Abstract
Chemoreception is essential for survival. Feeding, mating, and avoidance of predators depend on detection of sensory cues. Drosophila contains diverse families of chemoreceptors that detect odors, tastants, pheromones, and noxious stimuli, including receptors of the Or, Gr, IR, Ppk, and Trp families. We consider recent progress in understanding chemoreception in the fly, including the identification of new receptors, the discovery of novel biological functions for receptors, and the localization of receptors in unexpected places. We discuss major unsolved problems and suggest areas that may be particularly ripe for future discoveries, including the roles of these receptors in driving the circuits and behaviors that are essential to the survival and reproduction of the animal.
Keywords: Odor receptor (Or), Gustatory receptor (Gr), Ionotropic glutamate receptor (IR), Olfactory Receptor Neuron (ORN), Olfaction, Taste
The problem
Animals in their natural environments are immersed in a sea of chemical compounds. Some of these compounds signal the presence of nutrients, while others signify the danger of poisons. Some compounds indicate the proximity of a mating partner, while others warn of a predator. Animals must be able to detect and identify a wide variety of meaningful signals among the vast complexity of their chemical milieu.
In addition to chemical identity, chemical intensity can also be critical to an animal. The quantity of a sugar in a food source reflects its nutritive value, just as the quantity of a bitter compound such as strychnine may reflect its toxicity. Moreover, some stimuli are attractive at low concentrations and aversive at high concentrations. The temporal pattern of the stimulus is also important. For example, it may inform an animal of the proximity of an odor source.
This, then, is the problem: how to detect and interpret a wide variety of chemical signals amidst a cacophony of chemical noise. The signals are enormously diverse in chemical and temporal structure, and the ability to identify and quantitate them may be a matter of life and death.
This review discusses recent progress in understanding chemoreception, which is the foundation of all the perceptual processes and behavioral responses that follow. We focus on the chemoreceptors of Drosophila, which provides a powerful genetic model for the study of chemosensory reception. Although there has been great progress in the field, it is clear that critical problems remain to be solved and that major discoveries are in store.
The cellular context of chemoreception
Volatile compounds are sensed by olfactory receptor neurons (ORNs) of the olfactory system, whereas non-volatile compounds are detected by gustatory receptor neurons (GRNs) of the taste system. That said, the conceptual wall dividing olfaction and taste has been increasingly assaulted by a barrage of experimental results that establish new links between the two sensory modalities.
The adult olfactory system
The fly contains two olfactory organs, the antenna and the maxillary palp (Fig. 1). Both are covered with sensilla, sensory hairs that contain the dendrites of up to four ORNs (Fig. 2A) [1, 2]. The shafts of sensilla are perforated by numerous pores, or channels, through which odorants can pass. The ORN cell bodies lie below the sensillar shafts, adjacent to accessory cells. These cells secrete odorant binding proteins (OBPs) into the lymph that bathes the ORN dendrites [3]. OBPs are widely believed to carry odorants to odor receptors in the dendritic membranes, although other functions have been proposed (Box 1). ORNs project axons to the antennal lobe of the brain, where signals are processed and transmitted to higher-order centers [4].
Fig. 1.
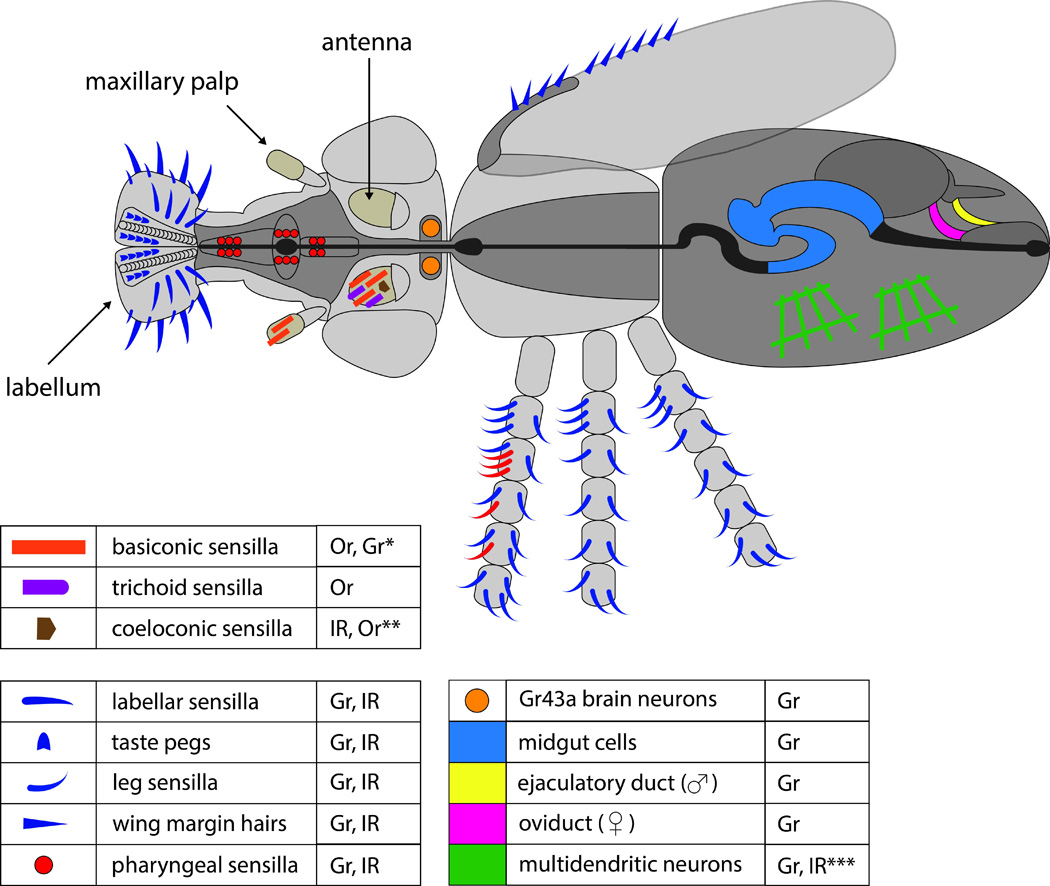
Expression of the three largest chemoreceptor families in Drosophila. Light gray coloring indicates the exterior of the fly; dark gray coloring indicates the interior. Tan highlights the antenna and maxillary palp, which primarily house olfactory neurons. Leg sensilla indicated in red are male specific. Gr expression in the gut occurs in enteroendocrine cells as opposed to neurons. Multidendritic neurons are subcuticular. Expression of Grs and IRs is based in most cases on expression of Gr-GAL4 and IR-GAL4 drivers. Expression of Ors is based on Or-GAL4 drivers and in situ hybridizations. Classes of receptors that have been identified in each corresponding sensillum type or tissue are indicated at right of labels. *Antennal Grs include Gr21a and Gr63a in basiconic sensilla neurons and others [62, 85]. **A single Or has been localized to one neuron of one coeloconic sensillum. ***A single IR has been localized to multidendritic neurons in the abdomen
Fig. 2.
