Abstract
RAF and MEK inhibitors are effective in BRAF-mutant melanoma but not in BRAF-mutant colorectal cancer. To gain additional insights into this difference, we performed a genome-scale, pooled shRNA enhancer screen in a BRAF-mutant, BRAF-inhibitor resistant colorectal cancer cell line exposed to the selective RAF inhibitor PLX4720. We identified multiple genes along the receptor tyrosine kinase (RTK)-mitogen-activated protein kinase (MAPK) signalling axis that, when suppressed, either genetically or pharmacologically, sensitized cells to the selective RAF inhibitor through sustained inhibition of MAPK signalling. Strikingly, CRAF was a key mediator of resistance that could be overcome by the use of pan-RAF inhibitors in combination with a MEK inhibitor. Furthermore, the combination of pan-RAF and MEK inhibitors displayed strong synergy in melanoma and colorectal cancer cell lines with RAS-activating events such as RTK activation, KRAS mutation or NF1 loss of function mutations. Combinations of selective RAF inhibitors such as PLX4720 or dabrafenib with MEK inhibitors did not incur such profound synergy, suggesting that inhibition of CRAF by pan-RAF inhibitors plays a key role in determining cellular response. Importantly, in contrast to the modest activity seen with single agent treatment, dual pan-RAF and MEK inhibition results in the induction of apoptosis, greatly enhancing efficacy. Notably, combined pan-RAF and MEK inhibition can overcome intrinsic and acquired resistance to single-agent RAF/MEK inhibition, supporting dual pan-RAF and MEK inhibition as a novel therapeutic strategy for BRAF- and KRAS-mutant cancers.
Keywords: BRAF inhibitor, BRAF, MEK, drug resistance, colorectal cancer
Introduction
Despite the impressive early successes with BRAF-targeted therapies in BRAF-mutant melanoma, intrinsic and acquired resistance presents an enormous challenge and ultimately leads to progressive disease (1). Furthermore, widespread, intrinsic resistance of BRAF-mutant colorectal cancers to selective RAF inhibitors has severely blunted the use of these agents in this context (2). This observation could be taken to suggest that colorectal cancers are indifferent to RAF inhibitors and that a BRAF mutation alone is not sufficient to inform drug selection in the clinic. However, BRAF has been shown to be a high-ranking dependency that discriminates between BRAF-mutant and BRAF-wild type cell lines, solidifying its status as a key oncogenic driver (3). These data are therefore consistent with a continued dependency on BRAF signalling for cell survival and validates it as a drug target in colorectal cancer. Furthermore, initial studies suggested that some cancers are able to evade the effects of BRAF inhibition and reactivate the MAPK pathway. Sustained MAPK pathway activity can be brought about by amplification of BRAF, overexpression of CRAF, mutation of NRAS, mutation of NF1, PDGFR_ signalling, IGF1R/PI3K activity, MEK1 mutation, expression of MAP3K8/COT or an altered transcriptional state (4–12). Surprisingly, second-site mutations of the BRAF gene have not yet been implicated in resistance to BRAF inhibition, despite demonstrating the potential to do so in preclinical models (13).
Therefore, to enable an unbiased and global assessment of potential RAF-inhibitor combination therapies for BRAF-mutant cancers, we performed a pooled, shRNA enhancer screen configured to identify genes, which when suppressed, sensitized cells to the effects of the selective RAF inhibitor PLX4720 (14). Candidate hits from the screen would then enable us to identify specific vulnerabilities in BRAF-mutant cancers that might indicate novel opportunities for combination therapies to maximize patient benefit.
Materials and Methods
Cell lines and reagents
Cell lines were obtained from the American Tissue Culture Collection (ATCC) or the National Cancer Institute (NCI) and cultured in DMEM (Cellgro), 10% FBS (Gemini Bio-Products). Cells were passaged for less than 6 months after receipt and authenticated by short tandem repeat profiling. PLX4720, AZD6244, AZ628, RAF265, GSK1120212, GSK2118436 were purchased from Selleck Chemicals. MLN2480 was purchased from Chemiscene. Expression constructs for LacZ, KRASG12V and MEKQ56P have been described previously (5, 11).
Cell proliferation assays
Cells were seeded into 96 well plates at a density permitting logarithmic growth for the duration of the experiment. The following day, compounds were added to the cells dissolved in DMSO/medium and incubated for 96 h. CellTiter-Glo reagent (Promega) was then added to the wells, the plates were shaken for 15 minutes, then luminescence was read using an Envision plate reader (Perkin-Elmer). GI50 values or % inhibition were calculated using GraphPad Prism. Where cell proliferation was determined relative to the starting cell titer, an untreated control plate was frozen at the time of treatment and inhibitor-treated plates were frozen following the 96 h incubation. Plates were then thawed and CellTiter-Glo reagent was used as described to determine cell proliferation. For colony formation assays, cells were seeded at low density into 12-well plates. The following day, cells were treated with compounds and the medium/compound was replaced every 5 d. Following 7–10 d of treatment, cells were washed with PBS, fixed in 4% formaldehyde for 30 min and then stained with 0.5% crystal violet solution. Unbound dye was removed by washing with excess water. Wells were photographed and the dye was resuspended in 10% acetic acid and absorbance measured at 595 nm. Relative cell growth was determined as a percentage of the DMSO-treated control. The Cytotox-Glo assay (Promega) was used as recommended by the supplier following a 72 h exposure to the inhibitors.
In-cell Western
Cell lines were seeded into clear bottomed, black walled 96 well plates. The next day, cells were treated with a 10-point titration of PLX4720 for 24 h. Cells were then fixed and permeabilised with 4% formaldehyde, 0.1% TX-100 for 30 minutes. Wells were blocked with LI-COR blocking buffer and then incubated with an antibody to phospho-ERK1/2 (Sigma) and to ERK2 (Santa Cruz Biotechnology) for 4 h. Cells were then washed with 0.1% Tween 20. Cells were then incubated with secondary antibodies conjugated to IR dye for 1 h. Cells were then washed with 0.1% Tween 20 and PBS was added to the wells. Plates were then read using the In-cell Western protocol on an Odyssey scanner (LI-COR). Phospho-ERK1/2 was normalized to total ERK2 and percent-inhibition determined relative to control wells.
Cell lysis and Western blotting
Cells were lysed in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris pH8.0). Protein concentration was determined using the BCA assay (Sigma). Proteins were resolved by SDS-PAGE and transferred to PVDF-Fl membranes (Millipore) using iBlot (Life Technologies). Blots were blocked with LI-COR blocking buffer and probed with the indicated antibodies overnight at 4°C. Detection of proteins was via IR-dye conjugated secondary antibodies and IR-fluorescence detection via the Odyssey system (LI-COR).
