Abstract
Background
Despite a similar histologic appearance, upper tract urothelial carcinoma (UTUC) and urothelial carcinoma of the bladder (UCB) tumors have distinct epidemiologic and clinicopathologic differences.
Objective
To investigate whether the differences between UTUC and UCB result from intrinsic biological diversity.
Design, setting, and participants
Tumor and germline DNA from patients with UTUC (n = 83) and UCB (n = 102) were analyzed using a custom next-generation sequencing assay to identify somatic mutations and copy-number alterations in 300 cancer-associated genes.
Outcome measurements and statistical analysis
We described co-mutation patterns and copy-number alterations in UTUC. We also compared mutation frequencies in high-grade UTUC (n = 59) and high-grade UCB (n = 102).
Results and limitations
Comparison of high-grade UTUC and UCB revealed significant differences in the prevalence of somatic alterations. Alterations more common in high-grade UTUC included fibroblast growth factor receptor 3 (FGFR3; 35.6% vs 21.6%; p = 0.065), Harvey rat sarcoma viral oncogene homolog (HRAS; 13.6% vs 1.0%; p = 0.001), and cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) (CDKN2B; 15.3% vs 3.9%; p = 0.016). Genes less frequently mutated in high-grade UTUC included tumor protein p53 (TP53; 25.4% vs 57.8%; p < 0.001), retinoblastoma 1 (RB1; 0.0% vs 18.6%; p < 0.001), and AT rich interactive domain 1A (SWI-like) (ARID1A; 13.6% vs 27.5%; p = 0.050). Because our assay was restricted to genomic alterations in a targeted panel, rare mutations and epigenetic changes were not analyzed.
Conclusions
High-grade UTUC tumors display a spectrum of genetic alterations similar to high-grade UCB. However, there were significant differences in the prevalence of several recurrently mutated genes including HRAS, TP53, and RB1. As relevant targeted inhibitors are being developed and tested, these results may have important implications for the site-specific management of patients with urothelial carcinoma.
Patient summary
Comparison of next-generation sequencing of upper tract urothelial carcinoma (UTUC) with urothelial bladder cancer identified that similar mutations were present in both cancer types but at different frequencies, indicating a potential need for unique management strategies. UTUC tumors were found to have a high rate of mutations that could be targeted with novel therapies.
Keywords: Upper tract urothelial carcinoma, Genomics, Bladder cancer, Targeted therapy
1. Introduction
Although most urothelial carcinomas originate in the bladder (urothelial carcinoma of the bladder [UCB]), in an estimated 5–8% of cases, the primary tumor site is the renal pelvis or ureter (upper tract urothelial carcinoma [UTUC]) [1]. Despite a common histologic appearance, epidemiologic and clinicopathologic differences suggest that UTUC and UCB may represent two disparate disease entities [2–7]. Upper tract tumors, for example, are more often invasive than bladder tumors at the time of surgery, indicating that UTUC may represent a more aggressive disease phenotype.
Molecular studies have documented some biological distinctions between UTUC and UCB. Microsatellite instability, for example, is more common in UTUC [8]. Promoter methylation has also been shown to be more common and, when present, more extensive in UTUC [9]. Such epigenetic differences may be related to variability in exposures to specific carcinogenic metabolites excreted in the urine (longer dwell time in the bladder) or to the different embryologic origins of the bladder and upper tract (urogenital sinus vs ureteric bud). Previous efforts at genetic characterization of UTUC utilized technologies based on polymerase chain reaction to assess the mutation status of relatively few genes [10]. In this study, we sought to genomically characterize UTUC more comprehensively using massively parallel next-generation sequencing with the goal of determining whether the clinicopathologic differences between UTUC and UCB are the result of differences in the spectrum of somatic genetic driver events.
2. Methods
2.1. Samples
Frozen tumor samples, normal renal parenchymal tissue, and blood were collected from 83 patients with UTUC treated with radical nephroureterectomy under a protocol approved by the institutional review board. For all specimens, representative hematoxylin and eosin slides from frozen tissue and formalin-fixed paraffin-embedded sections were reviewed by a board-certified genitourinary pathologist to confirm the histology, grade, and stage. All samples were primary urothelial carcinoma (no predominant variant histologies were included). DNA was extracted as previously described [11]. Clinical and demographic information was obtained from a prospectively maintained institutional database.
2.2. Targeted capture and sequencing
All protein-coding exons of 300 cancer-associated genes were sequenced using the Integrated Molecular Profiling of Actionable Cancer Targets (MSK-IMPACT) assay [12]. Briefly, an equimolar pool of bar-coded libraries generated from genomic DNA was subjected to exon capture using custom oligonucleotides and sequenced on an Illumina HiSeq 2500 (Illumina Inc, San Diego, CA, USA). Validation of mutations identified by the MSK-IMPACT assay in 10 frequently altered genes was performed using the Illumina MiSeq platform.
