspection of the crystallographic structure of STAT3 revealed that residues K551 and W546 are in close proximity to glutamic acid E434 in the DNA binding domain (Fig. 2A). We therefore mutated E434 to alanine to test a possible function of this predicted interaction. STAT3 E434A showed a 60% reduction in activity. Interestingly, the double mutant EE434/435AA had been described earlier in the literature as dominant negative STAT3 mutant (35). We confirmed that EE434/435AA has greatly impaired transcription activity (10% of wild-type STAT3; Fig. 1A) as well as faster off times from DNA (80% loss of DNA binding in 6 min; Fig. 3D), a phenotype that mirrors the K551A mutation. In line with a predicted cooperativity between residues K551 and W546, double mutation W546A/K551A caused a slightly greater loss in activity than K551A alone (5% of activity remaining; Fig. 1A). These results taken together strongly support K551:E434:W546 functional interactions within the linker domain, which are then communicated through the linker domain, perhaps also involving E435 to affect the STAT3 function via contacts with the DNA-binding domain.
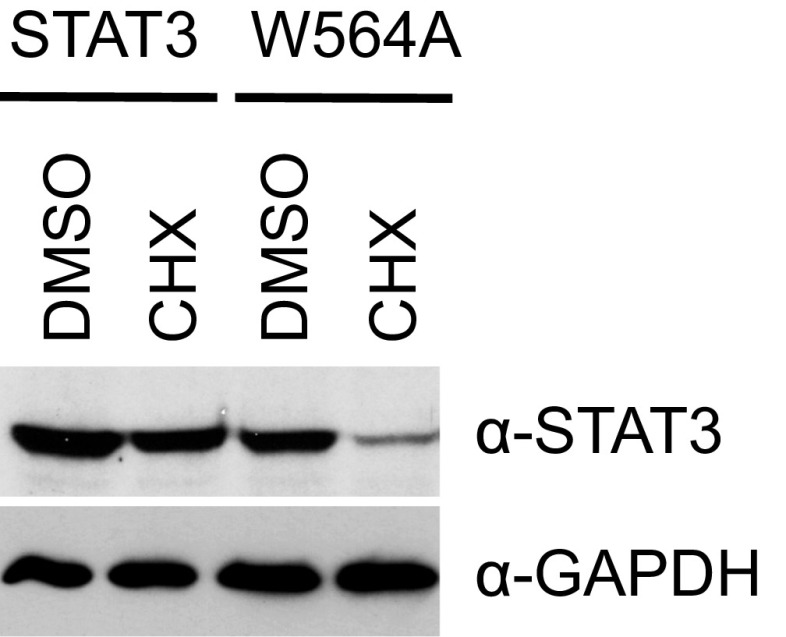
We next investigated the importance of conserved tryptophan residues in the last helix of the linker domain. Whereas the mutation STAT1 W555A had no effect on transcription, STAT3 W562A showed a 50% reduction in activity. Mutation of residues W557 (STAT1) and W564 (STAT3) impaired transcription by 90% and 75%, respectively. Of note, both W557 in STAT1 and W564 in STAT3 when changed to alanine produced less protein than wild type, potentially because the mutant protein was unstable. Stability of STAT3 W564A was tested after construct transfection comparing wild-type and mutant proteins in cells treated with cycloheximide. The W564A protein declined 70% within 4 h of cycloheximide treatment, whereas the wild-type protein was stable (Fig. S2). Disruption of a contact of the last helix in STAT3 by a single amino acid change appears to lead to protein instability.
Fig. S2.
W564A shows decreased protein stability in cycloheximide-treated cells. A4 cells in six-well plates were transfected with 100 ng of expression constructs for wild-type STAT3 or W564A. Twenty-two hours after transfection, cells were treated for 4 h with DMSO or 100 μg/mL cycloheximide (CHX). Whole-cell lysates were analyzed by Western blotting for total STAT3. Blots were reprobed with GAPDH antibody as loading control.
In the original STAT1 crystallography paper, the proximity and, therefore, possible interaction of residues in the last helix in the STAT linker domain with either the DNA binding domain or the SH2 domain, were pointed out (see figure 5 and accompanying discussion in ref. 3). Also, in STAT3, there is an unstructured loop in the SH2 domain just distal to the arginine-containing pocket to which the phosphotyrosine from the partner monomer is bound (Fig. S1A). This loop comes close to the residues in helix 8 of STAT3. In particular, S611 in the STAT3 SH2 loop is close to W564 (Fig. 2B). [This final helix (residues ∼561 to ∼574) is called helix 10 in STAT1 and helix 8 in STAT3 because other short (3- to 5-aa-long) helical regions received different numbering.]
The region of the final linker domain helix proximate to this SH2 domain loop contains several charged residues, D566, D570, K573, and K574 downstream from the tryptophan residues (W562 and W564) discussed earlier. In the crystal structure of STAT3, these residues are also close to the SH2 loop discussed above (Fig. 2B). We therefore made the constructs D566A, D570A, D570K, K573A, and K574A in STAT3 to test for effects on transcriptional activation and on dephosphorylation. All of these mutations had a drastic negative effect on transcription (Fig. 1A) with activities between −2% and 15% of wild-type STAT3. Unexpectedly D566A, D570A, and especially D570K showed increased background reporter activity and low levels of STAT3 tyrosine phosphorylation before IL6 induction despite serum starvation for 18 h that effectively removed traces of tyrosine phosphorylation of wild-type STAT3 and other linker domain mutants (Fig. 1B).