Pooled lentiviral shRNA screen
The conditions and procedure for pooled lentiviral shRNA screening have been described previously (11). Briefly, RKO cells were seeded into 12-well plates at a density of 3x106 cells/well, a total of 7.2x107 cells/replicate were infected with a 90,000 shRNA, pooled library at an MOI of 0.3–0.5. Cells were centrifuged at 2000 rpm for 2 h and medium changed immediately after. The next day, cells were pooled and selected in the presence of 1 μg/ml puromycin for 3 d. Cells were then passaged in the presence of either DMSO or 3 μM PLX4720 for 16 population doublings. Cells were harvested by trypsinization and stored in PBS at −80°C. Extraction of genomic DNA, amplification of shRNA sequences and screen deconvolution by massively parallel sequencing (Illumina) were performed as previously described (11). The depletion of shRNAs in the PLX4720-treated arm relative to the DMSO-treated arm was determined using the RIGER algorithm and this produced a statistically significant ranked list of genes according to the “second best shRNA” analysis in Gene-E (http://www.broadinstitute.org/cancer/software/GENE-E/).
shRNA/ORF lentiviral infection
Cells were seeded into 6 or 96 well plates as required. The following day, the medium was supplemented with 4 μg/ml polybrene (Millipore) and lentiviral particles were added to the cells to give an MOI of ~1. Plates were centrifuged at 2,000 rpm for 30 minutes and medium was changed immediately after. 72 h later, cells were treated with inhibitors as required. shRNA constructs used are as follows: shLuc TRCN0000072243; shMET TRCN0000000393, TRCN0000121233; shPTPN11 TRCN0000005002, TRCN0000005003; shRAF1 TRCN0000001066, TRCN0000197115, shSHOC2 TRCN0000151603, TRCN0000154502; shNF1 TRCN0000039717. Further details are available via The RNAi Consortium portal http://www.broadinstitute.org/rnai/public/.
Results
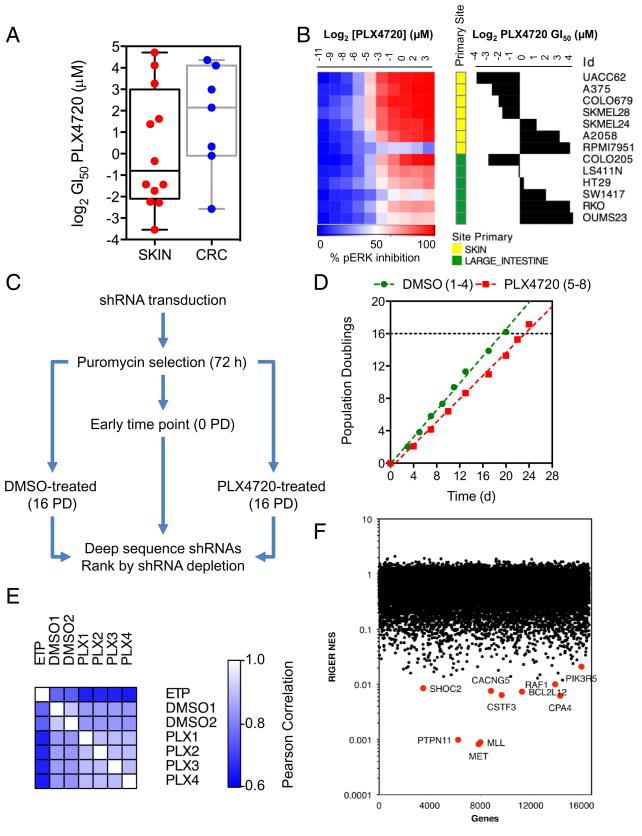
We profiled a panel of 20 BRAF-mutant melanoma and colorectal cancer (CRC) cell lines for sensitivity to PLX4720 (Figure 1A). The majority of CRC lines showed reduced sensitivity to the inhibitor compared to the melanoma lines. To determine if this was related to inhibition of the MAPK pathway, we measured ERK1/2 phosphorylation following a 24 h incubation with PLX4720 by in-cell Western (Figure 1B). Compared to the melanoma lines, only one CRC line achieved near-complete suppression of phospho-ERK (COLO205) and was highly sensitive to PLX4720. The other lines tested all failed to reach >75% inhibition of phospho-ERK and, as demonstrated in clinical trials of vemurafenib, complete (>90%) suppression of the pathway was required for efficacy (15). Interestingly, when RKO cells were treated with 3 μM PLX4720, ERK phosphorylation was inhibited by >90% at 1–2 h but this recovered during 4–24 h to approximately 50% of controls (Figure S1A). Furthermore, the cells continued to proliferate in the presence of the drug (Figure S1B).
Figure 1. A synthetic lethal shRNA screen identifies enhancers of BRAF inhibition in colorectal cancer.
(A) BRAF-mutant melanoma and colorectal cancer cell lines were profiled for sensitivity to PLX4720 following a 4 d exposure. GI50 values were determined using non-linear regression analysis in GraphPad Prism. Box and whisker plots represent the median values, 25th and 75th percentiles and min/max.
(B) Cell lines were treated with PLX4720 and ERK phosphorylation was determined by in-cell Western. Results are presented as a heat map with PLX4720 GI50 values plotted for comparison.
(C) Protocol for a synthetic lethal RNAi screen in RKO colorectal cancer cells for modifiers of sensitivity to PLX4720.
(D) Proliferation of RKO cells infected with the shRNA library and cultured in the presence of either DMSO or 3 μM PLX4720.
(E) Pearson correlation was determined between experimental replicates for each condition in the screen to assess reproducibility.
(F) Following 16 population doublings, the abundance of each shRNA was determined and used to generate a ranked list of shRNAs based upon the log-fold change between the DMSO- and PLX4720-treated replicates. RIGER analysis was employed to rank synthetic lethal genes according to the second best hairpin metric. The top ten ranking genes are indicated.
To understand what factors might permit reactivation of MAPK signalling in the presence of PLX4720, we conducted a pooled shRNA screen targeting >16,500 genes to identify which genes, when silenced, sensitized cells to the inhibitor (Figure 1C). The RKO CRC cell line for used this study as it was readily transduced with the lentiviral shRNA library, was highly resistant to PLX4720 (GI50: 17 μM) and had a short doubling time (<24 h). Moreover, these cells have previously been shown to possess a dependency on BRAF for proliferation (3). The cells were infected with a pooled library of ~90,000 shRNAs, then cultured in the presence of either DMSO vehicle or 3 μM PLX4720 (Figure 1D). shRNAs depleted only in the presence of PLX4720 were identified by Illumina sequencing (11). The Pearson correlation between replicate samples for each experimental condition confirmed reproducibility of the screen result (Figure 1E). RNAi Gene Enrichment (RIGER, (16)) was used to rank genes according to the second best scoring shRNA per gene (Figure 1F, Supplementary Table S1). The top-ranking gene was MET which encodes the receptor tyrosine kinase MET/HGFR, followed by MLL/KMT2A (encoding a lysine methyl-transferase) and PTPN11, which encodes the protein tyrosine phosphatase SHP2. We used the protein-interaction tool DAPPLE (17) to identify possible connections between the top 300 genes in the screen (Figure S2). Notably, a node encompassing genes for the RTKs MET, ERBB3 and NTRK2, adapter proteins GAB1 and CRKL, the protein tyrosine phosphatases PTPN6 and PTPN11 and several members of the MAPK pathway SHOC2, RAF1, MAP2K2 stood out due to the known functional relationships among the proteins encoded by these genes.
