Inturned-mediated complex formation of NPHP4 and DAAM1 is important for ciliogenesis and ciliary function in multiciliated cells, presumably because of its requirement for the local rearrangement of actin cytoskeleton.
Abstract
Motile cilia polarization requires intracellular anchorage to the cytoskeleton; however, the molecular machinery that supports this process remains elusive. We report that Inturned plays a central role in coordinating the interaction between cilia-associated proteins and actin-nucleation factors. We observed that knockdown of nphp4 in multiciliated cells of the Xenopus laevis epidermis compromised ciliogenesis and directional fluid flow. Depletion of nphp4 disrupted the subapical actin layer. Comparison to the structural defects caused by inturned depletion revealed striking similarities. Furthermore, coimmunoprecipitation assays demonstrated that the two proteins interact with each other and that Inturned mediates the formation of ternary protein complexes between NPHP4 and DAAM1. Knockdown of daam1, but not formin-2, resulted in similar disruption of the subapical actin web, whereas nphp4 depletion prevented the association of Inturned with the basal bodies. Thus, Inturned appears to function as an adaptor protein that couples cilia-associated molecules to actin-modifying proteins to rearrange the local actin cytoskeleton.
Introduction
Although most cells express nonmotile primary cilia, epithelial cells in the central nervous system, respiratory tract, and fallopian tubes of the uterus are equipped with multiple motile cilia (Brooks and Wallingford, 2014). Motile cilia are polarized in several ways to generate directional fluid or particle flows (Wallingford, 2010). The “rotational polarization” describes the polarity of cilia and their appendages within a single cell and enables the motile cilia to beat in a directional fashion. In Xenopus laevis epidermal cells, this type of polarity is established by an early positional bias and subsequent positive hydromechanical feedback mechanisms that reinforce the directional beating pattern (Mitchell et al., 2007); it is characterized by the orientation of the basal foot in the direction of the effective stroke and the striated rootlet, pointing to the opposite direction (Wallingford, 2010). Dishevelled (Dvl), which associates with the basal bodies at the apical membrane, appears to activate RhoA in multiciliated cells (Park et al., 2008). Ultimately, Dvl is asymmetrically localized within the center of the ciliary rootlet, and this asymmetry is essential for a normal rotational polarity (Park et al., 2008). The anaphase-promoting complex, which is found in the proximity of basal bodies and targets Dvl for degradation, may contribute to the confined Dvl localization in fully polarized cilia because knockdown of the anaphase-promoting complex/cyclosome disrupts ciliary polarity (Ganner et al., 2009). The planar cell polarity effector molecule Inturned is also required for normal ciliary polarization, and it recruits RhoA to the apical membrane. Depletion of Inturned prevents the association of RhoA with the apical membrane, resulting in a thinning of the apical actin cytoskeleton and abnormal ciliary polarization (Park et al., 2006, 2008). However, the precise function of Inturned remains unknown.
The role of the actin cytoskeleton for ciliogenesis, basal body docking, and ciliary polarization is well established (Park et al., 2006; Pan et al., 2007; Dawe et al., 2009; Bershteyn et al., 2010). The apical actin web consists of two layers: a dense net-like apical layer immediate underneath the plasma membrane and a subapical layer with short actin filaments that connect neighboring basal bodies (Werner et al., 2011). These short interconnecting actin fibers are particularly important in generating the metachronal wave of synchronously beating axonemes across multiciliated cells (Brooks and Wallingford, 2014). The actin-depolymerizing drug cytochalasin D disrupts these actin fibers in stage 29 Xenopus embryos and disturbs metachrony without reversing the rotational polarity (Werner et al., 2011). However, the molecular machinery that generates these specific subapical actin fibers is unknown.
Formation of actin filaments or branched actin polymers requires actin nucleation factors to overcome unfavorable initial association rates (Qualmann and Kessels, 2009). There are three classes of actin nucleation factors: (a) Arp2/3, a multimeric protein complex involved in actin branching; (b) formins (mDIA1-mDIA3, FRL1-2, DAAM1-2, FHOD1/3, FMN1-2, Delphin, and INF1-2); and (c) WH2-containing proteins (SPIRE1, SPIRE2, COBL, and LMOD1-LMOD3). Although these factors are critical for actin polymerization, dynamic changes of actin polymerization require the presence of additional actin nucleation-promoting factors (Campellone and Welch, 2010). Several actin-modifying proteins have been identified in the ciliary proteome, including the two Formins DAAM1 and FHOD3. Daam1 was identified through interaction with the PDZ domain of Dvl2 (Habas et al., 2001). Daam1 binds both Dvl2 and RhoA and is required for normal gastrulation of Xenopus embryos (Habas et al., 2001), a process accompanied by extensive cytoskeletal rearrangements.
Nephronophthisis (NPH), a heterogenetic autosomal recessive ciliopathy, is characterized primarily by cystic kidney disease with progression to end-stage renal disease, retinopathy, and varying cerebral defects (Benzing and Schermer, 2012). Proteins of the NPH gene family reside within the ciliary compartment but also colocalize with focal adhesions at the plasma membrane. Recently, several focal adhesion proteins were found to associate with the basal bodies of multiciliated cells and are required for normal basal body migration and docking (Antoniades et al., 2014). Interestingly, two NPH proteins, nephrocystin-1 (NPHP1) and nephrocystin-4 (NPHP4), interact with focal adhesion molecules and have been implicated in cell–cell contacts and directed cell migration (Benzing et al., 2001; Burcklé et al., 2011; Liebau et al., 2011; Slanchev et al., 2011). Mutations of NPH proteins also affect motile cilia. Whereas the fluid and particle flow across the ventral node is disrupted in NPHP2/Inversin-deficient mice (Okada et al., 1999), NPHP2, NPHP3, and NPHP16 mutations cause situs inversus (Otto et al., 2003; Bergmann et al., 2008; Hoff et al., 2013). In addition, patients with NPHP4 mutations display spermatogenesis defects (Alazami et al., 2013), and an increased susceptibility to respiratory infections, indicating a dysfunction of motile cilia in the respiratory tract, has been observed in patients with NPHP1 mutations (Fliegauf et al., 2006). NPHPs appear to form large protein networks to participate in defined cellular functions (Sang et al., 2011); these protein complexes have been extensively studied in Caenorhabditis elegans, where they participate in structural elements of the transition zone and control the access of molecules to the ciliary axoneme (Jauregui et al., 2008; Williams et al., 2010, 2011; Reiter et al., 2012; Warburton-Pitt et al., 2014). In contrast, the role of NPHPs in ciliary motility and polarization remains largely unknown. We report now that NPHP4 interacts with Inturned, which facilitated the formation of a tertiary complex between NPHP4 and the actin-nucleating protein Daam1. Knockdown of nphp4, inturned, or daam1 in Xenopus resulted in strikingly similar abnormalities of the subapical actin layer. Furthermore, NPHP4 was required for Inturned to localize to the basal bodies of motile cilia. Our findings suggest that the subapical actin web is controlled by interactions between cilia-associated molecules such as NPHP4, which can recruit Inturned, and actin-modifying proteins such as Daam1 to modify the local actin cytoskeleton.
