Summary
Whole exome sequencing of cutaneous melanoma has led to the detection of P29 mutations in RAC1 in 5–9% of samples, but the role of RAC1 P29 mutations in melanoma biology remains unclear. Using reverse phase protein array analysis to examine the changes in protein/phospho-protein expression, we identified cyclin B1, PD-L1, Ets-1, and Syk as being selectively upregulated with RAC1 P29S expression and downregulated with RAC1 P29S depletion. Using the melanoma patient samples in TCGA, we found PD-L1 expression to be significantly increased in RAC1 P29S patients compared to RAC1 WT as well as other RAC1 mutants. The finding that PD-L1 is upregulated suggests that oncogenic RAC1 P29S may promote suppression of the antitumor immune response. This is a new insight into the biological function of RAC1 P29S mutations with potential clinical implications as PD-L1 is a candidate biomarker for increased benefit from treatment with anti-PD1 or anti-PD-L1 antibodies.
Keywords: melanoma, RAC1, PD-L1, anti-PD-1, immune evasion
Introduction
New efforts in sequencing the exomes of cutaneous melanomas led to the detection of recurrent P29S mutations in Ras-related C3 botulinum toxin substrate (RAC1) in 5–9% of samples, making this the third most frequent activating mutation in sun-exposed melanoma after BRAF V600 and NRAS Q61 mutations (Hodis et al., 2012; Krauthammer et al., 2012). RAC1 belongs to the RHO family of small GTPases, which act as molecular switches that cycle between an active GTP-bound state and an inactive GDP-bound state. RAC proteins induce the formation of membrane ruffles and lamellipodia through their regulation of actin polymerization, making them essential in the maintenance of cell morphology and cell migration (Ridley, 2001). RAC1 also influences cell proliferation and gene transcription. Among the many effector proteins associated with RAC1 are scaffold proteins (Pard6 A,G; IQGAP1,2; Nck1; Cdc42SE1,2; IL1Rap1; Hspc121), serine/threonine kinases (Pak1-3, Map3K11, PrkcA), and the regulatory p85 subunit of PI3K (PIK3R1) (Bustelo et al., 2007).
The proline to serine substitution at codon 29 (RAC1 P29S) is a C>T transition (CCT>TCT), which is consistent with a molecular signature associated with ultraviolet radiation damage (Krauthammer et al., 2012). The RAC1 codon 29 is part of the switch I region and is distinct from the gain-of-function mutations found in RAS isoforms at codons 12 or 61, which lead to defective GTP hydrolysis (Davis et al., 2013). The P29S mutation in the RAC1 protein leads to a fast cycling GTPase, with increased inherent GDP/GTP nucleotide exchange (Davis et al., 2013). Clinically, melanomas with the RAC1 P29 mutation are associated with increased thickness, increased mitotic rate, ulceration, nodular subtype, and nodal disease at diagnosis (Mar et al., 2014). From a structural point of view, it is known that RAC1 effectors use residues within the switch I and switch II regions as the major docking/recognition sites (Bishop and Hall, 2000). Therefore, the P29S mutation in the switch I region may have other effects on signal transduction beyond the observed fast cycling phenotype. There have been limited biochemical studies showing RAC1 P29S to have increased binding to downstream effectors as well as enhanced migration and proliferation (Davis et al., 2013; Krauthammer et al., 2012).
Given the prevalence of RAC1 mutations in melanoma and the relative dearth of knowledge on the mechanism through which RAC1 P29S transforms murine melanocytes (Krauthammer et al., 2012), we examined the signaling pathways associated with RAC1 expression. Through reverse phase protein array (RPPA) analysis, we found that cyclin B1, PD-L1, Ets-1, and Syk were significantly upregulated with RAC1 P29S expression and downregulated with RAC1 P29S depletion. Western blot and flow cytometry analyses revealed a robust increase in PD-L1 specifically with RAC1 P29S expression. Using the Skin Cutaneous Melanoma (SKCM) database in The Cancer Genome Atlas (TCGA), we found PD-L1 expression to be significantly increased in RAC1 P29S compared to RAC1 WT melanoma patients. Thus, our data provide new insight into the biological function of RAC1 P29S mutations as being involved in suppressing the antitumor immune response. Clinically, immunotherapies that target the inhibitory ligand PD-L1 or its receptor PD-1 have shown high response rates of 30–50% in melanoma, many of which are durable (Atkins et al., 2014; Hamid et al., 2013; Topalian et al., 2012; Weber et al., 2013; Wolchok et al., 2013). As PD-L1 is a candidate biomarker for increased likelihood of benefit from these therapies, the RAC1 P29 subset of melanoma patients may derive increased benefit.
Results
RAC1 mutations in melanoma
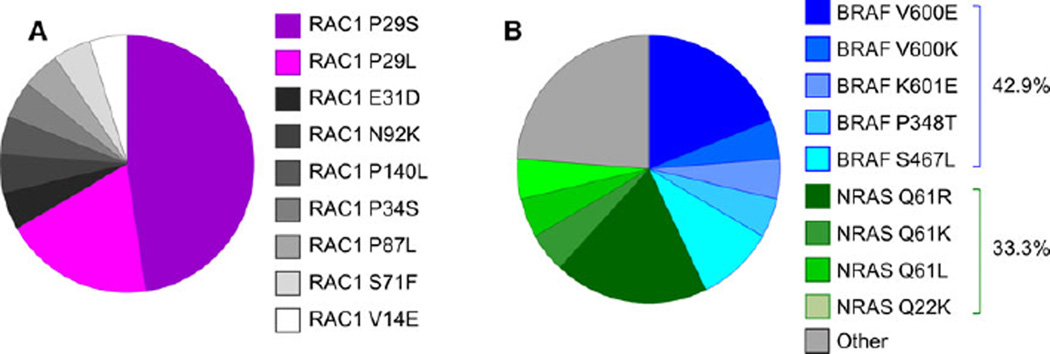
Previous studies have identified RAC1 mutations in 5–9% of cutaneous melanoma samples (Hodis et al., 2012; Krauthammer et al., 2012). In agreement with published studies, the rate of RAC1 mutations was 5.5% (21/382) in the SKCM database from TCGA Research Network (Cerami et al., 2012; Gao et al., 2013). Of these, 47.6% of the mutations are P29S and 19.0% of the mutations are P29L, making a total of 66.6% of the mutations occurring at the P29 codon (Figure 1A). The other mutations identified are V14E, E31D, P34S, S71F, P87L, N92K, and P140L. Interestingly, none of these mutations occur at the Q61 or G12 codon, which are known Ras family activating mutations seen in other cancer types. In the cutaneous melanoma dataset from the TCGA, we found the rate of co-existing BRAF mutation to be 42.9% (9/21) and that of co-existing NRAS mutation to be 33.3% (7/21) (Figure 1B). The rate of BRAF mutations in the entire dataset is 46.3% (177/382) and that of NRAS is 24.9% (95/382). Compared to all melanoma patients, those with the RAC1 mutation have 1.37 the odds of having a co-existing NRAS mutation but only 0.87 the odds of having a co-existing BRAF mutation. Therefore, there is significant overlap between NRAS and BRAF mutations with RAC1 mutations, and the rate of co-existing NRAS mutation is higher than expected.