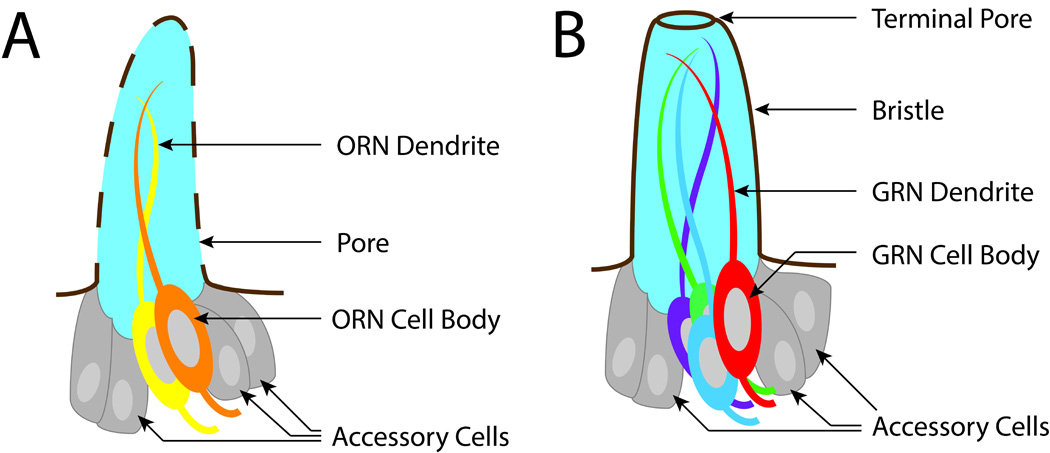
Chemosensory sensilla. (A) olfactory and (B) gustatory sensilla in adult flies. The dendrites are bathed in sensillar lymph (light blue). The shaft of the olfactory sensillum is perforated by small pores, while the shaft of the gustatory sensillum contains a single pore, located at the tip. Both olfactory and gustatory sensilla contain non-neuronal accessory cells, which secrete OBPs into the lymph.
Box 1. A large and diverse family of soluble chemosensory proteins.
Large families of soluble proteins called Odorant binding proteins (Obps) have been identified in many insects, originally in a moth [105, 106]. Obps are small proteins (~14kDa) that are highly divergent in sequence but that share six cysteines. They bind odorants, with different degrees of binding selectivity reported for different Obps [107, 108]. Obps are reminiscent of Ors in their number (52 genes in D. melanogaster), sequence divergence, and diversity of expression patterns. Intriguingly, some are expressed at exceedingly high levels [85, 109].
Obps have been proposed to bind and transport hydrophobic odorants across the hydrophilic sensillum lymph to odor receptors in the dendrites of ORNs [106]. However, Obps have also been proposed to act in the termination of odor response, perhaps by removing odorants from receptors or from the sensillar lymph [105, 106, 110]. Odor receptors expressed in heterologous systems such as Sf9 cells respond to odors in the absence of Obps. The response profiles of Ors appear not to depend on Obps, in some if not all cases [111]. Obps increase the sensitivity of responses in vitro [112, 113], but in one system the sensitivity could be increased equally well by the addition of a non-specific protein [114]. These results leave open the question of why olfactory organs express large families of diverse Obps, in diverse patterns.
A variety of studies support a role for Obps in olfactory perception in vivo. RNAi knockdown of Obps led to abnormalities in olfactory behavior in Drosophila [109] and decreased physiological responses in mosquito antennae [115, 116]. Mutation of an Obp reduced the response of an ORN to the pheromone cVA in Drosophila, although the mechanism has been controversial [117, 118]. Some Obps are also expressed in the taste system, and mutations of some of them altered responses to tastants; interestingly, one of the responses that was altered was inhibitory [68], rather than excitatory [117, 119].
In summary, enormous biological resources are devoted to the synthesis and regulation of these abundant and diverse proteins, but much remains to be learned about their mechanism of action and role in chemosensory coding.
Most olfactory sensilla fall into three morphological classes [2, 5, 6]. Basiconic sensilla are located on the antenna and maxillary palp, and detect many food odors, including esters, alcohols, and aldehydes. Trichoid sensilla are found on the antenna and detect fly odors, including pheromones. Coeloconic sensilla are found on the antenna and respond to many acids and amines.
The adult gustatory system
Drosophila contains GRNs in a variety of locations [1, 7] (Fig. 1). The labellum, on the proboscis, contains gustatory sensilla that have a single, large pore at the tip (Figure 2B). Many labellar sensilla contain four GRNs, and in some cases one neuron is sensitive to sugars, one to bitter compounds and high concentrations of salt, one to low concentrations of salt, and one to water, i.e. osmolarity. Shorter structures called taste pegs are also located on the labellum; they house a single sugar-sensitive GRN.
The legs also contain taste sensilla [7–9], allowing the fly to sample potential food sources before making contact with the mouthparts. Some leg sensilla contain a sugar-sensitive neuron but not a bitter-sensitive neuron, while others contain a bitter-sensitive neuron but not a sugar-sensitive neuron. Other sensilla have both; still others have neither, leaving a large number of “orphan neurons” whose sensitivities and ligands should be a fertile topic of future exploration.
The anterior margin of the wing contains chemosensory sensilla, which were found to respond to tastants in pioneering studies of Marion-Poll and others [5, 10]. Sensilla on the ovipositor of larger flies have been shown electrophysiologically to have gustatory function [11, 12]. In Drosophila, sensilla on the distal tip of the female abdomen exhibit morphology and innervation patterns suggestive of taste function [5, 13], but functional data to support this suggestion are sparse.
Flies also contain internal taste cells [14]. For example, the labral, ventral, and cibarial sense organs (LSO, VCSO, DCSO) of the adult pharynx contain sensilla that house up to eight neurons [5]. These neurons project dendrites into pits that open into the esophageal lumen. Post-ingestive nutrient monitoring also occurs in the brain [15, 16], and presumably in enteroendocrine cells of the midgut [17].
The larval chemosensory system
Larvae hatch on food sources and then burrow into them, emerging only in the final phase of larval life to pupate. During most of larval life, therefore, chemical stimuli seem likely to reach the larva primarily via fluids rather than air. In experimental paradigms, however, larvae of all instars respond to airborne odorants [18, 19]. The primary olfactory organ, the dorsal organ (DO), contains both ORNs (n~21) and GRNs (n~9)(Fig. 3). Two other external taste organs, the terminal organ (TO) and the ventral organ (VO) also contain GRNs [20, 21]. Internal chemosensory organs line the larval pharynx and contain small numbers of GRNs as well [20, 21]. Interestingly, some of the pharyngeal GRNs are among the few larval sensory neurons to survive remodeling during metamorphosis, being incorporated into some of the adult pharyngeal sensory organs [22].
Fig. 3.
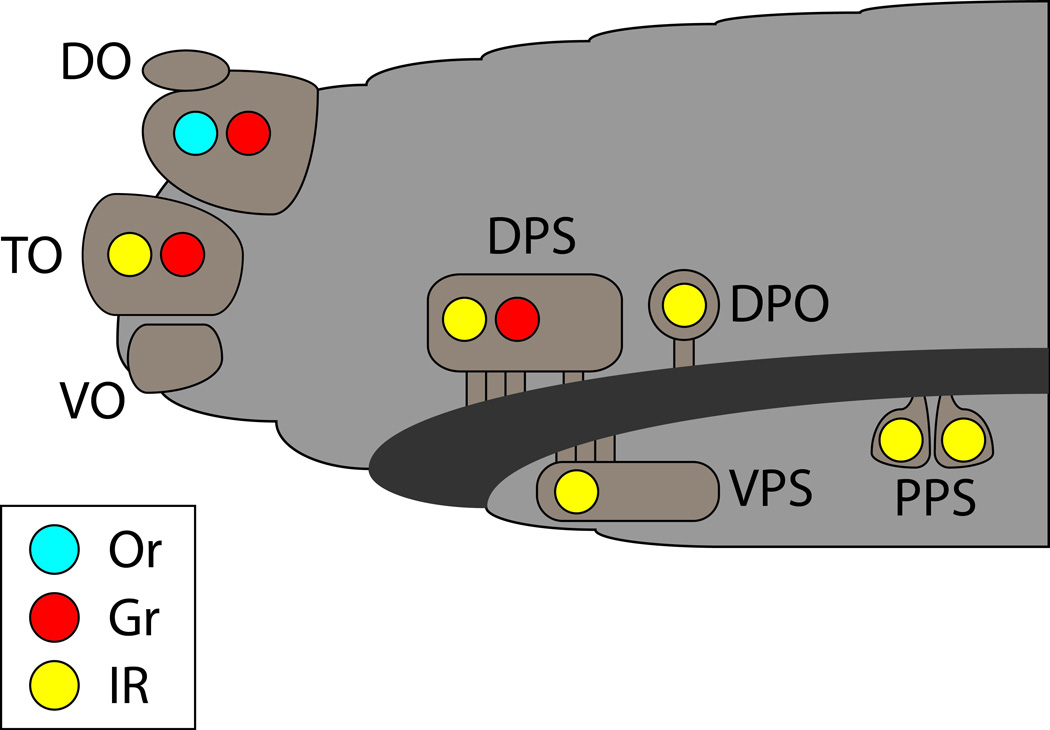
Chemosensory organs of the larval head. The dorsal organ (DO), terminal organ (TO), and ventral organ (VO) are external; the dorsal pharyngeal sensilla (DPS), ventral pharyngeal sensilla (VPS) and dorsal pharyngeal organ (DPO) are internal and line the gastrointestinal tract (dark gray) of the larva. Colored circles indicate the expression of at least one receptor of the indicated category in the indicated organ. Mapping of Grs and IRs is based primarily on analysis of GAL4 drivers.