Results and discussion
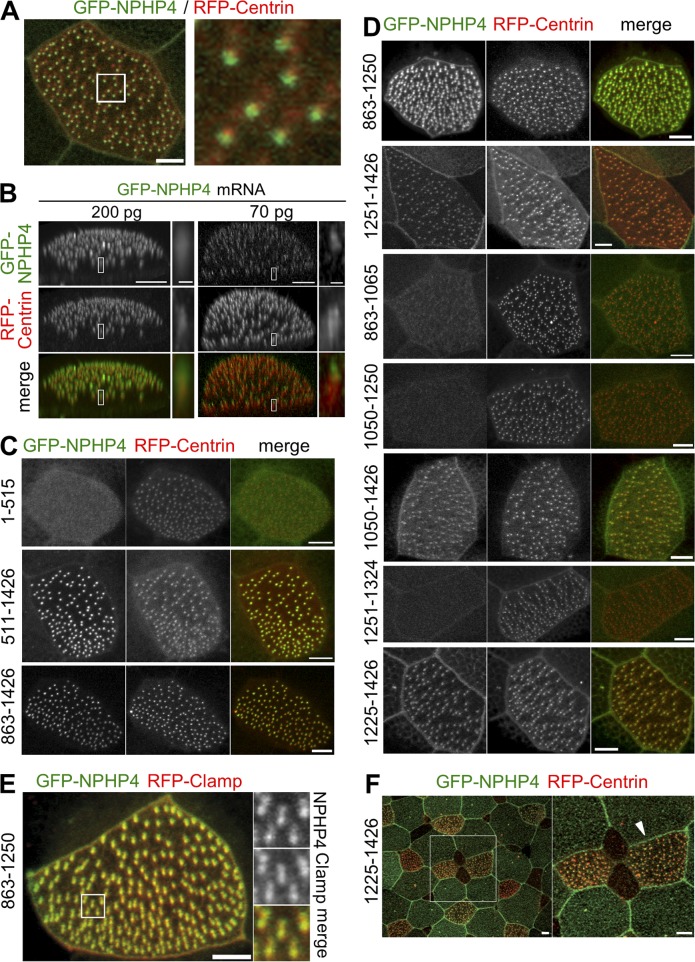
A C-terminal fragment localizes NPHP4 to the basal body/transition zone in multiciliated epidermal cells of Xenopus embryos
We found that Xenopus nphp4 is present in oocytes and during early stages of Xenopus development (stages 1–42; Fig. S1, A–C), preferentially in ciliated tissues such as the pronephric duct, olfactory placode, and multiciliated cells of the epidermis. To explore the epidermis of developing Xenopus embryos as a model system to study the molecular functions of NPHP4 in motile cilia, we targeted the expression of NPHP4 to the epidermis. Injection of mRNA encoding full-length GFP-tagged human NPHP4 (200 pg) revealed the localization of NPHP4 to the basal bodies, labeled by RFP-Centrin (Fig. 1 A and Fig. S1 D). 3D reconstruction of confocal images revealed that NPHP4 localized to a region extending from the basal body to a region immediately distal of the basal body referred to as the transition zone (Fig. 1 B). Lower doses (70 pg) of microinjected NPHP4 restricted the localization of fluorescently labeled NPHP4 almost exclusively to the presumptive transition zone. The association between the basal body and NPHP4 was already detectable at stage 19, well before nucleation of the ciliary axoneme (Fig. S1 E), suggesting essential functions of NPHP4 during early ciliogenesis and before basal body docking. We next determined whether NPHP4 contains a certain targeting sequence typically required for intraflagellar transport–dependent ciliary trafficking (Sung and Leroux, 2013). Analysis of GFP-tagged NPHP4 truncations revealed that the C-terminal half of NPHP4 (amino acids 863–1,426) is sufficient to recruit NPHP4 to the basal bodies, fully recapitulating the localization of full-length NPHP4 (Fig. 1, A and C; and Table S1). This C-terminal domain could be further divided into two distinct subdomains (Fig. 1 D). One is spanning amino acids 1,251–1,426 delineating a minimum necessary sequence for NPHP4 localization; smaller fragments (e.g., amino acids 1,251–1,324) failed to localize to the basal bodies. The other domain spans amino acids 863–1,250. This subdomain was not confined to the basal body but is also colocalized with the ciliary rootlets labeled by RFP-Clamp (Fig. 1 E). These findings suggested that targeting of NPHP4 is not mediated by a short linear recognition motif but rather by a more complex 3D structure. Interestingly, some of the C-terminal fragments (e.g., amino acids 1,225–1,426) showed an enhanced membrane association of NPHP4 (Fig. 1 F), consistent with the dual cellular localization reported for NPHP4.
Figure 1.
Localization of full-length and truncated NPHP4 in multiciliated cells of the Xenopus epidermis. At the four-cell stage, Xenopus embryos were injected with mRNA for GFP-NPHP4 (green in merge), together with mRNAs for RFP-Centrin or RFP-Clamp (red in merge) to label basal bodies and ciliary rootlets, respectively. At stage 32, confocal microscopy was performed to study the localization of full-length and truncated NPHP4 in the cells with multiple motile cilia of epidermal skin. Maximum intensity projection of obtained confocal datasets is shown, unless noted otherwise. (A) Colocalization of GFP-NPHP4 with RFP-Centrin. Area enclosed by the white box is magnified in the right panel (5.3× magnification; see Fig. S1 A for separate GFP and RFP channels). (B) 3D reconstruction of confocal datasets is shown. Injection of mRNA encoding for GFP-NPHP4 (200 pg) revealed the broad localization of GFP-NPHP4 extending from the basal body to the presumptive transition zone distal to the basal body, whereas lower-dose injection of mRNA (70 pg) revealed a more confined localization of GFP-NPHP4 to the presumptive transition zone. The insets are magnified on the right panels. (C–F) Localization of NPHP4 truncations is studied. Numbers on the left of the panels indicate the first and last amino acid of the NPHP4 truncations. Expression of all constructs was confirmed by Western blot. (C) The C-terminal half of NPHP4 (amino acids 863–1,426) is sufficient for localization to the basal body. (D–F) The C-terminal half of NPHP4 contains two distinct subdomains targeting NPHP4 to the basal bodies: amino acids 863–1,250 and amino acids 1,251–1,426. (E) The first subdomain (amino acids 863–1,250) showed additional localization to the ciliary rootlet, colocalizing with RFP-Clamp. The inset is magnified by 2.4× on the right panels. (F) The C-terminal fragment spanning amino acids 1,225–1,426 showed enhanced localization to the plasma membrane (arrowhead). Area enclosed by the white box is magnified in the right panel. Bars: (main) 5 µm; (B, magnified insets) 500 nm.
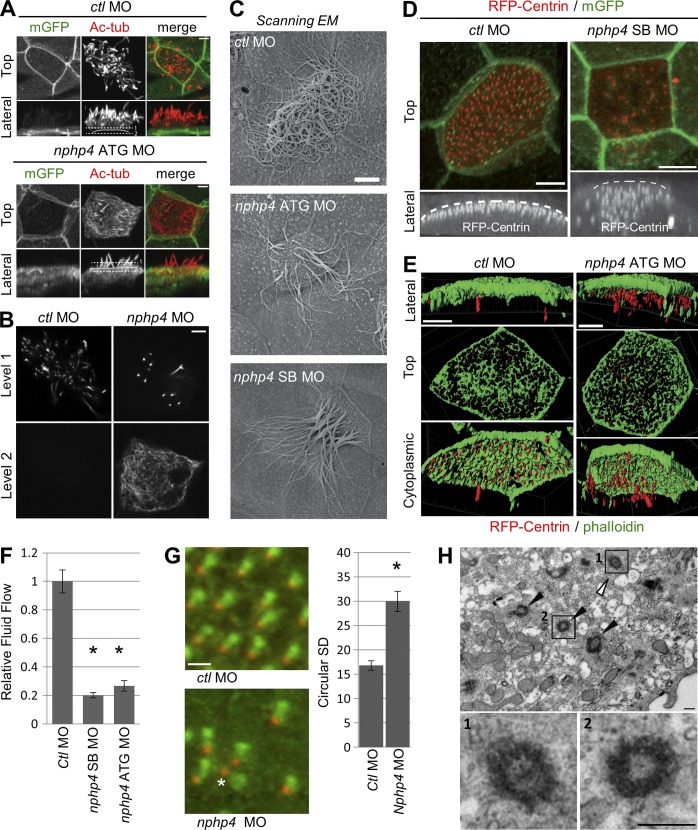
The role of NPHP4 in ciliogenesis
We explored the role of NPHP4 in ciliogenesis by using antisense morpholino oligonucleotides (MO) to deplete endogenous Xenopus Nphp4 protein. In control morpholino (ctl MO)-injected embryos, the apical surface of the multiciliated cells of the epidermis is decorated with more than 100 cilia (Fig. 2, A and C). These cilia are uniformly distributed across the apical cell surface as revealed by the distribution of basal bodies from which cilia nucleate (Fig. 2 D). Depletion of nphp4 by a translation-blocking MO (nphp4 ATG MO) targeting nphp4 mRNA resulted in a reduction of intact cilia that penetrated the surface of the cell (Fig. 2, A–C); instead, signal intensity for acetylated-tubulin staining was increased within the cytoplasm (Fig. 2 B, bottom). Lateral views of confocal images revealed that a large number of basal bodies were retained below the apical actin web in nphp4-depleted Xenopus embryos (Fig. 2 E). These basal bodies still nucleated microtubular structures positive for acetylated tubulin within the cytoplasm (Fig. 2 B). However, transmission electron microscopy revealed that only a few of these structures represented typical ciliary axonemes (Fig. 2 H), suggesting that basal bodies rarely nucleate a ciliary axoneme before reaching the apical membrane. The apical actin cytoskeleton also appeared slightly thinner in nphp4-deficient cells than in wild-type cells (Fig. 2 E). To quantify the functional consequences of nphp4 depletion, we measured the fluid flow speed across the ciliated-epidermal skin by tracking fluorescent particles, added to the culture media of Xenopus embryos. Embryos injected with nphp4 ATG MO showed a striking reduction in flow speed compared with ctl MO-injected embryos (Fig. 2 F), which was partially rescued by GFP-tagged nphp4 (Fig. S2 A). To further confirm that the observed defects were derived from depletion of nphp4, we designed a splice-blocking nphp4 MO (nphp4 SB MO; Fig. S2 B). We observed that injection of this MO caused identical defects elicited by nphp4 ATG MO (Fig. 2, C, D, and F). Although the reduction in cilia number appeared the main cause for the reduced flow speed, we examined ciliary polarization, using the combination of fluorescently labeled Clamp/Centrin to determine basal body polarity (Park et al., 2008). This analysis revealed a moderate but statistically significant disorientation in nphp4-deficient Xenopus depicted by an increase in circular standard deviation in comparison to control embryos (Fig. 2 G). To further characterize the function of NPHP4 in motile cilia, we monitored the beating of cilia by performing high-speed video microscopy and analyzed the beating pattern and frequency of cilia in nphp4-deficient embryos. Although the motile cilia of ctl MO–injected embryos showed a high-frequent directional pattern (Video 1), the cilia of nphp4 MO–treated embryos displayed a slow and uncoordinated beating pattern (Video 2). Given the relatively preserved basal body polarity, these observations suggested that the impaired function of motile cilia in nphp4-deficient cells resulted primarily from the loss of their connection to the actin cytoskeleton.
