Yeast Pex1- and Pex6-deficient cells harbor peroxisomal membrane ghosts that lack matrix proteins but contain all major peroxisomal membrane proteins and acquire matrix proteins to develop into normal peroxisomes upon Pex1 reintroduction.
Abstract
Pex1 and Pex6 are two AAA-ATPases that play a crucial role in peroxisome biogenesis. We have characterized the ultrastructure of the Saccharomyces cerevisiae peroxisome-deficient mutants pex1 and pex6 by various high-resolution electron microscopy techniques. We observed that the cells contained peroxisomal membrane remnants, which in ultrathin cross sections generally appeared as double membrane rings. Electron tomography revealed that these structures consisted of one continuous membrane, representing an empty, flattened vesicle, which folds into a cup shape. Immunocytochemistry revealed that these structures lack peroxisomal matrix proteins but are the sole sites of the major peroxisomal membrane proteins Pex2, Pex10, Pex11, Pex13, and Pex14. Upon reintroduction of Pex1 in Pex1-deficient cells, these peroxisomal membrane remnants (ghosts) rapidly incorporated peroxisomal matrix proteins and developed into peroxisomes. Our data support earlier views that Pex1 and Pex6 play a role in peroxisomal matrix protein import.
Introduction
Peroxisomes are ubiquitous and versatile cell organelles that are involved in a large variety of metabolic pathways. Conserved functions are hydrogen peroxide metabolism and fatty acid β-oxidation (Smith and Aitchison, 2013). Peroxisomes proliferate in response to various internal or external cues, thus ensuring that organelle abundance continuously adapts to cellular needs (Mast et al., 2015).
In higher eukaryotes, peroxisome deficiency is lethal (Fujiki et al., 2012; Hu et al., 2012). However, yeast mutants that show a defect in peroxisome biogenesis are normally viable and capable to grow on media containing glucose, but not on substrates that are metabolized by peroxisomal enzymes (e.g., oleic acid and methanol). This unique property enabled using simple yeast genetic screens to identify genes (PEX genes) that play a role in peroxisome formation (Erdmann and Kunau, 1992).
Upon reintroduction of the deleted PEX genes in yeast peroxisome-deficient (pex) mutants, peroxisomes invariably reappear. So far, different mechanisms of peroxisome reintroduction have been described. Deletion of a PEX gene encoding a protein involved in peroxisomal matrix protein import (e.g., Pex14) results in cells containing peroxisomal membrane remnant structures, designated ghosts, in conjunction with mislocalization of matrix proteins in the cytosol. Peroxisomal membrane proteins (PMPs) are normally present in these ghosts because sorting and insertion of PMPs is independent of matrix protein import. Upon reintroduction of the corresponding PEX gene, these preexisting ghosts develop into normal peroxisomes by importing matrix proteins.
For a long time, it was generally accepted that yeast mutants affected in peroxisomal membrane formation (i.e., pex3 or pex19 mutants) lack peroxisomal membrane remnants (Hettema et al., 2000). However, we recently showed that yeast pex3 and pex19 cells do contain small preperoxisomal vesicles (PPVs), which contain only a subset of PMPs, whereas other PMPs are mislocalized and very instable (Knoops et al., 2014). Upon reintroduction of the corresponding genes, the latter PMPs are also sorted to the PPVs, which results in the formation of a functional peroxisomal importomer and hence matrix protein import, thus leading to the maturation of PPVs into normal peroxisomes.
Recently, an alternative pathway of peroxisome reintroduction has been described for yeast pex1 and pex6 cells. According to this model, two types of ER-derived vesicles fuse upon reintroduction of Pex1 or Pex6, before the formation of normal peroxisomes (van der Zand et al., 2012). These vesicles each carry half a peroxisomal translocon complex, namely either proteins of the receptor docking complex (Pex13 and Pex14) or the RING complex (Pex2, Pex10, and Pex12) together with Pex11. This would imply that in yeast pex1 and pex6 cells, two types of biochemically distinct vesicles accumulate. Upon Pex1 or Pex6 reintroduction, heterotypical fusion of these vesicles would lead to the assembly of the full peroxisomal translocon, thus allowing PMP import.
Here we analyzed the ultrastructure of yeast pex1 and pex6 mutant cells and the mode of peroxisome reintroduction in depth using advanced, high-resolution microscopy techniques, i.e., electron tomography (ET), immunolabeling, and correlative light and electron microscopy (CLEM). The results of these studies are contained in this paper.
