Abstract
We present an updated and integrated version of our widely used protein-protein docking and binding affinity benchmarks. The benchmarks consist of non-redundant, high quality structures of protein-protein complexes along with the unbound structures of their components. Fifty-five new complexes were added to the docking benchmark, 35 of which have experimentally-measured binding affinities. These updated docking and affinity benchmarks now contain 230 and 179 entries, respectively. In particular, the number of antibody-antigen complexes has increased significantly, by 67% and 74% in the docking and affinity benchmarks, respectively.
We tested previously developed docking and affinity prediction algorithms on the new cases. Considering only the top ten docking predictions per benchmark case, a prediction accuracy of 38% is achieved on all 55 cases, and up to 50% for the 32 rigid-body cases only. Predicted affinity scores are found to correlate with experimental binding energies up to r=0.52 overall, and r=0.72 for the rigid complexes.
Keywords: Protein-protein complex structure, Antibody-antigen, Conformational change, Protein-protein interface, Binding free energy
Introduction
Protein-protein interactions are among the most important processes in biology, playing fundamental roles in the immune system, signaling pathways, and enzyme inhibition. Proteome-wide studies have revealed that most proteins interact with other proteins [1]. The experimental characterization of the structure of a protein-protein complex is, however, difficult and not always successful. To complement experimental approaches, computational techniques for the prediction of protein complexes have been developed over the years, stimulated by the CAPRI experiment (Critical Assessment of PRedicted Interactions) [2]. Computational approaches for modeling protein-protein complex structures include ab-initio docking methods [3,4], homology-based methods based on the experimental structures of similar complexes [5–11], and integrative, information-driven methods [12], These approaches typically attempt to predict the most likely structure of a complex, but are not designed to predict how strongly the proteins bind or whether they bind at all. Thus a more complete computational description of protein-protein interaction also requires algorithms that can predict binding affinities. Although energy functions for affinity prediction and the ranking of docking poses are related, they are often developed specifically for their respective purposes and so far have shown varying and rather limited performance [13]. Example areas where scoring functions can be improved are entropic contributions [14], solvent effects [15], and the optimal combination of terms [16].
Essential for the development of computational algorithms are training and test sets that are reliable and sufficiently large. It is computationally daunting to sift the Protein Data Bank for structures of protein-protein complexes; the experimental conditions and accuracies of these structures vary widely and are not always straightforward to assess, and neither is the definition of the biological unit. Recognizing this, various benchmarks were developed that attempt to collect a reliable and well-understood set of data. Our docking benchmark, which after its initial development [17] has seen three updates [18–20], is widely used for developing and assessing docking methods. Key features are the availability of both the complex structure and the unbound structures of the component proteins, non-redundancy, and reliability of the data. Other benchmarks include DOCKGROUND [21], which also focuses on protein-protein interactions, and benchmarks that contain complexes of proteins with nucleic acids [22,23].
More recently we used our protein-protein docking benchmark as a starting point for developing a structure-based affinity benchmark [24,25], which includes the entries from our docking benchmark for which experimental binding affinities were available. The affinity benchmark has been used for the development of algorithms for predicting protein-protein binding free energies, with a typical correlation coefficient of r=0.6 with experimentally measured binding free energies [26–28].
In this paper we present updates to our docking and affinity benchmarks, of which the development is tightly integrated. We added 55 new protein-protein complexes to the docking benchmark, for 35 of which experimental affinities could be found that were added to the affinity benchmark. These new additions to both benchmarks were then used, as an independent test set, to assess the performance of four docking algorithms and a large panel of affinity prediction algorithms that had been previously developed without seeing any of the new cases. This allowed us to assess the performance of docking and affinity predictions, both of which remained limited due to conformational changes, with an indication that low affinity complexes were also more challenging to dock.
Results and Discussion
Composition