Figure 2.
Xenopus Nphp4 is essential for building functional cilia. (A and B) Confocal microscopy of multiciliated cells of Xenopus epidermal skin. Cilia and apical cell surface are labeled with anti–acetylated tubulin (Ac-tub; red in merge) and membrane-targeted GFP (mGFP; green in merge). (A) Maximum intensity projection (top), and 3D reconstruction projected in the x-z plane (lateral) for the confocal datasets are shown. The apical surface is indicated by a solid white line. Embryos injected with 20 ng control morpholino (ctl MO) showed normal ciliogenesis, whereas 20 ng nphp4 ATG MO–injected embryos showed ciliogenesis defects. (B) Single optical sections comparing the signals for anti–acetylated tubulin at levels 1 and 2 in ctl MO– and nphp4 ATG MO–treated Xenopus embryos (see dashed white lines in the lateral views of A) revealed that ciliary microtubules are nucleated below the apical cell surface in nphp4-deficient, but not ctl MO–treated, Xenopus embryos. (C) Scanning EM of the Xenopus epidermis confirmed the ciliogenesis defect in 16 ng nphp4 ATG MO– and 8 ng nphp4 SB MO–injected Xenopus embryos. (D) Labeling of basal bodies and the apical cell surface with RFP-Centrin and mGFP revealed that basal bodies are aligned along the apical cell surface and distributed uniformly across the cell surface in ctl MO–injected Xenopus embryos. In contrast, basal bodies in nphp4 MO–injected Xenopus embryos failed to migrate to the apical cell surface and remained within the cytoplasm. (Top) Maximum intensity projection of confocal datasets. (Bottom) Serial confocal images projected in the x-z plane with the apical cell surface indicated by a dashed white line. (E) The multiciliated cells of the Xenopus epidermis formed a dense actin cytoskeleton (phalloidin, green) at the apical cortex. Apically localized basal bodies (RFP-Centrin, red) were embedded in this actin web (left). In nphp4-deficient Xenopus embryos, basal bodies remained within the cytoplasm below the apical actin web, which is also thinner than in control cells (right). Confocal 3D datasets were processed with Imaris software, and the lateral (top), top (middle), and cytoplasmic (bottom) views are shown. (F) Depletion of nphp4 resulted in a decreased rate of cilia-driven fluid flow as revealed by the movement of polypropylene beads across the epidermis. Shown is a summary of four independent experiments (n ≥ 25). Error bars, SEM; t test; *, P < 0.002. (G) nphp4-deficient Xenopus embryos showed a mild perturbation of basal body polarization, revealed by the relative position of the basal bodies (RFP-Centrin) and the ciliary rootlets (GFP-Clamp). A moderate dose of 6.8 ng nphp4 SB MO was injected to allow apical docking of basal bodies, a prerequisite for polarization. The right panels show the magnified images for the regions enclosed by white boxes. Only a small number of basal bodies were incorrectly polarized (asterisk) in nphp4-deficient cells. The polarization was quantified by angular measurements of Clamp/Centrin pairs. Depletion of nphp4 resulted in a moderate increase of circular standard deviation as compared with the control. Error bars, SEM. (H) Transmission electron microscopy detected occasional ciliary axonemes (white arrowhead, box 1), and a large number of basal bodies (black arrowheads, box 2) in the cytoplasm of nphp4-depleted Xenopus multiciliated cells. Bars: (A–E) 5 µm; (G) 1 µm; (H) 200 nm.
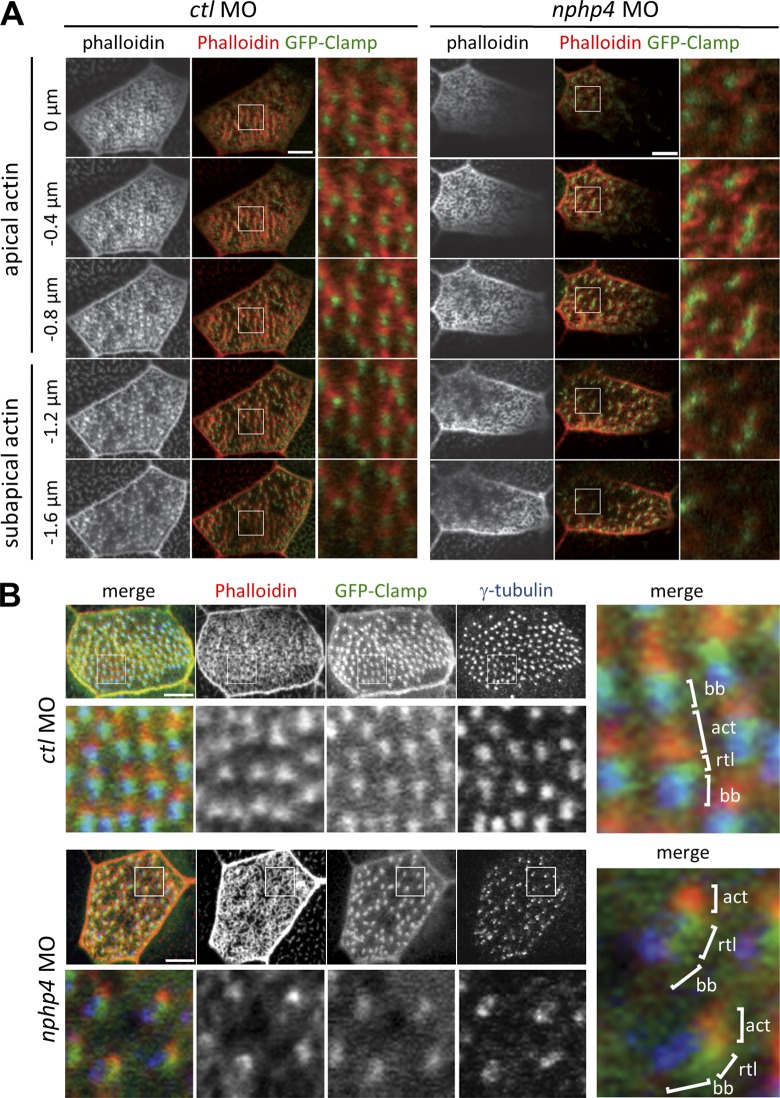
NPHP4 is required for a normal subapical actin network formation
Lifeact is a 17-amino-acid peptide derived from the budding yeast protein ABP-140 (Riedl et al., 2008). Colocalization of GFP-fused Lifeact (Lifeact-GFP) with the phalloidin-labeled F-actin structure in fixed Xenopus embryos (Fig. S2 C) prompted us to use Lifeact-GFP for studying the actin dynamics in multiciliated cells. Lifeact-GFP in combination with time-lapse imaging revealed that actin bundles continuously formed and dissolved during the initial stages of ciliogenesis (stage 20; Fig. S2 D and Video 3). Basal bodies seemed to stabilize actin bundles because areas devoid of basal bodies contained fewer actin bundles, suggesting a direct impact of basal bodies on actin cytoskeleton formation (Fig. S2, D and E). By late tailbud stage, ciliogenesis was completed and fully functional cilia were formed. In these mature multiciliated cells, basal bodies were embedded in a dense actin network at the apical surface of the cell (Fig. S2 F). Consistent with recently reported findings (Werner et al., 2011), we observed that the most apical actin layer formed a meshwork-like structure immediately below the apical plasma membrane, whereas the subapical actin layer contained evenly distributed filament-like structures (Fig. S2 G). The subapical actin layer developed ∼1 µm below the apical layer with filaments connecting neighboring basal bodies along the stroke direction (Fig. 3). Upon depletion of nphp4, the subapical actin pool became irregular, whereas the apical actin remained unaffected (Fig. 3 A). Triple labeling using γ-tubulin to mark the basal bodies, clamp to identify the rootlets, and phalloidin to identify the subapical actin bundles revealed an orderly sequence among these three structures in ctl MO–treated cells (Fig. 3 B). In contrast, nphp4-depleted cells showed a reduced nucleation of subapical actin from the tip of the rootlet. The basal bodies of nphp4-deficient cells were irregularly distributed, and the weakly nucleated subapical actin failed to connect neighboring basal bodies (Fig. 3 B); however, the basal bodies and rootlets remained correctly polarized (Fig. 2 G). These observations suggested that NPHP4 contributes to the organization of the actin cytoskeleton, which has been previously hypothesized based on the interaction partners of NPHP4 (Sang et al., 2011).
