Abstract
Many microbial communities contain organized patterns of cell types, yet relatively little is known about the mechanism or function of this organization. In colonies of the budding yeast Saccharomyces cerevisiae, sporulation occurs in a highly organized pattern, with a top layer of sporulating cells sharply separated from an underlying layer of nonsporulating cells. A mutant screen identified the Mpk1 and Bck1 kinases of the cell-wall integrity (CWI) pathway as specifically required for sporulation in colonies. The CWI pathway was induced as colonies matured, and a target of this pathway, the Rlm1 transcription factor, was activated specifically in the nonsporulating cell layer, here termed feeder cells. Rlm1 stimulates permeabilization of feeder cells and promotes sporulation in an overlying cell layer through a cell-nonautonomous mechanism. The relative fraction of the colony apportioned to feeder cells depends on nutrient environment, potentially buffering sexual reproduction against suboptimal environments.
Keywords: cell-wall integrity, cell permeability, cell-cell signaling, Saccharomyces cerevisiae, sporulation
AS embryos develop, cells of different fates organize into patterns [reviewed in Kicheva et al. (2012) and Perrimon et al. (2012)] . Intriguingly, even unicellular microbial species form communities in which different cell types are organized into patterns [reviewed in Kaiser et al. (2010), Honigberg (2011), Vachova et al. (2012), and Loomis (2014)]. For example, colonies of the budding yeast Saccharomyces cerevisiae form an upper layer of larger cells (U cells) overlying a layer of smaller cells (L cells). U and L cells differ in their metabolism, gene expression, and resistance to stress, and U and L layers are separated by a strikingly sharp boundary (Cap et al. 2012; Vachova et al. 2013). Patterns are also observed in yeast biofilms, where cells closest to the plastic surface grow as ovoid cells, whereas cells further from the surface differentiate into hyphae for Candida species [reviewed in Finkel and Mitchell (2011)] or pseudohyphae and eventually asci for S. cerevisiae (White et al. 2011).
Sporulation also occurs in patterns within yeast colonies. Specifically, a narrow horizontal layer of sporulated cells forms through the center of the colony early during colony development. As colonies continue to mature, this layer progressively expands upward to include the top of the colony; this wave is driven by progressive alkalization and activation of the Rim101 signaling pathway (Piccirillo et al. 2010). In contrast, cells at the bottom of the colony, i.e., directly contacting the agar substrate, also sporulate at early stages of colony development, but this narrow cell layer does not expand as the colony matures (Piccirillo et al. 2010). The same colony sporulation pattern is observed in a range of laboratory yeasts as well as in S. cerevisiae and S. paradoxus isolated from the wild. Indeed, in these wild yeasts, the same colony sporulation pattern forms on a range of fermentable and nonfermentable carbon sources (Piccirillo and Honigberg 2010).
The mechanism of sporulation patterning and its function remain mysterious. Colony sporulation patterns may reflect differences in nutrient environment across the community as well as cell-to-cell signals within communities [reviewed in Honigberg (2011)]. One function of sporulation patterning may be to localize sporulated cells to the surfaces of colonies to maximize their dispersal; spores are resistant to environmental stress and may be largely dispersed by insect vectors that feed at the surfaces of these microbial communities [reviewed in Neiman (2011)]. A second possible function of sporulation patterning is to efficiently distribute limited nutrients within the community. Although sporulation is a response to nutrient starvation, paradoxically, it is also a program that requires a sizable energy investment (Ray et al. 2013). Energy is required for chromosomes to replicate, pair, recombine, and segregate in meiosis; for spore-wall formation [reviewed in Kupiec et al. (1997)]; and for the induction of hundreds of gene products, some to very high levels (Chu et al. 1998; Primig et al. 2000; Grassl et al. 2010). In laboratory sporulation cultures, respiration of acetate provides the energy for sporulation, but it is less clear how wild yeast communities obtain this energy.
To investigate the mechanism and function of colony sporulation patterning, we identified genes specifically required for efficient sporulation in colonies. Characterization of the role of one such gene, RLM1, in sporulation patterning led to the discovery that the nonsporulating layer contains “feeder cells” that promote meiosis in the overlying cell layer through a cell-nonautonomous mechanism. The fraction of cells in the colony apportioned to the feeder layer depends on environment, and this “differential partitioning” may serve to buffer efficient sexual reproduction against suboptimal environments.
Materials and Methods
Strains
All strains used in this study are shown in Supporting Information, Table S1. The genetic screen was performed on the Homozygous Yeast Knockout Collection (Open Biosystems), and all other experiments (except for Figure 1, A and B) were conducted on mutants constructed in the W303 strain background (SH3881). Deletion mutants in the W303 strain background were constructed using PCR fragments to delete >90% of the ORF, and these alleles were verified by diagnostic PCR using primers flanking the targeted region (Gray and Honigberg 2001; Gray et al. 2005).
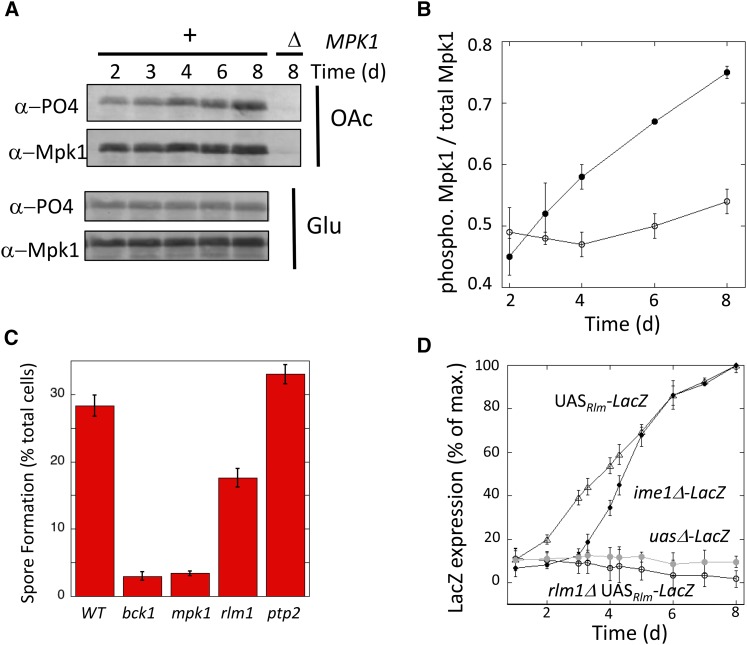
Figure 1.
CWI pathway induced in sporulating colonies. (A) Western blot of phosphorylated Mpk1 (α-PO4) and total Mpk1 (α-Mpk1) levels at the indicated times on either acetate (OAc) or glucose (Glu) growth medium for MPK1+ (lanes 1–5, SH2081) and mpk1Δ (lane 6, SH 4704) strains. (B) Ratio of phosphorylated Mpk1 to total Mpk1 in colonies on acetate (filled circles) or glucose (open circles) growth medium (n = 3). (C) Sporulation in 4-day spot colonies of indicated genotype. Mutants constructed in the W303 background: wild type (SH3881), bck1Δ (SH4770), mpk1Δ (SH4324), rlm1Δ (SH4767), and ptp2Δ (SH4502 and SH4503) (n =6). (D) Expression of UASRlm1-lacZ in RLM1+ (triangles, SH4838) and rlm1Δ spot colonies (open circles, SH4848) and expression of ime1Δ-lacZ (solid circles, SH4924) and uasΔ-lacZ in RLM1+ strains (shaded circles, SH4839) (n = 3).
Media and growth
The medium used for the mutant screen in the S288C background (SPO) contains 1% potassium acetate, 0.1% yeast extract, and 0.05% glucose and amino acids/nucleic acids to balance auxotrophies. For assays of colony behavior, except as noted, spot colonies were inoculated with 1 × 105 cells in 0.5 μl of water on YNA-2 plates (Piccirillo et al. 2010) or on the same medium containing 40 μg/ml X-gal (Fisher Scientific). Alternatively (e.g., for colony sections), ∼300 cells were spread on YA plates (2% potassium acetate, 0.5% yeast extract, and 2% agar, pH 6.0). Other media used in this study have been described previously (Piccirillo and Honigberg 2010; Rose et al. 1990).