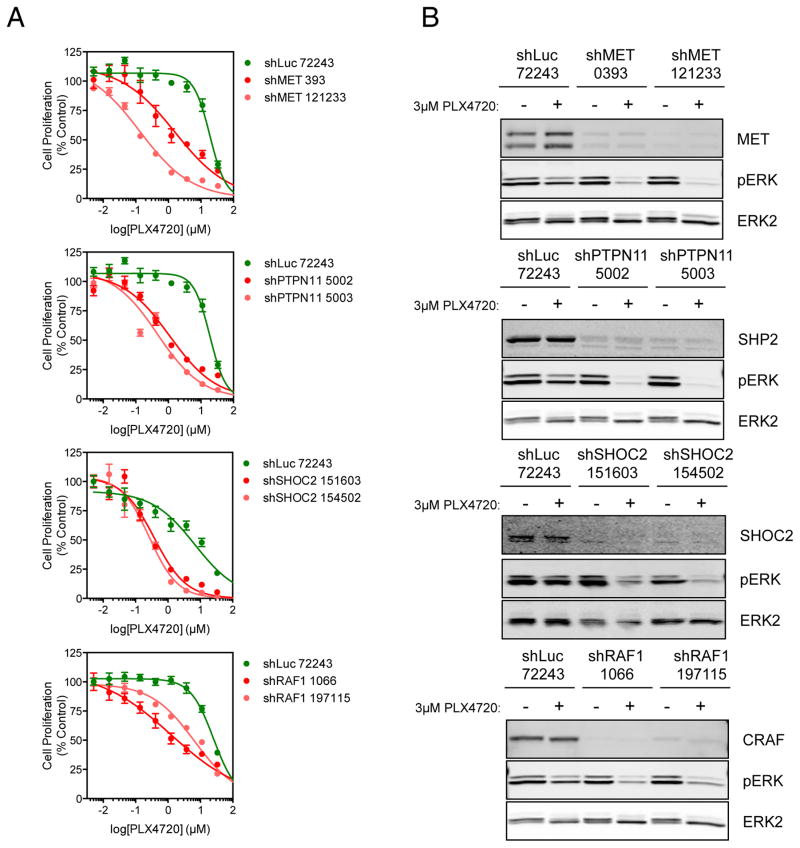
We therefore decided to validate key genes in this node as mediators of resistance to BRAF inhibition. Compared to a control shRNA targeting luciferase, knockdown of MET/HGFR, PTPN11/SHP2, SHOC2 and RAF1/CRAF all sensitized RKO cells to PLX4720 (Figure 2A) and permitted greater sustained suppression of ERK1/2 phosphorylation at 24 h following treatment with 3 μM PLX4720 (Figure 2B). Suppression of most of these genes sensitized cells to BRAF inhibition in other CRC cell lines SW1417, LS411N and WIDR (Figure S3); the exception to this was MET, whose sensitization effects were restricted to the RKO cell line. The sensitization of the RKO line by MET suppression is likely due to high expression of HGF by these cells, which activates MET signalling, thus creating a dependency on MET in the absence of signalling by oncogenic BRAF (18). We confirmed that inhibition of MET using crizotinib, SGX523 or foretinib in combination with PLX4720 resulted in near-complete inhibition of ERK1/2 phosphorylation and synergistic anti-proliferative activity as determined using the Bliss independence model (Figure S4) (19). Interestingly, inhibition of SHP2 using the tool compounds NSC87877 (20) or PHPS1 (21) in combination with PLX4720 also yielded greater anti-proliferative activity than single-agent treatment and greater suppression of ERK1/2 phosphorylation, resulting in modest synergy (Figure S5). Thus it seems likely that other RTKs, such as EGFR that has been demonstrated to confer resistance to BRAF inhibition in colorectal cancer cell lines (4, 22), also depend upon SHP2 to signal to the MAPK pathway and drive resistance. We confirmed that combined inhibition of BRAF and EGFR was synergistic in WiDr and SW1417 cells (data not shown).
Figure 2. Validation of candidate synthetic lethal genes.
(A) RKO cells were infected with individual lentiviral shRNA expression vectors targeting high-ranking genes from the primary screen or a control shRNA targeting luciferase. Cells were treated with increasing concentrations of PLX4720 for 4 d. Cell proliferation was determined using the CellTiter-Glo assay. GI50 values were determined using GraphPad Prism.
(B) RKO cells were infected as in (A) then, 72 h after infection, were treated with 3 μM PLX4720 for 18 h and protein lysates were analysed by Western blotting for the indicated proteins.
