Abstract
Sexual reproduction and the exchange of genetic information are essential biological processes for species across all branches of the tree of life. Over the last four decades, biochemists have continued to identify many of the factors that facilitate reproduction, but the molecular mechanisms that mediate this process continue to elude us. However, a recurring observation in this research has been the rapid evolution of reproductive proteins. In animals, the competing interests of males and females often result in arms race dynamics between pairs of interacting proteins. This phenomenon has been observed in all stages of reproduction, including pheromones, seminal fluid components, and gamete recognition proteins. In this article, we review how the integration of evolutionary theory with biochemical experiments can be used to study interacting reproductive proteins. Examples are included from both model and non-model organisms, and recent studies are highlighted for their use of state-of-the-art genomic and proteomic techniques.
Significance
Despite decades of research, our understanding of the molecular mechanisms that mediate fertilization remain poorly characterized. To date, molecular evolutionary studies on both model and non-model organisms have provided some of the best inferences to elucidating the molecular underpinnings of animal reproduction. This review article details how biochemical and evolutionary experiments have jointly enhanced the field for 40 years, and how recent work using high-throughput genomic and proteomic techniques have shed additional insights into this crucial biological process.
Keywords: Sexual selection, Reproduction, Pheromones, Fertilization, Evolution
1. Introduction
Sexual reproduction, while prevalent along every branch of the tree of life, remains a challenge for evolutionary biologists to explain [1]. Asexual reproduction offers the advantages of propagating twice the genetic material, lacks the costs associated with finding mates, and can more rapidly establish favorable epistatic effects [2]. Early mathematical models also supported asexual reproduction as the optimal reproductive strategy. However, in more realistic scenarios of dynamic ecosystems with changing environments and co-evolving symbiotes, frequent recombination is needed and natural selection favors sexual reproduction [3,4]. Given the breadth and diversity of sexually reproducing organisms, it is no surprise that various strategies have evolved to improve reproductive success. In animals, males often perform various courtship displays [5–7], deliver pheromones that affect female behavior and physiology [8–10], and regulate the contents of their ejaculate based on female quality [11,12]. Similarly, to improve mate fitness and quality, females must be able to discriminate between these cues for honest or dishonest signals of fitness [13]. Both the male and female characteristics involved can be modified by sexual selection. Under sexual reproduction, mates must be procured to provide complementary genetic material, much like a predator capturing prey for energy and nutrient acquisition. Just as predators and prey often evolve through arms race dynamics, the continual adaptation between elaborate male traits and female perception represents one of the most well characterized examples of rapid, exacerbated co-evolution [14].
The literature is rich with examples of co-evolving sexually selected traits [15]. For historical reasons, the majority of study systems have been visible characteristics such as body size, coloration, mating behaviors, and secondary sexual traits [16]. In recent decades, as molecular biology and biochemistry have advanced, research on sexually selected traits has broadened to include the study of reproductive proteins [14], which we broadly define as any polypeptide directly involved in reproduction. While all reproductive proteins may be subject to sexual selection, the most interesting examples are likely those that directly bind molecules derived from the other sex: examples include pheromones and their cognate receptors [17], interacting egg and sperm surface proteins [18], and seminal proteins that alter female physiology [19]. A recurring theme among reproductive proteins is rapid evolution. As sexual reproduction is an essential biological process for most animals, one might expect that the majority of the reproductive proteins would be under strong negative selection to maintain compatibility. However, the recurring pattern of rapidly evolving reproductive proteins has been observed in both vertebrates and invertebrates at several stages of reproduction [14]. Because selection is most likely to act on functionally important residues in a protein, signatures of positive Darwinian selection can often guide further investigation into their underlying biochemical mechanisms [20], with studies of reproductive proteins serving as exemplars of applying molecular evolutionary techniques to characterize protein function [21–25].
Near the turn of the century and following the completion of the human genome project, a surge of high throughput technologies emerged which have altered the size and scope of questions that biologists can now ask. Various next-generation sequencing (NGS) platforms permit de novo analysis of whole genomes and transcriptomes for both model and non-model organisms [26,27]. Likewise, advances in mass spectrometry (MS) now provide the opportunity to qualitatively and quantitatively characterize whole proteomes [3,28,29,30]. These techniques have additionally been adapted to a wide array of other specific “omic” applications (e.g., metabolomics, phosphoproteomics, pharmacogenomics), but both NGS- and MS-based approaches are quickly becoming the standard for the initial characterization of any biological system [23]. Here we review the biochemical investigations of many reproductive proteins that span various levels of reproduction: pre-copulatory behavior (pheromones), copulation (seminal proteins), and fertilization (sperm/egg proteins). Recent studies in the field have employed NGS- and MS-based approaches, and we discuss how applying such “omics” techniques to reproductive systems may be further integrated with detailed mechanistic and theoretical evolutionary models.
2. Molecular evolution and models of sexual selection
The molecular evolution of any given trait is shaped by neutrality or some form of selection (balancing, directional, or disruptive) (Fig. 1A). Balancing selection reduces genetic diversity and stabilizes a trait at some optimum phenotype. Disruptive selection is the opposite of balancing selection and favors individuals with extreme phenotypes. Finally, directional selection shifts a trait towards a single extreme. Two suites of statistical tests which use either allele frequencies or nucleotide substitutions have been developed, and each tests for selection on relatively different time scales. In the first suite of analyses, assumptions are made concerning the rates at which specific mutations are accumulated and distributed among alleles, often within and between populations. These tests are particularly valuable for identifying recent selection following selective sweeps, but are also heavily influenced by population demographics and bottlenecks (for more thorough review, see [31]). The second set of analyses compares the frequency of nucleotide substitutions at codons within genes – usually between species – and describes trends on relatively longer time scales. In the absence of selection, most nucleotide substitutions (and amino acid substitutions) are free to accumulate at the basal mutation rate. The rate of synonymous substitutions (dS) provides an estimate of this mutation rate, and under neutrality, non-synonymous substitutions (dN) should similarly occur, yielding a ratio of dN/dS ≈ 1. Because most non-synonymous substitutions alter the tertiary structure of a protein and negatively impact function, non-synonymous substitutions should occur more rarely (dN/dS < 1). Residues where non-synonymous substitutions are disfavored are described as under negative or purifying selection [20,32–34]. Unsurprisingly, the average dN/dS across the protein coding sequence for most genes is less than one, and in humans, the genome-wide average dN/dS ∼ 0.25 [35]. The purging of deleterious mutations by purifying selection often results in stabilizing selection of a trait. However, under situations where rapid mutation may be adaptive, non-synonymous substitutions can accumulate more quickly than the mutation rate (dN/dS > 1) and the trait is described as under positive selection [32]. The forces leading to positive selection often generate directional selection, but disruptive selection is also possible when nearly any deviation from the mean is similarly favorable.
Fig. 1.
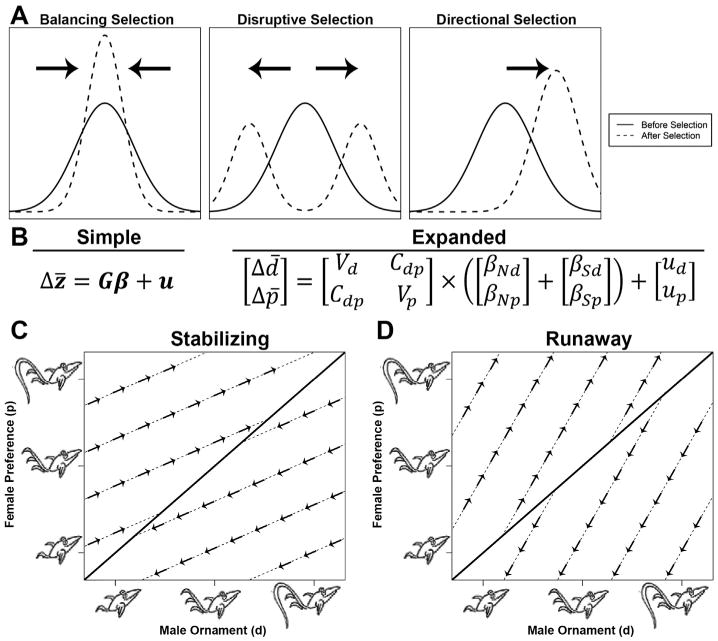
An introduction to trait selection and a summary of the G matrix model of sexual selection described by Lande [41] (and graphics adapted from Mead and Arnold [210]). (A) When a trait is under selection, three different modes are used to describe the direction that the mean moves: stabilizing selection (same mean, less variation), disruptive selection (moves towards the extremes), or directional selection (moves away from the mean along a single trajectory). (B) A mathematical representation of the G matrix, where in the simplified form a change in the mean of a trait (Δz̄) is equal to its genetic structure (G) times some selective force (β) plus a mutation constant (u). In the expanded form, the two traits being observed are a male display (d) and the corresponding female preference for the display (p). The G matrix is composed of the genetic variation for both d and p (Vd and Vp), and covariance (or genetic coupling) between the two traits (Cdp). Both natural (βN) and sexual (βS) selective forces can act on both the male display (βNd, βSd) and the female preference (βNp, βSp), and each trait is subject to independent mutations (ud and up). When there is weak or no genetic coupling (Cdp ∼ 0), the matrix simplifies such that the change in male display is only dependent on the direct selection acting on the display, and similarly for female preference. Therefore, male traits will evolve until female preferences are matched, represented by different evolutionary trajectories (dashed lines) converging towards an optimum (solid diagonal line) (C). However, when there is genetic coupling (Cdp > 0), selection on the male display can “pull” the female preference in a particular direction (indicated by the cross multiplication of Cdp and βd in B), and likewise for selection on the preference. Because of this deviation from independent selection towards the typical optimum, it is possible for male displays and female preferences to undergo “runaway selection” and evolve indefinitely, yielding ever increasingly more elaborate ornaments (D).
Since each residue differentially contributes to a given protein's structure and function, and it is likely that all three forces of selection are simultaneously acting on protein-coding genes to different degrees. For example, with serine proteases, most of the protein surface is covered in polar residues that are functionally neutral and highly interchangeable. Within the active site, purifying selection preserves the catalytic triad of serine, histidine, and aspartate that are critical for enzymatic activity, and mutations are rare save for cases of atypical function [36]. However, for select serine proteases that are involved in apoptosis, adaptive response to pathogens which cause cell death has promoted positive selection on active site residues that mediate inhibitor binding and substrate specificity [37]. While the identification of neutrally evolving sites and those under purifying selection can be advantageous for understanding protein function, both forces are common in maintaining protein function and do not necessarily reflect adaptation to specific stimuli. Hence, greater interest is often placed on sites under positive selection, and various statistical packages exist to compute dN/dS scores along phylogenetic trees for both whole genes and specific residues [20].
While positive selection and rapid evolution have been documented in a range of systems [33,38–40], they are practically hallmarks of interacting reproductive proteins [14]. Over the last few decades, various quantitative genetic models have been developed to address how elaborate male traits and female preferences may evolve. While qualitatively described by Fisher in the 1930s, Lande [41] was the first to formalize the theory using a genetic correlation matrix (Fig. 1B–D). To illustrate this, assume that females of a given species have a preference for some male ornament, such as large, colorful peacock tails. If there is some heritable component to both the male ornament and the female preference, female peacocks with strong preferences and males with bright tails should produce daughters and sons who carry both traits, leading to genetic correlation and linkage disequilibrium. This process may continue iteratively through generations, which can lead to greater genetic association, stronger preferences, and more pronounced ornaments as part of a model of runaway selection [16]. This has alternatively been dubbed “the sexy son” hypothesis [42]. However, various factors may restrain the characteristics from evolving indefinitely. Elaborate ornaments may be energetically costly, and the ornament and preference may evolve to an upper limit until they are balanced by natural selection (“sensory bias”) [43]. There are also instances where choosing to mate with males with large ornaments may confer a cost to the female, such as reduced viability and/or increased risk of predation, leading to antagonism between mating preference and survivorship (“sexual conflict”) [44]. Alternatively, the male ornament may itself be indicative of some underlying adaptive trait, such as increased offspring survival or fecundity (direct benefits) [45,46]. In this example, both male peacock tails and female perception are complex, polygenic traits. Techniques such as high resolution quantitative trait loci (QTL) mapping can approximate the relative contribution of individual genes to these traits; however, these methods are not sensitive to low-impact genes, require high quality genome sequences, and are highly labor intensive.
The archetype of male ornaments and female preferences provides a framework that also applies to interacting reproductive proteins. Elaborate male anatomical displays and their perception within the female nervous system are almost certainly pairs of complex, polygenic traits. However, reproductive protein dynamics may be as simple as a single male-derived molecule docking with a female receptor through protein–protein interactions [47,48]. This one-to-one stoichiometry permits a more tractable environment to study co-evolution, theoretically at single amino acid resolution given sufficient phylogenetic data [49]. The use of NGS and MS-based proteomics are rapidly increasing the feasibility of such studies, as will be described below.
3. Protein pheromones in vertebrates
One of the largest barriers to mating is often the search for potential mates, particularly when population densities are low. Volatile chemoattractants in the form of pheromone signals are widely used by both invertebrates and vertebrates. The term “pheromone” was originally coined by Karlson and Luscher in 1954, derived from the Greek words pherin (“to attract”) and hormon (“to stimulate”) [50]. The first described pheromone was bombykol, a long chain alcohol released by female silkmoths to attract conspecific males [51]. Since this original discovery, a wide range of pheromone molecules have been described that not only attract potential mates, but also serve as territory markers [52], signal species identity [53], coordinate migratory behavior [54], and prime the nervous system to sense additional pheromone signals [55]. While volatile odorants are the most well-documented and characterized type of pheromones, many vertebrates also utilize water-soluble peptide or protein pheromones [17,56]. In contrast to their volatile counterparts, which are generally synthesized as part of complex enzymatic cascades, peptide/protein pheromones are direct gene products whose evolutionary histories can be more readily probed by sequencing and other standard molecular biological techniques.