Mutant screen
Cultures from the yeast deletion collection were pinned from 96-well thawed glycerol stocks to microtiter plates containing 200 μl of YPDA yeast medium per well. Plates were sealed, incubated for 40 hr at 30°, and then used to inoculate plates containing 200 μl of YPA medium per well. After incubation for 72 hr at 30°, these latter cultures were pinned to single-well microtiter plates containing SPO agar medium. Plates were sealed and incubated for 2 weeks at 30° and then assayed for sporulation levels.
Of the 450 candidate mutants identified in the initial screen, ∼250 had not been annotated previously as sporulation defective in cultures. These 250 mutant strains were consolidated into a 96-well-format secondary collection, which also contained multiple isolates of the isogenic wild-type control strain, and rescreened as earlier. Candidates identified in this rescreen that sporulated at <6% of the wild type were consolidated into a tertiary collection (22 mutants) that contained multiple isolates of each candidate along with control strains, and these tertiary collections were rescreened as earlier. Tertiary collections also were screened for sporulation efficiency in cultures as follows: 300 μl of YPDA cultures were grown overnight at 22° with shaking in 24-well microtiter plates, then harvested, washed twice in water, resuspended in 300 μl of SPO medium, and incubated as earlier for 72 hr.
Assays for Mpk1 phosphorylation
To extract protein from colonies, ∼1000 cells per plate were spread on SPO plates. After incubation, all colonies on the plate were pooled and suspended in 1 ml of sterile water. Approximately, 5 × 107 cells from these suspensions were harvested, washed, and the protein extracted as described by von der Haar (2007). Western blot analysis was performed with lanes containing protein from equivalent numbers of cells. Mpk1 primary Ab (1:600; Santa Cruz Biotechnology) was used with anti-goat IgG-HRP secondary Ab (1:30,000; Santa Cruz Biotechnology). Phosph-p44/42 MAPK (Thr202, Tyr206) Ab (1:2000; Cell Signaling Technology) was used with anti-rabbit IgG-HRP secondary Ab (1:2000; Cell Signaling Technology). Signal was detected using ECL Plus reagents (Amersham), and all incubations and washes after transfer were adapted from the protocol provided by the manufacturer. Western blot images were captured using a Typhoon 9400 imager and quantified using ImageQuant software (National Institutes of Health).
Colony sectioning
Colonies grown on YA medium were embedded in Spurr’s medium, stained (omitting OsO4), sectioned, and the distribution of spores quantified as described previously (Piccirillo and Honigberg 2011; Piccirillo et al. 2011). Images of embedded sections were adjusted for brightness and contrast. Colonies were visualized for LacZ or permeability to propidium iodide (PI) by modifying a previously described method (Cap et al. 2012). In brief, the colony plus an underlying 3-mm square of agar were removed to a glass slide and overlaid with 2% agar containing 120 μg/ml of X-gal, 0.1% SDS, and 6% dimethylformamide (DMF; except where noted) or 16 μg/ml PI. After X-gal overlay, colonies were incubated at 30° for 24 hr (except where noted). To section the colonies, a razor blade attached to a micromanipulator was used to cleave the colony and block in half from top to bottom. The cleaved blocks then were trimmed to an ∼3-mm cube, and the exposed side of the colony was placed face down in mounting medium and examined by fluorescence microscopy. Images of GFP, RFP, LacZ, and PI samples were adjusted for brightness only.
Assays for colony sporulation efficiency, viability, gene expression, cell autonomy, osmosensitivity, and cell permeability
To determine sporulation efficiency, colonies were scraped from a plate, resuspended in 1 M sorbitol or water, and examined by microscope. For the mutant screen, ∼100–200 cells were viewed in the microscope to calculate sporulation efficiency; for all other experiments, 250–300 cells were viewed.
Determination of gene expression in pure and chimeric colonies was as described previously (Piccirillo et al. 2010), except that in addition to measuring LacZ expression, in the current study we also measured expression of GFP and RFP (mCherry) fusion genes. For this purpose, digital images of single colonies were captured using an Olympus SZX12 stereomicroscope equipped with a fluorescence source, and these images were quantified using ImageJ software.
To determine the ratio of reporter to signal cells after growth is complete, 2- to 3-day colonies were resuspended in 1 M sorbitol and plated on YPD medium. Colonies from this plate then were patched on a master plate and replica plated to MPX medium to identify ime2-lacZ isolates or to SP3+ medium for 4 days and examined under a blue-light lamp (Dark Reader, Clare Chemical Research) to identify ime2-GFP isolates. Expression of the reporter in chimeric colonies was standardized to the average fraction of reporter cells in the colonies (n=3).
To measure the viability of nonsporulated cells, spot colonies were suspended and diluted in 1 M sorbitol and plated on synthetic medium containing 0.5 M sorbitol. To ensure that only diploid cells could grow into colonies, the medium lacked both histidine and lysine (Lee and Honigberg 1996). To measure osmosensitivity of nonsporulated diploids, the preceding cell suspensions also were diluted 100-fold in distilled water and plated on His− Lys− medium lacking sorbitol. Osmosensitivity is calculated as colony-forming units (cfu) per cells plated for colony suspensions diluted and plated in the absence of sorbitol divided by cfu per cells plated for the same suspensions diluted and plated in the presence of sorbitol. To measure cell permeability, spot colonies were completely scraped from the plate and resuspended in a 2 M sorbitol solution containing 10 μg/ml PI and examined under UV illumination.
Statistics and reproducibility
All quantitative data in the study are expressed as the mean ± SEM of at least three biological replicas, with error bars representing the SEM. P-values are from unpaired Student’s t-tests (raw). All experiments were replicated on at least two separate dates, and all experiments comparing the wild-type strain to mpk1Δ or rlm1Δ mutants in the W303 background were performed with at least two independently derived mutant isolates. In contrast, experiments with mutants from the yeast deletion collection were performed on the single isolates present in the collection. All experiments comparing two or more strains that were based on scoring cells (e.g., spore formation, GFP expression, and PI permeability) were performed as double-blind experiments, and at least 250 cells were counted for each sample.
Data availability
All strains used in this study and all data gathered from the mutant screen and rescreens are available on request.
Results
Screen for mutants defective in sporulation specifically in colonies
Because sporulation in colonies displays properties not seen in cultures, such as sporulation patterns, we reasoned that some genes would be specifically required for sporulation in colonies. To identify these genes, we screened the yeast homozygous diploid deletion (YKO) series (Giaever et al. 2002) for mutants defective in colony sporulation, and we then rescreened these candidates for any that had normal sporulation levels in suspended cultures of the same medium (see Materials and Methods). Of the seven mutants identified (Table S2), three (MPK1/SLT2, BCK1, and SMI1) are deleted for genes encoding enzymes in the cell-wall integrity (CWI) pathway.
To confirm that the CWI pathway is required for sporulation in colonies, we performed two additional experiments. First, we measured sporulation in other mutants deleted for genes encoding core/nonredundant components of this pathway. In addition to Mpk1 (the MAPK) and Bck1 (the MEKK), Wsc1, a receptor that activates the pathway; Tus1, a GEF that positively regulates the pathway; and Rlm1, a transcription factor phosphorylated and activated by Mpk1, are all required for sporulation in colonies. Furthermore, these genes were required for colony sporulation to a much greater extent than they were required for sporulation in cultures (Table S3). Second, because the CWI pathway responds to cell-wall damage, we tested the possibility that the sporulation defect in cwiΔ colonies simply resulted from lower viability in these mutants. However, most cwiΔ mutants that were defective in colony sporulation retained at least as high viability in colonies as the wild-type strain (Table S3).
