Abstract
Background and Objectives:
Staphylococcus aureus is one of the main causatives of bovine mastitis. Resistance of some strains to methicillin, can complicate the treatment of its infections. On the other hand, enterotoxin production is also important. Therefore, the aim of our study was to investigate the methicillin resistance and enterotoxin production in S. aureus isolates caused bovine mastitis.
Materials and Methods:
Four hundred and fifty milk samples were collected. After isolation of Staphylococcus aureus, MRSA strains were detected by cefoxitin disc diffusion and oxacillin agar screening methods. DNA was extracted by phenol – chloroform method and PCR was applied for mecA, sea and seb genes. SCCmec types of mecA gene were identified using multiplex-PCR.
Results:
Fifty-four (12%) S. aureus were isolated. Out of these, 10 and 9 MRSA strains identified by cefoxitin disc diffusion and oxacillin agar screening methods, respectively. All 10 MRSA isolates identified by cefoxitin disc diffusion, were positive for mecA gene and all of them belonged to SCCmec type IV. The sea genes were detected in 19 isolates and only two isolates were positive for seb genes. One isolate possessed both sea and seb genes.
Conclusion:
Findings of this study indicated that results of cefoxitin disc diffusion test is in concordance with the PCR for mecA gene and has a higher sensitivity compared to oxacillin agar screening method. Finally, Our findings suggest that enterotoxin A is the dominant type.
Keywords: Bovine mastitis, Staphylococcus aureus, MRSA, Staphylococcal enterotoxins
INTRODUCTION
Mastitis is defined as an inflammation of the mammary gland (1). One of the most common organism causing mastitis is Staphylococcus aureus (2). The majority of intramammary infections due to S. aureus are subclinical and hence, the response of this infection to treatment is comparatively poor. Progression of disease due to delayed diagnosis and treatment in such cases, often leads to elimination of cows from herds (3, 4).
On the other hand, resistance of some S. aureus strains to methicillin, can complicate treatment of its infections (5).
Methicillin resistance in S. aureus isolates, obtained from bovine mastitis cases, was first reported in Belgium durin 1972 to 1975 (6).
It is caused by the acquisition of an exogenous gene, mecA, that encodes an additional penicillin-binding protein (PBP), termed PBP 2a or PBP2′, which possesses reduced affinities for binding to beta-lactam antibiotics (6).
mecA gene is carried by a large mobile genetic element, staphylococcal cassette chromosome mec (SCCmec). Each SCCmec element has an intact copy of mecA, a copy of insertion sequence IS431, and, when present, complete or truncated mec regulatory genes mecI and mecR1.
Although MRSA strains isolated from humans have been well characterized and examined by the analyses of their SCCmec complex, there have been few studies about MRSA isolated from animals or livestock products and their SCCmec complex characteristics (7).
Infected cows can shed large numbers of S. aureus in their milk and are probably the main source of contamination of raw milk with S. aureus strains. Milk is a good substrate for S. aureus growth and enterotoxin production. Although pasteurization, kills S. aureus cells, thermostable SEs generally retain their biological activity. They are recognized as being agents of staphylococcal food poisoning that is of major concern in public health programs worldwide.
MATERIALS AND METHODS
Isolation of S. aurus from milk samples.
A total of 450 milk samples were collected from dairy cows with mastitis in industrial dairy herds of Isfahan. To detect S. aureus, 1ml of milk sample was inoculated on Baird – Parker agar. After 24–48 h of incubation at 37°C, suspected colonies that formed grey-black, shiny and convex colonies with clear zones around, were sub-cultured on blood agar plate and incubated for a further 24 h at 37°C. Staphylococcus aureus was identified based on colony morphology, beta hemolysis on blood agar, observation of Gram-positive cluster under microscope after Gram staining, positive catalase test, tube coagulase and DNase (11, 12).
Determination of methicillin resistance by cefoxitin disk diffusion method.