We added 55 cases to the docking benchmark (Table 1). PDB entries 3AAD and 3P57 show two and three distinct binding modes, respectively. As in the previous versions of the benchmark, the complexes that display multiple binding modes were split into different cases. This represents an increase of 31% over the previous 175 cases. We could find binding affinity data for 35 of the cases, which brought the total number of cases in the affinity benchmark to 179, a 24% increase. In Table 2 we show the composition of the updated benchmarks compared with the previous versions. The most noticeable increase is for antibody-antigen complexes: from 24 cases to 40 cases in the docking benchmark and from 19 cases to 33 cases in the affinity benchmark, which reflects a surging interest in antibody-based therapeutics.
Table 1.
New cases in the docking benchmark 5 and affinity benchmark 2.
| Cat.a | PDB ID 1b | Protein 1 | PDB ID 2b | Protein 2 | I-RMSD (Å) |
ΔASAc (Å2) |
Kd (M) |
ΔGd (kcal/mol) |
T (°C) |
|
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| Rigid body | ||||||||||
| 2VXT_HL:I | A | 2VXU_HL | Murine reference antibody 125-2H Fab | 1J0S_A(6) | Interleukin-18 | 1.33 | 2163 | 5.33e-10 | −12.65 | |
| 2W9E_HL:A | A | 2W9D_HL | ICSM 18 Fab fragment | 1QM1_A | Prion protein fragment | 1.13 | 1677 | 1.3e-10 | −13.49 | |
| 3EOA_LH:I | A | 3EO9_LH | Efalizumab Fab fragment | 3F74_A | Integrin alpha-L I domain | 0.39 | 1272 | 2.2e-9 | −11.81 | 25 |
| 3HMX_LH:AB | A | 3HMW_LH | Ustekinumab Fab | 1F45_AB | Interleukin-12 | 0.73 | 1841 | |||
| 3MXW_LH:A | A | 3MXV_LH | Anti-Shh 5E1 chimera Fab fragment | 3M1N_A | Sonic Hedgehog N-terminal domain | 0.48 | 1696 | 7e-9 | −11.31 | 30 |
| 3RVW_CD:A | A | 3RVT_CD | 4C1Fab | 3F5VA | DER P 1 allergen | 0.50 | 1383 | 1.9e-8 | −10.53 | 25 |
| 4DN4_LH:M | A | 4DN3_LH | CNTO888 Fab | 1DOL_A | MCP-1 | 0.81 | 1317 | 3.8e-ll | −14.22 | 25 |
| 4FQI_HL:ABEFCD | A | 4FQH_HL | CR9114 Fab | 2FK0_ABCDEF | H5N1 influenza virus hemagglutinin | 1.08 | 1459 | 9e-10 | −12.55 | 30 |
| 4G6J_HL:A | A | 4G5Z_HL | Canakinumab antibody fragment | 4I1B_A | Interleukin-1 beta | 0.61 | 1893 | 4.1e-9 | −11.44 | 25 |
| 4G6M_HL:A | A | 4G6K_HL | Gevokizumab antibody fragment | 4I1B_A | Interleukin-1 beta | 0.49 | 1673 | 2.9e-10 | −13.01 | 25 |
| 4GXU_MN:ABEFCD | A | 4GXV_HL | 1F1 antibody | 1RUZ_HIJKLM | 1918 HI Hemagglutinin | 0.78 | 1830 | 6.2e-9 | −11.2 | |
| 1JTD_B:A | EI | 3QI0_A | BLIP-II | 1BTL_A | TEM-1 beta-lactamase | 0.44 | 2180 | 2.72e-ll | −14.41 | 25 |
| 2A1A_B:A | ES | 3UIU_A | Eukayotic translation initiation factor 2-alpha kinase 2 | 1Q46_A | eIF2 alpha subunit | 1.35 | 1186 | |||
| 2GAF_D:A | ER | 3OWG_A | Poly(A) polymerase VP55 | 1VPT_A | Vaccinia protein VP39 | 0.69 | 3368 | 1.2e-9 | −12.17 | |
| 2YVJ_A:B | ER | 2YVF_A | Ferredoxin reductase BPHA4 | 2E4P_A | Biphenyl dioxygenase ferredoxin subunit | 0.60 | 1377 | |||
| 3A4S_A:D | EI | 1A3S_A | SUMO-conjugating enzyme UBC9 | 3A4R_A | NFATC2-interacting protein SLD2 ubiquitin-like domain | 0.72 | 1116 | 2.81e-6 | −7.57 | 25 |
| 3K75_D:B | ER | 1BPB_A | DNA polymerase beta | 3K77_A | Reduced XRCC1, N-terminal domain | 0.64 | 1195 | 1.1e-7 | −9.49 | |
| 3LVK_AC:B | ER | 3LVM_AB | Cysteine desulfurase IscS | 1DCJ_A(12) | Sulfurtransferase tusA | 0.81 | 1609 | 3.04e-7 | −8.89 | 25 |
| 3PC8_A:C | ER | 3PC6_A | DNA repair protein XRCC1 | 3PC7_A | DNA ligase III-alpha BRCT domain | 0.50 | 1240 | 1.02e-7 | −9.54 | |
| 3VLB_A:B | EI | 3VLA_A | EDGP | 3VL8_A | Xyloglucan-specific endo-beta-1,4-glucanase A | 0.51 | 2020 | |||
| 4HX3_BD:A | EI | 4HWX_AB | Neutral proteinase inhibitor ScNPI | 1C7K_A | Zinc endoprotease | 0.90 | 2086 | 6e-6 | −7.41 | 37 |
| 4H03_A:B | ES | 1GIQ_A | Iota toxin component IA | 1IJJ_A | Alpha actin | 0.68 | 1474 | |||
| 1EXB_ABDC:EGFH | OX | 1QRQ_ABCD | KV beta2 protein beta subunit | 1QDV_ABCD | KV1.2 potassium channel N-terminal domain | 0.62 | 3558 | |||
| 1M27_AB:C | OX | 1D4T_AB | SAP-SLAM Complex | 3UA6_A | Fyn kinase SH3 domain | 1.22 | 799 | 3.45e-6 | −7.45 | 25 |
| 2GTP_A:D | OG | 1GFI_A | Alpha-1 subunit Guanine nucleotide-binding protein G(I) | 2BV1_A | RGS1 | 0.54 | 1442 | |||
| 2X9A_D:C | OR | 1S62_A(8) | TolA C-terminal domain | 2X9B_A | G3P TolA binding domain | 1.33 | 1571 | 4.4e-6 | −7.31 | 25 |
| 3BIW_A:E | OX | 3BIX_A | Neuroligin-1 | 2R1D_A | Neuroligin-1-beta | 0.39 | 1191 | 9.7e-8 | −9.41 | 20 |
| 3H2V_A:E | OX | 3MYI_A | Vinculin tail domain | 1WI6_A(8) | Raverl RRM1 domain | 0.80 | 1263 | 2.21e-5 | −6.31 | 23 |
| 3P57_AB:P | OX | 3KOV_AB | MEF2A | 3IO2_A | p300 TAZ2 domain | 0.53 | 1291 | |||
| 3P57_CD:P | OX | 3KOV_AB | MEF2A | 3IO2_A | p300 TAZ2 domain | 0.74 | 1177 | |||
| 3P57_IJ:P | OX | 3KOV_AB | MEF2A | 3IO2_A | p300 TAZ2 domain | 0.91 | 1126 | |||
| 4M76_A:B | OR | 1C3D_A | C3D | 1M1U_A | Integrin alpha-M CD11B A-domain | 0.43 | 1046 | 4.5e-7 | −8.66 | 25 |
| Medium | ||||||||||
| 3EO1_AB:CF | A | 3EO0_AB | GC-1008 Fab fragment | 1TGJ_AB | Transforming Growth Factor-Beta 3 | 1.37 | 1630 | |||
| 3G6D_LH:A | A | 3G6A_LH | CNTO607 Fab | 1IK0_A(10) | Interleukin-13 | 1.86 | 1793 | 1.84e-11 | −14.65 | 25 |
| 3HI6_XY:B | A | 3HI5_HL | AL-57 Fab fragment | 1MJN_A | Integrin alpha-L I domain | 1.65 | 1871 | 4.7e-6 | −7.27 | |
| 3L5W_LH:I | A | 3L7E_LH | C836 Fab | 1IK0_A(11) | Interleukin-13 | 0.48 | 1138 | 5.4e-11 | −14.01 | 25 |
| 3V6Z_AB:F | A | 3V6F_AB | FabE6 | 3KXS_F | Capsid protein assembly domain | 1.83 | 1922 | 3.3e-9 | −11.57 | |
| 4FZA_A:B | ER | 1UPL_A | MO25 alpha | 3GGF_A | Serine/threonine-protein kinase MST4 | 2.04 | 1695 | |||
| 4IZ7_A:B | EI | 1ERK_A | Non-phosphorylated ERK | 2LS7_A(1) | PEA-15 Death Effector Domain | 1.56 | 1202 | 1.33e-7 | −9.44 | 27 |
| 4LW4_AB:C | ES | 4LW2_AB | Cysteine desulfurase CsdA | 1NI7_A(8) | Cysteine desulfuration protein CsdE | 1.60 | 1610 | |||