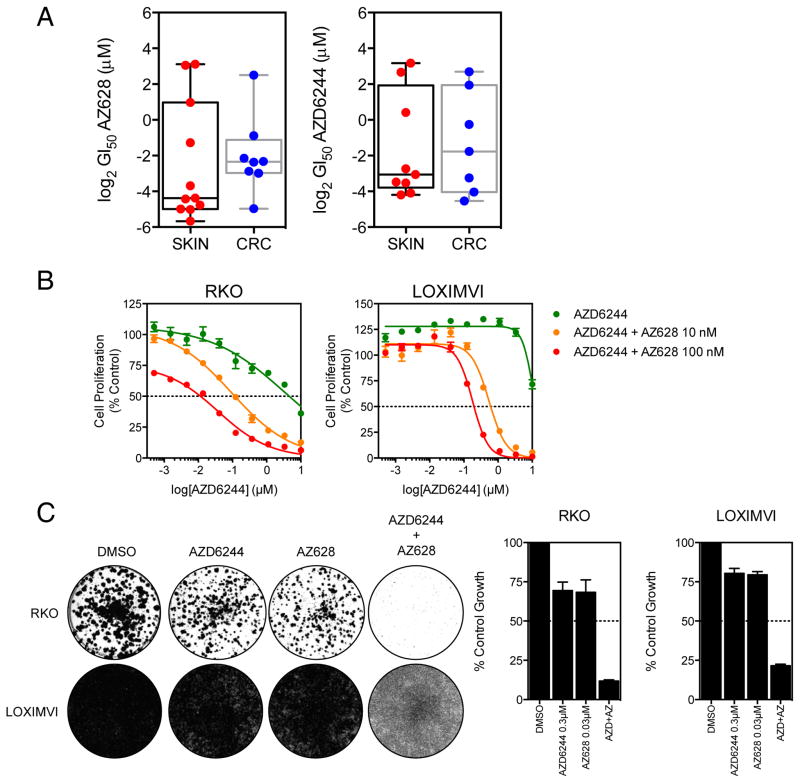
Near-complete inhibition of ERK1/2 phosphorylation seemed essential to elicit an anti-proliferative response in the PLX4720-resistant cell lines. Hence, we sought to examine drugs that were more likely to extinguish RAF-MEK-ERK signalling to this degree. We selected AZ628 for these studies, which is an inhibitor of BRAFV600E, BRAF and CRAF (23) (a so-called “pan-RAF” inhibitor). Notably, whilst PLX4720 is technically a pan-RAF inhibitor based on enzymatic assays, in cells the functional outcome is selective for mutant BRAF inhibition (14, 23). Alternatively, we reasoned that profound MAP kinase pathway inhibition downstream of RAF (e.g., using a MEK inhibitor) might overcome CRAF-mediated resistance. Consistent with this, both melanoma and CRC cell lines were generally more sensitive to AZ628 or the MEK inhibitor AZD6244 than PLX4720, although some lines still exhibited resistance (Figure 3A). Therefore, we explored combination strategies using AZ628 and AZD6244 in PLX4720-resistant lines. The RKO cell line has a GI50 of 0.5 ± 0.04 μM for AZ628 and 4.7 ± 0.9 μM for the MEK inhibitor AZD6244 (Figure 3B). However, when 10 nM AZ628 was added to a titration of AZD6244, the GI50 for AZD6244 dropped considerably to 240 ± 131 nM and if the concentration of AZ628 was increased to 100 nM, the GI50 further decreased to 25 ± 9 nM. Similarly, LOXIMVI cells, which have a GI50 of 8.3 ± 3.6 μM for AZ628, were sensitized to AZD6244 when treated with 10 and 100 nM AZ628, resulting in the GI50 decreasing from 8.2 ± 1.1 μm to 384 ± 129 nM and 128 ± 39 nM respectively. Furthermore, in longer-term colony formation assays, treatment of RKO and LOXIMVI cell lines with 300 nM AZD6244 and 30 nM AZ628 potently inhibited cell proliferation only when used in combination (Figure 3C).
Figure 3. PLX4720-resistant cells are exquisitely sensitive to combined pan-RAF and MEK inhibition.
(A) The sensitivity of BRAF mutant melanoma and colorectal cancer cell lines treated with either AZ628 or AZD6244 was assessed following a 96 h treatment with a 10-point titration of the drugs. CellTiter-glo data was used to generate GI50 values.
(B) RKO and LOXIMVI cells were treated with a titration of AZD6244 in the presence of either DMSO vehicle, 10 nM AZ628 or 100 nM AZ628 for 96 h. Cell proliferation was assessed by CellTiter-glo.
(C) RKO and LOXIMVI cells were treated with 300 nM AZD6244, 30 nM AZ628 alone or in combination. Cells were cultured for 7–10 days and then stained with crystal violet. Cell proliferation was quantified by measuring the absorbance of solubilized crystal violet stain.
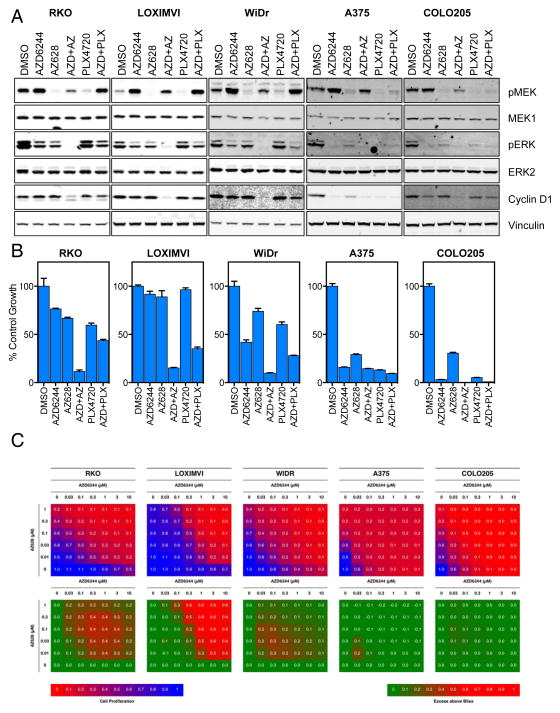
Next, we investigated the effects of combining AZD6244 and AZ628 on MAPK pathway activity. PLX4720-resistant RKO, LOXIMVI and WIDR cells and PLX4720-sensitive A375 and COLO205 cells were treated with 1 μM AZD6244 or 100 nM AZ628 either alone or in combination for 16 h (Figure 4A). In the PLX4720-resistant lines single agent treatment failed to completely inhibit ERK1/2 phosphorylation and did not suppress the expression of cyclin D1, a MAPK-regulated protein. However, when the two compounds were combined, ERK1/2 phosphorylation was completely inhibited and cyclin D1 expression was markedly reduced. Strikingly, treatment with the relatively high concentration of 10 μM PLX4720 did not inhibit either ERK1/2 phosphorylation or cyclin D1 expression and even in combination with AZD6244 did not appreciably inhibit MAPK pathway activity. PLX4720-sensitive cell lines showed good suppression of the MAPK pathway by single agents and the combination. Notably, the effects on MAPK signalling correlated well with the inhibition of cell proliferation (Figure 4B) whereby in the PLX4720-resistant cell lines, only the combination of AZD6244 and AZ628 resulted in robust inhibition. Using the Bliss independence model, this combination proved to be highly synergistic across a broad range of concentrations and was greatest in lines resistant to single agent treatment (Figure 4C). For comparison the combination of AZD6244 and PLX4720 did not potently inhibit cell proliferation nor did it produce a strong synergistic effect in PLX4720-resistant lines (Figure S6).
Figure 4. Combinatorial inhibition of BRAF, CRAF and MEK is synergistic in cell lines displaying intrinsic resistance to PLX4720.
(A) RKO, LOXIMVI, WiDr, A375 and COLO205 cancer cell lines were treated with 1 μM AZD6244, 0.1 μM AZ628, 10 μM PLX4720 or a combination of AZD6244 with either AZ628 or PLX4720 for 18 h. Cell lysates were analyzed by Western blotting for the indicated proteins.
(B) Cell lines were treated as in (A) for 96 h and cell proliferation was assessed by CellTiter-Glo assay.
(C) Cells were treated with a titration of AZD6244 versus a titration of either AZ628 or PLX4720 in combination for 96 h. Cell proliferation was assessed using CellTiter-Glo and presented in the blue-red heat maps. The green-red heat maps present the degree of synergistic interaction between the compounds, as determined using the ‘Bliss Independence model’ where values greater than 0 indicate an effect greater than the combined fractional inhibition of the single agents, indicative of synergy.