CWI pathway induced in wild-type colonies
As an independent test of the hypothesis that the CWI pathway is required for colony sporulation, we determined whether this pathway is activated as colonies mature. One measure of CWI pathway activation is phosphorylation of Mpk1 (de Nobel et al. 2000; Martin et al. 2000). Colonies were grown on medium containing either acetate, which promotes both colony growth and subsequent sporulation, or glucose, which only promotes growth, not subsequent sporulation. Samples were removed at the indicated times from 2 to 8 days and assayed for total levels of Mpk1 and for the activated Mpk1 phospho-isoform by Western blot. On acetate medium, phosphorylation of Mpk1 increased as colonies matured (Figure 1, A and B, filled circles). In contrast, on the otherwise-identical glucose medium, colonies failed to induce Mpk1 phosphorylation over the same time period (Figure 1, A and B, open circles).
We next asked whether the CWI pathway regulated colony sporulation in the W303 strain background; colony sporulation is more rapid and efficient in W303 than in the S288C background used for the mutant screen. We constructed mpk1Δ, bck1Δ rlm1Δ, and ptp2Δ mutants in the W303 background and measured spore formation in 4-day colonies (Figure 1C). The mpk1Δ, bck1Δ, and rlm1Δ mutants all had much lower colony sporulation than the wild-type strain. The rlm1Δ mutant had significantly higher sporulation than the other two mutants, perhaps reflecting that Rlm1 is only one of the targets regulated by the Bck1-Mek1/2-Mpk1 MAP kinase pathway. In addition, the ptp2Δ mutant displayed a small but likely significant (P = 0.048) increase in colony sporulation relative to the wild-type strain, consistent with the function of Ptp2 as a protein phosphatase that inactivates Mpk1 (Mattison et al. 1999). Based on the high levels of colony sporulation in the W303 strain background and the involvement of multiple components of the CWI pathway in this sporulation, the W303 background was used for all remaining experiments in this study.
As a second test of CWI pathway activation in colonies, we monitored Rlm1 activity as colonies matured. Rlm1 activation was detected using a hybrid promoter (UASRlm1-CYC1-lacZ) (Jung et al. 2002), and LacZ expression in colonies was quantified as described by Piccirillo et al. (2010). This reporter gene was induced several hundred-fold as colonies matured, and this induction depended on both RLM1 and UASRlm1 (Figure 1D). Interestingly, colonies activate Rlm1 1–2 days prior to the transcription of IME1, as measured using an IME1-promoter/lacZ fusion gene. IME1 is one of the first genes activated in meiosis; this gene encodes a transcriptional activator that serves as a master regulator for meiotic initiation.
Rlm1 is required for early meiotic gene expression
Because Rlm1 is activated in colonies before IME1 is expressed, we asked whether Rlm1 was required for transcription of this master regulator. Indeed, we found significantly higher ime1Δ-lacZ expression in RLM1+ colonies than in rlm1Δ colonies (Figure 2A, left). Similarly, we determined the effect of rlm1Δ on expression of a second regulator of meiosis, IME2. IME2 encodes a protein kinase that activates multiple targets during early and middle stages of meiosis [reviewed in Honigberg (2004)]. IME2 transcription is directly activated by binding of Ime1 to the IME2 promoter, but IME2 transcription is also regulated via IME1-independent pathways [reviewed in Kassir et al. (2003)]. Consistent with the ime1Δ-lacZ results, we found that ime2Δ-GFP/IME2+ strain expressed the fusion gene to higher levels in wild-type (RLM1+) colonies than in rlm1Δ colonies (Figure 2A, right). Thus, Rlm1 promotes transcription of both IME1 and IME2 in colonies.
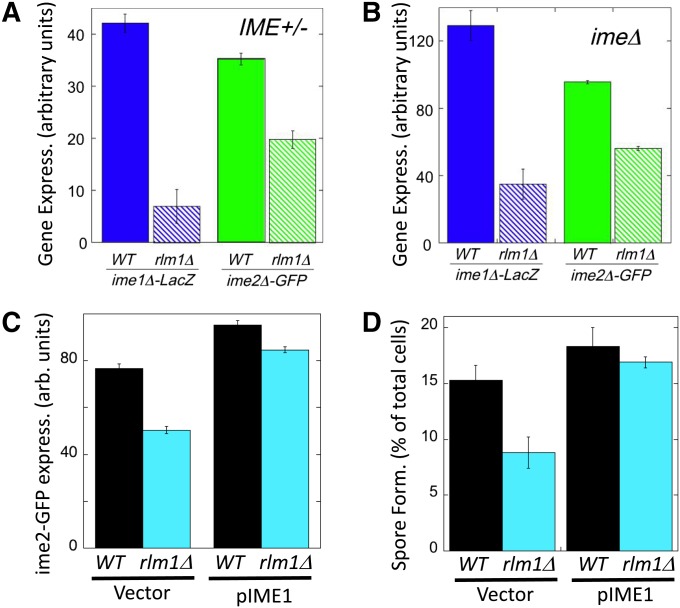
Figure 2.
An rlm1Δ mutant affects IME1 and IME2 expression and is suppressed by IME1 overexpression. (A) Effect of rlm1Δ on IME expression in IME+ colonies. ime1Δ-lacZ/IME1+ and ime2Δ-GFP/IME2+ expression in 6-day spot colonies that are either RLM1+ (wild type = solid bars, SH3827 and SH5105, respectively) or rlm1Δ (hatched bars, SH4799 and SH4789, respectively) (n = 4). (B) Effect of rlm1Δ on IME expression in ime1Δ colonies. ime1Δ-lacZ/ime1Δ and ime2Δ-GFP/ime2Δ expression in 6-day spot colonies that are either RLM1+ (wild type = solid bars, SH3830 and SH5071, respectively) or rlm1Δ (hatched bars, SH4800 and SH5072, respectively) (n =4). (C) Effect of IME1 overexpression on ime2-GFP expression in rlm1Δ colonies. GFP expression levels in RLM1 ime2-GFP and rlm1Δ ime2-GFP spot colonies containing only the control vector (left, SH 5280 and SH 5276) or containing a high-copy IME1 plasmid (right, SH5282 and SH5278) (n = 4). (D) Effect of IME1 overexpression on spore formation in rlm1Δ colonies. RLM1+ and rlm1Δ spot colonies contain only the control vector (SH5074 and SH5094) or contain a high-copy IME1 plasmid (SH5073 and SH5085) (n = 3).
IME1 and IME2 are regulated through both positive- and negative-feedback loops (Rubinstein et al. 2007; Ray et al. 2013). To determine whether Rlm1 acts entirely upstream of IME1 vs. via feedback controls, we measured ime1Δ-lacZ expression in strains lacking functional Ime1 (i.e., ime1Δ-lacZ/ime1Δ), and we also measured ime2Δ-GFP expression in strains lacking functional Ime2 (i.e., ime2Δ-GFP/ime2Δ). We found that expression of either IME gene is still significantly decreased in rlm1Δ mutants when the corresponding Ime protein is absent (Figure 2B). Thus, consistent with the relative timing of Rlm1 and IME1 activation in colonies, Rlm1 acts upstream of both IME1 and IME2 to stimulate colony sporulation.
To confirm that Rlm1 stimulates spore formation in colonies largely by activating IME1 transcription, we overexpressed IME1 in a rlm1Δ colony using a high-copy-number plasmid (Lee and Honigberg 1996; Honigberg and Lee 1998) and measured both ime2-GFP expression and spore formation. Overexpression of IME1 increased ime2-GFP expression (Figure 2C) and spore formation (Figure 2D) in both RLM1+ and rlm1Δ colonies; thus, IME1 expression limits sporulation in both strains. Furthermore, IME1 overexpression in rlm1Δ colonies caused this mutant to both express ime2-GFP and form spores at levels comparable to RLM1+ colonies (Figure 2, C and D). Thus, IME1 is a major target of Rlm1 in regulating colony sporulation.