| 3AAA_AB:C | OX | 3AA7_AB | Actin capping protein | 1MYO_A(30) | Myotrophin | 1.78 | 1686 | 2.1e-8 | −10.3 | 20 |
| 3AAD_A:D | OX | 1EQF_A | Double bromodomain | 1TEY_A(13) | Histone chaperone ASF1 | 2.00 | 1461 | |||
| 3BX7_A:C | OX | 3BX8_A | Lipocalin 2 | 3OSK_A | CTLA-4 extracellular domain | 1.63 | 2349 | 9e-9 | −10.98 | 25 |
| 3DAW_A:B | OX | 1IJJ-A | Alpha actin | 2HD7_A(5) | Twinfilin-1 C-terminal domain | 1.49 | 2323 | 2e-5 | −6.41 | |
| 3R9A_AC:B | OR | 1H0C_AB | Alanine-glyoxylate aminotransferase | 2C0M_A | PEX5P TPR repeat domain | 1.91 | 1926 | 3.5e-6 | −7.44 | 25 |
| 3SZK_DE:F | OX | 3ODQ_AB | MetHaemoglobin | 2H3K_A | ISDH-N1 | 2.10 | 1263 | 9.01e-8 | −9.45 | 20 |
| 3S9D_B:A | OR | 1N6U_A(15) | IFNAR2 | 1ITF_A(9) | IFNa2 | 1.69 | 1841 | 3e-9 | −11.63 | |
| 4JCV_ADBC:E | OX | 1VDD_ABCD | Recombinational repair protein RecR | 1W3S_A | DNA repair protein RecO | 1.62 | 1949 | |||
| Difficult | ||||||||||
| 3FN1_B:A | ER | 2EDI_A(5) | UQ_con domain from NEDD8-conjugating enzyme UBE2F | 2LQ7_A | NEDD8-activaating enzyme E1 catalytic subunit | 3.65 | 1897 | |||
| 3H11_BC:A | ER | 4JJ7_AB | Caspase-8 | 3H13_A | c-FLIPL protease-like domain | 3.79 | 3169 | |||
| 4GAM_AFBGCH:D | ER | 1XVB_ABCDEF | Methane monooxygenase hydroxylase | 1CKV_A(9) | Methane monooxygenase regulatory protein B | 5.79 | 6671 | |||
| 1RKE_A:B | OX | 1SYQ_A | Vinculin head | 3MYI_A | Vinculin tail | 4.25 | 2614 | |||
| 3AAD_A:B | OX | 1EQF_A | Double bromodomain | 1TEY_A(4) | Histone chaperone ASF1 | 4.37 | 1654 | |||
| 3F1P_A:B | OX | 1P97_A(9) | HIF2 alpha C-terminal PAS domain | 1X0O_A(5) | ARNT C-terminal PAS domain | 2.52 | 1919 | 1.4e-6 | −7.85 | 20 |
| 3L89_ABC:M | OR | 3L88_ABC | Ad21 fiber knob | 1CKL_A | CD46 SCR1 and SCR2 domains | 2.51 | 2167 | 2.84e-7 | −8.93 | 25 |
Categories: antibody-antigen (A); enzyme-inhibitor (EI); enzyme-substrate (ES); enzyme complex with a regulatory or accessory chain (ER); others, G-protein containing (OG); others, receptor containing (OR); others, miscellaneous (OX).
Numbers in parentheses denote the NMR model that was chosen as the unbound structure.
Change in solvent accessible surface area upon complex formation, calculated using the NACCESS program (see methods).
Calculated using ΔG = RT ln Kd, where R is the gas constant and T the absolute temperature, with T set to 298.15 K when unknown.
Table 2.
Composition of the updated docking and affinity benchmarks (in parentheses are values for the previous versions of the benchmarks, docking v4 and affinity v1).
| Docking | Affinity | |||
|---|---|---|---|---|
|
| ||||
| N | % | N | % | |
| All | 230 (175) | 179 (144) | ||
|
| ||||
| Enzyme containing | 88 (71) | 38% (41%) | 69 (61) | 39% (42%) |
| Antibody-antigen | 40 (24) | 17% (14%) | 33(19) | 18% (13%) |
| Others | 102 (80) | 45% (45%) | 77 (64) | 43% (45%) |
|
| ||||
| Rigid-bodya | 151(119) | 65% (68%) | ||
| Mediuma | 45 (29) | 20% (17%) | ||
| Difficulta | 34 (27) | 15% (15%) | ||
|
| ||||
| Rigid (I-RMSD<1.0Å)a | 93(75) | 52% (52%) | ||
| Flexible (I-RMSD>1.0Å)a | 86 (69) | 48% (48%) | ||
See Methods for definition
In the previous versions of the benchmarks, some categories are underrepresented, most notably the antibody-antigen cases (14%) and difficult cases (15%), while rigid-body cases are overrepresented (68%). Although there still is overrepresentation and underrepresentation in the updated benchmark, the newly added cases do not worsen the representation of any category, and achieve a more balanced composition for most categories. We examined the new cases on various properties related to size and flexibility of the component proteins, but only found the total solvent accessible surface area of the component proteins to be significantly smaller in docking benchmark 4 than the 55 new cases (p-value=0.05; Kolmogorov-Smirnov test), with average total surface areas of ~24,000 Å2 and ~29,000 Å2, respectively. It is not clear, however, to what extent this difference reflects changes in the content of the PDB. Finally, the cases in the docking benchmark that involve NMR structures increased from 16 cases (9%) in version 4 to 32 cases (14%) in version 5.
Performance of docking algorithms
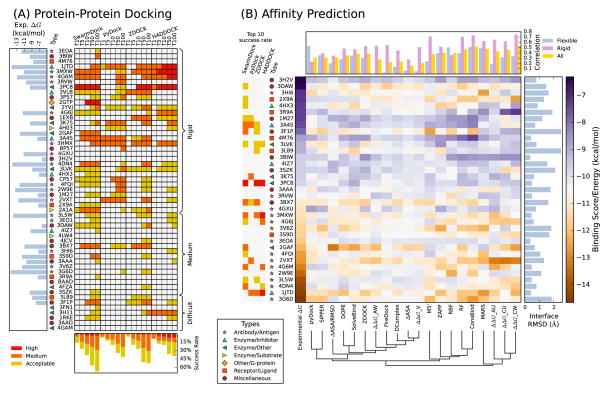
Four docking algorithms (see Material and Methods) we applied to the new cases and their results are shown in Figure 1A. SwarmDock [29,30], PyDock [31], and ZDOCK [32,33] are ab-initio methods, whereas HADDOCK uses bioinformatics predictions to drive the docking [34], in this particular case it uses CPORT to predict interface residues [35] and PARATOME [36] to identify CDR loops of antibodies (see Methods). Overall the success rates (at least one acceptable prediction for a benchmark case) ranged between 5–16% for the top prediction, 20–38% for the top 10 predictions, and 40–67% for the top 100 predictions, comparable to the success rates on version 4 of the docking benchmark using SwarmDock and ZDOCK [37,38]. As expected, the success rate was much higher for the rigid-body category, with the success rates for the top 10 predictions at 31–50%, compared to 4–22% for the medium and difficult cases. The success rates also varied according to biological category, highest for enzyme containing complexes (29–41%) followed by the antibody/antigen complexes (13–38%) and finally the other complexes (5–36%).
Figure 1.