Results and discussion
Components of the docking and RING complex colocalize in pex1 and pex6 cells
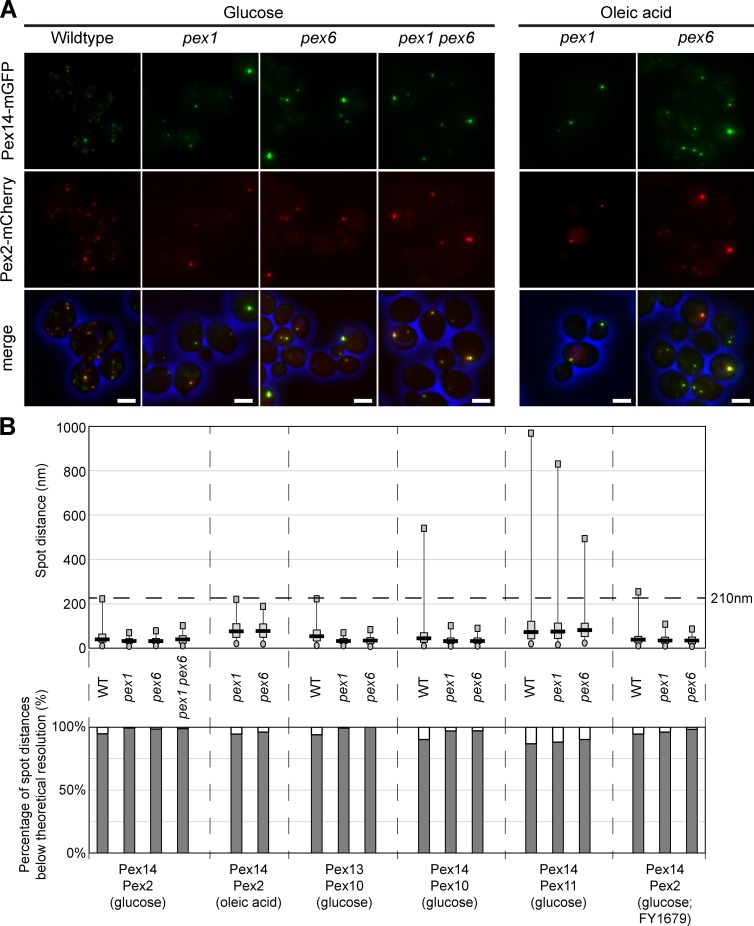
We first analyzed the localization of PMPs of the docking and RING complex by fluorescence microscopy (FM). PMPs were chromosomally tagged to create endogenously expressed C-terminal fusions with the monomeric red fluorescent protein mCherry (Pex2 and Pex10) or monomeric green fluorescent protein mGFP (Pex13 and Pex14). FM revealed that the fluorescent spots of the docking and RING proteins overlapped in Saccharomyces cerevisiae BY4742 pex1 and pex6 cells, similar as observed in wild-type (WT) controls (Fig. 1 A and S1 A). In addition, the spots of Pex11-mCherry, a PMP involved in peroxisome fission, coincided with Pex14-mGFP spots (Fig. S1 A).
Figure 1.
Pex2 and Pex14 colocalize in pex1 and pex6 cells. (A) FM analysis of BY4742 WT, pex1, pex6, or pex1 pex6 cells producing Pex14-mGFP and Pex2-mCherry, grown on glucose (4 h) or oleic acid media (16 h). Cells were fixed with formaldehyde and embedded in agarose before image acquisition at the Deltavision microscope. Bars, 2.5 µm. (B, top) The object based colocalization was determined by calculating the distance between the centers of gravity of the mGFP and mCherry spots of the different indicated PMPs. The dashed horizontal line at 210 nm indicates the lateral resolution limit that theoretically can be achieved with our setup. The circles represent the best fifth percentile and the squares the worst fifth percentile. The gray box is limited by the 25th and 75th percentile, whereas the black line represents the median. For all measurements, n was at least 250 measurements. (bottom) Percentages of spots below the resolution limit.
To seek further support for this PMP colocalization, we performed quantitative FM analysis. All mCherry spots present in 25 randomly acquired FM images were selected, and their distance to the closest mGFP spot was measured (Fig. S1 B). High colocalization values were obtained for all PMP pairs tested in glucose-grown cells of BY4742 pex1, pex6, and WT controls (Fig. 1 B). Similar results (Figs. 1 B and S1 A) were obtained for a pex1 pex6 double-deletion strain, for cells grown on oleic acid, or when WT and mutant strains of the FY1679 parental strain were used (as was used by van der Zand et al., 2012; Fig. S1 A). These data suggest that all PMPs analyzed colocalize in the absence of Pex1 or Pex6 as they do in WT control cells.
Quantification of the number of Pex14-GFP spots revealed a mean number of 1.5 ± 0.04 (n = 441) per cell in glucose-grown pex1 cells, which is similar to what was previously reported (see Fig. 3 A in van der Zand et al., 2012). This number did not increase significantly upon incubation of cells in oleic acid medium (mean number of 1.6 ± 0.20 Pex14-mGFP spots per cell; n = 540).
pex1 and pex6 cells contain peroxisomal membrane ghosts
To identify the nature of the structures to which the fluorescent PMPs were localized, we performed CLEM using cryosections of pex1 cells. CLEM of oleic acid-grown pex1 cells producing Pex10-mCherry revealed that at the site of a single red fluorescent spot, clusters of small membrane structures were present (Fig. 2 A). Immunocytochemistry using specific antibodies against Pex14 revealed that these membranes contained Pex14, suggesting that they represented peroxisomal membranes (Fig. 3 E). Similar Pex14-containing structures were observed in oleic acid–induced pex6 cells (Fig. 3 F). These structures were not detected in WT controls, in which α-Pex14–specific label was confined to peroxisomes (Fig. 3 D). To test whether the Pex14-labeled structures also contained the RING protein Pex2 or the fission protein Pex11, we performed double-labeling experiments using cryosections of pex1 cells producing either HA-tagged Pex2 or Pex11. These experiments revealed that in pex1 cells producing Pex2-HA, both α-HA– and α-Pex14–dependent specific labeling was present at the same membrane structures (Fig. 3 G), indicating that the cells did not contain two different types of vesicles. Similarly, Pex14 colocalized with Pex11-HA (Fig. 3 H) at the same membrane structures in pex1 cells. Specific α-Pex14 and α-HA label was invariably confined to the membrane structures and never observed at any other structure in the cell. Double-labeling experiments revealed that peroxisomal ghosts containing both Pex11 and Pex14 were also present in glucose-grown pex1 cells (Fig. S2 A) but were difficult to detect because of their low abundance. These data suggest that the ghosts proliferate upon incubation of cells in oleic acid media, similar to peroxisomes in WT cells. Proliferation most likely occurs by fission because in cells of a pex6 pex11 double-deletion strain, invariably a single ghost structure was observed (Fig. S2 C).