Rlm1 activation in colonies occurs in a separate population of cells than meiotic initiation
Although Rlm1 is required for IME1 expression in colonies, the IME1 promoter lacks obvious Rlm1 binding sites, so we asked whether Rlm1 is activated in the same cells in colonies as IME1 is transcribed. For this purpose, we used the UASRlm1-lacZ and ime2Δ-GFP alleles described earlier. Colonies were grown for 6 days and then covered with agar containing X-gal, sectioned, and examined by fluorescent and reflected-light microscopy. As expected from previous studies (Piccirillo et al. 2010), ime2Δ-GFP was expressed in a band of cells extending from the middle to the top of the colony (Figure 3A, center-left panel). In contrast, the UASRlm1-lacZ allele was expressed in a separate horizontal band of cells extending from the bottom to the middle of the colony (Figure 3A, top left panel). The GFP and LacZ expression patterns were adjoining but not overlapping (Figure 3A, bottom panel). To quantify the populations of cells in colonies expressing ime2Δ-GFP and/or UASRlm1-lacZ, we resuspended cells from 6-day colonies and measured the frequency of cells in the overall population that express one or both fusion genes. Whereas most of the cells in the colony expressed one or the other gene, very few cells expressed both genes (Figure 3, B and C).
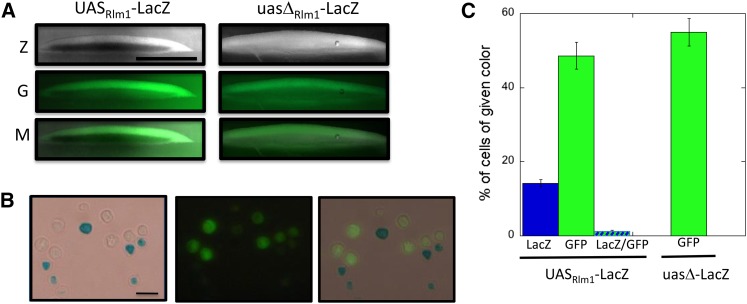
Figure 3.
ime2-GFP expression and Rlm1activation occur in separate colony regions and populations. (A) Section of 6-day ime2Δ-GFP UASRlm-lacZ colony (left, SH5065) and ime2Δ-GFP uasΔ-lacZ colony (right, SH5067) overlaid with X-gal agar (see Materials and Methods). Z = LacZ expression (dark region in left image corresponds to blue region of colony); G = GFP; and M = merge of Z and G images. Scale bar, 500 μm. (B) Cells resuspended from ime2Δ-GFP UASRlm-lacZ spot colony as in A except with 230 μg/ml X-gal and DMF and SDS omitted from overlay agar. LacZ (left), GFP (center), and overlays (right). Scale bar, 10 μm. (C) Quantification of percentage of cells in colonies as in B expressing GFP and/or LacZ in the same strains; cells were scored by light/fluorescence microscopy (n = 3).
To control for possible interference on the GFP signal from the blue X-gal metabolite, we compared the fraction of cells expressing GFP in a strain carrying the UASRlm1-lacZ allele to a control strain carrying a variant lacZ allele lacking this UAS (Figure 3C). We found that GFP expression was not significantly different in the two strains. Similarly, colony sections from this control strain displayed a similar pattern of ime2-GFP expression limited to a top layer in the colony (Figure 3A, right side). Thus, IME2 is expressed in a cell population within colonies that is largely separate from the cell population in which Rlm1 is active.
Rlm1 regulates colony sporulation via a cell-nonautonomous mechanism
Taken together, the preceding experiments indicate that Rlm1 is activated in an underlying layer of cells in colonies, but Rlm1 stimulates sporulation in a separate overlying layer of cells. These results suggest that Rlm1 promotes sporulation through a cell-nonautonomous mechanism. To test cell autonomy directly, we employed the chimeric colony assay (Piccirillo et al. 2010). In brief, this assay (Figure 4A; see also Materials and Methods) uses spot colonies containing approximately equal concentrations of a “reporter strain” and a “signal strain.” The reporter strain carries a reporter gene, in the present case the ime1Δ-RFP allele. The signal strain lacks this reporter gene but contains either a wild-type or mutant allele (in the present case, RLM1 or rlm1Δ). Reporter gene expression is compared in chimeric colonies containing either RLM1 or rlm1Δ signal strains. If the genotype of the signal strain affects expression of the reporter gene, then it must be doing so by a cell-nonautonomous mechanism (Figure 4A).
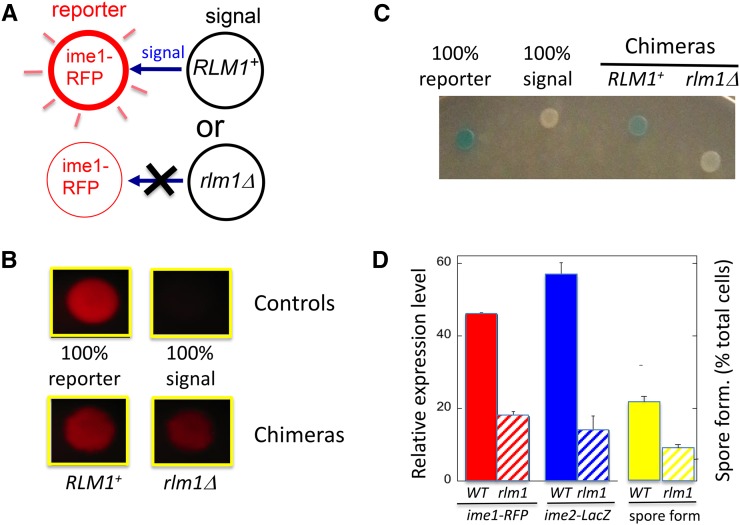
Figure 4.
Rlm1 promotes IME expression and spore formation by a cell-nonautonomous mechanism. (A) Diagram of chimeric colony assay for testing cell autonomy. Chimeric colonies contain equal numbers of reporter cells (left), which carry a reporter allele (e.g., ime1Δ-RFP), and signal cells (right), which lack the reporter. If the genotype of the signal cell affects the response from the reporter cells, the corresponding gene must regulate the response via a nonautonomous mechanism. (B) Effect of rlm1Δ allele on ime1Δ-RFP expression in chimeric colonies. Top two control colonies contain either only the RFP reporter strain (left, SH4414) or only the RLM1+ signal strain (right, SH5071). Bottom two colonies contain chimeras with a mixture of the reporter strain and either RLM1+ (left, SH5071) or rlm1Δ (right, SH5072) signal strains. (C) Effect of rlm1Δ allele on ime2Δ-lacZ expression in chimeric colonies. (From left to right) The colonies contain only the ime2Δ-lacZ reporter strain (SH3825), only the RLM1+ signal strain (SH3883), an RLM1+ chimera (equal mixture of SH3825 and SH3883), and an rlm1Δ chimera (equal mixture of SH3825 and SH4783) (n =4). (D) Quantification of response in RLM1+ (solid bars) or rlm1Δ (hatched bars) chimeric colonies: reporter cells measure either ime1Δ-RFP expression as in B (left, red; n = 5), ime2Δ-lacZ expression as in C (center, blue) or spore formation (right, yellow; n = 3), with the SPO+ reporter strain being the RLM1+ IME2+ strain (SH3881) and the signal strains being the SPO− strains SH3883 (ime2Δ RLM1+) and SH4783 (ime2Δ rlm1Δ) (n = 3).
Our results support the idea that Rlm1 promotes sporulation through a cell-nonautonomous mechanism. That is, we found that chimeric colonies containing an rlm1Δ signal strain expressed ime1-RFP in the reporter strain to a much lower extent than when the signal strain was RLM1+ (Figure 4, B and D, left). As a second test of autonomy, we also measured expression of an ime2-lacZ reporter strain in chimeras containing either an RLM1+ or rlm1Δ signal strain. As with the ime1-RFP reporter, the ime2-lacZ reporter was expressed to significantly lower levels in rlm1Δ chimeras than in RLM1+ chimeras (Figure 4, C and D, center). In addition, we observed similar results when the reporter strain is rlm1Δ rather than RLM1+ (Figure S1). Finally, we used a variation of the chimeric colony assay in which the signal strains were ime1Δ (as well as either RLM1 or rlm1Δ), allowing us to specifically measure spore formation (rather than expression) in the IME+ reporter strain. In this case, the reporter strain formed spores to significantly lower levels in chimeras containing the rlm1Δ signal strain than when the signal strain was RLM1+ (Figure 4D, right; P = 0.015).