2.3. Sequence analysis
Sequence reads were aligned to the reference human genome (hg19) using the CASAVA pipeline (http://support.illumina.com/sequencing/sequencing_software/casava.html), the Burrows-Wheeler alignment tool v.0.6.2 (SourceForge; Slashdot Media, San Jose, CA, USA), and GATK [13]. Single-nucleotide variants were called using MuTect v.1.0.27783 (Broad Institute, Cambridge, MA USA), and insertions and deletions (indels) were called using the SomaticIndelDetector tool [14]. All candidate mutations and indels were reviewed manually.
The accumulated sequence coverage for each exon was compared in tumor and matched germline samples, after performing samplewide LOESS normalization for GC percentage across exons and normalizing for global differences in “on-target” sequence coverage. Increases and decreases in the tumor-to-germline coverage ratios were used to infer copy-number alterations. Coverage ratios ≥3 were defined as amplifications, and ratios ≤0.3 were defined as deletions, whereas ratios ≥2x and ≤0.5x were defined as gains and losses, respectively. To detect somatic structural aberrations, we filtered for duplicates using the Picard tools (Broad Institute) java package (SAMtools; SourceForge) and searched for candidate structural rearrangements using DELLY [15]. Read pairs from both tumor and matched germline samples were used. All candidate structural aberrations were filtered, annotated, and reviewed manually.
2.4. Statistical analysis
Prespecified genomic alterations deemed to have functional significance were analyzed. For known oncogenes, we included recurrent point mutations and amplifications. For putative tumor suppressors, we included truncating mutations (nonsense, frameshift indels) and deletions. Demographic characteristics were compared using the Wilcoxon rank-sum test or Fisher exact test. Bivariate comparisons of individual mutation frequencies by cohort were performed using the Fisher exact test. We performed a multivariable logistic regression analysis for the association between mutation and cohort, adjusted for T stage. Counts of gains, losses, and total copy-number alterations were analyzed using negative binomial regression to account for overdispersion in the data. Although this study was exploratory, we also adjusted p values for multiple comparisons using the Benjamini-Hochberg method to control the false discovery rate. The p values <0.05 were considered statistically significant. All analyses were conducted using R v.3.1.1 (R Foundation, Vienna, Austria) including the survival and cmprsk packages.
2.5. Data availability
Genomic and associated clinicopathologic data are publicly available through the Memorial Sloan Kettering Cancer Center cBioPortal for Cancer Genomics [16].
3. Results
3.1. Genomic characterization of upper tract urothelial carcinoma
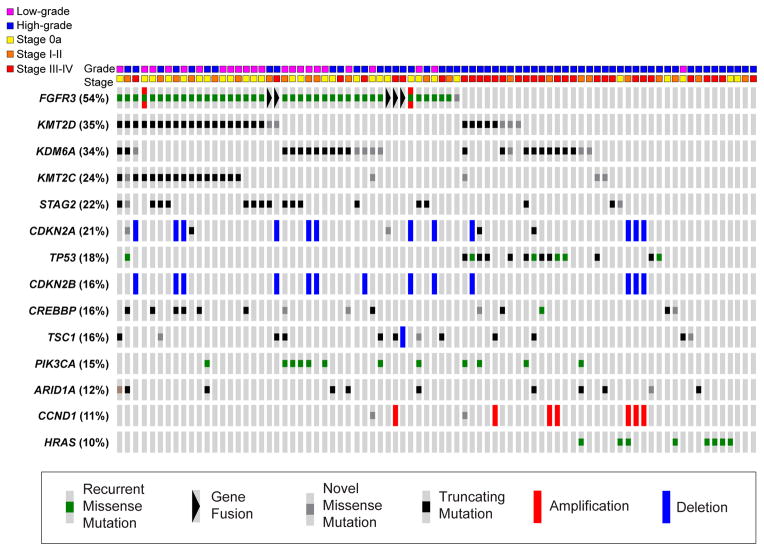
With the goal of defining the spectrum of somatic genetic alterations in UTUC, we analyzed 83 UTUC tumors (60 high grade and 23 low grade) using a capture-based, massively parallel, next-generation sequencing assay (MSK-IMPACT). The median number of mutated genes per sample was five. Of note, one tumor was identified to be ultramutated, harboring 422 somatic mutations and no focal copy-number alterations (CNAs) (Supplementary Fig. 1). Analysis identified a hotspot mutation of the exonuclease domain of polymerase (DNA directed), epsilon, catalytic subunit (POLE), V411L, previously reported to be associated with an ultramutated phenotype in endometrial and colorectal cancers [17,18]. To our knowledge, this is the first report of an ultramutated urothelial tumor associated with a POLE mutation. Because most of the genes examined in this tumor harbored somatic mutations, most of which presumably represent passenger events, this sample was excluded from subsequent statistical analyses.