Differences in pheromone chemistry have resulted in the evolution of specialized anatomy for their detection. The olfactory system of terrestrial vertebrates includes two discrete sets of neuronal epithelia: the main olfactory epithelia (MOE) and vomeronasal epithelia (VNE) [57,58]. In general, the MOE senses generic odorants while the VNE is specialized to detect pheromones and similar heterospecific cues (although there is evidence for significant crosstalk between these systems and a few exceptions to the rule [59]). There is further partitioning within the VNE such that neurons express one of two main types of G protein coupled receptors (GPCRs): vomeronasal type 1 receptors (V1Rs) that detect volatile pheromones [60], or vomeronasal type 2 receptors (V2Rs) that bind peptide or protein pheromones [61]. One of the goals of modern pheromone research has been to precisely characterize the co-evolution between specific protein pheromones and their cognate V2Rs.
Despite significant advancements towards independently characterizing pheromone and receptor profiles, pairing specific receptors with their ligands has remained a challenge. Protein pheromones are generally purified using various chromatographic separations, with their identity ascertained through joint sequencing and mass spectral characterization. Receptor identification, by comparison, has relied predominantly on molecular and immunohistochemical techniques. Both V1R and V2R receptor types were identified in rodents through the preparation of single-cell cDNA libraries, subtractive hybridization against MOE cDNA to remove genes common to all olfactory neurons, and localization verified by in situ hybridization [60,61]. Thanks in part to these early studies, the majority of V2R sequences have been mined by homology search from completed genome sequences. Within the mouse and rat genomes, there are at least 209 and 168 V2R genes respectively [62]. As GPCRs, V2Rs contain seven helical transmembrane domains that anchor them to the plasma membrane, but there are also large N terminal extracellular domains that were postulated to be the site of pheromone binding [58]. The dN/dS ratio for the N-terminal extracellular domain is approximately three times higher than the mean dN/dS for the whole protein, and 27 positions were specifically detected to be under positive selection [62]. V2Rs do not readily express in cell culture and require specific chaperones to translocate to the plasma membrane [63]. The current approach to identifying receptor:ligand pairs involves co-labeling neurons with probes for specific V2Rs and for signals of activation upon pheromone treatment. Riboprobes with varying degrees of degeneracy can be designed to detect one or more V2R sequences by in situ hybridization [64]. Neuronal activation can be detected using a number of methods: calcium imaging (with neurons pre-loaded with fluorescent Ca2+probes suchasFura-2/AMorendogenously expressing transgenic reporters) [65], immediate-early gene activation (such as c-Fos or Ebr1, with either mRNA detected by in situ hybridization [64] or protein by immunohistochemistry [66]), or the amino acid derivative agmatine (which selectively enters activated neurons and can later be detected by immunohistochemistry [67]). In a recent study examining mouse MOE and VNE transcriptomes, there were few differences between male and female receptor profiles, suggesting that other neuronal features or changes in the central nervous system are responsible for sex-specific pheromone effects [68].
Only a few murine protein families have been demonstrated to activate V2R-containing neurons, including major urinary proteins (MUPs) [69], exocrine secreted peptides (ESPs) found in tears [70], and select MHC peptides [71]. Both the MUPs and ESPs are highly duplicated, multigene families that display variable isoform expression depending on genetic background, sex, and age. MUPs are part of the lipocalin superfamily of binding proteins, and the NMR solution structure revealed a large hydrophobic pocket which binds secreted metabolites [72]. While mouse urine has long been known to contain a repertoire of pheromones, the original sole function of MUPs was thought to be binding volatile pheromones and preventing their rapid evaporation upon release into the environment, extending the longevity of the signal. However, bioassays testing bacterially expressed recombinant MUPs (which lack the volatile component of mouse urine) revealed that the proteinaceous component itself can act as a pheromone; specifically, exposure to recombinant MUPs induced male aggressive behavior [69]. MUP expression is highly variable between different inbred mice strains; for example, mass spectral analysis identified MUP17 as highly expressed in both wild and C57BL/6 male mice, yet BALB/C males only expressed trace levels. MUP17 plays a key role in female sexual behavior, and is also referred to as “darcin,” in reference to the amorous male protagonist of Pride and Prejudice. Recombinant darcin alone increased female mating receptivity, but also promoted learning of associated volatile compounds. While females were not innately attracted to many volatile compounds in male urine, if darcin was exogenously added to the volatile mixture when applied to females, repeat exposure to the volatile components alone similarly increased receptivity. Hence, MUP bioactivity may rely on both the lipocalin-like binding site and additional protein–protein interactions with V2R receptors [73,74]. It is noteworthy that hamster vaginal secretions contain a similar lipocalin molecule, aphrodisin, which is also hypothesized to serve as a carrier for volatile compounds to the male VNE [75].
Similar to the MUP family, the exocrine secreted peptide (ESP) family is a highly duplicated multigene family that performs an array of pheromone activities in mice. The first identified member of the family, ESP1, was discovered in the extraorbital lacrimal gland as a male-specific component that stimulated female sexual behaviors. The bioactive component of this gland secretion was isolated by anion exchange chromatography, and the identity of ESP1 ascertained by protein sequencing using Edman degradation and mass spectrometry. A subsequent genome-wide search localized ESP1 (along with the other 23 tandemly repeated genes) to chromosome 17 [76]. Co-labeling of ESP1-activated neurons with specific V2R probes revealed V2Rp5 to be the specific receptor for ESP1 [70]. The 3D structure of ESP1 was determined by NMR, and when supplemented with biochemical data from both mutagenesis and affinity chromatography experiments, charge interactions were suggested to drive binding between ESP1 and V2Rp5 [77]. Surprisingly, the predominantly alpha helical structure of ESP1 resembles that of the sea hare pheromone attractin [78]. At least ten of the ESP genes (ESP2-11) are expressed in both male and female mice, usually in more than one type of tear gland [76]; however, ESP36 is at least one example of a female-specific isoform, and ESP22 is a juvenile-specific isoform [79]. When recombinant ESP22 was applied to female mice, males were less sexually active, and it was hypothesized that the function of ESP22 may be to discourage mounting attempts on juveniles [80]. The specific receptors and roles of the other ESP family members remain unknown, but presumably perform additional pheromone functions as part of rodent communication [81].
In addition to mammals, protein pheromones have also been well studied in salamanders [47,82,83]. As basal tetrapods, amphibians are particularly well-suited to understanding the early evolution of pheromones in terrestrial vertebrates. Amphibian protein pheromones evolved at least 350 million years ago [84], and have been studied in two different salamander families: Salamandridae (newts) and Plethodontidae (lungless salamanders). The first characterized peptide pheromone was the decapeptide sodefrin, which is released from the cloaca of male firebelly newts (Cynops pyrroghaster) to attract gravid females during the mating season. Sodefrin was originally identified by separating male abdominal gland extract using reversed phase liquid chromatography, screening the chromatographic fractions for ability to attract female newts, and analyzing the single bioactive fraction by gas–liquid phase protein sequencing [85]. The closely related species Cynops ensicauda uses a similar peptide termed silefrin that varies from sodefrin by only two substitutions (P3L and L8Q), yet the activity of each peptide is species-specific [86]. Recently, a variant of sodefrin was found in a single population of C. pyrroghaster with a highly conservative L8V mutation, and it was only effective at attracting local females [87]. These two natural sodefrin variants suggest that residue eight likely plays a critical role in receptor binding. Subsequent cDNA sequencing revealed that the decapeptide was cleaved from a larger ∼20 kDa precursor using a dibasic cleavage site, similar to many peptide hormones such as insulin and glucagon [88]. Homologs of the precursor known as sodefrin precursor-like protein (SPF) have been identified in additional newt species [89], and in Lissotriton helveticus, the precursor similarly facilitates mating by increasing female nudging of the male cloaca [84]. Phylogenetic comparison of SPF sequences revealed that the cleaved sodefrin decapeptide is unique to the Cynops genus, such that a second pheromone independently evolved from a previously co-opted pheromone gene [89]. Sodefrin signals through the female newt olfactory system, but the receptor(s) remain unidentified.
While adult newts are primarily terrestrial animals, their life cycle begins with an aquatic larval stage and they return to freshwater habitats to reproduce. Consequently, the release of water-soluble proteins and peptides into the aqueous environment is analogous to the release of volatile odorants into the air. In contrast, lungless salamanders of the family Plethodontidae are purely terrestrial, with females laying clutches of eggs on land and juveniles hatching already in their adult morph [47]. Plethodontid salamanders contain various skin glands that secrete pheromones which provide information on species [90, 91], body size [92], sex [90], female gravidity [92,93], diet [94,95], and parasite load [96]. The most well-characterized of these glands is a male-specific chin gland – the mental gland – that seasonally hypertrophies in response to elevated plasma androgens that coincide with the courtship season [97,98]. Previously termed the “hedonic gland” [99], the primary function of this gland seems to be the production of proteinaceous pheromones that modify female courtship behavior and reduce the length of courtship [24,100–103]. Once the gland fully develops, nearly all of its transcriptional and translational capacity is diverted to pheromone synthesis, such that >70% of the glandular mRNA codes for pheromone [104]. In contrast to the mammalian examples where pheromones are exclusively sensed by olfaction, female plethodontid salamanders receive pheromone by one of two delivery mechanisms. In the majority of plethodontid salamanders (∼300 species), mental gland development is accompanied by the development of hypertrophied premaxillary teeth which are used to “scratch” the dorsum of the female, and pheromones diffuse transdermally into the bloodstream. However, in a single clade of large eastern Plethodon salamanders (28 species), mental gland morphology has transitioned to a large pad-like structure that is “slapped” to the female nares and pheromones are delivered to the olfactory system [47].
To date, three major pheromone families have been transcriptionally, proteomically, and functionally characterized: SPF (a homolog of the newt pheromone), Plethodontid Receptivity Factor (PRF), and Plethodontid Modulating Factor (PMF). Found in both newts and plethodontid salamanders, the SPF family is likely >300 million years old and is possibly the oldest vertebrate pheromone family [89]. All three proteins are derived from separate gene families: SPF is related to phospholipase A2 inhibitors, PRF is related to the IL-6 helical cytokines, and PMF is a member of the three-finger protein (TFP) superfamily. RTPCR based analyses revealed that SPF and PMF are present in all plethodontid species, while PRF is a more recently derived family found only in eastern Plethodon species, including those with olfactory delivery (Fig. 2A). Determination of dN/dS ratios revealed strong signatures of positive selection and rapid evolution in all three pheromone families [105–108]. In addition to rapid evolution, the three gene families have been subjected to extensive gene duplication such that they are generally maintained as multi-isoform blends in the pheromone extract.
Fig. 2.
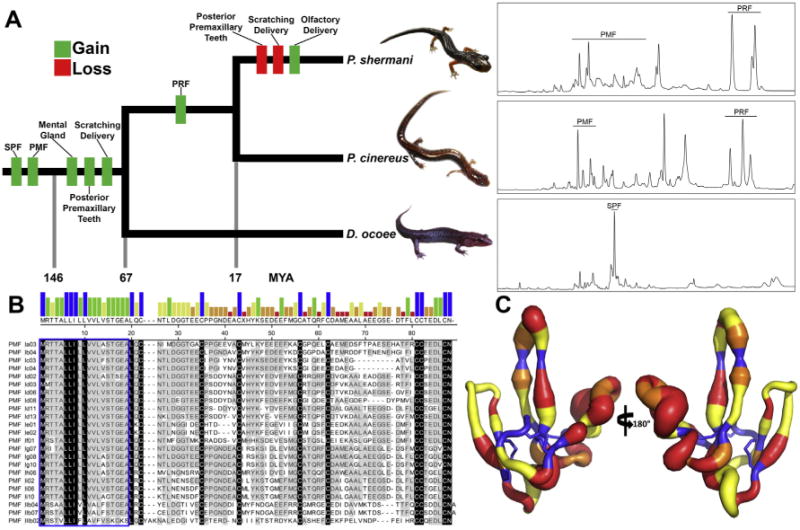
Summary of plethodontid pheromone evolution. (A) A reduced phylogenetic tree emphasizing species for which proteomic characterizations have been performed (P. shermani, P. cinereus, and D. ocoee). Approximate times in millions of years (MYA) for gain and loss of key morphological, behavioral, or genetic trait acquisitions are included, as well as representative reverse phase chromatograms for the pheromones of each species (using 70 minute gradients at 1% acetonitrile/min). While PMF is one of the oldest pheromone gene families, its expression varies dramatically between the three species, with D. ocoee expressing a single PMF isoform at extremely low abundance (<1% total pheromone, not visible by HPLC), P. cinereus expressing ∼4–6 isoforms, and P. shermani expressing >30 isoforms. (B) Alignment of PMF isoforms identified in the P. shermani pheromone extract by HPLC and MS-based proteomics (adapted from [109]). The bar graph shows consensus strength, the blue box highlights the signal peptide, and the few absolutely conserved residues (mostly the conserved eight cysteine core) are colored in black. (C) A putty model of the PMF 3D structure from [25], with backbone diameter representing residue heterogeneity (Shannon Weaver diversity index), and color signifying the mode of evolution (blue, purifying selection; yellow, neutral selection; orange, positive selection at >95% confidence; red, positive selection at >99% confidence).
