Abstract
Purpose
To spatially map human lens Aquaporin-0 (AQP0) protein modifications, including lipidation, truncation, and deamidation, from birth through middle age using matrix-assisted laser desorption ionization (MALDI) imaging mass spectrometry (IMS).
Methods
Human lens sections were water-washed to facilitate detection of membrane protein AQP0. We acquired MALDI images from eight human lenses ranging in age from 2 months to 63 years. In situ tryptic digestion was used to generate peptides of AQP0 and peptide images were acquired on a 15T Fourier transform ion cyclotron resonance (FTICR) mass spectrometer. Peptide extracts were analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) and database searched to identify peptides observed in MALDI imaging experiments.
Results
Unmodified, truncated, and fatty acid–acylated forms of AQP0 were detected in protein imaging experiments. Full-length AQP0 was fatty acid acylated in the core and cortex of young (2- and 4-month) lenses. Acylated and unmodified AQP0 were C-terminally truncated in older lens cores. Deamidated tryptic peptides (+0.9847 Da) were mass resolved from unmodified peptides by FTICR MS. Peptide images revealed differential localization of un-, singly-, and doubly-deamidated AQP0 C-terminal peptide (239–263). Deamidation was present at 4 months and increases with age. Liquid chromatography–MS/MS results indicated N246 undergoes deamidation more rapidly than N259.
Conclusions
Results indicated AQP0 fatty acid acylation and deamidation occur during early development. Progressive age-related AQP0 processing, including deamidation and truncation, was mapped in human lenses as a function of age. The localization of these modified AQP0 forms suggests where AQP0 functions may change throughout lens development and aging.
Keywords: ocular lens, MALDI IMS, deamidation, truncation, posttranslational modifications, in situ digestion, aging
The ocular lens is a transparent optical element, focusing light onto the retina for clear vision. Tight cellular packing, lack of light-scattering organelles, and a careful balance of protein and water concentration ensure lens transparency and provide focusing properties.1–3 Epithelial cells on the lens anterior surface differentiate into lens fiber cells, such that new fiber cells are continuously added to the lens cortex atop older fiber cells in the lens core, creating concentric rings that reflect cellular age.4 Lens fibers mature through a process of elongation, cellular compaction, and removal of organelles. Mature fiber cells in the core experience no cellular turnover and they are devoid of light-scattering nuclei as well as other organelles.5 Without machinery for cellular turnover or protein synthesis, long-lived cells and their contents survive for decades in the lens.6 Moreover, aging lens proteins are subject to modification over time. Some posttranslational modifications (PTMs), like phosphorylation, are programmed to occur in a specific region of the lens, while truncation accumulates in a time-dependent manner with age.7 In the lens, PTMs may alter protein function by modulating existing protein function when no new protein is produced.
One essential, yet extensively modified, protein is the lens major intrinsic protein (MIP), or aquaporin-0 (AQP0). Aquaporin-0 is a dual-function water channel and adhesion molecule comprising approximately 50% of lens membrane protein.8 The central pore of each AQP0 monomer shuttles water across the plasma membrane.9 Aquaporin-0 also is involved in junction formation and cell-to-cell adhesion.10–16 Mutations or loss of AQP0 lead to cataract, highlighting the importance of this protein for lens function.17–28
Aquaporin-0 is a 28 kD protein comprised of six transmembrane domains and an intracellular C-terminal tail. The highly modified tail regulates AQP0 permeability via calmodulin binding.29–31 This region of AQP0 also binds cytoskeletal proteins, including filensin and phakinin,32 and may have a role in establishing fiber cell organization. Numerous PTMs decorate the AQP0 C-terminus over time, some of which have been functionally characterized. Phosphorylation of Ser-235 reduces the binding affinity of calmodulin, thereby boosting the permeability of the channel.31,33,34 Cleavage of the C-terminal tail removes protein–protein binding regions and may encourage AQP0 to form cell–cell junctions.7,13 More recently, fatty acid modifications were discovered on lysine 238 and on the N-terminal methionine, which may target the protein to detergent-resistant lipid raft regions of the membrane.35,36 Deamidation and isomerization of asparagine residues have been reported but have not been fully characterized.37,38 The localization of these PTMs within the lens can provide clues of the changing physiological role of AQP0 over time.
Matrix-assisted laser desorption ionization (MALDI) imaging mass spectrometry (IMS) has been used to map the spatial localization of modified AQP0 within the lens.35,39 In MALDI IMS, proteins are detected by their mass-to-charge (m/z) value and each m/z value can be mapped across a tissue section with intensity encoded as a heat map.40 Protein modifications are characterized by specific shifts in m/z values.
Previous MALDI experiments showed the localization of truncated and lipid-modified AQP0.35,41 Although informative, these molecular images captured a single time point in the complex aging process. Furthermore, the spatial distribution and accumulation of deamidated AQP0 is unknown.
The aim of this study was to characterize the localization of modified AQP0 in human lens from birth through middle age using MALDI IMS. We optimized methods to image modified AQP0, revealing unprecedented images of deamidated and truncated forms of the functionally-important AQP0 C-terminal tail. Understanding the localization of these modifications in young or old fiber cells can provide clues on how AQP0 function changes during development, aging, and cataract formation.
Materials and Methods
Materials
Proteomics-grade porcine trypsin, methanol, high-performance liquid chromatography (HPLC)-grade acetonitrile, formic acid, sinapinic acid, and α-cyano-4-hydroxycinnamic acid (CHCA) were purchased from Sigma-Aldrich Corp. (St. Louis, MO, USA). Acetic acid, ammonium bicarbonate, and Triton X-100 were from Fisher Scientific (Pittsburgh, PA, USA). Gold-coated MALDI targets were from AB Sciex (Framingham, MA, USA). Frozen human lenses were obtained from the National Disease Research Interchange (Philadelphia, PA, USA) and stored at −80°C until further use. Lenses used in this study were from individuals with no reported ocular pathology. Eight lenses were analyzed in total: 2 (male [M]) and 4 (female [F]) months, and 21 (M), 23 (F), 39 (M), 47 (M), 56 (M), and 63 (F) years old. All other reagents were purchased from Sigma-Aldrich Corp. unless otherwise stated. Human tissues were acquired in compliance with the Declaration of Helsinki.