Figure 3.
Nphp4 is essential for the organization of the subapical actin layer. (A) Serial confocal images for actin (phalloidin) and ciliary rootlets (GFP-Clamp) of cells with multiple motile cilia revealed that the subapical actin layer became irregular in nphp4-deficient Xenopus embryos, whereas apical actin pool remained unaffected. The depth of each optical section from the first section is shown on the left. A magnified image of the inset is shown on the right side (4× magnification). (B) Triple staining for actin (phalloidin, red in merge), ciliary rootlets (GFP-Clamp, green in merge), and basal bodies (γ-tubulin, blue in merge) revealed that actin filaments in the subapical actin layer connect a basal body with the ciliary rootlet of the neighboring basal body. In nphp4-deficient cells, the subapical actin layer was poorly nucleated and failed to provide the connection between basal bodies. Top panels show the maximum intensity projection of serial confocal images. Single optical sections at the level of the subapical actin pool (boxed area) are magnified and shown in the bottom panels (4.5× magnification). Right panels are higher magnification images of the single optical sections depicting the subapical actin organization (10.5× magnification). act, actin; bb, basal body; rtl, ciliary rootlet. Bars, 5 µm.
Functional interactions among NPHP4, INTURNED, and DAAM1
Because NPHP4 does not harbor any enzymatic domain, NPHP4 likely influences the actin cytoskeleton through interaction with other proteins. To obtain further insight, we decided to identify potential binding partners through a candidate approach based on two criteria: (1) mutant proteins displaying an overlapping ciliary phenotype, and (2) actin-modifying proteins that have been identified in cilia. Immunoprecipitation assays in HEK 293T cells revealed that the Drosophila melanogaster actin-nucleating factor cappuccino as well as human INTURNED interacted with NPHP4; other actin-nucleating factors did not or only weakly bound to NPHP4 (Fig. S3 A). Cappuccino preferentially interacted with the C-terminal domain of NPHP4, although a reduced interaction was observed between cappuccino and the N-terminal 515 amino acids of NPHP4 (Fig. S3 B), suggesting an indirect mode of interaction between both proteins. Cappuccino is a Drosophila Formin family member; more than a dozen Formins have been described in vertebrates (Chesarone et al., 2010). A previous proteomic study of mouse photoreceptors identified three Formins within the ciliary compartment, DAAM1, FORMIN-2, and FHOD3 (Liu et al., 2007). DAAM1 interacted only weakly with NPHP4 but revealed a strong interaction with INTURNED (Fig. S3, C and D). Because NPHP4 immobilized INTURNED, we next tested if INTURNED can mediate the formation of a ternary complex between NPHP4 and DAAM1. NPHP4 failed to interact with DAAM1 when coexpressed with DAAM1 (Fig. S3 E). However, when INTURNED was added, DAAM1 precipitated NPHP4, suggesting that NPHP4, INTURNED, and DAAM1 indeed form heteromeric complexes (Fig. S3 E).
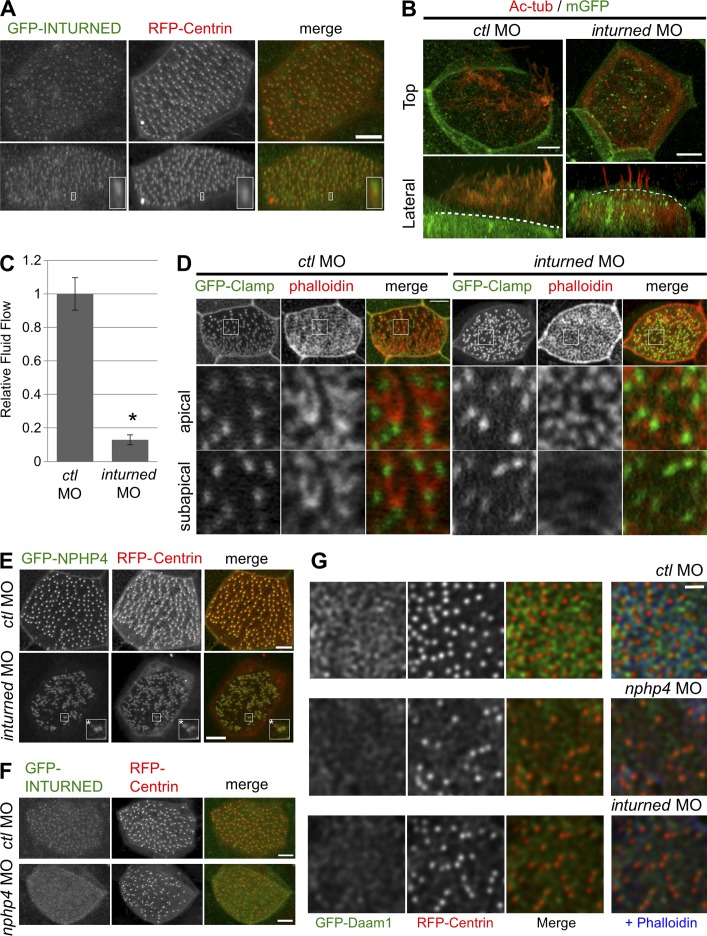
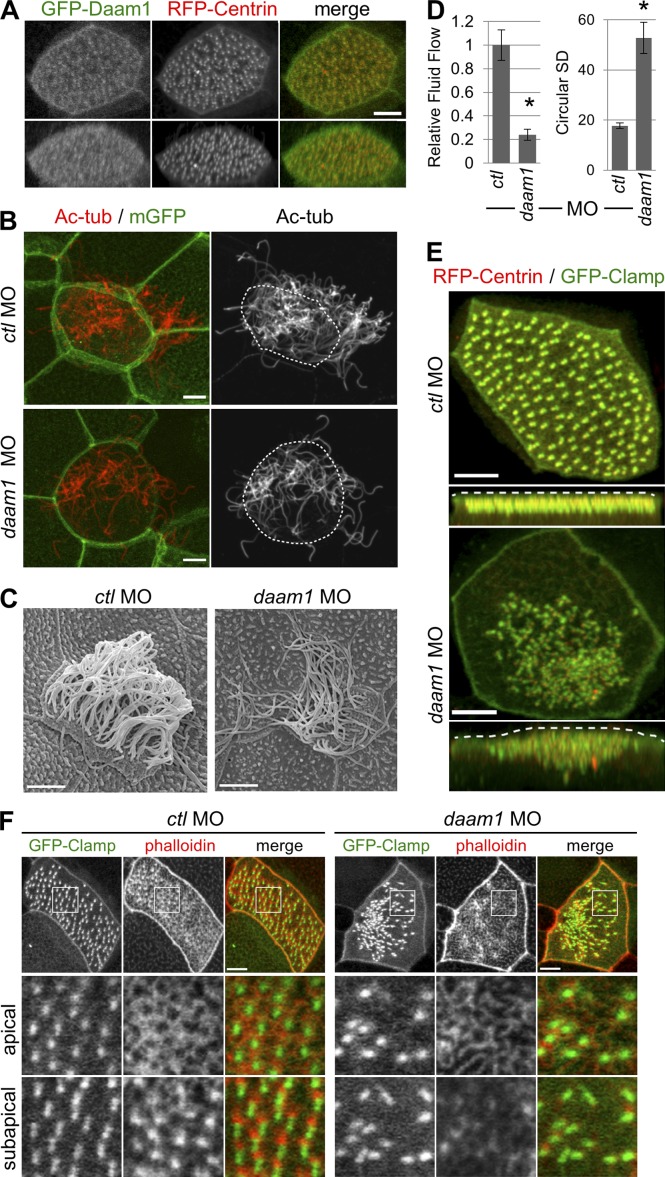
Because we identified INTURNED and DAAM1 as binding partners of NPHP4, we next examined the effects of inturned and daam1 depletion in the multiciliated cells of the Xenopus epidermis. Consistent with the previously reported role of Inturned (Park et al., 2006), knockdown of inturned caused ciliogenesis defects accompanied by reduced flow speed of fluorescent particles across the epidermis (Fig. 5, B and C). Because a reduced density of cortical actin web in inturned-deficient cells has been reported (Park et al., 2006), we analyzed the actin organization in detail and found that the formation of the subapical actin network was similarly altered as in nphp4-deficient cells (Fig. 5 D); however, inturned depletion also affected the apical actin network. Cells from daam1-deficient embryos recapitulated the ciliary defects observed in nphp4-deficient cells, including defective basal body docking and decreased ciliogenesis and ciliary flow as well as reduced nucleation of the subapical actin network (Fig. 4); the fluid flow defect was partially rescued by GFP-tagged Daam1 (Fig. S2 H). FORMIN-1 and -2 are the closest homologs to cappuccino, and FORMIN-2 is part of the ciliary proteome (Liu et al., 2007). To determine whether the observed role is unique for DAAM1, we tested whether the knockdown of formin-1 and formin-2 cause similar ciliary defects. Although depletion of formin-1 was only partially effective, RT-PCR revealed a substantial reduction in formin-2. However, knockdown of formin-2 did not reduce ciliary fluid flow, suggesting that Formin-2, in contrast to Daam1, does not play a major role in the formation of the subapical actin web and the function of motile cilia in the Xenopus epidermis (Fig. S3 F).