Figure 1.
Characterization of RAC1 mutants in melanoma. (A) The distribution of RAC1 mutations in the Skin Cutaneous Melanoma (SKCM) database from the TCGA Research Network. (B) The distribution of mutant RAC1 melanomas with co-mutations in BRAF or NRAS.
RAC1 mutants have limited effects on growth
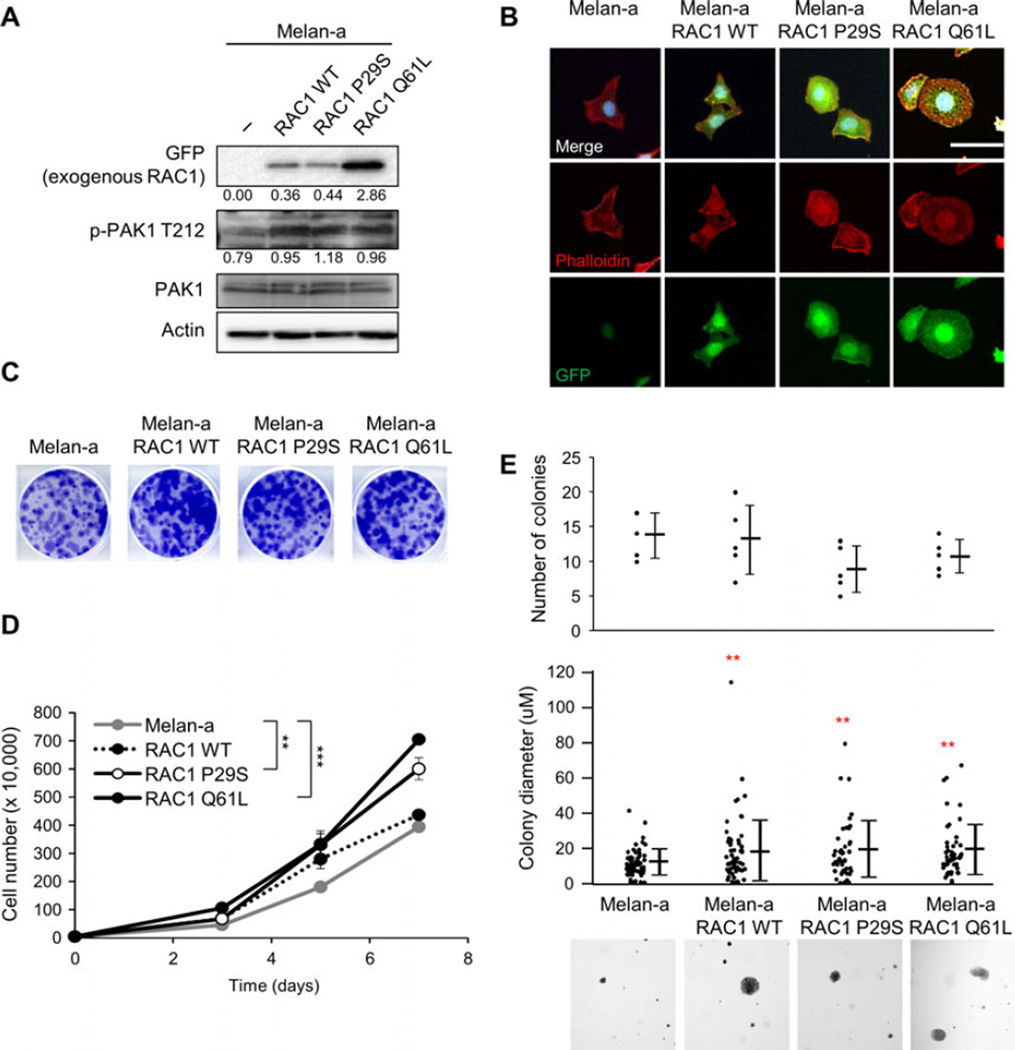
To study the effects of RAC1 expression on cell growth independent of other transforming mutations, we utilized melan-a cells, an immortal murine melanocyte cell line that possesses characteristics of normal melanocytes (Bennett et al., 1987). We used site-directed mutagenesis to introduce a P29S mutation in wild-type (WT) RAC1 with an eGFP tag (Figure S1). The RAC1 WT construct was used as a control for RAC1 overexpression and RAC1 Q61L as a constitutively active form of RAC1. Western blotting showed that the RAC1 constructs were expressed in melan-a (Figure 2A). To determine RAC1 activity, we looked at p21-activated kinase 1 (Pak1), a downstream mediator of RAC1 (Bid et al., 2013). Expression of RAC1 increased levels of phospho-Pak1. Fluorescence staining of the actin cytoskeleton with phalloidin indicated that the expression of RAC1 P29S and Q61L led to more cells with membrane ruffling (Figures 2B and S2).
Figure 2.
RAC1 expression increases growth. (A) Western blot analysis of melan-a cell lines expressing RAC1 WT-eGFP, RAC1 P29S-eGFP, and RAC1-Q61L-eGFP. Quantitation is normalized to actin control. (B) Immunofluorescence image of melan-a, melana RAC1 WT-eGFP, melan-a RAC1 P29S-eGFP, and melan-a RAC1 Q61L-eGFP cells stained for DAPI (blue), phalloidin (red), and GFP (green). Scale bar = 100 µm. (C) Melan-a, melan-a RAC1 WT, melan-a RAC1 P29S, and melan-a RAC1 Q61L cells were plated at low density and cultured for 1 week. Cells were stained with crystal violet, representative images shown. (D) Melan-a, melan-a RAC1 WT (P = 0.069), melan-a RAC1 P29S, and melan-a RAC1 Q61L cell proliferation over 7 days (n = 3; errors bars, SE; **P < 0.01; *** P < 0.001). (E) Soft agar growth assay for melan-a, melan-a RAC1 WT, melan-a RAC1 P29S, and melan-a RAC1 Q61L cells. The number of colonies and size of colonies (in triplicate fields of view) was determined after 4 weeks (n = 3; mean ± SE; **P < 0.01). Representative 4 × images are shown.