An inhibition zone diameter of ≤19 mm was reported as resistant and ≥20 mm was considered as sensitive. S. aureus ATCC 25923 and S. aureus ATCC 33591 were used as methicillin sensitive and resistant controls, respectively.
Determination of methicillin resistance by oxacillin screen agar method.
Mueller-Hinton agar (MHA) plates containing 4% NaCl and 6 μg/ml of oxacillin were prepared. To calculate the amount of oxacillin powder, following formula was used:
S. aureus ATCC 25923 and S. aureus ATCC 33591 were used as methicillin sensitive and resistant controls, respectively.
Determination of minimum inhibitory concentration for oxacillin.
Oxacillin MIC were determined by using Agar dilution method. The lowest concentration of oxacillin that completely inhibits growth, reported as oxacillin MIC. According to CLSI guidelines, concentrations of ≥ 4 and ≥ 256 were considered as the lowest and the highest oxacillin MIC for MRSA strains, respectively.
Determination the sensitivity and specificity of cefoxitin disc diffusion and oxacillin agar screening methods methods compared to PCR.
The following formulas was used to determine sensitivity and specificity of methods.
DNA extraction.
Phenol-chloroform method was used to extract DNA. The extracted DNA were dissolved in 50 μl sterile distilled water and stored in −20°C.
PCR for identification of mecA gene.
The PCR mastermix contained 2.5 μl 10x Buffer, 0.6 μl MgCl2, 0.4 μl dNTP, 0.3 μl TaqDNA polymerase, 0.5 μl of each primer (10 pmol/ μl) and 5 μl template DNA. The final volume was adjusted to 25 μl by adding 15.2 μl sterile ultrapure water and was placed on thermal cycler. Amplification was carried out using following conditions:
An initial denaturation at 94°C for 5 min, 35 cycles of denaturation at 94°C for 15 s, annealing at 55°C for 15s and extension at 72°C for 20 s, followed by a final extension at 72°C for 5 min (13, 14).
SCCmec typing by multiplex-PCR.
Mastermix contained 2.5 μl 10x Buffer, 0.75μl MgCl2, 0.5μl dNTP, 0.25μl TaqDNA polymerase, 0.5 μl of α3R1, βF1, ccrCF and ccrCR primers (10 pmol/ μl), 0.3 μl of 1272F1, 1272R1, 5RmecA, 5R431 primers (10 pmol/ μl) and 3μl template DNA. The final volume was adjusted to 25 μl by adding 14.8μl sterile ultrapure water and was placed on thermal cycler. Amplification was carried out through following conditions:
An initial denaturation at 94°C for 5 min, 35 cycles of denaturation at 94°C for 15 s, annealing at 55°C for 15 s and extension at 72°C for 20 s, followed by a final extension at 72°C for 5 min (15).
Multiplex-PCR for identification of sea and seb genes.
The PCR Mastermix contained 2.5 μl 10x Buffer, 0.6 μl MgCl2, 0.5μl dNTP, 0.25μl Taq DNA polymerase, 0.75μl of each primer (10 pmol/ μl) and 3μl template DNA. The final volume was adjusted to 25 μl by adding 15μl sterile ultrapure water and was placed on thermal cycler. Amplification was carried out using following conditions:
An initial denaturation at 94°C for 5 min, 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min and extension at 72°C for 1 min, followed by a final extension at 72°C for 7 min.
All primers are listed in Table 1.
Table 1.