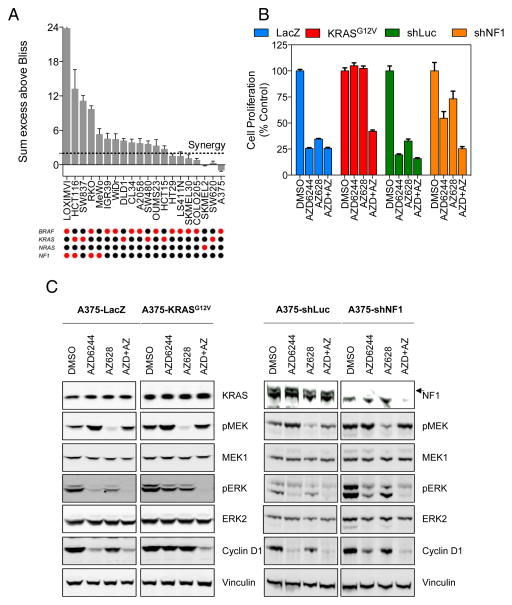
Given that the combination of a pan-RAF inhibitor and a MEK inhibitor could drive a profound response, we sought to establish what factors were responsible for this. Accordingly, we screened a panel of melanoma and colorectal cancer cell lines and determined the sum excess over Bliss score across the range of concentrations tested. Strikingly, we observed that some of the cell lines that exhibited resistance to the single agents but were synergistically inhibited by the combination also had an activating mutation in either KRAS (SW480, SW837, DLD1, HCT116, HCT15) or loss of function mutations in NF1 (LOXIMVI, HCT116, RKO, MeWo) (Figure 5A) (24). All of these events could promote elevated RAS signalling, which may confer resistance to the aforementioned single agents. We did however observe KRAS and NRAS mutant cell lines (SW620 and SKMEL2 respectively) that did not display synergy, likely because they were more sensitive to single agent treatment alone. RAS activity would be expected to promote CRAF activation (25) and potentially lead to sustained signalling in the presence of a selective RAF inhibitor such as PLX4720 (23, 26, 27). In this case, a greater dependency may exist on CRAF, which would be more efficiently targeted by AZ628 (7).
Figure 5. RAS activation is associated with sensitivity to combined pan-RAF and MEK inhibition.
(A) Synergy between AZD6244 and AZ628 was determined using the Bliss independence model in colorectal and melanoma cell lines with genetic lesions in BRAF, NF1, KRAS or NRAS (red dots indicate mutation, black indicate wild type). The sum of excess over Bliss was calculated across the matrix of concentrations tested. Values in excess of 2, indicated by the dotted red line, indicated a synergistic response.
(B) A375 cells were infected with lentiviral expression vectors encoding ORFs for LacZ and KRASG12V or shRNAs for luciferase and NF1. After 72 h, cells were treated with 1 μM AZD6244, 0.1 μM AZ628 or a combination of both compounds for 96 h. Cell proliferation was assessed using CellTiter-Glo.
(C) A375 cells were infected with lentiviral expression vectors as in (B). After 72 h, cells were treated with 1 μM AZD6244, 0.1 μM AZ628 or a combination of both compounds for 18 h. Cell lysates were analyzed for the indicated proteins.
Therefore, to test the involvement of RAS in the response to pan-RAF/MEK inhibition, we introduced KRASG12V or shRNA targeting the RAS-GAP NF1 into the A375 melanoma cell line, which confer >200-fold or >30-fold resistance to PLX4720 respectively (11). The cells were then treated with AZD6244 and AZ628 alone or in combination and cell proliferation was assessed after 96 h (Figure 5B). In the control LacZ or shLuc cells, AZD6244 and AZ628 potently inhibited proliferation alone and in combination. In contrast, expression of KRASG12V or knockdown of NF1 conferred resistance to single agent treatment but retained sensitivity to the combination of both drugs. The cells were also treated as above for 16 h and MAPK pathway activity was assessed by Western blotting (Figure 5C). In the control cells expressing LacZ or shLuc, both inhibitors reduced ERK1/2 phosphorylation and decreased the expression of cyclin D1. However, cells expressing KRASG12V or shRNA targeting NF1 maintained ERK1/2 phosphorylation and the expression of cyclin D1 when treated with the single agents but were acutely sensitive to the combination of both agents, resulting in loss of ERK1/2 phosphorylation and cyclin D1 expression.
Given that the majority of resistance mechanisms to BRAF inhibitors involve either RAS activation by mutation, amplification or upstream activation of RTKs we posited that cells showing acquired resistance to PLX4720 would be sensitive to the combination of MEK and pan-RAF inhibition. We generated a PLX4720-resistant cell line by exposing A375 cells to 1 μM PLX4720 for approximately 1 month and isolated resistant clones. This A375R10 clone was 23-fold less sensitive to PLX4720 than its parental pre-treatment A375 cells (Figure S7A) and displayed a greater degree of CRAF S338 phosphorylation, a marker of CRAF activation promoted by RAS (28). Furthermore, the levels of phospho-MEK1/2, phospho-ERK1/2 and cyclin D1 were far less affected by PLX4720-treatment than compared to the sensitive, parental cells (Figure S7B). Whilst resistant to treatment with single agent AZD6244 or AZ628, the combination of these two compounds markedly inhibited proliferation of the cells (Figure S7C) and this correlated with greater suppression of MAPK pathway activity in the A375R10 cell line (Figure S7D).
To test the possibility that the anti-proliferative effect of the drug combination was due to off-target effects of either inhibitor, we tested combinations of different pan-RAF and MEK inhibitors. The combination of RAF265 (29) and AZD6244, MLN2480 (30) and AZD6244 and of AZ628 and trametinib (GSK1120212) (31) all resulted in highly synergistic activity in lines that were resistant to the single agents and this again correlated well with near complete MAPK pathway inhibition (Figures S8–10). We also tested the combination of AZD6244 and dabrafenib (GSK2118436) (32) or trametinib and dabrafenib. Strikingly, whilst some modest synergy was observed between the MEK inhibitors and dabrafenib, the concentrations required to achieve a substantial reduction in proliferation were considerably higher compared to AZ628 and the degree of synergy was lower (Figure S11A). Interestingly, when we treated RKO cells with AZ628 and an inhibitor of ERK2, VTX-11e (33), little or no synergy was observed, despite potent inhibition of proliferation (Figure S11B). This is consistent with ERK2 activity being the critical integrator of RAS-RAF-MEK upstream signalling, and that single agent ERK2 inhibition may overcome the resistance exhibited to single agent pan-RAF or MEK inhibition. This also underscores the specificity of the pan-RAF/MEK combination for its effect on the MAPK pathway. Importantly, we could rescue cells from the combination of AZD6244 and AZ628 by expression of the drug-resistant mutant MEKQ56P (5), which permitted sustained ERK1/2 phosphorylation and cyclin D1 expression in the presence of the inhibitors (Figure S12A) and restored cell proliferation (Figure S12B) consistent with MAPK pathway inhibition being the key effector of response to the combination.