More detailed proteomic analyses have been performed for three plethodontid species representing cases of transdermal delivery with no PRF (Desmognathes ocoee), transdermal delivery with PRF (Plethodon cinereus), and olfactory delivery (Plethodon shermani). Initial pheromone characterization was performed for P. shermani using multiple types of high performance liquid chromatography (HPLC) to isolate individual proteins, which were then identified by MS analysis using a shotgun EST library as a reference [109,110]. Subsequent studies with D. ocoee and P. cinereus instead used de novo transcriptomes as reference databases, with amplification of identified candidates by RT-PCR and validation by Sanger sequencing [111,112]. Despite detection of SPF mRNA by RT-PCR in all three species, it was only observed proteomically in D. ocoee, where it comprised ∼30% of the total pheromone. While D. ocoee may express up to ∼70 different SPF isoforms, six major classes of highly divergent isoforms shared <50% sequence identity. Protein abundance estimates by spectral counting suggest that isoform expression is relatively stable between individual male D. ocoee [112]. When applied to females during courtship, purified SPF decreased courtship time [102]. Proteomic analyses of the D. ocoee pheromone extract also led to the identification of several peptide hormone paralogs such as glucagon, insulin, leptin, and relaxin. It was hypothesized that when delivered to the female bloodstream, these hormone-like pheromones may directly influence female physiology by exploiting pre-existing endocrine receptors [112]. In contrast, both P. cinereus and P. shermani with transdermal and olfactory delivery, respectively, secrete pheromone mixtures of predominantly PRF and PMF, but with multiple notable differences. First, PRF was heavily glycosylated in P. cinereus and not P. shermani, with the loss of glycosylation resulting from mutation of two NXS/NXT N-glycosylation motifs and an alternative signal peptidase cleavage position which removes an O-glycan near the N-terminus. While glycosylation can affect various aspects of protein solubility, stability, and receptor specificity, its functional significance in this pheromone context remains to be determined [111]. Second, P. shermani expresses ∼5–10 times more PMF isoforms compared to P. cinereus (Fig. 2A). Specifically, individual P. shermani males express more than 30 different PMF isoforms. Most, if not all, of this diversity appears to be from gene duplication, and 99 different putative isoforms were identified through cDNA sequencing of a single population. HPLC-based analyses suggest that each male expresses a unique combination of isoforms [110], and behavioral studies show that female behavior changes upon exposure to different combination of isoforms [24]. Mass spectral characterization of P. shermani PMFs confirmed that at least 27 different PMFs are expressed, and are highly divergent with an average amino acid conservation of ∼30% (Fig. 2B) [109].
One possible hypothesis as to why olfactory delivery evolved as a replacement to transdermal delivery is signal amplification. Akin to normal endocrine cues, pheromones supplied to the bloodstream likely bind to their receptors in a concentration-dependent manner. Hence, male salamanders with transdermal delivery must deliver some critical dosage of pheromone to elicit a response. However, application of pheromone to the olfactory system offers the opportunity for signal amplification via neuronal activation. In P. shermani, V2Rs are highly expressed by VNE neurons [113], with different neurons separately detecting PRF and PMF [114,115]. Because V2Rs are tuned to recognize signals at nanomolar levels or lower [71], there is likely limited concentration dependence for olfactory pheromones, and less restriction on gene duplication, such that plethodontid males benefit from the increased PMF isoform diversity. More isoforms increases the likelihood of stimulating females with any possible receptor profile, and improves the chances of mating success. In a recent study, the 3D structure of PMF was determined by multidimensional NMR, and NMR relaxation analysis revealed that the residues under positive selection tended to be on the most flexible regions of the molecule (that result from a novel disul fide bonding pattern compared to all other three-finger proteins; Fig. 2C) [25]. Evolved flexibility may be yet another way to create pheromone diversity and stimulate additional receptors. However, as with the case of sodefrin, the receptors for plethodontid pheromones are unknown, and it remains difficult to explicitly test models of sexual selection.
4. Seminal fluid proteins
For animals that utilize internal fertilization, male ejaculate is a complex and highly variable mixture, containing cells (gametes), mucins, small metabolites (sugars and amino acids to nutrify spermatozoa), hormones, and various peptides and proteins — this last category simply being termed seminal fluid proteins (SFPs). The types and functions of SFPs vary tremendously between species, including structural proteins (mucins [116] and/or aggregate-forming proteins that result in copulatory plugs [116,117]), antimicrobial peptides [118], and proteins that directly affect female physiology and behavior by modulating female receptivity, longevity, egg production, feeding, and sperm storage potential [119]. Classical biochemical methods are generally not well-suited to studying SFP interactions. Since they are delivered as part of the male ejaculate only during mating, it is often difficult to purify compounds and artificially apply them within the appropriate biological context (with a few notable exceptions [120]). In contrast to the simpler case of pheromones being synthesized by single glands, seminal fluid is the cumulative product of several organ secretions (in humans, this includes the testes, prostate, seminal vesicle, and bulbourethral glands [121]), and disentangling the relative contributions from each of these tissues can be challenging. However, advances in gene expression analysis and proteomic methods have driven a recent explosion in our broader understanding of seminal fluid composition and its effects on female physiology.
The majority of SFP studies have utilized insect models (for thorough review, see [119]). A number of microarray, EST, and proteomic studies have been performed on honey bees [122], beetles [123], mosquitoes [124–126], butterflies [127,128], ticks [129], bed bugs [130], and field crickets [131,132];unsurprisingly as an established laboratory model, fruit flies (Drosophila spp.) have been the most extensively characterized [133]. Sperm competition is well-documented in Drosophila, and it was accurately predicted that SFPs would play a significant role in this process. A particular focus has been on accessory gland proteins (Acps), which constitute the majority of seminal fluid proteins [133]. Genome analysis, whole sperm proteomics, and tissue-specific microarray analysis from Drosophila melanogaster males allowed prediction of 112 Acps [134]. Two key Acps have been the subject of extensive biological and molecular characterization: sex peptide (SP, or Acp70Aa) and ovulin (Acp26Aa). Both proteins increase female egg production, but through different mechanisms: SP stimulates oocyte maturation and accumulation of yolk proteins [135,136], while ovulin stimulates ovulation by triggering octopamine signaling in the central nervous system (through an unknown receptor) [137]. SP also decreases female receptivity to re-mating, increases feeding behavior, and changes dietary preferences from carbohydrate-rich to protein-rich food. Energy acquisition and mating have been extensively linked in many systems [138,139], and this change in feeding behavior is likely a secondary response to increased yolk production [140]. SP-induced effects are observable less than 24 h after mating and can last up to two weeks. This long-term response results from SP binding to sperm and being released gradually over several days following proteolytic cleavage [141,142]. The SP receptor (SPR) was identified by screening D. melanogaster females generated as part of a genome-wide series of RNA-interference (RNAi) lines, with SPR knockdown females laying fewer eggs and re-mating at similar frequencies to virgin females. SPR is a GPCR expressed on neurons in both the female reproductive tract and central nervous system [143].
The evolution and interactions between SP and SPR are atypical compared to other studied reproductive proteins. First, there is minimal evidence that SP is rapidly evolving [144]. Second, in contrast to pheromone examples where there is likely one-to-one relationships between ligands and receptors, SPR is a promiscuous receptor both in expression profile and ligand specificity. Specifically, SPR is also expressed in the brains of larvae and adult males, and has similar affinity for both SP and myoinhibitory peptides (MIPs) [145,146]. A recent study showed that MIP-SPR interactions play a critical role in Drosophila sleep behavior [147]. Third, there is little evidence for co-evolution between the two genes. Evolutionary rate covariation (ERC) is a recently developed metric for correlating the normalized number of substitutions between protein-coding genes along a phylogenetic tree, such that continual co-evolution between proteins can be inferred from strong positive correlations [148]. When this metric was applied to annotated Drosophila reproductive genes, there was weak ERC between SP and SPR; however, 111 other genes had significant ERC with SP or one of five other SFPs that cooperate with SP in a network. Twenty-one candidates were selected from this list for RNAi knockdown, and five genes had a significant effect on female re-mating behavior [149]. SP and SPR seem to have evolved through sexual conflict, where males prioritize their own mating success over female fitness [16]. Because the binding constant between SP and SPR is a proxy for female preference, it is influenced by both the sequence and expression level of each protein. SPR sequence evolution has presumably experienced purifying selection to maintain its non-reproductive functions (e.g., MIP binding); conversely, SP has likely evolved to an optimum to maximize binding affinity. Without a changing SPR sequence, SP cannot “chase” SPR, leading to it also being under stabilizing selection (Fig. 1C). With respect to the gene expression and the Lande model (Fig. 1B), SP is likely under positive sexual selection (increases mating success) and negative natural selection (costly to produce, given that males have limited Acps that they strategically allocate during multiple matings [150]). Similarly, SPR is also probably under negative natural selection, as females suffer reduced longevity from increased mating and receiving excess Acps [151–153]. Sexual selection on SPR is difficult to infer, because of the challenges in ascertaining if reduced mating frequency in response to sex peptide is purely a byproduct of reduced fitness [154] (zero or weakly negative sexual selection), or also in part due to mate choice and increased female receptivity (positive sexual selection). Even in the absence of sexual selection, SPR expression could be maintained in the female reproductive tract through genetic coupling with SP (with regard to Fig. 1B, this implies Vp × — βNp ≈ Cdp × (βSd — βNd)), leading to a stable sexual conflict scenario. Testing such hypotheses is likely difficult and would require estimating female mating receptivity, likely using carefully controlled mate choice experiments and/or sophisticated neurophysiological approaches.
While more than 100 putative Acps have been identified through male-specific tissue screens, identifying expression of these proteins and their actual transmission to the female reproductive tract remains challenging. In one study [22], the novel proteomics strategy of whole fly isotopic labeling was employed to selectively identify male versus female proteins. Flies were fed yeast grown on 15N labeled media, and after one full generation, no unlabeled peptides were detectable. Male flies fed unlabeled yeast were then mated with 15N labeled females, and soluble proteins extracted from the female reproductive tract for MS analysis performed with searches tuned to only identify unlabeled peptides. This design provided a zero background to detect only male-derived peptides, resulting in identification of 138 SFPs, only 75 of which were previously predicted based on EST sequencing. Highly conservative tests for molecular evolution showed that at least nine of the genes were rapidly evolving. Repeating the experiment on two additional Drosophila species (Drosophila simulans and Drosophila yakuba) revealed only 63 SFPs were commonly expressed in all three species, likely suggesting lineage-specific gene expression.
The same isotopic labeling strategy was adapted to the study of seminal fluid proteins from the house mouse (Mus domesticus) [155]. Again, females were isotopically labeled with a 15N-based diet to minimize background, and the female reproductive tract was investigated for unlabeled proteins post- mating. A total of 69 proteins were identified, 62 of which belong to the copulatory plug, and seven copulatory plug structural proteins constituted ∼37% of the total protein. A previous proteomic study documented 506 proteins secreted from six distinct regions of the male M. domesticus reproductive tract, and only a few of these genes contained an elevated dN/dS ratio relative to the genomewide average of ∼0.12 (when compared to the rat genome) [156]. In contrast, the 69 SFPs had a significantly higher mean dN/dS of 0.27 (with the non-SFP reproductive tract proteins having a mean dN/dS of ∼0.06). In addition to structural proteins, there was an enrichment for proteins related to protection from oxidative stress and protease inhibition. When the MS search parameters were adjusted to focus on proteins with 15N incorporation to identify female proteins expressed in response to mating, six proteins were identified including two proteases: kallikrein-related peptidase 14 and lactotransferrin. Interestingly, both proteins had elevated dN/dS ratios (0.32 and 0.74, respectively), and may be rapidly co-evolving with SFP protease inhibitors [155]. Notably, the same categories of genes are enriched among human SFPs [121], and further suggest that many protein families are common to the male ejaculates of most species, with variation in copy number, sequence, and expression level driven by female sexual selection [157].
5. Gamete recognition proteins
The birth of sexual selection arguably initiated with the differentiation of sperm and egg, and each gamete benefiting from a different reproductive strategy [16]. Consequently, the interaction of egg and sperm are one of the most essential biological processes, and has been of major research interest, due in part to the likely role such interactions play in human infertility [14,158]. The process of fertilization can be divided into various stages, and while the particular molecules vary between organisms, the core process is highly conserved. Oocytes are surrounded by glycoproteinaceous egg coat that serves as an initial barrier to sperm, termed the zona pellucida (ZP) in mammals and the vitel line envelope (VE) in other animals. Sperm are attracted to oocytes by chemotaxis, and upon reaching the ZP/VE, form an acrosomal process: an extension of the sperm head that is coated in proteins secreted from an acrosome granule. The acrosomal proteins create a hole in the ZP/VE, permitting sperm passage to the oocyte plasma membrane, where a second set of molecular interactions fuse the two membranes and result in zygote formation.
The most well-characterized fertilization systems are those of marine invertebrates, including sea urchins [159,160], abalone [161], sea snails [162], oysters [163], and sea stars [164]. Early biochemical studies were facilitated by the ample availability of gametes from such external fertilizers to prepare various biochemical fractions. In particular, sea urchins have been a model of fertilization for over a century [165], and the first fertilization protein was identified in sea urchins nearly forty years ago [160]. Early ultrastructural studies of sea urchin sperm suggested that the contents of the acrosomal process were associating with the oocyte membrane [166], and biochemical analysis revealed that the acrosomal granule contained a single protein, termed bindin [160]. Agglutination experiments using bindin from different species of sea urchin revealed greater efficacy when bindin was paired with conspecific oocytes, suggesting species-specificity [159]. It was approximately a decade later before bindin was first cloned from the species Strongylocentrotus purpuratus [167], and has since been documented in several orders of sea urchin [165]. The size of sea urchin bindin varies between 193–418 residues, but includes a highly conserved ∼60 residue domain with 29 invariant residues [168]. Part of this domain is an 18 residue peptide (B18) that promotes fusion of the gamete plasma membranes [169]. Much of the variation in bindin length stems from repeats of short glycine-rich motifs that may be either N- or C-terminal to the conserved core [165].