The most frequently mutated genes in the 82 UTUC tumors analyzed included those identified as commonly altered in previous studies of UCB including fibroblast growth factor receptor 3 (FGFR3), lysine (K)specific demethylase 6A (KDM6A), lysine (K)-specific methyltransferase 2D (KMT2D), cyclin-dependent kinase inhibitor 2A (CDKN2A), and tumor protein p53 (TP53) [19–21]. Within the top 14 most frequently altered genes, we found 67 recurrent missense mutations previously reported in the Catalogue of Somatic Mutations in Cancer (COSMIC), 31 novel missense mutations, 114 truncating mutations, 7 amplifications, and 26 homozygous deletions (Fig. 1). All mutations identified in FGFR3, tuberous sclerosis 1 (TSC1), AT rich interactive domain 1A (SWI-like) (ARID1A), CREB binding protein (CREBBP), Harvey rat sarcoma viral oncogene homolog (HRAS), KDM6A, KMT2D, TP53, inositol polyphosphate-4-phosphatase, type I, 107kDa (INPP4A), and phosphatidylinositol-4,5-biphosphate 3-kinase, catalytic subunit alpha (PIK3CA) were subsequently confirmed using an orthogonal MiSeq-based assay with 100% concordance. We also identified five intrachromosomal FGFR3-transforming, acidic coiled-coil containing protein 3 (TACC3) translocations predicted to result in activating gene fusions [22,23]. The breakpoints were located in intron 17 (four cases) or exon 18 (one case) of FGFR3 and intron 10 (four cases) or exon 7 (one case) of TACC3. All five translocations were subsequently confirmed using Sanger sequencing (Supplementary Fig. 2). Consistent with prior studies, we identified a predominantly mutually exclusive pattern of alterations in the RTK/RAS/MAPK pathway and p53/MDM2 (Supplementary Fig. 3).
Fig. 1.
Representation of the 14 most frequently altered genes in a series of 82 upper tract urothelial carcinoma tumors. Mutations are categorized as missense mutations reported in COSMIC (green), gene fusions (black triangle), novel missense mutations (gray), truncating nonsense mutations or indels (black), amplifications (red bar), and deletions (blue bar).
3.2. Comparison of genomic profiles of high-grade upper tract urothelial carcinoma and urothelial carcinoma of the bladder
To investigate potential differences in the mutational landscape between upper and lower tract tumors, we compared the 59 high-grade UTUC tumors in our cohort with 102 high-grade UCB tumors from patients without a history of UTUC also sequenced using MSK-IMPACT [24]. Patient demographics and tumor characteristics were similar between the two cohorts (Table 1). The median age of the UTUC and UCB cohorts was 70 yr (interquartile range [IQR]: 63–76 yr) and 68 yr (IQR: 63–76 yr), respectively.
Table 1.
Comparison of patient and tumor characteristics of the cohorts with high-grade upper tract urothelial carcinoma and high-grade urothelial carcinoma of the bladder
| All UTUC, 83 tumors, n (%) | High-grade UTUC, 59 tumors, n (%) | High-grade UCB, 102 tumors, n (%) | p value | |
|---|---|---|---|---|
| Age at surgery, yr, median (IQR) | 68 (63–75) | 70 (63–76) | 68 (63–76) | 0.584 |
| Sex | ||||
| Male | 55 (66.3) | 40 (67.8) | 77 (75.5) | 0.359 |
| Female | 28 (33.7) | 19 (32.2) | 25 (24.5) | |
| Smoking status | 0.351 | |||
| Never | 21 (25.3) | 13 (22.0) | 30 (29.4) | |
| Former | 46 (55.4) | 39 (66.1) | 55 (53.9) | |
| Current | 16 (19.3) | 7 (11.9) | 17 (16.7) | |
| pT stage | 0.106 | |||
| pTa/pT1 | 44 (53.0) | 22 (37.3) | 25 (24.5) | |
| pT2/pT3/pT4 | 39 (57.0) | 37 (62.7) | 77 (75.5) | |
| pN stage | 0.313 | |||
| Negative | 45 (54.2) | 30 (50.8) | 55 (53.9) | |
| Positive | 17 (20.5) | 17 (28.8) | 35 (34.3) | |
| No LN dissection | 21 (25.3) | 12 (20.3) | 12 (11.8) |
IQR = interquartile range; LN = lymph node; UCB = urothelial carcinoma of the bladder; UTUC = upper tract urothelial carcinoma.