Tissue Preparation
Frozen human lenses were sectioned to 20-μm thickness using a CM 3050 Cryostat (Leica Microsystems, Bannockburn, IL, USA). Sections were transferred to gold-coated MALDI target plates via methanol soft landing.39 Using a modified protocol,39 sections were pipette washed with double distilled water (ddH2O) for 1 minute, followed by desiccation to dryness after each wash. After 10 water washes, sections were washed three times with 1% formic acid (1 minute), with 30-second water washes in between. Sections were desiccated before matrix application.
MALDI IMS of Lens Membrane Protein AQP0
Matrix was applied using a Portrait Spotter (Labcyte, Sunnyvale, CA, USA). Arrays of matrix droplets were placed with 400-μm spacing and were interleaved for a final droplet spacing of 200 μm. Sinapinic acid matrix (20 mg/mL in 70:29.7:0.3 ACN:H2O:TFA with 0.025% Triton X-100) was spotted, one droplet per pass, for a total of 40 droplets per spot.
We collected MALDI protein imaging data on a Bruker Autoflex Speed II TOF instrument (Bruker Daltonics, Billerica, MA, USA). External calibration was performed using a protein standard mix (equine cytochrome C, equine apomyoglobin, bovine trypsinogen, and rabbit carbonic anhydrase) before data collection. Methods were established in FlexControl software (1 kHz laser, medium spot size laser, 350 ns pulsed ion extraction, m/z 17,000–35,000, 300 laser shots per spot) and images were viewed in FlexImaging software. For images shown, all four human lens sections (4-month, and 21-, 39-, and 56-year) were imaged on one plate, in one data file, and normalized via total ion current (TIC).42 Total ion current normalization calculates the normalized peak intensity for each spectral feature (peak) by dividing its intensity by the total ion current in each mass spectrum across an entire imaging data set, in this case across four lens images. Ion intensities are then displayed across the image by assigning the largest normalized signal for each m/z to a value of 100% and the lowest normalized signal as 0%. Therefore, each ion image is calculated independently from other ion images. For display purposes, data were interpolated and pixel intensities were rescaled using “brightness optimization” in FlexImaging software. This feature optimizes the brightness of each individual signal to use the entire dynamic range, as indicated by the color scale bars. The integrated intensity for each m/z signal (±0.3% m/z units) was plotted as a normalized TIC value. A total of three technical replicates were run to ensure reproducibility of molecular patterns. Triplicate images also were acquired from a second sample set, including 2-month, and 23-, 47-, and 63-year lenses.
In Situ Digestion and MALDI IMS of AQP0 Tryptic Peptides
For peptide imaging, lens sections were washed with water and 1% formic acid as described above and sprayed with trypsin (0.04 μg/μL in 100 mM ammonium bicarbonate, pH 8, with 10% ACN). Trypsin was applied in eight passes by a TM Sprayer (HTX Technologies, Carrboro, NC, USA) modified with a syringe pump at 7.5 μL/min (Harvard Apparatus, Holliston, MA, USA). Digestion was done in a humidified petri dish at 37°C with 0.2 mL ammonium bicarbonate for 2 hours. Incubation conditions were optimized to minimize delocalization and no drying was required before matrix application. We applied CHCA matrix in 4 passes using the TM sprayer (5 mg/mL CHCA in 50:49.7:0.3 ACN:H2O:TFA, 700 mm/min velocity, 2-mm track spacing).
Peptide imaging data were collected on a Bruker SolariX 15T FTICR MS (Bruker Daltonics). Images over the entire peptide mass range (m/z 600–10,000) were collected with 1000 laser shots per pixel with a raster step size of 100 μm. Images were acquired using a Smartbeam II 2kHz Nd:YAG (355 nm) laser using the small laser setting (estimated spot size ∼60 μm). Continuous accumulation of selected ions (CASI)43 imaging also was used to achieve better sensitivity for low-abundance species. For CASI images, a narrow mass range around the signal of interest was selected (1475–3000 for m/z 2550) with a Q1 mass of 2555 and an isolation window of m/z 20. An ICR transient length of 3.35 seconds was used to achieve a resolving power of 250,000 at m/z 2550. The raster step size was 125 μm; 5000 laser shots were acquired per pixel. Ion images were generated in Bruker FlexImaging software. Full mass range data were normalized by TIC and each m/z signal was plotted ± 0.005 m/z units.
Microextraction and nanoLC-MS/MS Analysis for Peptide Identification
Serial lens sections were washed and sprayed with trypsin as described above. Spatially-directed extraction of tryptic peptides was accomplished as described by Schey et al.44 Extracts were collected from the outer and inner cortex of four lenses (4-month, and 21-, 39-, and 56-year). Using a gel-loading pipet tip, 1 μL solvent (20% ACN with 0.1% formic acid, HPLC-grade) was pipetted onto a lens region, aspirated repeatedly, and collected. This microextraction procedure was repeated, and the pooled peptide extract was diluted 10-fold with the same solvent. Half of the extract was bomb-loaded onto a reverse-phase 360 μm outer diameter (o.d.) × 150 μm inner diameter (i.d.) capillary trap column (3 cm length/5 μm Jupiter C18 beads, 300Å; Phenomenex, Torrence, CA, USA) in-line with a 360 μm o.d. × 100 μm i.d. reverse-phase analytical column equipped with a laser-pulled emitter tip and packed with 20 cm of Jupiter C18 beads (3 μm, 300Å; Phenomenex). Using an Eksigent nanoHPLC, peptides were eluted at a flow rate of 500 nL/min over a 90-minute gradient of 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B). The gradient consisted of 2% to 40% B in 64 minutes, 40% to 90% B in 8 minutes, 90% B for 2 minutes, followed by equilibration at 2% B. Gradient-eluted peptides were mass analyzed on an LTQ Velos Orbitrap mass spectrometer with a nanoelectrospray ionization source (Thermo Fisher Scientific, San Jose, CA, USA). The