Models of molecular evolution for bindin were variable between genera, such that species from four of the six genera tested showed signatures of positive selection (dN/dS > 1). An interesting quality about the genera with rapidly evolving bindin is that they include sympatric species with overlapping habitats, while the two non-rapidly evolving genera contain only allopatric species [170]. These data are in support of the reinforcement hypothesis: overlapping habitat creates risk of species hybridization, and if hybrids are not viable and/or less fit, selection should favor mechanisms to reinforce species boundaries. However, some allopatric species still display signatures of rapid bindin evolution such that reinforcement alone is not sufficient explanation. Another attractive hypothesis is one of sexual conflict and polyspermy. Following fertilization, the sea urchin VE depolarizes and becomes impermeable to additional sperm; however, before this depolarization, if eggs are penetrated by multiple sperm they will become nonviable. Hence, natural selection may counter sexual selection by favoring oocytes whose VEs have lower affinity for bindin [171]. Population genetics and field experiments with wild sea urchin populations support this hypothesis, as higher fertilization rates have been observed for uncommon bindin genotypes under high population densities where polyspermy risk is increased [172]. It was not until ∼25 years after the discovery of bindin that its receptor EBR1 was cloned from two Strongylocentrotus spp. by using subtractive cDNA hybridization. EBR1 is a large ∼350 kDa protein that includes an N-terminal ADAMTS-like domain and ∼19 species-specific repeats [173]. Similar to bindin, EBR1 evolved under positive selection in Stronglyocentrotus spp., although fewer residues were under selection compared to bindin [174]. Only EBR1 sequences from Stronglyocentrotus are deposited in GenBank. The large transcript size and presumably high sequence variability have made amplifying full-length EBR1 difficult; however, the sea star EBR1 was recently characterized using a de novo transcriptome and contained a similar architecture to the sea urchin homolog [164]. More sea urchin genera will need to be sampled to assess how reinforcement versus sexual conflict may contribute to the co-evolutionary chase between bindin and EBR1.
In addition to sea urchin and related echinoderms, the marine gastropod abalone has been another valuable model for characterizing interacting gamete proteins. Early studies with abalone acrosome extract revealed its ability to “unravel” purified egg VEs, creating an ∼3 μm hole for sperm passage. This activity was attributed to a single component: the 16 kDa protein lysin [175]. Similar to bindin, lysin displayed species-specificity in that its more effective at dissolving con-specific VEs compared to those of other species [21]. Estimating dN/dS ratios for lysin revealed strong positive selection [161], and it has since become a textbook example of rapid evolution [176]. Crystal structures were determined for lysin from two species (red abalone, Haliotis rufescens, and green abalone, Haliotis fulgens), and the two proteins have nearly identical 3D structures, with backbone atoms varying on average by ∼0.28 Å (Fig. 3A) [177]. Modification of crystallization conditions and accompanying FRET-based experiments revealed that lysin predominantly exists as a dimer [178]. Lysin monomers consist of a disjointed triple helix whose surface is polarized and has two faces of concentrated with either basic or hydrophobic residues, with the hydrophobic face defining the dimerization interface (Fig. 3B). Notably, the majority of residues under positive selection were concentrated on the N- and C termini, which form a nexus on one side of the monomer (Fig. 3C). These regions were also seemingly flexible and not well-resolved in the crystal structure. Chimeric recombinant lysins were prepared by substituting subsequences of lysin from red and pink abalone (Haliotis corrugata), and exchanging the N- and C-termini from each species was sufficient to invert the species specificity of VE dissolution [179]. In contrast to bindin, which mediates both VE dissolution and membrane fusion, lysin seems to only interact with the VE. A second molecule, sperm protein 18 kDa (Sp18), likely facilitates membrane fusion [180]. Similar to lysin, Sp18 has also been subjected to pervasive positive selection and exhibits species-specific activity [181]. However, while there is little discernable sequence similarity between lysin and Sp18, the crystal structure of Sp18 revealed a highly similar alpha helical topology [182], and the two genes share nearly identical intron–exon boundaries [181]. Hence, Sp18 and lysin are likely paralogous genes, duplicating from a common ancestral protein that mediated both VE dissolution and membrane fusion similar to bindin. Upon gene duplication of this ancestor, the new genes that now represent lysin and Sp18 were free to specialize in order to more efficiently facilitate fertilization.
Fig. 3.
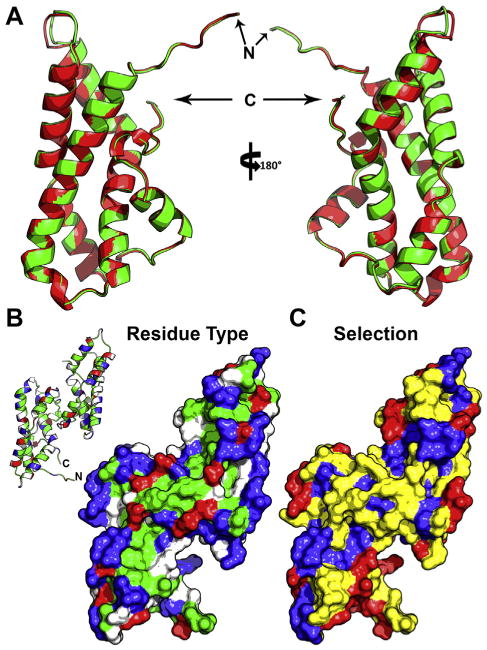
Structural models of abalone sperm lysin. (A) Backbone cartoon overlay of lysin crystal structures from two different species: red and green abalone (with ribbon color matching the species common name, and the N- and C-termini noted). The average backbone rmsd between the two models is ∼0.28Å.(B) Surface model of the red abalone lysin dimer, colored based on residue type (blue, basic; red, acidic; green, hydrophobic; white, polar), with an inlay of the backbone structure for reference. The majority of hydrophobic residues cluster on or near the dimer interface, with the two exposed monomeric faces containing long tracts of basic residues. (C) The same surface model overlaid with the estimated mode of selection (blue, purifying; yellow, neutral; red, positive; from [211]). The residues under positive selection centralize on the edges of the basic tract and the nexus at the N- and C-termini.
Analogous to EBR1, the vitelline envelope receptor for lysin (VERL) is a giant, highly repetitive glycoprotein of ∼1000 kDa that was identified through lysin affinity purification [21]. Combined sequencing of RACE cDNA and a genomic cosmid library revealed that the VERL gene codes for a 3722 residue protein (∼410 kDa), ∼90% of which results from 22 repeats of ∼153 residues [183]. This repetitive domain is homologous to the vertebrate ZP-N polymerization domain (which will be described in further detail below) [184]. As part of a long fibrous molecule, these repetitive polymerization domains are thought to form an interlocking network of hydrogen bonds that stabilize the VE supramolecular structure. Binding curves between lysin and VERL are consistent with positive cooperativity, and suggest a stoichiometry of ∼2–3 lysin molecules per VERL repeat. Lysin presumably dissolves the VE by competing for hydrogen bonds between VERL repeats, resulting in the splaying of the fibers that allows sperm passage [21]. The majority of repeats (3–22, ordered N-terminus to C-terminus) are ∼99% identical within a given species, and are hypothesized to be homogenized through unequal crossing over and concerted evolution [185]. In contrast, repeats 1 and 2 have evolved independently under positive selection [186], and the dN/dS ratios between lysin and these first two VERL repeats are correlated along branches of the phylogenetic tree [187]. The cumulative result of these findings is that lysin and VERL evolve by chase dynamics: the first two repeats of VERL move away from lysin's optimum, and lysin quickly adapts to maintain high affinity interactions. These N-terminal repeats are likely on the outermost surface of the VE, making them the first repeats lysin encounters upon secretion from the acrosome. After binding repeats 1 and 2, affinity for the homogenized repeats would increase by cooperativity. However, in contrast to the pheromone system where olfaction provides signal amplification, non-enzymatic VE dissociation requires that lysin have an extremely low KA for VERL and/or be in large stoichiometric excess. Sexual selection seems to have favored both, as lysin binds VERL at nanomolar concentrations [21], and individual sperm produce tremendous quantities of lysin (up to one gram of lysin can be purified from a single male abalone; W.J. Swanson, personal observation).
In addition to the trio of lysin, VERL, and Sp18, recent proteomic studies have suggested greater complexity in both the male and female sides of abalone fertilization. Joint transcriptome sequencing and MS proteomics of abalone sperm revealed two isoforms of a small acrosomal protein (termed sperm protein 6 kDa, or Sp6) which together are more abundant than either lysin or Sp18. The two isoforms of Sp6 are highly similar, differing primarily in a polyaspartate region near the N-terminus of four or eight residues, such that the two isoforms are referred to as Sp6_4D and Sp6_8D. Comparison of Sp6 sequences between abalone species was again indicative of rapid evolution by positive selection [23]. The function of Sp6 remains to be determined, but given its high net negative charge in contrast to the positive charges of lysin and Sp18, one hypothesis is that it stabilizes one or both of the molecules (with positive selection as a consequence of lysin/Sp18 rapid co-evolution with female receptors). Sp6 may also interact with additional VE proteins. While VERL was the first VE protein characterized through lysin affinity chromatography, deep cDNA sequencing and shotgun proteomics permitted identification of more than a dozen additional ZP domain-containing proteins in the abalone VE (Fig. 4A) [188,189]. The ZP domainis an ∼250 residue motif common to all vertebrate egg coat proteins, consisting of two disulfide bonded halves which independently adopt modified immunoglobulin-like folds. The separate halves of this domain are termed ZP-N and ZP-C, respective to their orientation in the protein. While polymerization between ZP domains involves intermolecular contacts between both ZP-N and ZP-C [190], biochemical studies have demonstrated ZP-N alone is sufficient for polymerization [191,192]. Most vertebrates have three ZP proteins – simply numbered ZP1, ZP2, and ZP3. The ZP primarily consists of ZP2 and ZP3 in approximately equal quantities, with both proteins under positive selection [193]. In abalone, 32 genes expressed in the ovary were identified that contained full ZP domains, with expression of 29 validated by tandem MS. One of these proteins was VERL, such that its structure includes 22 separate ZP-N repeats followed by a complete ZP domain (Fig. 4C). Approximate protein quantification using the normalized spectral abundance factor (NSAF) suggested that VERL is a relatively minor component (∼1% by stoichiometry; Fig. 4B) [188,189]; however, it is the only domain with multiple tandem ZP-N repeats, and may be acting as a scaffolding protein that interlocks with the other ZP-containing proteins. This crosslinking function would also explain why it is the target of lysin and is key for disrupting VE supramolecular structure. Interestingly, vertebrate ZP2 has a similar architecture of serial ZP-N repeats before a complete ZP domain(Fig. 4E) and may be providing an analogous scaffolding function. Phylogenetic analysis of the abalone ZP proteins identified one component, VEZP14, as a more recent paralog of VERL, although without the multiple tandem ZP-N repeats [189]. Co-evolutionary rate analysis revealed the surprising pattern of inverted rates of evolution between VERL and VEZP14: when lysin and VERL were most rapidly evolving, VEZP14 was evolving slowly, and vice versa (Fig. 4D). Lysin binds VEZP14 with similar affinity to VERL, such that VEZP14 may have evolved as a competitive inhibitor to slow and/or restrict lysin-mediated dissolution of the VE in certain contexts (analogous to strategies which attenuate polyspermy risk in sea urchins) [194]. The role of molecular decoys is well-established in immunology and pathogen response (where rapid evolution is also common) [195], but VEZP14 is the first instance that this has been proposed for a sexually selected system. This potentially suggests that there may be a small subset of “tools” that have evolved multiple times for systems under arms race dynamics, regardless of the source of selection.
Fig. 4.
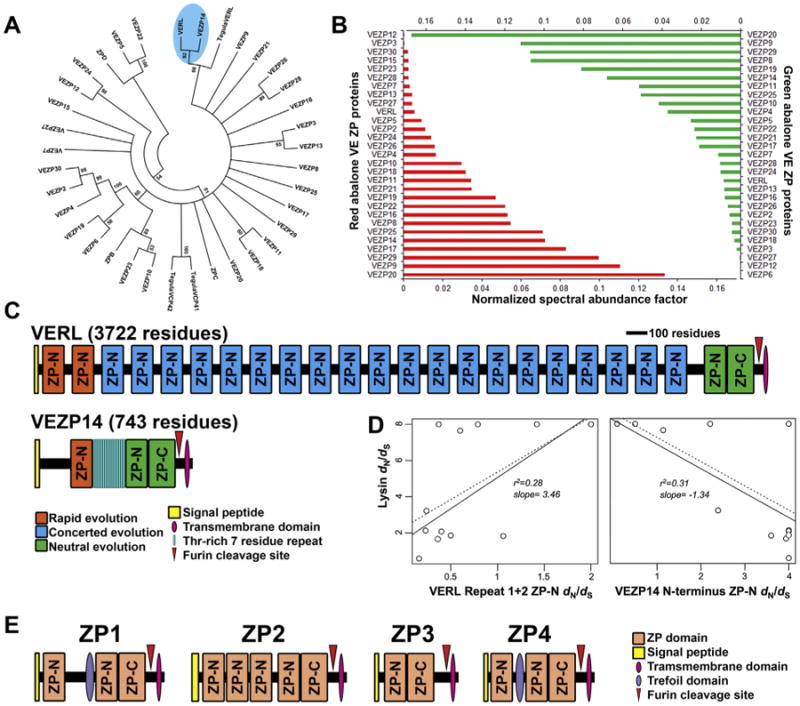
Evolution and proteomics of ZP-domain containing proteins in abalone. (A) Phylogenetic tree of the 32 ZP-domain containing proteins from red abalone (H. rufescens), with three Tegula ZP domains included as outgroups. Notably, VEZP14 and VERL are well-supported on a separate branch from Tegula VERL, suggesting that they result from a more recent gene duplication. (B) Estimate of protein expression for ZP domain-containing proteins based on normalized spectral abundance factor (NSAF), determined by multidimensional protein identification (MUDPit). Both red and green abalone have relatively similar expression levels of homologous genes, with VERL representing ∼1% expression (adapted from [189]). (C) The gene architecture of VERL and VEZP14. (D) Co-evolutionary rates of VERL and VEZP14 relative to lysin, with the positive correlation between lysin and VERL suggesting that as VERL rapidly evolves, so does lysin, with the inverse relationship for lysin and VEZP14 (adapted from [194]). (E) Gene architecture of human ZP proteins, adapted from [193]. It is noteworthy that ZP2 contains a shorter, but similar, tandem array of ZP-N domains like VERL.
