Analysis of Pex1 and dynamin-related protein function indicates peroxisomes multiply mainly by growth and division in Saccharomyces cerevisiae, whereas no evidence was found for the previously proposed role for Pex1 in peroxisome formation by fusion of ER-derived preperoxisomal vesicles.
Abstract
A recent model for peroxisome biogenesis postulates that peroxisomes form de novo continuously in wild-type cells by heterotypic fusion of endoplasmic reticulum–derived vesicles containing distinct sets of peroxisomal membrane proteins. This model proposes a role in vesicle fusion for the Pex1/Pex6 complex, which has an established role in matrix protein import. The growth and division model proposes that peroxisomes derive from existing peroxisomes. We tested these models by reexamining the role of Pex1/Pex6 and dynamin-related proteins in peroxisome biogenesis. We found that induced depletion of Pex1 blocks the import of matrix proteins but does not affect membrane protein delivery to peroxisomes; markers for the previously reported distinct vesicles colocalize in pex1 and pex6 cells; peroxisomes undergo continued growth if fission is blocked. Our data are compatible with the established primary role of the Pex1/Pex6 complex in matrix protein import and show that peroxisomes in Saccharomyces cerevisiae multiply mainly by growth and division.
Introduction
For many years, peroxisomes were thought to be autonomous organelles that multiply by growth and division and that import membrane and matrix proteins posttranslationally from the cytosol (Lazarow, 2003). Most peroxisomal matrix proteins contain a C-terminal peroxisomal targeting signal type 1 (PTS1; Gould et al., 1987). PTS1-containing proteins (cargo) are recognized in the cytosol by a soluble receptor (Pex5), which delivers its cargo by binding the docking complex (Pex13/14/17) on the peroxisomal membrane (Otera et al., 2002; Agne et al., 2003). The receptor and its cargo dissociate, and the receptor is recycled to the cytosol (Liu et al., 2012). Recycling requires the receptor to be monoubiquitinated (Platta et al., 2007; Okumoto et al., 2011) by the RING finger complex (Pex2/10/12; Williams et al., 2008; Platta et al., 2009) and extracted from the peroxisomal membrane by the AAA− ATPases Pex1 and Pex6 (Platta et al., 2004, 2005; Miyata and Fujiki, 2005). The docking complex and RING finger complex are physically linked via Pex8 and together form the importomer (Agne et al., 2003). After deubiquitination, the receptor is ready for another round of import (Debelyy et al., 2011; Miyata et al., 2012).
Targeting and insertion of peroxisomal membrane proteins (PMPs) does not require the machinery used for matrix protein import. Targeting of most PMPs (class 1) depends on the predominantly cytoplasmic Pex19, which has a chaperone function that helps it to function as a targeting signal receptor; Pex19 binds targeting signals in newly synthesized PMPs and delivers them to the peroxisomal membrane by docking onto Pex3 (Sacksteder et al., 2000; Fang et al., 2004; Rottensteiner et al., 2004; Pinto et al., 2006; Yagita et al., 2013; Chen et al., 2014). Some PMPs (class 2), including Pex3, contain targeting signals that are not recognized by Pex19, and these proteins follow an alternative route to peroxisomes (Jones et al., 2004; Hoepfner et al., 2005; Tam et al., 2005; Kim et al., 2006; Matsuzaki and Fujiki, 2008; Halbach et al., 2009; Fakieh et al., 2013; Knoops et al., 2014).
Most yeast mutants that lack functional peroxisomes (i.e., that are unable to import PTS1 proteins) contain peroxisomal membranes. However, two mutants, pex3 and pex19, appear to lack peroxisomal membranes altogether (Hettema et al., 2000; Koek et al., 2007). Upon complementation of these mutants, peroxisomes form from the ER (Fig. 1; Hoepfner et al., 2005; Tam et al., 2005). This process was visualized by inducing the expression of Pex3-GFP in Saccharomyces cerevisiae pex3 cells. Pex3 was first observed in ER-associated puncta, which subsequently dissociated from the ER and matured into peroxisomes (Hoepfner et al., 2005). Since then, the involvement of the ER in de novo peroxisome formation has been confirmed in various experimental setups (Haan et al., 2006; Toro et al., 2009).
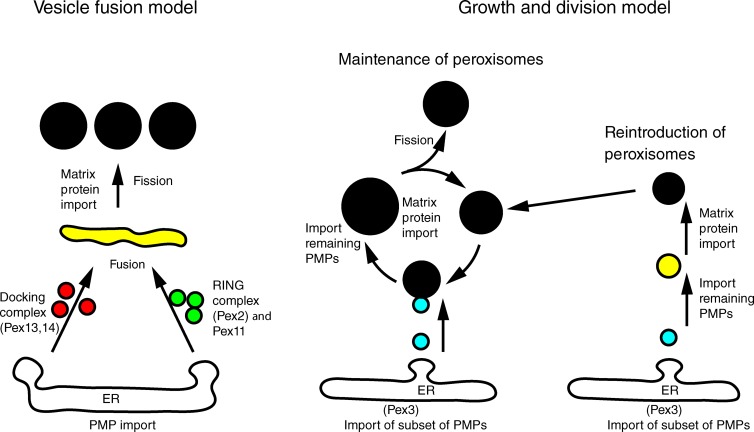
Two models for peroxisome multiplication in wild-type (WT) yeast cells have been proposed (Fig. 1). In the first, peroxisomes multiply predominantly by growth and division, with the ER providing membrane lipids and a subset of PMPs, including Pex3 and Pex22 (Motley and Hettema, 2007; Halbach et al., 2009; Hettema and Motley, 2009; Nuttall et al., 2011; Fakieh et al., 2013), via vesicles that fuse with existing peroxisomes. Other PMPs are inserted directly into the peroxisomal membrane (Fang et al., 2004). The finding that newly synthesized PMPs are transported to existing peroxisomes supports this model (Motley and Hettema, 2007; Fakieh et al., 2013; Menendez-Benito et al., 2013). Fission of peroxisomes is mediated by the direct action of the dynamin-related proteins (DRPs) Vps1 and Dnm1 (Hoepfner et al., 2001; Kuravi et al., 2006; Vizeacoumar et al., 2006; Motley and Hettema, 2007; Motley et al., 2008; Williams et al., 2015). Most vps1/dnm1 cells contain a single enlarged peroxisome (Kuravi et al., 2006; Motley and Hettema, 2007; Nagotu et al., 2008). The elongated peroxisome of vps1/dnm1 cells passes through the bud neck and divides upon cytokinesis, resulting in efficient segregation between mother and daughter cells (Hoepfner et al., 2001; Kuravi et al., 2006; Motley and Hettema, 2007). DRPs are not required for the reintroduction of peroxisomes in cells that temporarily lack them (Motley and Hettema, 2007).
Figure 1.
Schematic representation of models for peroxisome multiplication in S. cerevisiae. The vesicle fusion model proposes all PMPs traffic via the ER and exit in distinct vesicles containing Pex11 and RING finger proteins (Pex2/10/12; green) or docking complex proteins (Pex13/14/17; red). Heterotypic fusion between these vesicles requires the Pex1/6 complex and results in an intermediate compartment (yellow) in which the importomer is fully functional and matrix protein import commences. Peroxisomes form continuously, regardless of whether peroxisomes are already present. According to the growth and division model, preexisting peroxisomes receive newly synthesized membrane and matrix proteins and multiply by DRP-dependent fission. Pex1 and Pex6 are required for matrix protein import by the recycling of the PTS receptors. Only a subset of membrane proteins traffic via the ER (cyan), the remainder being inserted directly into peroxisomes (black). Peroxisomes form de novo only if no peroxisomes are present (reintroduction of peroxisomes): Pex3 localizes first to ER-associated puncta, which subsequently lose ER association (cyan) and acquire other PMPs (yellow), eventually importing matrix (PTS1 containing) proteins (black). Once a cell has formed peroxisomes de novo, they continue to multiply by growth and division.
An alternative model postulates that all PMPs insert first into the ER (van der Zand et al., 2010), where docking complex proteins (Pex13/14) are sorted away from Pex11 and RING finger complex proteins (Pex2/10/12) before the exit of these complexes in distinct vesicles (Fig. 1; van der Zand et al., 2012). Heterotypic vesicle fusion is proposed to result in the formation of an active translocon, after which the import of matrix proteins can occur (van der Zand et al., 2012). According to this model, vesicle fusion requires the AAA+ ATPases Pex1 and Pex6 and gives rise to a continuous stream of new peroxisomes in WT cells that add to the existing population as well as being the mechanism of peroxisome formation in cells lacking peroxisomes (van der Zand et al., 2012; Tabak et al., 2013; van der Zand and Tabak, 2013). DRPs are proposed to act after the Pex1/Pex6–mediated vesicle fusion event. Pex1 and Pex6 have also been suggested to mediate membrane fusion reactions of preperoxisomal structures during the maturation of peroxisomes in Yarrowia lipolytica (Titorenko and Rachubinski, 2000).
Studies in plants, yeast, and mammals have revealed that peroxisomes do not fuse homotypically (Arimura et al., 2004; Motley and Hettema, 2007; Bonekamp et al., 2012). However, both models described above require delivery of membrane material (lipids and proteins). Whereas the vesicle fusion model proposes heterotypic fusion of distinct ER-derived vesicles, the growth and division model proposes that ER-derived vesicles fuse with peroxisomes.
In this study, we reexamined the role of Pex1, Pex6, and the DRPs Vps1 and Dnm1 in peroxisome biogenesis. We found that depletion of Pex1 rapidly blocks matrix protein import but does not affect membrane protein delivery to peroxisomes. We show by genetic analysis that peroxisomal membranes are not maintained by a linear pathway whereby Pex1 acts upstream of DRPs and that reintroduction of peroxisomal membranes does not require Pex1. We find markers previously reported to be present in distinct vesicles localize to the same membranes in pex1 and pex6 cells. We show that peroxisomes undergo continued growth if fission is blocked and do not form de novo if peroxisomes are already present. These data support a model whereby peroxisomes multiply mainly by growth and division, and whereby Pex1/Pex6 has a direct role in matrix protein import and not in PMP biogenesis.
To understand the discrepancy between our conclusions and those of van der Zand et al. (2012), we replicated their experimental (bimolecular fluorescence complementation [BiFC]) setup. We found an increased turnover of peroxisomal membranes in pex1 and pex6 cells caused by pexophagy, and BiFC-positive peroxisomes divide asymmetrically. We discuss the implications of these new findings.