As one caveat regarding chimeric colony assays, these assays cannot exclude the possibility that Rlm1 stimulates sporulation through both cell-autonomous and cell-nonautonomous mechanisms. However, the population studies described in the preceding section indicate that IME2 expression and Rlm1 activation occur in largely different subpopulations of the colony, and this result is more consistent with an entirely nonautonomous mechanism. Taken together, all three chimeric colony assays, along with the pattern of Rlm1 activation, suggest that Rlm1 activity is required in the underlying cell layer to stimulate sporulation in the overlying cell layer (i.e., cell nonautonomy).
Rlm1 is required for sporulation pattern in colonies
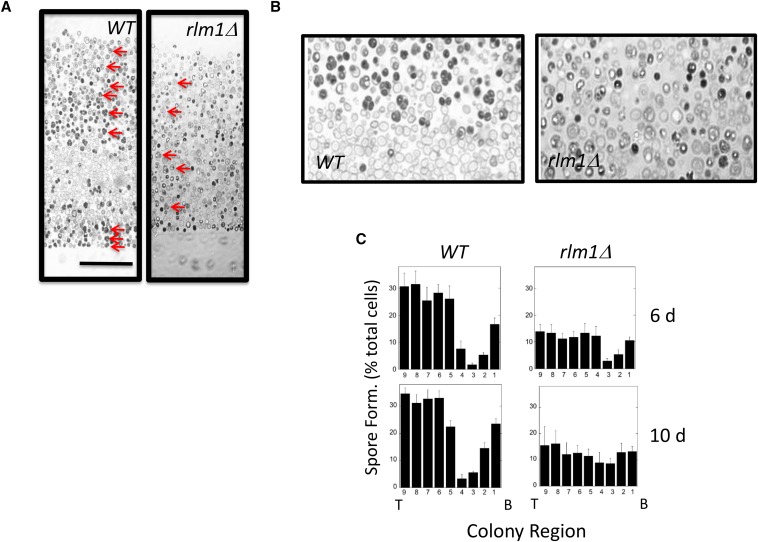
Because Rlm1 stimulates colony sporulation through a cell-nonautonomous mechanism, we asked whether Rlm1 is involved in colony sporulation patterning. For this purpose, we embedded and sectioned RLM1+ and rlm1Δ colonies. As observed previously (Piccirillo and Honigberg 2010; Piccirillo et al. 2010), wild-type colonies incubated for 6 days displayed two sharply defined layers of sporulating cell separated by a layer of nonsporulating cells (Figure 5, A and B, left). In contrast, in rlm1Δ colonies, no clear sporulation pattern was observed, with low levels of asci distributed throughout the colony (Figure 5, A and B, right). We confirmed this difference by quantifying the distribution of asci in multiple RLM1+ and rlm1Δ colonies incubated for either 6 or 10 days (Figure 5C). Even in rlm1Δ colonies, a region of less efficient sporulation was sometimes observed in the central region of the colony at early times (Figure 5C), but this layer was always narrower and less defined than in wild-type colonies. These results indicate that Rlm1 is required for the sharp boundary between nonsporulated and sporulated layers formed in wild-type colonies.
Figure 5.
Patterns of sporulation and RLM1 expression in colonies. (A) Central region of a section from 10-day RLM1+ (wild type, SH3881) and rlm1Δ (SH4767) colonies. Arrows point out representative asci in both sections. Scale bar, 50 μm. (B) Higher-magnification image of the central region in the image shown in A. (C) Distribution of sporulated cells in RLM1+ and rlm1Δ colonies at the indicated times. To determine this distribution, a grid containing nine equal-sized rectangles stacked on top of one the other (long side horizontal) was superimposed on each section and scaled to just cover the central region of the colony from bottom to top. Distributions show the percentage of the population recognized as asci in each region of the colony from the bottom (B, left) to top (T, right) of the colony. Each distribution shows mean percentage sporulation for four or more colonies.
We previously reported that in wild-type colonies many cells in the nonsporulated layer are characteristically enlarged and lightly stained relative to other cells in the colony (Piccirillo and Honigberg 2010) (Figure 5, A and B, left). In this study, we observed that these enlarged cells were largely absent from rlm1Δ colonies (Figure 5, A and B, right). Because Rlm1 is specifically expressed in an underlying layer of cells in the colony, and because Rlm1 acts nonautonomously to promote sporulation in the overlying layer (and hence sporulation patterning), we term the cells of the underlying layer feeder cells.
Nonsporulated cells become osmosensitive as colonies develop
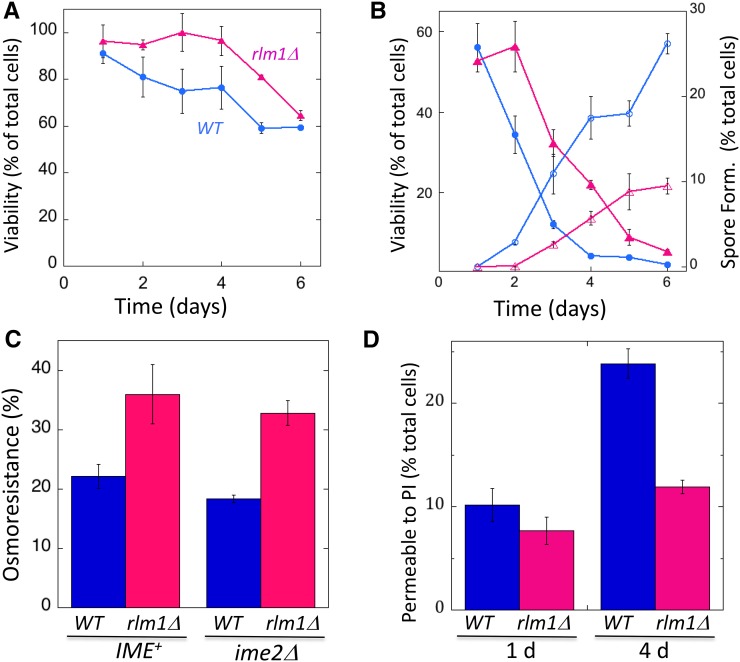
The distinctive size and appearance of wild-type feeder cells in embedded colonies prompted us to investigate their viability and osmosensitivity. Because spores retain high viability under a variety of stresses (Neiman 2011), we used a genetic assay to measure viability specifically in the nonsporulated (i.e., diploid) cells (see Materials and Methods).
As an initial experiment, we resuspended cells from RLM1+ or rlm1Δ colonies in a high-osmolarity solution (1 M sorbitol) used to provide osmostability to the cells and then plated these cells on medium adjusted to the same osmolarity. Under these osmolarity-stabilizing conditions, wild-type diploids showed a relatively constant viability over 6 days (Figure 6A), at which time colony sporulation was largely complete (Figure 6B). Consistent with the relatively high viability of most cwiΔ mutants in colonies (Table S2), rlm1Δ diploids displayed at least the same level of viability as wild-type diploids (Figure 6A).
Figure 6.
RLM1 increases the permeability of nonsporulated cells in colonies. (A) Viability of nonsporulated (diploid) cells under osmotically stabilized conditions. Wild-type colonies, blue circles (SH3881); rlm1Δ colonies, magenta triangles (SH4708) (n =3). (B) Viability of nonsporulated cells from same colonies as in A after resuspension in water and plating on standard osmolarity medium. Symbols are the same as in A, and fractions of cells from colonies that have formed spores are shown by corresponding open symbols (n =3). (C) Effect of rlm1Δ on osmoresistance. Four-day RLM1+ colonies (blue bars) compared to rlm1Δ colonies (magenta bars) in both IME2+ (left side, SH3881 and SH4767, respectively) and ime2Δ (right side, SH3883 and SH5072) backgrounds. All colonies were resuspended in sorbitol, and the suspension was split and then diluted and plated either in the presence or absence of sorbitol (n = 3). Osmoresistance = 100% × cfu (absence of sorbitol) per cfu (presence of sorbitol). (D) Permeability to PI. Fraction of cells from colonies at the indicated times that are permeable to PI after resuspension in 2 M sorbitol. (C and D) RLM1+ (wild type, blue bars, SH3830) and rlm1Δ (magenta bars, SH4800) colonies were assayed (n = 3).