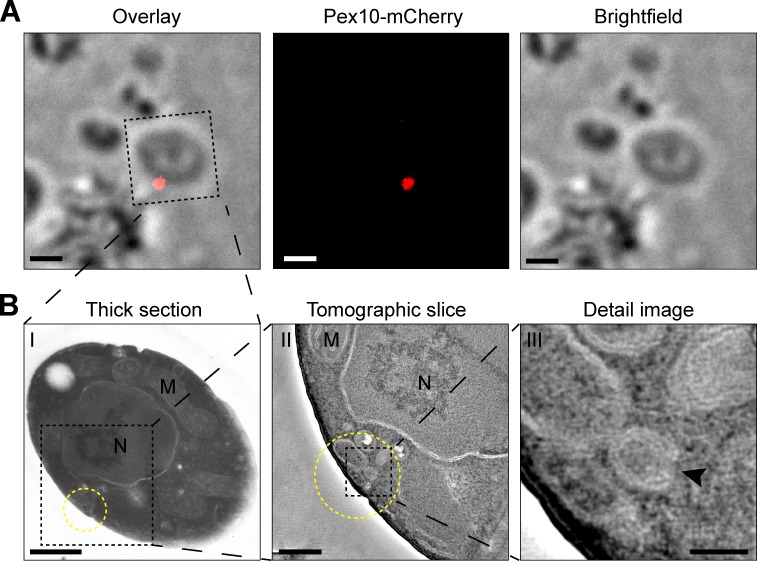
Figure 2.
CLEM revealed the presence of clusters of membrane structures at Pex10-mCherry spots. (A) FM image and brightfield image of a 160-nm-thin cryosection cut from an oleic acid–grown pex1 cell producing Pex10-mCherry (left, overlay). (B) Electron micrographs of the same cell where at the site of the fluorescent spot (yellow circle), a cluster of membrane structures is present. (B, II and III) Electron tomography was used to enhance the contrast, showing the morphology of the membrane structures at 10-nm-thick Z-planes through the reconstructed volume. The black arrowhead indicates an opening in the structure toward the cytosol. Bars: (A) 1 µm; (B, I) 500 nm; (B, II) 200 nm; (B, III) 50 nm.
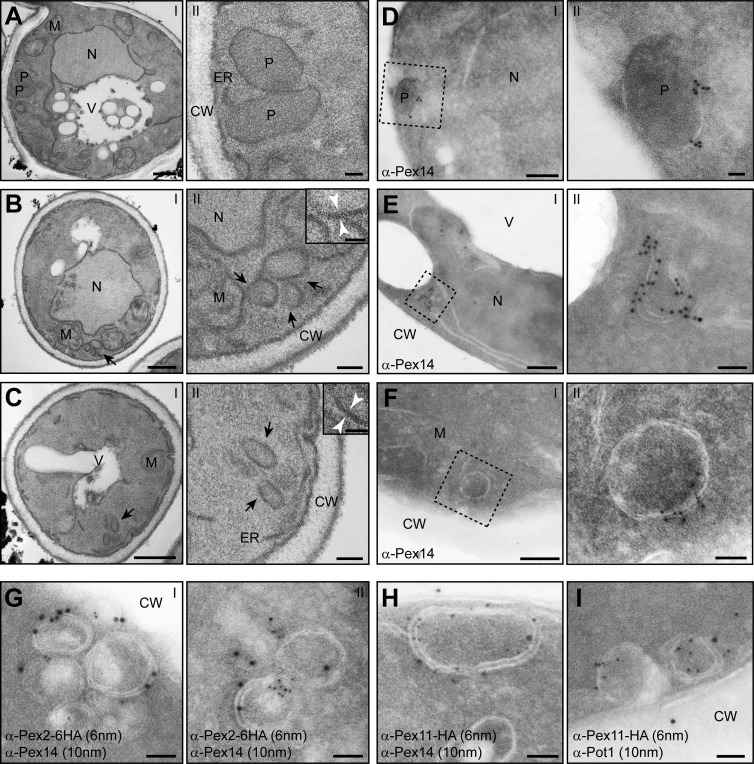
Figure 3.
pex1 and pex6 cells harbor large peroxisomal ghosts that contain Pex2, Pex11, and Pex14 but lack thiolase. (A–C) EM analysis of ultrathin sections of KMnO4-fixed BY4742 WT (A, I and II), pex1 (B, I and II), or pex6 cells (C, I and II). Cells were grown for 16 h in the presence of oleic acid. The insets in panel II of B and C show higher magnifications, which reveal the double membranes in cross sections. (D–F) Immunolabeling of cryosections of aldehyde-fixed, oleic acid–grown cells labeled with α-Pex14 antibodies (WT: D, I and II; pex1: E, I and II; pex6: F, I and II). (G and H) Ultrathin sections of aldehyde-fixed, oleic acid–grown pex1 cells, which produce C-terminal HA-tagged Pex2 (G) or Pex11 (H), were labeled using rabbit α-Pex14 and mouse α-HA antibodies and detected using α-rabbit 10-nm and α-mouse 6-nm gold particles. (I) Double immunolabeling using α-thiolase antibodies (Pot1), showing that the Pex11-labeled structures do not accumulate thiolase. CW, cell wall; M, mitochondrion; N, nucleus; P, peroxisome; V, vacuole. Bars: (A–C, I) 500 nm; (D–F, I) 200 nm; (A–C, II) 100 nm; (B, II, inset; C, II, inset; D, II; E, II; F, II; and G–I) 50 nm.
The peroxisomal matrix protein thiolase (Pot1) did not localize to the membrane structures in oleic acid–induced pex1 cells (Fig. 3 I), which suggests that the remnants are defective in matrix protein import. Therefore, we considered the remnants to represent peroxisomal ghosts that contain all PMPs and can proliferate but are unable to import matrix proteins.