While biomedical implications have led to great interest in characterizing mammalian and human fertilization, our understanding of the molecular components and their interactions remains limited. As previously mentioned, ZP2 and ZP3 are the major constituents of the mammalian ZP, comprising ∼80% of the total protein [193]. Humans and other primates contain a fourth ZP protein (ZP4) that is a recent gene duplication of ZP1 [196]. Both ZP2 and ZP3 are within the top 10% most rapidly evolving genes in the human genome [197], and similar trends were found for field mice and birds [198,199]. Mouse oocytes with gene knockouts for ZP2 or ZP3 fail to form a proper ZP, with knockouts of ZP1 having a loose, fibrous ZP (suggesting a possible crosslinking role for ZP1, likely through its extra ZP-N motif) [200]. It was initially postulated that the sperm binding motif was exclusively on ZP3 [201], but the current consensus is that the interactions are more complex and involve carbohydrate recognition, the ZP supramolecular structure, or both [190,193,202,203]. Additional gene deletion studies have identified sperm factors important for ZP adhesion, but no clear molecular mechanism has been proposed [204]. However, significant advancements were recently made towards understanding mammalian membrane fusion. Using a high-throughput ELISA-like screen for membrane protein/ligand interactions [205], folate receptor 4 (FolR4, now renamed “Juno”) was characterized as the egg cell surface receptor for sperm Izumo1 and determined to be critical for membrane fusion. Immunocytochemistry experiments also demonstrated that upon sperm binding, Juno is shed from the oocyte membrane through vesicle formation and exocytosis, acting as a block to polyspermy. This pathway seems to be conserved in all mammals [206], but the co-evolutionary dynamics of these proteins remain uncharacterized.
6. Conclusions and future considerations
The rapid co-evolution of male proteins and female receptors through arms race-like dynamics is a phenomenon that permeates all levels of complex animal reproduction, from pre-copulatory events until a sperm fuses with an egg, and is not exclusive to any specific lineage. The case has been presented here that studies of reproductive systems are strongly enhanced by jointly characterizing the molecular underpinnings through classical biochemistry and the likely targets of selection through models of molecular evolution. The availability of high-throughput next-generation sequencing and mass spectral proteomics allow simultaneous identification of both. In the plethodontid salamander system, proteomics permitted identification of the subset of translated PMF isoforms behind the dozens of gene duplication events [109]. In Drosophila, whole animal isotopic labeling and shotgun proteomics dramatically expanded the range of identified seminal fluid proteins, for which evolutionary rates could be rapidly calculated from whole genome sequence data [22]. In abalone, joint transcriptomics and proteomics together permitted identification of a highly abundant, rapidly evolving protein that – because of atypical biochemical properties – had gone unnoticed despite more than 20 years of research [23]. As argued in the introduction, the application of these methods will continue to expand the number of organisms for which reproductive proteins are characterized, and allow more broad range phylogenetic comparisons. At the present, these high-throughput technologies are most well-suited for candidate identification, and still do not have the precision to fully replace biochemical structure/function-driven experiments. Structures determined by NMR and X-ray crystallography are limited for reproductive protein, and nearly all examples were discussed here. The NMR-derived model of ESP1 and homology model of V2Rp5 are the current best example of any interacting pair of reproductive proteins, but these data do not directly address the challenge of how particular mutations in a receptor might drive adaptive co-evolution in a male ornament. Sequencing based methods also provide limited information on post-translational modifications, such as glycosylation, which are functionally critical in many reproductive proteins [207]. While time, cost, and labor may preclude future structural studies for many reproductive proteins, improved in silico docking protocols, sequencing of multiple homologs for residue covariance between interacting partners [208], and advancements in NGS-based high-throughput mutational screens [209] will permit more rapid cross-validation of molecular evolutionary models. Such studies should drive the testing of many of the available hypotheses regarding sexual selection, providing a unification between theory and empiricism that currently does not exist.
Acknowledgments
We would like to thank Dr. Jan Aagaard, Jennifer McCreight, Emily Killingbeck, and Andrew Knight, as well as two anonymous reviewers for their comments on the manuscript. This work was supported by NIH grant R01HD076862 (to W.J.S.) and NIH training grant T32HG000035 (to D.B.W.).
Footnotes
Conflicts of interest: The authors have no conflicts of interest to disclose.
References
- 1.Otto S. Sexual reproduction and the evolution of sex. Nat Educ. 2008;1:182. [Google Scholar]
- 2.De Visser JAGM, Elena SF. The evolution of sex: empirical insights into the roles of epistasis and drift. Nat Rev Genet. 2007;8:139–149. doi: 10.1038/nrg1985. [DOI] [PubMed] [Google Scholar]
- 3.Felsenstein J. The evolutionary advantage of recombination. Genetics. 1974;78:737–756. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otto SP, Lenormand T. Resolving the paradox of sex and recombination. Nat Rev Genet. 2002;3:252–261. doi: 10.1038/nrg761. [DOI] [PubMed] [Google Scholar]
- 5.Fusani L, Day LB, Canoine V, Reinemann D, Hernandez E, Schlinger BA. Androgen and the elaborate courtship behavior of a tropical lekking bird. Horm Behav. 2007;51:62–68. doi: 10.1016/j.yhbeh.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Arnold SJ. The Reproductive Biology of Amphibians. Springer; 1977. The Evolution of Courtship Behavior in New World Salamanders with Some Comments on Old World Salamandrids; pp. 141–183. [Google Scholar]
- 7.Masonjones HD, Lewis SM. Courtship behavior in the dwarf seahorse, Hippocampus zosterae. Copeia. 1996:634–640. [Google Scholar]
- 8.Melrose DR, Reed HC, Patterson RL. Androgen steroids associated with boar odour as an aid to the detection of oestrus in pig artificial insemination. Br Vet J. 1971;127:497–502. doi: 10.1016/s0007-1935(17)37337-2. [DOI] [PubMed] [Google Scholar]
- 9.Albone ES. Mammalian Semiochemistry. J Wiley; New York: 1984. [Google Scholar]
- 10.Johnston RE. Chemical communication and pheromones: the types of chemical signals and the role of the vomeronasal system. In: Finger TE, Silver WL, Restrepo D, editors. The Neurobiology of Taste and Smell. Wiley-Liss; New York: 2000. pp. 101–127. [Google Scholar]
- 11.Wedell N, Gage MJ, Parker GA. Sperm competition, male prudence and sperm-limited females. Trends Ecol Evol. 2002;17:313–320. [Google Scholar]
- 12.Simmons LW, Kvarnemo C. Ejaculate expenditure by malebush crickets decreases with sperm competition intensity. Proc R Soc Lond Ser B Biol Sci. 1997;264:1203–1208. [Google Scholar]
- 13.Heinze J, d'Ettorre P. Honest and dishonest communication in social Hymenoptera. J Exp Biol. 2009;212:1775–1779. doi: 10.1242/jeb.015008. [DOI] [PubMed] [Google Scholar]
- 14.Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- 15.Andersson M. Sexual Selection. Princeton Univ. Press; Princeton, NJ: 1994. [Google Scholar]
- 16.Kokko H, Jennions MD, Brooks R. Unifying and testing models of sexual selection. Annu Rev Ecol Evol Syst. 2006;37:43–66. [Google Scholar]
- 17.Wyatt T. Pheromones and Animal Behavior: Chemical Signals and Signatures. Cambridge Univeristy Press; 2014. [Google Scholar]
- 18.Vacquier VD. Evolution of gamete recognition proteins. Science. 1998;281:1995–1998. doi: 10.1126/science.281.5385.1995. [DOI] [PubMed] [Google Scholar]
- 19.Wolfner M. The gifts that keep on giving: physiological functions and evolutionary dynamics of male seminal proteins in Drosophila. Heredity. 2002;88:85–93. doi: 10.1038/sj.hdy.6800017. [DOI] [PubMed] [Google Scholar]
- 20.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 21.Swanson WJ, Vacquier VD. The abalone egg vitelline envelope receptor for sperm lysin is a giant multivalent molecule. Proc Natl Acad Sci. 1997;94:6724–6729. doi: 10.1073/pnas.94.13.6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Findlay GD, Yi X, MacCoss MJ, Swanson WJ. Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS Biol. 2008;6:e178. doi: 10.1371/journal.pbio.0060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmer MR, McDowall MH, Stewart L, Ouaddi A, MacCoss MJ, Swanson WJ. Mass spectrometry and next-generation sequencing reveal an abundant and rapidly evolving abalone sperm protein. Mol Reprod Dev. 2013;80:460–465. doi: 10.1002/mrd.22182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilburn DB, Eddy SL, Chouinard AJ, Arnold SJ, Feldhoff RC, Houck LD. Pheromone isoform composition differentially affects female behaviour in the red-legged salamander, Plethodon shermani. Anim Behav. 2015;100:1–7. [Google Scholar]
- 25.Wilburn DB, Bowen KE, Doty KA, Arumugam S, Lane AN, Feldhoff PW, et al. Structural insights into the evolution of a sexy protein: novel topology and restricted backbone flexibility in a hypervariable vertebrate pheromone. PLoS ONE. 2014;9:e96975. doi: 10.1371/journal.pone.0096975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koboldt Daniel C, Steinberg Karyn M, Larson David E, Wilson Richard K, Mardis ER. The next-generation sequencing revolution and its impact on, genomics. Cell. 2013;155:27–38. doi: 10.1016/j.cell.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Dijk EL, Auger H, Jaszczyszyn Y, Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30:418–426. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Bensimon A, Heck AJR, Aebersold R. Mass spectrometry-based proteomics and network biology. Annu Rev Biochem. 2012;81:379–405. doi: 10.1146/annurev-biochem-072909-100424. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Fonslow BR, Shan B, Baek MC, Yates JR. Protein analysis by shotgun/bottom-up proteomics. Chem Rev. 2013;113:2343–2394. doi: 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacCoss MJ, Yates JR., III Proteomics: analytical tools and techniques. Curr Opin Clin Nutr Metab Care. 2001;4:369–375. doi: 10.1097/00075197-200109000-00006. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen R. Molecular signatures of natural selection. Annu Rev Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- 32.Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- 33.Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- 34.Wong WSW, Yang Z, Goldman N, Nielsen R. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics. 2004;168:1041–1051. doi: 10.1534/genetics.104.031153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oleksyk TK, Smith MW, O'Brien SJ. Genome-wide scans for footprints of natural selection. Philos Trans R Soc B. 2010;365:185–205. doi: 10.1098/rstb.2009.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carter P, Wells JA. Dissecting the catalytic triad of a serine protease. Nature. 1988;332:564–568. doi: 10.1038/332564a0. [DOI] [PubMed] [Google Scholar]
- 37.da Fonseca RR, Kosiol C, Vinař T, Siepel A, Nielsen R. Positive selection on apoptosis related genes. FEBS Lett. 2010;584:469–476. doi: 10.1016/j.febslet.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Rosenberg HF, Nei M. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fitch WM, Leiter J, Li X, Palese P. Positive Darwinian evolution in human influenza A viruses. Proc Natl Acad Sci. 1991;88:4270–4274. doi: 10.1073/pnas.88.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodríguez-Trelles F, Tarrío R, Ayala FJ. Convergent neofunctionalization by positive Darwinian selection after ancient recurrent duplications of the xanthine dehydrogenase gene. Proc Natl Acad Sci. 2003;100:13413–13417. doi: 10.1073/pnas.1835646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lande R. Models of speciation by sexual selection on polygenic traits. Proc Natl Acad Sci. 1981;78:3721–3725. doi: 10.1073/pnas.78.6.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weatherhead PJ, Robertson RJ. Offspring quality and the polygyny threshold: “the sexy son hypothesis”. Am Nat. 1979;113:201–208. [Google Scholar]
- 43.Fuller RC, Houle D, Travis J. Sensory bias as an explanation for the evolution of mate preference. Am Nat. 2005;166:437–446. doi: 10.1086/444443. [DOI] [PubMed] [Google Scholar]
- 44.Rowe L, Cameron E, Day T. Escalation, retreat, and female indifference as alternative outcomes of sexually antagonistic coevolution. Am Nat. 2005;165:S5–S18. doi: 10.1086/429395. [DOI] [PubMed] [Google Scholar]
- 45.Hoezler GA. The good parent process of sexual selection. Anim Behav. 1989;38:1067–1078. [Google Scholar]