Results
Peroxisomes grow in the absence of DRP-dependent fission
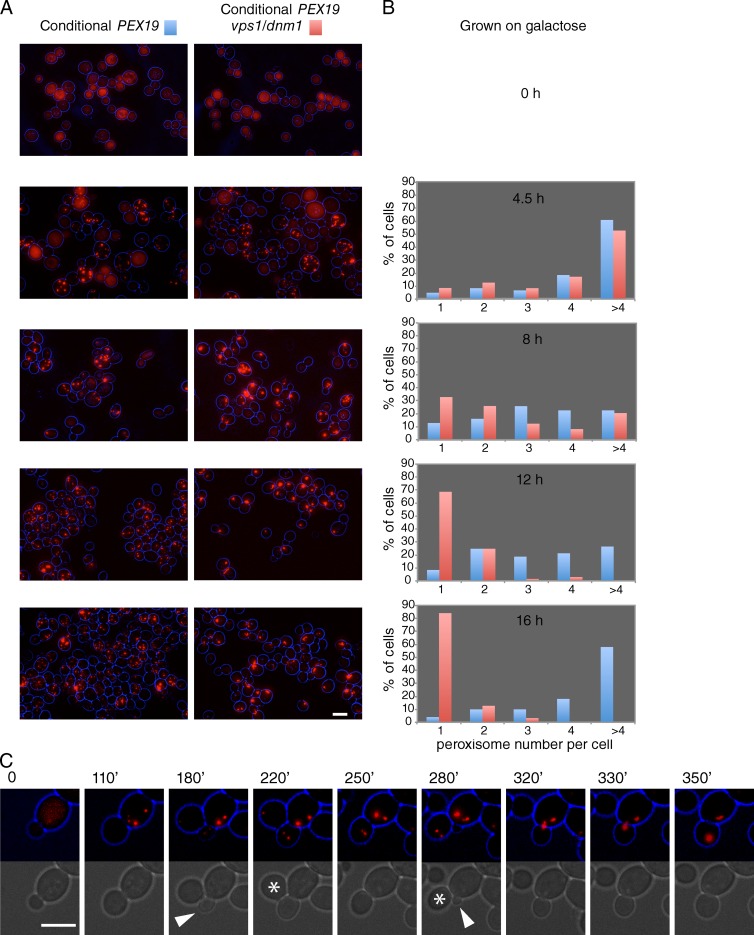
According to the growth and division model, DRPs are required for fission of peroxisomes that grow as they continue to receive PMPs (and other membrane constituents). To test this, we forced cells lacking DRPs to form peroxisomes de novo (Fig. 2). We replaced the PEX19 promoter with the GAL1 promoter (in WT and vps1/dnm1 backgrounds) so that de novo formation is driven by conditional Pex19 expression. When cells are grown on glucose, Pex19 is not expressed, and the peroxisomal matrix marker HcRed-PTS1 is cytosolic. Pex19 expression is induced by switching cells to galactose medium. Import of HcRed-PTS1 first becomes detectable after ∼3.5 h on this carbon source, the minimum time required to form peroxisomes de novo (Hoepfner et al., 2005; Tam et al., 2005; Motley and Hettema, 2007). At the early time points of de novo formation, both strains form multiple small peroxisomes per cell. The cultures were kept under conditions of exponential growth, with images being captured at intervals (shown for 4.5 h onwards). The frequency of vps1/dnm1 cells with more than four peroxisomes decreased with time. After 16 h, >80% of vps1/dnm1 cells contained a single enlarged peroxisome. That this reduction in peroxisome number is not a consequence of fusion of peroxisomes in vps1/dnm1 cells is shown in Video 1, Fig. 2 C, and Fig. S1. In the video, we show that the newly formed peroxisomes in vps1/dnm1 cells segregate until there is just one peroxisome, which becomes elongated as both mother and daughter try to inherit it. In Fig. S1, we pulse labeled peroxisomes in Matα or MatA cells with HcRed-PTS1 or GFP-PTS1, respectively. After mating, the red and green peroxisomes remained separate, in contrast to mitochondria, which fuse readily (Fig. S1; Nunnari et al., 1997). We have previously shown that peroxisomes in vps1/dnm1 cells do not fuse (Motley et al., 2008). We conclude that the reduction of peroxisome number in vps1/dnm1 cells is caused by dilution of peroxisomes by segregation in the absence of fission. We have tested a variety of PMPs (Pex3, Pex11, and Pex13-GFP) in this de novo assay, and as for the PTS1 protein, the number of puncta decreases until there is just a single peroxisome per cell in the vps1/dnm1 background at the later time points (Fig. S2). We conclude that peroxisomes in dividing vps1/dnm1 cells, once formed, proceed to grow into enlarged structures. The presence of a single enlarged peroxisome per vps1/dnm1 cell strongly suggests that new peroxisomes do not form if a peroxisome is already present and is in line with our previous observations (Motley and Hettema, 2007).
Figure 2.
De novo–formed peroxisomes grow into elongated peroxisomes in DRP-deficient cells. (A) Galactose-controllable PEX19 strains transformed with constitutive HcRed-PTS1 were grown on raffinose (top) or galactose medium for the times indicated in B. (B) Peroxisome number per cell was determined for at least 200 cells for each time point. Blue, conditional pex19 expression in WT background. Red, conditional pex19 expression in vps1Δ/dnm1Δ background. (C) Stills of a time-lapse video (Video 1): peroxisomes formed de novo in vps1/dnm1 cells expressing HcRed-PTS1 segregate on cell division, until a single enlarged peroxisome remained. Asterisks indicate cells that move away. Unit of measure is in minutes. Arrows indicate new buds forming. Bars, 5 µm.
Genetic analysis shows Pex1 does not act upstream of DRPs in peroxisomal membrane biogenesis
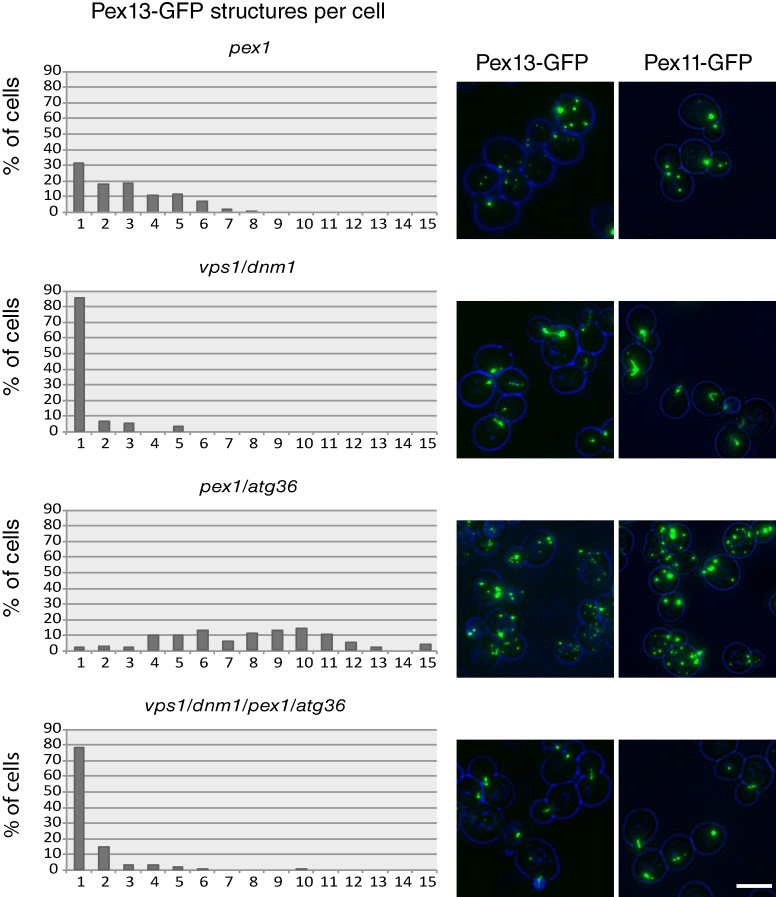
The vesicle fusion model proposes a role for Pex1/6-mediated vesicle fusion before fission of peroxisomes by DRPs (Fig. 1). To test this, we determined the number of peroxisomal membrane structures in various mutant backgrounds (Fig. 3). Because pex1 cells are deficient in matrix protein import, we used the peroxisomal membrane markers Pex11- and Pex13-GFP.
Figure 3.
Quantitation of membrane structures in pex1Δ, pex1Δ/atg3Δ6, vpsΔ1/dnm1Δ, and vps1Δ/dnm1Δ/pex1Δ/atg3Δ6 cells. Mutants expressing Pex11- or Pex13-GFP from their endogenous promoters (on plasmids) were imaged by epifluorescence microscopy. Cells were kept in a log phase on glucose-containing medium for 18 h before imaging. The doubling time of WT, vps1Δ/dnm1Δ, pex1Δ/atg36Δ, and vps1Δ/dnm1Δ/pex1Δ/atg36Δ strains under this condition was 101 min, 100 min, 106 min, and 115 min, respectively. The number of Pex13-GFP puncta per cell was determined for at least 200 cells per strain. Bar, 5 µm.
pex1 cells have a reduced number of peroxisomal membrane puncta (>70% have one to three puncta; Fig. 3) caused by increased turnover (Nuttall et al., 2014). Peroxisomal membrane structures are more abundant in pex1 cells lacking the pexophagy receptor Atg36 (>90% of pex1/atg36 cells have four or more fluorescent puncta; Motley et al., 2012). Cells with quadruple deletion of vps1/dnm1/pex1/atg36 have strongly reduced numbers of membrane structures (i.e., their phenotype resembles that of vps1/dnm1 and not pex1/atg36 cells; Fig. 3). This cannot be explained by a linear model whereby Pex1 acts upstream of DRPs.
Pex1 is not required for de novo formation of peroxisomal membranes
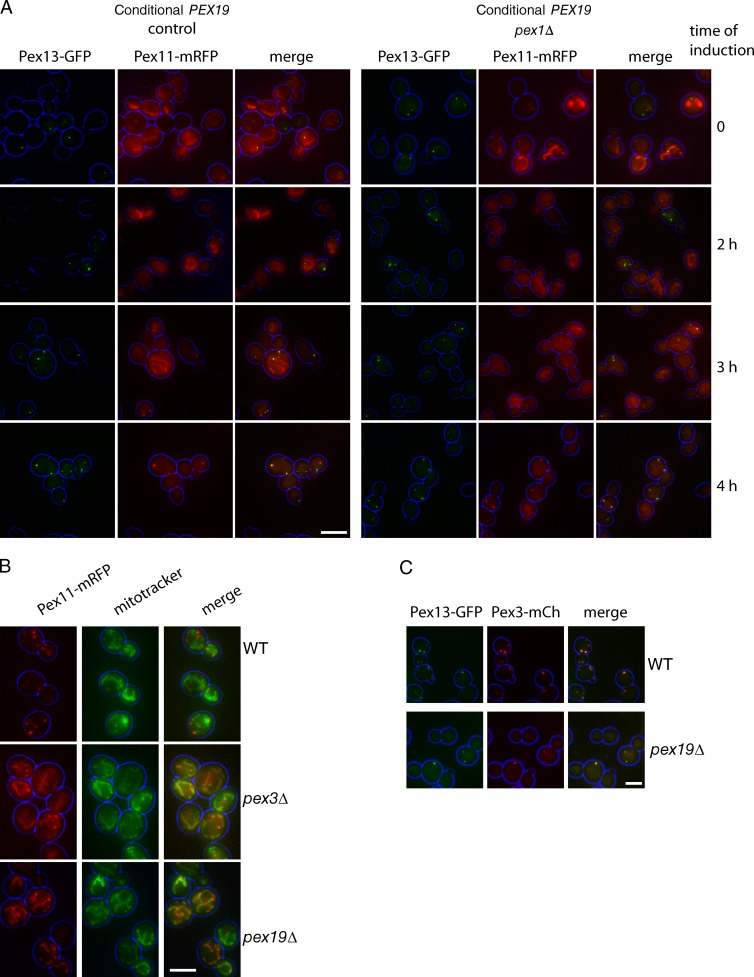
We tested the role of Pex1 in the formation of peroxisomal membranes using an approach similar to that described above for Fig. 2, in which we forced cells to form peroxisomes de novo by replacing the PEX19 promoter with the GAL1 promoter. We used Pex13-GFP and Pex11–monomeric RFP (mRFP) as markers for the distinct vesicles (van der Zand et al., 2012), which should persist in the absence of Pex1. Both PMPs show very faint signals at early time points, as PMPs are unstable in the absence of peroxisomes (Hettema et al., 2000), with Pex11-mRFP faintly labeling a tubular network and Pex13-GFP labeling puncta (Fig. 4 A). In pex19 cells, these puncta also contain Pex3 (Fig. 4 C). Pex3-GFP has previously been seen in the peroxisomal ER (pER) in cells forming peroxisomes de novo (Hoepfner et al., 2005; Tam et al., 2005). The tubular network faintly labeled by Pex11-mRFP in peroxisome-deficient cells colocalizes with MitoTracker (Fig. 4 B). After 3-h growth on a galactose medium, Pex11-mRFP becomes detectable in the Pex13-GFP puncta both in the presence or the absence of Pex1. Therefore, during de novo peroxisome formation (peroxisome reintroduction), peroxisomal membrane structures containing both Pex11-mRFP and Pex13-GFP are formed independently of Pex1.