To monitor osmosensitivity, we resuspended colonies in water and then plated these cells on standard medium not adjusted for osmolarity. Surprisingly, resistance of diploid cells to low-osmolarity conditions dropped dramatically as wild-type colonies matured. Interestingly, this decreased osmoresistance occurred at the same time or slightly before sporulation levels increased in the colony (Figure 6B). These results suggest that nonsporulated diploids become more permeable as the colonies mature.
Rlm1 affects timing of osmosensitivity/permeability in developing colonies
We next measured osmosensitivity in rlm1Δ colonies. Interestingly, whereas rlm1Δ diploids also became osmosensitive as colonies matured, this transition was delayed by 1–2 days in rlm1Δ colonies relative to wild-type colonies (Figure 6B). As for the wild-type colonies, rlm1Δ colonies become osmosensitive at approximately the same time or slightly before spore formation (Figure 6B).
To directly compare osmoresistance in wild-type and rlm1Δ colonies, 4-day colonies were resuspended in sorbitol, and aliquots from this cell suspension were either diluted in sorbitol and plated on medium containing sorbitol or diluted in water and plated in medium lacking sorbitol. Osmoresistance of diploid cells is defined as the fraction of diploids forming colonies in the absence of sorbitol divided by the fraction of diploids forming colonies in the presence of sorbitol. We found that osmoresistance of rlm1Δ diploids at 4 days was significantly higher in RLM1+ diploids (Figure 6C), and this is true for both in IME+ colonies (P = 0.048, n = 6) and ime2Δ colonies (P = 0.003, n = 3).
As an independent test of the effect of Rlm1 on cell permeability, we resuspended cells from 1- and 4-day colonies in a sorbitol solution containing the nucleic acid stain PI and determined the fraction of the cell population permeable to this dye. Because permeability to PI increases in sporulating cells (Figure S2), we performed these experiments in an ime1Δ background, i.e., in the absence of sporulation. In RLM1+ colonies, the permeability to PI increased two- to threefold from 1 to 4 days (Figure 6D, left). In contrast, in rlm1Δ colonies, there was little or no increase in PI permeability from 1 to 4 days (Figure 6D, right). Thus, PI permeability increases and osmoresistance decreases as colonies mature, and Rlm1 contributes to both transitions.
Distinct patterns of permeable and meiotic cells in colonies
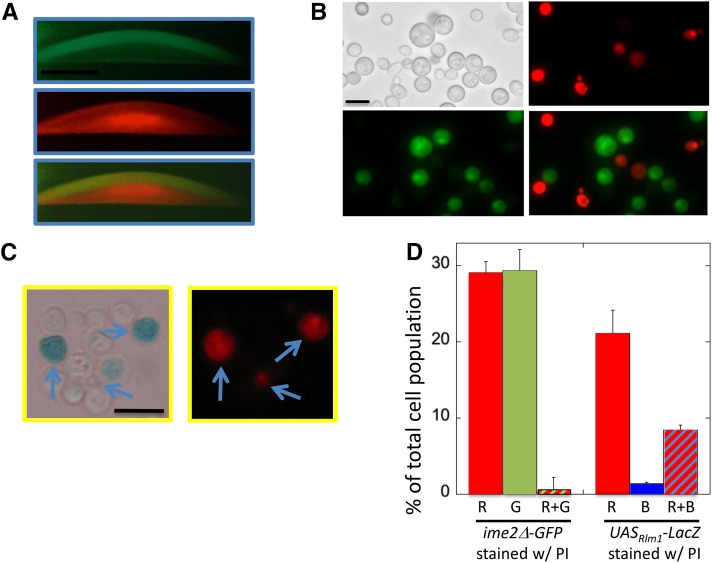
To further investigate the link between cell permeability and sporulation, we examined the localization of these two processes within colonies. Specifically, we compared the pattern of PI permeability to the pattern of expression of an ime2Δ-GFP allele in sectioned colonies. All colonies were ime2Δ for the reason stated for the preceding experiment. Interestingly, after 4 days of incubation, a sharp band of PI-permeable cells directly underlies the broad layer of ime2Δ-GFP expression (Figure 7A).
Figure 7.
PI-permeable feeder cells underlie the sporulation layer in colonies and colocalize in colonies with active Rlm1. (A) Sections of 4-day colonies containing the ime2Δ-GFP allele (SH5071) overlaid with agar containing PI (see Materials and Methods): (top) GFP, (middle) PI, and (bottom) overlay. (B) Six-day colonies as in A resuspended in PI/sorbitol and visualized by bright-field microscopy (top left), red fluorescence microscopy (top right), green fluorescence microscopy (bottom left), and merge of green and red images (bottom right). (C) Six-day spot colonies containing the UASRlm-lacZ allele (SH5065) were overlaid with agar containing 120 μg/ml X-gal and 0.1% SDS and incubated for 2 days; then they were resuspended as in B and visualized by bright-field (left) and fluorescent microscopy (right). Arrows indicate representative cells that are both blue and red. (D) Quantification of cell populations in B and C. (Left) Cells as in B that fluoresce only red (R), only green (G), or both colors (R + G) (n = 4). (Right) Cells as in C, except that incubation after overlay was only 1 day, staining only red (R), only blue (B), or both colors (R + B) (n = 3).
To quantify the overlap between PI-permeable and ime2Δ-GFP-expressing cell populations, we resuspended ime2Δ-GFP colonies in an osmotically stabilized solution, stained the cells with PI, and examined them by fluorescence microscopy. We found that 20–30% of cells stained with PI and a similar percentage expressed detectable levels of ime2Δ-GFP, but almost no cells both expressed ime2Δ-GFP and stained with PI (Figure 7, A and B, left). Thus, as expected, PI-permeable (feeder) cells were largely a distinct population from ime2Δ-GFP-expressing (meiotic) cells.
UASRlm1 is expressed in the same cells that become permeable
Complementary to the preceding experiments, we examined the possible overlap between PI-permeable cells and Rlm1-active cells in colonies because both are characteristic of feeder cells. It was not possible to measure localization of these two populations in the same colony section because PI interferes with LacZ activity. As an alternative approach, colonies containing the UASRlm1-lacZ allele were first incubated in X-gal overlay agar and then resuspended in PI. We found that almost all cells that express UASRlm1-lacZ also were permeable to PI (Figure 7, C and D, right). Interestingly, some cells that were permeable to PI did not detectably express UASRlm1-lacZ. These cells might have lost the plasmid-borne UASRlm1-lacZ allele, or Rlm1 may be inactive in some permeable cells. In summary, population studies of Rlm1 activity, PI permeability, and ime2Δ-GFP expression in colonies demonstrate that Rlm1-active feeder cells are permeable to PI and remain distinct from the meiotic cell population.
Colony partitioning may buffer dependence of sporulation on environment
To investigate the biological function of colony partitioning, we asked whether the relative partitioning of a colony into meiotic and feeder cell fates depends on the colony environment. We found that in wild-type colonies, lowering the concentration of acetate (from 2 to 0.5%; compare rows 1 and 2 in Table 1) led to a higher fraction of the cell population becoming feeder cells, as determined by permeability to PI (Table 1, column 3) or by activation of Rlm1 (Table 1, column 4). Similarly, yeast grows optimally at 30°, but when colonies were grown instead at 34°, these colonies displayed a higher fraction of feeder cells (Table 1; compare rows 1 and 3). Not surprisingly, both of these deviations from optimal sporulation conditions resulted in decreased sporulation (Table 1, column 5). Strikingly, sporulation under suboptimal conditions displayed a much greater dependence on Rlm1 than colonies grown under optimal sporulation conditions (Table 1, columns 5–7). Thus, depending on the environment, yeast colonies vary in partitioning between feeder cells and germline cells. Under stressful/suboptimal conditions, a higher fraction of feeder cells in the colony might increase the overall efficiency of sporulation in the colony.