- 46.Price T, Schluter D, Heckman NE. Sexual selection when the female directly benefits. Biol J Linn Soc Lond. 1993;48:187–211. [Google Scholar]
- 47.Houck LD, Arnold SJ. Courtship and mating behavior. In: Sever DM, editor. Phylogeny and Reproductive Biology of Urodela (Amphibia) Science Publishers; Enfield, New Hampshire: 2003. pp. 383–424. [Google Scholar]
- 48.Clark NL, Aagaard JE, Swanson WJ. Evolution of reproductive proteins from animals and plants. Reproduction. 2006;131:11–22. doi: 10.1530/rep.1.00357. [DOI] [PubMed] [Google Scholar]
- 49.McLaughlin RN, Jr, Poelwijk FJ, Raman A, Gosal WS, Ranganathan R. The spatial architecture of protein function and adaptation. Nature. 2012;491:138–142. doi: 10.1038/nature11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karlson P, Luscher M. “Pheromones”: a new term for a class of biologically active substances. Nature. 1959;183:55–56. doi: 10.1038/183055a0. [DOI] [PubMed] [Google Scholar]
- 51.Butenandt A, Beckmann R, Hecker E. Über den sexuallockstoff des seidenspinners, i. der biologische test und die isolierung des reinen sexuallockstoffes bombykol. Hoppe Seylers Z Physiol Chem. 1961;324:71–83. doi: 10.1515/bchm2.1961.324.1.71. [DOI] [PubMed] [Google Scholar]
- 52.Jaeger RG. Pheromonal markers as territorial advertisement by terrestrial salamanders. In: Duvall D, Muller-Schwarze D, Silverstein RM, editors. Chemical Signals in Vertebrates. Vol. 4. Plenum Press; New York: 1986. pp. 191–203. [Google Scholar]
- 53.Levesque H, Scaffidi D, Polkinghorne C, Sorensen P. A multi-component species identifying pheromone in the goldfish. J Chem Ecol. 2011;37:219–227. doi: 10.1007/s10886-011-9907-6. [DOI] [PubMed] [Google Scholar]
- 54.Fine J, Sorensen P. Isolation and biological activity of the multi-component sea lamprey migratory pheromone. J Chem Ecol. 2008;34:1259–1267. doi: 10.1007/s10886-008-9535-y. [DOI] [PubMed] [Google Scholar]
- 55.Conte YL, Hefetz A. Primer pheromones in social Hymenoptera. Annu Rev Entomol. 2008;53:523–542. doi: 10.1146/annurev.ento.52.110405.091434. [DOI] [PubMed] [Google Scholar]
- 56.Wyatt T. Pheromones and signature mixtures: defining species-wide signals and variable cues for identity in both invertebrates and vertebrates. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2010;196:685–700. doi: 10.1007/s00359-010-0564-y. [DOI] [PubMed] [Google Scholar]
- 57.Luo M, Fee MS, Katz LC. Encoding pheromonal signals in the accessory olfactory bulb of behaving mice. Science. 2003;299:1196e201. doi: 10.1126/science.1082133. [DOI] [PubMed] [Google Scholar]
- 58.Dulac C, Torello AT. Molecular detection of pheromone signals in mammals: from genes to behaviour. Nat Rev Neurosci. 2003;4:551–562. doi: 10.1038/nrn1140. [DOI] [PubMed] [Google Scholar]
- 59.Baum MJ, Kelliher KR. Complementary roles of the main and accessory olfactory systems in mammalian mate recognition. Annu Rev Physiol. 2009;71:141–160. doi: 10.1146/annurev.physiol.010908.163137. [DOI] [PubMed] [Google Scholar]
- 60.Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 61.Herrada G, Dulac C. A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell. 1997;90:763–773. doi: 10.1016/s0092-8674(00)80536-x. [DOI] [PubMed] [Google Scholar]
- 62.Yang H, Shi P, Zhang YP, Zhang J. Composition and evolution of the V2r vomeronasal receptor gene repertoire in mice and rats. Genomics. 2005;86:306–315. doi: 10.1016/j.ygeno.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 63.Loconto J, Papes F, Chang E, Stowers L, Jones EP, Takada T, et al. Functional expression of murine V2R pheromone receptors involves selective association with the M10 and M1 families of MHC class Ib molecules. Cell. 2003;112:607–618. doi: 10.1016/s0092-8674(03)00153-3. [DOI] [PubMed] [Google Scholar]
- 64.Isogai Y, Si S, Pont-Lezica L, Tan T, Kapoor V, Murthy VN, et al. Molecular organization of vomeronasal chemoreception. Nature. 2011;478:241–245. doi: 10.1038/nature10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boschat C, Pélofi C, Randin O, Roppolo D, Lüscher C, Broillet MC, et al. Pheromone detection mediated by a V1r vomeronasal receptor. Nat Neurosci. 2002;5:1261–1262. doi: 10.1038/nn978. [DOI] [PubMed] [Google Scholar]
- 66.Papes F, Logan DW, Stowers L. The vomeronasal organ mediates interspecies defensive behaviors through detection of protein pheromone homologs. Cell. 2010;141:692–703. doi: 10.1016/j.cell.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Michel WC, Steullet P, Cate HS, Burns CJ, Zhainazarov AB, Derby CD. High-resolution functional labeling of vertebrate and invertebrate olfactory receptor neurons using agmatine, a channel-permeant cation. J Neurosci Methods. 1999;90:143–156. doi: 10.1016/s0165-0270(99)00077-1. [DOI] [PubMed] [Google Scholar]
- 68.Ibarra-Soria X, Levitin MO, Saraiva LR, Logan DW. The olfactory transcriptomes of mice. PLoS Genet. 2014;10:e1004593. doi: 10.1371/journal.pgen.1004593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chamero P, Marton TF, Logan DW, Flanagan K, Cruz JR, Saghatelian A, et al. Identification of protein pheromones that promote aggressive behaviour. Nature. 2007;450:899–902. doi: 10.1038/nature05997. [DOI] [PubMed] [Google Scholar]
- 70.Haga S, Hattori T, Sato T, Sato K, Matsuda S, Kobayakawa R, et al. The male mouse pheromone ESP1 enhances female sexual receptive behaviour through a specific vomeronasal receptor. Nature. 2010;466:118–122. doi: 10.1038/nature09142. [DOI] [PubMed] [Google Scholar]
- 71.Leinders-Zufall T, Ishii T, Mombaerts P, Zufall F, Boehm T. Structural requirements for the activation of vomeronasal sensory neurons by MHC peptides. Nat Neurosci. 2009;12:1551–1558. doi: 10.1038/nn.2452. [DOI] [PubMed] [Google Scholar]
- 72.Lucke C, Franzoni L, Abbate F, Lohr F, Ferrari E, Sorbi RT, et al. Solution structure of a recombinant mouse major urinary protein. Eur J Biochem/FEBS. 1999;266:1210–1218. doi: 10.1046/j.1432-1327.1999.00984.x. [DOI] [PubMed] [Google Scholar]
- 73.Roberts S, Simpson D, Armstrong S, Davidson A, Robertson D, McLean L, et al. Darcin: a male pheromone that stimulates female memory and sexual attraction to an individual male's odour. BMC Biol. 2010;8:75. doi: 10.1186/1741-7007-8-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roberts SA, Davidson AJ, McLean L, Beynon RJ, Hurst JL. Pheromonal induction of spatial learning in mice. Science. 2012;338:1462–1465. doi: 10.1126/science.1225638. [DOI] [PubMed] [Google Scholar]
- 75.Briand L, Trotier D, Pernollet JC. Aphrodisin, an aphrodisiac lipocalin secreted in hamster vaginal secretions. Peptides. 2004;25:1545–1552. doi: 10.1016/j.peptides.2003.10.026. [DOI] [PubMed] [Google Scholar]
- 76.Kimoto H, Haga S, Sato K, Touhara K. Sex-specific peptides from exocrine glands stimulate mouse vomeronasal sensory neurons. Nature. 2005;437:898–901. doi: 10.1038/nature04033. [DOI] [PubMed] [Google Scholar]
- 77.Yoshinaga S, Sato T, Hirakane M, Esaki K, Hamaguchi T, Haga-Yamanaka S, et al. Structure of the mouse sex peptide pheromone ESP1 reveals a molecular basis for specific binding to the class C G-protein-coupled vomeronasal receptor. J Biol Chem. 2013;288:16064–16072. doi: 10.1074/jbc.M112.436782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garimella R, Xu Y, Schein CH, Rajarathnam K, Nagle GT, Painter SD, et al. NMR solution structure of attractin, a water-borne protein pheromone from the mollusk Aplysia californica. Biochemistry. 2003;42:9970–9979. doi: 10.1021/bi0274322. [DOI] [PubMed] [Google Scholar]
- 79.Kimoto H, Sato K, Nodari F, Haga S, Holy TE, Touhara K. Sex- and strain-specific expression and vomeronasal activity of mouse ESP family peptides. Curr Biol. 2007;17:1879–1884. doi: 10.1016/j.cub.2007.09.042. [DOI] [PubMed] [Google Scholar]
- 80.Ferrero DM, Moeller LM, Osakada T, Horio N, Li Q, Roy DS, et al. A juvenile mouse pheromone inhibits sexual behaviour through the vomeronasal system. Nature. 2013;502:368–371. doi: 10.1038/nature12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abe T, Touhara K. Structure and function of a peptide pheromone family that stimulate the vomeronasal sensory system in mice. Biochem Soc Trans. 2014;42:873–877. doi: 10.1042/BST20140051. [DOI] [PubMed] [Google Scholar]
- 82.Woodley SK. Pheromone communication in amphibians. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2010;196:713–727. doi: 10.1007/s00359-010-0540-6. [DOI] [PubMed] [Google Scholar]
- 83.Woodley SK. Chemosignals, hormones, and amphibian reproduction. Horm Behav. 2015;68:3–13. doi: 10.1016/j.yhbeh.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 84.Van Bocxlaer I, Treer D, Maex M, Vandeberg W, Janssenswillen S, Stregen G, et al. Side-by-side secretion of late Palaeozoic diverged courtship pheromones in an aquatic salamander. Proc R Soc Lond B. 2015;282 doi: 10.1098/rspb.2014.2960. 20142960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kikuyama S, Toyoda F, Ohmiya Y, Matsuda K, Tanaka S, Hayashi H. Sodefrin: a female-attracting peptide phermone in newt cloacal glands. Science. 1995;267:1643–1645. doi: 10.1126/science.7886452. [DOI] [PubMed] [Google Scholar]
- 86.Yamamoto K, Kawai Y, Hayashi T, Ohe Y, Hayashi H, Toyoda F, et al. Silefrin, a sodefrin-like pheromone in the abdominal gland of the sword-tailed newt, Cynops ensicauda. FEBS Lett. 2000;472:267–270. doi: 10.1016/s0014-5793(00)01455-1. [DOI] [PubMed] [Google Scholar]
- 87.Nakada T, Toyoda F, Iwata T, Yamamoto K, Conlon JM, Kato T, et al. Isolation, characterization and bioactivity of a region-specific pheromone, [Val8]sodefrin from the newt Cynops pyrrhogaster. Peptides. 2007;28:774–780. doi: 10.1016/j.peptides.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 88.Kikuyama S, Toyoda F. Sodefrin: a novel sex pheromone in a newt. Rev Reprod. 1999;4:1–4. doi: 10.1530/ror.0.0040001. [DOI] [PubMed] [Google Scholar]
- 89.Janssenswillen S, Vandebergh W, Treer D, Willaert B, Maex M, Van Bocxlaer I, et al. Origin and diversification of a salamander sex pheromone system. Mol Biol Evol. 2015;32:472–480. doi: 10.1093/molbev/msu316. [DOI] [PubMed] [Google Scholar]
- 90.Dawley EM. Recognition of individual, sex, and species odours by salamanders of the Plethodon glutinosus–P. jordani complex. Anim Behav. 1984;32:352–361. [Google Scholar]
- 91.Dawley EM. Evolution of chemical signals as a premating isolating mechanism in a complex of terrestrial salamanders. In: Duvall D, Muller-Schwarze D, Silverstein RM, editors. Chemical Signals in Vertebrates. Vol. 4. Plenum Press; New York: 1986. pp. 221–224. [Google Scholar]
- 92.Marco A, Chivers DP, Kiesecker JK, Blaustein AR. Mate choice by chemical cues in western redback (Plethodon vehiculum) and Dunn's (P. dunni) salamanders. J Chem Ecol. 1998;27:1333–1344. [Google Scholar]
- 93.Danzter BJ, Jaeger RG. Male redbacked salamanders can determine the reproductive status of conspecific females through volatile chemical signals. Herpetologica. 2007;63:176–183. [Google Scholar]
- 94.Walls SC, Mathis A, Jaeger RG, Gergits WF. Male salamanders with high quality diets have faeces attractive to females. Anim Behav. 1989;38:546–548. [Google Scholar]
- 95.Chouinard AJ. Rapid onset of mate quality assessment via chemical signals in a woodland salamander (Plethodon cinereus) Behav Ecol Sociobiol. 2012;66:765–775. [Google Scholar]
- 96.Maksimowich DS, Mathis A. Pheromonal markers as indicators of parasite load: parasite-mediated behavior in salamanders (Plethodon angusticlavius) Acta Ethol. 2001;3:83–87. [Google Scholar]
- 97.Woodley SK. Plasma androgen levels, spermatogenesis, and secondary sexual characteristics in two species of Plethodontid salamanders with dissociated reproductive patterns. Gen Comp Endocrinol. 1994;96:206–214. doi: 10.1006/gcen.1994.1175. [DOI] [PubMed] [Google Scholar]
- 98.Sever DM. Induction of secondary sex characters in Eurycea quadridigitata. Copeia. 1976:830–833. [Google Scholar]
- 99.Sever DM. Morphology and seasonal variation of the mental hedonic glands of the dwarf salamander, Eurycea quadridigitata (Holbrook) Herpetologica. 1975;31:241–251. [Google Scholar]
- 100.Houck LD, Bell AM, Reagan-Wallin NL, Feldhoff RC. Effects of experimental delivery of male courtship pheromones on the timing of courtship in a terrestrial salamander, Plethodon jordani (Caudata: Plethodontidae) Copeia. 1998:214–219. [Google Scholar]
- 101.Feldhoff RC, Rollmann SM, Houck LD. Chemical analyses of courtship pheromones in a Plethodontid salamander. In: Johnston RE, Műller-Schwarze D, Sorensen P, editors. Advances in Chemical Signals in Vertebrates. Kluwer Academic/Plenum; New York: 1999. pp. 117–125. [Google Scholar]
- 102.Houck LD, Mead LS, Watts RA, Arnold SJ, Feldhoff PW, Feldhoff RC. A candidate vertebrate pheromone, SPF, increases female receptivity in a salamander. In: Hurst J, Beynon R, Muller-Schwarze D, editors. Chemical Signals in Vertebrates. Vol. 11. Springer; New York: 2007. pp. 213–221. [Google Scholar]