Figure 4.
Pex1 is not required for de novo formation of peroxisomal membranes. (A) Galactose-controllable PEX19 strains expressing Pex13-GFP and Pex11-mRFP from their endogenous promoters (on plasmids) were grown on raffinose (top) or galactose medium for the times indicated. The Pex11-mRFP signal was weak and was enhanced strongly to show localization. (B) WT, pex3Δ, and pex19Δ cells expressing Pex11-mRFP from its endogenous promoter on plasmid were grown to log phase and stained with MitoTracker green. (C) WT and pex19Δ cells expressing Pex13-GFP and Pex3-mCherry (Pex3-mCH) from endogenous promoters (on plasmids) were grown to log phase. Bars, 5 µm.
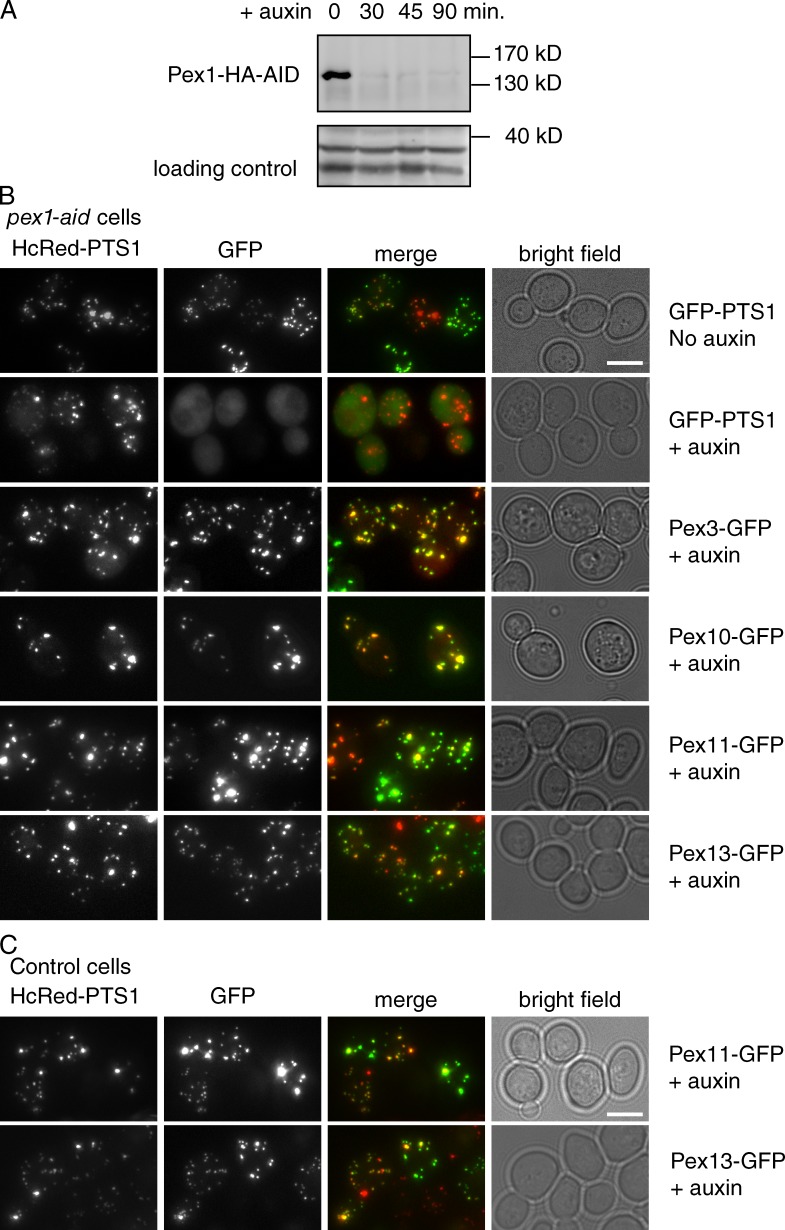
Induced degradation of Pex1 confirms its role in matrix protein import
We depleted Pex1 using an auxin-inducible degron tag (Nishimura et al., 2009; Nuttall et al., 2014). Degron-tagged Pex1 is undetectable 60 min after the addition of auxin (Fig. 5 A). We induced fluorescent reporter proteins after Pex1 depletion and examined their localization. 90 min after auxin addition, newly synthesized PMPs were transported to preexisting peroxisomes (labeled with HcRed), whereas the import of newly synthesized GFP-PTS1 was blocked. This shows that PMPs still reach peroxisomes in Pex1-depleted cells and are not trapped in the ER or a preperoxisomal compartment. A role for Pex1 and Pex6 in PTS1 import has been well documented (Miyata and Fujiki, 2005; Platta et al., 2005) and is confirmed by our depletion experiment. We noticed occasional PMP-GFP puncta that did not contain detectable matrix marker in both WT and Pex1-depleted cells (Fig. 5 C). We think this arises because of asymmetric fission of peroxisomes (see Fig. 6).
Figure 5.
Short-term depletion of Pex1 affects matrix protein import but not PMP transport. (A) Western blot analysis of Pex1-HA-AID levels in cells as grown in <0–90 min after addition of 0.5-mM auxin. (B) Pex1-HA-AID cells expressing the peroxisomal matrix marker HcRed-PTS1 were transformed with various PMP-GFP expression plasmids driven by the GAL1 promoter. Cells were grown to log phase on raffinose medium, and auxin was added to 0.5 mM. 45 min later, cells were resuspended in a galactose medium + auxin for 15 min (Pex3, Pex11, and Pex13) or 45 min (Pex10) to induce PMP-GFP expression and imaged by epifluorescence microscopy. (C) WT cells expressing HcRed-PTS1 constitutively and Pex11- or Pex13-GFP from the GAL1 promoter were grown as described in B in the presence of auxin. Bars, 5 µm.
Figure 6.
Colocalization of Pex11 and Pex13 in pex1 and pex6 cells. (A) pex1, pex6, pex1/pex6, and WT cells expressing Pex11-RFP and Pex13-GFP (from endogenous promoters on plasmids) were imaged after 3-h growth in log phase. (B) The distance between the center of each fluorescent Pex11-RFP and Pex13-GFP spot was calculated as described in the Materials and methods subsection Image acquisition and processing and is depicted as a box plot. n > 400 spots for each strain. The pale green line across each box indicates the median. (C) vps1/dnm1/pex1/atg36 cells expressing Pex11-RFP and Pex13-GFP (from endogenous promoters on plasmid) were imaged after 18 h in exponential growth phase. (D) WT and pex1/pex6 cells expressing plasmid-based Pex13-mCherry constitutively and Pex11-GFP from the GAL1 promoter were given a short pulse of Pex11-GFP as indicated. Bars, 5 µm.
Colocalization of PMPs in pex1 and pex6 cells
The experiments in Fig. 4 indicate that Pex1 is not required for de novo formation of peroxisomal membranes or for PMPs to reach existing peroxisomes. To investigate this further, we quantified colocalization of Pex11-mRFP and Pex13-GFP by comparing the coordinates of the center of each fluorescent spot in pex1, pex6, pex1/pex6, and WT cells (see Materials and methods subsection Image acquisition and processing; Fig. 6). We found that the absence of Pex1 or Pex6, or both, does not significantly decrease the degree of colocalization of these markers (Fig. 6, A and B), with the median distance between them being 113, 113, 160, and 160 nm for pex1, pex6, pex1/pex6, and WT, respectively. This is below the resolution of our setup. Furthermore, we show that Pex11-mRFP and Pex13-GFP clearly colocalize in the extended membranes of vps1/dnm1/pex1/atg36 cells (i.e., the absence of Pex1 does not prevent their trafficking to the same membrane; Fig. 6 C) and that a short (15 min) pulse of Pex11-GFP reaches Pex13-mCherry–labeled peroxisomal membranes in pex1/pex6 cells (Fig. 6 D).
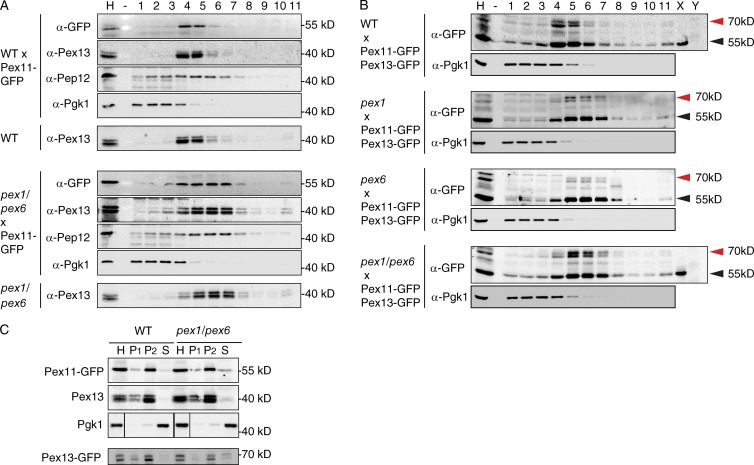
Homogenates of WT and pex1, pex6, and pex1/pex6 cells were analyzed by flotation equilibrium density gradient centrifugation (Fig. 7). Tagged Pex11 and Pex13 cofractionated with each other in all strains analyzed. Endogenous Pex13 floats to the same density as Pex13-GFP and Pex11-GFP in transformed and untransformed cells, indicating that tagging Pex11 and Pex13 does not affect the fractionation of peroxisomal membranes. The distribution of PMPs within the gradients is different in WT cells compared with mutants: the bulk of the PMPs (tagged or untagged) are present in fractions 4 + 5 in WT cells and in fractions 4–7 in pex1, pex6, and pex1/pex6 cells (Fig. 7). The shift to lower density fractions of peroxisomal membranes (ghosts) has been reported before (Santos et al., 1988; Gärtner et al., 1991; van Roermund et al., 1991; Motley et al., 1994; Hettema et al., 2000). Furthermore, differential centrifugation experiments (Fig. 7) show that the 25,000 g pellet contains the bulk of Pex11-GFP, Pex13-GFP, and Pex13 in both WT and pex1/pex6 cells, whereas the bulk of the cytoplasmic marker (Pgk1) is present in the 25,000 g supernatant. A small but reproducible amount of Pex13, Pex11-GFP, and Pex13-GFP was observed in the supernatant fraction of pex1/pex6 cells. The subcellular fractionation experiments support the fluorescence microscopy observations that Pex11 and Pex13 are present in the same membrane structures in cells lacking Pex1 and/or Pex6.
Figure 7.