Peroxisomal ghosts in pex1 cells are empty peroxisomal membranes that form a rounded membrane structure
Further morphological analysis revealed that the ghosts were rounded in shape, which in ultrathin cross sections appeared as double-membrane rings (Fig. 2 B and Fig. 3, B and C). These ghosts measured up to 100 nm in diameter. In longitudinal sections, a single hole was observed in the structure (Fig. 2 B, III, black arrowhead). Our data support a model in which pex1 cells contain empty peroxisomes, which flatten and curve into rounded structures. The structures do not represent autophagosomes, as they were also observed in cells of a pex1 atg1 double-deletion strain (Fig. S2 B). The ghosts strongly resemble those previously described in S. cerevisiae (Hettema et al., 2000) and Hansenula polymorpha pex1 and pex6 cells (Koek et al., 2007), as well as in Pex6-deficient CHO cells (Hashiguchi et al., 2002).
Pex1 reintroduction results in the import of matrix proteins into preexisting peroxisomal ghosts
To study peroxisome reintroduction in pex1 cells, we created a conditional allele of Pex1 by tagging it with the yeast-optimized auxin-inducible degron (AID*) 6HA tag (AID*-6HA; Morawska and Ulrich, 2013). Control experiments confirmed that upon growth of Pex1-AID* cells in media lacking auxin, Pex1-AID*-6HA protein was present (Fig. 4 A) in conjunction with normal import of DsRed-SKL into peroxisomes (Fig. 4 B). However, when cells were grown in the presence of auxin, Pex1-AID*-6HA protein was not detectable and DsRed-SKL mislocalized to the cytosol (Fig. 4, A and C).
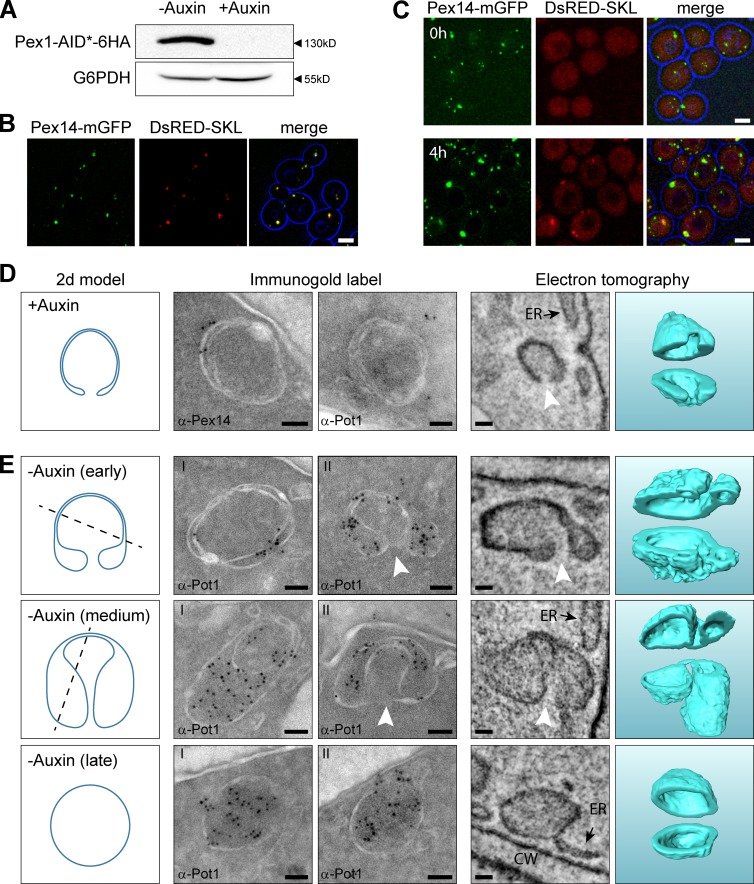
Figure 4.
Peroxisomal ghosts import matrix proteins upon reintroduction of Pex1 in Pex1-deficient cells and develop into peroxisomes. (A) Western blot analysis of total cell extracts of Pex1-AID* cells using α-HA antibodies, to demonstrate the presence of the Pex1-AID*-6HA fusion protein. Cells were precultivated for 16 h on glucose medium followed by 4 h on oleic acid medium in the presence or absence of 1 mM auxin. Equal amounts of protein were loaded per lane. Glucose 6-phosphate dehydrogenase (G6PDH) was used as loading control. (B and C) Confocal laser scanning microscopy (CLSM) analysis of Pex1-AID* cells grown for 16h on glucose medium followed by incubation for 4h on oleic acid medium in the absence (B) or presence (C, top; T = 0 h) of auxin. (C–E) Pex1-AID* cells, precultivated for 16 h on glucose and subsequently for 4 h on oleic acid medium in the presence of auxin, were shifted to oleic acid medium without auxin (T = 0 h). CLSM analysis revealed that after 4 h of incubation in the absence of auxin (C, bottom), DsRed-SKL spots became detectable that colocalized with Pex14-mGFP. (D) Immunogold labeling and ET showing that at T = 0 h rounded structures are present in the cells. In cross sections, the structures are visualized as double rings. The structures lack thiolase (α-Pot1) but contain Pex14. ET revealed that these structures are rounded and contained a single opening to the cytosol (white arrowheads). (E) After 2 h of growth on oleic acid in the absence of auxin, thiolase accumulated at the structures as evident from immunolabeling using α-Pot1 antibodies. The left panels in D and E show 2D models of common sections that were observed in both iEM and ET. The dashed lines illustrate cutting planes that explain the left iEM image, whereas the 2D model represents the right iEM image as well as the left ET panel, which shows a 10-nm-thick section through the ET volume. The ghost and the different stages of peroxisome formation are 3D modeled in the right ET panel and cut open for visualization purposes. Bars: (B and C) 2.5 µm; (D and E) 50 nm. CW, cell wall.