The largest families of chemoreceptors
Chemoreceptors of the fly are numerous and sundry (Fig. 4). A recurrent source of excitement in the field has been the discovery of new kinds of chemoreceptors. Below we describe the largest classes of chemoreceptors. We also introduce some additional classes of chemoreceptors in Box 2. Other kinds of receptors may well lurk in the genome of the fly, awaiting discovery.
Fig. 4.
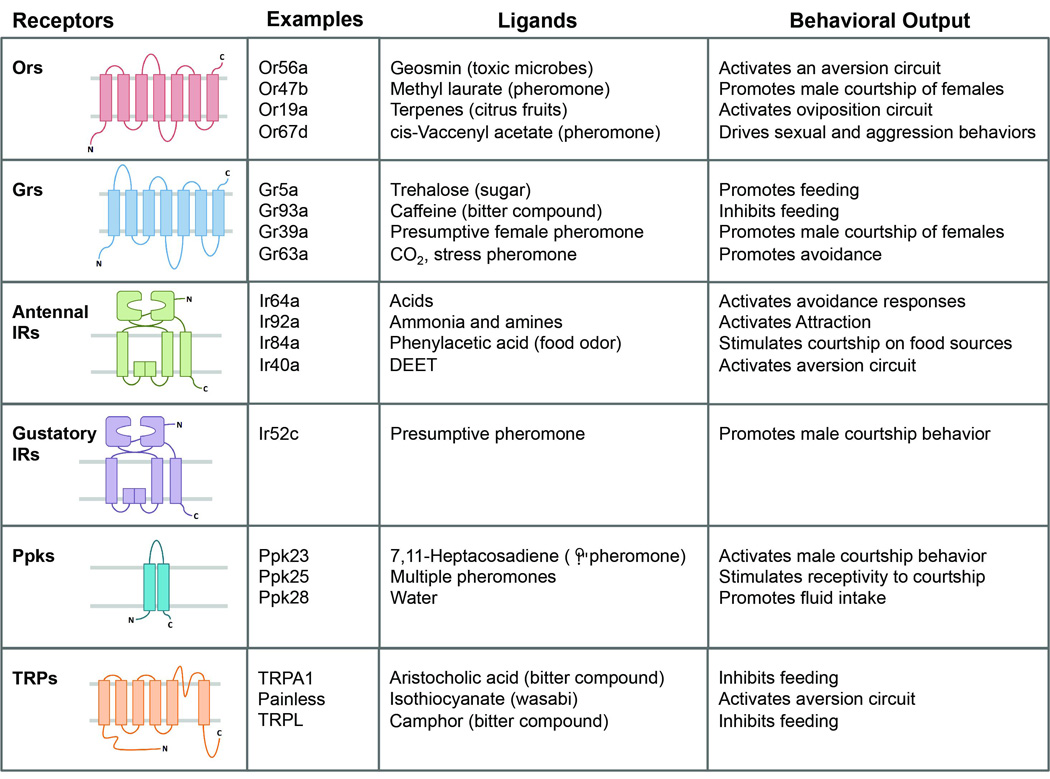
Classes of chemoreceptors. The predicted topologies of the different receptor classes are indicated. The topology of Grs is not well-established and may vary among Grs. Examples of receptors in each class are provided, along with presumptive stimuli and behaviors activated by each receptor. We note that TRPA1 also responds to heat and acts in thermotaxis behavior, while TRPL responds to light and acts in phototaxis behavior.
Box 2. Other chemoreceptors in the fly: Ppks and Trps.
Pickpocket (Ppk)/Degenerin-Epithelial sodium Channels (Deg/ENaCs) also play interesting roles in chemoreception. There are 31 ppk genes in Drosophila, and they are predicted to encode two-transmembrane-domain proteins [120]. Ppk28 is required for the sensing of water and osmolarity, and misexpression of ppk28 in bitter-sensitive neurons confers water sensitivity to them [121, 122]. ppk23 and ppk29 are expressed in male forelegs, among other chemosensory tissues, and are required for male courtship behavior toward females [123–125]. ppk23 mutant males also showed abnormally high courtship toward other males, suggesting that the gene can function in both excitatory and inhibitory circuits. Both ppk genes are required for responses to identified fly pheromones.
Trp proteins are six-transmembrane-domain proteins that act in a variety of sensory modalities. A number of Trps are expressed in taste and olfactory organs of the fly [7, 85]. Two of them, TrpA1 and Painless, act in thermosensation [126] but are also expressed in taste organs. TRPA1 is required for the response to noxious electrophiles, and is activated by electrophiles when expressed in Xenopus oocytes [127]. TRPA1 also functions in the response to aristolochic acid, a bitter compound [128], as does TRPL in the response to camphor [129] and Painless in the behavioral aversion to isothiocyanate, the pungent ingredient of wasabi [130]. Interestingly, the mechanisms of action in these three latter cases differ from that by which TRPA1 detects electrophiles.
Outstanding Questions Box
What are the 3D structures of Ors and Grs? What are their multimeric compositions? Where do ligands bind? What are the transduction mechanisms?
How do individual neurons select which chemoreceptor genes to express, from an enormous repertoire?
What circuits and behaviors are driven by individual Ors, Grs, IRs, and other chemoreceptors?
Do IRs and Grs detect different classes of tastants or different features of the same gustatory stimuli?
How are signals from IRs and Grs integrated in the brain?
How are olfactory and taste signals integrated with each other and with other sensory inputs?
Does combinatorial coding of taste information by IRs and Grs enhance discrimination or memory of taste stimuli?
How is signaling via chemoreceptors modulated by experience, and how is it shaped through evolution?
Odor receptors (ORs)
The Drosophila melanogaster genome contains 60 Or genes, which are predicted to encode 62 seven-transmembrane-domain proteins via alternative splicing [23]. These proteins have little if any sequence similarity to the odor receptors of vertebrates or C. elegans, which are G protein-coupled receptors (GPCRs). Moreover, the membrane topology of Ors is distinct from that of GPCRs [24]. Heterologous expression studies have provided evidence that Ors can function as ligand-gated ion channels and can transduce olfactory information independent of G proteins [25–27]; however, a role for G proteins in olfactory signaling has been supported by several studies, and the mechanism of Or-mediated transduction remains an active topic of investigation [28–30].
Most ORNs of basiconic and trichoid sensilla express a single member of the Or family, which confers the odorant response profile of the neuron [31, 32]. These neurons also express a co-receptor, Orco, which heterodimerizes with the Or and is essential for the targeting of the complex to the dendritic membrane [33].
The response profiles of Ors have been characterized in detail by expressing them in an in vivo expression system called the “empty neuron” system [32, 34]. This system is based on a mutant neuron that lacks an endogenous Or and does not respond to odorants. An individual Or may be expressed in this neuron via the GAL4-UAS system and the odorant responses that it confers are measured by singleunit electrophysiology. The response profiles conferred by many Ors were found to match closely with the response profiles of specific ORN classes of the wild type antenna, which supported the fidelity of the expression system and allowed construction of a receptor-to-neuron map of the antenna [32]. This map was confirmed and extended by molecular analysis, providing a near-complete map of Or expression in the fly’s olfactory organs [31, 35].