Cofractionation of Pex11-GFP with Pex13 and Pex13-GFP in WT, pex1, pex6, and pex1/pex6 cells. (A and B) Homogenates (H; 800 g postnuclear supernatant) adjusted to 60% sucrose were separated by flotation analysis through sucrose equilibrium density gradient centrifugation. Fractions were collected from the bottom, and equal volumes were analyzed by Western blotting. Homogenates were prepared from glucose-grown cells of the strains as indicated. Cytosolic (Pgk1; A and B) and endosomal (Pep12; A) markers were included as the control for separation of membranes from cytosol. Pex13 is detected as a doublet, as Pex13 is prone to partial breakdown by proteolysis during subcellular fractionation (Elgersma et al., 1996). The samples in B were TCA precipitated and concentrated fourfold, as Pex13-GFP signals were weak. X and Y indicate control samples containing a P2 fraction of WT or pex1/pex6 cells expressing Pex11-GFP only and untransformed WT or pex1/pex6 cells. Red arrowheads indicate doublets of Pex13-GFP. Black arrowheads indicate Pex11-GFP. (C) Homogenates from WT and pex1/pex6 cells transformed with either Pex11-GFP or Pex13-GFP were separated by centrifugation first at 2,500 g and then at 25,000 g into a 2,500 g pellet (P1), a 25,000 g pellet (P2), and a 25,000 g supernatant fraction (S). Equivalent portions of each fraction were analyzed by Western blotting. Black lines indicate that intervening lanes have been spiced out.
As our conclusions contrast with those of a previous study (van der Zand et al., 2012), we replicated their experimental setup by reconstructing some of their strains and BiFC Venus tags. We generated identical strains (same tags, linker length, and parental strains) and used the same experimental conditions. To detect interactions between peroxins as a measure of importomer assembly, van der Zand et al. (2012) used BiFC after mating haploid yeast tagged in the genome with Venus GFP N- and C-terminal halves (VN and VC). BiFC occurs when nonfluorescent GFP halves are brought together by interaction between the proteins they are fused to. The presence of a signal was interpreted as the presence of complex formation (i.e., that the proteins are present in the same membrane structure). The absence of a signal was interpreted as the proteins being present in distinct membrane structures (van der Zand et al., 2012).
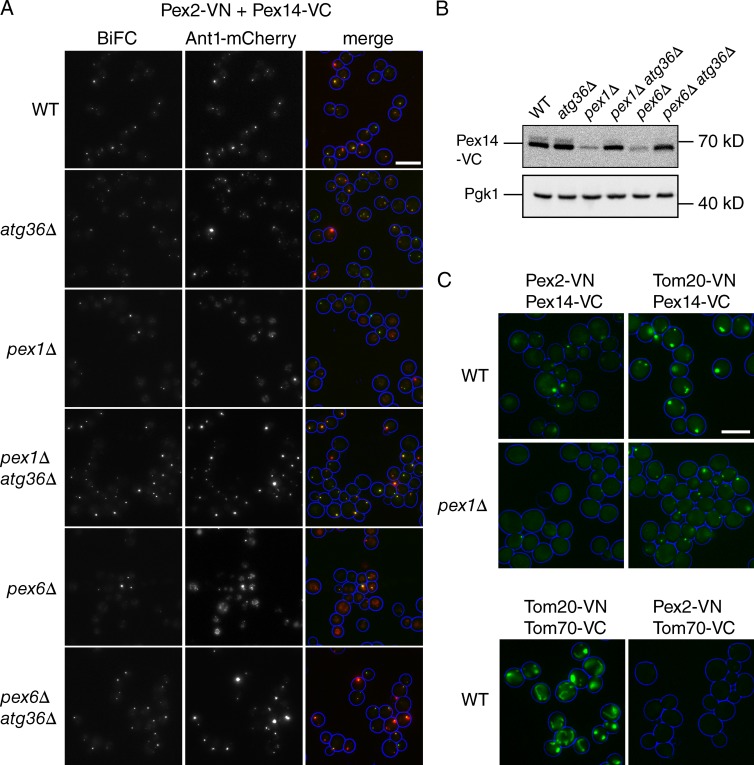
As markers for the docking complex, we used Pex13-VN and Pex14-VC, and for the RING finger complex, we used Pex2-VN. As shown in Fig. S3, peroxisomes formed within 3 h of mating pex1 with pex6 cells, but the BiFC signals took longer to develop. Similar to the observations of van der Zand et al. (2012), 24 h after mating, BiFC was evident in all combinations of Pex13-VN and Pex14-VC (pex1 × 6, 6 × 6, and 1 × 1), and a weak signal between Pex2-VN and Pex14-VC was observed in the pex1 × pex6 mating combination, although, in our hands, only in a minority (<10%) of mating cells. This signal is very weak and varies between experiments. The lack of Pex2-VN/Pex14-VC BiFC in pex1 × pex1 and pex6 × pex6 mating combinations was previously interpreted as an indication that Pex2 and Pex14 are present in distinct vesicles. However, the lack of an already faint signal could also be a consequence of enhanced pexophagy that occurs in pex1 and pex6 cells (Nuttall et al., 2014). Furthermore, the significance of BiFC signals forming from constitutively expressed tags after such long incubations (24–72 h; van der Zand et al., 2012) in cells that have peroxisomes after 3 h is not clear. We therefore tested for these interactions in haploid pex1 and pex6 cells.
Fig. 8 A shows BiFC between Pex2-VN and Pex14-VC in haploid cells. This BiFC is readily detectable in WT cells, but interestingly, we also see a signal in pex1 and pex6 cells, although only in a minority of cells (∼25%). Double labeling with the PMP marker Ant1-mCherry stains the vacuole in many pex1 and pex6 cells, which we would expect as a result of enhanced pexophagy. When the pexophagy receptor is disrupted in these haploid strains, vacuolar labeling of Ant1 is prevented and Pex14-VC is stabilized to near-WT levels (by Western blotting; Fig. 8 B). BiFC between Pex2-VN and Pex14-VC now becomes evident in most pex1/atg36 and pex6/atg36 cells. We conclude that the weak signal of Pex2/Pex14 BiFC in pex1 and pex6 cells is a consequence of increased peroxisome turnover.
Figure 8.
A block in pexophagy restores BiFC signals in pex1 and pex6 cells. (A) BiFC between Pex2-VN and Pex14-VC (genomically tagged) in haploid cells. Cells were transformed with a plasmid expressing Ant1-mCherry and grown on a glucose-containing medium to allow BiFC signals to develop. Mean number of Ant1-mCherry puncta per cell + SD were as follows: WT, 2.5 ± 1.6; atg36, 3.5 ± 2.3; pex1, 0.55 ± 0.6; pex1/atg36, 1.4 ± 0.7; pex6, 0.6 ± 0.6; and pex6/atg36, 1.6 ± 0.9. Percentage of cells showing colocalization between BiFC and Ant1 were as follows: WT, 84%; atg36, 89%; pex1, 95%; pex1/atg36, 93%; pex6, 98%; and pex6/atg36, 90%. At least 100 cells were analyzed. (B) Western blot showing Pex14-VC in strains as indicated. PGK1 was used as a loading control. (C) BiFC between mitochondrial and peroxisomal proteins in WT and pex1 cells as indicated. A singe slice of each sample was captured so that the fluorescence intensity in the images reflects the BiFC signal strength in the samples. Bars, 5 µm.
To control for specificity, we tested for BiFC in haploid cells between all combinations of Pex2-VN, Pex14-VC, and mitochondrial outer membrane proteins Tom20-VN and Tom70-VC. As shown in Fig. 8 C, the Tom20/Tom70 pair shows mitochondrial BiFC, and as expected, the Tom70/Pex2 pair is negative. Surprisingly, however, the Pex14/Tom20 pair gives a signal. Furthermore, this BiFC is stronger than that between Pex14 and Pex2. Whether this signal is meaningful is not clear. BiFC signals between proteins in different membranes have been previously reported (Pu et al., 2011; Mattiazzi Ušaj et al., 2015). The strength of the BiFC signal correlates with the level of expression of the proteins tested: quantification of C-tagged proteins indicates the number of molecules per cell is 339 for Pex2, 2,570 for Pex14, 5,680 for Tom20, and 45,300 for TOM70 (Ghaemmaghami et al., 2003). The strength of the BiFC signal may be influenced more by the relative abundance of the proteins tested than by an interaction between them or even their presence in the same membrane.
Asymmetric segregation of BiFC signal with matrix marker
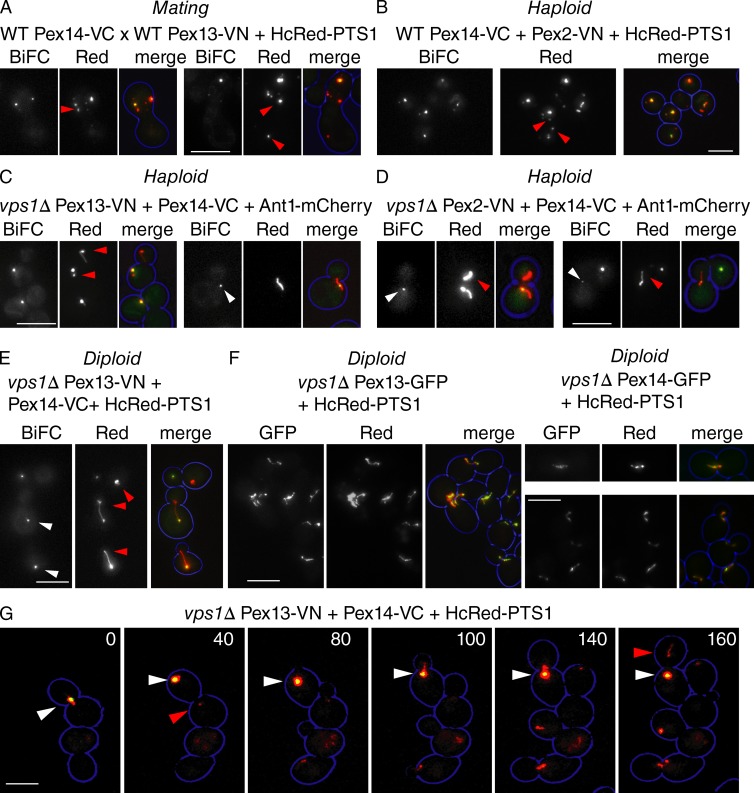
In agreement with van der Zand et al. (2012), we noticed that some peroxisomes lack a BiFC signal when mating WT VC- and VN-tagged strains (Fig. 9 A and their Figs. 1 and 6). Although the formation of BiFC puncta without matrix content may reflect de novo formation (van der Zand et al., 2012; and their Fig. 6), an alternative explanation is that BiFC and peroxisomal matrix marker do not segregate equally. In support of this, red-only peroxisomes are evident in haploid WT cells expressing VN and VC tags and HcRed-PTS1 (Fig. 9 B), which is striking because we and others have shown that newly synthesized PMPs traffic to existing peroxisomes (Motley and Hettema, 2007; Fakieh et al., 2013; Menendez-Benito et al., 2013). If unequal segregation of BiFC and red-PTS1 content explains the occurrence of red-only peroxisomes, we would expect such a segregation defect to be more evident on the elongated peroxisomes of vps1 cells, and this is what we saw: BiFC signals appear as puncta on the extended peroxisomes of haploid (Fig. 9, C and D) and diploid (Fig. 9 E) vps1 cells. In contrast, Pex13-GFP and Pex14-GFP label the whole peroxisomal structure (Fig. 9 F), although high magnification reveals that this labeling is not evenly spread over the peroxisomal membrane, particularly in the case of Pex14-GFP.