Figure 5.
Inturned plays a role similar to Nphp4 for normal ciliogenesis, and basal body localization of Inturned requires Nphp4. (A) GFP-tagged INTURNED (green in merge) preferentially localized to a region distal to the basal body labeled by RFP-Centrin (red in merge). Maximum intensity projection (top) and 3D reconstruction (bottom) of confocal images are shown. The representative localization pattern is shown magnified in the inset (4.3× magnification). (B) Confocal imaging of cilia stained with anti–acetylated tubulin (Ac-tub). Reduced number of cilia formed above the apical cell surface after injection of 17 ng inturned MO. Maximum intensity projection (top) and the projection in the x-z plane (bottom) of confocal images are shown. Dashed line indicates the apical surface of cells as revealed by membrane-associated GFP (mGFP; green). (C) 8 ng inturned MO–injected embryos presented with a decreased rate of ciliary fluid flow across the epidermis. Shown is the summary of four independent experiments (n ≥ 39; t test; *, P = 0.0014). (D) Injection of 8 ng inturned MO resulted in a reduced nucleation of the subapical actin pool as well as fragmentation of apical actin web. Actin and ciliary rootlet were stained with phalloidin (red in merge) and GFP-Clamp (green in merge), respectively. (Top) Maximum intensity projection of serial confocal images. The area enclosed by white boxes was magnified (5.6×); single optical sections at the level of apical actin pool (middle) and subapical actin pool (bottom) are shown. Bars, 5 µm. (E) Localization of GFP-NPHP4 (green in merge) to the basal body (RFP-Centrin, red in merge) was only slightly affected by the depletion of inturned (7 ng MO). Areas enclosed by the white boxes are magnified in the insets (2.5×). Only a small subset of basal bodies in the inturned-deficient cells revealed a reduced accumulation of NPHP4 (asterisks). (F) Depletion of nphp4 (8 ng MO) resulted in a reduced colocalization of GFP-INTURNED (green in merge) with the basal bodies, and GFP-INTURNED revealed a more homogenous distribution in nphp4-deficient cells. (G) Whereas GFP-Daam1 was closely associated with the actin filaments in Xenopus epidermal cells microinjected with a control MO (ctl MO; Fig. S3 F), the depletion of nphp4 or inturned in Xenopus epidermal skin cells decreased both actin and GFP-Daam1 in the subapical region. The image depicts three z sections at a −600 nm in cells as indicated. Bars, 1 µm.
Figure 4.
daam1-deficient multiciliated cells recapitulate the ciliary phenotypes of nphp4-deficient cells. (A) GFP-tagged Daam1 (green in merge) preferentially localized to a region distal to the basal body (RFP-Centrin, red in merge) at the apical surface. Maximum intensity projection (top) or 3D reconstruction (bottom) of confocal images is shown. (B) Confocal imaging of cilia (Ac-tub, red). As compared with the control, reduced number of cilia formed above the apical cell surface (membrane-associated GFP [mGFP], green) after injection of 12 ng daam1 MO. The apical cell surface is depicted by dashed white lines on the right panels. (C) The ciliogenesis defect was confirmed by scanning electron microscopy. (D) daam1-deficient Xenopus embryos show a decreased rate of cilia-driven fluid flow across the epidermis (four independent experiments with n > 45; error bars, SEM; t test; *, P = 0.0014). Depletion of daam1 was also associated with a polarity defect and an increase in circular SD (t test; *, P < 0.001). (E) Basal bodies in daam1-deficient multiciliated cells failed to migrate to the apical cell surface and remained in the cytoplasm. Basal bodies and ciliary rootlets were labeled with RFP-Centrin and GFP-Clamp. Maximum intensity projection (top) or projection in the x-z plane (bottom) of serial confocal images are shown. The apical cell surface is indicated by dashed white line. (F) The subapical actin layer displayed irregularities in daam1–deficient multiciliated cells. Ciliary rootlets were labeled with GFP-Clamp (green in merge) and actin stained with phalloidin (red in merge). (Top) Maximum intensity projection of serial confocal images. Single optical sections at the level of apical and subapical actin layer of the boxed area are shown magnified in the middle and bottom panels (4× magnification). Bars, 5 µm.
To obtain further support for a functional link between NPHP4, Inturned, and Daam1, we examined the subcellular localization of INTURNED and Daam1 in the multiciliated cells by generating GFP-tagged version of these proteins. GFP-tagged INTURNED showed a speckled cytoplasmic distribution; colabeling of the basal bodies with RFP-Centrin revealed that INTURNED colocalized with the basal bodies (Fig. 5 A). GFP-tagged Daam1 also accumulated at the apical surface of the multiciliated cells (Fig. 4 A). Colabeling with RFP-Centrin and phalloidin revealed that GFP-Daam1 decorated the actin filaments and in some instances was found in close proximity of the basal bodies (Fig. S3 G). Thus, the subcellular localization of these three proteins supports the idea that Nphp4, Inturned, and Daam1 form ternary complexes at the basal body to modulate the subapical actin network.
To determine the hierarchy among these three proteins, we examined which component of the Nphp4–Inturned–Daam1 complex plays a pivotal role in recruiting this complex to the basal body/ciliary compartment. In inturned-deficient embryos, we observed no striking effect on the accumulation of GFP-NPHP4 at the basal bodies; most of the basal bodies were decorated with the GFP-NPHP4 signal, and only a small subset of basal bodies was devoid of the GFP signal (Fig. 5 E). In contrast, the staining pattern of GFP-INTURNED at the apical membrane became homogenous, and the accumulation of GFP-INTURNED at the basal bodies was strongly reduced in nphp4-deficient embryos (Fig. 5 F), whereas depletion of nphp4 or inturned reduced both actin and Daam1 accumulation in the subapical actin layer (Fig. 5 G). Thus, Nphp4 appears to recruit Inturned to the basal bodies of motile cilia, interaction of Inturned with Daam1 can then modify the actin cytoskeleton in a spatially restricted manner. Because NPHP4 associates with basal bodies already during early developmental stages (Fig. S1 E), the protein complex could modify the actin cytoskeleton before basal bodies reach the apical plasma membrane. The Inturned–Daam1 interaction likely plays a role in other cellular programs. Although both proteins have been implicated as downstream effectors of the planar cell polarity (PCP) pathway (Habas et al., 2001; Park et al., 2006), the role of Inturned in mammalian convergent extension as a downstream effector molecule of Frizzled (Fz)/Dvl signaling has remained unclear. For example, whereas Daam1 is recruited by the PCP core proteins Celsr1/Fz/Dvl to adherent junctions and is essential for convergent extension movements during neural tube closure (Nishimura et al., 2012), Inturned is not required for this process, but rather seems to function in neural tube patterning, requiring the presence of cilia (Heydeck and Liu, 2011). Cilia are linked to PCP signaling; however, the hierarchical relationship between PCP core proteins and cilia is not fully understood (Brooks and Wallingford, 2014). Although genetic analysis has placed Inturned downstream of Fz during Drosophila wing development (Lee and Adler, 2002; Adler et al., 2004), Inturned in association with other PCP effector molecules can also act upstream of PCP core proteins to instruct their localization (Wang et al., 2014). Furthermore, Inturned physically interacts with “Multiple wing hairs” (Mwh), a Drosophila protein that contains a Diaphanous GTPase-binding and an FH3 domain (Lu et al., 2010). Although Inturned affects the localization of Mwh (Lu et al., 2010), recent results demonstrate that Mwh can directly regulate the actin cytoskeleton (Lu et al., 2015). Collectively, these observations suggest an evolutionary conserved pathway, where Inturned interacts with site-specific proteins to assume a distinct subcellular localization and then recruits actin-modifying proteins to alter the local actin cytoskeleton. Because NPHP4 can target Dvl for degradation (Burcklé et al., 2011), NPHP4 could antagonize Dvl2/Daam1-mediated Rho activation to redirect the function of Daam1 in a spatially restricted manner.
Materials and methods
Embryo manipulations
Xenopus embryos were manipulated as described previously (Lienkamp et al., 2010). In brief, in vitro–fertilized embryos were cultured in 0.3× Marc’s modified Ringer’s medium (MMR) and staged according to Nieuwkoop and Faber. For most experiments, microinjections were made into the two ventral blastomeres at the four-cell stage to target the epidermis. Each blastomere was injected with 10-nl solution containing 8–20 ng of antisense MOs (GeneTools) and various amounts of mRNAs using a time- and pressure-triggered microinjection system (Narishige). Embryos were cultured until stage 32–34 unless noted otherwise. The institutional animal committee approved all experiments.