Ectopic expression of Or genes revealed that Ors dictate several properties of ORN response [32]. The odor response spectrum, the mode of response (excitation v. inhibition), the termination kinetics, and the level of spontaneous activity all depend on the receptor. An individual receptor can mediate both excitatory and inhibitory responses to different odors in the same cell. Systematic functional analysis of the Or repertoire revealed that some receptors were narrowly tuned and others broadly tuned with respect to a panel of 110 diverse odorants [36]. Tuning depends on concentration: receptors that respond broadly to high concentrations of odorants respond to a narrower range of odorants when tested at lower concentrations. Coding is combinatorial, in that different odors elicit responses from different subsets of receptors.
A particularly exciting area of recent research has extended this primary analysis of odor sensitivity to include compounds of special biological significance to the fly. One receptor, Or56a, is narrowly tuned to geosmin, a compound that alerts flies to the presence of toxic microbes and that activates an aversion circuit [37]. Or19a detects terpenes present in citrus fruits, and activates an oviposition circuit [38]. Or67d, Or47b, Or88a, and Or65a detect fly pheromones that act in sexual, aggression, or social aggregation behaviors [39–44]. These findings illustrate an interesting philosophical problem that transcends the entire field of chemoreception: given the vast dimension of chemical space and our ability to sample only an infinitesimal fraction of it, it is difficult to be certain what ligand any receptor has evolved to bind. That said, it is particularly compelling when low concentrations of a compound from the natural environment of the fly are found to activate a particular receptor, a dedicated circuit, and an adaptive behavior.
The receptor-to-neuron map of the olfactory system raises an intriguing developmental problem. How do individual ORNs select the Ors that they express, from a family of 60 genes? A combinatorial code of transcription factors, including the POU domain transcription factor Acj6 and the Kruppel-like transcription factor Rotund, underlies much of the process [45, 46]; epigenetic mechanisms also contribute to receptor regulation in ORNs [47]. A code of regulatory elements upstream of Or genes acts in specifying their expression patterns [48]. Another puzzle is posed by the spatial distribution of expression. ORNs that express the same receptor are restricted to particular regions of the antenna, but within a given region, ORNs that express one individual receptor are intermingled with ORNs that express other receptors [49]. Intriguingly, this patterning arises in part through mobility: sensilla are initially specified during early pupal development, and then move beneath the surface of the antennal disc during late pupal development to give rise to a scattered pattern [50].
Larvae express 25 Ors, of which 13 are larval-specific [19, 31, 51]. For 19 receptors, an odorant that activates it strongly has been identified [52]. Silencing of neurons that express different receptors reduces behavioral response to different odorants [19]. Mutational analysis of two receptors showed that they mediate responses to different concentrations of ethyl acetate [53].
Gustatory receptors (Grs)
The D. melanogaster genome contains 60 Gr genes, which are predicted to encode 68 proteins via alternative splicing [23]. These proteins are extremely divergent, containing as little as 8% amino acid identity. Grs are weakly related to Ors, which can be viewed as a single lineage within a larger insect chemoreceptor superfamily [23]. The topology of insect Gr proteins is not as clearly established as that of Or proteins [54].
Grs are expressed in the labellum, legs, and pharynx of the adult fly [9, 55–59], the larval taste organs [20], and in a variety of other adult tissues, including the antenna, maxillary palp, enteroendocrine cells of the gut, multidendritic cells of the abdominal body wall, neurons innervating reproductive organs, and the brain [15, 17, 60–63]. Gr expression studies have been constrained by the inability to visualize most tested Gr transcripts by in situ hybridization. Localization of Gr expression has thus primarily been carried out directly using RT-PCR, RNA-Seq, or microarray analysis, or indirectly using transgenic and knock-in reporter techniques, all of which are subject to limitations. That said, expression studies of labellar and leg Grs have in some cases been confirmed by electrophysiological studies of individual taste sensilla [9, 59].
Unlike ORNs, some GRNs coexpress many receptors. One class of bitter-sensitive neurons in the labellum expresses reporter constructs representing 29 different Grs [59], a bitter neuron in the leg coexpresses 18 [9], and a neuron of the larval taste system coexpresses 17 [20]. Expression of Grs in bitter neurons is combinatorial, although some receptors are expressed broadly. Gr33a, for example, is expressed ubiquitously in bitter neurons of the labellum, leg, and larva. Sugar-sensitive neurons also coexpress several receptors [64, 65]. Strikingly, the sets of Grs expressed in bitter and sugar neurons are mutually exclusive [66]. Despite this division of sweet and bitter sensing into separate cells, there is evidence that sugar neurons are both inhibited by bitter compounds and activated by sugars, raising new and exciting questions about how the fly detects and integrates information from stimuli of opposing valence [67, 68].
Loss-of-function experiments have shown that some Grs, such as Gr5a, Gr61a and members of the Gr64a-f cluster are required for responses to various sugars [62, 64, 65, 69]. Reciprocally, all of these genes conferred a sugar response when ectopically expressed in an antennal neuron that has no endogenous sugar responses [70]. All of these genes lie in one clade of the Gr family [23].
Bitter responses also depend on Grs. For example, Gr93a is required for electrophysiological response of one class of taste sensilla to caffeine, but not to several other bitter compounds [71]. Gr8a is required for response to the aversive compound L-canavanine, but not other compounds [72]. Mutation of Gr33a, by contrast, reduced electrophysiological response to a variety of structurally diverse bitter compounds [73]. The breadth of this phenotype and its expression pattern suggested the possibility that Gr33a acts as a co-receptor for other Grs. However, Gr33a differs from Orco in that it is not required for trafficking of other tested Grs to the membrane. Moreover, normal electrophysiological response to caffeine also depends on a third broadly expressed Gr, Gr66a [74], raising the possibility of a signaling complex consisting of three or more Gr subunits.
Some Grs are required for sexual behavior. Mutation or knockdown of Gr39a in males led to reduced courtship of females, consistent with the notion that these receptors signal the presence of an excitatory female pheromone [75]. Mutation of Gr32a also led males to court other males and previously mated females, as if this receptor signals the presence of an inhibitory pheromone [76]. Remarkably, Gr32a also acts in males of Drosophila melanogaster to prevent them from mating with females of different species [77]. We note that mutation of the broadly expressed Gr33a produced elevated male-male courtship [73], consistent with the notion that Gr33a acts as a coreceptor for Gr32a. One study found that knockdown of Gr68a, or silencing of Gr68a-expressing neurons in males, reduced male courtship towards females, suggesting that Gr68a is a receptor for a female pheromone and is required for efficient courtship. [78]. However, a recent investigation found that Gr68a-expressing male neurons sense an anti-aphrodisiac compound produced by males that can inhibit male courtship toward females [79], illustrating that there are still numerous exciting puzzles to be solved regarding the role of Grs in pheromone detection and sexual behavior in Drosophila.
Early studies revealed expression of a few Grs in the antenna [60, 63]. Gr21a and Gr63a are coexpressed in one class of antennal ORNs, where together they confer response to CO2 [80, 81], which is an important cue used in detecting fermenting food sources and is a stress signal in Drosophila [82]. Mosquito ORNs that express orthologs of these Ors respond to CO2 and odorants from human skin, informing mosquitoes of the proximity of human hosts [83]. Gr28b.d is expressed in a specialized structure of the antenna called the arista and plays a critical role in thermosensation [84].
Surprisingly, recent RNA-seq analysis revealed that 12 Grs are expressed in the antenna [85], a larger number than previously detected using transgenic GAL4 reporters or in situ hybridization. RNA-seq examines RNA levels directly, unlike transgenic GAL4 reporters, and its sensitivity is greater than that of conventional in situ hybridization. Intriguingly, five members of the Gr64a-f cluster of sugar receptor genes are expressed in the antenna, where their function remains enigmatic [62, 85].