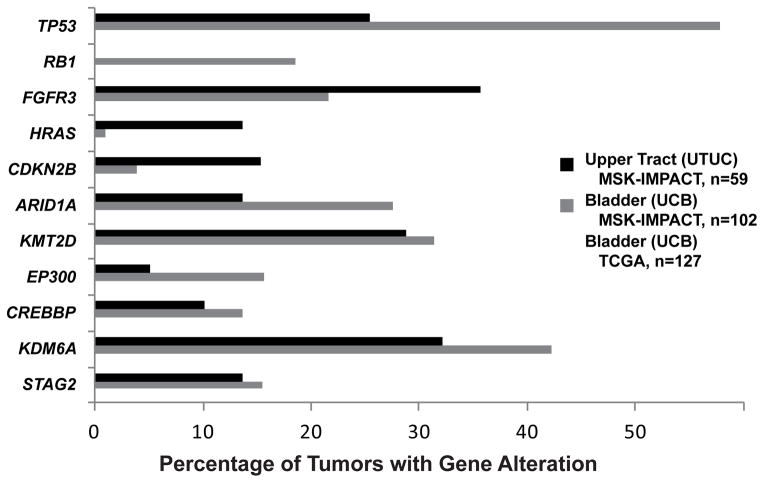
Mean sequencing coverage across all targeted exons for the UTUC and UCB cohorts was 650 times and 579 times, respectively. The mutation frequencies in our UCB cohort were similar to those reported by the Cancer Genome Atlas (TCGA) [19]. Overall, the landscape of alterations in the UTUC and UCB cohorts was similar, but the prevalence of mutations differed. FGFR3, HRAS, and cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) (CDKN2B), for example, were more frequently altered in UTUC (35.6% vs 21.6%, p = 0.065; 13.6% vs 1.0%, p = 0.001; and 15.3% vs 3.9%, p = 0.016, respectively), whereas TP53 and ARID1A were more frequently altered in UCB (57.8% vs 25.4%, p < 0.001 and 27.5% vs 13.6%, p = 0.050, respectively) (Fig. 2). A higher frequency of FGFR3-TACC3 fusions was identified in the UTUC cohort (8.5% vs 2.0%). Alterations in the Rb and p53 pathways were previously reported as commonly altered in high-grade UCB. Strikingly, we detected no retinoblastoma 1 (RB1) mutations in the UTUC cohort compared with an 18.6% frequency in UCB tumors (p < 0.001). We also identified fewer TP53/MDM2 proto-oncogene, E3 ubiquitin protein ligase (MDM2) alterations in the UTUC cohort (35.6% vs 62.7%; p = 0.001). The previously mentioned differences, except for FGFR3, remained statistically significant when adjusted for the effects of tumor stage (Supplementary Table 1).
Fig. 2. Significant differences in prevalence of mutations identified between the high-grade upper tract urothelial carcinoma (black bars) and high-grade urothelial carcinoma of the bladder (gray bars) cohorts. Mutation frequencies of the Cancer Genome Atlas high-grade bladder cohort (white bars) are displayed for comparison.
TCGA = The Cancer Genome Atlas; UCB = urothelial carcinoma of the bladder; UTUC = upper tract urothelial carcinoma.
We also identified a trend toward differences between the UTUC and UCB cohorts in the frequency of alterations in several additional driver oncogenes and tumor suppressors including TSC1 (11.9% vs 3.9%; p = 0.100) and PIK3CA (10.2% vs 21.6%; p = 0.084). Consistent with recent next-generation sequencing analyses of UCB tumors, we also identified frequent mutations in chromatin-modifying genes (CMGs) including the histone demethylase KDM6A, the histone methyltransferases KMT2A (MLL), KMT2D (MLL2), and lysine (K)-specific methyltransferase 2C (KMT2C) (MLL3), the histone acetyltransferases CREBBP and E1A binding protein p300 (EP300), and the SWI/SNF complex genes ARID1A and SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 (SMARCA4) [19–21]. With the exception of ARID1A, there was no significant difference in the frequency of mutations in these CMGs in UTUC versus UCB tumors.
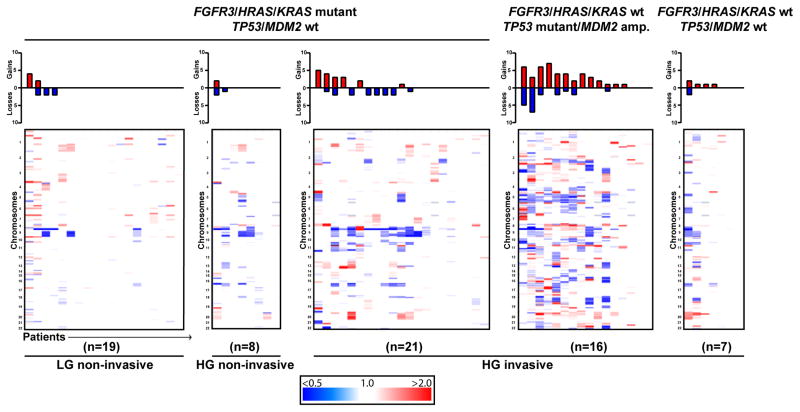
When examining the landscape of CNAs in UTUC, we found that TP53/MDM2-altered UTUC tumors possessed a high frequency of CNAs, with all but three tumors harboring at least one copy-number gain or loss. When compared with FGFR3/HRAS/Kirsten sarcoma viral oncogene homolog (KRAS) mutant high-grade invasive UTUC tumors (n = 21), TP53/MDM2-altered high-grade invasive UTUC tumors (n = 16) had significantly more copy-number gains and total CNAs (rate ratio [RR]: 2.91 [95% confidence interval (CI), 1.13–7.71; p = 0.028] vs 2.49 [95% CI, 1.19–5.33; p = 0.017], respectively) (Fig. 3). These results in UTUC are consistent with previously reported data suggesting an association between aneuploidy and TP53 mutation in UCB [25]. As might be expected, high-grade tumors had more CNAs than low-grade tumors (p = 0.004), and invasive tumors had more CNAs than noninvasive tumors (p < 0.001).