RNA extraction and RT-PCR
RNA was isolated from Xenopus embryos using the RNeasy kit (QIAGEN), and cDNA synthesis was performed using the RevertAid H Minus kit (Fermentas) or the First-Strand cDNA Synthesis kit (Invitrogen). Primer sequences used for RT-PCR are provided in Table S1.
Whole-mount in situ hybridization
For probe synthesis, Xenopus nphp4 was subcloned into pGEM-T system (Promega) and in vitro transcription was performed using SP6 (sense probe) or T7 (antisense probe) polymerase and DIG-labeling mix (Roche). Xenopus wild-type or albino embryos were fixed with MEMFA (0.1 M Mops, 2 mM EGTA, 1 mM MgSO4, and 3.7% formaldehyde, pH7.4) for 1 h at RT and stored at −20°C in methanol until required. Whole-mount in situ hybridization was performed as described previously, with some modifications (Lienkamp et al., 2010). In brief, fixed embryos were rehydrated in PBST (PBS with 0.1% Tween 20), treated with 10 µg/ml Proteinase K for 10 min, washed with triethanolamine with and without 0.25% acetic anhydride, and refixed with 4% PFA for 20 min at RT. After washing with PBST, embryos were prehybridized with hybridization buffer (50% formamide, 5× SSC, 100 µg/ml heparin, 1× Denhart’s, 0.1% Tween 20, 10 mM EDTA, 1 mg/ml Torula RNA, and 0.1% CHAPS) at 60°C for 5–6 h and incubated with 1–2 µg/ml probe in hybridization buffer at 60°C overnight. After a wash with hybridization buffer followed by washes with 2× SSCT (2× SSC with 0.1% Tween 20) at 60°C, embryos were treated with RNases (100 μ/ml RNase T [Sigma-Aldrich] and 20 µg/ml RNase A [QIAGEN]) in 2× SSCT for 30 min at 37°C and washed with 2× SSCT, 0.2× SSCT, and maleic acid buffer (MAB; 100 mM maleic acid and 150 mM NaCl, pH 7.5). After sequential blocking with 2% blocking reagent (Roche) in MAB for 15 min at RT and 2% blocking reagent/20% sheep serum in MAB for 1 h at RT, embryos were incubated with a 1:2,000 dilution of alkaline phosphatase–conjugated sheep anti-DIG antibody (Roche) at 4°C overnight. After washes in MAB and alkaline phosphatase buffer (100 M Tris, 50 mM MgCl2, 100 mM NaCl, 0.1% Tween20, and 5 mM levamisole, pH 9.5), embryos were stained with BM purple (Roche). After the desired signal strength was obtained, reaction was stopped with MAB and refixation was performed with 10% formaldehyde/5% glacial acetic acid at 4°C overnight followed by washes in 70% ethanol/30% PBS and 1× SSC. Wild-type embryos were cleared in bleaching solution (0.5× SSC, 2% formaldehyde, and 1.2% hydrogen peroxide) for 1–2 h at RT. Double staining for acetylated tubulin was performed after in situ hybridization. Embryos in PBS were imaged at RT using an MZ16F stereomicroscope (Leica) equipped with a Plan-Apochromat 1.0× objective and a DFC450C camera (Leica). LAS V4.3 software (Leica) was used for image acquisition. Acquired images were exported as TIFF files and imported into Photoshop software CS5 (Adobe) to arrange figures.
Immunofluorescence staining and microscopy
Xenopus embryos were fixed for 1 h at RT with MEMFA and washed with PBST (antibody staining). After blocking for 30–60 min in PBST containing 10% FBS and 5% DMSO, the embryos were incubated with mouse anti–acetylated-tubulin antibody (1:500 dilution; T6793; Sigma-Aldrich), or with a mouse anti–γ-tubulin antibody (1:2,500 dilution; T6557; Sigma-Aldrich,) at 4°C overnight. The embryos were then washed three times with PBST, incubated for 2–5 h at RT with Alexa Fluor 546– or 647–labeled secondary antibodies (1:500 dilution; Invitrogen), and washed three times with PBST. For F-actin staining, embryos were washed three times with TBSN (TBS with 0.05% NP-40) after MEMFA fixation, incubated for 2–5 h at RT with Alexa Fluor 488– or Alexa Fluor 568–phalloidin (1:300 dilution; Invitrogen), and then washed three times with TBSN. Confocal fluorescence microscopy was performed at RT with an Axiovert 200 LSM 510 Duo-Live microscope equipped with an LCI Plan Neofluar 63×/1.3 NA water immersion objective (Carl Zeiss). The excitation of fluorophores (GFP/Alexa Fluor 488, RFP/Alexa Fluor 568, and Alexa Fluor 647) was performed at 488, 561, and 633 nm, respectively. For detection of the emission signal at specified ranges, the spectral metal detector or normal photomultiplier channels were used with band-pass filter 505–550, long-pass (LP) 575, and LP 650 nm. Confocal pinhole diameters were always adjusted to 0.8-µm sections (according to 1 airy unit at 488 nm excitation) and z stacks (∼60 z planes, 0.4 µm z distance, pixel time 1.21 µs, line average 16) were recorded. Pictures were taken with scan speed 9 and 8 bit. Zen 2010 software (Carl Zeiss) was used for image acquisition. Confocal fluorescence microscopy for the Daam1 localization analysis in Fig. S3 G was performed at RT with a TCS SP8 microscope equipped with an HC PL APO 63×/1.3 glycerine objective (Leica). The excitation of fluorophores (GFP, RFP, and Alexa Fluor 647) was performed sequentially at 488, 561 and 633 nm. The spectral emission detection unit was set to the following wavelength ranges: 492–581, 566–658, and 640–738 nm, accordingly. Confocal pinhole diameter was always adjusted to 0.8-µm z sections (according to 1 airy unit at 488 nm excitation), and z stacks (∼40 planes, 0.2 µm distance, pixel time 975 ns, frame accumulation 2, and line average 2) were recorded. Pictures were taken with a 400-Hz scan speed and 8 bit. Leica Application Suite X was used for image acquisition. The acquired images were deconvolved with Huygens essential software (Scientific Volume Imaging), applying the classic maximum likelihood estimation mode and using a theoretical point spread function. Scanning electron microscopy was performed as described previously (Hoff et al., 2013). In brief, Xenopus embryos were fixed using Bouin solution. Dehydration with 70% EtOH, 80% EtOH, 90% EtOH, and 100% EtOH was followed by incubation in 50:50 EtOH/HMDS. After incubation in 100% HMDS, the solvent was allowed to evaporate. All samples were coated with gold using a Cool Sputter Coater E 5100 (Polaron). Samples were imaged using a Leo 1450 VP electron microscope (Carl Zeiss). Transmission electron microscopy was performed as described previously (Epting et al., 2015). In brief, Xenopus embryos were fixed with 4% PFA/1% glutaraldehyde in PBS overnight at 4°C and postfixed with 1% osmium tetroxide for 30 min at RT, dehydrated with two 10-min ethanol (50% and 60%) washings, and incubation in 1% uranyl acetate in 70% ethanol overnight at 4°C. Embryos were dehydrated in a graded ethanol series. After washing in propylene oxide, embryos were embedded in Durcupan (Fluka). Transverse ultrathin sectioning (60 nm) was performed using a Leica EM UC6. The sections were mounted on copper grids and examined in an electron microscope (LEO 906E; Carl Zeiss). For analyzing ciliary flow, embryos were placed in 0.3× MMR containing the anesthetic drug MS222 (0.02%; Sigma-Aldrich) and small fluorescent beads (Invitrogen). Time-lapse microscopy was performed at RT with a SPOT Insight FireWire system (Diagnostic Instruments) at 10× on a Leica MZ16 stereomicroscope. Images were analyzed with Imaris software (Bitplane) and the mean speed of beads was calculated for each embryo. For analyzing ciliary beating, embryos were injected at the four-cell stage with MO, together with mRNA for membrane-targeted GFP to label cilia, placed in MS222-containing 0.3× MMR at stage 32, and the GFP fluorescence was recorded at RT using an Axiovert 200 LSM 5 high-speed line scanning microscope equipped with an LCI Plan Neofluar 63×/1.3 NA water immersion objective (Carl Zeiss). LP495 filter was used for detection of the emission signal. Scanning area was 256 × 256 pixels, 108 frames/s, pixel size was 0.42 µm, and pixel dwell time was 15.4 µs. The confocal slit diameter was fully opened, resulting to an optical section thickness of 5.8 µm. Zen 2010 software was used for image acquisition. Pictures were digitized with 8 bit. Acquired images were exported as TIFF files and imported into Photoshop software CS5 to arrange figures.