Compared to RAC1 WT, both RAC1 P29S and RAC1 Q61L have been shown to increase proliferation (Hodis et al., 2012; Krauthammer et al., 2012). In colony proliferation assays, we noted a similar increase in cell growth with RAC1 Q61L (P < 0.001) > RAC1 P29S (P = 0.001) > RAC1 WT (P = 0.069) compared to parental cells (Figure 2C, D. We examined the effect of RAC1 expression on growth in soft agar, an indicator of the ability of tumor cells to escape the requirement for interactions with the extracellular matrix. There was no difference in the number of colonies that formed, but there was a significant increase in the average colony size with RAC1 expression compared to parental melan-a cells (Figure 2E). In our system, expression of RAC1 led to an increase in cell growth in both anchorage-dependent and anchorage-independent conditions.
RAC1 mutants do not confer resistance to MEK inhibitors
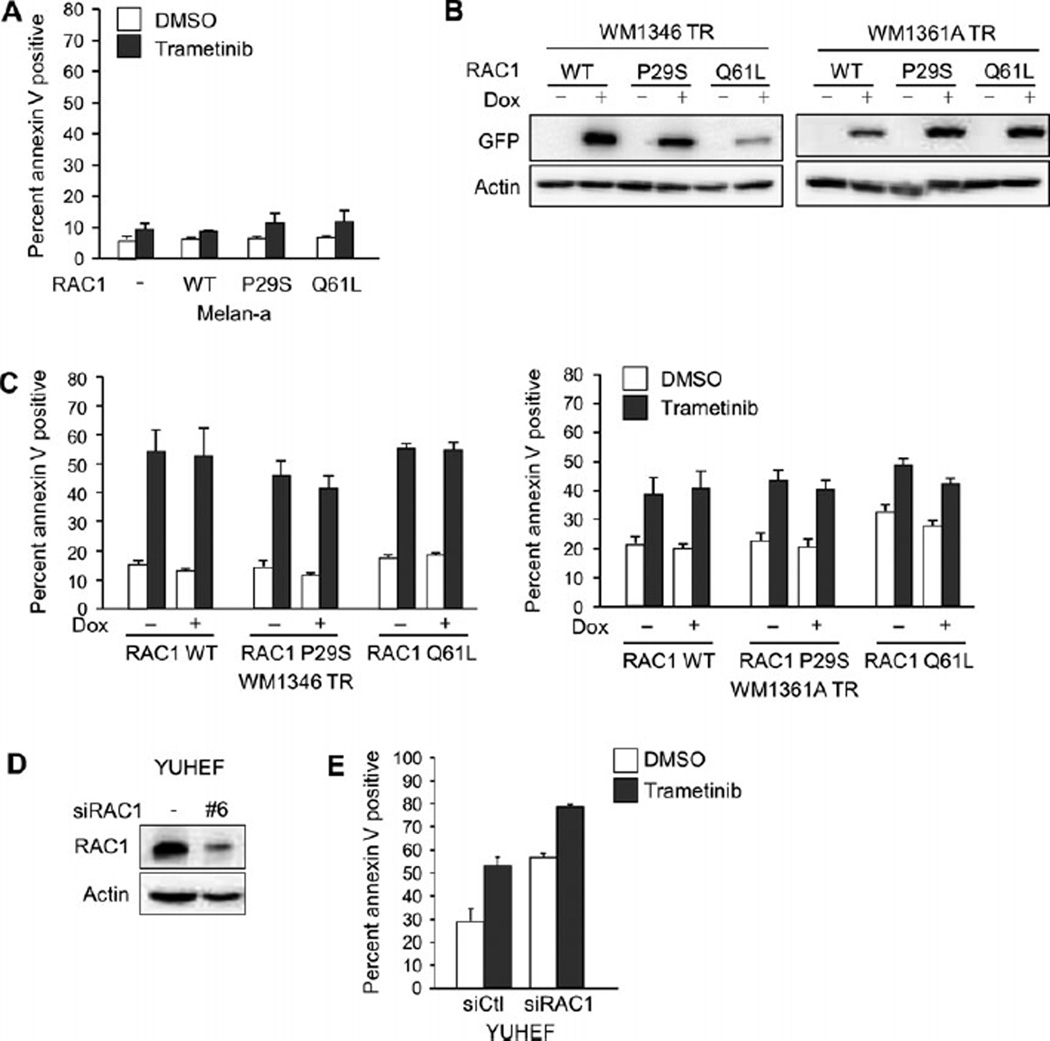
Studies from other groups indicate that RAC1 P29S is a mode of resistance to RAF inhibitors in mutant BRAF melanoma (Watson et al., 2014). It is important to understand how RAC1 mutations alter the response to targeted therapies in subsets of melanomas. We examined the effect of RAC1 expression in response to the MEK inhibitor, trametinib. In 3D culture systems that mimic the dermal microenvironment of melanoma, melan-a cells were resistant to the effects of trametinib, with no significant increase in apoptosis, as measured by levels of the early apoptotic marker annexin V (Figure 3A). The expression of RAC1, either wild type or mutant, did not affect the level of apoptosis induced by trametinib.
Figure 3.
RAC1 expression does not affect sensitivity to MEK inhibition. (A) Melan-a RAC1 WT, melan-a RAC1 P29S, and melan-a RAC1 Q61L cells were cultured in 3D collagen and treated with trametinib (90 nm) for 48 h. Cells were stained with annexin V and analyzed by flow cytometry (n = 3; errors bars, SE). (B) WM1346 TR and WM1361A TR cells with tetracycline-inducible expression of RAC1 WT, RAC1 P29S, and RAC1 Q61L were treated with dox for 72 h. Cells were lysed and lysates analyzed by Western blot. (C) WM1346 TR and WM1361A TR cells with inducible expression of RAC1 WT, RAC1 P29S, and RAC1 Q61L cells were pretreated with dox for 72 h. Cells were then cultured in 3D collagen and treated as in (A). (D) YUHEF cells were transfected with non-targeting or RAC1-targeting siRNA, and after 72 h, cells were lysed and lysates analyzed by Western blot. (E) YUHEF cells were transfected as in (D), cultured in 3D collagen, and treated as in (A).
Given that melan-a cells were insensitive to trametinib, we were not able to test the protective effects of RAC1 mutants in cell death assays. Mutant NRAS melanoma cells have varied response to MEK inhibition (Solit et al., 2006; Vu and Aplin, 2014). Therefore, we generated two mutant NRAS cell lines with inducible expression of RAC1 WT, RAC1 P29S or RAC1 Q61L (Figure 3B). In 3D culture systems, the two mutant NRAS human melanoma cell lines, WM1346 TR and WM1361A TR, were sensitive to trametinib, as indicated by increased annexin V positivity following trametinib treatment (Figure 3C). Inducible expression of RAC1 WT, P29S, or Q61L did not significantly change the degree of apoptosis induced by trametinib (Figure 3C). In YUHEF cells, a melanoma cell line wild type for both BRAF and NRAS with an endogenous RAC1 P29S mutation, we depleted endogenous RAC1 by siRNA transfection (Figure 3D) to determine whether RAC1 loss sensitized the cells to MEK inhibition. In response to trametinib, YUHEF cells had a 23.9% increase in apoptosis (Figure 3E). RAC1 depletion increased apoptosis in YUHEF cells, and this was further enhanced by 21.8% following addition of trametinib. The similar increase in trametinib-induced apoptosis in the absence and presence of RAC1 depletion indicated that RAC1 depletion did not increase sensitivity to trametinib. In sum, RAC1 does not confer resistance to MEK inhibition.