Grs are also expressed in pharyngeal organs and in enteroendocrine cells of the gut [17, 55–58]. These cells may regulate food intake and other functions. Gr43a is expressed in the brain, where it functions as a fructose receptor and controls feeding responses [15].
The mechanism of Gr-mediated transduction is in great need of further investigation. Of 11 Grs of the silkmoth Bombyx mori expressed in Xenopus oocytes, one, BmGr-9, conferred response to a tastant, D-fructose [86]. This receptor and its Drosophila ortholog, Gr43a, also conferred responses when expressed in cultured cells. Analysis of these responses was consistent with the hypothesis that BmGr-9 is a ligand-gated ion channel; evidence to support a role for G protein-mediated signaling was not found. By contrast, in vivo studies of taste responses to sugars have found a requirement for G protein signaling, but different studies implicated different G proteins (Goα, Gqα, or Gsα, as well as Gγ1) [7], and the roles of some G proteins may be modulatory or indirect. Moreover, different Gr proteins may signal through different mechanisms.
Ionotropic receptors (IRs)
The D. melanogaster genome also contains ~60 IR genes, which are related to ionotropic glutamate receptor genes (iGluRs) [87]. IR genes are predicted to encode ligand-gated cation channels with three transmembrane domains [88].
Antennal IRs
Approximately 17 IRs are expressed in the antenna, mostly in ORNs of coeloconic sensilla [85, 87, 89], which typically do not express Ors [31]. These IRs confer response to many organic acids and amines [89, 90]. IR92a is required for response to particular amines [91], whereas IR64a is acid-sensitive [92]. Like Ors, the trafficking and function of these IRs depend on the expression of widely expressed co-receptors, including IR8a or IR25a [93]. For example, IR64a and IR8a are physically associated in vivo and form a functional channel when coexpressed in Xenopus oocytes [94].
Some antennal IRs play interesting behavioral roles. IR84a is activated by food odors, and this activation increases levels of male courtship behavior [95]. Evidently the ORN expressing IR84a influences a male courtship circuit. D. melanogaster mates primarily on food sources, and thus IR84a appears to provide a neural link between food and sex. Another intriguing role for an IR concerns the avoidance of the insect repellent DEET: in Drosophila this aversion is mediated in part via IR40a, which is expressed in neurons within a three-chambered pit on the antennal surface [87, 96]. Like Gr28b.d, IR21a expresses in the arista of the Drosophila antenna [87]; it will be interesting to investigate whether IR21a also plays a role in thermosensation.
Gustatory IRs
Some IRs are expressed in gustatory organs of adult or larval Drosophila [88, 97, 98], where investigations into their roles in taste perception have only recently been initiated. A large clade of IRs, the IR20a clade, consists of ~35 members that are more distantly related to the iGluRs. GAL4 drivers representing genes of the IR20a clade were recently found to be expressed in taste neurons of all gustatory organs of the fly, including the labellum, the legs, the pharynx, and the anterior wing margin [98]. Neurons expressing these drivers project to taste centers in the CNS. Eleven drivers are expressed in the larva, mostly in single pairs of pharyngeal neurons [97].
Some gustatory IR drivers are coexpressed with Gr drivers in bitter- or sugar-sensitive neurons of the labellum, suggesting that some of these IRs function in detection of aversive or appetitive stimuli, respectively [98]. The IRs may detect stimuli not recognized by Grs; alternatively, activation of one class of receptor may lead to modulation of another.
Other gustatory IR drivers are expressed in “orphan” taste neurons that do not express Grs or other known receptors, and that do not respond to canonical food sources [9, 98]. Analysis of IR52c and IR52d has shown sexually dimorphic expression in such neurons of the male foreleg, which makes contact with females during courtship behavior, and genetic analysis has verified that IR52c and IR52d play roles in sexual behavior [98]. The neurons in which they are expressed are activated by exposure to virgin D. melanogaster females, but not by exposure to males or virgin females of a sibling species, D. simulans, suggesting a role in recognition of a species-specific female pheromone.
Another IR that is not a member of the IR20a clade, IR76b, has been proposed to be a detector of appetitive, low concentrations of salt, based on molecular, genetic, and physiological evidence [99]. IR76b has also been proposed to be a co-receptor with other IRs [93]. IR25a may be a co-receptor in both olfactory and taste systems [88, 93, 97], providing another interesting link between the two sensory modalities.
Concluding Remarks and Future Perspectives
Great progress has been made in understanding the receptors that constitute the basis of all of chemosensory perception. However, critical boxes remain black, key principles remain controversial, and major topics remain unexplored. It is as if a new continent has been discovered but only the coastline has been mapped.
Ors and Grs are currently represented as squiggles through cartoon membranes, with much uncertainty in the case of Grs. A 3D structure of Ors and Grs and their co-receptors, with and without ligands, is urgently needed. Although experimental determination of Ors and Grs has remained elusive, a recent 3D modeling study provided evidence that the packing of Or transmembrane helices is distinct from that of canonical GPCRs [100]. In the case of Grs, we need to know whether receptors assemble as complex multimers, possibly of three or more subunits. We also need to know whether any of these receptors interact directly with G proteins, and to clarify the potential roles of various G proteins in signaling.
It seems clear that new pheromone receptors await identification. The pheromonal profile of the fly has recently been found to be much richer than previously thought and may include 58 hydrocarbons [101, 102]. Many of these compounds appear to be transferred from males to females, and some in the opposite direction, during sexual behavior. The number of pheromones thus appears to greatly exceed the number of known pheromone receptors. It has also become clear that larvae signal to each other via pheromones [103]. Beyond pheromones, the natural environment is teeming with signals – from predators, a cornucopia of food sources, and the chemical arsenal of microbes and plants that do not want to be eaten. Receptors for many signals await identification.
In addition to these “orphan ligands” for which receptors have not been identified, there are “orphan receptors” for which no ligands have been identified. The future may well see some happy unions between these ligands and receptors; in any case there are clearly many inviting opportunities for ligand and receptor de-orphanization. We note also that there are many “orphan neurons”, such as in the legs and the larva, for which neither ligands nor receptors have been identified.
The establishment of neural circuits depends critically on the expression of the receptors that drive them. Receptors that signal mates, nutrients, and toxins must be expressed in neurons that drive mating, consumption, and avoidance. Much remains to be learned about the genetic and epigenetic mechanisms by which individual neurons select the Ors they express, and virtually nothing is known of how neurons select Grs or IRs.
Finally, how does the pattern of receptor expression serve the behavior of the animal? Does the combinatorial pattern of Gr and IR expression in taste organs allow for combinatorial coding of taste information and thereby enhance the specificity or discriminatory power of the system? How does it allow a single bitter compound to have both a positive and a negative valence, i.e. to suppress feeding but activate egg-laying [57, 104]? The behavior of Drosophila is governed by the function of more than 180 receptors, and an understanding of how behaviors are activated depends critically on an understanding of the receptors that activate them.
Trends Box.
Odorant receptors (Ors) have been found to activate an increasing number of behavioral circuits.
Gustatory receptors (Grs) are expressed in a wide diversity of organs. Emerging results reveal roles in an expanding repertoire of functions, extending beyond chemoreception.
Ionotropic receptors (IRs) are expressed not only in olfactory organs but in taste organs. A large clade has recently been found to be expressed in all taste organs of the fly.
Ors, Grs, and IRs all have roles in the sexual behavior of the fly.