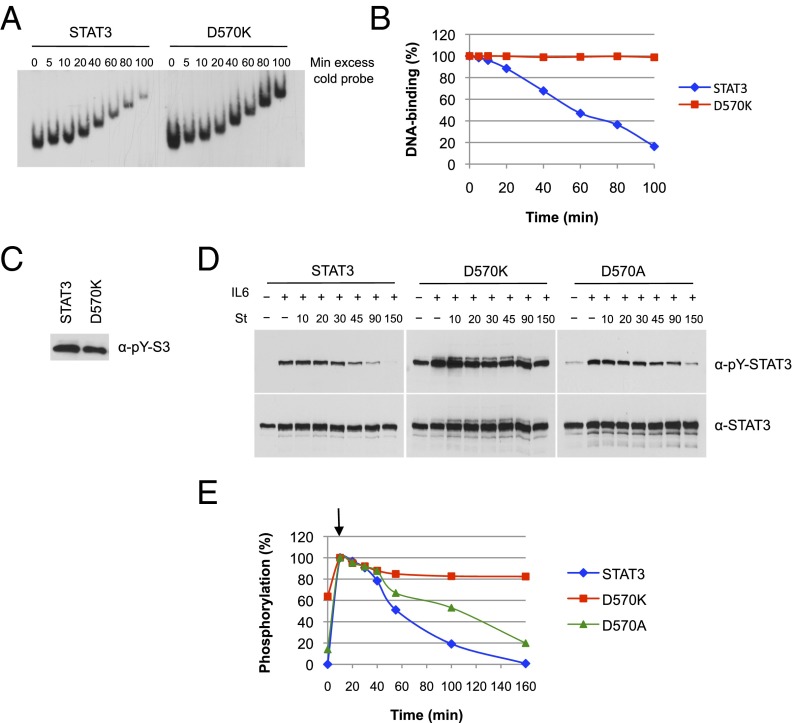
Because STAT3 can be activated by many tyrosine kinases other than receptor-bound JAKs (11), we tested to see whether the “constitutive” STAT3 Y705 phosphorylation activation was due to either src, a constitutive kinase for STAT3, or to the tec kinase that is activated in PI3K-transformed cells (36). Inhibitors specific for each of these kinases did not affect the low but distinct constitutive Y705 phosphorylation in the transfected D570K mutants. Mutant D570K produced a striking phenotype that requires a more detailed description. Despite high levels of induced tyrosine phosphorylation (Fig. 1B), the low level of background transcriptional activity of D570K consistently decreased after initial IL-6 stimulation.
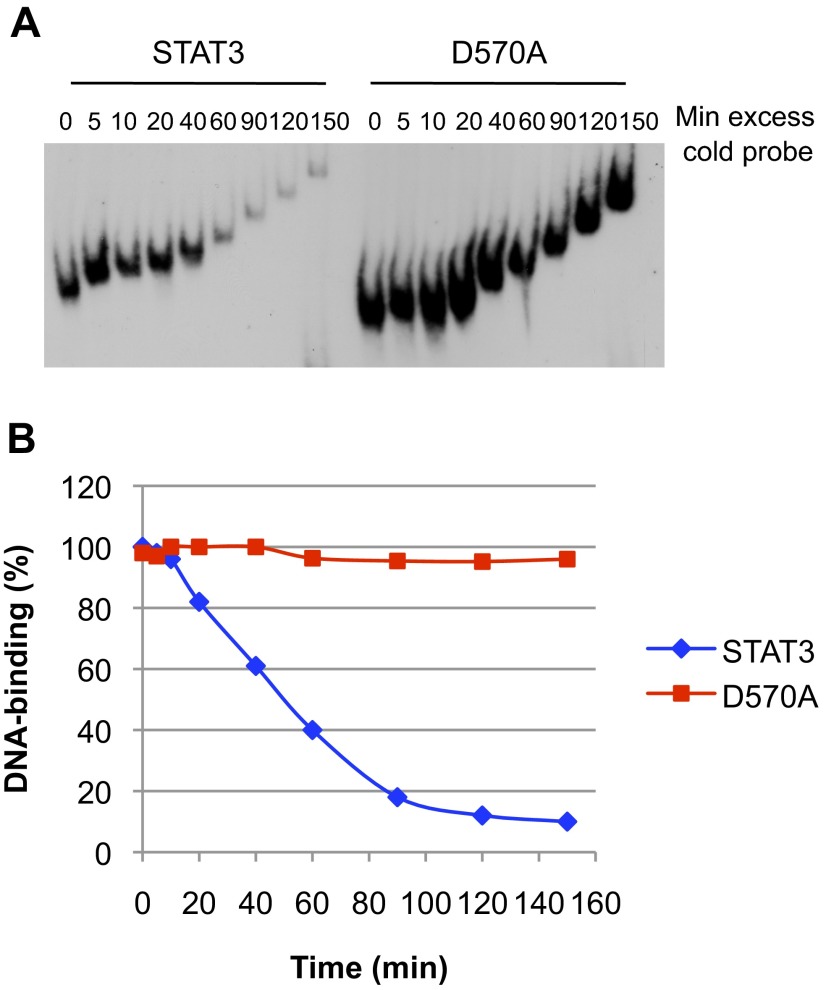
In addition to not responding to IL-6–induced transcription, D570K remained persistently bound to DNA. In off-time experiments with purified protein and radiolabeled oligonucleotides, D570K remained bound to DNA in the presence of unlabeled competitor oligonucleotides for 2.5 h (Fig. 4 A and B show time course over 100 min). The same long-term binding was obtained for the D570A mutant that remained bound to DNA for 2.5 h although stimulating transcription only 5% as well as wild type (Fig. S3).