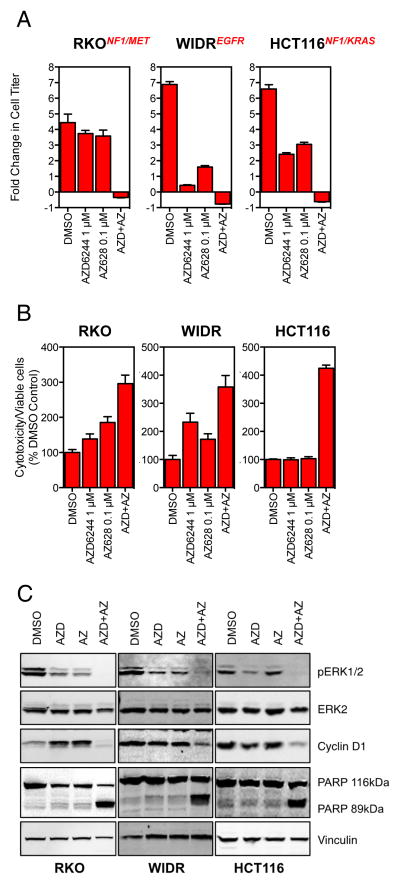
To further characterize the strong synergistic response produced by the combination of the pan-RAF and MEK inhibitors, we measured proliferation relative to the number of cells present at the commencement of treatment. When treated with single agents, cells were still able to proliferate, albeit at a reduced rate (Figure 6A). Strikingly, when used in combination, we observed a net loss of cells relative to the starting number consistent with cell death. We therefore assessed cytotoxicity using the Cytotox-glo assay which measures protease activity associated with cell death. The combination of AZ628 and AZD6244 led to significantly increased cytotoxicity compared to the single agents (Figure 6B and Figure S13). The induction of apoptosis by the drug combination was further confirmed by the detection of cleaved PARP following a 72 h exposure to the inhibitors (Figure 6C). Therefore, while exposure to single agent pan-RAF/MEK inhibition was able to reduce, but not stop, cell proliferation; the combination of both agents resulted in significant apoptosis.
Figure 6. Pan-RAF and MEK inhibition is cytotoxic and induces apoptosis.
(A) RKO, WIDR and HCT116 cells were treated with 1 μM AZD6244, 0.1 μM AZ628 or a combination of the two for 96 h. Cell proliferation was determined relative to the starting number of cells prior to treatment. Genes known to play a role in resistance to BRAF inhibition are indicated in red for each cell line.
(B) RKO, WIDR and HCT116 cells were treated as in (A) and after 72 h cytotoxicity was determined relative to the proportion of viable cells using the Cytotox-Glo assay.
(C) Cells were treated as in (B) and after 72 h cell lysates were generated and analyzed by Western blotting for the stated proteins.
Discussion
Although BRAF is a clear driver of melanoma and colorectal cancers, attempts at targeting this oncogene have delivered mixed responses due to intrinsic or acquired resistance mechanisms (34). Our shRNA screening approach identified multiple potential candidates for RAF inhibitor-based combinatorial therapies. Strikingly, our analysis of genes that promote resistance to PLX4720 indicated enrichment for genes within the RTK-MAPK pathway, including MET, ERBB3, NTRK2, GAB1, CRKL, SHP2, SHOC2, MAP2K2 and CRAF. These combinations have clear mechanistic rationales; for example MET inhibition overcomes signalling induced by overexpression of the MET ligand HGF, as others and we have observed (18, 35). Similarly, inhibition of SHP2 is consistent with blocking RTK-mediated signalling that may compensate for inhibition of BRAF, through relief of negative feedback (36, 37). Furthermore, inhibition of SHP2 may have wider utility than inhibition of specific RTKs, as multiple RTKs are known to depend upon SHP2 to mediate mitogenic signalling and have been implicated in resistance to BRAF inhibition eg. EGFR (WIDR, SW1417 and LOXIMVI cells) (4, 22, 38) and MET (RKO cells) (18, 35, 39). However, more potent and selective inhibitors of the SHP2 phosphatase are needed before this strategy can be tested clinically. We also identified SHOC2, a protein that mediates the activation of CRAF by RAS as a candidate target to block RTK- or RAS-driven resistance to BRAF inhibition (40, 41). Targeting protein-protein interactions with small molecules is technically challenging but may be an alternative to targeting specific RAS mutants directly as recently described (42). Interestingly, our data also show that loss of CRAF sensitizes cells to inhibition of BRAF, consistent with CRAF being activated by selective RAF inhibitors either through relief of negative feedback mechanisms, so-called ‘paradoxical activation’ or relief of auto-inhibition (26, 36, 43). Notably, RKO cells have multiple loss of function mutations in NF1 and have activated MET due to high expression of the MET ligand HGF; which could cooperate to increase RAS activation and promote activation of CRAF. Taken together, the data suggest that reactivation of the MAPK pathway in a CRAF-dependent manner plays a major role in the manifestation of resistance to BRAF inhibition.
In light of these observations, we targeted the MAPK pathway with the combination of a pan-RAF inhibitor and a MEK inhibitor to fully suppress signalling. Combined treatment with AZD6244 and AZ628 resulted in a profound, synergistic inhibition of cell proliferation. In comparison the combination of AZD6244 and PLX4720 or trametinib and dabrafenib failed to show strong synergy. We hypothesized that the synergistic activity of combinations with AZ628 was due to inhibition of both BRAF and CRAF (7). Pan-RAF inhibitors such as AZ628 reportedly retain potency against CRAF in cells whereas inhibitors such as PLX4720 do not (23). This is because kinase assays conducted at low ATP concentrations in vitro suggest that PLX4720 is equipotent against BRAFV600E and CRAF. However, in the cellular environment with physiologically relevant ATP concentrations of 1 mM, the potency of PLX4720 against CRAF is dramatically reduced because the Km(app) for ATP is much higher for BRAFV600E compared to wild type CRAF (14, 23). Thus, in cells, the wild type proteins are relatively resistant to PLX4720 when compared to the BRAFV600E mutant. Furthermore, both PLX4720 and AZ628 can promote RAF dimerization, but only PLX4720 causes paradoxical MAPK pathway activation that is only partially reversed by MEK inhibition, consistent with our observation that PLX4720 in combination with AZD6244 displays no synergy and has a limited antiproliferative effect (23, 26). Other mechanisms have recently been described whereby MEK is less susceptible to MEK inhibitors when activated by CRAF than when it is activated by BRAFV600E (44). This may also explain why despite being downstream of both BRAF and CRAF, the MEK inhibitor AZD6244 was only able to reduce ERK1/2 phosphorylation by about 50% when used as a single agent, perhaps also in part due to relief of negative feedback (36). Consistent with this, we have previously shown that moderate RAS activation by NF1 loss can confer resistance to AZD6244 in BRAF-mutant melanoma (11). Therefore, AZ628 inhibits BRAF and CRAF, prevents MEK activation and maintains sensitivity to the MEK inhibitor, resulting in greater pathway inhibition and synergistic effects on proliferation. It should be noted that AZ628, RAF265 and MLN2480 are all type II inhibitors, whereas PLX4720, vemurafenib and dabrafenib are type I inhibitors. It is conceivable that the inactive confirmation induced by type II inhibitor binding is less favourable for transactivation of RAF dimers, as correct positioning of the DFG motif is required to form the ‘regulatory hydrophobic spine’ as found in the active conformation (45, 46). Thus a reduced ability to transactivate RAF dimers translates into more complete pathway inhibition with type II inhibitors versus type I inhibitors. Overall, our complimentary pharmacological and genetic approaches support the concept that more effective targeting of wild type CRAF drives the synergy observed between pan-RAF and MEK inhibitors.