Figure 9.
Failure of BiFC to segregate with peroxisomes in mating cells, haploid cells, and diploid cells. (A) WT MatA Pex14-VC cells containing red-PTS1 on a plasmid were mated with WT Matα Pex13-VN cells and imaged 24 h after mating. Two examples are shown. (B) BiFC between Pex2-VN and Pex14-VC in WT haploid cells expressing HcRed-PTS1 from a plasmid. (C and D) VPS1 was disrupted in haploid cells double tagged as indicated, and peroxisomes were visualized using Ant1-mCherry. Two examples of each are shown. (E and F) Pex13-VN/14-VC BiFC (E) versus Pex13-GFP and Pex14-GFP (F) in diploid vps1 cells expressing HcRed-PTS1 from a plasmid. (G) Video stills from Video 2 of Pex13-VN/Pex14-VC diploid vps1 cells expressing HcRed-PTS1 from a plasmid. VN, VC, and GFP tags were genomically integrated (i.e., a WT copy in addition to tagged protein was present in diploid cells). Red arrowheads indicate peroxisomes or parts of peroxisomes without a BiFC signal. White arrowheads indicate punctate BiFC signals colocalizing with the elongated peroxisomes. White arrowheads in G indicate peroxisome with BiFC signal from which a peroxisome without BiFC signal splits off (red arrowheads). Bars, 5 µm.
We imaged diploid vps1 cells expressing Pex13-VN and Pex14-VC, as this BiFC pair resulted in the strongest signal and allowed for time-lapse microscopy over several cell divisions (Fig. 9 G and Video 2). The time-lapse analysis illustrates that BiFC signal frequently fails to segregate when the HcRed-PTS1–labeled peroxisome divides on cytokinesis. We conclude that red-only peroxisomes arise because the BiFC complex often fails to segregate when peroxisomes divide.
Discussion
By studying the role of DRPs and AAA+ ATPases, we have tested two models of peroxisome multiplication. The first model proposes that peroxisomes multiply by growth and division. According to this model, peroxisome growth is the result of delivery from the ER of vesicles carrying lipids and a subset of PMPs. Other PMPs and matrix proteins are imported directly into peroxisomes, and division is mediated by DRPs. When peroxisomes are absent, they can be reintroduced by de novo formation from the ER (Fig. 1).
The second model proposes that peroxisomes form de novo regardless of whether peroxisomes are already present. All PMPs enter the ER and exit in distinct preperoxisomal vesicles that undergo heterotypic fusion, bringing together components of the import machinery. Fusion is mediated by the AAA ATPases Pex1 and Pex6. After fusion and assembly of the importomer, matrix protein import commences, resulting in a new peroxisome. This linear maturation model ends with fission of peroxisomes by DRPs (Fig. 1; van der Zand et al., 2012; Tabak et al., 2013; van der Zand and Tabak, 2013).
The results reported here indicate that peroxisomes multiply mainly by growth and division in S. cerevisiae. We found that peroxisomes can grow in size and receive newly synthesized PMPs, and that membrane growth and delivery of PMPs occurs independent of Pex1 and Pex6. Our data support previous findings of the direct involvement of Pex1 and Pex6 in matrix protein import. We do not find evidence to support the proposal that Pex1 and Pex6 are required for the formation of new peroxisomal membranes by fusion of ER-derived vesicles.
Most cells lacking the DRPs Vps1 and Dnm1 contain a single peroxisome. This phenotype is difficult to explain if new peroxisomes form continuously, unless these would form de novo as large structures that neither divide nor segregate between mother and daughter cell during cell division. Our results show this is not the case: multiple small peroxisomes appear initially in vps1/dnm1 cells forced (by conditional Pex19 expression) to form peroxisomes de novo. These new peroxisomes increase in size but decrease in number during several rounds of cell division as they distribute between mother and daughter cell until a single peroxisome per cell is observed. Upon cytokinesis, this single peroxisome is split into two, allowing segregation between mother and daughter cell. The single elongated peroxisome phenotype of vps1/dnm1 cells is stably inherited over many cell generations (Video 2; Hoepfner et al., 2001; Kuravi et al., 2006; Motley and Hettema, 2007), implying that peroxisomes continue to receive new membrane and matrix constituents. This is consistent with pulse chase experiments showing that peroxisomal markers segregate between mother and daughter peroxisomes on fission (Motley and Hettema, 2007; Knoblach et al., 2013; Menendez-Benito et al., 2013). Not all proteins distribute equally on peroxisome fission, however (Cepińska et al., 2011; Knoblach et al., 2013), and this is observed to some extent in vps1/dnm1 cells for Pex14-GFP (Fig. 9 F). The asymmetric distribution in the membrane of Pex13/Pex14 BiFC is much greater than that of Pex13-GFP or Pex14-GFP (Fig. 9, E and F). Asymmetric fission most likely underlies the occasional occurrence of PMP-GFP puncta that lack detectable matrix content (Figs. 5 and 6), as PMP-GFP puncta without content are not observed in vps1/dnm1 cells (Fig. 9 F). Distinct peroxisome populations are generated by asymmetric distribution of PMPs and content followed by membrane fission during Woronin body biogenesis (Managadze et al., 2007; Liu et al., 2008). Asymmetric segregation also underlies the removal of intraorganellar aggregates from peroxisomes (Manivannan et al., 2013). In accordance with van der Zand et al. (2012), we found that BiFC signals fail to label all peroxisomes. However, we show that this is not a consequence of de novo formation of peroxisomes, but of asymmetric fission of BiFC-positive peroxisomes.
A recent modeling study (Mukherji and O’Shea, 2014) proposed that peroxisomes form mainly de novo when S. cerevisiae cells are grown on glucose, whereas under conditions of peroxisome proliferation, the DRPs contribute to multiplication by peroxisome fission. In that study, the effect of disruption of DRPs on peroxisome abundance in cells grown on glucose was not tested. We (Motley and Hettema, 2007; Motley et al., 2008) and others (Kuravi et al., 2006) have shown that peroxisome abundance is severely reduced in DRP-deficient cells grown on glucose (Fig. S4). Therefore, at least under our experimental conditions, peroxisomes multiply mainly by growth and division. In plants and mammals, there is support for the multiplication of peroxisomes by growth and division (Huybrechts et al., 2009; Delille et al., 2010; Barton et al., 2013), although both forms of multiplication have been reported to occur simultaneously in mammalian cells (Kim et al., 2006).
Although we have not detected de novo peroxisome formation in S. cerevisiae cells containing peroxisomes, we may have missed low levels of this. There may be conditions whereby de novo formation is induced in WT cells. A complex of ER reticulons and Pex30 has been reported to form a site of close contact between ER and peroxisomes. The absence of this complex enhances the rate at which new peroxisomes form during peroxisome reintroduction. This suggests a link between regulation of de novo peroxisome formation and ER morphology (Yan et al., 2007; David et al., 2013).
That the ER plays a central role in peroxisome biogenesis is supported by studies in many different organisms (Dimitrov et al., 2013; Agrawal and Subramani, 2015; Kim and Hettema, 2015). In S. cerevisiae, there is strong support for a role for the ER during reintroduction of peroxisomes in mutants temporarily lacking them (Hoepfner et al., 2005; Kragt et al., 2005; Tam et al., 2005; Haan et al., 2006). When Pex3-GFP expression is induced in pex3-deficient yeast cells, new peroxisomes form from a subdomain of the ER, the pER. In S. cerevisiae, the pER was reported to be ER associated (Bascom et al., 2003; Hoepfner et al., 2005), although no continuation with the ER was found in Hansenula polymorpha (Knoops et al., 2014). Other PMPs localized to this compartment in pex3 or pex19 cells, including Pex3, Pex13, Pex14, and Pex22 (Figs. 4 and 6; Faber et al., 2002; Bascom et al., 2003; Hoepfner et al., 2005; Kragt et al., 2005; Tam et al., 2005; Haan et al., 2006; Kim et al., 2006; Toro et al., 2009; van der Zand et al., 2010; Fakieh et al., 2013; Knoops et al., 2014). Some PMPs do not enter the pER, but insert late when new peroxisomes are formed in H. polymorpha (Knoops et al., 2014). We observe this for Pex11-GFP in S. cerevisiae. In the absence of peroxisomes, Pex11 is unstable (Hettema et al., 2000; Motley et al., 2012), and the low levels of Pex11-GFP that remain mislocalize to mitochondria (Fig. 4; Mattiazzi Ušaj et al., 2015). Upon peroxisome reintroduction, Pex11-GFP appears in puncta with Pex13 close to the time that GFP-PTS1 import commences (3–4 h; Figs. 2 and 4). These observations are compatible with a previously proposed model of de novo formation from the ER (during peroxisome reintroduction) by a process of maturation: a subset of PMPs traffic via the ER to the pER, preperoxisomes form from this specialized part of the ER, other PMPs are inserted, and a final maturation step is the import of matrix proteins (Hoepfner et al., 2005; Knoops et al., 2014).
In cells multiplying peroxisomes by growth and division (for example, in WT S. cerevisiae cells), we favor a model whereby peroxisomes receive lipids and a subset of PMPs via vesicular transport from the ER, whereas other PMPs are inserted into peroxisomes directly (Fig. 1). This model is supported by the following findings: that Pex15 appended with a glycosylation site is fully glycosylated in WT cells (Lam et al., 2010), that pER-trapped Pex3 can be transported to existing peroxisomes (Motley and Hettema, 2007), and that signals for Pex3 insertion into the ER, its sorting within the ER, and its subsequent sorting from pER to peroxisomes are required both during reintroduction of peroxisomes (de novo formation from the ER) and for transport of Pex3 to peroxisomes in WT cells (Fakieh et al., 2013). Many PMPs are recognized by Pex19 and could, after docking onto Pex3, be directly inserted into the peroxisomal membrane as has been shown to occur in mammalian cells and Neurospora crassa (Pinto et al., 2006; Matsuzaki and Fujiki, 2008; Yagita et al., 2013; Chen et al., 2014). Besides the direct route to peroxisomes, there is also a PMP trafficking route via the ER in mammals and plants (Kim et al., 2006; Karnik and Trelease, 2007; Toro et al., 2009; Aranovich et al., 2014).
The triple ATPases, Pex1 and Pex6, are important for matrix protein import. A series of studies have uncovered a role for these proteins in the recycling of PTS1 and PTS2 targeting receptors (Platta et al., 2004, 2005; Miyata and Fujiki, 2005; Debelyy et al., 2011; Miyata et al., 2012). Additional roles for these proteins have been proposed, including maturation of precursor peroxisomes in Y. lipolytica (Titorenko et al., 2000) and heterotypic fusion of distinct ER-derived vesicles in S. cerevisiae (van der Zand et al., 2012). Our data are compatible with a role for Pex1 and Pex6 in matrix protein import. We did not find evidence to support a role for these proteins in membrane protein biogenesis as proposed in the vesicle fusion model (van der Zand et al., 2012). This model is in part based on the observation that markers for the distinct ER exit routes label separate structures in pex1 and pex6 cells. We could not reproduce this observation (Figs. 4, 6, and 7). We found that these markers colocalize and cofractionate in WT, pex1, pex6, and pex1/pex6 cells. We expressed bright fluorescent proteins and fixed cells to reduce exposure times and movement. This may explain the discrepancy.