Fig. 4.
Activated D570K persistently binds to DNA and resists dephosphorylation. (A) DNA binding of D570K. Equal amounts of purified phospho-STAT3 and D570K were incubated for 15 min with 32P-labeled m67SIE probe. Excess of cold probe was then added at time 0, and the protein–DNA complexes were loaded onto the running gel at the indicated times. (B) Quantification of the data shown in A. (C) Purified phospho-STAT3 (4 ng) and D570K (4 ng) used in A were analyzed for Y705 phosphorylation (pY) by immunoblot. (D) Dephosphorylation kinetics of D570K and D570A. A4 cells were transfected with expression constructs for wild-type STAT3, D570K, or D570A and serum starved for 18 h. Parallel samples of cells were left untreated or treated with IL-6 (20 ng/mL) and soluble IL-6 receptor (25 ng/mL) for 10 min, and IL-6/IL-6 receptor for 10 min followed by the tyrosine kinase inhibitor staurosporine (St, 1 μM) for the indicated times (10, 20, 30, 45, 90, and 150 min). Nuclear extracts were prepared and analyzed by Western blotting for phospho-Y705 STAT3 (Top) and total STAT3 (Bottom). (E) Phospho-STAT3 signals were quantified by ImageJ software and normalized for total STAT3 signals. The graph represents the experiment shown in (D). Arrow denotes start of staurosporine treatment after 10 min of IL-6.
Fig. S3.
Activated D570A persistently binds to DNA. (A) Equal amounts of purified phospho-STAT3 and D570A were incubated for 15 min with 32P-labeled m67SIE probe. Excess of cold probe was then added at time 0, and the protein–DNA complexes were loaded onto the running gel at the indicated times. (B) Quantification of the data shown in A.
Effect of STAT3 Linker Domain Mutants on Phosphorylation, Nuclear Entry, and Dephosphorylation
The closure of the STAT activation cycle involves dephosphorylation and return of the monomer molecules to the cytoplasm. Currently this dephosphorylation event is thought not to occur on DNA but only after departure from DNA. Vinkemeier and colleagues investigated these events by using a clever strategy with STAT1 (37). Positively charged residues were placed within the contact sites in the DNA binding domain known from the crystal structure. This mutant produced a phosphorylated STAT1 dimer that binds tightly to both specific STAT binding sites and nonspecific sites but does not leave or become dephosphorylated, supporting the conclusion that dephosphorylation requires departure from the template. It is known that the TC45 phosphatase is the responsible protein in the cell nucleus for dephosphorylation and that the purified enzyme will dephosphorylate DNA-free molecules in a cell-free context and the in vitro reaction can be inhibited by furnishing specific DNA-binding sites (37, 38).
To complete the investigation of the effect of the STAT3 mutants described in this work in the whole cycle of phosphorylation and dephosphorylation, we next determined whether cytoplasmic activation and accumulation in the nucleus was affected.
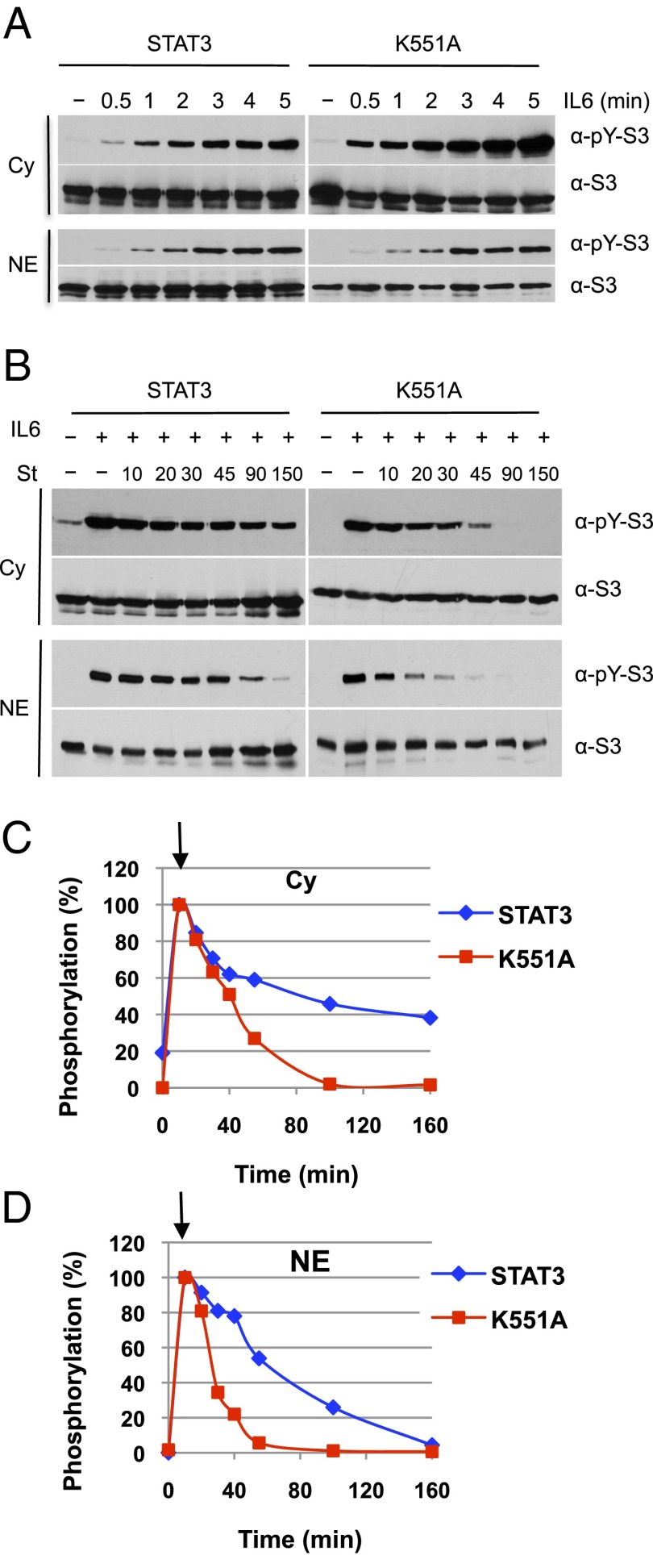
No experiments of which we are aware have begun observation of this cycle in short times after exposure to an activating ligand. Fig. 5A shows that nonphosphorylated STAT3 in these transfected cells is partitioned approximately equally between nucleus and cytoplasm, as has been ascribed in other cells (39) to nuclear shuttling of nonphosphorylated STAT3 (and STAT1; ref. 37). Within 30 s of IL-6 treatment, cytoplasmic pSTAT3 is detectable compared with untreated cell extracts with a gradual rise to a maximum in 5–10 min (Fig. 5A and first sample in Fig. 5B in which cells were treated with IL-6 for 10 min). Nuclear entry of pSTAT3 is evident in 1 min after IL-6 treatment and steadily increases for the 5 min of observation (Fig. 5A) to a total of approximately twofold less than in the cytoplasm. The estimated transit time of a dimeric STAT3 from cytoplasm to nucleus from this data are on the order of a few minutes at most.
Fig. 5.