PD-L1 expression is specific to RAC1 P29S signature
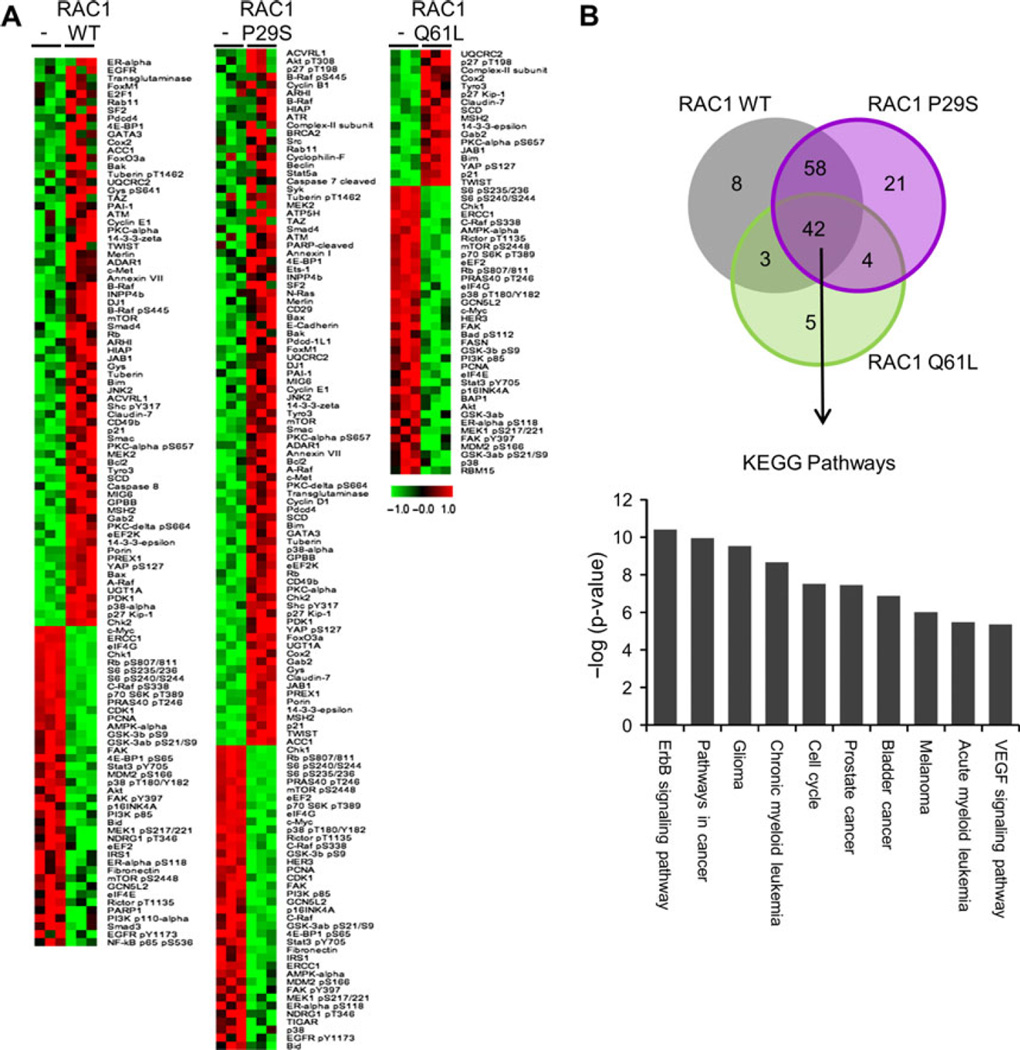
Next, we utilized a high-throughput antibody-based RPPA platform (Tibes et al., 2006) to identify proteins and signaling pathways regulated by RAC1 expression. The RPPA analysis utilized 218 validated antibodies. A significance analysis of microarrays (SAM) identified several proteins that were differentially expressed with each RAC1 construct (Figure 4A). With RAC1 WT expression, there were 71 proteins that were upregulated and 40 that were downregulated; with RAC1 P29S expression, 87 were upregulated and 38 downregulated; and with RAC1 Q61L expression, 17 were upregulated and 36 were downregulated. There were 42 different proteins and phospho-proteins that were similarly regulated with the expression of the RAC1 WT, RAC1 P29S, and RAC1 Q61L compared to parental melan-a cells (Figure 4B). KEGG pathway analysis (DAVID software) of the up- and downregulated proteins showed an enrichment of pathways involved in various cancers, including melanoma, and cell cycle regulation (Figure 4C). The enrichment of pathways involved in cell cycle and cancer signaling likely reflects the increase in cell growth seen with RAC1 expression (Figure 2C–E). The majority of the up- or downregulated proteins were shared in common between or among RAC1 WT, RAC1 P29S, and RAC1 Q61L. However, several proteins were uniquely regulated, specifically 8 with RAC1 WT, 21 with RAC1 P29S, and 5 with RAC1 Q61L (Figure 4B).
Figure 4.
Signaling pathways modulated by RAC1 mutants. (A) Significance analysis of microarray for RPPA of parental melan-a cells and melan-a cells expressing RAC1 WT, RAC1 P29S, and RAC1 Q61L. Expression was normalized to the mean. (B) Venn diagram depicting number of proteins significantly regulated in, between, and among melan-a cells expressing RAC1 WT, RAC1 P29S, and RAC1 Q61L. (C) Top ten KEGG pathways enriched in the 42 proteins similarly regulated by RAC1 WT, RAC1 P29S, and RAC1 Q61L.