Table 1. Colony partitioning and Rlm1 dependence in suboptimal sporulation environments.
| Condition | Spore formation (%)c | |||||
|---|---|---|---|---|---|---|
| % KOAc | Temp. (°C) | % Permeablea | % lacZ+b | Wild type | rlm1Δ | Wild type/rlm1Δ |
| 2 | 30° | 31 ± 1 | 6 ± 0.2 | 50 ± 0.2 | 35 ± 2 | 1.4 |
| 0.5 | 30° | 66 ± 1 | 30 ± 2 | 24 ± 2 | 5 ± 2 | 4.8 |
| 2 | 34° | 48 ± 1 | 31 ± 1 | 31 ± 2 | 10 ± 1 | 3.1 |
Percentage of cells in colony permeable to PI after suspension in 2 M sorbitol (n = 3).
Percentage of cells in UASRlm1-lacZ colony producing visible level of X-gal metabolite (n = 3).
Percentage of cells in colony that have formed spores (n = 3).
Discussion
Yeast evolved to proliferate and differentiate within multicellular communities. However, only recently has it become clear that these communities partition into regions sharply defined by differences in gene expression and cell differentiation (see earlier). In this study, we show that Rlm1, a target of the cell-wall integrity MAPK pathway, was activated specifically in an underlying layer of feeder cells in the colony. Rlm1 acts nonautonomously to stimulate meiotic gene expression and spore formation in the overlying germline cell layer and is required for the sharp boundary between feeder and sporulation layers in colonies. One possible explanation for these results is that the function of feeder cells is to supply signals/nutrients that stimulate sporulation in the overlying layer.
Several additional lines of evidence support this hypothesis for feeder-cell function. In the first place, as colonies mature, the nonsporulated cells undergo a transition in which they sharply increase in osmosensitivity and also sharply increase in permeability to PI, consistent with the idea that they release compounds that stimulate sporulation in the overlying layer. In the second place, this transition occurs at the same time or shortly before sporulation in colonies, and rlm1Δ colonies are both delayed in this transition and defective in initiating sporulation. Finally, these PI-permeable cells localize to the feeder-cell layer. Taken together, these results suggest that Rlm1 promotes colony sporulation by promoting the permeability of the feeder-cell layer.
One caveat regarding the idea that Rlm1 stimulates colony sporulation by activating the permeabilization of feeder cells is that whereas nonsporulated cells in rlm1Δ colonies are delayed in the timing of increased osmosensitivity, these colonies eventually become as osmosensitive as wild-type colonies. Thus, if feeder-cell permeabilization stimulates colony sporulation, then the timing of this permeabilization must critical to the stimulation.
A role for Rlm1 in feeder-cell development is consistent with the known activities of the CWI pathway. In addition to responding to cell-wall damage, this pathway is required to remodel the cell wall during several types of differentiation, e.g., during formation of mating projections and polarized cell growth (Zarzov et al. 1996; Buehrer and Errede 1997; van Drogen and Peter 2002). Because the CWI pathway induces cell-wall synthesis, the increased permeability of feeder cells is more likely to result from cell-wall remodeling than from cell-wall damage.
Feeder cells in sporulating colonies may be more analogous to L cells in haploid colonies than to stationary-phase cells in suspended cultures. L cells are found in an underlying layer of cells in haploid colonies and differ in a number of properties from the overlying layer of upper (U) cells. For example, L cells exhibit greater sensitivity to zymolyase treatment (removal of the cell wall) than U cells and greater expression of transporter genes (Cap et al. 2012; Vachova et al. 2013)—both of these properties are consistent with increased permeability. In contrast, stationary-phase cell walls thicken and strengthen (Smith et al. 2000), and as cell volume and turgor pressure increase (Martinez de Maranon et al. 1996), permeability of these stationary-phase cells to water eventually decreases (Suh et al. 2003).
If feeder cells in colonies represent a distinct, mutually exclusive differentiation fate from meiosis and spore formation, this may help to explain the sharp boundary between sporulating and nonsporulating cell layers in colonies—namely, that feeder cells have lost the ability to sporulate. On a similar note, we observed that as colonies mature, an initially narrow layer of sporulating cells in the center of the colony expands upward to eventually include the top of the colony but that this layer mostly does not expand downward (Piccirillo et al. 2010). The unidirectional nature of this expansion would be explained if cells that have already differentiated into feeder cells are blocked from sporulating.
The increased permeability of feeder cells could represent an alternative to programmed cell death (PCD) for redistributing nutrients within yeast communities. It has been proposed that PCD in a subpopulation of a microbial community could provide nutrients necessary for the remaining members of the community to proliferate, both in yeast communities (Fabrizio et al. 2004; Herker et al. 2004; Vachova and Palkova 2005) and in bacterial communities (Tanouchi et al., 2013; Allocati et al. 2015). Such a mechanism is related to but distinct from the findings reported in this study. Rlm1-dependent permeabilization of nonsporulated cells does not rapidly result in cell death; indeed, these cells remain viable for many days after sporulation is complete. We speculate that within the environment of sporulating colonies, the increased permeability of feeder cells could be a more controlled method of transferring nutrients between yeast cells than PCD.
Partitioning of colonies into distinct populations of feeder cells and germline cells may help to resolve the “energy paradox” described earlier. In particular, feeder cells may supply nutrients necessary for the overlying layer of cells to sporulate efficiently. In the wild, communities of yeast are largely clonal; thus, a genotype that promotes more efficient sporulation in the overall population through a range of environments would be subject to positive selection. Indeed, evolutionary selection for partitioning is consistent with colony patterning being observed in multiple laboratory and wild strains of S. cerevisiae and wild S. paradoxus strains (Piccirillo and Honigberg 2010).
Across all species, cells have evolved to be exquisitely responsive to certain extracellular signals. However, the opposite side of the coin may be equally fundamental—cells evolve mechanisms to maximize efficiency in a wide range of environments. One well-known example of this “environmental buffering” is the heat-shock response. Although the influence of diet on reproductive capability has been well demonstrated in Drosophila melanogaster, Caenorhabditis elegans, and mammals (Ashworth et al. 2009; Ables et al. 2012; Hubbard et al., 2013), to our knowledge, mechanisms by which gametogenesis efficiency is maintained under suboptimal environments have not been explored. Our study suggests the hypothesis that differential partitioning of cell fate within a yeast community provides environmental buffering—here termed the DPEB hypothesis. According to the DPEB hypothesis, under suboptimal conditions for sporulation, a larger portion of the entire community is apportioned to feeder cells. This increased feeder-cell population then compensates for the poor environment by providing the energy necessary for the remaining nonfeeder cells in the community to sporulate efficiently. We suggest that DPEB may prove ubiquitous in a wide range of cell types and species, including microbial communities and metazoan development.
Supplementary Material
Acknowledgments
We are grateful to David Levin (Boston University) for the gift of the UASRlm1 plasmids and to Jin-Yuan Price (University of Missouri–Kansas City) for advice on experiments. This research was funded by National Institutes of General Medicine grant R15-GM-094770.