K551A has similar activation kinetics as wild-type STAT3 but shows faster dephosphorylation in the nucleus. (A) Immediate activation of STAT3 and K551A by IL-6 and flow from the cytoplasm to the nucleus. A4 cells transfected with expression constructs for wild-type STAT3 or K551A were left untreated or treated with IL-6 (80 ng/mL) and soluble IL-6 receptor (100 ng/mL) for the indicated times (0.5, 1, 2, 3, 4, and 5 min) after serum starvation. Cytoplasm (Cy) and nuclear fractions (NE) were extracted from identical samples and equivalent amounts of both fractions analyzed by immunoblotting with antibodies against phospho-Y705 and total STAT3. (B) Dephosphorylation kinetics of K551A. A4 cells expressing wild-type STAT3 or K551A were serum starved for 18 h. Parallel samples of cells were left untreated or treated with IL-6 (20 ng/mL) and soluble IL-6 receptor (25 ng/mL) for 10 min, and IL-6/IL-6 receptor for 10 min followed by staurosporine (1 μM) for the indicated times in minutes. Cytoplasmic (Cy) and nuclear extracts (NE) were prepared and equivalent amounts were analyzed by Western blotting for phospho-Y705 STAT3 and total STAT3. (C and D) Quantification of phospho-STAT3 signals in cytoplasm (C) and nuclear extracts (D). Data were normalized for total STAT3. Graphs represent the experiments shown in B. Arrow marks start of staurosporine treatment after 10 min of IL-6.
Fig. 5B uses the quick-acting general kinase inhibitor staurosporine, at 10 min, to stop further pSTAT3 formation, allowing the examination of the dephosphorylation of nuclear pSTAT3 (40). Dephosphorylation of nuclear STAT3 in these transfected A4 cells after IL-6 activation is not nearly as rapid as is STAT1 after IFN-γ activation in HeLa cells where the cytoplasmic pSTAT1 is gone 15 min after staurosporine treatment and all nuclear pSTAT1 is gone in 45 min (40). In the present experiments, cytoplasmic pSTAT3 declines slowly in staurosporine, and the decline in the nucleus is only approximately two- to fourfold in 45 and 90 min. In the same analysis, the K551A mutant with the rapid off rate from DNA and 5–10% of wild-type STAT3 transcriptional stimulation was dephosphorylated completely within 45 min in the nucleus and within 90 min in the cytoplasm. Because TC45, the phosphatase responsible for dephosphorylation, is nuclear, nuclear entry has to occur for dephosphorylation to be completed (38).
We also tested the effect of staurosporine on the D570K mutant. During in vitro off time measurements, D570K was much more stable than wild-type pSTAT3 (Fig. 4 A and B). Treatment with staurosporine left nuclear pSTAT3 D570K at the same level for 2.5 h (Fig. 4 D and E). This result differs radically from wild-type protein, which has a nuclear pSTAT3 half decay time of ∼40 min, and in staurosporine it becomes completely dephosphorylated. These results imply all the D570K molecules remain bound to DNA. We do not know at this point whether all molecules are bound to true STAT target sites. Further, as noted above, the K551A mutant, which has a faster than normal off time, is dephosphorylated much faster than wild-type protein.
All these results agree with the Vinkemeier laboratory’s conclusion (37) that there is no discernible dephosphorylation of a mutant that persistently binds DNA compared with wild-type protein and support the conclusion that disengagement from DNA is required before dephosphorylation. So we have, by accident, engineered a single amino acid mutant, D570K, that fails to drive transcription, remains bound to DNA for hours, and is not dephosphorylated in vivo as is wild-type protein.
Interpretations of STAT3 Linker Mutants Through Examination of Linker Domain Structure
Table S1 gives the biochemical phenotype of the mutants discussed in this paper. Fig. 2 shows portions of the crystallographic structure of STAT3 (principally the linker domain) and the residues of wild-type protein where mutations and/or potential contacts were made (4). Here, we make an attempt based on plausible expectations from proximity of potentially interactive amino acids to provide a possible structural explanation of the functional effect of some of the mutants.