Plasmids, MOs, and mRNA synthesis
For expression in mammalian cells, full-length fly cappuccino, human CD2AP, human DAAM1, human INTURNED, human NPHP4, human SPIRE, and truncated versions of human NPHP4 (residues 1–515, 511–1,426, and 863–1,426) were PCR amplified, cloned, sequenced, and subcloned into Flag-, Myc-, or V5-tagged pcDNA6 vectors (Invitrogen). To identify full-length cDNA of X. laevis nphp4 and daam1, BLASTN searches against X. laevis genome ver.5 were performed, using full-length X. tropicalis nphp4 identified in the U.S. Department of Energy Joint Genome Institute genome assembly v4.1 (JGI XT Genome 4.1) Scaffold 300 or X. tropicalis daam1 (XM_004917213.1), respectively. Scaffold 2903 contained all the coding exons for nphp4 except for the last one, whereas scaffold 94,422 contained the last coding exon and scaffold 14,125 contained a full-length daam1. Primers were designed based on these genomic sequences and were used to amplify the coding region of nphp4 and daam1. The PCR product was cloned into pGEM-T vector (Promega) and sequenced. For expression in Xenopus embryos, human INTURNED, Xenopus nphp4, daam1, centrin4, and clamp were subcloned into VF10 vectors with GFP or RFP at the N terminus. Lifeact DNA was generated by annealing a pair of complementary oligonucleotides, restriction digestion, and subcloning into pCS2+ vector carrying C-terminal GFP. After linearization, these plasmids were transcribed in vitro into mRNA using mMessage Machine kit (Ambion). To design MOs, Xenopus genomic sequences were obtained by performing BLASTN search against X. laevis genome ver.5 using X. tropicalis cDNA sequences. The genomic DNA was amplified by PCR, cloned, and sequenced. The sequence of MOs is as follows: control MO (ctl MO), 5′-CCTCTTACCTCAGTTACAATTTATA-3′; nphp4 ATG MO, 5′-ATATTCTCCCAGTCCTTCATCTTGA-3′; nphp4 SB MO, 5′-TAGGGTTCAGGTCGTCTTACCTCGT-3′; formin-1 MO, 5′-TCCAAAAATCGGTACATACCTTGTT-3′; formin-2 MO1, 5′-GCTTGACAACCTGACAAAATGAACA-3′; formin-2 MO2, 5′-CTCTTGCACAAAATACTCACATTGC-3′; inturned MO, 5′-TCCATATCCCCCAGCAGCACTTCTA-3′; daam1 MO (Habas et al., 2001), 5′-GCCGCAGGTCTGTCAGTTGCTTCTA-3′. Detailed information on these MOs is also provided in Table S2.
Accession numbers
NCBI RefSeq accession numbers: human SPIRE, NM_020148; fly cappuccino, NM_057618; human DAAM1, NM_014992; human NPHP4, NM_015102, human INTURNED, BC130611. UniGene accession numbers: Xenopus centrin4, Xl.50473; Xenopus clamp, Xl.26316.
Cell culture, coimmunoprecipitation, and Western blots
Human embryonic kidney (HEK 293T) cells were grown in DMEM supplemented with 10% FBS. Transient transfection and coimmunoprecipitation were performed as described previously (Ganner et al., 2009). In brief, transfections were performed using the calcium phosphate method; cells were washed with cold PBS and lysed in 1% Triton X-100, 20 mM Tris, pH 7.5, 50 mM NaCl, 50 mM NaF, 15 mM Na4P2O7, 0.1 mM EDTA, and 2 mM Na3VO4 supplemented with protease inhibitor mix (Roche). Lysates were incubated with anti-Flag M2-agarose beads (Sigma-Aldrich) for 2 h and washed with lysis buffer. For Western blots, proteins were fractionated by SDS-PAGE and detected by mouse monoclonal antibody to Flag (Sigma-Aldrich) and GFP (Santa Cruz Biotechnology) and rabbit polyclonal antibody to Flag (Sigma-Aldrich) and V5 (Millipore).
Online supplemental material
Fig. S1 demonstrates the spatial and temporal expression of nphp4 and localization of its protein product in Xenopus. Fig. S2 depicts the actin cytoskeleton in Xenopus multiciliated cells and contains the specificity control for nphp4 and daam1 MOs and the analysis of actin cytoskeleton in Xenopus multiciliated cells. Fig. S3 identifies binding partners of NPHP4, demonstrates the effect of formin-1 and formin-2 knockdown on fluid flow, and depicts the Daam1 localization in multiciliated Xenopus epidermal cells at high resolution. Table S1 summarizes the results of the subcellular localization of NPHP4 truncations in multiciliated Xenopus epidermal cells. Table S2 lists the primers used for Xenopus nphp4 and odc RT-PCR. Table S3 contains the sequence information of the MOs used in the report. Video 1 depicts the beating pattern of cilia in ctl- and MO-injected Xenopus embryo. Video 2 shows the beating pattern of cilia in nphp4 MO-injected Xenopus embryo. Video 3 shows the dynamic rearrangement of the actin cytoskeleton in multiciliated Xenopus epidermal cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201502043/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201502043.dv.
Supplementary Material
Acknowledgments
We would like to thank all members of the Renal Division for helpful discussions and for critically reading early versions of the manuscript. Special thanks goes to R. Nitschke and the staff of the Life Imaging Center in the Center for Systems Biology, Albert-Ludwigs-University Freiburg for excellent support in image acquisition as well as to B. Joch for the support of EM sample preparation.
G. Walz is supported by the Deutsche Forschungsgemeinschaft SFB 746 and 1140. T. Huber and O. Ronneberger are supported by SFB 1140. S. Lienkamp is supported by Deutsche Forschungsgemeinschaft Emmy Noether program and by SFB 1140.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- Dvl
- Dishevelled
- Fz
- Frizzled
- MAB
- maleic acid buffer
- MMR
- Marc’s modified Ringer’s medium
- MO
- morpholino oligonucleotides
- NPH
- nephronophthisis
- NPHP
- nephrocystin
- PCP
- planar cell polarity
References
- Adler P.N., Zhu C., and Stone D.. 2004. Inturned localizes to the proximal side of wing cells under the instruction of upstream planar polarity proteins. Curr. Biol. 14:2046–2051. 10.1016/j.cub.2004.11.007 [DOI] [PubMed] [Google Scholar]
- Alazami A., Alshammari M., Baig M., Salih M., Hassan H., and Alkuraya F.. 2013. NPHP4 mutation is linked to cerebello-oculo-renal syndrome and male infertility. Clin. Genet. 85:371–375. 10.1111/cge.12160 [DOI] [PubMed] [Google Scholar]
- Antoniades I., Stylianou P., and Skourides P.A.. 2014. Making the connection: ciliary adhesion complexes anchor basal bodies to the actin cytoskeleton. Dev. Cell. 28:70–80. 10.1016/j.devcel.2013.12.003 [DOI] [PubMed] [Google Scholar]
- Benzing T., and Schermer B.. 2012. Clinical spectrum and pathogenesis of nephronophthisis. Curr. Opin. Nephrol. Hypertens. 21:272–278. 10.1097/MNH.0b013e3283520f17 [DOI] [PubMed] [Google Scholar]
- Benzing T., Gerke P., Höpker K., Hildebrandt F., Kim E., and Walz G.. 2001. Nephrocystin interacts with Pyk2, p130(Cas), and tensin and triggers phosphorylation of Pyk2. Proc. Natl. Acad. Sci. USA. 98:9784–9789. 10.1073/pnas.171269898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann C., Fliegauf M., Brüchle N.O., Frank V., Olbrich H., Kirschner J., Schermer B., Schmedding I., Kispert A., Kränzlin B., et al. 2008. Loss of nephrocystin-3 function can cause embryonic lethality, Meckel-Gruber-like syndrome, situs inversus, and renal-hepatic-pancreatic dysplasia. Am. J. Hum. Genet. 82:959–970. 10.1016/j.ajhg.2008.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershteyn M., Atwood S.X., Woo W.M., Li M., and Oro A.E.. 2010. MIM and cortactin antagonism regulates ciliogenesis and hedgehog signaling. Dev. Cell. 19:270–283. 10.1016/j.devcel.2010.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks E.R., and Wallingford J.B.. 2014. Multiciliated cells. Curr. Biol. 24:R973–R982. 10.1016/j.cub.2014.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burcklé C., Gaudé H.M., Vesque C., Silbermann F., Salomon R., Jeanpierre C., Antignac C., Saunier S., and Schneider-Maunoury S.. 2011. Control of the Wnt pathways by nephrocystin-4 is required for morphogenesis of the zebrafish pronephros. Hum. Mol. Genet. 20:2611–2627. 10.1093/hmg/ddr164 [DOI] [PubMed] [Google Scholar]
- Campellone K.G., and Welch M.D.. 2010. A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11:237–251. 10.1038/nrm2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone M.A., DuPage A.G., and Goode B.L.. 2010. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat. Rev. Mol. Cell Biol. 11:62–74. 10.1038/nrm2816 [DOI] [PubMed] [Google Scholar]