Acknowledgments
We thank Karen Menuz and Zhe He for helpful comments. We apologize to those whose work has not been cited due to space constraints or amnesia. Our work is supported by an NRSA (R.J.) and grants (J.C.) from the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annu Rev Neurosci. 2007;30:505–533. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- 2.Su CY, Menuz K, Carlson JR. Olfactory perception: receptors, cells, and circuits. Cell. 2009;139(1):45–59. doi: 10.1016/j.cell.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shanbhag SR, et al. Expression mosaic of odorant-binding proteins in Drosophila olfactory organs. Microsc Res Tech. 2001;55(5):297–306. doi: 10.1002/jemt.1179. [DOI] [PubMed] [Google Scholar]
- 4.Wilson RI. Early olfactory processing in Drosophila: mechanisms and principles. Annu Rev Neurosci. 2013;36:217–241. doi: 10.1146/annurev-neuro-062111-150533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stocker RF. The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res. 1994;275(1):3–26. doi: 10.1007/BF00305372. [DOI] [PubMed] [Google Scholar]
- 6.Martin F, et al. Elements of olfactory reception in adult Drosophila melanogaster. Anat Rec (Hoboken) 2013;296(9):1477–1488. doi: 10.1002/ar.22747. [DOI] [PubMed] [Google Scholar]
- 7.Liman ER, Zhang YV, Montell C. Peripheral coding of taste. Neuron. 2014;81(5):984–1000. doi: 10.1016/j.neuron.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meunier N, et al. Peripheral coding of bitter taste in Drosophila. J Neurobiol. 2003;56(2):139–152. doi: 10.1002/neu.10235. [DOI] [PubMed] [Google Scholar]
- 9.Ling F, et al. The molecular and cellular basis of taste coding in the legs of Drosophila. J Neurosci. 2014;34(21):7148–7164. doi: 10.1523/JNEUROSCI.0649-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yanagawa A, Guigue AM, Marion-Poll F. Hygienic grooming is induced by contact chemicals in Drosophila melanogaster. Front Behav Neurosci. 2014;8:254. doi: 10.3389/fnbeh.2014.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolbarsht ML, Dethier VG. Electrical activity in the chemoreceptors of the blowfly. I. Responses to chemical and mechanical stimulation. J Gen Physiol. 1958;42(2):393–412. doi: 10.1085/jgp.42.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rice MJ. Blowfly ovipositor receptor neurone sensitive to monovalent cation concentration. Nature. 1977;268:747–749. [Google Scholar]
- 13.Taylor BJ. Sexually dimorphic neurons of the terminalia of Drosophila melanogaster: II. Sex-specific axonal arborizations in the central nervous system. J Neurogenet. 1989;5(3):193–213. doi: 10.3109/01677068909066208. [DOI] [PubMed] [Google Scholar]
- 14.Nayak SV, Singh RN. Sensilla on the tarsal segments and mouthparts of adult Drosophila melanogaster Meigen (Diptera: Drosophilidae) Int. J. Insect Morphol. Embryol. 1983;12(5–6):273–291. [Google Scholar]
- 15.Miyamoto T, et al. A fructose receptor functions as a nutrient sensor in the Drosophila brain. Cell. 2012;151(5):1113–1125. doi: 10.1016/j.cell.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dus M, et al. Nutrient Sensor in the Brain Directs the Action of the Brain-Gut Axis in Drosophila. Neuron. 2015 doi: 10.1016/j.neuron.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park JH, Kwon JY. Heterogeneous expression of Drosophila gustatory receptors in enteroendocrine cells. PLoS One. 2011;6(12):e29022. doi: 10.1371/journal.pone.0029022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monte P, et al. Characterization of the larval olfactory response in Drosophila and its genetic basis. Behav Genet. 1989;19(2):267–283. doi: 10.1007/BF01065910. [DOI] [PubMed] [Google Scholar]
- 19.Fishilevich E, et al. Chemotaxis behavior mediated by single larval olfactory neurons in Drosophila. Curr Biol. 2005;15(23):2086–2096. doi: 10.1016/j.cub.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 20.Kwon JY, et al. Molecular and cellular organization of the taste system in the Drosophila larva. J Neurosci. 2011;31(43):15300–15309. doi: 10.1523/JNEUROSCI.3363-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerber B, Stocker RF. The Drosophila larva as a model for studying chemosensation and chemosensory learning: a review. Chem Senses. 2007;32(1):65–89. doi: 10.1093/chemse/bjl030. [DOI] [PubMed] [Google Scholar]
- 22.Gendre N, et al. Integration of complex larval chemosensory organs into the adult nervous system of Drosophila. Development. 2004;131(1):83–92. doi: 10.1242/dev.00879. [DOI] [PubMed] [Google Scholar]
- 23.Robertson HM, Warr CG, Carlson JR. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2003;100(Suppl 2):14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benton R, et al. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4(2):e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato K, et al. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452(7190):1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- 26.Wicher D, et al. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452(7190):1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- 27.Smart R, et al. Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochem Mol Biol. 2008;38(8):770–780. doi: 10.1016/j.ibmb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Kain P, et al. Reduced odor responses from antennal neurons of G(q)alpha, phospholipase Cbeta, and rdgA mutants in Drosophila support a role for a phospholipid intermediate in insect olfactory transduction. J Neurosci. 2008;28(18):4745–4755. doi: 10.1523/JNEUROSCI.5306-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng Y, et al. The stimulatory Galpha(s) protein is involved in olfactory signal transduction in Drosophila. PLoS One. 2011;6(4):e18605. doi: 10.1371/journal.pone.0018605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ignatious Raja JS, et al. Role of Go/i subgroup of G proteins in olfactory signaling of Drosophila melanogaster. Eur J Neurosci. 2014;39(8):1245–1255. doi: 10.1111/ejn.12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Couto A, Alenius M, Dickson BJ. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol. 2005;15(17):1535–1547. doi: 10.1016/j.cub.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 32.Hallem EA, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117(7):965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 33.Larsson MC, et al. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43(5):703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 34.Dobritsa AA, et al. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37(5):827–841. doi: 10.1016/s0896-6273(03)00094-1. [DOI] [PubMed] [Google Scholar]
- 35.Fishilevich E, Vosshall LB. Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol. 2005;15(17):1548–1553. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- 36.Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125(1):143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 37.Stensmyr MC, et al. A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell. 2012;151(6):1345–1357. doi: 10.1016/j.cell.2012.09.046. [DOI] [PubMed] [Google Scholar]
- 38.Dweck HK, et al. Olfactory preference for egg laying on citrus substrates in Drosophila. Curr Biol. 2013;23(24):2472–2480. doi: 10.1016/j.cub.2013.10.047. [DOI] [PubMed] [Google Scholar]
- 39.Kurtovic A, Widmer A, Dickson BJ. A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature. 2007;446(7135):542–546. doi: 10.1038/nature05672. [DOI] [PubMed] [Google Scholar]
- 40.Dweck HK, et al. Pheromones mediating copulation and attraction in Drosophila. Proc Natl Acad Sci U S A. 2015;112(21):E2829–E2835. doi: 10.1073/pnas.1504527112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Anderson DJ. Identification of an aggression-promoting pheromone and its receptor neurons in Drosophila. Nature. 2010;463(7278):227–231. doi: 10.1038/nature08678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ha TS, Smith DP. A pheromone receptor mediates 11-cis-vaccenyl acetate-induced responses in Drosophila. J Neurosci. 2006;26(34):8727–8733. doi: 10.1523/JNEUROSCI.0876-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu W, et al. Social regulation of aggression by pheromonal activation of Or65a olfactory neurons in Drosophila. Nat Neurosci. 2011;14(7):896–902. doi: 10.1038/nn.2836. [DOI] [PubMed] [Google Scholar]