Table S1.
Summary of effects of STAT3 linker domain mutants
| Mutant | Transcription activation, % | DNA off time | Intramolecular contact |
| K551A | <10 | t1/2,<1 min | E434 |
| W546A | 25–30 | t1/2, 20 min | E434 |
| W546A+K551A | 5 | ND | E434 |
| E434A | 40 | WT | K551, W546 |
| E434A/E435A | 10 | t1/2, 3 min | K551, W546, H437, R382 |
| W562A | 40–50 | ND | P471 |
| W564A | 20–25 | ND* | S611 |
| W566A | 5 | ND | — |
| D570A | 5–8 | Stable 2.5 h | K574 |
| D570K | −2 | Stable 2.5 h | K574 |
| K573A | 15 | NA | — |
| K574A | <10 | NA | D570 |
| WT | 100 | t1/2, 40 min | — |
All mutants are in linker domain except E434A (DBD), which contacts linker domain residues.
ND W564A is a less stable protein than wild type; see Fig. S2.
First, in the K551 region, the positively charged epsilon NH2 group of the lysine side chain is close enough (∼2.8–3.1 Å) to the negatively charged glutamic acid E434 in a loop within but not contacting DNA in the DNA-binding domain to engage in hydrogen bonding. Likewise, the pyrrole ring of the tryptophan residue 546 is also close enough to hydrogen bond with E434 (Fig. 2A). Recall that mutagenesis of either K551 or W546 to A resulted in less stable binding to DNA (faster off time). The alanine substitutions, K551A and W546A, would disrupt these potential H bonds. The crystal structures of STAT3 with these mutations are required to provide further insight into why there is such a dramatic effect on the stability of DNA binding with what might have been anticipated to be a small physical change.
Next, we considered the position of the side chains of the negatively charged residues D570 and the positively charged K573 and K574 in the last helix of the linker. The D570 could interact with either K573 or K574. However, none of the side chains of these residues seems close to residues in the DNA-binding domain or to the SH2 domain. Why these potential interactions would cause the strong DNA binding of D570K, is unclear. Even in its most extended conformation, the lysine side chain of the D570K mutant would be at least ∼3.5 Å from the DNA backbone. A crystal structure would be most valuable to understand this mutant (D570K).
Finally, as noted earlier, the proximity of the last helix of the linker domain in STAT1 was noted to be quite close to a loop in the SH2 domain. In STAT3, the same SH2 loop region exists close to the last helix in the linker domain. The SH2 loop is ∼10–15 residues downstream from the pocket containing residue R609, which is crucial for the pY705 on a tail from the opposite STAT monomer to bind, producing the phosphorylated dimer. The W564A mutant in the last helix lowered the transcription rate three- to fourfold, and the side chain of the W564 hydrogen bonds with the backbone carbonyl group of S611. Transfection of the W564A produced decreased transcription but apparently did so by destabilizing the protein (Fig. S2).
Discussion
Upon completion of the structure of STATs 1, 3, and 5 and the initial and growing demonstration of the widespread importance of STAT3 as a driver in human cancer (33), great effort has been applied to blocking STAT3 function by interrupting the interlocking SH2 Y705 interaction that renders STAT3 an active dimeric transcription factor (reviewed in ref. 41). Using excellent wet chemistry and computer-assisted methods, many compounds have been created that attack this target, several successfully, reducing tyrosine 705 phosphorylation, inhibiting tumor cells in culture, and even effecting a modest inhibition of human tumor xenografts in mice (20, 42–45). However, no molecule has been devised or unearthed that has adequate specificity and pharmacologic efficacy to provide hope that this obvious anticancer target will soon yield an effective anti-STAT3 drug in humans.
As has been known for more than 20 years, effective DNA binding of activated STATs and subsequent transcriptional activation depends on residues in the DNA-binding domain (46), but the Yang et al. results with STAT1 suggested and the present results provide conclusive evidence that effective DNA binding also depends on residues in the linker domain. The logical implication is that the linker domain has a previously insufficiently appreciated structural role in maintaining an effective DNA-binding domain. Here, we show that linker domain mutants reveal key residues that enforce the presumptive structural role of the linker domain. If small molecules can be found that effect the same structural changes that the presently described mutants cause, they would be of great interest. The STAT3-destabilizing effects of the K551A mutant could well rest on the K551-E434 contact, which provides a possible target for disruption. Likewise, the transcriptional inhibition of W564A mutation affords another target, the W564–S611 interaction, which destabilizes the STAT3 structure (Fig. S2).
Perhaps the most intriguing but admittedly puzzling lead comes from the D570K mutation. That this abnormally tight DNA binding mutant that does not drive transcription and resists dephosphorylation says a mutation can cause what appears to be not only a disabling mutation but suggests a molecule that permanently disables not only a pSTAT3 molecule but for as long as binding persists, disables a STAT3 site on a chromosome. Further work (crystal structure first) is definitely in order for this puzzling result, plus a determination of whether this tight association persists through, or even affects, DNA replication.
Finally, the usefulness of these results may be to focus attention on the fact that residues relatively distant from what has been the well-attended main arena of interest—STAT3 DNA binding at defined sites and SH2-phosphotyrosine interchange—suggests that STAT3 functions (and STAT function in general) might be affected through subtle interactions of neighboring STAT domains on each other.