(A) Performance of four docking algorithms on the new cases in the benchmarks, showing whether acceptable/medium/high quality structures evaluated using the CAPRI criteria were present in the top 1/5/10/50/100 predictions for each case (denoted by T1, T5, T10, T50, and T100, respectively). Also shown are the overall success rates (bottom), complex type (left) and binding energy where available (far left). The complexes are ordered first by the difficulty category, then by I-RMSD. (B) Evaluation of affinity prediction methods. Complexes are ordered by increasing experimental affinities, to which the predicted affinities were fitted using linear regression in order to compare the performance of various prediction methods. The performances are grouped using a weighted average linkage agglomerative clustering algorithm (bottom). Correlations against the experimental data are shown at the top, for all the new benchmark cases as well as for the flexible complexes (I-RMSD ≥ 1.0 Å) only or for the rigid complexes (I-RMSD < 1.0Å) only. Also shown are the I-RMSD values (right), complex type (left), and the docking success rate at top 10 predictions (far left).
We observed that the performances of the different docking algorithms were correlated; for 25% of the rigid-body cases, not a single acceptable solution was found in the top 10 predictions by any of the algorithms, and for 22% cases all four methods succeeded. These figures are much higher than would be expected if the complexes with correct predictions were randomly distributed amongst the rigid-body cases (16% and 2%, respectively). Some insight into why some interactions were inherently easier to dock than others, even within the rigid-body category, can be gleaned by focusing on the cases for which affinities are available. When all the docking algorithms failed to find an acceptable solution in the top 10 predictions, the affinity predictors also predicted weak binding energies (3EOA, 3BIW, 4M76, 3RVW, 4GXU, 3H2V). This is either because the complexes are indeed of low affinity, or due to deficiencies in the energy functions used in both docking and affinity prediction. The success rates were higher for enzyme containing and antibody-antigen complexes than for other complexes, as the latter tend to form weaker interactions.
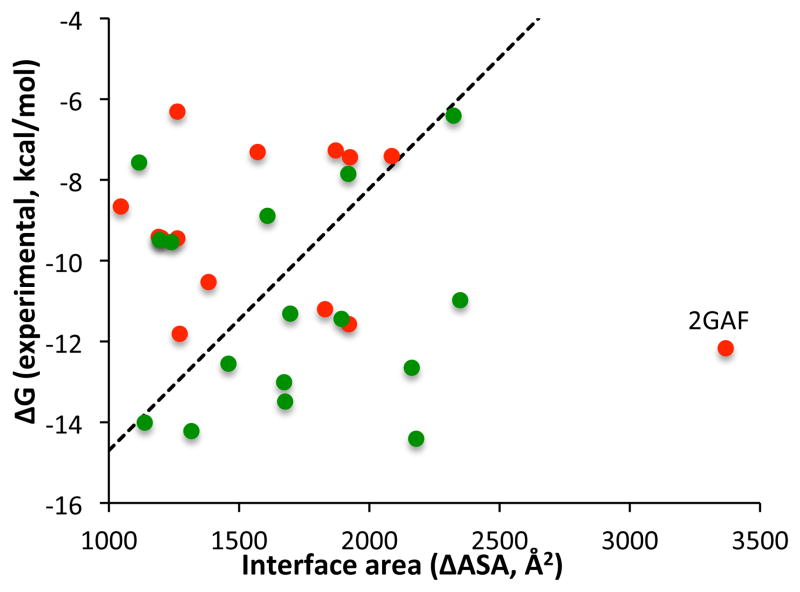
We searched for features indicative of a successful docking outcome. We define a successful run as a benchmark case for which at least three out of four docking algorithms yielded an acceptable or better prediction in the top 100 predictions, while an unsuccessful docking run had at most one algorithm with an acceptable prediction in the top 100 predictions. We asked which features could separate the cases with successful docking runs from the cases with unsuccessful docking runs. Because a major driving force in many protein-protein docking algorithms is the desolvation of the protein components [28], we computed the buried interface area (ΔASA) upon complex formation, which is a good measure for desolvation. We further hypothesized that strong binders were easier to dock than weak binders. Indeed ΔASA and experimentally measured binding free energy achieved a good separation of the two sets of cases with successful and unsuccessful docking runs (Figure 2). Note that the correlation between ΔASA and the experimental binding energy is low, as reported in Figure 1B and discussed below. These two features were individually mildly predictive of docking success (for example, the seven strongest binders all resulted in successful docking runs), the combination of them could almost cleanly separate the successful and unsuccessful docking runs. Below the separating line, 79% docking runs were successful, and above the line the docking performance drops to 31%. The outlier 2GAF [39] has the largest interface area of all the cases and a binding energy stronger than any of the other cases with unsuccessful docking runs. Below we discuss this complex in more detail.
Figure 2.
Interface area vs. experimental binding energy of the benchmark cases with successful docking runs (green; at least three docking protocols yielding acceptable predictions in the top 100) or unsuccessful docking runs (red; at most one docking protocol yielding acceptable predictions in the top 100).
Performance of affinity prediction algorithms
The change in buried surface area, ΔASA, does not correlate well with binding energy (r=−0.16), even for the rigid complexes (I-RMSD < 1.0Å, r=−0.28), due to complexes with large ΔASA but low affinity, such as the snpA protease/inhibitor complex (4HX3), as well as high affinity complexes with low surface area such as the C836 (3L5W) and carlumab (4DN4) antibodies, which are highly optimized for cytokine binding. Similarly, the binding energy does not correlate highly with I-R-SD (r=−0.24), and only a small improvement is found using a minimal linear model combining ΔASA and I-RMSD (r=0.31) [40]. We further evaluated a number of prediction methods that include the specific geometry and composition of the interaction (Figure 1B). This yielded overall correlations of up to r=0.53, with a predictive power much higher for rigid complexes, up to r=0.75, than for the flexible cases, up to r=0.53. The best performing methods were trained using either the first version of the affinity benchmark [25] or using changes in affinity upon mutation [41], yet these functions yielded lower correlations on the new benchmark cases than the best correlation of r=0.63 previously reported for the original affinity benchmark [26,27,42]. The correlations were lower for the statistical potentials and docking scores.
For some of the complexes, the predictions were consistently poor across all methods. All methods underestimated the affinities for the antibody/hemagglutinin complex (4GXU), which features a glycosylated asparagine at the periphery of the interface, the C3D/integrin α-M complex (4M76), for which the interaction is mediated via a Ca2+ ion at the core of the interface, and the efalizumab/integrin α-L complex (3EOA), which is the most rigid interaction in the benchmark (I-RMSD = 0.39 Å). On the other hand, all methods overestimated the affinities for the actin/twinfilin (3DAW), ALM57/integrin α-L (3HI6), TolA/G3P (2X9A) and HIF2/ARNT (3F1P) complexes, all of which have high flexibility, for which the energy penalty of conformational rearrangement may not be well estimated.