- 44.Ejima A, et al. Generalization of courtship learning in Drosophila is mediated by cis-vaccenyl acetate. Curr Biol. 2007;17(7):599–605. doi: 10.1016/j.cub.2007.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bai L, Carlson JR. Distinct functions of acj6 splice forms in odor receptor gene choice. J Neurosci. 2010;30(14):5028–5036. doi: 10.1523/JNEUROSCI.6292-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Q, et al. Combinatorial rules of precursor specification underlying olfactory neuron diversity. Curr Biol. 2013;23(24):2481–2490. doi: 10.1016/j.cub.2013.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sim CK, et al. Epigenetic regulation of olfactory receptor gene expression by the Myb-MuvB/dREAM complex. Genes Dev. 2012;26(22):2483–2498. doi: 10.1101/gad.201665.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ray A, van der Goes van Naters W, Carlson JR. A regulatory code for neuron-specific odor receptor expression. PLoS Biol. 2008;6(5):e125. doi: 10.1371/journal.pbio.0060125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vosshall LB, et al. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96(5):725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- 50.Song E, et al. Determinants of the Drosophila odorant receptor pattern. Dev Cell. 2012;22(2):363–376. doi: 10.1016/j.devcel.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 51.Kreher SA, Kwon JY, Carlson JR. The molecular basis of odor coding in the Drosophila larva. Neuron. 2005;46(3):445–456. doi: 10.1016/j.neuron.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 52.Mathew D, et al. Functional diversity among sensory receptors in a Drosophila olfactory circuit. Proc Natl Acad Sci U S A. 2013;110(23):E2134–E2143. doi: 10.1073/pnas.1306976110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kreher SA, et al. Translation of sensory input into behavioral output via an olfactory system. Neuron. 2008;59(1):110–124. doi: 10.1016/j.neuron.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang HJ, et al. Topological and functional characterization of an insect gustatory receptor. PLoS One. 2011;6(8):e24111. doi: 10.1371/journal.pone.0024111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dunipace L, et al. Spatially restricted expression of candidate taste receptors in the Drosophila gustatory system. Curr Biol. 2001;11(11):822–835. doi: 10.1016/s0960-9822(01)00258-5. [DOI] [PubMed] [Google Scholar]
- 56.Thorne N, et al. Taste perception and coding in Drosophila. Curr Biol. 2004;14(12):1065–1079. doi: 10.1016/j.cub.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 57.Joseph RM, Heberlein U. Tissue-specific activation of a single gustatory receptor produces opposing behavioral responses in Drosophila. Genetics. 2012;192(2):521–532. doi: 10.1534/genetics.112.142455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.LeDue EE, et al. Pharyngeal sense organs drive robust sugar consumption in Drosophila. Nat Commun. 2015;6:6667. doi: 10.1038/ncomms7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weiss LA, et al. The molecular and cellular basis of bitter taste in Drosophila. Neuron. 2011;69(2):258–272. doi: 10.1016/j.neuron.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scott K, et al. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell. 2001;104(5):661–673. doi: 10.1016/s0092-8674(01)00263-x. [DOI] [PubMed] [Google Scholar]
- 61.Park JH, Kwon JY. A systematic analysis of Drosophila gustatory receptor gene expression in abdominal neurons which project to the central nervous system. Mol Cells. 2011;32(4):375–381. doi: 10.1007/s10059-011-0128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fujii S, et al. Drosophila sugar receptors in sweet taste perception, olfaction, and internal nutrient sensing. Curr Biol. 2015;25(5):621–627. doi: 10.1016/j.cub.2014.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thorne N, Amrein H. Atypical expression of Drosophila gustatory receptor genes in sensory and central neurons. J Comp Neurol. 2008;506(4):548–568. doi: 10.1002/cne.21547. [DOI] [PubMed] [Google Scholar]
- 64.Dahanukar A, et al. Two Gr genes underlie sugar reception in Drosophila. Neuron. 2007;56(3):503–516. doi: 10.1016/j.neuron.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiao Y, Moon SJ, Montell C. A Drosophila gustatory receptor required for the responses to sucrose, glucose, and maltose identified by mRNA tagging. Proc Natl Acad Sci U S A. 2007;104(35):14110–14115. doi: 10.1073/pnas.0702421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Z, et al. Taste representations in the Drosophila brain. Cell. 2004;117(7):981–991. doi: 10.1016/j.cell.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 67.French AS, et al. Dual mechanism for bitter avoidance in Drosophila. J Neurosci. 2015;35(9):3990–4004. doi: 10.1523/JNEUROSCI.1312-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jeong YT, et al. An odorant-binding protein required for suppression of sweet taste by bitter chemicals. Neuron. 2013;79(4):725–737. doi: 10.1016/j.neuron.2013.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jiao Y, et al. Gr64f is required in combination with other gustatory receptors for sugar detection in Drosophila. Curr Biol. 2008;18(22):1797–1801. doi: 10.1016/j.cub.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Freeman EG, Wisotsky Z, Dahanukar A. Detection of sweet tastants by a conserved group of insect gustatory receptors. Proc Natl Acad Sci U S A. 2014;111(4):1598–1603. doi: 10.1073/pnas.1311724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee Y, Moon SJ, Montell C. Multiple gustatory receptors required for the caffeine response in Drosophila. Proc Natl Acad Sci U S A. 2009;106(11):4495–4500. doi: 10.1073/pnas.0811744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee Y, et al. Gustatory receptors required for avoiding the insecticide L-canavanine. J Neurosci. 2012;32(4):1429–1435. doi: 10.1523/JNEUROSCI.4630-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moon SJ, et al. A Drosophila gustatory receptor essential for aversive taste and inhibiting male-to-male courtship. Curr Biol. 2009;19(19):1623–1627. doi: 10.1016/j.cub.2009.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moon SJ, et al. A taste receptor required for the caffeine response in vivo. Curr Biol. 2006;16(18):1812–2817. doi: 10.1016/j.cub.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 75.Watanabe K, et al. Gr39a, a highly diversified gustatory receptor in Drosophila, has a role in sexual behavior. Behav Genet. 2011;41(5):746–753. doi: 10.1007/s10519-011-9461-6. [DOI] [PubMed] [Google Scholar]
- 76.Wang L, et al. Hierarchical chemosensory regulation of male-male social interactions in Drosophila. Nat Neurosci. 2011;14(6):757–762. doi: 10.1038/nn.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fan P, et al. Genetic and neural mechanisms that inhibit Drosophila from mating with other species. Cell. 2013;154(1):89–102. doi: 10.1016/j.cell.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bray S, Amrein H. A putative Drosophila pheromone receptor expressed in male-specific taste neurons is required for efficient courtship. Neuron. 2003;39(6):1019–1029. doi: 10.1016/s0896-6273(03)00542-7. [DOI] [PubMed] [Google Scholar]
- 79.Shankar S, et al. The neuropeptide tachykinin is essential for pheromone detection in a gustatory neural circuit. Elife. 2015;4 doi: 10.7554/eLife.06914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jones WD, et al. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. 2007;445(7123):86–90. doi: 10.1038/nature05466. [DOI] [PubMed] [Google Scholar]
- 81.Kwon JY, et al. The molecular basis of CO2 reception in Drosophila. Proc Natl Acad Sci U S A. 2007;104(9):3574–3578. doi: 10.1073/pnas.0700079104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Suh GS, et al. A single population of olfactory sensory neurons mediates an innate avoidance behaviour in Drosophila. Nature. 2004;431(7010):854–859. doi: 10.1038/nature02980. [DOI] [PubMed] [Google Scholar]