Our data show the strongest synergy in cell lines that were resistant to the single agents and also harboured RAS activating events such as mutation of KRAS (SW480, SW837, DLD1, HCT116, HCT15) or mutation/activation of upstream regulators such as NF1 (RKO, LOXIMVI, HCT116, MEWO), MET (RKO) or EGFR (LOXIMVI, WIDR). We confirmed this in A375 cells, made resistant to RAF or MEK inhibition by the expression of KRASG12V or knockdown of NF1, where the combination of AZ628 and AZD6244 potently suppressed proliferation. These data further demonstrate the ability of pan-RAF and MEK inhibition to treat common resistance mechanisms to BRAF inhibition (4, 11, 18, 22). Excitingly, the combination of a pan-RAF inhibitor and a MEK inhibitor appears to drive significant apoptosis. This is potentially transformative in terms of rapidly reducing tumour burden and thus reducing the opportunity for resistant populations to emerge.
This study set out to discover new approaches to treat BRAF mutant colorectal cancer, but this strategy may also have utility in melanoma and in tumors driven by oncogenic RAS. Our data is in agreement with recent reports describing synergistic activity following combined genetic or pharmacological suppression of BRAF, CRAF and MEK1/2 in RAS-mutant contexts (44, 47–49). This is attributed to the inhibition of BRAF and CRAF activities induced by relief of negative feedback following MEK inhibition. Overall, these studies underline the continued dependence on MAPK signalling in RAF inhibitor-resistant cancers and nominate potential therapeutic options for patients exhibiting resistance to vemurafenib or dabrafenib-based therapies. The synergistic activity observed in models of resistance driven by RTK activation suggests this strategy may be an alterative to dual BRAF and EGFR targeting (50), or triple BRAF, EGFR and PI3K inhibition which have shown response rates of up to 32% (50, 51). However, the tolerability of pan-RAF inhibitors has been questioned based on a phase I study of RAF265 that suggested intermittent dosing might be required, although this is possibly due to the drug’s exceptionally prolonged half-life of 11 d (52). As the latest generation of pan-RAF inhibitors progress into clinical testing (49, 53, 54), it will be important to do so in combination with MEK inhibitors to see if such treatment is well tolerated and whether it yields the striking responses described herein.
Supplementary Material
Acknowledgments
Financial support
This study was supported by grants to L.A. Garraway from the National Cancer Institute (5P50CA127003-05) STARR Cancer Consortium (I4-A448), the NIH New Innovator Award (DP2OD002750) and the Melanoma Research Alliance.
Footnotes
Conflicts of Interest
Dr. Garraway is a consultant to Novartis, Millenium/Takeda, and Boehringer Ingelheim, and a recipient of a grant from Novartis.
References
- 1.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. The New England journal of medicine. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kopetz S, Desai J, Chan E, Hecht JR, O’Dwyer PJ, Lee RJ, et al. PLX4032 in metastatic colorectal cancer patients with mutant BRAF tumors. ASCO Meeting Abstracts. 2010;28:3534. [Google Scholar]
- 3.Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12372–7. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer discovery. 2012;2:227–35. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20411–6. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–72. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montagut C, Sharma SV, Shioda T, McDermott U, Ulman M, Ulkus LE, et al. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer research. 2008;68:4853–61. doi: 10.1158/0008-5472.CAN-07-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:3085–96. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whittaker SR, Theurillat JP, Van Allen E, Wagle N, Hsiao J, Cowley GS, et al. A Genome-Scale RNA Interference Screen Implicates NF1 Loss in Resistance to RAF Inhibition. Cancer discovery. 2013;3:350–62. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konieczkowski DJ, Johannessen CM, Abudayyeh O, Kim JW, Cooper ZA, Piris A, et al. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer discovery. 2014;4:816–27. doi: 10.1158/2159-8290.CD-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whittaker S, Kirk R, Hayward R, Zambon A, Viros A, Cantarino N, et al. Gatekeeper mutations mediate resistance to BRAF-targeted therapies. Science translational medicine. 2010;2:35ra41. doi: 10.1126/scitranslmed.3000758. [DOI] [PubMed] [Google Scholar]
- 14.Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3041–6. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–9. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, et al. Highly parallel identification of essential genes in cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20380–5. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS genetics. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–4. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keith CT, Borisy AA, Stockwell BR. Multicomponent therapeutics for networked systems. Nature reviews Drug discovery. 2005;4:71–8. doi: 10.1038/nrd1609. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Sung SS, Yip ML, Lawrence HR, Ren Y, Guida WC, et al. Discovery of a novel shp2 protein tyrosine phosphatase inhibitor. Molecular pharmacology. 2006;70:562–70. doi: 10.1124/mol.106.025536. [DOI] [PubMed] [Google Scholar]
- 21.Hellmuth K, Grosskopf S, Lum CT, Wurtele M, Roder N, von Kries JP, et al. Specific inhibitors of the protein tyrosine phosphatase Shp2 identified by high-throughput docking. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7275–80. doi: 10.1073/pnas.0710468105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–3. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 23.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–5. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 24.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dumaz N, Hayward R, Martin J, Ogilvie L, Hedley D, Curtin JA, et al. In melanoma, RAS mutations are accompanied by switching signaling from BRAF to CRAF and disrupted cyclic AMP signaling. Cancer research. 2006;66:9483–91. doi: 10.1158/0008-5472.CAN-05-4227. [DOI] [PubMed] [Google Scholar]