Fig. 3. Comparison of copy-number alterations in upper tract urothelial carcinoma stratified by pathologic grade, stage, and FGFR3/HRAS and TP53/MDM2 alteration status. Copy-number gains (red) and losses (blue) are quantified in bar graphs at the top.
HG = high grade; LG = low grade.
3.3. Comparison of low-grade and high-grade upper tract urothelial carcinoma
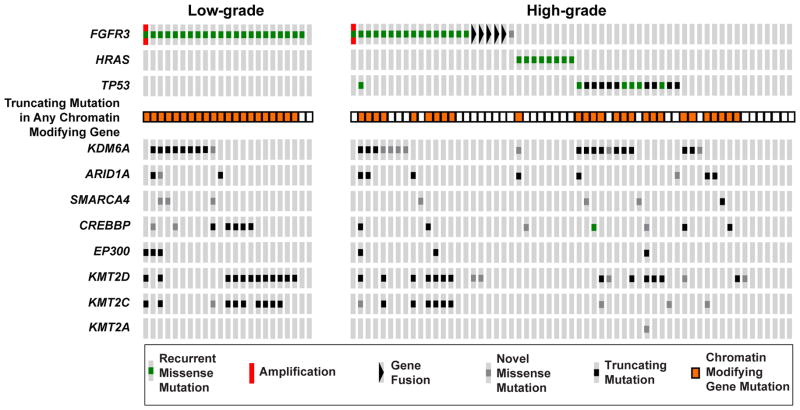
Given the known association between FGFR3 mutations and low-grade histology in UCB, we sought to define their prevalence in low-grade UTUC. Overall, 22 of 23 low-grade UTUC tumors were found to harbor known recurrent activating mutations in FGFR3 (Fig. 4). Analysis of the high-grade UTUC tumors identified a mutually exclusive mutation pattern among FGFR3, HRAS, and TP53. Notably, TP53 mutations were found exclusively in high-grade tumors, whereas mutations in CMGs were common in both low-grade and high-grade tumors. These latter results suggest that CMG alterations are likely an early event in the pathogenesis of UTUC rather than drivers of progression to high-grade disease.
Fig. 4.
OncoPrint comparing mutations in FGFR3, HRAS, and TP53 between low-grade and high-grade upper tract urothelial carcinoma. FGFR3 mutations were detected in 22 of 23 low-grade tumors, and a pattern of mutual exclusivity between FGFR3, HRAS and TP53 was seen in the high-grade tumors. Alterations in chromatin-modifying genes were common in both the low-grade and high-grade tumors.
To determine whether the FGFR3 mutations identified were present in morphologically and spatially distinct components of the tumors, we performed microdissection of four UTUC cases to separate tumor components for additional sequencing. We used a customized MiSeq assay to sequence 10 genes, including FGFR3, within spatially and histologically distinct portions of these four tumors including areas of low- versus high-grade disease, noninvasive versus invasive disease, or primary tumor versus metastatic lymph node. There was complete concordance in FGFR3 status between the tumor components as well as the matched lymph node metastasis (Table 2). Mutations in CMGs (KDM6A and KMT2D) were also concordant, supporting the hypothesis that such mutations occur early in the pathogenesis of this disease.
Table 2.
Concordance of mutations in FGFR3 and chromatin-modifying genes (KDM6A and KMT2D) in the various pathologic components (low vs high grade, noninvasive vs invasive, primary tumor vs metastatic lymph node) of four upper tract urothelial carcinoma tumors
| ID | Tumor sample | Type | Mutation status | ||
|---|---|---|---|---|---|
|
| |||||
| FGFR3 | KDM6A | KMT2D | |||
|
|
|||||
| Tumor 24 | Primary tumor Microdissection | Frozen | S249C | C1181Y | Q4687* |
| Tumor 24 | low-grade component Microdissection | FFPE | S249C | C1181Y | Q4687* |
| Tumor 24 | high-grade component | FFPE | S249C | C1181Y | Q4687* |
|
| |||||
| Tumor 47 | Primary tumor Microdissection | Frozen | S249C | wt | wt |
| Tumor 47 | low-grade component Microdissection | FFPE | S249C | wt | wt |
| Tumor 47 | high-grade component | FFPE | S249C | wt | wt |
|
| |||||
| Tumor 49 | Primary tumor | Frozen | S249C | wt | Q3994* |
| Tumor 49 | Microdissection noninvasive component | FFPE | S249C | wt | Q3994* |
| Tumor 49 | Microdissection invasive component | FFPE | S249C | wt | Q3994* |
|
| |||||
| Tumor 68 | Primary tumor | Frozen | S249C, F591L | P966R | wt |
| Tumor 68 | Primary tumor | FFPE | S249C, F591L | P966R | wt |
| Tumor 68 | Lymph node metastasis | FFPE | S249C, F591L | P966R | wt |
FFPE = formalin-fixed paraffin-embedded; wt = wild type.