instrument was operated using a data-dependent method. Full scan (m/z 300–2000) spectra were acquired with the Orbitrap (resolution 60,000) using an MS1 AGC target value of 1 × 106 with 100 ms maximum injection time. Dynamic exclusion settings allowed for a repeat count of 1 within a repeat duration of 10 seconds, and the exclusion duration time was set to 15 seconds. The top 16 most abundant ions in each MS scan were selected for fragmentation via collision-induced dissociation (CID) in the LTQ trap. An isolation width of 2 m/z, activation time of 10 ms, and 35% normalized collision energy were used to generate MS2 spectra. Tandem mass spectra were acquired using an MS2 AGC target value of 1 × 104 with 100 ms maximum injection time. An average of 30,000 MS2 spectra were generated from each 90-minute run. For selected LC-MS/MS analyses, the LTQ Orbitrap Velos was operated using a method consisting of targeted scan events, for which specific m/z values corresponding to AQP0 peptides were provided in the data acquisition method to facilitate collection of targeted MS/MS spectra despite the low intensity of peptide precursors. Spectra acquired of AQP0 peptides of interest, including those with nontryptic termini, were inspected using Xcalibur 3.0.63 Qual Browser software (Thermo Fisher Scientific). For manual peptide assignment, peaks in MS/MS spectra with a precursor mass within 5 parts per million (ppm) of the calculated mass were compared to theoretical b- and y-ions. Other considerations included overall signal intensity, good coverage of the entire peptide sequence including modified residue(s), labeling of a majority of fragment ions, and few unattributed peaks of high intensity. For additional peptide identification, tandem mass spectra were converted into DTA files using Scansifter and searched using a custom version of Sequest (Thermo Fisher Scientific) operating on the Vanderbilt ACCRE computing cluster. We searched MS/MS spectra against a concatenated forward and reverse (decoy) database containing the Homo sapiens subset of UniprotKB Sprot protein database (20,360 proteins, available in the public domain at www.uniprot.org). Additional search parameters included: trypsin enzyme specificity, monoisotopic masses were used for searching product ions, and oxidation of methionine and phosphorylation of serine, threonine, and tyrosine were allowed as variable modifications. Scaffold 4.3.4 (Proteome Software, Portland, OR, USA) was used to summarize and validate search results, where a minimum probability threshold of 95% was required for peptide identifications and data were filtered to a false-discovery rate (FDR) of <1% at the protein level. Peptide signals from MALDI IMS (acquired on the FTICR instrument) and LC-MS/MS (acquired on the Orbitrap instrument) experiments were matched by accurate mass (<5 ppm error required) to identify peptides from imaging experiments.
Lens Homogenization and LC-MS/MS for Identifying Lipid-Modified Peptides
Lipid modification of AQP0 C-terminal peptide in 4-month lens was confirmed by LC-MS/MS, using a modified protocol.35 Soluble peptides were extracted from the digested pellet with 0.1% formic acid, then 100% ACN was used to extract hydrophobic peptides from the pellet. The ACN peptide extract was dried down and reconstituted with 5% ACN/0.1% formic acid before LC-MS/MS analysis as described above.
Results
MALDI IMS of Lens Membrane Protein AQP0
We acquired MALDI images of endogenous protein from young and older human lenses to spatially map age-related processing of AQP0. The procedure of Grey et al.39 and Thibault et al.45 was modified by adding a series of 1% formic acid washes to remove crystallins to facilitate AQP0 imaging. The matrix solution also was optimized to include 70% ACN with 0.025% Triton X-100 detergent. This matrix solution generated uniform spots and provided excellent extraction and co-crystallization of AQP0 with matrix (data not shown).
Representative spectra from two human lenses highlight the accumulation of truncated AQP0 with age (Fig. 1). Full-length AQP0 1-263 (m/z 28,127 ± 0.3%) is abundant in the 2-month lens and very few soluble crystallin protein signals (19–23 kDa) are present due to extensive tissue washing. Fatty acid–acylated AQP0 also is observed (m/z 28,386 ± 0.15%). In a 21-year lens, full-length AQP0 signal decreases, while lower m/z signals appear due to C-terminal cleavage. Truncated AQP0 corresponding to 1-259 (m/z 27,704 ± 0.15%), 1-246 (m/z 26,364 ± 0.3%), and 1-243 (m/z 26,056 ± 0.15%) could be detected in all lenses except 2- and 4-month. Ion images reveal that AQP0 1 to 263 is present throughout the 4-month lens but only in the very outer cortex of the older lenses (Fig. 2A; Supplementary Fig. S1). In the older lens cores, major C-terminal cleavage products are abundant (Figs. 2B–D). In agreement with previous reports, truncation at residue 259 is most intense in the outer cortex, while truncated products 1-246 and 1-243 are most abundant in the central core. The absence of signal from the 56-year lens core is likely due to further truncation and other posttranslational modifications, resulting in a heterogeneous population of AQP0 forms. We repeated MALDI IMS experiments on another set of human lenses, ranging from 2 months to 63 years, and the observed truncation patterns are very similar to these results (data not shown).
Figure 1.
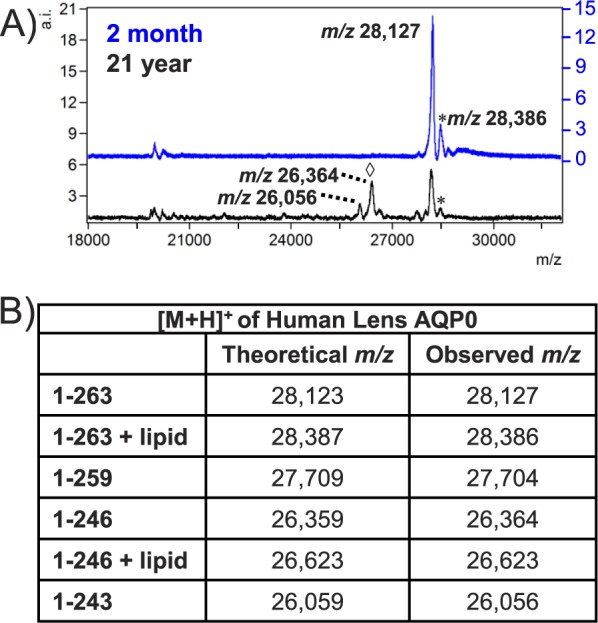
Representative MALDI-TOF spectra from 2-month and 21-year human lens. (A) The major signal in the 2-month lens is full-length AQP0 1-263. Lipid-modified AQP0 (*) is present in both lenses, while major truncation peaks (◊) are apparent only in the 21-year lens. Spectra were generated from regions of interest including the cortex and core of each lens. (B) Theoretical and observed m/z values for modified forms of AQP0. Putative identifications listed are based on accurate mass and previous publications.39 a.i., arbitrary intensity.