Highlighted case: Poly(A) polymerase VP55/Vaccinia protein VP39 (2GAF)
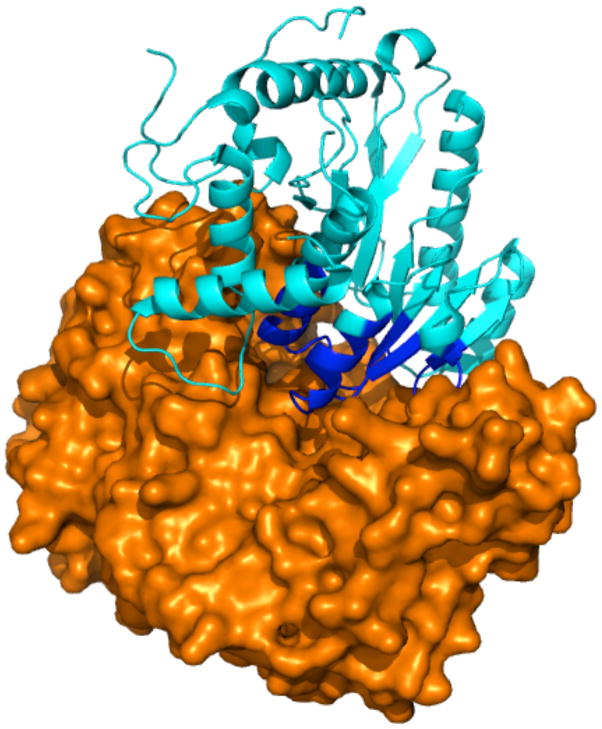
Figure 2 shows that the combination of experimentally measured binding energy and buried surface area forms a good indicator for a successful docking run. The complex of Poly (A) polymerase VP55 and Vaccinia protein VP39 (2GAF) [39], however, is a striking outlier. Only a single docking protocol was successful, despite 2GAF having the largest buried surface area of all complexes and stronger binding than any of the other complexes that had at most one successful docking run. Furthermore, this complex belongs to the rigid-body category, with an I-RMSD of 0.69 Å, and we did not find co-factors or other aspects that might complicate the docking. We studied 2GAF in more detail to understand the poor docking performance. Inspection of the structure (Figure 3) suggests that the difficulty may be related to the deep cavity of the receptor being completely filled by the ligand. To quantify this, we calculated the degree of encapsulation of a protein by its binding partner using Cα atoms, and performed the same calculation for all the benchmark cases in Figure 2. We found that 39 residues of the vaccinia protein VP39 are within the cavity of the Poly(A) polymerase VP55 (indicated in blue in Figure 3). This is the highest number observed in the set of proteins considered for Figure 2; 4FQI and 3BX7 have 25 and 12 residues encapsulated, respectively, while all other proteins have fewer than ten residues within the cavities (39 proteins show zero resides). Presumably the tight fit seen in 2GAF renders the mouth of the energy funnel narrow, which may impact the ability of docking algorithms to find and enter the energy funnel. In addition, the tight fit may cause difficulty for grid-based methods (ZDOCK, PyDock), because even small deviations from the ideal position, resulting from the discreet rigid-body conformational parameters, may cause clashes that prevent favorable scores. Indeed, for a run with a finer rotational sampling (6° vs. the default of 15°), ZDOCK found a high-accuracy prediction at rank 23. SwarmDock was able to find a solution in the top 5. Small conformational changes allowed by SwarmDock, which may have alleviated steric clashes at the funnel entrance, could have facilitated a smoother entry to the binding funnel. Indeed, the lowest frequency normal mode corresponds to the opening of the binding cavity, allowing ligand insertion. In the case of HADDOCK, it was the low quality of the bioinformatics predictions for the ligand binding site (recall of 7%) that prevented the sampling of near-native solutions. Docking with center-of-mass or random ambiguous interaction restraints (two ab-initio docking modes of HADDOCK) does generate acceptable solutions in the top 50 (data not shown). In general, it appears that the poor performance of the docking algorithms for 2GAF is caused by the inability to correctly sample or find the native orientation of the ligand within the receptor cavity. This makes 2GAF an exception to the general consensus in the field that failures of docking protocols are caused either by inaccuracies of the scoring functions (including explicit solvation and entropy effects) or the difficulty of modeling protein conformational changes [43,44].
Figure 3.
Crystal structure (2GAF) of the complex of Poly(A) polymerase (orange) VP55 and Vaccinia protein VP39 (blue and cyan). Vaccinia protein VP39 residues that are within the Poly(A) polymerase cavity are colored blue, while the residues outside the cavity are colored cyan.
Conclusions
We have presented updated versions to our widely used protein-protein docking and affinity benchmarks with respectively 55 and 35 new entries. This represents relative increases of 31% and 24% cases, respectively compared with the previous versions. The updated benchmarks have slightly improved the balance with respect to both complex types and the range of conformational changes between bound and unbound forms. They are available from the following sites: http://zlab.umassmed.edu/benchmark (docking benchmark) and http://bmm.cancerresearchuk.org/~bmmadmin/Affinity (affinity benchmark).
We analyzed the performance of four different docking methods and a comprehensive set of state-of-the-art protein-protein complex affinity prediction methods. We found that the newly added complexes provide a challenging test set for both docking and affinity prediction algorithms: Structure predictions success rates and correlations with experimentally obtained affinities are lower than reported using previous versions of the benchmark. These updated benchmarks will aid the community in improving these algorithms and increasing our understanding of biomolecular interactions.
Methods
Benchmark construction
We collected new structures for our benchmarks from the Protein Data Bank (PDB) [45] using a semiautomatic pipeline. We first used the BLAST sequence homology search tool [46] to find protein-protein complexes for which the experimental structures of both the complex and the unbound component proteins were available. We also used the SACS resource [47] to collect a candidate list of antibody-antigen complexes. These complexes were then filtered using various quality criteria: (1) the complex structure needed to be determined by X-ray crystallography, the unbound structures by either X-ray crystallography or nuclear magnetic resonance (NMR); (2) the sequence identity between bound and unbound chains needed to be at least 96% with an alignment coverage larger than 80%; (3) the X-ray resolution needed to be 3.25 Å or better; (4) chains needed to consist of at least 30 residues.
While constructing the previous versions of our docking benchmark [17–20], we deemed two complexes redundant when the pairs of interacting domains were the same at the SCOP [48] family level. Antibody-antigen complexes were considered redundant only when the SCOP families of the antigens were identical, and at least 80% of the antigen interface residues were shared between the two complexes. We used SCOPe 2.03 [49] (previously named SCOP 1.75C), which represented a limited update with respect to the 1.75 release used for the first four versions of the docking benchmark. To further compensate for the lack of SCOP coverage for the most recently solved PDB structures, we inferred their SCOP family level assignments using the older PDB entries with identical sequences and known SCOP IDs.
We manually investigated the candidate complexes extensively, consulting the literature associated with the PDB entries. We checked whether any residues were missing or mutated in the interface (allowing such residues only if binding would not be affected), and whether co-factors that affect binding were present or compatible in both bound and unbound forms. The starting point for the manual step was the first biological assembly listed in the PDB, although in a number of cases these were not accurate and an alternative assembly had to be used. When multiple entries were available for a complex or a component protein, we chose the entry that had the best overall structure quality. This was to some extent a subjective criterion, as we had to balance all the aforementioned features in the decision. For component proteins with NMR structures we chose the model that had the lowest interface root mean square deviation (I-RMSD) from the bound structure. Finally, we prepared structure files that included the fewest protein chains that correctly reflected the binding process, aligned the bound and unbound structures, and retained only those HETATM fields that we deemed biologically relevant.