Indicates nonsense mutation.
3.4. Clinicopathologic genomic associations in upper tract urothelial carcinoma
We found that mutations in TP53 (p = 0.008), FGFR3 (p < 0.001), CREBBP (p = 0.04), KMT2C (p = 0.02), and stromal antigen 2 (STAG2) (p = 0.006) were significantly associated with grade. Tumors with TP53 mutations were more frequently high grade, whereas those with FGFR3, CREBBP, and STAG2 mutations were more frequently low grade.
TP53 (p = 0.002), cyclin D1 (CCND1) (p = 0.046), FGFR3 (p < 0.001), erb-b2 receptor tyrosine kinase 2 (ERBB2) (p = 0.046), erb-b2 receptor tyrosine kinase 3 (ERBB3) (p = 0.046), KRAS (p = 0.016), and STAG2 (p = 0.013) were significantly associated with T stage. Tumors with TP53, CCND1, ERBB2, ERBB3, and KRAS mutations were more frequently pT3/pT4, whereas those with FGFR3 mutations were more frequently pTa/pT1/pT2.
4. Discussion
Cancers are increasingly classified and, when possible, treated based on mutational status. The heterogeneity of clinical outcomes within tumor types has been hypothesized to result, at least partly, from variation in the underlying genetic profiles of individual tumors. Urothelial tumors arising from the upper tract are more frequently invasive at diagnosis than those originating in the bladder [2–7]. Anatomic differences may account for much of this disparity because the thinner smooth muscle covering of the upper tract may allow for more rapid progression to non–organ-confined disease. By comparing the mutational profiles of UTUC and UCB, we sought to determine whether differences in somatic mutation patterns account, at least in part, for the clinicopathologic differences noted between these tumor types.
Consistent with the genetic profile of high-grade UCB reported by the TCGA and others, we found that mutations in FGFR3, HRAS, TP53, and CMGs are common in UTUC [19–21,25]. Our comparison of high-grade UTUC with UCB identified more frequent mutations in FGFR3 and HRAS and less prevalent mutations in TP53 and RB1. To exclude the possibility that the higher fraction of high-grade UTUC tumors harboring FGFR3 and HRAS mutations was an artifact resulting from intratumoral heterogeneity [26], we compared multiple histologically and spatially distinct areas of four tumors. In all cases, the FGFR3 and CMG mutations were completely concordant. Also of note, CMG mutations were found at high frequency in low-grade UTUC tumors and in the noninvasive components of two high-grade UTUC tumors, suggesting such events could occur early as one form of disease pathogenesis in some tumors.
Two distinct mechanisms of urothelial carcinogenesis have been proposed [27–29]. In the first, low-grade tumors, which typically harbor mutations in FGFR3 and HRAS, acquire additional genetic and/or epigenetic alterations leading to a high-grade morphology, invasion, and metastasis. Alternatively, high-grade tumors may arise de novo, associated with mutations in TP53 and RB1. Consistent with this model, we identified activating FGFR3 mutations in 22 of 23 low-grade UTUC tumors. All 23 low-grade tumors were the TP53 and RB1 wild type. Unexpectedly, activating missense mutations of FGFR3 and HRAS were found to be more common in high-grade UTUC (27.1% and 13.6%) compared with high-grade UCB. All five FGFR3-TACC3 translocations in the UTUC cohort were present in high-grade tumors. These FGFR3- and HRAS-altered high-grade UTUC tumors were, with one exception, the TP53 wild type. In sum, the results are consistent with a model in which low-grade FGFR3 and HRAS mutant tumors more frequently progress to high-grade invasive disease when they arise in the upper tract versus the bladder. This may simply reflect lead-time bias resulting from the greater difficulty in both detection and complete endoscopic resection of UTUC, which may allow for the accumulation of additional genetic or epigenetic changes not identified by our analyses that mediate the transition to high-grade histology.
Our study had several limitations. Although this is the largest series to date of UTUC tumors profiled using next-generation sequencing, a larger study may have revealed other significant patterns of co-mutated genes. Furthermore, although our capture-based assay included all genes highly mutated in the recent TCGA study of high-grade UCB, less frequent mutational events and structural alterations not included in the MSK-IMPACT assay would have been missed. Finally, epigenetic differences and/or differences in gene expression may be more important drivers of disease progression than genomic alterations, a possibility not addressed by our targeted DNA sequencing approach.