Figure 2.
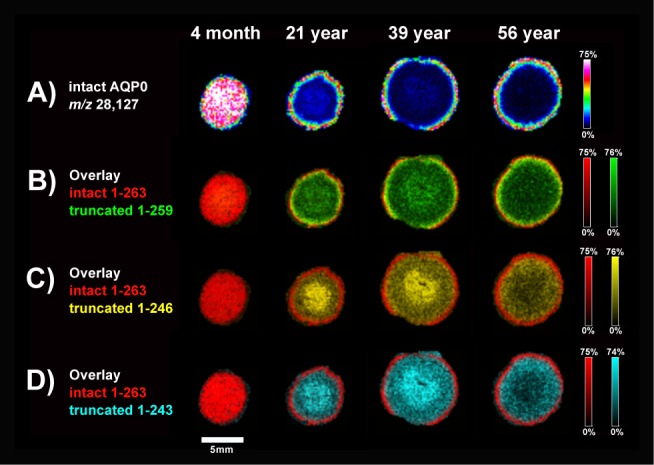
Imaging of AQP0 protein demonstrates age-related truncation. (A) Full-length AQP0 1-263 signal is lost due to extensive C-terminal truncation in older lens cores. Accumulation of 1-259 (B), 1-246 (C), and 1-243 (D) truncated AQP0 is apparent in older lens cores. Images were acquired with 200 μm raster step size on a Bruker Autoflex TOF. Ion intensities are normalized to the TIC for each ion across the tissue. Color scale bars indicate the range of intensities plotted.
Fatty acid–acylated AQP0 was detected in all lenses (Fig. 3; Supplementary FiSupplementary Fig. S2). In the 4-month lens, full-length, fatty acid–acylated AQP0 was present in the lens cortex and core. This finding represents the youngest lens reported to contain lipid-modified AQP0 and suggests that AQP0 lipidation is an early processing event. The presence of acylated AQP0 was confirmed by LC-MS/MS in the 4-month lens (Supplementary Fig. S3). Older lenses contained full-length (1-263 + lipid) and truncated acylated AQP0 (1-246 + lipid), depending on fiber cell age. The lipid modification detected by MALDI is observed as a fairly broad peak, likely containing a combination of oleic acid (C18:1, +264 Da) and palmitic acid (C16:0, +238 Da) as reported previously.35
Figure 3.
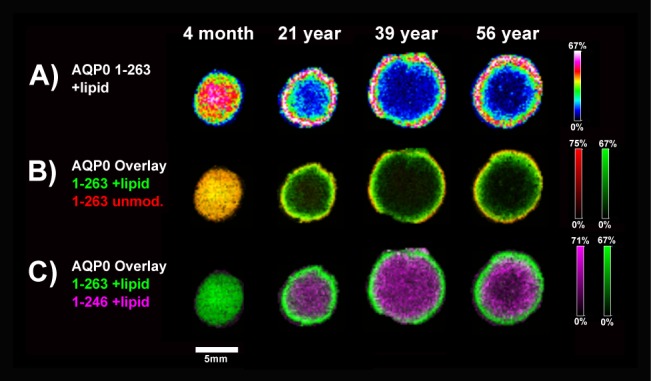
Fatty acid–acylated AQP0 protein. Aquaporin-0 is modified with a fatty acid in young fiber cells (A), with a narrow ring of unmodified AQP0 before the lipid is added (B). The fatty acylated form also is C-terminally truncated, predominantly at residue 246 (C). Images were acquired with 200 μm raster step size on a Bruker Autoflex TOF. Ion intensities are normalized to the TIC for each ion across the tissue. Color scale bars indicate the range of intensities plotted.
In Situ Digestion and MALDI IMS of AQP0 Tryptic Peptides
After extensive washing to remove soluble proteins, lens sections were digested with spray-coated trypsin to generate spatially-localized peptides that reflect endogenous protein distribution. Using a high mass resolution FTICR MS instrument operated in CASI mode, peptides were analyzed by MALDI IMS. Continuous accumulation of selected ions afforded increased sensitivity during high mass resolution (200,000) experiments, enabling resolution of isotopic clusters of deamidated and undeamidated AQP0 peptides. Mass defect differences were used to determine deamidation of AQP0 peptides, as shown for lens crystallins at the peptide level.46
The C-terminal tryptic peptide of AQP0 (239–263) can be deamidated at two asparagine residues, N246 and N259.37 Figure 4B shows the CASI spectrum for this AQP0 peptide from a 4-month lens. The undeamidated 239-263 AQP0 peptide (m/z 2550.274 ± 0.005) was only detected in the outer cortex of a 4-month human lens with low intensity (Fig. 4A). Singly- and doubly-deamidated AQP0 (m/z 2551.257 ± 0.005 and 2552.238 ± 0.005, respectively) could be detected even in a young lens, suggesting age-related deamidation can occur early in developing humans. Note the observed isotopic distribution matches the predicted isotopic distribution at a mass resolving power of approximately 250,000. In older lenses, undeamidated AQP0 cannot be detected by MALDI, and the doubly-deamidated peptide is most abundant (Fig. 5; Supplementary Fig. S4). Overlaid ion images show concentric rings of singly- and doubly-deamidated AQP0, while truncated AQP0 is localized more centrally in older lenses, suggesting deamidation precedes truncation. These unprecedented MALDI images acquired at 250,000 resolving power show early and progressive deamidation and truncation of AQP0.
Figure 4.
Deamidation of C-terminal AQP0 peptide 239-263. (A) Undeamidated AQP0 is present in the very young outer cortical fiber cells of a 4-month lens. Deamidation is more abundant in the lens core. Images were acquired with 125-μm raster step size using FTICR MS. All m/z values are plotted ± 0.005 m/z. Ion intensities are normalized to the TIC for each ion across the tissue. Color scale bars indicate the range of intensities plotted. (B) Continuous accumulation of selected ions FTICR spectrum showing un-, singly-, and doubly-deamidated AQP0 peptide. (C) Calculated and observed m/z values for AQP0 C-terminal peptide deamidation. Reported full scan and CASI m/z values were acquired on a Bruker SolariX 15T FTICR MS.