Materials and Methods
Cell culture and reagents are described in detail in SI Materials and Methods. All experiments were performed with STAT3 null A4 colon cancer cells derived by homologous recombination, a gift of G. Stark. Luciferase reporter assays, transfection assays, and cell extracts were all standard procedures, and purification of tyrosoine phosphorylated STAT3 proteins are fully described in SI Materials and Methods.
Note.
While completing this work, we learned from a private confidential communication that the compound described by Y. Li et al. (47) as a STAT3 inhibitor binds in a crystal structure in a pocket formed at the junction between the DNA binding domain and the linker domain as the basis for STAT3 inhibition.
Materials and Methods
Cell Culture and Reagents.
STAT3-null A4 cells were derived from human DLD1 colon cancer cells by homologous recombination (48) and were kindly provided by G. R. Stark. Cells were grown in McCoy’s 5A medium with l-glutamine (HyClone), supplemented with 10% (vol/vol) FBS, penicillin, and streptomycin (100 μg/mL) (Gibco-BRL). Before experimental treatment, cells were serum starved (1% or no FBS) overnight, and all treatments were carried out in the absence of serum. Human IL-6 (BioLegend) and the recombinant soluble form of the human IL-6 receptor (Sigma) were used at concentrations of 20 ng/mL or 80 ng/mL (IL-6) and 25 ng/mL or 100 ng/mL (IL-6-R) as indicated. Staurosporine (Abcam) was used at a concentration of 1 μM, cycloheximide (Sigma) at a concentration of 100 μg/mL. STAT3, and phospho-Y705 STAT3 antibodies were purchased from Cell Signaling Technology. M2 agarose and Flag-peptide were purchased from Sigma. Extraction buffers were complemented with protease inhibitor mixture (Halt; Thermo Scientific) and phosphatase inhibitor (Simple Stop; Gold Biotechnology) before use.
Site-Directed Mutagenesis.
Mammalian expression constructs for STAT3 mutants were generated by QuikChange site-directed mutagenesis (Stratagene) using mouse STAT3α in vector pcDNA3.1 as template. All mutants were sequenced over the entire reading frame. Expression constructs for protein purification from mammalian cells were generated by inserting a Flag-tag between amino acid residues 197 and 198 in the coiled-coil domain of STAT3α. The internally Flag-tagged wild-type STAT3 was then used as template for consecutive QuikChange site-directed mutagenesis to introduce point mutations in linker and DNA binding domain.
Luciferase Reporter Assays.
Transient transfections for reporter assays were carried out in 24-well plates with 1.0 × 105 cells per well by using 100 ng of α2-M luciferase reporter (−200 to −100; ref. 49), 4 ng of Renilla plasmid and 100 ng of STAT3 expression constructs. Cells were transfected with Lipofectamine 2000 Reagent (Invitrogen) and grown overnight in serum-free medium. Twenty-two hours past transfection, cells were left untreated or treated with IL-6 and soluble IL-6-R for 4 h. Luciferase assays were performed by using the Dual Luciferase Reporter Assay System (Promega) according to manufacturer’s instructions. Luciferase activities were normalized against Renilla luciferase activity of the cotransfected internal control plasmid. Each transfection was performed in duplicate, and results were confirmed in three to five independent experiments.
Purification of Tyrosine Phosphorylated STAT3 Proteins.
A4 cells were grown in 100-mm dishes and transfected with 15 μg of internally Flag-tagged STAT3 expression constructs. Transfected cells were serum-starved overnight (1% FBS) and kept for an additional 2–3 h in serum-free medium before induction with IL-6 and IL-6-R for 30 min. Whole-cell lysates [20 mM Hepes pH 7.9, 280 mM KCl, 1 mM EDTA, 10% (vol/vol) glycerol, 0.5% Nonidet P-40, 1 mM DTT and 0.5 mM PMSF] were prepared from two dishes and incubated with 20 μL of M2-resin for 4 h at 4 °C. Beads were washed three times with BC buffer [20 mM Hepes pH 7.9, 20% (vol/vol) glycerol, 0.5 mM EDTA, 1 mM DTT, 0.5 mM PMSF] containing 300 mM KCl and 0.1% Nonidet P-40 (BC300–0.1% Nonidet P-40) followed by one wash with buffer BC100–0.1% Nonidet P-40. Bound Flag-tagged proteins were eluted with two consecutive incubations with 25 μg/mL Flag-peptide in BC100–0.1% Nonidet P-40. Elutions were carried out at room temperature (RT) for 15 min each in a total volume of 100 μL. Elution fractions were analyzed side by side with input and supernatant fractions by SDS/PAGE with phospho Y705-STAT3 antibody. The amount of purified phospho STAT3 was estimated by SDS/PAGE and silver staining (Pierce) by comparing signal intensity of STAT3 bands to known amounts of BSA. Typical yields were ∼300 ng of purified tyrosine phosphorylated STAT3.
EMSA.
DNA binding activity of STAT3 mutants was assayed with 32P-labeled m67 probe (5′-CATTTCCCGTAAATCGTCGA-3′). Probe was prepared by end-labeling of double-stranded oligonucleotides with [γ-32P] ATP and T4 polynucleotide kinase (NEB). Purified in vivo phosphorylated STAT3 proteins (2–6 ng) were incubated with 0.019 pmol of probe in binding buffer (20 μL) containing 15 mM Hepes pH 7.9, 82 mM NaCl, 1 mM DTT, 0.43 mM EDTA, and 10% (vol/vol) glycerol. Reaction mixtures were incubated for 15 min at RT. To measure dissociation rates from DNA, a 40-fold excess of unlabeled oligonucleotides was then added for the indicated times. STAT3–DNA complexes were loaded onto a running 4% nondenaturing polyacrylamide gel (acrylamide/bisacrylamide 37.5:1) in 0.5× Tris-borate-EDTA, and electrophoresed at 130 V for 2.5–4 h at RT. The gel was dried and exposed to X-ray film. Bands were quantified by using ImageJ software.