BiFC studies were used to show that Pex1 and Pex6 are required for assembly of the importomer (van der Zand et al., 2012). However, increased pexophagy in pex1 and pex6 cells (Nuttall et al., 2014) may have precluded detection of BiFC: we show by blocking pexophagy that importomer subunits Pex2 and Pex14 do give BiFC in pex1 and pex6 cells. This is in line with previous studies that indicate importomer assembles in pex1 and pex6 cells (Agne et al., 2003; Kiel et al., 2004; Platta et al., 2004; Rosenkranz et al., 2006; Hensel et al., 2011). We show that depletion of Pex1 blocks matrix protein import (despite the presence of importomer in the membrane) but does not affect PMP delivery to peroxisomes. Furthermore, we find no role for Pex1 in the assembly of peroxisomal membranes during their reintroduction to conditional pex19 cells. We conclude that, within the boundaries of our experimental framework, peroxisomes multiply mainly by growth and division, and that the AAA+ ATPases Pex1 and Pex6 are involved in matrix protein import.
Materials and methods
Yeast strains used are shown in Table S1, and the oligonucleotides used are shown in Table S2. Yeast strains used for Figs. 2, 3, 4, 5, 6, 7, S1, S2, and S4 and Video 1 were derivatives of BY4741 (MATA his3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0) or BY4742 (MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0) and for Figs. 8, 9, and S3 and Video 2 were derivatives of FY1679-08A (MATA) and FY1679-06C (MATα), all obtained from the EUROSCARF (European Saccharomyces Cerevisiae Archive for Functional Analysis) consortium. Gene deletions were made by replacing the entire coding sequence with Schizosaccharomyces pombe HIS5 or Klebsiella pneumoniae hygromycin B phosphotransferase gene cassette that confers resistance to hygromycin B (Goldstein and McCusker, 1999); BiFC or split GFP plasmids used as templates to genomically C-terminal tag Pex2- and 13-VN and Pex14-VC were pFA6a-VN173-HIS3MX6, pFA6a-VN155-HIS3MX6, and pFA6a-VN173-TRP1 (Bioneer Corporation). URA3 and LEU2 centromere plasmids were derived from Ycplac33 and Ycplac111 (Gietz and Sugino, 1988). GFP-PTS1 and RFP-PTS1 are peroxisomal luminal marker proteins appended with the well-characterized PTS1 (Gould et al., 1988). The constitutive expression of HcRed-PTS1 was under the control of the HIS3 promoter (Figs. 2, 4, 6, and 8). The majority of constructs used in this study were generated by homologous recombination in yeast (Uetz et al., 2000). The ORF of interest was amplified by PCR. The 5′ ends of the primers included 18-nt extensions homologous to plasmid sequences flanking the intended insertion site, to enable repair of gapped plasmids by homologous recombination. For expression of genes under control of their endogenous promoter, 500 nt upstream from the ORF were included. Galactose-inducible constructs contained the GAL1 and GAL10 intragenic region. All yeast constructs contain the PGK1 terminator. Conditional expression constructs contained the GAL1 promoter.
Growth conditions and mating assay
Cells were grown overnight in selective glucose medium and diluted to OD 0.1 in either selective glucose medium (Figs. 3, 6 [A–C], 7, 8, 9, and S4 and Video 1) or in selective raffinose medium for 4 h followed by resuspension in galactose medium (Figs. 2, 4, 5, S1, and S2 and Video 1) for the times indicated. For mating, 107 cells of each mating type grown to logarithmic phase on yeast peptone dextrose (YPD) were mixed, pelleted, and spotted onto a prewarmed YPD plate and incubated at 30°C for the times indicated. For each experiment, >100 (mating) cells were examined, and images are representative.
Image acquisition and processing
Cells were analyzed with a microscope (Axiovert 200M; Carl Zeiss) equipped with an Exfo X-cite 120 excitation light source, band pass filters (Carl Zeiss and Chroma Technology Corp.), an α Plan-Fluar 100× 1.45 NA, Plan-Apochromat 63× 1.4 NA, or a-Plan 40× 0.65 NA Ph2 objective lens (Carl Zeiss) and a digital camera (Orca ER; Hamamatsu Photonics). Image acquisition was performed using Volocity software (PerkinElmer). Fluorescence images were collected as 0.5-µm Z stacks using exposures of up to 300 ms, merged into one plane in Openlab (PerkinElmer), and processed further in Photoshop (Adobe). Brightfield images were collected in one plane and processed where necessary to highlight circumference of the cells.
For quantitation of colocalization, images were acquired using an Axio Observer (Carl Zeiss) microscope with a 100× 1.45 NA α Plan-Fluar objective with an electron-multiplying charge-coupled device camera (EM-C2; Rolera) using ZEN software. Single slices were taken of Pex13-GFP and Pex11-mRFP. Colocalization analysis was performed computionally using a Jython script for FIJI (http://fiji.sc/Fiji). Images were processed with a Fast-Fourier transform bandpass filter before subtracting the background and converting to 8-bit images. Per channel, the center of each fluorescent spot was then located by finding maxima in the image. The coordinates of the center of each fluorescent spot in the green channel was then compared with the coordinates of spots in the red channel. The distance between these coordinates was then found using a measure of 0.08 microns/pixel. The green and red coordinates with the minimum distance between them was then recorded, and they were both removed from the list of coordinates to be compared. More than 400 spots were counted in this manner.
For the live cell imaging, images for video were acquired using an Axio Observer microscope with a 100× 1.45 NA α Plan-Fluar objective with an EM-C2 electron-multiplying charge coupled device camera using ZEN software. Stacks were taken every 20 min at 27°C. Images were then processed to give extended depth of field and manually thresholded, and a Gaussian filter with a kernel of 3 × 3 pixels was applied to remove noise using ZEN. Videos run at one frame per second. Cells were grown in a CellASIC Microfluidic system in 2% galactose (Video 1) or 2% glucose (Video 2) for 14 h. Stills of Video 1 are shown in Fig. 2 C of peroxisomes forming de novo in vps1/dnm1 cells (expressing HcRed-PTS1 from a plasmid) and segregating to daughter cells. Stills of Video 2, showing Pex13-VN/Pex14-VC in diploid vps1 cells expressing HcRed-PTS1 from a plasmid, are shown in Fig. 9 G.
Subcellular fractionation
Cells grown overnight on selective glucose medium (OD600 = 4–5) were converted to spheroplasts with Zymolyase 20T (5 mg/g cells). The spheroplasts were washed twice in 1.2-M sorbitol, 5-mM MES, pH 6, 1-mM EDTA, 1-mM KCl before resuspension in 0.65-M sorbitol, 5-mM MES, pH 6, 1-mM EDTA, and 1-mM KCl (fractionation buffer) containing 1-mM PMSF and protease inhibitor cocktail. Cell breakage was achieved by 10 strokes with a tight-fitting douncer. Intact cells and nuclei were removed by two centrifugation steps (800 g for 10 min). 1 ml homogenate was mixed with 3 ml of 80% sucrose in a fractionation buffer. The sample was loaded on the bottom of an SW41 tube over which a sucrose step gradient was loaded consisting of 1-ml fractions of 50, 45, 40, 35, 32.5, 30, and 25% sucrose (wt/vol). These gradients were centrifuged for 18 h at 100,000 g in an SW41 rotor at 4°C. 1-ml fractions were collected from the bottom of the tube and analyzed by SDS-PAGE and immunoblotting. All sucrose solutions were made in fractionation buffer.
3 ml homogenate was fractionated by sequential differential centrifugation, from which we obtained a 2,500 g pellet, a 25,000 g pellet, and a 25,000 g supernatant. Pellet fractions were resuspended in a 3-ml fractionation buffer. Equivalent volumes of these fractions were analyzed by SDS-PAGE and immunoblotting.
Immunoblotting
For preparation of extracts by alkaline lysis, cells were centrifuged and pellets resuspended in 0.2-M NaOH and 0.2% β-mercaptoethanol and left on ice for 10 min. Soluble protein was precipitated by addition of 5% TCA for a further 10 min. After centrifugation (13,000 g for 5 min at 4°C), soluble protein was resuspended in 10 µl of 1-M Tris-HCl, pH 9.4, and boiled in 90 µl of 1× SDS-PAGE sample loading buffer for 10 min. Samples (0.25–1 OD600 equivalent) were resolved by SDS-PAGE followed by immunoblotting. Monoclonal anti-GFP antibody was obtained from Roche (11814460001), and secondary antibody was HRP-linked anti–mouse polyclonal (1706516; Bio-Rad Laboratories). Pex13 antiserum was provided by R. Erdmann (Ruhr University, Bochum, Germany). Blots were blocked in 2% (wt/vol) fat-free Marvel milk in TBS–Tween 20 (50-mM Tris-HCl, pH 7.5, 150-mM NaCl, and 0.1% [vol/vol] Tween 20). Tagged proteins were detected using enhanced chemiluminescence (Biological Industries) and chemiluminescence imaging.
Online supplemental material
Fig. S1 presents a mating assay revealing that peroxisomes in vps1/dnm1 cells do not fuse. Fig. S2 presents additional data using peroxisomal membrane markers showing that peroxisomes in vps1/dnm1 cells grow into elongated structures. Fig. S3 presents additional data showing that upon mating of pex1 and pex6 cells, matrix protein import is restored much faster than the development of BiFC signals. Fig. S4 shows quantitation of peroxisome number in WT, vps1, and vps1/dnm1 cells grown on glucose. Video 1 presents a time-lapse analysis of peroxisome dynamics in vps1/dnm1 cells. Video 2 presents time-lapse analysis of asymmetric segregation of a peroxisomal membrane BiFC signal in support of Fig. 9. Table S1 shows yeast strains used in this study. Table S2 shows oligonucleotides used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201412066/DC1.
Supplementary Material
Acknowledgments
We would like to thank Jeremy Craven for discussion throughout the project. We would also like to thank various members of the Hettema lab (University of Sheffield, Sheffield, England, UK), including Pawel Gardzielewski for construction of the conditional pex19 strains, Joanne Lacey for construction of Gal-inducible Pex13 and Pex14-GFP plasmids, and Marzieh Radi for the running of Western blots. We would also like to thank Ralf Erdmann for Pex13 antiserum. We would also like to thank Kevin Knoops and Ida van der Klei for their constructive comments on the manuscript and for sharing unpublished data.