Figure 5.

Age-related accumulation of deamidation and truncation. Singly- and doubly-deamidated AQP0 peptide are visible in concentric rings in older lenses. Slightly older fiber cells contain deamidated peptide that has been truncated at residue 259. Images were acquired with 100-μm raster step size on a Bruker SolariX 15T FTICR MS. Ion intensities are normalized to the TIC for each ion across the tissue. Color scale bars indicate the range of intensities plotted.
Microextraction and nanoLC-MS/MS Analysis for Peptide Identification
Liquid chromatography-MS/MS analysis was performed to ensure the correct identification of AQP0 peptides detected in MALDI imaging experiments. Using high mass resolution instrumentation, un-, singly-, and doubly-deamidated AQP0 239-263 peptides can be distinguished by a shift in molecular weight. High-resolution tandem mass spectra were acquired to confirm the deamidation sites. Figure 6 shows a labeled MS2 spectrum for the doubly-charged, singly-deamidated peptide, m/z 1276.13, confirming N246 as the deamidation site. Both N246 and N259 were modified in the doubly-deamidated peptide (data not shown). The MALDI images show the singly-deamidated peptide is present in younger fiber cells, followed by the doubly-deamidated peptide in slightly older fiber cells. Together with LC-MS/MS data, these results indicated N246 is deamidated earlier than N259. This finding agrees with predicted rates of deamidation based on primary sequence; N246 is followed by a small glycine residue, whereas N259 is adjacent to a larger threonine residue.47 Supplementary Fig. S5 shows the accumulation of deamidated and truncated AQP0 with fiber cell age, plotted as peak areas from LC-MS/MS data of a 21-year lens. These results agree with imaging data (Fig. 5), revealing early deamidation followed by truncation.
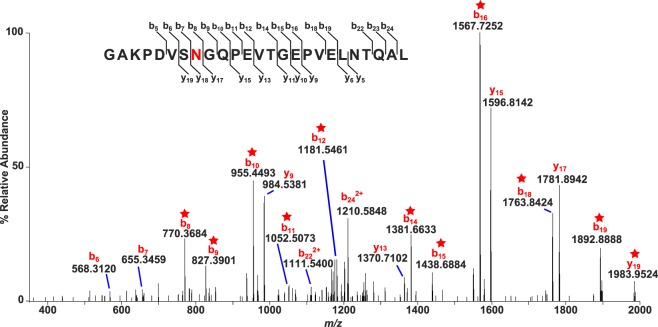
Figure 6.
Tandem mass spectrum identifying deamidation site at N246. High mass resolution CID spectrum of doubly-charged AQP0 peptide 239-259, m/z 1276.13, acquired on a Thermo LTQ Velos Orbitrap. The observed mass corresponds to one deamidated residue on the peptide; starred peaks confirmed deamidation at residue 246. The amino acid sequence showing fragmentation coverage is included above.
Discussion
The aim of this study was to image human AQP0 from birth through middle age to provide a spatially accurate depiction of AQP0 aging over time. We imaged four lenses in one experiment, ranging from 2 months to 63 years, which allowed a comparison of protein changes within old cells in the lens core (which is formed during embryonic development and contains cells as old as the individual) and within newly-differentiated fiber cells in the lens cortex. Protein imaging revealed fatty acid–acylated and truncated AQP0 patterns. In situ digestion and peptide imaging enabled unprecedented high mass resolution imaging of deamidated AQP0 peptides. Together with LC-MS/MS analysis, these data reveal time-dependent deamidation as it relates to truncation in the human lens.
Extensive AQP0 truncation is detected in the inner cortex and nucleus of all lenses except the youngest, 2- and 4-month lenses (Figs. 1, 2). Our results agree with published data, indicating approximately half of AQP0 is truncated by a cellular age of 24 years.36 Cleavage of AQP0 is expected to be nonenzymatic, as there is little enzymatic activity in mature fiber cells and truncation at asparagine residues likely proceeds via a well-studied backbone cleavage reaction.48 Furthermore, in vitro studies demonstrated spontaneous cleavage at N246 and N259 after peptide incubation at 37°C in ammonium bicarbonate buffer.38 Although most AQP0 is fated for truncation, the functional consequences of this modification are disputed. Crystals formed from nuclear AQP0, which is mostly truncated, tended to form bilayers.13 Treatment of AQP0 with proteolytic enzymes generates more junctional AQP013 and structural characterization suggests the truncated junctional form has a closed water pore.49 However, functional studies demonstrated equal permeability of full-length and truncated AQP0,11,50 and showed full-length AQP0 has adhesive properties.12 Another possible consequence of C-terminal truncation is that key residues involved in AQP0 protein–protein interactions are removed. Perhaps AQP0 binding with cytoskeletal proteins filensin, phakinin, and ezrin are no longer required in deep cortical cells since the fiber cell architecture already has been established in the outer cortex.7,32,51 Removal of the AQP0 C-terminus could also permit binding of increasingly insoluble proteins and complexes to the plasma membrane, potentially contributing to the lens permeability barrier and hardening around age 50.7,52 Recent studies conclude that truncation of AQP0 termini could help establish a proper refractive index gradient across the lens.11,53 Although the precise function of AQP0 truncation remains to be determined, it is remarkable that such extensive cleavage occurs even in healthy lenses at relatively young ages. Labile asparagine residues could serve as a molecular clock for predictable age-related truncation, as a means of modulating AQP0 function in older fiber cells.