Primers used in this study.
| Target | Primer | Sequence | Product size (bp) |
|---|---|---|---|
| mecA | F mecA | 5′TGGCTATCGTGTCACAATCG 3′ | 310 |
| R mecA | 5′CTGGAACTTGTTGAGCAGAG 3′ | ||
| SCCmec | F β | 5′- ATTGCCTTGATAATAGCCYTCT- 3′ | 937 |
| R α3 | 5′- TAAAGGCATCAATGCACAAACACT-3′ | ||
| F ccrC | 5′-CGTCTATTACAAGATGTTAAGGATAAT-3′ | 518 | |
| R ccrC | 5′- CCTTTATAGACTGGATTATTCAAAATAT-3′ | ||
| F 1272 | 5′- GCCACTCATAACATATGGAA-3′ | 415 | |
| R1272 | 5′- CATCCGAGTGAAACCCAAA-3′ | ||
| F5RmecA | 5′-TATACCAAACCCGACAACTAC -3′ | 359 | |
| R 5R431 | 5′-CGGCTACAGTGATAACATCC-3′ | ||
| sea | F sea | 5′-CCTTTGGAAACGGTTAAAACG-3′ | 127 |
| R sea | 5′-TCTGAACCTTCCCATCAAAAAC-3′ | ||
| seb | F seb | 5′-TCGCATCAAACTGACAAACG-3′ | 477 |
| R seb | 5′-GCAGGTACTCTATAAGTGCCTGC-3′ | ||
RESULTS
Out of 450 milk samples, 54 were positive for S. aureus. Among 54 S. aureus isolates, 10 (18.51%) MRSA strains were identified. Growth of MRSA was observed from the 11.11% (1/9) and 20% (9/45) of clinical and subclinical mastitis samples. Of 9 (16.67%) MRSA strains that were identified in this method, 11.11% (1/9) and 17.78% (8/45) were isolated from clinical and subclinical mastitis samples.
Among 9 MRSA isolated by Oxacillin screen agar method, Oxacillin MIC for 7 MRSA strains was 128 μg/ml and for 2 other strains was 256 μg/ml.
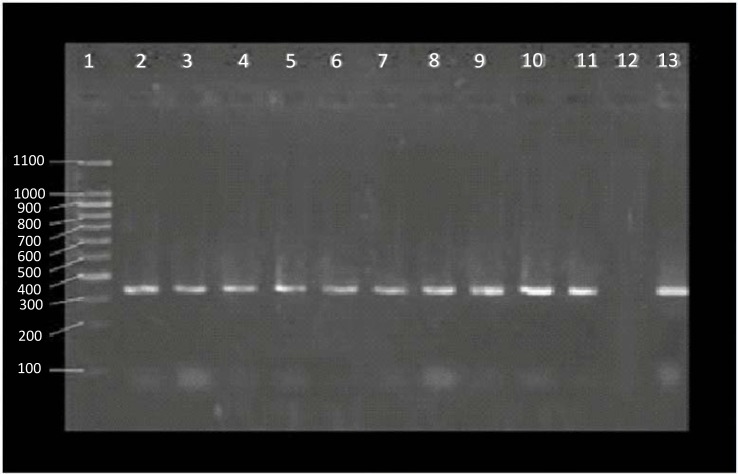
PCR was performed for all 54 S. aureus isolates. Out of this number, 10 isolates were positive regarding mecA gene (Fig. 1).
Fig. 1.
Results of mecA gene PCR. Lane 1: 100 bp marker, Lane 2 – 11: mecA positive strains isolated form milk samples, Lane 12: negative control, Lane 13: positive control
The sensitivity and specificity of the two phenotypic tests as compared with PCR are demonstrated in Table 2.
Table 2.
Determination the sensitivity and specificity of cefoxitin disc diffusion and oxacillin agar screening methods methods compared to PCR.