For reintroduction experiments, Pex1-AID* cells were first precultivated in glucose media in the presence of auxin and subsequently incubated for another 4 h in oleic acid media supplemented with auxin to induce peroxisomal proteins. Next, the cells were washed twice and further cultivated in oleic acid media lacking auxin. FM revealed that after 4 h of further cultivation in the absence of auxin DsRed-SKL, spots had reappeared, indicative of peroxisome formation (Fig. 4 C).
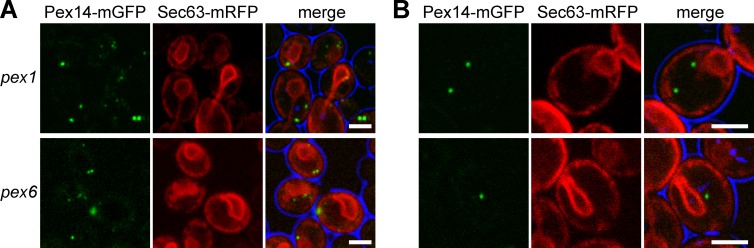
We then analyzed the cells at different stages of Pex1 reintroduction by EM. Before the removal of auxin, the cells contained the typical membrane ghosts to which Pex14 was localized (Fig. 4 D), similar to those observed in pex1 cells (compare with Fig. 3 B). Also, thiolase did not accumulate in these structures (Fig. 4 D). ET indicated that the ghosts in Pex1-depleted cells represented rounded structures with tightly opposed membranes. As in pex1 cells, these ghosts contained a single opening connecting the lumen of the structure with the cytosol (Fig. 4 D). Generally, these structures were localized in close vicinity of the plasma membrane and cortical ER (Fig. 5 A). ET analysis indicated that the membranes of the ghosts were not continuous with the ER (Fig. 4, D and E), which was underscored by FM analysis, which showed that Pex14-mGFP spots exist that do not colocalize with the ER marker Sec63-mRFP (Fig. 5 B).
Figure 5.
Peroxisomal ghosts are not continuous with the ER in pex1 and pex6 cells. (A and B) CLSM analysis of glucose-grown cells of pex1 and pex6 strains producing Pex14-mGFP and Sec63-mRFP. (A) Most Pex14-mGFP spots are present at the periphery of the cell, where the cortical ER is localized. (B) However, Pex14-mGFP spots that do not colocalize with the ER marker Sec63-mRFP signal are present as well. Bars, 2.5 µm.
Already 2 h after shifting the Pex1-AID* cells to oleic acid media lacking auxin, most ghosts had undergone a morphological change and different intermediate stages toward normal peroxisomes could be distinguished (Fig. 4 E). First, the space in-between the membranes expanded, particularly near the opening, because of the import of matrix protein, as was evident from immuno-labeling experiments using antibodies against thiolase (α-Pot1; Fig. 4 E, early). Next, structures were observed containing globular extensions that are likely the result of further import of matrix proteins (Fig. 4 E, medium). Still, these large extensions were connected to the initial membranes by a sheet of tightly opposed membranes and reached similar diameters as the peroxisomes that were ultimately generated (Fig. 4 E, medium). This indicates that the peroxisomes (Fig. 4 E, late) most likely are formed by fission of the ghost structure.
In summary, our results demonstrate that S. cerevisiae pex1 and pex6 cells contain peroxisomal membrane ghosts that harbor the docking proteins Pex13 and Pex14 as well as the RING proteins Pex2 and Pex10 together with Pex11. We were not able to detect the two types of biochemically distinct vesicles harboring either the docking or the RING proteins and Pex11 (van der Zand et al., 2012) using our fluorescence and immunocytochemical methods.
The structures most likely proliferate from existing ghosts by fission. These ghosts are defective in matrix protein import because of depletion of Pex1, but not in PMP insertion. Also, organelle fission is most likely not affected based on the finding that their proliferation depends on PEX11.
Upon reintroduction of Pex1, the ghosts imported matrix proteins and developed directly into peroxisomes essentially as described before for Pex6-deficient CHO cells upon genetic complementation (Hashiguchi et al., 2002; for a hypothetical model, see Fig. S3). Our data therefore support a model in which Pex1 and Pex6 play a role in matrix protein import (Platta et al., 2005) but cannot exclude an additional role for these proteins in vesicular fusion during peroxisome biogenesis (Titorenko and Rachubinski, 2000; van der Zand et al., 2012).
Materials and methods
Organisms and growth
The yeast strains used in this study are listed in Table S1. Cells were grown in selective medium (0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate and 1% casamino acids) containing 2% glucose (YM2) or 0.1% oleic acid and 0.2% Tween-80 (Sigma-Aldrich; YMO). Amino acids and uracil were added when needed.
DNA manipulations, cloning procedures, and strain constructions
S. cerevisiae BY4742 WT, pex1, and pex6 strains and the FY1679 WT strain were obtained from the Euroscarf collection (Table S1). All plasmids used in this study are described in Table S2. Gene fusions were made by PCR-based methods using the primers listed in Table S3. Correct introduction of the fusion genes was checked by colony PCR, Western blotting, and/or Southern blotting. All gene deletions were confirmed by PCR or Southern blotting.