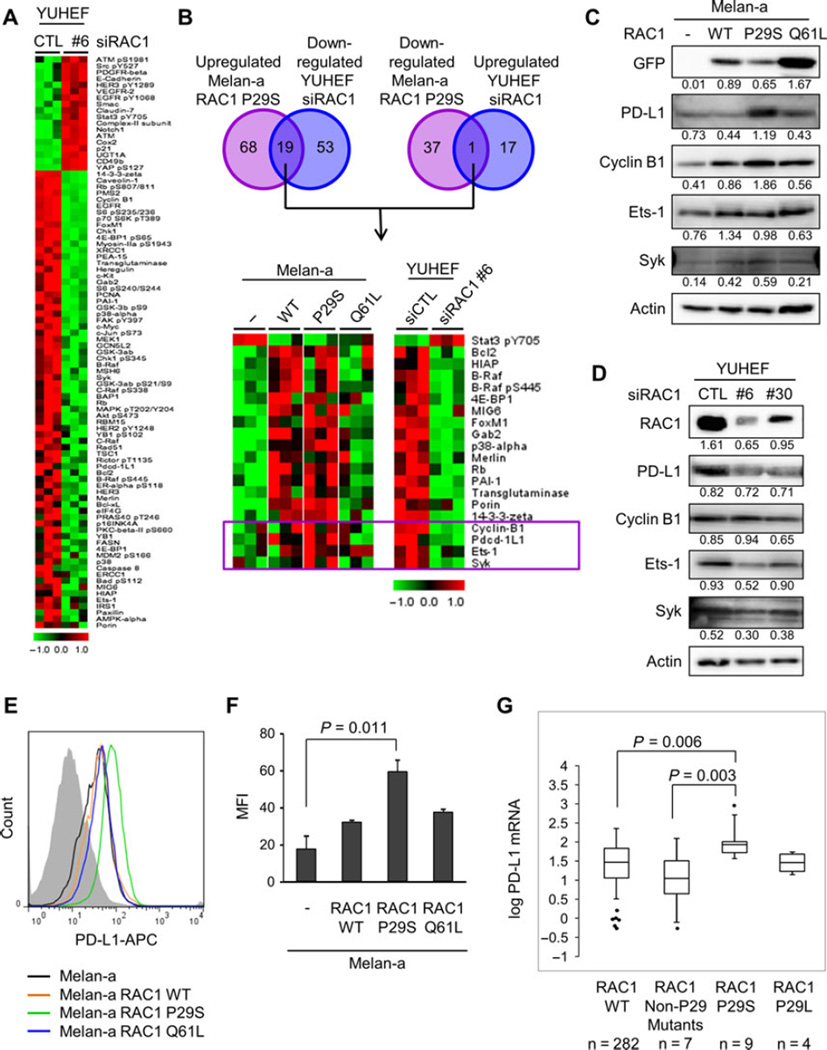
To define a protein signature specific to RAC1 P29S, we compared the changes with RAC1 P29S overexpression in melan-a cells and the changes with RAC1 depletion in YUHEF melanoma cells, which harbor an endogenous RAC1 P29S mutation. SAM showed 18 proteins that were upregulated and 72 that were downregulated with RAC1 depletion in YUHEF cells (Figure 5A). Nineteen proteins were upregulated with RAC1 P29S expression and downregulated with RAC1 depletion, and one protein was downregulated with RAC1 P29S expression and upregulated with RAC1 depletion (Figure 5B). Of the 20 proteins differentially regulated, four are part of the 21 protein subset that were uniquely changed with the expression of RAC1 P29S: cyclin B1, Pdcd-1L1 (aka PD-L1, CD274), Ets-1, and Syk (boxed in Figure 5B).
Figure 5.
Signaling pathways distinct in RAC1 P29S mutation. (A) Significance analysis of microarray for RPPA of YUHEF cells transfected with non-targeting siRNA or RAC1-targeting siRNA #6. Expression was normalized to the mean. (B) Venn diagram depicting number of proteins significantly upregulated in melan-a RAC1 P29S and downregulated in YUHEF cells transfected with RAC1-targeting siRNA #6 (left) and proteins significantly downregulated in melan-a RAC1 P29S and upregulated in YUHEF cells transfected with RAC1-targeting siRNA #6 (right). Heat map of differentially regulated proteins (bottom). Expression was normalized to the mean. The four boxed proteins are uniquely upregulated in melan-a cells expressing RAC1 P29S. (C) Western blot analysis of melan-a cells and melan-a cells expressing RAC1 WT, RAC1 P29S, or RAC1 Q61L. Quantitation is normalized to actin control. (D) Western blot analysis of YUHEF cells transfected with non-targeting or RAC1-targeting siRNAs, #6 and #30, for 72 h. Quantitation is normalized to actin control. (E) Representative FACS analysis of surface PD-L1 expression on melan-a cells with or without RAC1 expression. (F) Mean fluorescence intensity (MFI) of surface PD-L1-APC on melan-a cells with or without RAC1 expression. MFI was determined by gating on live cells and subtracting background (isotype) MFI (n = 3; mean ± S.E.). (G) Box and whisker plot of log PD-L1 (CD274) mRNA in RAC1 WT, RAC1 non-P29 mutants, RAC1 P29S, and RAC1 P29L patients in the SKCM database of TCGA Box.
We performed Western blot analysis to validate the findings of the RPPA. We found a modest increase in levels of cyclin B1, Ets-1, and Syk with RAC1 expression (Figure 5C). The level of PD-L1 was dramatically increased with RAC1 P29S expression. To evaluate the levels of the proteins of interest in YUHEF cells after depletion of RAC1, we utilized an additional siRNA targeting RAC1 (Figure 5D). With RAC1 depletion, there were decreases in PD-L1, Ets-1, and Syk that correlate with the degree of RAC1 knockdown. The specificity of the PD-L1 antibodies used for Western blot analysis was validated with PD-L1 knockdown in both a human melanoma cell line and melan-a cells (Figure S3A,B). We examined the surface expression of PD-L1 utilizing flow cytometry, after validating the specificity of the antibody with PD-L1 knockdown (Figure S3C). As was seen with Western blot analysis, surface staining of PD-L1 in melan-a cells expressing RAC1 WT and RAC1 Q61L was similar to that of parental cells melan-a, but expression of RAC1 P29S led to a significant increase in surface PD-L1 expression (Figure 5E, F.
To extend our studies to human patient samples, we evaluated the mutant RAC1 subset for PD-L1 expression in the SKCM database in the TCGA (Cerami et al., 2012; Gao et al., 2013). There was no significant difference in PD-L1 expression between the wild-type RAC1 and mutant RAC1 patients when accounting for the full spectrum of RAC1 mutations (P = 0.292). We next evaluated PD-L1 expression in patients with a RAC1 P29 mutation (Figure 5G). For the RAC1 P29S subset, we found a significant increase in PD-L1 mRNA (P = 0.005) compared to wild-type RAC1 patients. In the RAC1 P29L subset, we did not see an increase in PD-L1 expression (P = 0.824). With both exogenous expression of RAC1 P29S in vitro and endogenous expression of RAC1 P29S in melanoma patients, we found that RAC1 P29S expression is correlated with an increase in PD-L1 expression.
Discussion
In recent years, advances in DNA sequencing have led to the identification of many new mutations in melanomas. The current challenge lies in understanding the biological effects of these mutations to identify clinically significantly events. In agreement with published studies, the rate of RAC1 mutations was 5.5% (21/382) in the SKCM database from TCGA Research Network. Previously, it was found that in RAC1 mutant melanomas, the rate of co-existing NRAS mutation (30%) is higher than expected, whereas the rate of co-existing BRAF mutation was only 26% (Mar et al., 2014). In the cutaneous melanoma dataset from the TCGA, we found the rate of co-existing BRAF mutation to be 42.9% (9/21) and that of co-existing NRAS mutation to be 33.3% (7/21). Thus, there is a high degree of overlap between melanomas with mutations in RAC1 and those with mutations in BRAF or NRAS.