| Method | Detected as MRSA | Detected as MSSA | sensitivity (%) | Specificity (%) |
|---|---|---|---|---|
| Cefoxitin disc diffusion | 10 | 44 | 100 | 100 |
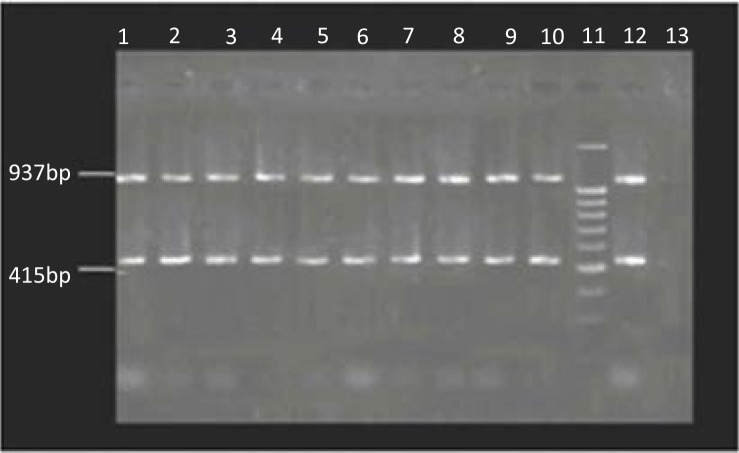
Electrophopresis of PCR products indicated that all 10 MRSA strains carried SCCmec type IV and yielded two 415 bp and 937 bp bands in the multiplex PCR (Fig. 2).
Fig. 2.
Results of SCCmec typing by Multiplex PCR. Lane 1–10: MRSA strains with SCCmec type IV, Lane 11: 100 bp marker, Lane 12: positive control, Lane 13: negative control
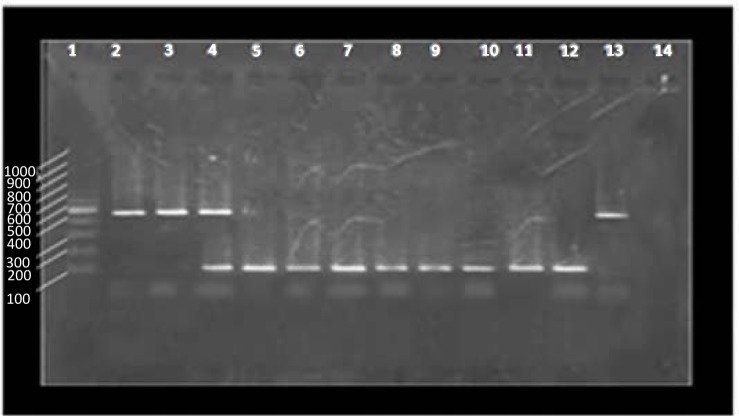
Twenty two (74/40%) enterotoxigenic strains were isolated. sea genes were detected in 19 (86.36%) isolates and only two isolates (9.1%) were positive for seb genes. One isolate (4.54%) possessed both sea + seb genes (Fig. 3).
Fig. 3.
Results of sea and seb genes amplification. Lane 1: 100 bp marker, Lane 2 and 3: seb positive strains (477 bp), Lane 4: sea + seb positive strains, Lane 5 to 11: sea positive strains (127 bp), Lane 12: positive control for sea gene, Lane 13: positive control for seb gene, Lane 14: negative control.
DISCUSSION
Our findings suggests a low prevalence of bovine mastitis caused by S. aureus in the study area. This result confirms the findings of Ebrahimi et al in, Shahrekord, Iran (12). So that, in his study, only 8 S. aureus (16.8%) were isolated from 98 milk samples collected from cows with mastitis (12). In another study conducted by Khan-nazar et al in 2002 in Shiraz, Iran, out of 1352 milk samples collected from bovine mastitis cases, 118 (8.73%) S. aureus were isolated (17). Also, according to findings of Atyabi et al from 2002 to 2006, the prevalence of S. aureus in bovine mastitis cases in Iran was very low, so that only 84 S. aureus (2.89%) were isolated from 2904 milk samples, in her study (3).
Like many other studies, the largest number of S. aureus isolates in this study were related to subclinical mastitis and 45 S. aureus (15.52%) were isolated out of the 290 milk samples which were collected from subclinical mastitis. While only 9 isolates (5.62%) of the 160 milk samples from clinical mastitis were isolated. It is because of the fact that the majority of intramammary infections due to S. aureus are subclinical (4). Findings of Abera et al in Ethiopia in 2010 also confirm these results and showed that out of 110 subclinical and 30 clinical mastitis milk samples, 49 (44.5%) and 10 (33.3%) S. aureus were isolated, respectively (9).