- 26.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–21. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–30. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diaz B, Barnard D, Filson A, MacDonald S, King A, Marshall M. Phosphorylation of Raf-1 serine 338-serine 339 is an essential regulatory event for Ras-dependent activation and biological signaling. Molecular and cellular biology. 1997;17:4509–16. doi: 10.1128/mcb.17.8.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su Y, Vilgelm AE, Kelley MC, Hawkins OE, Liu Y, Boyd KL, et al. RAF265 inhibits the growth of advanced human melanoma tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:2184–98. doi: 10.1158/1078-0432.CCR-11-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasco DW, Olszanski AJ, Patnaik A, Espino G, Neuwirth R, Faucette S, et al. MLN2480, an investigational oral pan-RAF kinase inhibitor, in patients (pts) with relapsed or refractory solid tumors: Phase I study. ASCO Meeting Abstracts. 2013;31:2547. [Google Scholar]
- 31.Gilmartin AG, Bleam MR, Groy A, Moss KG, Minthorn EA, Kulkarni SG, et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:989–1000. doi: 10.1158/1078-0432.CCR-10-2200. [DOI] [PubMed] [Google Scholar]
- 32.Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–65. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 33.Aronov AM, Tang Q, Martinez-Botella G, Bemis GW, Cao J, Chen G, et al. Structure-guided design of potent and selective pyrimidylpyrrole inhibitors of extracellular signal-regulated kinase (ERK) using conformational control. Journal of medicinal chemistry. 2009;52:6362–8. doi: 10.1021/jm900630q. [DOI] [PubMed] [Google Scholar]
- 34.Lito P, Rosen N, Solit DB. Tumor adaptation and resistance to RAF inhibitors. Nature medicine. 2013;19:1401–9. doi: 10.1038/nm.3392. [DOI] [PubMed] [Google Scholar]
- 35.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–9. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lito P, Pratilas CA, Joseph EW, Tadi M, Halilovic E, Zubrowski M, et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer cell. 2012;22:668–82. doi: 10.1016/j.ccr.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montero-Conde C, Ruiz-Llorente S, Dominguez JM, Knauf JA, Viale A, Sherman EJ, et al. Relief of feedback inhibition of HER3 transcription by RAF and MEK inhibitors attenuates their antitumor effects in BRAF-mutant thyroid carcinomas. Cancer discovery. 2013;3:520–33. doi: 10.1158/2159-8290.CD-12-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun C, Wang L, Huang S, Heynen GJ, Prahallad A, Robert C, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508:118–22. doi: 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- 39.Scott LM, Lawrence HR, Sebti SM, Lawrence NJ, Wu J. Targeting protein tyrosine phosphatases for anticancer drug discovery. Current pharmaceutical design. 2010;16:1843–62. doi: 10.2174/138161210791209027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li W, Han M, Guan KL. The leucine-rich repeat protein SUR-8 enhances MAP kinase activation and forms a complex with Ras and Raf. Genes & development. 2000;14:895–900. [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez-Viciana P, Oses-Prieto J, Burlingame A, Fried M, McCormick F. A phosphatase holoenzyme comprised of Shoc2/Sur8 and the catalytic subunit of PP1 functions as an M-Ras effector to modulate Raf activity. Molecular cell. 2006;22:217–30. doi: 10.1016/j.molcel.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 42.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–51. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holderfield M, Merritt H, Chan J, Wallroth M, Tandeske L, Zhai H, et al. RAF inhibitors activate the MAPK pathway by relieving inhibitory autophosphorylation. Cancer cell. 2013;23:594–602. doi: 10.1016/j.ccr.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 44.Lito P, Saborowski A, Yue J, Solomon M, Joseph E, Gadal S, et al. Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer cell. 2014;25:697–710. doi: 10.1016/j.ccr.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu J, Stites EC, Yu H, Germino EA, Meharena HS, Stork PJ, et al. Allosteric activation of functionally asymmetric RAF kinase dimers. Cell. 2013;154:1036–46. doi: 10.1016/j.cell.2013.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor SS, Kornev AP. Protein kinases: evolution of dynamic regulatory proteins. Trends in biochemical sciences. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Atefi M, Titz B, Avramis E, Ng C, Wong D, Lassen A, et al. Combination of Pan-RAF and MEK inhibitors in NRAS mutant melanoma. Molecular cancer. 2015;14:27. doi: 10.1186/s12943-015-0293-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lamba S, Russo M, Sun C, Lazzari L, Cancelliere C, Grernrum W, et al. RAF suppression synergizes with MEK inhibition in KRAS mutant cancer cells. Cell reports. 2014;8:1475–83. doi: 10.1016/j.celrep.2014.07.033. [DOI] [PubMed] [Google Scholar]
- 49.Nakamura A, Arita T, Tsuchiya S, Donelan J, Chouitar J, Carideo E, et al. Antitumor activity of the selective pan-RAF inhibitor TAK-632 in BRAF inhibitor-resistant melanoma. Cancer research. 2013;73:7043–55. doi: 10.1158/0008-5472.CAN-13-1825. [DOI] [PubMed] [Google Scholar]
- 50.Yaeger R, Cercek A, O’Reilly EM, Reidy DL, Kemeny N, Wolinsky T, et al. Pilot trial of combined BRAF and EGFR inhibition in BRAF-mutant metastatic colorectal cancer patients. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:1313–20. doi: 10.1158/1078-0432.CCR-14-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schellens JHM, van Geel RBJC, Spreafico A, Schuler M, Yoshino T, Delord J-P, Yamada Y, Lolkema MP, Faris JE, Eskens FALM, Sharma S, Yaeger R, Lenz H-J, Wainberg ZA, Avsar E, Chatterjee A, Jaeger S, Demuth T, Tabernero J. Final biomarker analysis of the phase I study of the selective BRAF V600 inhibitor encorafenib (LGX818) combined with cetuximab with or without the α-specific PI3K inhibitor alpelisib (BYL719) in patients with advanced BRAF-mutant colorectal cancer. Proceedings of the Annual Meeting of the American Association for Cancer Research; 2015; Philadelphia: AACR; 2015. [Google Scholar]
- 52.Sharfman WHFSH, Lawrence DP, Flaherty KT, Amaravadi RK, Kim KB, Dummer R, Gobbi S, Puzanov I, Sosman JA, Dohoney K, Lam LP, Kakar S, Tang Z, Krieter O, Atkins MB. Results from the first-in-human (FIH) phase I study of the oral RAF inhibitor RAF265 administered daily to patients with advanced cutaneous melanoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:abstr 8508. [Google Scholar]
- 53.Girotti MR, Lopes F, Preece N, Niculescu-Duvaz D, Zambon A, Davies L, et al. Paradox-breaking RAF inhibitors that also target SRC are effective in drug-resistant BRAF mutant melanoma. Cancer cell. 2015;27:85–96. doi: 10.1016/j.ccell.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okaniwa M, Hirose M, Arita T, Yabuki M, Nakamura A, Takagi T, et al. Discovery of a selective kinase inhibitor (TAK-632) targeting pan-RAF inhibition: design, synthesis, and biological evaluation of C-7-substituted 1,3-benzothiazole derivatives. Journal of medicinal chemistry. 2013;56:6478–94. doi: 10.1021/jm400778d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.