Imaging results show fatty acid–acylated AQP0 in all lenses, including young (2-month) cortex and core (Fig. 3). Previous MALDI profiling and imaging experiments revealed full-length (1-263) and truncated (1-246) fatty acid–acylated AQP0 in an 11-year lens,35,36 but other studies did not detect lipidated AQP0 in fetal lens.7 Our discovery of lipid-modified AQP0 in a very young lens suggests acylation may be developmentally programmed in younger fiber cells and not simply a consequence of AQP0 coexisting with membrane lipids for years. Our results suggested AQP0 is acylated in primary fiber cells of the fetal nucleus and in newly-synthesized AQP0 of secondary fibers. Over time, the fatty acid modification is observed on truncated AQP0, suggesting fatty acylation is an irreversible posttranslational modification in lens. The mechanism for fatty acid acylation in lens is unknown and no acyl transferases have been discovered that attach lipids to lysine residues. Lipid-modified AQP0 was shown to purify with detergent-resistant, or lipid-raft, regions of the plasma membrane.35 We hypothesize that the presence of a large fatty acid on the N- or C-terminus of AQP0 could help anchor the protein to the plasma membrane or target the protein to lipid raft regions for specific protein–protein interactions. Localization of AQP0 within the plasma membrane changes based on distance into the lens, starting with even distribution throughout the plasma membrane and eventually forming ribbon-like structures on the broad sides of fiber cells.54 The mechanism of AQP0 redistribution remains to be determined.
For the first time to our knowledge, deamidated forms of a protein were resolved and imaged by MALDI FTICR IMS (Figs. 4, 5). Deamidation is a common protein modification, resulting in an extremely small mass shift (+0.9847 Da), and is considered a molecular clock that reflects protein age. If deamidated and undeamidated peptides are present in a sample, it is challenging to mass resolve the monoisotopic form of the deamidated peptide from the first 13C isotope (+1.0034 Da) of the undeamidated peptide. To overcome this issue, we performed in situ digestion directly on lens sections to generate spatially-localized tryptic peptides and used a high mass resolution FTICR for imaging analysis (∼250,000 resolving power at m/z 2550). Tryptic digestion on tissue sections has been used to profile and image peptides from formalin-fixed, paraffin-embedded (FFPE) and frozen tissue sections.55–57
Based on AQP0 peptide images, differentially deamidated AQP0 forms have distinct, age-specific spatial localization within the lens. As shown in Figure 4, the 4-month lens contained un-, singly, and doubly-deamidated AQP0 peptides. These peptides form concentric rings in the lens as deamidation accumulates with increasing fiber cell age. Undeamidated AQP0 was detected only in the 4-month lens by MALDI. Figure 5 highlights the accumulation of deamidation and then truncation in older lenses. Loss of truncated, doubly-deamidated AQP0 peptide is likely due to further truncation, as observed in AQP0 protein images. It is noteworthy that the major truncation sites of AQP0 occur at residues 246 and 259 in AQP0; asparagine residues known to be deamidated in AQP0 and consistent with a nonenzymatic mechanism of cleavage. Singly-deamidated peptides contain N246D, indicating deamidation occurs more rapidly at N246 than at N259. Furthermore, it is unlikely that the observed deamidation is due to sample preparation because undeamidated AQP0 still is detected in the outer cortex of the 4-month lens and deamidated forms of AQP0 have unique spatial localization.
Asparagine residues can spontaneously form a cyclic succinimide intermediate, which can produce iso- or aspartic acid through loss of NH3.48 This pathway also can lead to nonenzymatic truncation, as described for crystallins58 and for AQP0 in vitro.38 Deamidation rates can be reliably predicted based on protein primary and secondary structure and other factors, including pH, temperature, and time, also dictate the rate of formation. Asparagine residues followed by small amino acids are rapidly deamidated, whereas larger residues provide steric hindrance and slow deamidation rates considerably.47 In AQP0, we observed N246 (sequence SNG) to be deamidated more rapidly than N259 (sequence LNT). Previous work also indicates truncation is more common at slowly-deamidating residues.59 In AQP0, residue 259 undergoes deamidation more slowly than 246 and truncation at residue 259 is the first to accumulate based on our imaging results.
Conclusions
We combined protein imaging and tryptic peptide imaging of a wide age range of human lenses to spatially characterize AQP0 processing over time. To our knowledge, our results are the first to show that AQP0 is fatty acid acylated in a very young lens nucleus. For the first time, we imaged deamidated tryptic peptides and showed the time course of AQP0 deamidation in human lens, which begins at a very young age. Spatial mapping of modified forms of AQP0 shows where AQP0 function is likely to change across the human lens.
Supplementary Material
Acknowledgments
The authors thank Salisha Hill, MS, and Amanda Hachey, MS, for their technical support.
Supported by National Institutes of Health (NIH; Bethesda, MD, USA) Grants T32-EY07135, EY-13462, P30-EY08126, 5P41-GM103391-05, 1S10OD012359-01, and S10RR027714. Tissues were procured by the National Disease Research Interchange (NDRI) with support from NIH Grant 3U42OD011158.