For construction of the FY1679 pex1 and pex6 strains, the PEX1 and PEX6 deletion cassettes were PCR amplified from BY4742 pex1 and pex6 strains using the primer pairs pKEK123/pKEK124 or pKEK127/pKEK128, respectively. The resulting PCR fragments of 2.6 and 2.3 kbp were transformed into FY1679 WT cells that were plated on YPD with geneticin selective antibiotics (GE Healthcare). For construction of the pex1 atg1 strain, the ATG1 deletion cassette was PCR amplified from the pAG25 plasmid using the primers pTER208/pTER209, resulting in a PCR fragment of 2.7 kbp, which was transformed into the BY4742 pex1 strain, which was plated on YPD containing nourseothricin selective antibiotics (GE Healthcare). For construction of the pex1.pex6 double-deletion strain, the PEX6 deletion cassette was PCR amplified from pHyg-AID*-6HA using the primer pair pKEK229/pKEK230, resulting in a PCR fragment of 1.7 kbp, which was transformed into a pex1 strain already containing Pex2-mCherry and Pex14-mGFP. In the same way, PEX6 was deleted in the Euroscarf pex11 strain to construct the pex11.pex6 double-deletion strain.
For construction of Pex14-mGFP strains, the PEX14-mGFP gene with downstream HIS marker was PCR amplified from strain WT PEX14-mGFP (Invitrogen) using the primers pKEK036/pKEK041. The resulting PCR fragment of 2.3 kbp was integrated in yeast cells using standard transfection protocols. For construction of Pex13-mGFP strains, the PEX13-mGFP gene with downstream HIS marker was PCR amplified from strain WT PEX13-mGFP (Invitrogen) using the primers pKEK164/pKEK165, resulting in a PCR fragment of 2.4 kbp.
For the construction of plasmid pRSA01, a PCR fragment of 700 bp was obtained by primers RSA10fw and RSA11rev on pCDNA3.1mCherry. The resulting BglII–SalI fragment was inserted between the BglII and SalI of pANL31. For construction of Pex2-mCherry strains, the mCherry gene and Zeocin selection marker of the pRSA01 plasmid were amplified with primers pKEK182/pKEK183, thus yielding a PCR fragment of 2.3 kbp with 5′ overlap on the PEX2 gene and 3′ overlap on the PEX2 terminator. For construction of Pex10-mCherry strains, the mCherry gene and Zeocin selection marker of the pRSA01 plasmid were amplified with primers pKEK038/pKEK039, thus yielding a PCR fragment of 2.3 kbp with 5′ overlap on the PEX10 gene and 3′ overlap on the PEX10 terminator. For construction of Pex11-mCherry strains, the mCherry gene and Zeocin selection marker of the pRSA01 plasmid were amplified with primers pKEK184/pKEK185, thus yielding a PCR fragment of 2.3 kbp with 5′ overlap on the PEX11 gene and 3′ overlap on the PEX11 terminator.
For C-terminal tagging of PEX11 with the HA tag, first the Zeocin marker was PCR amplified with 5′ overhang of the HA sequence (GYPYDVPDYASG) using the primers pKEK131/pKEK136, yielding a PCR fragment of 1.5 kbp. This PCR product was then amplified using 5′ overhang of the Pex11 C terminus and PEX11 terminator using the primers pKEK137/pKEK138, thus yielding a fragment of 1.6 kbp, which was transformed in pex1 cells. For C-terminal tagging of PEX2 with the HA tag, the pHyg-Pex2-6HA plasmid was constructed by SmaI and EheI digestion of the pHyg-AID*-6HA plasmid resulting in two fragments of 4,355 and 168 bp. The C-terminal part of Pex2 was amplified using primers pKEK202/pKEK203, yielding a product of 720 bp, which was assembled into the pHyg backbone using the Gibson Assembly Master Mix (New England Biolabs) and transformed into DH5α, which was then plated on LB containing ampicillin as selection marker. The final pHyg-Pex2-6HA plasmid was linearized with ApaI and transformed into yeast.
To obtain constitutive expression of a gene encoding DsRed containing a peroxisomal targeting signal 1, first the TDH3 promotor was amplified with primers TDH3_NotI.F/TDH3_BamHI.R, thus yielding a PCR fragment of 716 bp with 5′ NotI and 3′ BamHI digestion sites. The vector pHIPX7 GFP-SKL was digested with NotI and BamHI to excise the TEF promotor and filled with the digested TDH3 promotor, thus resulting in the pPtdh3 GFP-SKL plasmid. The plasmid pHIPZ4 DsRed–SKL was digested with BamHI and SalI and the obtained DsRed–SKL fragment was subsequently ligated into the pPtdh3 GFP-SKL which was digested with BamHI and SalI, finally resulting in the pPtdh3 DsRed-SKL plasmid. The plasmid was linearized with DraI and integrated into a TIR1 strain already containing Pex14-mGFP. The plasmid for integration of a C-terminal AID*-tag for Pex1 was created by Pex1 amplification with primers pKEK178/pKEK179, resulting in a product of 1697 bp, which was digested by HindIII and SalI and ligated in pHyg-AID*-6HA plasmid that was digested with the same enzymes. The resulting plasmid, pHyg-Pex1-AID*-6HA, was linearized with BstBI for integration in the TIR1 strain containing Pex14-mGFP, as well as DsRed-SKL. The TIR1 strain and AID* plasmid were provided by H. Ulrich, Institute of Molecular Biology, Mainz, Germany.
In general, S. cerevisiae cells were transformed using PCR-amplified DNA. 2 µg PCR-purified DNA and 0.1 µg carrier DNA were added to 60 µl cells. Carrier DNA was denatured at 100°C for 10 min. A solution of PEG/LiAc/DTT was added with a volume of 300 µl every 100 µl of cell-DNA mixture. Heat shock was performed for 15 min at 42°C, followed by 2 min on ice. Then, the cells were centrifuged for 1 min at 5,000 rpm, the supernatant discarded, the pellet suspended in 5 ml YPD, and incubated for 3 h at 30°C with 200 rpm shaking. Again, the cells were centrifuged for 1 min at 5,000 rpm and the pellet suspended in 100 µl YPD or YND and plated on selective YPD/YND plates.