We evaluated several properties from the structure files. The change in solvent accessible surface area (ΔASA) upon complex formation was calculated using the NACCESS algorithm [50]. The I-RMSD was calculated by superposing the unbound component proteins onto their bound forms, using the Cα atoms for residues that had any atom within 10 Å of any atom of the binding partner. We also assessed the expected difficulty of a benchmark entry for protein-protein docking algorithms [17–20]. Complexes with I-RMSD > 2.2 Å were considered difficult, and complexes with I-RMSD < 1.5 Å were considered rigid-body if their fnon-nat [51] were < 0.40. All other complexes were considered to be of medium docking difficulty.
We then used the set of complexes as a starting point for extending the structural affinity benchmark. For many entries, affinities were reported multiple times either by different groups or using different techniques. These measurements were mostly in mutual accordance with one another, typically within one order of magnitude in terms of equilibrium constant. When selecting the value to include in the benchmark, priority was given to affinities reported for samples matching the sequences of the reported structures of the complexes. When this criterion could not be met or still resulted in multiple values preference was based on sequence similarity and the measurement method. As in the first version of the affinity benchmark, most affinities were measured using surface plasmon resonance, isothermal titration calorimetry, or spectroscopic methods. The affinities of four new cases were measured using the more recent thermophoresis and bio-layer interferometry technologies. We also collected experimental conditions and additional thermodynamic and kinetic data whenever available. Affinities were measured at a pH in the 7–8 range, typically within the 20–25°C temperature range, and with an ionic strength of around 150 mM. In the context of affinity prediction we consider complexes with I-RMSD < 1.0 Å as rigid-body and the remaining complexes flexible.
Docking algorithms
ZDOCK is an FFT-based rigid-body docking algorithm that performs a grid-based exhaustive search with a 15° or 6° rotational sampling in three-dimensional (3D) rotational space and a 1.2 Å sampling in the 3D translational space [32,33,38,52]. For each combination of the three rotational angles, the best scoring prediction in the translational space is retained, yielding 3600 or 54000 predictions for the 15° and the 6° sampling respectively. Here we report results obtained using the 15° sampling. We used ZDOCK version 3.0.2 that uses the IFACE [53] scoring function and the advanced 3D convolution library [54].
SwarmDock is a flexible docking method employing a population-based memetic algorithm that combines a modified particle swarm optimization global search with an adaptive random local search [29,30]. Elastic network normal mode analysis is used to model flexibility, and the algorithm simultaneously optimizes translational, quaternion and normal coordinates, using the DComplex statistical potential as objective function [55]. The algorithm was run at the SwarmDock server [37]; swarms are initialized around ca. 120 points surrounding the receptor and the algorithm was run four times from each starting point for 600 iterations. The lowest energy solutions found in each run were ranked using the centroid potential of Tobi [56] and clustered, retaining only the lowest energy member of each cluster.
PyDock [31] is a protein-protein docking protocol built upon FTDock [57], an FFT based method that searches for geometrically complementary rigid-body poses in the translational and rotational space. FTDock predicts 10,000 poses which are then scored using an empirical potential composed of electrostatic interaction (Coulombic energy with a distance-dependent dielectric constant ε = 4.0r and charges specified by the AMBER94 force field [58], truncated to be in between 1.0 and −1.0 kcal/mol), desolvation (based on atomic solvation parameters optimized for rigid-body docking), and a limited (10%) contribution from the van der Waals energy (6–12 Lennard-Jones potential with atomic parameters from the AMBER94 force field, truncated to be below 1.0 kcal/mol).
HADDOCK (High Ambiguity Driven DOCKing) [34] is a semi-flexible docking protocol that uses bioinformatics predictions and biochemical/biophysical interaction data to drive the docking process. It uses CNS (Crystallography and NMR system) [59] as its structure calculation engine. The protocol consists of three steps: i) randomization of orientation and rigid-body docking via energy minimization driven by interaction restraints (it0), ii) semi-flexible refinement in the torsional angle space in which side-chain and backbone atoms of the interface residues are allowed to move (it1) and iii) Cartesian dynamics refinement in explicit solvent, typically water. The final structures are clustered using the pairwise backbone ligand interface RMSD and the resulting clusters ranked according to the HADDOCK score (weighted sum of the restraint energy, the van der Waals and electrostatic energies based on OPLS parameters [60] and a desolvation energy term [61]). Note that in the docking performance analysis presented here, no clustering was performed and individual models were selected based on their HADDOCK score.
We used the HADDOCK web server [62], outputting 10000/400/400 models for the three stages of the protocol. Restraints to drive the docking were derived from bioinformatics predictions by CPORT [35], except for the antibody-antigen complexes for which complementarity-determining regions (CDRs) identified with PARATOME [36] were defined as active, and all solvent-accessible residues of the antigen were used as passive residues to define ambiguous interaction restraints to drive the docking. The predicted interfaces (and their recall and precision) used for docking are available at the SBGRid Data Bank, along with all docking decoys and HADDOCK input files from the deposited HADDOCK docking set [63].
Affinity prediction algorithms
ZAPP predicts protein-protein binding free energies using a linear combination of nine energy terms and a constant [26]. Only one term uses the unbound structures in addition to the complex structures, while the other eight terms only require the complex structure.
ConsBind is an affinity prediction method based on machine learning in which the predicted affinity is a consensus of four learners [42]: multivariate adaptive regression splines (MARS), random forest regression (RF), radial basis function (RBF) interpolation, and an M5′ regression tree (M5′). The learners were trained using 143 of the 144 affinities in the previous affinity benchmark [25] with all 108 features extracted from the bound structures using the CCharPPI web server [64]. Information from the unbound structures was not used. The final consensus score is the arithmetic mean of the four learners.
Solvebind is a binding affinity prediction method based on the global surface model of Kastritis et al. [27], combining the number of atoms in the interface (NAtomsINT) and the percentages of charged and polar residues in the non-interacting surface (%AAcharNIS and %AApolNIS):
with α = 0.0857, β = −0.0685, γ = 0.0262, and δ = 3.0125 (obtained after four-fold cross-validation based on the rigid-body complexes of the previous affinity benchmark [25]). Properties of the non-interacting surface were found to correlate with affinity [13,27] and may regulate solvation and electrostatic contributions to binding affinity [27,65].
Besides the aforementioned binding affinity prediction methods developed in our groups, we also assessed the minimal affinity model of Janin (ΔASA/RMSD) [40], buried surface area (ΔASA), the DOPE [66] and DComplex [55] statistical potentials, the PyDock [31], SIPPER [67], ZDOCK [68] and FireDock [69] docking scores, as well as contact potentials (ΔΔG_AW, ΔΔG_AU, ΔΔG_CW, ΔΔG_CU) [41] and a surface energy model (ΔΔG_V) [70] derived from mutation data.