Another important issue in cases of bovine mastitis caused by S. aureus, is the resistance of some strains to methicillin which can complicate the treatment of its infections and also increases the risk of transmission of resistant strains to human population (2, 5).
The first report of mecA in MRSA from bovine milk appeared as recently as 2003: In a Korean study describing 265 S. aureus isolates from 894 milk samples, 12 (1.3%) were mecA positive (18).
In this study, three methods were applied to detect MRSA strains including: cefoxitin disc diffusion, oxacillin agar screening methods and detection of mecA gene using PCR.
Ten MRSA strains (18.52%) were identified with cefoxitin disc diffusion test and in these 10 isolates mecA gene was detected by PCR while oxacillin agar screening method detected 9 MRSA strains (16.66%). This finding is in accordance with data reported by Turkyilmaz et al. from 2002 to 2006 in Turkey. Among 93 S. aureus isolated in Turkyilmaz study, 16 (17.2%) were MRSA (19).
Like other studies, results of cefoxitin disc diffusion test is completely in agreement with the PCR for mecA gene. Since detection of mecA gene is considered the gold standard for identification of MRSA strains. We can say that, sensitivity and specificity of cefoxitin disc diffusion method in our study were 100%. The sensitivity and specificity of Oxacillin agar screening in our study were 90% and 100% respectively. Despite the low sensitivity of oxacillin agar screening, this test may be advocated in diagnostic laboratories due to its reproducibility and cost-effectiveness (20, 21). Recent studies indicate that cefoxitin disc diffusion testing is far superior to most of the currently recommended phenotypic methods like oxacillin screen agar testing and is now an accepted method for detection of MRSA by many reference groups including CLSI (22).
Since it is expected that, the prevalence of S. aureus, especially MRSA in herds, considering the importance of health and pollution control, be too low; even the very low incidence reported in this study can make many problems and increases the risk of transmission to other cows and human population.
SCCmec typing results of mecA genes in our study provide an evidence that one of the most frequent SCCmec types in bovine mastitis cases is type IV and all 10 mecA-positive strains harbored type IV of SCCmec. This finding is in accordance with many other studies in this field. For example, Nam et al, in 2005 during an investigation on the 9055 milk samples collected from cows infected with mastitis in Korea, showed that all 14 MRSA isolates, belong to SCCmec type IV(7). Findings of Witte et al. and Van Den Ede et al, also confirm this (23, 24).
Moreover, our results suggest enterotoxin A is the dominant type and were detected in 19 isolates. Two isolates were positive for seb genes and only 1 possessed both sea + seb genes.
According to Ghasemian Safaie and Rahimi study in 2009, out of 390 milk samples collected from subclinical bovine mastitis cases in Isfahan, Iran, 91(23.4%) S. aureus were isolated. Among these, 29 enterotoxigenic strains were identified which is similar to our results (25). SEA was the most frequent enterotoxin in their study. In another study that was conducted by Boynukara and colleagues in 2008, among 27 Enterotoxigenic strains that isolated from 480 bovine mastitis milk samples in Turkey, 25 and 2 isolates were positive for sea and seb genes, respectively (26). In all listed studies, the most frequent enterotoxin is SEA. Staphylococcal enterotoxin A is actually the main cause of food poisoning in the world, and most studies have been conducted about it.
CONCLUSION
Cefoxitin disc diffusion test can be an alternative to PCR for detection of MRSA, when use of PCR is not possible. PCR is the best method for identification of MRSA and enterotoxigenic strains because of its ability to detect silent genes that can not be identified by phenotypic methods. On the other hand, it should be noted that all MRSA strains isolated in this study, in terms of SCCmec, possess similar characteristics with community acquired methicillin resistant S. aureus (CA-MRSA), Since all of them belong to SCCmec type IV.