FM
Cells were fixed in 1% formaldehyde in PHEM buffer, consisting of 60 mM Pipes, 25 mM Hepes, 8 mM MgCl2, and 40 mM EGTA, pH 6.9, for 1 h and embedded in 1% low-melting-point agarose in PHEM buffer to prevent cell movement during imaging.
To assess PMP colocalization, single plain images were acquired through the middle of the cells for bright-field, mGFP and mCherry on a Personal Deltavision (GE Healthcare) using a Photometrics Coolsnap HQ2 digital camera and SoftWorks 5.5.1 software. All images were made at room temperature using 100× 1.30 NA Plan-Neofluar objectives (Carl Zeiss), resulting in a pixel size of 64 nm. The mGFP signal was visualized with a 470⁄40 nm band pass excitation filter, a 495-nm dichromatic mirror, and a 525/50-nm band-pass emission filter. mCherry fluorescence was visualized with a 572/35-nm band-pass excitation filter and a 632/60-nm band-pass emission filter. Because the mCherry spots displayed lower fluorescence intensities relative to the mGFP spots, all mCherry spots present in 25 randomly acquired fluorescence microscopy images were selected and their distance to the closest mGFP spot was measured (Fig. S1 B). To extract the fluorescent spots from the original 16-bit TIF images, the coordinates of mCherry spots were determined semiautomatically using IMOD (Kremer et al., 1996), after which they were automatically boxed out for both channels resulting in images with a frame size of 32 × 32 pixels. The signal was enhanced and normalized with a fast-Fourier transform band-pass filter (between 16 and 4 pixels) in ImageJ (National Institutes of Health). Per channel, all processed images were corrected equally for background signal and converted to 8-bit TIF images. For the object-based colocalization analysis, spots were masked and labeled using the Dipimage toolbox in MATLAB (MathWorks). The center of gravity was determined in both mGFP and mCherry channels and used to calculate the distance between the mostly centered spots.
Sec63-mRFP and DsRed localization images were captured with a confocal microscope (LSM510; Carl Zeiss), equipped with photomultiplier tubes (Hamamatsu Photonics) and Zen 2009 software. All images were made at room temperature using 100× 1.30 NA Plan-Neofluar objectives (Carl Zeiss). For Sec63-mRFP, the cells were fixed in 1% formaldehyde in PHEM buffer and embedded in 1% LMP agarose in PHEM buffer. mGFP fluorescence was analyzed by excitation of the cell with a 488-nm argon ion laser (Lasos), and emission was detected using a 500–550-nm band-pass emission filter. The mRFP and DsRed signals were visualized by excitation with a 543-nm helium neon laser (Lasos), and emission was detected using a 565- to 615-nm band-pass emission filter. To reduce possible bleed through of mGFP into the mRFP/DsRed channel, the fluorescence images were acquired sequentially. The resulting 3D confocal stacks were median filtered using a 3D 2 × 2 × 2 kernel and merged in Z-direction by averaging.
EM
For morphological analysis, S. cerevisiae cells were fixed in 1.5% potassium permanganate, poststained with 0.5% uranyl acetate, and embedded in epon 812 (Serva, 21045). Ultrathin sections were viewed in a Philips CM12 TEM. For ET, 10-nm gold beads were layered on top of 400-nm-thick sections and acted as fiducial markers for ET. Two single-axis tilt series, each containing 141 images with 1° tilt increments, were acquired with a pixel size of 1.16 nm on a FEI Tecnai20 at 200 kV using the FEI automated tomography software and a cooled slow-scan charge-coupled device camera (4k Eagle; FEI Company) in 2 × 2 binned mode. The tilt series were aligned and reconstructed using the IMOD software package (Kremer et al., 1996) and analyzed using the AMIRA visualization package (TGS Europe). To generate 3D surface-rendered models in AMIRA, masks of organelles were first drawn manually and afterward improved by thresholding.
For immunogold labeling, cells were washed twice in PHEM buffer, then fixed overnight in a mixture of 0.2% glutaraldehyde and 2% formaldehyde in PHEM buffer at 4°C and subsequently incubated for 15 min in a solution of 0.4% sodium periodate and for 30 min in 1% ammonium chloride. Upon embedding in 12% gelatin in PHEM buffer, ∼0.5-mm3 cubes were infiltrated overnight in 2.3 M sucrose in the same buffer. Cryosections of 70 nm were cut using a cryo diamond knife (Diatome) at −120°C using a Reichert Ultracut and mounted on carbon-coated formvar nickel grids. Immunolabeling of Pex14 and thiolase were performed using rabbit polyclonal antibodies followed by goat anti–rabbit antibodies conjugated to 6 or 10 nm gold (Aurion). Pex11-HA and Pex2-6HA were localized using mouse monoclonal antibodies raised against HA (H9658; Sigma-Aldrich,) and goat-anti-mouse antibodies conjugated to 6 nm gold (Aurion). Labeled sections were first stained for 2 min with 2% uranyl oxalate, pH 7, at room temperature and after a quick rinse on three drops of double distilled water stained and embedded in a mixture of 0.5% uranyl acetate and 0.5% methylcellulose (25 centipoise; Sigma-Aldrich) for 10 min on ice. Excess staining solution was drained, and the grids were left to dry before viewing them in a CM12 TEM (Philips).
For CLEM, the sample was prepared similarly as for immunogold labeling; however, 180-nm thin sections were cut and mounted on carbon-coated formvar copper grids that were over layered with 10-nm gold particles (Sigma 752584) for alignment of the tomograms. The grids were placed with section side facing the objective in a droplet of water on a coverslip. Fluorescence imaging was performed at room temperature using AxioVision 4.8.2 software on an Observer Z1 (Carl Zeiss) equipped with a 100× 1.30 NA Plan-Neofluar objective (Carl Zeiss) and an AxioCAM MRm camera (Carl Zeiss). mCherry fluorescence was visualized with a 587/25-nm band pass excitation filter and a 647/70-nm band-pass emission filter. After fluorescence imaging, the grid was poststained and embedded in a mixture of 0.5% uranyl acetate and 0.5% methylcellulose. The area of interest was found back in the electron microscope using the bright-field images as maps. Low-magnification EM was used to align the EM images on the bright-field FM images. Acquisition of the double-tilt tomography series was performed manually in a CM12 TEM running at 80 kV and included a tilt range of 40° to −40° with 5° increments. Reconstruction of the tomograms was performed using the IMOD software package (Kremer et al., 1996).