Footnotes
Communicating editor: A. P. Mitchell
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.180919/-/DC1
Literature Cited
- Ables E. T., Laws K. M., Drummond-Barbosa D., 2012. Control of adult stem cells in vivo by a dynamic physiological environment: diet-dependent systemic factors in Drosophila and beyond. Wiley Interdiscip. Rev. Dev. Biol. 1: 657–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allocati N., Masulli M., Di Ilio C., De Laurenzi V., 2015. Die for the community: an overview of programmed cell death in bacteria. Cell Death Dis. 6: e1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashworth C. J., Toma L. M., Hunter M. G., 2009. Nutritional effects on oocyte and embryo development in mammals: implications for reproductive efficiency and environmental sustainability. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364: 3351–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehrer B. M., Errede B., 1997. Coordination of the mating and cell integrity mitogen-activated protein kinase pathways in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 6517–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cap M., Stepanek L., Harant K., Vachova L., Palkova Z., 2012. Cell differentiation within a yeast colony: metabolic and regulatory parallels with a tumor-affected organism. Mol. Cell 46: 436–448. [DOI] [PubMed] [Google Scholar]
- Chu S., DeRisi J., Eisen M., Mulholland J., Botstein D., et al. , 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699–705. [DOI] [PubMed] [Google Scholar]
- de Nobel H., Ruiz C., Martin H., Morris W., Brul S., et al. , 2000. Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance and thermotolerance. Microbiology 146: 2121–2132. [DOI] [PubMed] [Google Scholar]
- Fabrizio P., Battistella L., Vardavas R., Gattazzo C., Liou L. L., et al. , 2004. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J. Cell Biol. 166: 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel J. S., Mitchell A. P., 2011. Genetic control of Candida albicans biofilm development. Nat. Rev. Microbiol. 9: 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., Chu A. M., Ni L., Connelly C., Riles L., et al. , 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. [DOI] [PubMed] [Google Scholar]
- Grassl J., Scaife C., Polden J., Daly C. N., Iacovella M. G., et al. , 2010. Analysis of the budding yeast pH 4–7 proteome in meiosis. Proteomics 10: 506–519. [DOI] [PubMed] [Google Scholar]
- Gray M., Honigberg S. M., 2001. Effect of chromosomal locus, GC content and length of homology on PCR-mediated targeted gene replacement in Saccharomyces. Nucleic Acids Res. 29: 5156–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M., Piccirillo S., Honigberg S. M., 2005. Two-step method for constructing unmarked insertions, deletions and allele substitutions in the yeast genome. FEMS Microbiol. Lett. 248: 31–36. [DOI] [PubMed] [Google Scholar]
- Herker E., Jungwirth H., Lehmann K. A., Maldener C., Frohlich K. U., et al. , 2004. Chronological aging leads to apoptosis in yeast. J. Cell Biol. 164: 501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigberg S. M., 2004. Ime2p and Cdc28p: co-pilots driving meiotic development. J. Cell. Biochem. 92: 1025–1033. [DOI] [PubMed] [Google Scholar]
- Honigberg S. M., 2011. Cell signals, cell contacts, and the organization of yeast communities. Eukaryot. Cell 10: 466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigberg S. M., Lee R. H., 1998. Snf1 kinase connects nutritional pathways controlling meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 4548–4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard E. J., Korta D. Z., Dalfo D., 2013. Physiological control of germline development. Adv. Exp. Med. Biol. 757: 101–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung U. S., Sobering A. K., Romeo M. J., Levin D. E., 2002. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 46: 781–789. [DOI] [PubMed] [Google Scholar]
- Kaiser D., Robinson M., Kroos L., 2010. Myxobacteria, polarity, and multicellular morphogenesis. Cold Spring Harb. Perspect. Biol. 2: a000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassir Y., Adir N., Boger-Nadjar E., Raviv N. G., Rubin-Bejerano I., et al. , 2003. Transcriptional regulation of meiosis in budding yeast. Int. Rev. Cytol. 224: 111–171. [DOI] [PubMed] [Google Scholar]
- Kicheva A., Cohen M., Briscoe J., 2012. Developmental pattern formation: insights from physics and biology. Science 338: 210–212. [DOI] [PubMed] [Google Scholar]
- Kupiec M., Byers B., Esposito R. E., Mitchell A. P., 1997. Meiosis and sporulation in Saccharomyces cerevisiae, pp. 889–1036 in The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology, edited by Pringle J. R., Broach J. R., Jones E. W. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Lee R. H., Honigberg S. M., 1996. Nutritional regulation of late meiotic events in Saccharomyces cerevisiae through a pathway distinct from initiation. Mol. Cell. Biol. 16: 3222–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis W. F., 2014. Cell signaling during development of Dictyostelium. Dev. Biol. 391: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin H., Rodriguez-Pachon J. M., Ruiz C., Nombela C., Molina M., 2000. Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275: 1511–1519. [DOI] [PubMed] [Google Scholar]
- Martinez de Maranon I., Marechal P. A., Gervais P., 1996. Passive response of Saccharomyces cerevisiae to osmotic shifts: cell volume variations depending on the physiological state. Biochem. Biophys. Res. Commun. 227: 519–523. [DOI] [PubMed] [Google Scholar]
- Mattison C. P., Spencer S. S., Kresge K. A., Lee J., Ota I. M., 1999. Differential regulation of the cell wall integrity mitogen-activated protein kinase pathway in budding yeast by the protein tyrosine phosphatases Ptp2 and Ptp3. Mol. Cell. Biol. 19: 7651–7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman A. M., 2011. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 189: 737–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N., Pitsouli C., Shilo B. Z., 2012. Signaling mechanisms controlling cell fate and embryonic patterning. Cold Spring Harb. Perspect. Biol. 4: a005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo S., Honigberg S. M., 2010. Sporulation patterning and invasive growth in wild and domesticated yeast colonies. Res. Microbiol. 161: 390–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo S., Honigberg S. M., 2011. Yeast colony embedding method. J. Vis. Exp. 49: pii: 2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo S., White M. G., Murphy J. C., Law D. J., Honigberg S. M., 2010. The Rim101p/PacC pathway and alkaline pH regulate pattern formation in yeast colonies. Genetics 184: 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo S., Wang H. L., Fisher T. J., Honigberg S. M., 2011. GAL1-SceI directed site-specific genomic (gsSSG) mutagenesis: a method for precisely targeting point mutations in S. cerevisiae. BMC Biotechnol. 11: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primig M., Williams R. M., Winzeler E. A., Tevzadze G. G., Conway A. R., et al. , 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26: 415–423. [DOI] [PubMed] [Google Scholar]
- Ray D., Su Y., Ye P., 2013. Dynamic modeling of yeast meiotic initiation. BMC Syst. Biol. 7: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P., 1990. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Rubinstein A., Gurevich V., Kasulin-Boneh Z., Pnueli L., Kassir Y., et al. , 2007. Faithful modeling of transient expression and its application to elucidating negative feedback regulation. Proc. Natl. Acad. Sci. USA 104: 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. E., Zhang Z., Thomas C. R., Moxham K. E., Middelberg A. P., 2000. The mechanical properties of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97: 9871–9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh K. J., Hong Y. S., Skirda V. D., Volkov V. I., Lee C. Y., et al. , 2003. Water self-diffusion behavior in yeast cells studied by pulsed field gradient NMR. Biophys. Chem. 104: 121–130. [DOI] [PubMed] [Google Scholar]
- Tanouchi Y., Lee A. J., Meredith H., You L., 2013. Programmed cell death in bacteria and implications for antibiotic therapy. Trends Microbiol. 21: 265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachova L., Palkova Z., 2005. Physiological regulation of yeast cell death in multicellular colonies is triggered by ammonia. J. Cell Biol. 169: 711–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachova L., Cap M., Palkova Z., 2012. Yeast colonies: a model for studies of aging, environmental adaptation, and longevity. Oxid. Med. Cell. Longev. 2012: 601836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachova L., Hatakova L., Cap M., Pokorna M., Palkova Z., 2013. Rapidly developing yeast microcolonies differentiate in a similar way to aging giant colonies. Oxid. Med. Cell. Longev. 2013: 102485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Drogen F., Peter M., 2002. Spa2p functions as a scaffold-like protein to recruit the Mpk1p MAP kinase module to sites of polarized growth. Curr. Biol. 12: 1698–1703. [DOI] [PubMed] [Google Scholar]
- von der Haar T., 2007. Optimized protein extraction for quantitative proteomics of yeasts. PLoS One 2: e1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. G., Piccirillo S., Dusevich V., Law D. J., Kapros T., et al. , 2011. Flo11p adhesin required for meiotic differentiation in Saccharomyces cerevisiae minicolonies grown on plastic surfaces. FEMS Yeast Res. 11: 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarzov P., Mazzoni C., Mann C., 1996. The SLT2(MPK1) MAP kinase is activated during periods of polarized cell growth in yeast. EMBO J. 15: 83–91. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All strains used in this study and all data gathered from the mutant screen and rescreens are available on request.