Finally, our results suggest that enterotoxin production in S. aureus strains, isolated from bovine mastitis, in Iran, is very high and enterotoxin A is the dominant type.
Acknowledgments
We are thankful to all Members of Microbiology Department of Medicine School in Isfahan University of Medical Sciences.
REFERENCES
- 1. Nelson Philpot W, Nickerson SC. (2000). Winning the fight against mastitis. 1nd ed Westfalia Surge. [Google Scholar]
- 2. Holmes MA, Zadoks RN. Methicillin resistant S. aureus in human and bovine mastitis. J Mammary Gland Biol Neoplasia 2011; 16: 373– 382. [DOI] [PubMed] [Google Scholar]
- 3. Atyabi N, Vodjgani M, Gharagozloo F, Bahonar A. Prevalence of bacterial mastitis in cattle from the farms around Tehran, Iran. J Vet Res 2006; 7: 76– 79. [Google Scholar]
- 4. Ghorbanpoor M, Seyfiabadshapouri M, Moatamedi H, Jamshidian M, Gooraninejad S. Comparison of PCR and bacterial culture methods for diagnosis of dairy cattle's subclinical mastitis caused by Staphylococcus aureus. J Vet Res 2007; 62: 87– 91. [Google Scholar]
- 5. Vanderhaeghen W, Cerpentier T, Adriaensen C, Vicca J, Hermans K, Butaye P. Methicillin-resistant Staphylococcus aureus (MRSA) ST398 associated with clinical and subclinical mastitis in Belgian cows. Vet Microbiol 2010; 144: 166– 171. [DOI] [PubMed] [Google Scholar]
- 6. Devriese LA, Van Damme LR, Fameree L. Methicillin (cloxacillin)-resitant Staphylococcus aureus strains isolated from bovine mastitis cases. ZentralblVeterinarmed B 1972; 19: 598– 605. [DOI] [PubMed] [Google Scholar]
- 7. Nam HK, Kun TP, Jin SM, Woo KJ, So HK, Jun MK, et al. Staphylococcal cassette chromosome mec (SCCmec) characterization and molecular analysis for methicillin-resistant Staphylococcus aureus and novel SCCmec subtype IVg isolated from bovine milk in Korea. J Antimicrob Chemother 2005; 56: 624– 632. [DOI] [PubMed] [Google Scholar]
- 8. Hashemi M, Kafi M, Safdarian M. The prevalence of clinical and subclinical mastitis in dairy cows in the central region of Fars province, south of Iran. Iran J Vet Res 2011; 12: 236– 241. [Google Scholar]
- 9. Abera M, Demie B, Aragaw K, Regassa F, Regassa A. Isolation and identification of Staphylococcus aureus from bovine mastitic milk and their drug resistance patterns in Adama town, Ethiopia. J Vet MedAnimHealth 2010; 2: 29– 34. [Google Scholar]
- 10. Ghorbanpoor M, Seyfiabadshapouri M, Moatamedi H, Jamshidian M, Gooraninejad S. Comparison of PCR and bacterial culture methods for diagnosis of dairy cattle's subclinical mastitis caused by Staphylococcus aureus. J Vet Res 2007; 62: 87– 91. [Google Scholar]
- 11. Vojgani M, Gharagozloo F, Bahonar A, Darabi M, Jafari H. Evaluation of therapeutic effects of topical application of eucalyptus essential oil (essence) on experimental mastitis caused by Streptococcus agalactiae. Iran Vet J 2006; 12: 5– 14. [Google Scholar]
- 12. Ebrahimi A, AkhavanTaheri M. Characteristics of staphylococci isolated from clinical and subclinical mastitis cows in Shahrekord. Iran J Vet Res 2009; 10: 273– 277. [Google Scholar]
- 13. Klevens R, Morisson M, Nadle J, Petit S. Invasive Methicillin-Resistant Staphylococcus aureus Infections in the United States. J Amer Med Assoc 2007; 298: 15– 20. [DOI] [PubMed] [Google Scholar]