Disclosure: J.L. Wenke, None; K.L. Rose, None; J.M. Spraggins, None; K.L. Schey, None
References
- 1. Mathias RT,, Rae JL,, Baldo GJ. Physiological properties of the normal lens. Physiol Rev. 1997; 77: 21–50. [DOI] [PubMed] [Google Scholar]
- 2. Jacob TJ. The relationship between cataract cell swelling and volume regulation. Prog Retin Eye Res. 1999; 18: 223–233. [DOI] [PubMed] [Google Scholar]
- 3. Wride MA. Lens fibre cell differentiation and organelle loss: many paths lead to clarity. Philos Trans R Soc Lond B Biol Sci. 2011; 366: 1219–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Taylor VL,, al-Ghoul KJ,, Lane CW,, Davis VA,, Kuszak JR,, Costello MJ. Morphology of the normal human lens. Invest Ophthalmol Vis Sci. 1996; 37: 1396–1410. [PubMed] [Google Scholar]
- 5. Bassnett S. Lens organelle degradation. Exp Eye Res. 2002; 74: 1–6. [DOI] [PubMed] [Google Scholar]
- 6. Lynnerup N,, Kjeldsen H,, Heegaard S,, Jacobsen C,, Heinemeier J. Radiocarbon dating of the human eye lens crystallines reveal proteins without carbon turnover throughout life. PLoS One. 2008; 3: e1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Korlimbinis A,, Berry Y,, Thibault D,, Schey KL,, Truscott RJW. Protein aging: truncation of aquaporin 0 in human lens regions is a continuous age-dependent process. Exp Eye Res. 2009; 88: 966–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alcala J,, Lieska N,, Maisel H. Protein composition of bovine lens cortical fiber cell membranes. Exp Eye Res. 1975; 21: 581–595. [DOI] [PubMed] [Google Scholar]
- 9. Kushmerick C,, Rice SJ,, Baldo GJ,, Haspel HC,, Mathias RT. Ion water and neutral solute transport in Xenopus oocytes expressing frog lens MIP. Exp Eye Res. 1995; 61: 351–362. [DOI] [PubMed] [Google Scholar]
- 10. Costello MJ,, McIntosh TJ,, Robertson JD. Distribution of gap junctions and square array junctions in the mammalian lens. Invest Ophthalmol Vis Sci. 1989; 30: 975–989. [PubMed] [Google Scholar]
- 11. Sindhu Kumari S, Varadaraj K. Intact and N- or C-terminal end truncated AQP0 function as open water channels and cell-to-cell adhesion proteins: end truncation could be a prelude for adjusting the refractive index of the lens to prevent spherical aberration. Biochim Biophys Acta. 2014; 1840: 2862–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kumari SS,, Varadaraj K. Intact AQP0 performs cell-to-cell adhesion. Biochem Biophys Res Commun. 2009; 390: 1034–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gonen T,, Cheng Y,, Kistler J,, Walz T. Aquaporin-0 membrane junctions form upon proteolytic cleavage. J Mol Biol. 2004; 342: 1337–1345. [DOI] [PubMed] [Google Scholar]
- 14. Fotiadis D,, Hasler L,, Muller DJ,, Stahlberg H,, Kistler J,, Engel A. Surface tongue-and-groove contours on lens MIP facilitate cell-to-cell adherence. J Mol Biol. 2000; 300: 779–789. [DOI] [PubMed] [Google Scholar]
- 15. Colom A,, Casuso I,, Boudier T,, Scheuring S. High-speed atomic force microscopy: cooperative adhesion and dynamic equilibrium of junctional microdomain membrane proteins. J Mol Biol. 2012; 423: 249–256. [DOI] [PubMed] [Google Scholar]
- 16. Simon SA,, Zampighi G,, McIntosh TJ,, Costello MJ,, Ting-beall HP,, Robertson JD. The structure of junctions between lens fiber cells. Biosci Rep. 1982; 2: 333–341. [DOI] [PubMed] [Google Scholar]
- 17. Kumari SS,, Gandhi J,, Mustehsan MH,, Eren S,, Varadaraj K. Functional characterization of an AQP0 missense mutation R33C, that causes dominant congenital lens cataract, reveals impaired cell-to-cell adhesion. Exp Eye Res. 2013; 116: 371–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Varadaraj K,, Kumari SS,, Patil R,, Wax MB,, Mathias RT. Functional characterization of a human aquaporin 0 mutation that leads to a congenital dominant lens cataract. Exp Eye Res. 2008; 87: 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu S,, Wang B,, Qi Y,, Lin H. The Arg233Lys AQP0 mutation disturbs aquaporin0-calmodulin interaction causing polymorphic congenital cataract. PLoS One. 2012; 7: e37637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shiels A,, Bassnett S,, Varadaraj K,, et al. Optical dysfunction of the crystalline lens in aquaporin-0-deficient mice. Physiol Genomics. 2001; 7: 179–186. [DOI] [PubMed] [Google Scholar]
- 21. Shiels A,, Mackay D,, Bassnett S,, Al-Ghoul K,, Kuszak J. Disruption of lens fiber cell architecture in mice expressing a chimeric AQP0-LTR protein. FASEB J. 2000; 14: 2207–2212. [DOI] [PubMed] [Google Scholar]
- 22. Shentu X,, Miao Q,, Tang X,, Yin H,, Zhao Y. Identification and functional analysis of a novel MIP gene mutation associated with congenital cataract in a Chinese family. PLoS One. 2015; 10: e0126679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Song Z,, Wang L,, Liu Y,, Xiao W. A novel nonsense mutation in the MIP gene linked to congenital posterior polar cataracts in a Chinese family. PLoS One. 2015; 10: e0119296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu Y,, Chen P,, Li J,, Zhu Y,, Zhai Y,, Yao K. A novel MIP gene mutation associated with autosomal dominant congenital cataracts in a Chinese family. BMC Med Genet. 2014; 15: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Francis P,, Berry V,, Bhattacharya S,, Moore A. Congenital progressive polymorphic cataract caused by a mutation in the major intrinsic protein of the lens, MIP (AQP0). Br J Ophthalmol. 2000; 84: 1376–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Okamura T,, Miyoshi I,, Takahashi K,, et al. Bilateral congenital cataracts result from a gain-of-function mutation in the gene for aquaporin-0 in mice. Genomics. 2003; 81: 361–368. [DOI] [PubMed] [Google Scholar]
- 27. Gu F,, Zhai H,, Li D,, et al. A novel mutation in major intrinsic protein of the lens gene (MIP) underlies autosomal dominant cataract in a Chinese family. Mol Vis. 2007; 13: 1651–1656. [PubMed] [Google Scholar]
- 28. Senthil KumarG,, Kyle JW,, Minogue PJ,, et al. An MIP/AQP0 mutation with impaired trafficking and function underlies an autosomal dominant congenital lamellar cataract. Exp Eye Res. 2013; 110: 136–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reichow SL,, Clemens DM,, Freites JA,, et al. Allosteric mechanism of water-channel gating by Ca2+-calmodulin. Nat Struct Mol Biol. 2013; 20: 1085–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reichow SL,, Gonen T. Noncanonical binding of calmodulin to aquaporin-0: implications for channel regulation. Structure. 2008; 16: 1389–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rose KML,, Wang Z,, Magrath GN,, Hazard ES,, Hildebrandt JD,, Schey KL. Aquaporin 0-calmodulin interaction and the effect of aquaporin 0 phosphorylation. Biochemistry. 2008; 47: 339–347. [DOI] [PubMed] [Google Scholar]