5. Conclusions
We performed targeted next-generation sequencing of 300 cancer-associated genes to define the genomic landscape of UTUC and to compare these results with UCB. Although the spectrum of genes mutated in UTUC and UCB was similar, we identified differences in the prevalence of alterations in several recurrently mutated genes including FGFR3, HRAS, TP53, and RB1. These data provide an important reference for developing multimodal strategies for the management of UTUC. The high prevalence of potentially actionable genetic events in UTUC suggests that routine genomic profiling may accelerate the development of novel personalized therapeutic approaches for this disease.
Supplementary Material
Scatter plot of mutation count and copy number alterations in 83 upper tract urothelial carcinoma tumors profiled with MSK-IMPACT. Ultra-mutated case (422 somatic mutations) highlighted in red.
(A) Schematic illustrating breakpoints in intron 17 of FGFR3 and intron 10 of TACC3, (B) Confirmation of translocation breakpoint using PCR and Sanger sequencing, (C) Predicted mRNA transcript and FGFR3-TACC3 fusion protein.
Oncoprint of the (A) RTK/RAS/MAPK and (B) p53 pathways showing mutual exclusivity.
Supplementary Table 1 — Comparison of frequencies of gene alterations in high-grade upper tract urothelial carcinoma (UTUC) and high-grade urothelial carcinoma of the bladder (UCB)
Take-home message.
We performed next-generation sequencing of 83 upper tract urothelial carcinoma (UTUC) tumors to characterize the landscape of genomic alterations. Comparison with high-grade bladder cancer identified more frequent FGFR3, HRAS, and CDKN2B alterations and fewer TP53 and RB1 mutations in high-grade UTUC.
Acknowledgments
Funding/Support and role of the sponsor: This work was supported by the National Institutes of Health, the Michael and Zena Wiener for Therapeutics Program in Bladder Cancer, Cycle for Survival, the Thompson Foundation, and the Urology Care Foundation Research Scholars Program (design and conduct of the study, and collection, analysis, and interpretation of the data). John P. Sfakianos was a research fellow in urologic oncology supported by NIH T32-CA82088.
We thank Kety Huberman, Igor Dolgalev, Olga Aminova, Sabrena Thomas, and Nathalie Lallier from the Geoffrey Beene Translational Oncology Core for assistance with the MiSeq studies; Nancy Bouvier from the Center for Molecular Oncology for assistance with MSK-IMPACT; and Maria Corazon Mariano, Katrina Allen, Priscilla McNeil, Daniel Navarrete, Anas Idelbi, and Anupama Gandhi from the Pathology Core for assistance with tissue collection and processing.
Footnotes
Author contributions: Eugene K. Cha had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Sfakianos, Cha, Iyer, Kim, Solit, Coleman.
Acquisition of data: Sfakianos, Cha, Iyer, Scott, Ren, Kim, Hakimi, Sun, Bajorin, Reuter, Bochner, Al-Ahmadie.
Analysis and interpretation of data: Sfakianos, Cha, Iyer, Scott, Zabor, Shah, Bagrodia, Ramirez, Hanrahan, Desai, Pinciroli, Schultz, Berger, Al-Ahmadie, Solit.
Drafting of the manuscript: Sfakianos, Cha.
Critical revision of the manuscript for important intellectual content: Sfakianos, Cha, Zabor, Solit, Coleman.
Statistical analysis: Zabor, Ostrovnaya.
Obtaining funding: Bochner, Solit, Coleman.
Administrative, technical, or material support: None.
Supervision: Rosenberg, Dalbagni, Bajorin, Reuter, Berger, Bochner, Solit, Coleman.
Other (specify): None.