RAC1 is known to positively regulate cell proliferation, and the P29S mutant has been shown to enhance cell growth (Davis et al., 2013; Krauthammer et al., 2012). We found that RAC1 expression led to an increase in both anchorage-dependent and anchorage-independent growth in immortalized melanocytes. Morphologically, expression of RAC1 P29S and RAC1 Q61L led to an increase in membrane ruffling, a phenotype commonly attributed to RAC1 activating mutations due to their regulation of F-actin reorganization (Davis et al., 2013). Given the proliferative advantage conferred by RAC1 mutations, the role of RAC1 in response to targeted therapies has been evaluated. While some have found melanoma cell lines with RAC1 P29S mutation to be resistant to RAF and MEK inhibitors (Watson et al., 2014), others have found that RAC1 mutation status does not predict sensitivity to RAF or MEK inhibitors (Halaban, 2015). Given the heterogeneity of responses, we analyzed the response of mutant NRAS melanoma to MEK inhibitors and found that RAC1 expression does not decrease cell death in the presence of the MEK inhibitor, trametinib. Conversely, RAC1 depletion in a melanoma cell line with an endogenous RAC1 P29S mutation does not lead to an increase in cell death upon MEK inhibition.
We utilized RPPA analysis to dissect the molecular pathways regulated by RAC1 expression. Of the 218 validated antibodies, 42 were similarly regulated with RAC1 WT, RAC1 P29S, and RAC1 Q61L expression. Twenty-one of the proteins were uniquely regulated by RAC1 P29S expression. Of these 21, four upregulated proteins (cyclin B1, PD-L1, Ets-1, and Syk) were also downregulated by RAC1 depletion in YUHEF melanoma cells, which harbor an endogenous RAC1 P29S mutation. Western blot and FACS analysis revealed a strong increase in levels of PD-L1 with RAC1 P29S expression. PD-L1 is the ligand for PD-1, and the two proteins form an inhibitory receptor/ligand axis within the immune system. Persistent PD-1 expression on T-cells induces T-cell exhaustion and loss of effector function (Barber et al., 2006). PD-1 has two binding ligands, PD-L1 and PD-L2, with PD-L1 having a broader expression pattern (Ohaegbulam et al., 2015). Mutations in melanoma likely contribute to the immunogenicity of this cancer, and the expression of mutated antigens in melanoma cells can be recognized by tumor infiltrating lymphocytes (Robbins et al., 2013). Inhibition of the MEK-ERK1/2 pathway has been shown to decrease the production of immunosuppressive soluble factors such as IL-10, IL-6, and IL-1 in mutant BRAF melanoma (Khalili et al., 2012; Sumimoto et al., 2006). Additionally, BRAF V600E has been shown to suppress the expression of melanocyte differentiation antigens (MDAs) such as MART-1 and gp100 (Boni et al., 2010) and MHC-1 surface expression (Bradley et al., 2015), which are critical components of the immunologic response to melanoma. Inhibition of the ERK1/2 pathway in BRAF V600E melanoma leads to an increase in expression of MDAs and MHC-1 (Boni et al., 2010; Bradley et al., 2015). To the best of our knowledge, our observation that PD-L1 is increased with RAC1 P29S is the first of a role for the RAC1 P29S oncogene in modulating the immunogenicity of melanoma. The exact mechanism through which RAC1 P29S regulates PD-L1 expression remains to be elucidated.
In melanoma, PD-L1 marks a subset of patients with decreased overall survival (Hino et al., 2010; Massi et al., 2014). Clinical trials of PD-1- or PD-L1-blocking monoclonal antibodies (mAb) in melanoma patients have shown high response rates of 30–50% (Atkins et al., 2014; Hamid et al., 2013; Topalian et al., 2012; Weber et al., 2013; Wolchok et al., 2013). Remarkably, these responses are durable with many lasting more than a year. Expression of PD-L1 by tumor cells may correlate with response to anti-PD-1 and PD-L1 therapies. In three Phase I studies of nivolumab, a fully human IgG4 mAb to PD-1, the relationship between tumor PD-L1 expression and response to therapy is unclear. Two trials showed a response in some PD-L1-positive patients and no response in PD-L1-negative patients (Brahmer et al., 2012; Topalian et al., 2012), while another did not find a correlation between response and PD-L1 positivity (Wolchok et al., 2013). In these three studies, PD-L1 positivity was defined as 5% of tumor cells exhibiting membranous staining on IHC. More recently, a Phase I clinical trial of melanoma patients receiving pembrolizumab, a humanized IgG4 PD-1-blocking mAb, PD-L1 expression, as defined by PD-L1 positivity in 1% of stained cells in IHC, was significantly associated with progression-free survival and response rate (Daud et al., 2014). Given the correlation between RAC1 P29S expression and PD-L1 expression, it would be interesting to evaluate the response to anti-PD-L1 or anti-PD-1 therapies in this subgroup.
In summary, we show a novel role for RAC1 P29S in modulating the expression of PD-L1 in melanoma. The expression of PD-L1 is a compensatory measure by melanomas to evade the immune system, which may explain the enrichment of RAC1 P29S-harboring tumor cells. Additionally, given the role of anti-PD-1 and PD-L1 antibodies, RAC1 P29S may be a useful predictor of response to such therapies.
Methods
Cell culture
WM1346 and WM1361A melanoma lines were kindly donated by Dr. Meenhard Herlyn (Wistar Institute, Philadelphia, PA, USA). Melan-a cells were provided by Dr. Dorothy Bennett (St. George’s University of London, UK). YUHEF cells were provided by Dr. Ruth Halaban (Yale University, New Haven, CT, USA). Melan-a cells were maintained in RPMI-1640 with 10% fetal bovine serum (FBS) and 200 nm TPA. YUHEF cells were maintained in OptiMEM media (Invitrogen, Carlsbad, CA, USA) supplemented with 5% FBS. WM cell lines were cultured in MCDB 153 medium containing 20% Leibovitz L-15 medium, 2% FBS, and 0.2% sodium bicarbonate.
Cloning and stable cell line generation
RAC1 WT-eGFP and RAC1 Q61L-eGFP cDNA were purchased from Addgene (Cambridge, MA). RAC1 P29S-eGFP cloned from RAC1 WT-eGFP using the following primers for site-directed mutagenesis: forward 5′-ATATTCTCCAGAAAATGCATT-3′ and reverse 5′-AATGCATTTTCTGGAGAATAT-3′. All DNA constructs were sequence-verified. Lentiviral particles and melan-a and tetracycline repressor-expressing (TR expressing) sublines WM1346 TR and WM1361A TR expressing RAC1 WT-eGFP and RAC1 P29S-eGFP and RAC1 Q61L-eGFP were generated, as previously described (Abel and Aplin, 2010). Transgene expression was induced with 0.1 µg/ml doxycycline in the cell culture medium.