Pex1 reintroduction
Cells producing Pex1 containing a C-terminal AID*-6HA-tag were precultivated for 16 h on YM2 medium containing 1 mM indole-3-acetic sodium salt (Sigma-Aldrich) and subsequently inoculated to OD600 = 0.1 in YMO medium containing 1 mM indole-3-acetic sodium salt. After 4 h of growth, cells were washed twice in sterile demi-water and resuspended in the same volume of YMO medium without indole-3-acetic sodium salt.
Online supplemental material
Fig. S1 shows FM analysis of colocalization of different PMPs in WT and mutant strains. Fig. S2 shows electron micrographs illustrating the presence of peroxisomal ghosts in glucose-grown pex1 cells as well as in oleic acid grown pex1 atg1 and pex6 pex11 double-deletion strains. Fig. S3 shows a hypothetical model of peroxisome reintroduction in yeast pex1 and pex6 cells. Tables S1, S2, and S3 contain the S. cerevisiae strains, plasmids, and primers used in this study, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201506059/DC1.
Supplementary Material
Acknowledgments
We thank Mohammed Zahir and Arjen Krikken for their valuable contributions and discussions and Leonard Bosgraaf for designing and preparing Fig. S3. We thank Egbert Boekema and Jan-Willem Veening for making their microscopy facilities available.
This work was supported by an EU Marie Curie IEF grant (FP7-330150) and a NWO Veni (863.14.003) to K. Knoops. I.J. van der Klei is supported by the Marie Curie ITN grant PERFUME (Grant Agreement Number 316723).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AID
- auxin-inducible degron
- CLEM
- correlative light and electron microscopy
- CLSM
- confocal laser scanning microscopy
- ET
- electron tomography
- FM
- fluorescence microscopy
- iEM
- immuno-EM
- mGFP
- monomeric GFP
- PMP
- peroxisomal membrane protein
- PPV
- preperoxisomal vesicle
- WT
- wild-type
References
- Erdmann R., and Kunau W.H.. 1992. A genetic approach to the biogenesis of peroxisomes in the yeast Saccharomyces cerevisiae. Cell Biochem. Funct. 10:167–174. 10.1002/cbf.290100306 [DOI] [PubMed] [Google Scholar]
- Fujiki Y., Yagita Y., and Matsuzaki T.. 2012. Peroxisome biogenesis disorders: molecular basis for impaired peroxisomal membrane assembly: in metabolic functions and biogenesis of peroxisomes in health and disease. Biochim. Biophys. Acta. 1822:1337–1342. 10.1016/j.bbadis.2012.06.004 [DOI] [PubMed] [Google Scholar]
- Hashiguchi N., Kojidani T., Imanaka T., Haraguchi T., Hiraoka Y., Baumgart E., Yokota S., Tsukamoto T., and Osumi T.. 2002. Peroxisomes are formed from complex membrane structures in PEX6-deficient CHO cells upon genetic complementation. Mol. Biol. Cell. 13:711–722. 10.1091/mbc.01-10-0479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema E.H., Girzalsky W., van Den Berg M., Erdmann R., and Distel B.. 2000. Saccharomyces cerevisiae pex3p and pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 19:223–233. 10.1093/emboj/19.2.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Baker A., Bartel B., Linka N., Mullen R.T., Reumann S., and Zolman B.K.. 2012. Plant peroxisomes: biogenesis and function. Plant Cell. 24:2279–2303. 10.1105/tpc.112.096586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Manivannan S., Cepinska M.N., Krikken A.M., Kram A.M., Veenhuis M., and van der Klei I.J.. 2014. Preperoxisomal vesicles can form in the absence of Pex3. J. Cell Biol. 204:659–668. 10.1083/jcb.201310148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koek A., Komori M., Veenhuis M., and van der Klei I.J.. 2007. A comparative study of peroxisomal structures in Hansenula polymorpha pex mutants. FEMS Yeast Res. 7:1126–1133. 10.1111/j.1567-1364.2007.00261.x [DOI] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., and McIntosh J.R.. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Mast F.D., Rachubinski R.A., and Aitchison J.D.. 2015. Signaling dynamics and peroxisomes. Curr. Opin. Cell Biol. 35:131–136. 10.1016/j.ceb.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska M., and Ulrich H.D.. 2013. An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast. 30:341–351. 10.1002/yea.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta H.W., Grunau S., Rosenkranz K., Girzalsky W., and Erdmann R.. 2005. Functional role of the AAA peroxins in dislocation of the cycling PTS1 receptor back to the cytosol. Nat. Cell Biol. 7:817–822. 10.1038/ncb1281 [DOI] [PubMed] [Google Scholar]
- Smith J.J., and Aitchison J.D.. 2013. Peroxisomes take shape. Nat. Rev. Mol. Cell Biol. 14:803–817. 10.1038/nrm3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko V.I., and Rachubinski R.A.. 2000. Peroxisomal membrane fusion requires two AAA family ATPases, Pex1p and Pex6p. J. Cell Biol. 150:881–886. 10.1083/jcb.150.4.881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zand A., Gent J., Braakman I., and Tabak H.F.. 2012. Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell. 149:397–409. 10.1016/j.cell.2012.01.054 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.