Supplementary Material
Footnotes
CDR definition used for docking antibody-antigen complexes with HADDOCK, predicted affinities listed by benchmark entry, experimental conditions of the affinities measurements, and the full references to the experimentally measured affinities.
The docking benchmark is hosted at http://zlab.umassmed.edu/benchmark, and the affinity benchmark at http://bmm.cancerresearchuk.org/~bmmadmin/Affinity
References
- 1.Wodak SJ, Vlasblom J, Turinsky AL, Pu S. Protein-protein interaction networks: the puzzling riches. Curr Opin Struc Biol. 2013;23:941–53. doi: 10.1016/j.sbi.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Janin J, Henrick K, Moult J, Eyck LT, Sternberg MJE, Vajda S, et al. CAPRI: a Critical Assessment of PRedicted Interactions. Proteins. 2003;52:2–9. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- 3.Ritchie DW. Recent progress and future directions in protein-protein docking. Curr Protein Pept Sci. 2008;9:1–15. doi: 10.2174/138920308783565741. [DOI] [PubMed] [Google Scholar]
- 4.Smith GR, Sternberg MJE. Prediction of protein-protein interactions by docking methods. Curr Opin Struc Biol. 2002;12:28–35. doi: 10.1016/s0959-440x(02)00285-3. [DOI] [PubMed] [Google Scholar]
- 5.Lu L, Lu H, Skolnick J. MULTIPROSPECTOR: An algorithm for the prediction of protein-protein interactions by multimeric threading. Proteins. 2002;49:350–64. doi: 10.1002/prot.10222. [DOI] [PubMed] [Google Scholar]
- 6.Mukherjee S, Zhang Y. Protein-protein complex structure predictions by multimeric threading and template recombination. Structure. 2011;19:955–66. doi: 10.1016/j.str.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szilagyi A, Zhang Y. Template-based structure modeling of protein–protein interactions. Curr Opin Struc Biol. 2014;24:10–23. doi: 10.1016/j.sbi.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogmen U, Keskin O, Aytuna AS, Nussinov R, Gursoy A. PRISM: protein interactions by structural matching. Nucleic Acids Research. 2005;33:W331–226. doi: 10.1093/nar/gki585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuncbag N, Gursoy A, Nussinov R, Keskin O. Predicting protein-protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nat Protoc. 2011;6:1341–54. doi: 10.1038/nprot.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinha R, Kundrotas PJ, Vakser IA. Docking by structural similarity at protein-protein interfaces. Proteins. 2010;78:3235–41. doi: 10.1002/prot.22812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vreven T, Hwang H, Pierce BG, Weng Z. Evaluating template-based and template-free protein-protein complex structure prediction. Brief Bioinformatics. 2014;15:169–76. doi: 10.1093/bib/bbt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodrigues JPGLM, Bonvin AMJJ. Integrative computational modeling of protein interactions. Febs J. 2014;281:1988–2003. doi: 10.1111/febs.12771. [DOI] [PubMed] [Google Scholar]
- 13.Kastritis PL, Bonvin AMJJ. Molecular origins of binding affinity: seeking the Archimedean point. Curr Opin Struc Biol. 2013;23:868–77. doi: 10.1016/j.sbi.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Kastritis PL, Bonvin AMJJ. On the binding affinity of macromolecular interactions: daring to ask why proteins interact. J R Soc Interface. 2013;10:20120835. doi: 10.1098/rsif.2012.0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kastritis PL, Visscher KM, van Dijk ADJ, Bonvin AMJJ. Solvated protein-protein docking using Kyte-Doolittle-based water preferences. Proteins. 2013;81:510–8. doi: 10.1002/prot.24210. [DOI] [PubMed] [Google Scholar]
- 16.Moal IH, Torchala M, Bates PA, Fernandez-Recio J. The scoring of poses in protein-protein docking: current capabilities and future directions. Bmc Bioinformatics. 2013;14:286. doi: 10.1186/1471-2105-14-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen R, Mintseris J, Janin J, Weng Z. A protein-protein docking benchmark. Proteins. 2003;52:88–91. doi: 10.1002/prot.10390. [DOI] [PubMed] [Google Scholar]
- 18.Mintseris J, Wiehe K, Pierce B, Anderson R, Chen R, Janin J, et al. Protein-Protein Docking Benchmark 2. 0: an update. Proteins. 2005;60:214–6. doi: 10.1002/prot.20560. [DOI] [PubMed] [Google Scholar]
- 19.Hwang H, Pierce B, Mintseris J, Janin J, Weng Z. Protein-protein docking benchmark version 3. 0. Proteins. 2008;73:705–9. doi: 10.1002/prot.22106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang H, Vreven T, Janin J, Weng Z. Protein-protein docking benchmark version 4. 0. Proteins. 2010;78:3111–4. doi: 10.1002/prot.22830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douguet D, Chen H-C, Tovchigrechko A, Vakser IA. DOCKGROUND resource for studying protein-protein interfaces. Bioinformatics. 2006;22:2612–8. doi: 10.1093/bioinformatics/btl447. [DOI] [PubMed] [Google Scholar]
- 22.van Dijk M, Bonvin A. A protein–DNA docking benchmark. Nucleic Acids Research. 2008;36:e88. doi: 10.1093/nar/gkn386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez-Cano L, Jiménez-García B, Fernandez-Recio J. A protein-RNA docking benchmark (II): extended set from experimental and homology modeling data. Proteins. 2012;80:1872–82. doi: 10.1002/prot.24075. [DOI] [PubMed] [Google Scholar]
- 24.Kastritis PL, Bonvin AMJJ. Are Scoring Functions in Protein-Protein Docking Ready To Predict Interactomes? Clues from a Novel Binding Affinity Benchmark. J Proteome Res. 2010;9:2216–25. doi: 10.1021/pr9009854. [DOI] [PubMed] [Google Scholar]
- 25.Kastritis PL, Moal IH, Hwang H, Weng Z, Bates PA, Bonvin AMJJ, et al. A structure-based benchmark for protein-protein binding affinity. Protein Sci. 2011;20:482–91. doi: 10.1002/pro.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vreven T, Hwang H, Pierce BG, Weng Z. Prediction of protein-protein binding free energies. Protein Sci. 2012;21:396–404. doi: 10.1002/pro.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kastritis PL, Rodrigues JPGLM, Folkers GE, Boelens R, Bonvin AMJJ. Proteins feel more than they see: fine-tuning of binding affinity by properties of the non-interacting surface. J Mol Biol. 2014;426:2632–52. doi: 10.1016/j.jmb.2014.04.017. [DOI] [PubMed] [Google Scholar]
- 28.Moal IH, Moretti R, Baker D, Fernandez-Recio J. Scoring functions for protein-protein interactions. Curr Opin Struc Biol. 2013;23:862–7. doi: 10.1016/j.sbi.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 29.Moal IH, Bates PA. SwarmDock and the use of normal modes in protein-protein docking. Int J Mol Sci. 2010;11:3623–48. doi: 10.3390/ijms11103623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Moal IH, Bates PA. Detection and refinement of encounter complexes for protein-protein docking: taking account of macromolecular crowding. Proteins. 2010;78:3189–96. doi: 10.1002/prot.22770. [DOI] [PubMed] [Google Scholar]