Western blot analysis
Cells lysed in sample buffer were separated by SDS-PAGE, and proteins were transferred electrophoretically onto Immobilon P membranes (Millipore Corp., Bedford, MA, USA). Membranes were blocked with phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) and 0.1% Tween-20 for 1 h and subsequently incubated with primary antibody overnight at 4°C. Antibodies for GFP (sc-8334), RAC1 (sc-95), PD-L1 (sc-19095), cyclin B1 (sc-245), Syk (sc-929), and Ets-1 (sc-55581) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), actin (A2066) from Sigma-Aldrich (St. Louis, MO, USA), and PD-L1 (AF156) from R&D Systems (Minneapolis, MN, USA). Membranes were washed in PBS/Tween and incubated with anti-mouse or anti-rabbit IgG peroxidase conjugates (Calbiochem, San Diego, CA, USA) for 1 h at room temperature. Western blots were developed using SuperSignal chemiluminescent substrate (Pierce, Rockford, IL, USA). Immunoreactivity was detected and quantified using a Fluor-S Multi-Imager and Quantity-One software (Bio-Rad, Hercules, CA, USA).
Immunofluorescence
Melanoma cells growing on glass coverslips were washed with PBS and fixed with 3.7% formaldehyde for 20 min. Cells were then permeabilized with 0.2% Triton X-100 for 5 min and nonspecific staining blocked with 1% BSA/PBS for 2 h at room temperature. Coverslips were then incubated with primary antibodies diluted in 1% BSA/PBS overnight at 4°C. After primary antibody, coverslips were washed three times with PBS before incubation with appropriate Alexa Fluor-conjugated secondary antibodies (Invitrogen) for 1 h at room temperature. For phalloidin staining, cells were incubated with phalloidin (Sigma-Aldrich) for 10 min at room temperature. In some instances, the coverslips were incubated with DAPI to visualize nuclei. Coverslips were mounted and visualized on an Eclipse Ti inverted microscope with NIS-Elements AR 3.00 software (Nikon, Melville, NY, USA).
Colony formation assay
Mutant NRAS cells (4 × 103) were plated per six-well plate in complete medium with or without inhibitors, which were replenished every 2 days. After 10 days, cells were stained with crystal violet in formalin, plates were imaged by scanning, and colonies were imaged on a Nikon Eclipse Ti inverted microscope with nis-elements ar 3.00 software.
3D collagen gels and apoptosis assay
Collagen gels were cast and cells were isolated and stained with annexin V-APC as previously described (Kaplan et al., 2012). Apoptosis was analyzed by flow cytometry on FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Data were analyzed by FlowJo software (Tree Star Inc., Ashland, OR, USA).
RNA interference
YUHEF cells were transfected for 4 h with chemically synthesized short interfering RNA (siRNA) at a final concentration of 25 nm using Oligofectamine (Invitrogen). Cells were harvested after a further 68 h. The RAC1 siRNAs (#6: M-003560-06 and #30: D-003560-30) were purchased from Dharmacon Inc. (Lafayette, CO, USA).
RPPA analysis
Cells were lysed and proteins prepared, as previously described (Tibes et al., 2006). Lysates were run against 187 validated antibodies, and analyses were performed using normalized data. Analysis was performed with triplicate normalized RPPA data and SAM using MeV 4.9 (Dana-Farber Cancer Institute, Boston, MA, USA). KEGG pathway analysis of up- and downregulated proteins with RAC1 expression was performed using DAVID (Huang da et al., 2009a,b).
Reagents
Trametinib (GSK11202212) was purchased from Selleck Chemicals LLC (Houston, TX, USA).
PD-L1 cell surface staining
Melan-a parental cells and those expressing RAC1 were harvested. Cells were washed and fixed in 4% paraformaldehyde for 10 minutes. Cells were then stained with anti-PD-L1 for 1 h. Data acquisition was performed on a LSR II with FACSCalibur flow cytometer (BD Biosciences). Data were analyzed by FlowJo software (Tree Star Inc.). Mean fluorescence intensity (MFI) was determined by gating on live cells and subtracting background (isotype) MFI.
Statistical analysis
Statistical analyses were performed using spss v22.0 (IBM, Armonk, NY, USA). A P value of <0.05 was considered statistically significant. Tukey HSD was used to evaluate growth curves, and Student’s t-test was used to evaluate colony number and size in soft agar. A one-way anova analysis of PD-L1 mRNA expression levels was performed on 9 RAC1 P29S mutants, 4 RAC1 P29L mutants, 7 non-P29 RAC1 mutants, and 282 RAC1 WT. Log-transformed PD-L1 expression levels of each RAC1 type were found to be normally distributed using Shapiro–Wilk’s test (P > 0.05). There was homogeneity of variances among all groups, as determined by Levene’s test of equality of variances (P = 0.343). Dunnett’s one-tailed multiple comparison post hoc tests were performed to determine the statistical significance of the RAC1 P29S samples compared to the other groups.
Supplementary Material
Figure S1. Generation of RAC1 P29S.
Figure S2. RAC1 P29S and RAC1 Q61L expression increases membrane ruffling.
Figure S3. Validation of PD-L1 antibodies.
Appendix S1. PD-L1 knockdown.
Significance.
Recent efforts in exome sequencing of cutaneous melanoma samples identified a recurrent RAC1 P29S mutation. We utilized an unbiased protein array to determine proteins regulated by RAC1 P29S and identified a correlation between PD-L1, a negative immune modulator, and RAC1 P29S. This finding provides novel insight into the biology of RAC1 P29S, with a potential role in evading the antitumor immune response. Additionally, RAC1 P29S melanoma patients may derive increased benefit from immune therapies that target PD-L1 or its receptor, PD-1.
Acknowledgements
We are grateful to Dr. Meenhard Herlyn (Wistar Institute, Philadelphia, PA) for supplying the WM melanoma cell lines, Dr. Dorothy Bennett (St. George’s University of London, UK) for the melan-a cells, and Dr. Ruth Halaban (Yale University, New Haven, CT) for the YUHEF cells. Also, we thank Bridget King for helping with the initial RAC1 mutant generation.
Source of support
This work was supported by the National Institutes of Health Grant F31-CA174331 (H.L. Vu), an Established Investigator Award from the Melanoma Research Foundation (A.E. Aplin), and grants from the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (A.E. Aplin and M.A. Davies). The Sidney Kimmel Cancer Center at Thomas Jefferson University is funded by National Cancer Institute Support Grant 1P30CA56036. The Reverse Phase Protein Array Core Facility at M.D. Anderson is supported by the NCI Cancer Center Support Grant, CA-16672.