- Dawe H.R., Adams M., Wheway G., Szymanska K., Logan C.V., Noegel A.A., Gull K., and Johnson C.A.. 2009. Nesprin-2 interacts with meckelin and mediates ciliogenesis via remodelling of the actin cytoskeleton. J. Cell Sci. 122:2716–2726. 10.1242/jcs.043794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epting D., Slanchev K., Boehlke C., Hoff S., Loges N.T., Yasunaga T., Indorf L., Nestel S., Lienkamp S.S., Omran H., et al. 2015. The Rac1 regulator ELMO controls basal body migration and docking in multiciliated cells through interaction with Ezrin. Development. 142:1553 10.1242/dev.124214 [DOI] [PubMed] [Google Scholar]
- Fliegauf M., Horvath J., von Schnakenburg C., Olbrich H., Müller D., Thumfart J., Schermer B., Pazour G.J., Neumann H.P., Zentgraf H., et al. 2006. Nephrocystin specifically localizes to the transition zone of renal and respiratory cilia and photoreceptor connecting cilia. J. Am. Soc. Nephrol. 17:2424–2433. 10.1681/ASN.2005121351 [DOI] [PubMed] [Google Scholar]
- Ganner A., Lienkamp S., Schäfer T., Romaker D., Wegierski T., Park T.J., Spreitzer S., Simons M., Gloy J., Kim E., et al. 2009. Regulation of ciliary polarity by the APC/C. Proc. Natl. Acad. Sci. USA. 106:17799–17804. 10.1073/pnas.0909465106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habas R., Kato Y., and He X.. 2001. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell. 107:843–854. 10.1016/S0092-8674(01)00614-6 [DOI] [PubMed] [Google Scholar]
- Heydeck W., and Liu A.. 2011. PCP effector proteins inturned and fuzzy play nonredundant roles in the patterning but not convergent extension of mammalian neural tube. Dev. Dyn. 240:1938–1948. 10.1002/dvdy.22696 [DOI] [PubMed] [Google Scholar]
- Hoff S., Halbritter J., Epting D., Frank V., Nguyen T.M., van Reeuwijk J., Boehlke C., Schell C., Yasunaga T., Helmstädter M., et al. 2013. ANKS6 is a central component of a nephronophthisis module linking NEK8 to INVS and NPHP3. Nat. Genet. 45:951–956. 10.1038/ng.2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauregui A.R., Nguyen K.C., Hall D.H., and Barr M.M.. 2008. The Caenorhabditis elegans nephrocystins act as global modifiers of cilium structure. J. Cell Biol. 180:973–988. 10.1083/jcb.200707090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., and Adler P.N.. 2002. The function of the frizzled pathway in the Drosophila wing is dependent on inturned and fuzzy. Genetics. 160:1535–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebau M.C., Höpker K., Müller R.U., Schmedding I., Zank S., Schairer B., Fabretti F., Höhne M., Bartram M.P., Dafinger C., et al. 2011. Nephrocystin-4 regulates Pyk2-induced tyrosine phosphorylation of nephrocystin-1 to control targeting to monocilia. J. Biol. Chem. 286:14237–14245. 10.1074/jbc.M110.165464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienkamp S., Ganner A., Boehlke C., Schmidt T., Arnold S.J., Schäfer T., Romaker D., Schuler J., Hoff S., Powelske C., et al. 2010. Inversin relays Frizzled-8 signals to promote proximal pronephros development. Proc. Natl. Acad. Sci. USA. 107:20388–20393. 10.1073/pnas.1013070107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Tan G., Levenkova N., Li T., Pugh E.N. Jr., Rux J.J., Speicher D.W., and Pierce E.A.. 2007. The proteome of the mouse photoreceptor sensory cilium complex. Mol. Cell. Proteomics. 6:1299–1317. 10.1074/mcp.M700054-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Yan J., and Adler P.N.. 2010. The Drosophila planar polarity proteins inturned and multiple wing hairs interact physically and function together. Genetics. 185:549–558. 10.1534/genetics.110.114066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Schafer D.A., and Adler P.N.. 2015. The Drosophila planar polarity gene multiple wing hairs directly regulates the actin cytoskeleton. Development. 142:2478–2486. 10.1242/dev.122119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell B., Jacobs R., Li J., Chien S., and Kintner C.. 2007. A positive feedback mechanism governs the polarity and motion of motile cilia. Nature. 447:97–101. 10.1038/nature05771 [DOI] [PubMed] [Google Scholar]
- Nishimura T., Honda H., and Takeichi M.. 2012. Planar cell polarity links axes of spatial dynamics in neural-tube closure. Cell. 149:1084–1097. 10.1016/j.cell.2012.04.021 [DOI] [PubMed] [Google Scholar]
- Okada Y., Nonaka S., Tanaka Y., Saijoh Y., Hamada H., and Hirokawa N.. 1999. Abnormal nodal flow precedes situs inversus in iv and inv mice. Mol. Cell. 4:459–468. 10.1016/S1097-2765(00)80197-5 [DOI] [PubMed] [Google Scholar]
- Otto E.A., Schermer B., Obara T., O’Toole J.F., Hiller K.S., Mueller A.M., Ruf R.G., Hoefele J., Beekmann F., Landau D., et al. 2003. Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nat. Genet. 34:413–420. 10.1038/ng1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., You Y., Huang T., and Brody S.L.. 2007. RhoA-mediated apical actin enrichment is required for ciliogenesis and promoted by Foxj1. J. Cell Sci. 120:1868–1876. 10.1242/jcs.005306 [DOI] [PubMed] [Google Scholar]
- Park T.J., Haigo S.L., and Wallingford J.B.. 2006. Ciliogenesis defects in embryos lacking inturned or fuzzy function are associated with failure of planar cell polarity and Hedgehog signaling. Nat. Genet. 38:303–311. 10.1038/ng1753 [DOI] [PubMed] [Google Scholar]
- Park T.J., Mitchell B.J., Abitua P.B., Kintner C., and Wallingford J.B.. 2008. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat. Genet. 40:871–879. 10.1038/ng.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B., and Kessels M.M.. 2009. New players in actin polymerization--WH2-domain-containing actin nucleators. Trends Cell Biol. 19:276–285. 10.1016/j.tcb.2009.03.004 [DOI] [PubMed] [Google Scholar]
- Reiter J.F., Blacque O.E., and Leroux M.R.. 2012. The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep. 13:608–618. 10.1038/embor.2012.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607. 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang L., Miller J.J., Corbit K.C., Giles R.H., Brauer M.J., Otto E.A., Baye L.M., Wen X., Scales S.J., Kwong M., et al. 2011. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 145:513–528. 10.1016/j.cell.2011.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slanchev K., Pütz M., Schmitt A., Kramer-Zucker A., and Walz G.. 2011. Nephrocystin-4 is required for pronephric duct-dependent cloaca formation in zebrafish. Hum. Mol. Genet. 20:3119–3128. 10.1093/hmg/ddr214 [DOI] [PubMed] [Google Scholar]
- Sung C.H., and Leroux M.R.. 2013. The roles of evolutionarily conserved functional modules in cilia-related trafficking. Nat. Cell Biol. 15:1387–1397. 10.1038/ncb2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford J.B. 2010. Planar cell polarity signaling, cilia and polarized ciliary beating. Curr. Opin. Cell Biol. 22:597–604. 10.1016/j.ceb.2010.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yan J., Lee H., Lu Q., and Adler P.N.. 2014. The proteins encoded by the Drosophila Planar Polarity Effector genes inturned, fuzzy and fritz interact physically and can re-pattern the accumulation of “upstream” Planar Cell Polarity proteins. Dev. Biol. 394:156–169. 10.1016/j.ydbio.2014.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton-Pitt S.R., Silva M., Nguyen K.C., Hall D.H., and Barr M.M.. 2014. The nphp-2 and arl-13 genetic modules interact to regulate ciliogenesis and ciliary microtubule patterning in C. elegans. PLoS Genet. 10:e1004866 10.1371/journal.pgen.1004866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M.E., Hwang P., Huisman F., Taborek P., Yu C.C., and Mitchell B.J.. 2011. Actin and microtubules drive differential aspects of planar cell polarity in multiciliated cells. J. Cell Biol. 195:19–26. 10.1083/jcb.201106110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.L., Masyukova S.V., and Yoder B.K.. 2010. Normal ciliogenesis requires synergy between the cystic kidney disease genes MKS-3 and NPHP-4. J. Am. Soc. Nephrol. 21:782–793. 10.1681/ASN.2009060597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.L., Li C., Kida K., Inglis P.N., Mohan S., Semenec L., Bialas N.J., Stupay R.M., Chen N., Blacque O.E., et al. 2011. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J. Cell Biol. 192:1023–1041. 10.1083/jcb.201012116 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.