Transfection Assays and Extract Preparation.
Transient transfections for activation and dephosphorylation studies were carried out in 50-mm dishes. Approximately 9.0 × 105 A4 cells were seeded the day before and transfected with 1.5 μg or 3.0 μg of STAT3 expression constructs (1.5 μg of wild-type STAT3, D570K, or D570A, 3.0 μg of K551A). The DNA amount was adjusted empirically to yield approximately equal expression of different proteins. Cells were serum-starved overnight (1% FBS) and incubated for an additional 3–4 h in serum-free medium before induction with IL-6/IL-6-R. Activation studies used 80 ng/mL IL-6 and 100 ng/mL IL-6-R for indicated times (0.5–5 min). Dephosphorylation studies used 20 ng/mL IL-6 and 25 ng/mL IL-6-R for 10 min, followed by treatment with 1 μM staurosporine for the indicated times (10–150 min). Cells were collected on ice and resuspended in hypotonic buffer [20 mM Hepes pH 7.9, 10 mM KCL, 1 mM EDTA, 0.2% Nonidet P-40, 10% (vol/vol) glycerol]. After 10-min incubation on ice, cytoplasmic fractions were collected by centrifugation at 20,000 × g for 30 s. Pellets were washed with sucrose buffer (20 mM Tris/HCl pH 7.5, 1 mM MgCl2, 2 mM EDTA, 320 mM sucrose) and extracted with high salt buffer [20 mM Hepes pH 7.9, 420 mM NaCl, 1 mM EDTA, 20% (vol/vol) glycerol]. Nuclear extracts were collected after 30-min incubation on ice by centrifugation at 20,000 × g for 10 min.
Acknowledgments
We thank Dr. George R. Stark for providing STAT3-null A4 cells and Lois Cousseau for preparing the manuscript. This work was partly supported by a grant from the Robertson Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515876112/-/DCSupplemental.
References
- 1.Sehgal PB, Levy DE, Hirano T, editors. Signal Transducers and Activators of Transcription: Activation and Biology. Kluwer Academic Publishers; London/Boston: 2003. [Google Scholar]
- 2.Stark GR, Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36(4):503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X, et al. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93(5):827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 4.Becker S, Groner B, Müller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394(6689):145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 5.Neculai D, et al. Structure of the unphosphorylated STAT5a dimer. J Biol Chem. 2005;280(49):40782–40787. doi: 10.1074/jbc.M507682200. [DOI] [PubMed] [Google Scholar]
- 6.Vinkemeier U, Moarefi I, Darnell JE, Jr, Kuriyan J. Structure of the amino-terminal protein interaction domain of STAT-4. Science. 1998;279(5353):1048–1052. doi: 10.1126/science.279.5353.1048. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, et al. A reinterpretation of the dimerization interface of the N-terminal domains of STATs. Protein Sci. 2003;12(2):361–365. doi: 10.1110/ps.0218903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wen Z, Zhong Z, Darnell JE., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82(2):241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 9.Levine M, Cattoglio C, Tjian R. Looping back to leap forward: Transcription enters a new era. Cell. 2014;157(1):13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allis CD, Capparros M-L, Jenuwein T, Reinberg D, editors. Epigenetics. 2nd Ed Cold Spring Harbor Lab Press; New York: 2015. [Google Scholar]
- 11.Gough DJ, Levy DE, Johnstone RW, Clarke CJ. IFNgamma signaling-does it mean JAK-STAT? Cytokine Growth Factor Rev. 2008;19(5–6):383–394. doi: 10.1016/j.cytogfr.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Schindler C, Fu X-Y, Improta T, Aebersold R, Darnell JE., Jr Proteins of transcription factor ISGF-3: One gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc Natl Acad Sci USA. 1992;89(16):7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veals SA, et al. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol Cell Biol. 1992;12(8):3315–3324. doi: 10.1128/mcb.12.8.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horvath CM, Stark GR, Kerr IM, Darnell JE., Jr Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol Cell Biol. 1996;16(12):6957–6964. doi: 10.1128/mcb.16.12.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang JJ, et al. Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc Natl Acad Sci USA. 1996;93(26):15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wojciak JM, Martinez-Yamout MA, Dyson HJ, Wright PE. Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains. EMBO J. 2009;28(7):948–958. doi: 10.1038/emboj.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau JF, Nusinzon I, Burakov D, Freedman LP, Horvath CM. Role of metazoan mediator proteins in interferon-responsive transcription. Mol Cell Biol. 2003;23(2):620–628. doi: 10.1128/MCB.23.2.620-628.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang JJ, et al. Ser727-dependent recruitment of MCM5 by Stat1alpha in IFN-gamma-induced transcriptional activation. EMBO J. 1998;17(23):6963–6971. doi: 10.1093/emboj/17.23.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zakharova N, et al. Distinct transcriptional activation functions of STAT1alpha and STAT1beta on DNA and chromatin templates. J Biol Chem. 2003;278(44):43067–43073. doi: 10.1074/jbc.M308166200. [DOI] [PubMed] [Google Scholar]
- 20.McMurray JS. Structural basis for the binding of high affinity phosphopeptides to Stat3. Biopolymers. 2008;90(1):69–79. doi: 10.1002/bip.20901. [DOI] [PubMed] [Google Scholar]