This work was funded in part by a Wellcome Trust senior research fellowship in basic biomedical science awarded to E.H. Hettema (WT084265MA).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- BiFC
- bimolecular fluorescence complementation
- DRP
- dynamin-related protein
- mRFP
- monomeric RFP
- pER
- peroxisomal ER
- PMP
- peroxisomal membrane protein
- PTS
- peroxisomal targeting signal
- WT
- wild type
- YPD
- yeast peptone dextrose
References
- Agne B., Meindl N.M., Niederhoff K., Einwächter H., Rehling P., Sickmann A., Meyer H.E., Girzalsky W., and Kunau W.H.. 2003. Pex8p: an intraperoxisomal organizer of the peroxisomal import machinery. Mol. Cell. 11:635–646. 10.1016/S1097-2765(03)00062-5 [DOI] [PubMed] [Google Scholar]
- Agrawal G., and Subramani S.. 2015. De novo peroxisome biogenesis: Evolving concepts and conundrums. Biochim. Biophys. Acta. 10.1016/j.bbamcr.2015.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranovich A., Hua R., Rutenberg A.D., and Kim P.K.. 2014. PEX16 contributes to peroxisome maintenance by constantly trafficking PEX3 via the ER. J. Cell Sci. 127:3675–3686. 10.1242/jcs.146282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura S., Yamamoto J., Aida G.P., Nakazono M., and Tsutsumi N.. 2004. Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc. Natl. Acad. Sci. USA. 101:7805–7808. 10.1073/pnas.0401077101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton K., Mathur N., and Mathur J.. 2013. Simultaneous live-imaging of peroxisomes and the ER in plant cells suggests contiguity but no luminal continuity between the two organelles. Front. Physiol. 4:196 10.3389/fphys.2013.00196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bascom R.A., Chan H., and Rachubinski R.A.. 2003. Peroxisome biogenesis occurs in an unsynchronized manner in close association with the endoplasmic reticulum in temperature-sensitive Yarrowia lipolytica Pex3p mutants. Mol. Biol. Cell. 14:939–957. 10.1091/mbc.E02-10-0633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonekamp N.A., Sampaio P., de Abreu F.V., Lüers G.H., and Schrader M.. 2012. Transient complex interactions of mammalian peroxisomes without exchange of matrix or membrane marker proteins. Traffic. 13:960–978. 10.1111/j.1600-0854.2012.01356.x [DOI] [PubMed] [Google Scholar]
- Cepińska M.N., Veenhuis M., van der Klei I.J., and Nagotu S.. 2011. Peroxisome fission is associated with reorganization of specific membrane proteins. Traffic. 12:925–937. 10.1111/j.1600-0854.2011.01198.x [DOI] [PubMed] [Google Scholar]
- Chen Y., Pieuchot L., Loh R.A., Yang J., Kari T.M., Wong J.Y., and Jedd G.. 2014. Hydrophobic handoff for direct delivery of peroxisome tail-anchored proteins. Nat. Commun. 5:5790 10.1038/ncomms6790 [DOI] [PubMed] [Google Scholar]
- David C., Koch J., Oeljeklaus S., Laernsack A., Melchior S., Wiese S., Schummer A., Erdmann R., Warscheid B., and Brocard C.. 2013. A combined approach of quantitative interaction proteomics and live-cell imaging reveals a regulatory role for endoplasmic reticulum (ER) reticulon homology proteins in peroxisome biogenesis. Mol. Cell. Proteomics. 12:2408–2425. 10.1074/mcp.M112.017830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debelyy M.O., Platta H.W., Saffian D., Hensel A., Thoms S., Meyer H.E., Warscheid B., Girzalsky W., and Erdmann R.. 2011. Ubp15p, a ubiquitin hydrolase associated with the peroxisomal export machinery. J. Biol. Chem. 286:28223–28234. 10.1074/jbc.M111.238600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delille H.K., Agricola B., Guimaraes S.C., Borta H., Lüers G.H., Fransen M., and Schrader M.. 2010. Pex11pβ-mediated growth and division of mammalian peroxisomes follows a maturation pathway. J. Cell Sci. 123:2750–2762. 10.1242/jcs.062109 [DOI] [PubMed] [Google Scholar]
- Dimitrov L., Lam S.K., and Schekman R.. 2013. The role of the endoplasmic reticulum in peroxisome biogenesis. Cold Spring Harb. Perspect. Biol. 5:a013243 10.1101/cshperspect.a013243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y., Kwast L., Klein A., Voorn-Brouwer T., van den Berg M., Metzig B., America T., Tabak H.F., and Distel B.. 1996. The SH3 domain of the Saccharomyces cerevisiae peroxisomal membrane protein Pex13p functions as a docking site for Pex5p, a mobile receptor for the import PTS1-containing proteins. J. Cell Biol. 135:97–109. 10.1083/jcb.135.1.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber K.N., Haan G.J., Baerends R.J., Kram A.M., and Veenhuis M.. 2002. Normal peroxisome development from vesicles induced by truncated Hansenula polymorpha Pex3p. J. Biol. Chem. 277:11026–11033. 10.1074/jbc.M112347200 [DOI] [PubMed] [Google Scholar]
- Fakieh M.H., Drake P.J., Lacey J., Munck J.M., Motley A.M., and Hettema E.H.. 2013. Intra-ER sorting of the peroxisomal membrane protein Pex3 relies on its luminal domain. Biol. Open. 2:829–837. 10.1242/bio.20134788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Morrell J.C., Jones J.M., and Gould S.J.. 2004. PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. J. Cell Biol. 164:863–875. 10.1083/jcb.200311131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gärtner J., Chen W.W., Kelley R.I., Mihalik S.J., and Moser H.W.. 1991. The 22-kD peroxisomal integral membrane protein in Zellweger syndrome–presence, abundance, and association with a peroxisomal thiolase precursor protein. Pediatr. Res. 29:141–146. 10.1203/00006450-199102000-00007 [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O’Shea E.K., and Weissman J.S.. 2003. Global analysis of protein expression in yeast. Nature. 425:737–741. 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- Gietz R.D., and Sugino A.. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 74:527–534. 10.1016/0378-1119(88)90185-0 [DOI] [PubMed] [Google Scholar]
- Goldstein A.L., and McCusker J.H.. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 15:1541–1553. [DOI] [PubMed] [Google Scholar]
- Gould S.G., Keller G.A., and Subramani S.. 1987. Identification of a peroxisomal targeting signal at the carboxy terminus of firefly luciferase. J. Cell Biol. 105:2923–2931. 10.1083/jcb.105.6.2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S.J., Keller G.A., and Subramani S.. 1988. Identification of peroxisomal targeting signals located at the carboxy terminus of four peroxisomal proteins. J. Cell Biol. 107:897–905. 10.1083/jcb.107.3.897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haan G.J., Baerends R.J., Krikken A.M., Otzen M., Veenhuis M., and van der Klei I.J.. 2006. Reassembly of peroxisomes in Hansenula polymorpha pex3 cells on reintroduction of Pex3p involves the nuclear envelope. FEMS Yeast Res. 6:186–194. 10.1111/j.1567-1364.2006.00037.x [DOI] [PubMed] [Google Scholar]
- Halbach A., Rucktäschel R., Rottensteiner H., and Erdmann R.. 2009. The N-domain of Pex22p can functionally replace the Pex3p N-domain in targeting and peroxisome formation. J. Biol. Chem. 284:3906–3916. 10.1074/jbc.M806950200 [DOI] [PubMed] [Google Scholar]
- Hensel A., Beck S., El Magraoui F., Platta H.W., Girzalsky W., and Erdmann R.. 2011. Cysteine-dependent ubiquitination of Pex18p is linked to cargo translocation across the peroxisomal membrane. J. Biol. Chem. 286:43495–43505. 10.1074/jbc.M111.286104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema E.H., and Motley A.M.. 2009. How peroxisomes multiply. J. Cell Sci. 122:2331–2336. 10.1242/jcs.034363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema E.H., Girzalsky W., van Den Berg M., Erdmann R., and Distel B.. 2000. Saccharomyces cerevisiae pex3p and pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 19:223–233. 10.1093/emboj/19.2.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner D., van den Berg M., Philippsen P., Tabak H.F., and Hettema E.H.. 2001. A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155:979–990. 10.1083/jcb.200107028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner D., Schildknegt D., Braakman I., Philippsen P., and Tabak H.F.. 2005. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 122:85–95. 10.1016/j.cell.2005.04.025 [DOI] [PubMed] [Google Scholar]
- Huybrechts S.J., Van Veldhoven P.P., Brees C., Mannaerts G.P., Los G.V., and Fransen M.. 2009. Peroxisome dynamics in cultured mammalian cells. Traffic. 10:1722–1733. 10.1111/j.1600-0854.2009.00970.x [DOI] [PubMed] [Google Scholar]
- Jones J.M., Morrell J.C., and Gould S.J.. 2004. PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J. Cell Biol. 164:57–67. 10.1083/jcb.200304111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnik S.K., and Trelease R.N.. 2007. Arabidopsis peroxin 16 trafficks through the ER and an intermediate compartment to pre-existing peroxisomes via overlapping molecular targeting signals. J. Exp. Bot. 58:1677–1693. 10.1093/jxb/erm018 [DOI] [PubMed] [Google Scholar]
- Kiel J.A., Emmrich K., Meyer H.E., and Kunau W.H.. 2004. Ubiquitination of the peroxisomal targeting signal type 1 receptor, Pex5p, suggests the presence of a quality control mechanism during peroxisomal matrix protein import. J. Biol. Chem. 280:1921–1930. 10.1074/jbc.M403632200 [DOI] [PubMed] [Google Scholar]
- Kim P.K., and Hettema E.H.. 2015. Multiple pathways for protein transport to peroxisomes. J. Mol. Biol. 427:1176–1190. 10.1016/j.jmb.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P.K., Mullen R.T., Schumann U., and Lippincott-Schwartz J.. 2006. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J. Cell Biol. 173:521–532. 10.1083/jcb.200601036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblach B., Sun X., Coquelle N., Fagarasanu A., Poirier R.L., and Rachubinski R.A.. 2013. An ER-peroxisome tether exerts peroxisome population control in yeast. EMBO J. 32:2439–2453. 10.1038/emboj.2013.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Manivannan S., Cepinska M.N., Krikken A.M., Kram A.M., Veenhuis M., and van der Klei I.J.. 2014. Preperoxisomal vesicles can form in the absence of Pex3. J. Cell Biol. 204:659–668. 10.1083/jcb.201310148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koek A., Komori M., Veenhuis M., and van der Klei I.J.. 2007. A comparative study of peroxisomal structures in Hansenula polymorpha pex mutants. FEMS Yeast Res. 7:1126–1133. 10.1111/j.1567-1364.2007.00261.x [DOI] [PubMed] [Google Scholar]
- Kragt A., Voorn-Brouwer T., van den Berg M., and Distel B.. 2005. Endoplasmic reticulum-directed Pex3p routes to peroxisomes and restores peroxisome formation in a Saccharomyces cerevisiae pex3Δ strain. J. Biol. Chem. 280:34350–34357. 10.1074/jbc.M505432200 [DOI] [PubMed] [Google Scholar]
- Kuravi K., Nagotu S., Krikken A.M., Sjollema K., Deckers M., Erdmann R., Veenhuis M., and van der Klei I.J.. 2006. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J. Cell Sci. 119:3994–4001. 10.1242/jcs.03166 [DOI] [PubMed] [Google Scholar]
- Lam S.K., Yoda N., and Schekman R.. 2010. A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 107:21523–21528. 10.1073/pnas.1013397107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarow P.B. 2003. Peroxisome biogenesis: advances and conundrums. Curr. Opin. Cell Biol. 15:489–497. 10.1016/S0955-0674(03)00082-6 [DOI] [PubMed] [Google Scholar]