- 103.Houck LD, Palmer CA, Watts RA, Arnold SJ, Feldhoff PW, Feldhoff RC. A new vertebrate courtship pheromone that affects female receptivity in a terrestrial salamander. Anim Behav. 2007;73:315–320. [Google Scholar]
- 104.Kiemnec-Tyburczy KM, Watts RA, Gregg RG, von Borstel D, Arnold SJ. Evolutionary shifts in courtship pheromone composition revealed by EST analysis of plethodontid salamander mental glands. Gene. 2009;432:75–81. doi: 10.1016/j.gene.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 105.Watts RA, Palmer CA, Feldhoff RC, Feldhoff PW, Houck LD, Jones AG, et al. Stabilizing selection on behavior and morphology masks positive selection on the signal in a salamander pheromone signaling complex. Mol Biol Evol. 2004;21:1032–1041. doi: 10.1093/molbev/msh093. [DOI] [PubMed] [Google Scholar]
- 106.Palmer CA, Watts RA, Gregg RG, McCall MA, Houck LD, Highton R, et al. Lineage-specific differences in evolutionary mode in a salamander courtship pheromone. Mol Biol Evol. 2005;22:2243–2256. doi: 10.1093/molbev/msi219. [DOI] [PubMed] [Google Scholar]
- 107.Palmer CA, Watts RA, Houck LD, Picard AL, Arnold SJ. Evolutionary replacement of components in a salamander pheromone signaling complex: more evidence for phenotypic-molecular decoupling. Evolution. 2007;61:202–215. doi: 10.1111/j.1558-5646.2007.00017.x. [DOI] [PubMed] [Google Scholar]
- 108.Palmer CA, Picard AL, Watts RA, Houck LD, Arnold SJ. Rapid evolution of Plethodontid modulating factor (PMF), a hypervariable salamander courtship pheromone, is driven by positive selection. J Mol Evol. 2010;70:427–440. doi: 10.1007/s00239-010-9342-2. [DOI] [PubMed] [Google Scholar]
- 109.Wilburn DB, Bowen KE, Gregg RG, Cai J, Feldhoff PW, Houck LD, et al. Proteomic and UTR analyses of a rapidly evolving hypervaraible family of vertebrate pheromones. Evolution. 2012;66:2227–2239. doi: 10.1111/j.1558-5646.2011.01572.x. [DOI] [PubMed] [Google Scholar]
- 110.Chouinard AJ, Wilburn DB, Houck LD, Feldhoff RC. Individual variation in pheromone isoform ratios of the red-legged salamander, Plethodon shermani. In: East ML, Dehnhard M, editors. Chemical Signals in Vertebrates. Vol. 12. Springer; New York: 2013. pp. 99–115. [Google Scholar]
- 111.Wilburn DB, Bowen KE, Feldhoff PW, Feldhoff RC. Proteomic analyses of courtship pheromones in the redback salamander, Plethodon cinereus. J Chem Ecol. 2014;40:928–939. doi: 10.1007/s10886-014-0489-y. [DOI] [PubMed] [Google Scholar]
- 112.Doty KA, Wilburn DB, Bowen KE, Feldhoff PW, Feldhoff RC. Co-option and evolution of non-olfactory proteinaceous pheromones in a terrestrial lungless salamander. J Proteomics. 2015 doi: 10.1016/j.jprot.2015.09.019. Same issue as this article. [DOI] [PubMed] [Google Scholar]
- 113.Kiemnec-Tyburczy KM, Woodley SK, Watts RA, Arnold SJ, Houck LD. Expression of vomeronasal receptors and related signaling molecules in the nasal cavity of a caudate amphibian (Plethodon shermani) Chem Senses. 2012;37:335–346. doi: 10.1093/chemse/bjr105. [DOI] [PubMed] [Google Scholar]
- 114.Wirsig-Wiechmann CR, Houck LD, Feldhoff PW, Feldhoff RC. Pheromonal activation of vomeronasal neurons in Plethodontid salamanders. Brain Res. 2002;952:335–344. doi: 10.1016/s0006-8993(02)03369-3. [DOI] [PubMed] [Google Scholar]
- 115.Wirsig-Wiechmann CR, Houck LD, Wood JM, Feldhoff PW, Feldhoff RC. Male pheromone protein components activate female vomeronasal neurons in the salamander Plethodon shermani. BMC Neurosci. 2006;7:26. doi: 10.1186/1471-2202-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jensen-Seaman M, Li WH. Evolution of the hominoid semenogelin genes, the major proteins of ejaculated semen. J Mol Evol. 2003;57:261–270. doi: 10.1007/s00239-003-2474-x. [DOI] [PubMed] [Google Scholar]
- 117.Parga JA, Maga M, Overdorff DJ. High-resolution X-ray computed tomography scanning of primate copulatory plugs. Am J Phys Anthropol. 2006;129:567–576. doi: 10.1002/ajpa.20281. [DOI] [PubMed] [Google Scholar]
- 118.Lung O, Kuo L, Wolfner MF. Drosophila males transfer antibacterial proteins from their accessory gland and ejaculatory duct to their mates. J Insect Physiol. 2001;47:617–622. doi: 10.1016/s0022-1910(00)00151-7. [DOI] [PubMed] [Google Scholar]
- 119.Avila FW, Sirot LK, LaFlamme BA, Rubinstein CD, Wolfner MF. Insect seminal fluid proteins: identification and function. Annu Rev Entomol. 2011;56:21–40. doi: 10.1146/annurev-ento-120709-144823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen PS, Stumm-Zollinger E, Aigaki T, Balmer J, Bienz M, Bohlen P. A male accessory gland peptide that regulates reproductive behavior of female D. melanogaster. Cell. 1988;54:291–298. doi: 10.1016/0092-8674(88)90192-4. [DOI] [PubMed] [Google Scholar]
- 121.Pilch B, Mann M. Large-scale and high-confidence proteomic analysis of human seminal plasma. Genome Biol. 2006;7:R40. doi: 10.1186/gb-2006-7-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Collins AM, Caperna TJ, Williams V, Garrett WM, Evans JD. Proteomic analyses of male contributions to honey bee sperm storage and mating. Insect Mol Biol. 2006;15:541–549. doi: 10.1111/j.1365-2583.2006.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Parthansarthy R, Tan A, Sun Z, Chen Z, Rankin M, Palli S. Juvenile hormone regulation of male accessory gland activity in the red flour beetle, Tribolium castaneum. Mech Dev. 2009;126:563–579. doi: 10.1016/j.mod.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dottorini T, Nicolaides L, Ranson H, Rogers D, Crisanti A, Catteruccia F. A genome-wide analysis in Anopheles gambiae mosquitoes reveals 46 male accessory gland genes, possible modulators of female behavior. Proc Natl Acad Sci. 2007;104:16215–16220. doi: 10.1073/pnas.0703904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rogers D, Whitten M, Thailayil J, Soichor J, Levashina E, Catteruccia F. Molecular and cellular components of the mating machinery in Anopheles gambiae females. Proc Natl Acad Sci. 2008;105:19390–19395. doi: 10.1073/pnas.0809723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sirot LK, Poulson R, McKenna M, Girnary H, Wolfner MF. Identity and transfer of male reproductive gland proteins of the genue vector mosquito, Aedes aegypti: potential tools for female feeding and reproduction. Insect Biochem Mol Biol. 2008;38:176–189. doi: 10.1016/j.ibmb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Walters J, Harrison R. EST analysis of male accessory glands from Heliconius butterflies with divergent mating systems. BMC Genomics. 2008;9:592. doi: 10.1186/1471-2164-9-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Walters J, Harrison R. Combined EST and proteomic analysis identifies rapidly evolving seminal fluid proteins in Heliconius butterflies. Mol Biol Evol. 2010;27:2000–2013. doi: 10.1093/molbev/msq092. [DOI] [PubMed] [Google Scholar]
- 129.Weiss B, Stepczynski J, Wong P, Kaufman W. Identification of genes differentially expressed in the testis/vas deferns of the fed male tick, Amblyomma bedraeum. Insect Biochem Mol Biol. 2002;32:785–793. doi: 10.1016/s0965-1748(01)00161-8. [DOI] [PubMed] [Google Scholar]
- 130.Reinhardt K, Wong C, Georgiou A. Detection of seminal fluid proteins in the bed bug, Cimex lecturlarius, using two-dimensional gel electrophoresis and mass spectrometry. Parasitology. 2009;136:283–292. doi: 10.1017/S0031182008005362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Andres J, Maroja L, Bogdanowicz S, Swanson WJ, Harrison R. Molecular evolution of seminal proteins in field crickets. Mol Biol Evol. 2006;23:1574–1584. doi: 10.1093/molbev/msl020. [DOI] [PubMed] [Google Scholar]
- 132.Andres J, Maroja L, Harrison R. Searching for candidate speciation genes using a proteomic approach: seminal proteins in field crickets. Proc R Soc Lond B. 2007;275:427–445. doi: 10.1098/rspb.2008.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ram K, Wolfner MF. Seminal influences: Drosophila Acps and the molecular interplay between males and females during reproduction. Integr Comp Biol. 2007;47:427–445. doi: 10.1093/icb/icm046. [DOI] [PubMed] [Google Scholar]
- 134.Dorus S, Busby S, Gerike U, Shabanowitz J, Hunt D, Karr T. Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nat Genet. 2006;38:1440–1445. doi: 10.1038/ng1915. [DOI] [PubMed] [Google Scholar]
- 135.Moshitzky P, Fleischmann I, Chaimov N, Suadan P, Klauser S, Kubli E, et al. Sex peptide activates juvenile hormone biosynthesis in Drosophila melanogaster corpus allatum. Insect Biochem Mol Biol. 1996;32:363–374. doi: 10.1002/(SICI)1520-6327(1996)32:3/4<363::AID-ARCH9>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 136.Soller M, Bownes M, Kubli E. Mating and sex peptide stimulate the accumulation of yolk in oocytes of Drosophila melanogaster. Eur J Biochem. 1997;243:732–738. doi: 10.1111/j.1432-1033.1997.00732.x. [DOI] [PubMed] [Google Scholar]
- 137.Rubinstein CD, Wolfner MF. Drosophila seminal protein ovulin mediates ovulation through female octopamine neuronal signaling. Proc Natl Acad Sci. 2013;110:17420–17425. doi: 10.1073/pnas.1220018110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chapman T, Partridge L. Female fitness in Drosophila melanogaster: an interaction between the effect of nutrition and of encounter rate with mates. Proc Biol Sci. 1996;22:755–759. doi: 10.1098/rspb.1996.0113. [DOI] [PubMed] [Google Scholar]
- 139.Abrahams MV. The trade-off between foraging and courting in male guppies. Anim Behav. 1993;45:673–681. [Google Scholar]
- 140.Apger-Mc Glaughon J, Wolfner MF. Post-mating change in excretion by mated Drosophila melanogaster females is a long-term response that depends on sex peptide and sperm. J Insect Physiol. 2013;59:1024–1030. doi: 10.1016/j.jinsphys.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Peng J, Chen S, Busser S, Liu H. Gradual release of sperm bound sex-peptide controls female postmating behavior in Drosophila. Curr Biol. 2005;15:207–213. doi: 10.1016/j.cub.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 142.LaFlamme BA, Ravi Ram K, Wolfner MF. The Drosophila melanogaster seminal fluid protease “seminase” regulates proteolytic and post-mating reproductive processes. PLoS Genet. 2012;8:e1002435. doi: 10.1371/journal.pgen.1002435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yapici N, Kim YJ, Ribeiro C, Dickson BJ. A receptor that mediates the post-mating switch in Drosophila reproductive behavior. Nature. 2008;451:33–37. doi: 10.1038/nature06483. [DOI] [PubMed] [Google Scholar]
- 144.Singh RS, Xu J, Kulathinal RJ. Rapidly Evolving Genes and Genetic Systems. Oxford Univ. Press; Oxford, London: 2012. [Google Scholar]
- 145.Kim Y, Bartalska K, Audsley N, Yamanaka N, Yapici N, Lee J, et al. MIPs are ancestral ligands for the sex peptide receptor. Proc Natl Acad Sci. 2010;107:6520–6525. doi: 10.1073/pnas.0914764107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Poels J, Van Loy T, Vandersmissen H, Van Hiel B, Van Soest S, Nachman R, et al. Myoinhibiting peptides are the ancestral ligands of the promiscuous Drosophila sex peptide receptor. Cell Mol Life Sci. 2010;67:3511–3522. doi: 10.1007/s00018-010-0393-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Oh Y, Yoon SE, Zhang Q, Chae HS, Daubnerova I, Shafer OT, et al. A homeostatic sleep-stabilizing pathway in Drosophila composed of the sex peptide receptor and its ligand, the myoinhibitory peptide. PLoS Biol. 2014;12:e1001974. doi: 10.1371/journal.pbio.1001974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Clark NL, Alani E, Aquadro CF. Evolutionary rate covariation reveals shared functionality and coexpression of genes. Genome Res. 2012;22:714–720. doi: 10.1101/gr.132647.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Findlay GD, Sitnik JL, Wang W, Aquadro CF, Clark NL, Wolfner MF. Evolutionary rate covariation identifies new members of a protein network required for Drosophila melanogaster female post-mating responses. PLoS Genet. 2014;10:e1004108. doi: 10.1371/journal.pgen.1004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wigby S, Sirot LK, Linklater JR, Nuehner N, Calboli FCF, Bretman A, et al. Seminal fluid protein allocation and male reproductive success. Curr Biol. 2009;19:751–757. doi: 10.1016/j.cub.2009.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fowler K, Partridge L. A cost of mating in female fruit flies. Nature. 1989;338:760–761. [Google Scholar]
- 152.Kuijiper B, Stewart A, Rice W. The cost of mating rises nonlinearly with copulation frequency in a laboratory population of Drosophila melanogaster. J Evol Biol. 2006;19:1795–1802. doi: 10.1111/j.1420-9101.2006.01186.x. [DOI] [PubMed] [Google Scholar]
- 153.Chapman T, Liddle L, Kalb J, Wolfner M, Partridge L. Cost of mating in Drosophila melanogaster females is mediated by male accessory gland products. Nature. 1995;373:241–244. doi: 10.1038/373241a0. [DOI] [PubMed] [Google Scholar]
- 154.Wigby S, Chapman T. Sex peptide causes mating costs in female Drosophila melanogaster. Curr Biol. 2005;15:316–321. doi: 10.1016/j.cub.2005.01.051. [DOI] [PubMed] [Google Scholar]