Financial disclosures: Eugene K. Cha certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Munoz JJ, Ellison LM. Upper tract urothelial neoplasms: incidence and survival during the last 2 decades. J Urol. 2000;164:1523–5. [PubMed] [Google Scholar]
- 2.Olgac S, Mazumdar M, Dalbagni G, Reuter VE. Urothelial carcinoma of the renal pelvis: a clinicopathologic study of 130 cases. Am J Surg Pathol. 2004;28:1545–52. doi: 10.1097/00000478-200412000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Stewart GD, Bariol SV, Grigor KM, Tolley DA, McNeill SA. A comparison of the pathology of transitional cell carcinoma of the bladder and upper urinary tract. BJU Int. 2005;95:791–3. doi: 10.1111/j.1464-410X.2005.05402.x. [DOI] [PubMed] [Google Scholar]
- 4.Catto JW, Yates DR, Rehman I, et al. Behavior of urothelial carcinoma with respect to anatomical location. J Urol. 2007;177:1715–20. doi: 10.1016/j.juro.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 5.Margulis V, Shariat SF, Matin SF, et al. Outcomes of radical nephroureterectomy: a series from the Upper Tract Urothelial Carcinoma Collaboration. Cancer. 2009;115:1224–33. doi: 10.1002/cncr.24135. [DOI] [PubMed] [Google Scholar]
- 6.Cha EK, Shariat SF, Kormaksson M, et al. Predicting clinical outcomes after radical nephroureterectomy for upper tract urothelial carcinoma. Eur Urol. 2012;61:818–25. doi: 10.1016/j.eururo.2012.01.021. [DOI] [PubMed] [Google Scholar]
- 7.Rink M, Ehdaie B, Cha EK, et al. Stage-specific impact of tumor location on oncologic outcomes in patients with upper and lower tract urothelial carcinoma following radical surgery. Eur Urol. 2012;62:677–84. doi: 10.1016/j.eururo.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 8.Roupret M, Azzouzi AR, Cussenot O. Microsatellite instability and transitional cell carcinoma of the upper urinary tract. BJU Int. 2005;96:489–92. doi: 10.1111/j.1464-410X.2005.05671.x. [DOI] [PubMed] [Google Scholar]
- 9.Catto JW, Azzouzi AR, Rehman I, et al. Promoter hypermethylation is associated with tumor location, stage, and subsequent progression in transitional cell carcinoma. J Clin Oncol. 2005;23:2903–10. doi: 10.1200/JCO.2005.03.163. [DOI] [PubMed] [Google Scholar]
- 10.van Oers JM, Zwarthoff EC, Rehman I, et al. FGFR3 mutations indicate better survival in invasive upper urinary tract and bladder tumours. Eur Urol. 2009;55:650–7. doi: 10.1016/j.eururo.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 11.Kim PH, Cha EK, Sfakianos JP, et al. Genomic predictors of survival in patients with high-grade urothelial carcinoma of the bladder. Eur Urol. 2015;67:198–201. doi: 10.1016/j.eururo.2014.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–64. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Won HH, Scott SN, Brannon AR, Shah RH, Berger MF. Detecting somatic genetic alterations in tumor specimens by exon capture and massively parallel sequencing. J Vis Exp. 2013:e50710. doi: 10.3791/50710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rausch T, Zichner T, Schlattl A, Stutz AM, Benes V, Korbel JO. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–9. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–22. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gui Y, Guo G, Huang Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–8. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo G, Sun X, Chen C, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet. 2013;45:1459–63. doi: 10.1038/ng.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet. 2013;22:795–803. doi: 10.1093/hmg/dds486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu YM, Su F, Kalyana-Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636–47. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim PH, Cha EK, Sfakianos JP, et al. Genomic predictors of survival in patients with high-grade urothelial carcinoma of the bladder. Eur Urol. 2015;67:198–201. doi: 10.1016/j.eururo.2014.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyer G, Al-Ahmadie H, Schultz N, et al. Prevalence and co-occurrence of actionable genomic alterations in high-grade bladder cancer. J Clin Oncol. 2013;31:3133–40. doi: 10.1200/JCO.2012.46.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Ahmadie HA, Iyer G, Janakiraman M, et al. Somatic mutation of fibroblast growth factor receptor-3 (FGFR3) defines a distinct morphological subtype of high-grade urothelial carcinoma. J Pathol. 2011;224:270–9. doi: 10.1002/path.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bakkar AA, Wallerand H, Radvanyi F, et al. FGFR3 and TP53 gene mutations define two distinct pathways in urothelial cell carcinoma of the bladder. Cancer Res. 2003;63:8108–12. [PubMed] [Google Scholar]
- 28.Lopez-Knowles E, Hernandez S, Malats N, et al. PIK3CA mutations are an early genetic alteration associated with FGFR3 mutations in superficial papillary bladder tumors. Cancer Res. 2006;66:7401–4. doi: 10.1158/0008-5472.CAN-06-1182. [DOI] [PubMed] [Google Scholar]
- 29.Lindgren D, Sjodahl G, Lauss M, et al. Integrated genomic and gene expression profiling identifies two major genomic circuits in urothelial carcinoma. PLoS One. 2012;7:e38863. doi: 10.1371/journal.pone.0038863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scatter plot of mutation count and copy number alterations in 83 upper tract urothelial carcinoma tumors profiled with MSK-IMPACT. Ultra-mutated case (422 somatic mutations) highlighted in red.
(A) Schematic illustrating breakpoints in intron 17 of FGFR3 and intron 10 of TACC3, (B) Confirmation of translocation breakpoint using PCR and Sanger sequencing, (C) Predicted mRNA transcript and FGFR3-TACC3 fusion protein.
Oncoprint of the (A) RTK/RAS/MAPK and (B) p53 pathways showing mutual exclusivity.
Supplementary Table 1 — Comparison of frequencies of gene alterations in high-grade upper tract urothelial carcinoma (UTUC) and high-grade urothelial carcinoma of the bladder (UCB)