- 32. Lindsey Rose KM, Gourdie RG, Prescott AR, Quinlan Ra, Crouch RK, Schey KL. The C terminus of lens aquaporin 0 interacts with the cytoskeletal proteins filensin and CP49. Invest Ophthalmol Vis Sci. 2006; 47: 1562–1570. [DOI] [PubMed] [Google Scholar]
- 33. Varadaraj K,, Kumari S,, Shiels A,, Mathias RT. Regulation of aquaporin water permeability in the lens. Invest Ophthalmol Vis Sci. 2005; 46: 1393–1402. [DOI] [PubMed] [Google Scholar]
- 34. Nemeth-Cahalan KL, Hall JE. pH and calcium regulate the water permeability of aquaporin 0. J Biol Chem. 2000; 275: 6777–6782. [DOI] [PubMed] [Google Scholar]
- 35. Schey KL,, Gutierrez DB,, Wang Z,, Wei J,, Grey AC. Novel fatty acid acylation of lens integral membrane protein aquaporin-0. Biochemistry. 2010; 49: 9858–9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gutierrez DB,, Garland D,, Schey KL. Spatial analysis of human lens aquaporin-0 post-translational modifications by MALDI mass spectrometry tissue profiling. Exp Eye Res. 2011; 93: 912–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schey KL,, Little M,, Fowler JG,, Crouch RK. Characterization of human lens major intrinsic protein structure. Invest Ophthalmol Vis Sci. 2000; 41: 175–182. [PubMed] [Google Scholar]
- 38. Ball LE,, Garland DL,, Crouch RK,, Schey KL. Post-translational modifications of aquaporin 0 (AQP0) in the normal human lens: spatial and temporal occurrence. Biochemistry. 2004; 43: 9856–9865. [DOI] [PubMed] [Google Scholar]
- 39. Grey AC,, Chaurand P,, Caprioli RM,, Schey KL. MALDI imaging mass spectrometry of integral membrane proteins from ocular lens and retinal tissue. J Proteome Res. 2009; 8: 3278–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Caprioli RM,, Farmer TB,, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997; 69: 4751–4760. [DOI] [PubMed] [Google Scholar]
- 41. Grey AC,, Li L,, Jacobs MD,, Schey KL,, Donaldson PJ. Differentiation-dependent modification and subcellular distribution of aquaporin-0 suggests multiple functional roles in the rat lens. Differentiation. 2009; 77: 70–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Deininger SO,, Cornett DS,, Paape R,, et al. Normalization in MALDI-TOF imaging datasets of proteins: practical considerations. Anal Bioanal Chem. 2011; 401: 167–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shen M,, Xiang P,, Shi Y,, Pu H,, Yan H,, Shen B. Mass imaging of ketamine in a single scalp hair by MALDI-FTMS. Anal Bioanal Chem. 2014; 406: 4611–4616. [DOI] [PubMed] [Google Scholar]
- 44. Schey KL,, Anderson DM,, Rose KL. Spatially-directed protein identification from tissue sections by top-down LC-MS/MS with electron transfer dissociation. Anal Chem. 2013; 85: 6767–6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thibault DB,, Gillam CJ,, Grey AC,, Han J,, Schey KL. MALDI tissue profiling of integral membrane proteins from ocular tissues. J Am Soc Mass Spectrom. 2008; 19: 814–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Robinson NE,, Lampi KJ,, McIver RT,, et al. Quantitative measurement of deamidation in lens betaB2-crystallin and peptides by direct electrospray injection and fragmentation in a Fourier transform mass spectrometer. Mol Vis. 2005; 11: 1211–1219. [PMC free article] [PubMed] [Google Scholar]
- 47. Robinson NE,, Robinson AB. Deamidation of human proteins. Proc Natl Acad Sci U S A. 2001; 98: 12409–12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Robinson NE,, Robinson AB. Molecular Clocks. Cave Junction, OR: Althouse Press; 2004. [Google Scholar]
- 49. Gonen T,, Sliz P,, Kistler J,, Cheng Y,, Walz T. Aquaporin-0 membrane junctions reveal the structure of a closed water pore. Nature. 2004; 429: 193–197. [DOI] [PubMed] [Google Scholar]
- 50. Ball LE,, Little M,, Nowak MW,, Garland DL,, Crouch RK,, Schey KL. Water permeability of C-terminally truncated aquaporin 0 (AQP0 1-243) observed in the aging human lens. Invest Ophthalmol Vis Sci. 2003; 44: 4820–4828. [DOI] [PubMed] [Google Scholar]
- 51. Wang Z,, Schey KL. Aquaporin-0 interacts with the FERM domain of ERM proteins in the ocular lens. Invest Ophthalmol Vis Sci. 2011; 52: 5079–5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Heys KR,, Friedrich MG,, Truscott RJW. Presbyopia and heat: changes associated with aging of the human lens suggest a functional role for the small heat shock protein α-crystallin, in maintaining lens flexibility. Aging Cell. 2007; 6: 807–815. [DOI] [PubMed] [Google Scholar]
- 53. Kumari SS,, Varadaraj K. Aquaporin 0 plays a pivotal role in refractive index gradient development in mammalian eye lens to prevent spherical aberration. Biochem Biophys Res Commun. 2014; 452: 986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zampighi GA,, Eskandari S,, Hall JE,, Zampighi L,, Kreman M. Micro-domains of AQP0 in lens equatorial fibers. Exp Eye Res. 2002; 75: 505–519. [DOI] [PubMed] [Google Scholar]
- 55. Groseclose MR,, Andersson M,, Hardesty WM,, Caprioli RM. Identification of proteins directly from tissue: in situ tryptic digestions coupled with imaging mass spectrometry. J Mass Spectrom. 2007; 42: 254–262. [DOI] [PubMed] [Google Scholar]
- 56. Casadonte R,, Caprioli RM. Proteomic analysis of formalin-fixed paraffin-embedded tissue by MALDI imaging mass spectrometry. Nat Protoc. 2011; 6: 1695–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stauber J,, MacAleese L,, Franck J,, et al. On-tissue protein identification and imaging by MALDI-Ion mobility mass spectrometry. J Am Soc Mass Spectrom. 2010; 21: 338–347. [DOI] [PubMed] [Google Scholar]
- 58. Voorter CE,, de Haard-Hoekman WA,, van den Oetelaar PJ,, Bloemendal H,, de Jong WW. Spontaneous peptide bond cleavage in aging alpha-crystallin through a succinimide intermediate. J Biol Chem. 1988; 263: 19020–19023. [PubMed] [Google Scholar]
- 59. Geiger T,, Clarke S. Deamidation isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J Biol Chem. 1987; 262: 785–794. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.