- 14. Kim JS, Song W, Kim HS, Cho HC, Lee KM, Choi MS, et al. Association between the methicillin resistance of clinical isolates of Staphylococcus aureus, their staphylococcal cassette chromosome mec (SCCmec) subtype classification, and their toxin gene profiles. Diag Microbiol Infect Dis 2006; 56: 289– 95. [DOI] [PubMed] [Google Scholar]
- 15. Boy K, Bartels MD. A new multiplex PCR for easy screening of methicillin resistant Staphylococcus aureus SCCmec type 1–5. Clin Microbiol Infect 2007; 13: 725– 757. [DOI] [PubMed] [Google Scholar]
- 16. MohammadSadegh M, AskariBadouei M, Gorjidooz M, Daneshvar M, Koochakzadeh AA. study on the clinical Coliform mastitis of Holstein cows on Garmsar suburban dairy farms. Vet Microbiol 2012; 8: 137– 149. [Google Scholar]
- 17. KhanNazer AH, Sarmadi R. Prevalence of clinical and subclinical mastitis, antibioticresistance and determination of minimum inhibitory concentration (MIC) in Staphylococcus aureus and Escherichia coli isolated from cases of bovineMastitis. J Vet Res 2005; 60: 247– 252. [Google Scholar]
- 18. Lee JH. Methicillin (oxacillin) –resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl Environ Microbiol 2003; 69: 6489– 6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Turkyilmaz S, Tekbiyik S, Oryasin E, Bozdogan B. Molecular epidemiology and antimicrobial resistance mechanisms of methicillin-resistant Staphylococcus aureus isolated from bovine milk. Zoonoses Public Health 2008; 57: 197– 203. [DOI] [PubMed] [Google Scholar]
- 20. Shariati L, Validi M, Tabatabaiefar MA, Karimi A, Nafisi M. Comparison of Real-Time PCR with Disk Diffusion, Agar Screen and E-test Methods for Detection of Methicillin-Resistant Staphylococcus aureus. Curr Microbiol 2010; 61: 520– 524. [DOI] [PubMed] [Google Scholar]
- 21. Skov R, Smyth R, Larsen AR, Bolmstrôm A, Karlsson A, Mills K, et al. Phenotypic detection of methicillin resistance in Staphylococcus aureus by disk diffusion testing and etest on Mueller-Hinton agar. J Clin Microbiol 2006; 44: 4395– 4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Skov R, Smyth R, Clausen M, Larsen AR, Frimodt Møller N, Olsson Liljequist B, et al. Evaluation of a cefoxitin 30μg disc on Iso-Sensitest agar for detection of methicillin resistant Staphylococcus aureus. J Antimicrob Chemother 2003; 52: 204– 207. [DOI] [PubMed] [Google Scholar]
- 23. Witte W, Strommenger B, Stanek C, Cuny C. Methicillin-resistant Staphylococcus aureus ST398 in humans and animals, Central Europe. Emerg Infect Dis 2007; 13: 255– 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Van Den Eede A, Martens A, Lipinska U, Struelens M, Deplano A, Denis O, et al. High occurrence of methicillin-resistant Staphylococcus aureus ST398 in equine nasal samples. Vet Microbiol 2009; 133: 138– 144. [DOI] [PubMed] [Google Scholar]
- 25. GhasemianSafai H, Rahimi E. Detection of classical enterotoxins of Staphylococcus aureus strains isolated from bovine subclinical mastitis in Isfahan, Iran. Vet Microbiol 2010; 141: 393– 394. [DOI] [PubMed] [Google Scholar]
- 26. Boynukara B, Gulhan T, Alisarli M, Gurturk K, Solmaz H. Classical enterotoxigenic characteristics of Staphylococcus aureus strains isolated from bovine subclinical mastitis in Van, Turkey. Int J Food Microbiol 2008; 125: 209– 211. [DOI] [PubMed] [Google Scholar]