Footnotes
Disclosure of potential conflict of interests
There is no potential conflict of interests.
Additional Supporting Information may be found in the online version of this article:
References
- Abel EV, Aplin AE. FOXD3 is a mutant B-RAF-regulated inhibitor of G(1)-S progression in melanoma cells. Cancer Res. 2010;70:2891–2900. doi: 10.1158/0008-5472.CAN-09-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins MB, Kudchadkar RR, Sznol M, et al. Phase 2, multicenter, safety and efficacy study of pidilizumab in patients with metastatic melanoma. ASCO Annual Meeting. 2014 (abstract 9001). [Google Scholar]
- Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- Bennett DC, Cooper PJ, Hart IR. A line of non-tumorigenic mouse melanocytes, syngeneic with the B16 melanoma and requiring a tumour promoter for growth. Int. J. ancer. 1987;39:414–418. doi: 10.1002/ijc.2910390324. [DOI] [PubMed] [Google Scholar]
- Bid HK, Roberts RD, Manchanda PK, Houghton PJ. RAC1: An emerging therapeutic option for targeting cancer angiogenesis and metastasis. Mol. Cancer Ther. 2013;12:1925–1934. doi: 10.1158/1535-7163.MCT-13-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AL, Hall A. Rho GTPases and their effector proteins. Biochem. J. 2000;348(Pt 2):241–255. [PMC free article] [PubMed] [Google Scholar]
- Boni A, Cogdill AP, Dang P, et al. Selective BRAFV600E inhibition enhances T-cell recognition of melanoma without affecting lymphocyte function. Cancer Res. 2010;70:5213–5219. doi: 10.1158/0008-5472.CAN-10-0118. [DOI] [PubMed] [Google Scholar]
- Bradley SD, Chen Z, Melendez B, et al. BRAF(V600E) co-opts a conserved MHC class I internalization pathway to diminish antigen presentation and CD8 + T-cell recognition of melanoma. Cancer Immunol. Res. 2015;3:602–609. doi: 10.1158/2326-6066.CIR-15-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustelo XR, Sauzeau V, Berenjeno IM. GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. BioEssays. 2007;29:356–370. doi: 10.1002/bies.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daud AI, Hamid O, Ribas A, et al. Antitumor activity of the anti-PD-1 monoclonal antibody MK-3475 in melanoma (MEL): Correlation of tumor PD-L1 expression with outcome. AACR Annual Meeting. 2014 (abstr CT104). [Google Scholar]
- Davis MJ, Ha BH, Holman EC, Halaban R, Schlessinger J, Boggon TJ. RAC1P29S is a spontaneously activating cancer-associated GTPase. Proc. Natl Acad. Sci. USA. 2013;110:912–917. doi: 10.1073/pnas.1220895110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaban R. RAC1 and melanoma. Clin. Ther. 2015;37:682–685. doi: 10.1016/j.clinthera.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid O, Robert C, Daud A, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N. Engl. J. Med. 2013;369:134–144. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino R, Kabashima K, Kato Y, Yagi H, Nakamura M, Honjo T, Okazaki T, Tokura Y. Tumor cell expression of programmed cell death-1 ligand 1 is a prognostic factor for malignant melanoma. Cancer. 2010;116:1757–1766. doi: 10.1002/cncr.24899. [DOI] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kaplan FM, Kugel CH, III, Dadpey N, Shao Y, Abel EV, Aplin AE. SHOC2 and CRAF mediate ERK1/2 reactivation in mutant NRAS-mediated resistance to RAF inhibitor. J. Biol. Chem. 2012;287:41797–41807. doi: 10.1074/jbc.M112.390906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalili JS, Liu S, Rodriguez-Cruz TG, et al. Oncogenic BRAF(V600E) promotes stromal cell-mediated immunosuppression via induction of interleukin-1 in melanoma. Clin. Cancer Res. 2012;18:5329–5340. doi: 10.1158/1078-0432.CCR-12-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauthammer M, Kong Y, Ha BH, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat. Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mar VJ, Wong SQ, Logan A, Nguyen T, Cebon J, Kelly JW, Wolfe R, Dobrovic A, McLean C, McArthur GA. Clinical and pathological associations of the activating RAC1 P29S mutation in primary cutaneous melanoma. Pigment Cell Melanoma Res. 2014;27:1117–1125. doi: 10.1111/pcmr.12295. [DOI] [PubMed] [Google Scholar]
- Massi D, Brusa D, Merelli B, et al. PD-L1 marks a subset of melanomas with a shorter overall survival and distinct genetic and morphological characteristics. Ann. Oncol. 2014;25:2433–2442. doi: 10.1093/annonc/mdu452. [DOI] [PubMed] [Google Scholar]
- Ohaegbulam KC, Assal A, Lazar-Molnar E, Yao Y, Zang X. Human cancer immunotherapy with antibodies to the PD-1 and PD-L1 pathway. Trends Mol. Med. 2015;21:24–33. doi: 10.1016/j.molmed.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ. Rho family proteins: coordinating cell responses. Trends Cell Biol. 2001;11:471–477. doi: 10.1016/s0962-8924(01)02153-5. [DOI] [PubMed] [Google Scholar]
- Robbins PF, Lu YC, El-Gamil M, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat. Med. 2013;19:747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solit DB, Garraway LA, Pratilas CA, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumimoto H, Imabayashi F, Iwata T, Kawakami Y. The BRAF-MAPK signaling pathway is essential for cancer-immune evasion in human melanoma cells. J. Exp. Med. 2006;203:1651–1656. doi: 10.1084/jem.20051848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, Kornblau SM. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol. Cancer Ther. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu HL, Aplin AE. Targeting TBK1 inhibits migration and resistance to MEK inhibitors in mutant NRAS melanoma. Mol. Cancer Res. 2014;12:1509–1519. doi: 10.1158/1541-7786.MCR-14-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson IR, Li L, Cabeceiras PK, et al. The RAC1 P29S hotspot mutation in melanoma confers resistance to pharmacological inhibition of RAF. Cancer Res. 2014;74:4845–4852. doi: 10.1158/0008-5472.CAN-14-1232-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JS, Kudchadkar RR, Gallenstein D, et al. Safety, efficacy, and biomarkers of nivolumab with vaccine in ipilimumab-refractory or na€ıve melanoma. J. Clin. Oncol. 2013;31:4311–4318. doi: 10.1200/JCO.2013.51.4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolchok JD, Kluger H, Callahan MK, et al. Nivolumab plus ipilimumab in advanced melanoma. N. Engl. J. Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Generation of RAC1 P29S.
Figure S2. RAC1 P29S and RAC1 Q61L expression increases membrane ruffling.
Figure S3. Validation of PD-L1 antibodies.
Appendix S1. PD-L1 knockdown.