- 21.O’Shea JJ, Holland SM, Staudt LM. JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl J Med. 2013;368(2):161–170. doi: 10.1056/NEJMra1202117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang E, Wen Z, Haspel RL, Zhang JJ, Darnell JE., Jr The linker domain of Stat1 is required for gamma interferon-driven transcription. Mol Cell Biol. 1999;19(7):5106–5112. doi: 10.1128/mcb.19.7.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang E, Henriksen MA, Schaefer O, Zakharova N, Darnell JE., Jr Dissociation time from DNA determines transcriptional function in a STAT1 linker mutant. J Biol Chem. 2002;277(16):13455–13462. doi: 10.1074/jbc.M112038200. [DOI] [PubMed] [Google Scholar]
- 24.Yang E. 2002. Studies of transcriptional control by the STATs. Ph.D. thesis (The Rockefeller University, New York)
- 25.Bromberg JF, et al. Stat3 as an oncogene. Cell. 1999;98(3):295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 26.Li L, Shaw PE. Autocrine-mediated activation of STAT3 correlates with cell proliferation in breast carcinoma lines. J Biol Chem. 2002;277(20):17397–17405. doi: 10.1074/jbc.M109962200. [DOI] [PubMed] [Google Scholar]
- 27.Casanova JL, Holland SM, Notarangelo LD. Inborn errors of human JAKs and STATs. Immunity. 2012;36(4):515–528. doi: 10.1016/j.immuni.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conley ME, Casanova JL. Discovery of single-gene inborn errors of immunity by next generation sequencing. Curr Opin Immunol. 2014;30:17–23. doi: 10.1016/j.coi.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koskela HL, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366(20):1905–1913. doi: 10.1056/NEJMoa1114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jerez A, et al. STAT3 mutations indicate the presence of subclinical T-cell clones in a subset of aplastic anemia and myelodysplastic syndrome patients. Blood. 2013;122(14):2453–2459. doi: 10.1182/blood-2013-04-494930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flanagan SE, et al. Activating germline mutations in STAT3 cause early-onset multi-organ autoimmune disease. Nat Genet. 2014;46(8):812–814. doi: 10.1038/ng.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat Rev Cancer. 2009;9(11):798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat Rev Cancer. 2014;14(11):736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 34.Burley SK, Petsko GA. Weakly polar interactions in proteins. Adv Protein Chem. 1988;39:125–189. doi: 10.1016/s0065-3233(08)60376-9. [DOI] [PubMed] [Google Scholar]
- 35.Nakajima K, et al. A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J. 1996;15(14):3651–3658. [PMC free article] [PubMed] [Google Scholar]
- 36.Hart JR, Liao L, Yates JR, 3rd, Vogt PK. Essential role of Stat3 in PI3K-induced oncogenic transformation. Proc Natl Acad Sci USA. 2011;108(32):13247–13252. doi: 10.1073/pnas.1110486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer T, Marg A, Lemke P, Wiesner B, Vinkemeier U. DNA binding controls inactivation and nuclear accumulation of the transcription factor Stat1. Genes Dev. 2003;17(16):1992–2005. doi: 10.1101/gad.268003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.ten Hoeve J, et al. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol Cell Biol. 2002;22(16):5662–5668. doi: 10.1128/MCB.22.16.5662-5668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reich NC, Liu L. Tracking STAT nuclear traffic. Nat Rev Immunol. 2006;6(8):602–612. doi: 10.1038/nri1885. [DOI] [PubMed] [Google Scholar]
- 40.Haspel RL, Salditt-Georgieff M, Darnell JE., Jr The rapid inactivation of nuclear tyrosine phosphorylated Stat1 depends upon a protein tyrosine phosphatase. EMBO J. 1996;15(22):6262–6268. [PMC free article] [PubMed] [Google Scholar]
- 41.Furqan M, et al. STAT inhibitors for cancer therapy. J Hematol Oncol. 2013;6:90–101. doi: 10.1186/1756-8722-6-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Auzenne EJ, et al. A phosphopeptide mimetic prodrug targeting the SH2 domain of Stat3 inhibits tumor growth and angiogenesis. J Exp Ther Oncol. 2012;10(2):155–162. [PMC free article] [PubMed] [Google Scholar]
- 43.Huang W, et al. A small molecule compound targeting STAT3 DNA-binding domain inhibits cancer cell proliferation, migration, and invasion. ACS Chem Biol. 2014;9(5):1188–1196. doi: 10.1021/cb500071v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu L-J, et al. Identification of a natural product-like STAT3 dimerization inhibitor by structure-based virtual screening. Cell Death Dis. 2014;5:e1293. doi: 10.1038/cddis.2014.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siddiquee K, et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci USA. 2007;104(18):7391–7396. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horvath CM, Wen Z, Darnell JE., Jr A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 1995;9(8):984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- 47.Li Y, et al. Suppression of cancer relapse and metastasis by inhibiting cancer stemness. Proc Natl Acad Sci USA. 2015;112(6):1839–1844. doi: 10.1073/pnas.1424171112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang J, et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci USA. 2010;107(50):21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ginsberg M, et al. Amino acid residues required for physical and cooperative transcriptional interaction of STAT3 and AP-1 proteins c-Jun and c-Fos. Mol Cell Biol. 2007;27(18):6300–6308. doi: 10.1128/MCB.00613-07. [DOI] [PMC free article] [PubMed] [Google Scholar]