- 83.Tauxe GM, et al. Targeting a dual detector of skin and CO2 to modify mosquito host seeking. Cell. 2013;155(6):1365–1379. doi: 10.1016/j.cell.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ni L, et al. A gustatory receptor paralogue controls rapid warmth avoidance in Drosophila. Nature. 2013;500(7464):580–584. doi: 10.1038/nature12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Menuz K, et al. An RNA-seq screen of the Drosophila antenna identifies a transporter necessary for ammonia detection. PLoS Genet. 2014;10(11):e1004810. doi: 10.1371/journal.pgen.1004810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sato K, Tanaka K, Touhara K. Sugar-regulated cation channel formed by an insect gustatory receptor. Proc Natl Acad Sci U S A. 2011;108(28):11680–11685. doi: 10.1073/pnas.1019622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Benton R, et al. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136(1):149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Croset V, et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6(8):e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Silbering AF, et al. Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J Neurosci. 2011;31(38):13357–13375. doi: 10.1523/JNEUROSCI.2360-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yao CA, Ignell R, Carlson JR. Chemosensory coding by neurons in the coeloconic sensilla of the Drosophila antenna. J Neurosci. 2005;25(37):8359–8367. doi: 10.1523/JNEUROSCI.2432-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Min S, et al. Dedicated olfactory neurons mediating attraction behavior to ammonia and amines in Drosophila. Proc Natl Acad Sci U S A. 2013;110(14):E1321–E1329. doi: 10.1073/pnas.1215680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ai M, et al. Acid sensing by the Drosophila olfactory system. Nature. 2010;468(7324):691–695. doi: 10.1038/nature09537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Abuin L, et al. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69(1):44–60. doi: 10.1016/j.neuron.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ai M, et al. Ionotropic glutamate receptors IR64a and IR8a form a functional odorant receptor complex in vivo in Drosophila. J Neurosci. 2013;33(26):10741–10749. doi: 10.1523/JNEUROSCI.5419-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Grosjean Y, et al. An olfactory receptor for food-derived odours promotes male courtship in Drosophila. Nature. 2011;478(7368):236–240. doi: 10.1038/nature10428. [DOI] [PubMed] [Google Scholar]
- 96.Kain P, et al. Odour receptors and neurons for DEET and new insect repellents. Nature. 2013;502(7472):507–512. doi: 10.1038/nature12594. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 97.Stewart S, et al. Candidate ionotropic taste receptors in the Drosophila larva. Proc Natl Acad Sci U S A. 2015;112(14):4195–4201. doi: 10.1073/pnas.1503292112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koh TW, et al. The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron. 2014;83(4):850–865. doi: 10.1016/j.neuron.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang YV, Ni J, Montell C. The molecular basis for attractive salt-taste coding in Drosophila. Science. 2013;340(6138):1334–1338. doi: 10.1126/science.1234133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hopf TA, et al. Amino acid coevolution reveals three-dimensional structure and functional domains of insect odorant receptors. Nat Commun. 2015;6:6077. doi: 10.1038/ncomms7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yew JY, et al. A new male sex pheromone and novel cuticular cues for chemical communication in Drosophila. Curr Biol. 2009;19(15):1245–1254. doi: 10.1016/j.cub.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Everaerts C, et al. Drosophila cuticular hydrocarbons revisited: mating status alters cuticular profiles. PLoS One. 2010;5(3):e9607. doi: 10.1371/journal.pone.0009607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mast JD, et al. Evolved differences in larval social behavior mediated by novel pheromones. Elife. 2014;3:e04205. doi: 10.7554/eLife.04205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang CH, et al. Drosophila egg-laying site selection as a system to study simple decision-making processes. Science. 2008;319(5870):1679–1683. doi: 10.1126/science.1151842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vogt RG, Riddiford LM. Pheromone binding and inactivation by moth antennae. Nature. 1981;293(5828):161–163. doi: 10.1038/293161a0. [DOI] [PubMed] [Google Scholar]
- 106.Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- 107.Sandler BH, et al. Sexual attraction in the silkworm moth: structure of the pheromone-binding-protein-bombykol complex. Chem Biol. 2000;7(2):143–151. doi: 10.1016/s1074-5521(00)00078-8. [DOI] [PubMed] [Google Scholar]
- 108.Damberger FF, et al. Structural basis of ligand binding and release in insect pheromone-binding proteins: NMR structure of Antheraea polyphemus PBP1 at pH 4.5. J Mol Biol. 2007;373(4):811–819. doi: 10.1016/j.jmb.2007.07.078. [DOI] [PubMed] [Google Scholar]
- 109.Swarup S, Williams TI, Anholt RR. Functional dissection of Odorant binding protein genes in Drosophila melanogaster. Genes Brain Behav. 2011;10(6):648–657. doi: 10.1111/j.1601-183X.2011.00704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ziegelberger G. Redox-shift of the pheromone-binding protein in the silkmoth Antheraea polyphemus. Eur J Biochem. 1995;232(3):706–711. [PubMed] [Google Scholar]
- 111.Kiely A, et al. Functional analysis of a Drosophila melanogaster olfactory receptor expressed in Sf9 cells. J Neurosci Methods. 2007;159(2):189–194. doi: 10.1016/j.jneumeth.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 112.Syed Z, et al. Pheromone reception in fruit flies expressing a moth's odorant receptor. Proc Natl Acad Sci U S A. 2006;103(44):16538–16543. doi: 10.1073/pnas.0607874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Grosse-Wilde E, et al. Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur J Neurosci. 2007;25(8):2364–2373. doi: 10.1111/j.1460-9568.2007.05512.x. [DOI] [PubMed] [Google Scholar]
- 114.van den Berg MJ, Ziegelberger G. On the function of the pheromone binding protein in the olfactory hairs of Antheraea polyphemus. J. Insect Physi. 1991;37(1):79–85. [Google Scholar]
- 115.Biessmann H, et al. The Anopheles gambiae odorant binding protein 1 (AgamOBP1) mediates indole recognition in the antennae of female mosquitoes. PLoS One. 2010;5(3):e9471. doi: 10.1371/journal.pone.0009471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pelletier J, et al. An odorant receptor from the southern house mosquito Culex pipiens quinquefasciatus sensitive to oviposition attractants. PLoS One. 2010;5(4):e10090. doi: 10.1371/journal.pone.0010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xu P, et al. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron. 2005;45(2):193–200. doi: 10.1016/j.neuron.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 118.Gomez-Diaz C, et al. Ligands for pheromone-sensing neurons are not conformationally activated odorant binding proteins. PLoS Biol. 2013;11(4):e1001546. doi: 10.1371/journal.pbio.1001546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Matsuo T, et al. Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia. PLoS Biol. 2007;5(5):e118. doi: 10.1371/journal.pbio.0050118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zelle KM, et al. The genetic architecture of degenerin/epithelial sodium channels in Drosophila. G3 (Bethesda) 2013;3(3):441–450. doi: 10.1534/g3.112.005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cameron P, et al. The molecular basis for water taste in Drosophila. Nature. 2010;465(7294):91–95. doi: 10.1038/nature09011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen Z, Wang Q, Wang Z. The amiloride-sensitive epithelial Na+ channel PPK28 is essential for drosophila gustatory water reception. J Neurosci. 2010;30(18):6247–6252. doi: 10.1523/JNEUROSCI.0627-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lu B, et al. ppk23-Dependent chemosensory functions contribute to courtship behavior in Drosophila melanogaster. PLoS Genet. 2012;8(3):e1002587. doi: 10.1371/journal.pgen.1002587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Thistle R, et al. Contact chemoreceptors mediate male-male repulsion and male-female attraction during Drosophila courtship. Cell. 2012;149(5):1140–1151. doi: 10.1016/j.cell.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Toda H, Zhao X, Dickson BJ. The Drosophila female aphrodisiac pheromone activates ppk23(+) sensory neurons to elicit male courtship behavior. Cell Rep. 2012;1(6):599–607. doi: 10.1016/j.celrep.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 126.Barbagallo B, Garrity PA. Temperature sensation in Drosophila. Curr Opin Neurobiol. 2015;34C:8–13. doi: 10.1016/j.conb.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kang K, et al. Analysis of Drosophila TRPA1 reveals an ancient origin for human chemical nociception. Nature. 2010;464(7288):597–600. doi: 10.1038/nature08848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kim SH, et al. Drosophila TRPA1 channel mediates chemical avoidance in gustatory receptor neurons. Proc Natl Acad Sci U S A. 2010;107(18):8440–8445. doi: 10.1073/pnas.1001425107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang YV, et al. Food experience-induced taste desensitization modulated by the Drosophila TRPL channel. Nat Neurosci. 2013;16(10):1468–1476. doi: 10.1038/nn.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Al-Anzi B, Tracey WD, Jr, Benzer S. Response of Drosophila to wasabi is mediated by painless, the fly homolog of mammalian TRPA1/ANKTM1. Curr Biol. 2006;16(10):1034–1040. doi: 10.1016/j.cub.2006.04.002. [DOI] [PubMed] [Google Scholar]