- Liu F., Ng S.K., Lu Y., Low W., Lai J., and Jedd G.. 2008. Making two organelles from one: Woronin body biogenesis by peroxisomal protein sorting. J. Cell Biol. 180:325–339. 10.1083/jcb.200705049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Ma C., and Subramani S.. 2012. Recent advances in peroxisomal matrix protein import. Curr. Opin. Cell Biol. 24:484–489. 10.1016/j.ceb.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Managadze D., Würtz C., Sichting M., Niehaus G., Veenhuis M., and Rottensteiner H.. 2007. The peroxin PEX14 of Neurospora crassa is essential for the biogenesis of both glyoxysomes and Woronin bodies. Traffic. 8:687–701. 10.1111/j.1600-0854.2007.00560.x [DOI] [PubMed] [Google Scholar]
- Manivannan S., de Boer R., Veenhuis M., and van der Klei I.J.. 2013. Lumenal peroxisomal protein aggregates are removed by concerted fission and autophagy events. Autophagy. 9:1044–1056. 10.4161/auto.24543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki T., and Fujiki Y.. 2008. The peroxisomal membrane protein import receptor Pex3p is directly transported to peroxisomes by a novel Pex19p- and Pex16p-dependent pathway. J. Cell Biol. 183:1275–1286. 10.1083/jcb.200806062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiazzi Ušaj M., Brložnik M., Kaferle P., Žitnik M., Wolinski H., Leitner F., Kohlwein S.D., Zupan B., and Petrovič U.. 2015. Genome-wide localization study of yeast Pex11 identifies peroxisome–mitochondria interactions through the ERMES complex. J. Mol. Biol. 427:2072–2087. 10.1016/j.jmb.2015.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez-Benito V., van Deventer S.J., Jimenez-Garcia V., Roy-Luzarraga M., van Leeuwen F., and Neefjes J.. 2013. Spatiotemporal analysis of organelle and macromolecular complex inheritance. Proc. Natl. Acad. Sci. USA. 110:175–180. 10.1073/pnas.1207424110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata N., and Fujiki Y.. 2005. Shuttling mechanism of peroxisome targeting signal type 1 receptor Pex5: ATP-independent import and ATP-dependent export. Mol. Cell. Biol. 25:10822–10832. 10.1128/MCB.25.24.10822-10832.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata N., Okumoto K., Mukai S., Noguchi M., and Fujiki Y.. 2012. AWP1/ZFAND6 functions in Pex5 export by interacting with cys-monoubiquitinated Pex5 and Pex6 AAA ATPase. Traffic. 13:168–183. 10.1111/j.1600-0854.2011.01298.x [DOI] [PubMed] [Google Scholar]
- Motley A.M., and Hettema E.H.. 2007. Yeast peroxisomes multiply by growth and division. J. Cell Biol. 178:399–410. 10.1083/jcb.200702167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A., Hettema E., Distel B., and Tabak H.. 1994. Differential protein import deficiencies in human peroxisome assembly disorders. J. Cell Biol. 125:755–767. 10.1083/jcb.125.4.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A.M., Ward G.P., and Hettema E.H.. 2008. Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J. Cell Sci. 121:1633–1640. 10.1242/jcs.026344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A.M., Nuttall J.M., and Hettema E.H.. 2012. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 31:2852–2868. 10.1038/emboj.2012.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji S., and O’Shea E.K.. 2014. Mechanisms of organelle biogenesis govern stochastic fluctuations in organelle abundance. eLife. 3:e02678 10.7554/eLife.02678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagotu S., Krikken A.M., Otzen M., Kiel J.A., Veenhuis M., and van der Klei I.J.. 2008. Peroxisome fission in Hansenula polymorpha requires Mdv1 and Fis1, two proteins also involved in mitochondrial fission. Traffic. 9:1471–1484. 10.1111/j.1600-0854.2008.00772.x [DOI] [PubMed] [Google Scholar]
- Nishimura K., Fukagawa T., Takisawa H., Kakimoto T., and Kanemaki M.. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods. 6:917–922. 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- Nunnari J., Marshall W.F., Straight A., Murray A., Sedat J.W., and Walter P.. 1997. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell. 8:1233–1242. 10.1091/mbc.8.7.1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuttall J.M., Motley A., and Hettema E.H.. 2011. Peroxisome biogenesis: recent advances. Curr. Opin. Cell Biol. 23:421–426. 10.1016/j.ceb.2011.05.005 [DOI] [PubMed] [Google Scholar]
- Nuttall J.M., Motley A.M., and Hettema E.H.. 2014. Deficiency of the exportomer components Pex1, Pex6, and Pex15 causes enhanced pexophagy in Saccharomyces cerevisiae. Autophagy. 10:835–845. 10.4161/auto.28259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto K., Misono S., Miyata N., Matsumoto Y., Mukai S., and Fujiki Y.. 2011. Cysteine ubiquitination of PTS1 receptor Pex5p regulates Pex5p recycling. Traffic. 12:1067–1083. 10.1111/j.1600-0854.2011.01217.x [DOI] [PubMed] [Google Scholar]
- Otera H., Setoguchi K., Hamasaki M., Kumashiro T., Shimizu N., and Fujiki Y.. 2002. Peroxisomal targeting signal receptor Pex5p interacts with cargoes and import machinery components in a spatiotemporally differentiated manner: conserved Pex5p WXXXF/Y motifs are critical for matrix protein import. Mol. Cell. Biol. 22:1639–1655. 10.1128/MCB.22.6.1639-1655.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto M.P., Grou C.P., Alencastre I.S., Oliveira M.E., Sá-Miranda C., Fransen M., and Azevedo J.E.. 2006. The import competence of a peroxisomal membrane protein is determined by Pex19p before the docking step. J. Biol. Chem. 281:34492–34502. 10.1074/jbc.M607183200 [DOI] [PubMed] [Google Scholar]
- Platta H.W., Girzalsky W., and Erdmann R.. 2004. Ubiquitination of the peroxisomal import receptor Pex5p. Biochem. J. 384:37–45. 10.1042/BJ20040572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta H.W., Grunau S., Rosenkranz K., Girzalsky W., and Erdmann R.. 2005. Functional role of the AAA peroxins in dislocation of the cycling PTS1 receptor back to the cytosol. Nat. Cell Biol. 7:817–822. 10.1038/ncb1281 [DOI] [PubMed] [Google Scholar]
- Platta H.W., El Magraoui F., Schlee D., Grunau S., Girzalsky W., and Erdmann R.. 2007. Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J. Cell Biol. 177:197–204. 10.1083/jcb.200611012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta H.W., El Magraoui F., Bäumer B.E., Schlee D., Girzalsky W., and Erdmann R.. 2009. Pex2 and pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol. Cell. Biol. 29:5505–5516. 10.1128/MCB.00388-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J., Ha C.W., Zhang S., Jung J.P., Huh W.K., and Liu P.. 2011. Interactomic study on interaction between lipid droplets and mitochondria. Protein Cell. 2:487–496. 10.1007/s13238-011-1061-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenkranz K., Birschmann I., Grunau S., Girzalsky W., Kunau W.H., and Erdmann R.. 2006. Functional association of the AAA complex and the peroxisomal importomer. FEBS J. 273:3804–3815. 10.1111/j.1742-4658.2006.05388.x [DOI] [PubMed] [Google Scholar]
- Rottensteiner H., Kramer A., Lorenzen S., Stein K., Landgraf C., Volkmer-Engert R., and Erdmann R.. 2004. Peroxisomal membrane proteins contain common Pex19p-binding sites that are an integral part of their targeting signals. Mol. Biol. Cell. 15:3406–3417. 10.1091/mbc.E04-03-0188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacksteder K.A., Jones J.M., South S.T., Li X., Liu Y., and Gould S.J.. 2000. PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J. Cell Biol. 148:931–944. 10.1083/jcb.148.5.931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M.J., Imanaka T., Shio H., and Lazarow P.B.. 1988. Peroxisomal integral membrane proteins in control and Zellweger fibroblasts. J. Biol. Chem. 263:10502–10509. [PubMed] [Google Scholar]
- Tabak H.F., Braakman I., and van der Zand A.. 2013. Peroxisome formation and maintenance are dependent on the endoplasmic reticulum. Annu. Rev. Biochem. 82:723–744. 10.1146/annurev-biochem-081111-125123 [DOI] [PubMed] [Google Scholar]
- Tam Y.Y., Fagarasanu A., Fagarasanu M., and Rachubinski R.A.. 2005. Pex3p initiates the formation of a preperoxisomal compartment from a subdomain of the endoplasmic reticulum in Saccharomyces cerevisiae. J. Biol. Chem. 280:34933–34939. 10.1074/jbc.M506208200 [DOI] [PubMed] [Google Scholar]
- Titorenko V.I., and Rachubinski R.A.. 2000. Peroxisomal membrane fusion requires two AAA family ATPases, Pex1p and Pex6p. J. Cell Biol. 150:881–886. 10.1083/jcb.150.4.881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko V.I., Chan H., and Rachubinski R.A.. 2000. Fusion of small peroxisomal vesicles in vitro reconstructs an early step in the in vivo multistep peroxisome assembly pathway of Yarrowia lipolytica. J. Cell Biol. 148:29–44. 10.1083/jcb.148.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro A.A., Araya C.A., Cordova G.J., Arredondo C.A., Cardenas H.G., Moreno R.E., Venegas A., Koenig C.S., Cancino J., Gonzalez A., and Santos M.J.. 2009. Pex3p-dependent peroxisomal biogenesis initiates in the endoplasmic reticulum of human fibroblasts. J. Cell. Biochem. 107:1083–1096. [DOI] [PubMed] [Google Scholar]
- Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., et al. . 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 403:623–627. 10.1038/35001009 [DOI] [PubMed] [Google Scholar]
- van der Zand A., and Tabak H.F.. 2013. Peroxisomes: offshoots of the ER. Curr. Opin. Cell Biol. 25:449–454. 10.1016/j.ceb.2013.05.004 [DOI] [PubMed] [Google Scholar]
- van der Zand A., Braakman I., and Tabak H.F.. 2010. Peroxisomal membrane proteins insert into the endoplasmic reticulum. Mol. Biol. Cell. 21:2057–2065. 10.1091/mbc.E10-02-0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zand A., Gent J., Braakman I., and Tabak H.F.. 2012. Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell. 149:397–409. 10.1016/j.cell.2012.01.054 [DOI] [PubMed] [Google Scholar]
- van Roermund C.W., Brul S., Tager J.M., Schutgens R.B., and Wanders R.J.. 1991. Acyl-CoA oxidase, peroxisomal thiolase and dihydroxyacetone phosphate acyltransferase: aberrant subcellular localization in Zellweger syndrome. J. Inherit. Metab. Dis. 14:152–164. 10.1007/BF01800588 [DOI] [PubMed] [Google Scholar]
- Vizeacoumar F.J., Vreden W.N., Fagarasanu M., Eitzen G.A., Aitchison J.D., and Rachubinski R.A.. 2006. The dynamin-like protein Vps1p of the yeast Saccharomyces cerevisiae associates with peroxisomes in a Pex19p-dependent manner. J. Biol. Chem. 281:12817–12823. 10.1074/jbc.M600365200 [DOI] [PubMed] [Google Scholar]
- Williams C., van den Berg M., Geers E., and Distel B.. 2008. Pex10p functions as an E3 ligase for the Ubc4p-dependent ubiquitination of Pex5p. Biochem. Biophys. Res. Commun. 374:620–624. 10.1016/j.bbrc.2008.07.054 [DOI] [PubMed] [Google Scholar]
- Williams C., Opalinski L., Landgraf C., Costello J., Schrader M., Krikken A.M., Knoops K., Kram A.M., Volkmer R., and van der Klei I.J.. 2015. The membrane remodeling protein Pex11p activates the GTPase Dnm1p during peroxisomal fission. Proc. Natl. Acad. Sci. USA. 112:6377–6382. 10.1073/pnas.1418736112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita Y., Hiromasa T., and Fujiki Y.. 2013. Tail-anchored PEX26 targets peroxisomes via a PEX19-dependent and TRC40-independent class I pathway. J. Cell Biol. 200:651–666. 10.1083/jcb.201211077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Rachubinski D.A., Joshi S., Rachubinski R.A., and Subramani S.. 2007. Dysferlin domain-containing proteins, Pex30p and Pex31p, localized to two compartments, control the number and size of oleate-induced peroxisomes in Pichia pastoris. Mol. Biol. Cell. 19:885–898. 10.1091/mbc.E07-10-1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.