- 155.Dean MD, Findlay GD, Hoopmann M, Wu C, MacCoss M, Swanson WJ, et al. Identification of ejaculated proteins in the house mouse (Mus domesticus) via iso topic labeling. BMC Genomics. 2011;12:306. doi: 10.1186/1471-2164-12-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dean MD, Clark NL, Findlay GD, Karen RC, Yi X, Swanson WJ, et al. Proteomics and comparative genomic investigations reveal heterogeneity in evolutionary rate of male reproductive proteins in mice (Mus domesticus) Mol Biol Evol. 2009;26:1733–1743. doi: 10.1093/molbev/msp094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Clark NL, Swanson WJ. Pervasive adaptive evolution in primate seminal proteins. PLoS Genet. 2005;1:e35. doi: 10.1371/journal.pgen.0010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.de Kretser DM. Male infertility. Lancet. 1997;349:787–790. doi: 10.1016/s0140-6736(96)08341-9. [DOI] [PubMed] [Google Scholar]
- 159.Glabe CG, Vacquier VD. Species specific agglutination of eggs by bindin isolated from sea urchin sperm. Nature. 1977;267:836–838. doi: 10.1038/267836a0. [DOI] [PubMed] [Google Scholar]
- 160.Vacquier VD, Moy GW. Isolation of bindin: the protein responsible for adhesion of sperm to sea urchin eggs. Proc Natl Acad Sci. 1977;74:2456–2460. doi: 10.1073/pnas.74.6.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lee YH, Ota T, Vacquier VD. Positive selection is a general phenomenon in the evolution of abalone sperm lysin. Mol Biol Evol. 1995;12:231–238. doi: 10.1093/oxfordjournals.molbev.a040200. [DOI] [PubMed] [Google Scholar]
- 162.Hellberg ME, Dennis AB, Arbour-Reily P, Aagaard JE, Swanson WJ. The Tegula tango: a coevolutionary dance of interacting, positively selected sperm and egg proteins. Evolution. 2012;66:1681–1694. doi: 10.1111/j.1558-5646.2011.01530.x. [DOI] [PubMed] [Google Scholar]
- 163.Brandriff B, Moy GW, Vacquier VD. Isolation of sperm bindin from oyster (Crassostrea gigas) Gamete Res. 1978;1:89–99. [Google Scholar]
- 164.Hart MW. Structure and evolution of the sea star egg receptor for sperm bindin. Mol Ecol. 2013;22:2143–2156. doi: 10.1111/mec.12251. [DOI] [PubMed] [Google Scholar]
- 165.Zigler KS. The evolution of sea urchin sperm bindin. Int J Dev Biol. 2008;52:791–796. doi: 10.1387/ijdb.072521kz. [DOI] [PubMed] [Google Scholar]
- 166.Dan JC. Acrosome reaction and lysins. In: Metz CB, Monroy A, editors. Fertilization. Academic Press; New York: 1967. [Google Scholar]
- 167.Gao B, Klein LE, Britten RJ, Davidson EH. Sequence of mRNA coding for bindin, a species-specific sperm protein required for fertilization. Proc Natl Acad Sci. 1986;83:8634–8638. doi: 10.1073/pnas.83.22.8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zigler KS, Lessios HA. 250 million years of bindin evolution. Biol Bull. 2003;205:8–15. doi: 10.2307/1543440. [DOI] [PubMed] [Google Scholar]
- 169.Rocha S, Lucio M, Pereira MC, Reis S, Brezesinski G. The conformation of fusagenic B18 peptide in surfactant solutions. J Pept Sci. 2008;14:436–441. doi: 10.1002/psc.935. [DOI] [PubMed] [Google Scholar]
- 170.Lessios HA, Zigler KS. Rates of sea urchin bindin evolution. In: Singh RS, Xu J, Kulathinal RJ, editors. Rapidly Evolving Genes and Genetic Systems. Oxford University Press; London: 2012. [Google Scholar]
- 171.Vacquier VD, Swanson WJ. Selection in the rapid evolution of gamete recognition proteins in marine invertebrates. Cold Spring Harb Perspect Biol. 2011;3:a002931. doi: 10.1101/cshperspect.a002931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Levitan DR, Terhorst CP, Fogarty ND. The risk of polyspermy in three congeneric sea urchins and its implications for gametic incompatibility and reproductive isolation. Evolution. 2007;61:2007–2014. doi: 10.1111/j.1558-5646.2007.00150.x. [DOI] [PubMed] [Google Scholar]
- 173.Kamei N, Glabe CG. The species-specific egg receptor for sea urchin sperm adhesion is EBR1, a novel ADAMTS protein. Genes Dev. 2003;17:2502–2507. doi: 10.1101/gad.1133003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pujolar JM, Pogson GH. Positive Darwinian selection in gamete recognition proteins of Strongylocentrotus sea urchins. Mol Ecol. 2011;20:4968–4982. doi: 10.1111/j.1365-294X.2011.05336.x. [DOI] [PubMed] [Google Scholar]
- 175.Lewis CA, Talbot CF, Vacquier VD. A protein from abalone sperm dissolves the egg vitelline layer by a nonenzymatic mechanism. Dev Biol. 1982;92:227–239. doi: 10.1016/0012-1606(82)90167-1. [DOI] [PubMed] [Google Scholar]
- 176.Barton NH, Briggs DEG, Eisen JA, Goldstein DB, Patel NH. Evolution. Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]
- 177.Kresge N, Vacquier VD, Stout CD. The high resolution crystal structure of green abalone sperm lysin: implications for species-specific binding of the egg receptor. J Mol Biol. 2000;296:1225–1234. doi: 10.1006/jmbi.2000.3533. [DOI] [PubMed] [Google Scholar]
- 178.Shaw A, Fortes PA, Stout CD, Vacquier VD. Crystal structure and subunit dynamics of the abalone sperm lysin dimer: egg envelopes dissociate dimers, the monomer is the active species. J Cell Biol. 1995;130:1117–1125. doi: 10.1083/jcb.130.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lyon JD, Vacquier VD. Interspecies chimeric sperm lysins identify regions mediating species-specific recognition of the abalone egg vitelline envelope. Dev Biol. 1999;214:151–159. doi: 10.1006/dbio.1999.9411. [DOI] [PubMed] [Google Scholar]
- 180.Swanson WJ, Vacquier VD. Liposome fusion induced by a Mr 18 000 protein localized to the acrosomal region of acrosome-reacted abalone spermatozoa. Biochemistry. 1995;34:14202–14208. doi: 10.1021/bi00043a026. [DOI] [PubMed] [Google Scholar]
- 181.Vacquier V, Swanson W, Lee YH. Positive Darwinian selection on two homologous fertilization proteins: what is the selective pressure driving their divergence? J Mol Evol. 1997;44:S15–S22. doi: 10.1007/pl00000049. [DOI] [PubMed] [Google Scholar]
- 182.Kresge N, Vacquier VD, Stout CD. The crystal structure of a fusagenic sperm protein reveals extreme surface properties. Biochemistry. 2001;40:5407–5413. doi: 10.1021/bi002779v. [DOI] [PubMed] [Google Scholar]
- 183.Galindo BE, Moy GW, Swanson WJ, Vacquier VD. Full-length sequence of VERL, the egg vitelline envelope receptor for abalone sperm lysin. Gene. 2002;288:111–117. doi: 10.1016/s0378-1119(02)00459-6. [DOI] [PubMed] [Google Scholar]
- 184.Swanson WJ, Aagaard JE, Vacquier VD, Monné M, Sadat Al Hosseini H, Jovine L. The molecular basis of sex: linking yeast to human. Mol Biol Evol. 2011;28:1963–1966. doi: 10.1093/molbev/msr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Swanson WJ, Aquadro CF, Vacquier VD. Polymorphism in abalone fertilization proteins is consistent with the neutral evolution of the egg's receptor for lysin (VERL) and positive Darwinian selection of sperm lysin. Mol Biol Evol. 2001;18:376–383. doi: 10.1093/oxfordjournals.molbev.a003813. [DOI] [PubMed] [Google Scholar]
- 186.Gallindo BE, Vacquier VD, Swanson WJ. Positive selection in the egg receptor for abalone sperm lysin. Proc Natl Acad Sci U S A. 2003;100:4639–4643. doi: 10.1073/pnas.0830022100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Clark NL, Gasper J, Sekino M, Springer SA, Aquadro CF, Swanson WJ. Coevolution of interacting fertilization proteins. PLoS Genet. 2009;5:e1000570. doi: 10.1371/journal.pgen.1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Aagaard JE, Yi X, MacCoss MJ, Swanson WJ. Rapidly evolving zona pellucida domain proteins are a major component of the vitelline envelope of abalone eggs. Proc Natl Acad Sci. 2006;103:17302–17307. doi: 10.1073/pnas.0603125103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Aagaard JE, Vacquier VD, MacCoss MJ, Swanson WJ. ZP domain proteins in the abalone egg coat include a paralog of VERL under positive selection that binds lysin and 18-kDa sperm proteins. Mol Biol Evol. 2010;27:193–203. doi: 10.1093/molbev/msp221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Han L, Monné M, Okumura H, Schwend T, Cherry AL, Flot D, et al. Insights into egg coat assembly and egg–sperm interaction from the X-ray structure of full-length ZP3. Cell. 2010;143:404–415. doi: 10.1016/j.cell.2010.09.041. [DOI] [PubMed] [Google Scholar]
- 191.Jovine L, Janssen WG, Litscher ES, Wassarman PM. The PLACI-homology region of the ZP domain is sufficient for protein polymerisation. BMC Biochem. 2006;7:11. doi: 10.1186/1471-2091-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Monne M, Han L, Schwend T, Burendahl S, Jovine L. Crystal structure of the ZP-N domain of ZP3 reveals the core fold of animal egg coats. Nature. 2008;456:653–657. doi: 10.1038/nature07599. [DOI] [PubMed] [Google Scholar]
- 193.Claw KG, Swanson WJ. Evolution of the egg: new findings and challenges. Annu Rev Genomics Hum Genet. 2012;13:109–125. doi: 10.1146/annurev-genom-090711-163745. [DOI] [PubMed] [Google Scholar]
- 194.Aagaard JE, Springer SA, Soelberg SD, Swanson WJ. Duplicate abalone egg coat proteins bind sperm lysin similarly, but evolve oppositely, consistent with molecular mimicry at fertilization. PLoS Genet. 2013;9:e1003287. doi: 10.1371/journal.pgen.1003287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mantovani A, Locati M, Vecchi A, Sozzani S, Allavena P. Decoy receptors: a strategy to regulate inflammatory cytokines and chemokines. Trends Immunol. 2001;22:328–336. doi: 10.1016/s1471-4906(01)01941-x. [DOI] [PubMed] [Google Scholar]
- 196.Goudet G, Mugnier S, Callebaut I, Monget P. Phylogenetic analysis and identification of pseudogenes reveal a progressive loss of Zona pellucida genes during evolution of vertebrates. Biol Reprod. 2008;78:796–806. doi: 10.1095/biolreprod.107.064568. [DOI] [PubMed] [Google Scholar]
- 197.Bleil JD, Wassarman PM. Structure and function of the zona pellucida: identification and characterization of the proteins of the mouse oocyte's zona pellucida. Dev Biol. 1980;76:185–202. doi: 10.1016/0012-1606(80)90371-1. [DOI] [PubMed] [Google Scholar]
- 198.Turner LM, Hoekstra HE. Adaptive evolution of fertilization proteins within a genus: variation in ZP2 and ZP3 in deer mice (Peromyscus) Mol Biol Evol. 2006;23:1656–1669. doi: 10.1093/molbev/msl035. [DOI] [PubMed] [Google Scholar]
- 199.Calkins JD, El-Hinn D, Swanson WJ. Adaptive evolution in an avian reproductive protein: ZP3. J Mol Evol. 2007;65:555–563. doi: 10.1007/s00239-007-9034-8. [DOI] [PubMed] [Google Scholar]
- 200.Rankin T, Talbot P, Lee E, Dean J. Abnormal zonae pellucidae in mice lacking ZP1 result in early embryonic loss. Development. 1999;126:3847–3855. doi: 10.1242/dev.126.17.3847. [DOI] [PubMed] [Google Scholar]
- 201.Wassarman PM. Mammalian fertilization: molecular aspects of gamete adhesion, exocytosis, and fusion. Cell. 1999;96:175–183. doi: 10.1016/s0092-8674(00)80558-9. [DOI] [PubMed] [Google Scholar]
- 202.Gahlay G, Gauthier L, Baibakov B, Epifano O, Dean J. Gamete recognition in mice depends on the cleavage status of an egg's zona pellucida protein. Science. 2010;329:216–219. doi: 10.1126/science.1188178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Rankin TL, Coleman JS, Epifano O, Hoodbhoy T, Turner SG, Castle PE, et al. Fertility and taxon-specific sperm binding persist after replacement of mouse sperm receptors with human homologs. Dev Cell. 2003;5:33–43. doi: 10.1016/s1534-5807(03)00195-3. [DOI] [PubMed] [Google Scholar]
- 204.Claw KG, George RD, Swanson WJ. Detecting coevolution in mammalian sperm–egg fusion proteins. Mol Reprod Dev. 2014;81:531–538. doi: 10.1002/mrd.22321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kerr JS, Wright GJ. Avidity-based extracellular interaction screening (AVEXIS) for the scalable detection of low-affinity extracellular receptor–ligand interactions. J Vis Exp. 2012;3881 doi: 10.3791/3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Bianchi E, Doe B, Goulding D, Wright GJ. Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature. 2014;508:483–487. doi: 10.1038/nature13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Springer SA, Gagneux P. Glycan evolution in response to collaboration, conflict, and constraint. J Biol Chem. 2013;288:6904–6911. doi: 10.1074/jbc.R112.424523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ovchinnikov S, Kamisetty H, Baker D. Robust and accurate prediction of residue–residue interactions across protein interfaces using evolutionary information. eLife. 2014 doi: 10.7554/eLife.02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Fowler DM, Araya CL, Fleishman SJ, Kellogg EH, Stephany JJ, Baker D, et al. High-resolution mapping of protein sequence–function relationships. Nat Methods. 2010;7:741–746. doi: 10.1038/nmeth.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Mead LS, Arnold SJ. Quantitative genetic models of sexual selection. Trends Ecol Evol. 2004;19:264–271. doi: 10.1016/j.tree.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 211.Yang Z, Swanson WJ, Vacquier VD. Maximum-likelihood analysis of molecular adaptation in abalone sperm lysin reveals variable selective pressures among lineages and sites. Mol Biol Evol. 2000;17:1446–14. doi: 10.1093/oxfordjournals.molbev.a026245. [DOI] [PubMed] [Google Scholar]