- 31.Cheng TM-K, Blundell TL, Fernandez-Recio J. pyDock: electrostatics and desolvation for effective scoring of rigid-body protein-protein docking. Proteins. 2007;68:503–15. doi: 10.1002/prot.21419. [DOI] [PubMed] [Google Scholar]
- 32.Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52:80–7. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 33.Chen R, Weng Z. A novel shape complementarity scoring function for protein-protein docking. Proteins. 2003;51:397–408. doi: 10.1002/prot.10334. [DOI] [PubMed] [Google Scholar]
- 34.Dominguez C, Boelens R, Bonvin A. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. Journal of the American Chemical Society. 2003;125:1731–7. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 35.De Vries SJ, Bonvin AMJJ. CPORT: a consensus interface predictor and its performance in prediction-driven docking with HADDOCK. PLoS ONE. 2011;6:e17695. doi: 10.1371/journal.pone.0017695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kunik V, Ashkenazi S, Ofran Y. Paratome: an online tool for systematic identification of antigen-binding regions in antibodies based on sequence or structure. Nucleic Acids Research. 2012;40:W521–4. doi: 10.1093/nar/gks480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torchala M, Moal IH, Chaleil RAG, Fernandez-Recio J, Bates PA. SwarmDock: a server for flexible protein-protein docking. Bioinformatics. 2013;29:807–9. doi: 10.1093/bioinformatics/btt038. [DOI] [PubMed] [Google Scholar]
- 38.Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, Weng Z. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771–3. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moure CM, Bowman BR, Gershon PD, Quiocho FA. Crystal structures of the vaccinia virus polyadenylate polymerase heterodimer: insights into ATP selectivity and processivity. Mol Cell. 2006;22:339–49. doi: 10.1016/j.molcel.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 40.Janin J. A minimal model of protein-protein binding affinities. Protein Sci. 2014;23:1813–7. doi: 10.1002/pro.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moal IH, Fernandez-Recio J. Intermolecular contact potentials for protein–protein interactions extracted from binding free energy changes upon mutation. J Chem Theory Computation. 2013;9:3715–727. doi: 10.1021/ct400295z. [DOI] [PubMed] [Google Scholar]
- 42.Moal IH, Agius R, Bates PA. Protein-protein binding affinity prediction on a diverse set of structures. Bioinformatics. 2011;27:3002–9. doi: 10.1093/bioinformatics/btr513. [DOI] [PubMed] [Google Scholar]
- 43.Bonvin A. Flexible protein-protein docking. Curr Opin Struc Biol. 2006;16:194–200. doi: 10.1016/j.sbi.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 44.Zacharias M. Accounting for conformational changes during protein-protein docking. Curr Opin Struc Biol. 2010;20:180–6. doi: 10.1016/j.sbi.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Berman HM. The Protein Data Bank. Nucleic Acids Research. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul S. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allcorn LC, Martin ACR. SACS--Self-maintaining database of antibody crystal structure information. Bioinformatics. 2002;18:175–81. doi: 10.1093/bioinformatics/18.1.175. [DOI] [PubMed] [Google Scholar]
- 48.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–40. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 49.Fox NK, Brenner SE, Chandonia J-M. SCOPe: Structural Classification of Proteins—extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Research. 2014;42:D304–9. doi: 10.1093/nar/gkt1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hubbard SJ, Thornton JM. “NACCESS,” Computer Program. Department of Biochemistry and Molecular Biology, University College London; 1993. [Google Scholar]
- 51.Méndez R, Leplae R, De Maria L, Wodak SJ. Assessment of blind predictions of protein-protein interactions: current status of docking methods. Proteins. 2003;52:51–67. doi: 10.1002/prot.10393. [DOI] [PubMed] [Google Scholar]
- 52.Vreven T, Pierce BG, Hwang H, Weng Z. Performance of ZDOCK in CAPRI rounds 20–26. Proteins. 2013;81:2175–82. doi: 10.1002/prot.24432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mintseris J, Pierce B, Wiehe K, Anderson R, Chen R, Weng Z. Integrating statistical pair potentials into protein complex prediction. Proteins. 2007;69:511–20. doi: 10.1002/prot.21502. [DOI] [PubMed] [Google Scholar]
- 54.Pierce BG, Hourai Y, Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS ONE. 2011;6:e24657. doi: 10.1371/journal.pone.0024657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu S, Zhang C, Zhou H, Zhou Y. A physical reference state unifies the structure-derived potential of mean force for protein folding and binding. Proteins. 2004;56:93–101. doi: 10.1002/prot.20019. [DOI] [PubMed] [Google Scholar]
- 56.Tobi D. Designing coarse grained-and atom based-potentials for protein-protein docking. Bmc Struct Biol. 2010;10:40. doi: 10.1186/1472-6807-10-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gabb HA, Jackson RM, Sternberg MJ. Modelling protein docking using shape complementarity, electrostatics and biochemical information. J Mol Biol. 1997;272:106–20. doi: 10.1006/jmbi.1997.1203. [DOI] [PubMed] [Google Scholar]
- 58.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, et al. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. Journal of the American Chemical Society. 1995;117:5179–97. [Google Scholar]
- 59.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 60.Jorgensen WL, Tirado-Rives J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. Journal of the American Chemical Society. 1988;110:1657–66. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 61.Fernandez-Recio J, Totrov M, Abagyan R. Identification of protein-protein interaction sites from docking energy landscapes. J Mol Biol. 2004;335:843–65. doi: 10.1016/j.jmb.2003.10.069. [DOI] [PubMed] [Google Scholar]
- 62.De Vries SJ, van Dijk M, Bonvin AMJJ. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5:883–97. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 63.Vangone A, Bonvin AMJJ. SBGRid Data Bank, V1. 2015. HADDOCK decoys for 55 new entries in Docking Benchmark 5. [DOI] [Google Scholar]
- 64.Moal IH, Jiménez-García B, Fernandez-Recio J. CCharPPI web server: computational characterization of protein-protein interactions from structure. Bioinformatics. 2015;31:123–5. doi: 10.1093/bioinformatics/btu594. [DOI] [PubMed] [Google Scholar]
- 65.Visscher KM, Kastritis PL, Bonvin AMJJ. Non-interacting surface solvation and dynamics in protein-protein interactions. Proteins. 2015;83:445–58. doi: 10.1002/prot.24741. [DOI] [PubMed] [Google Scholar]
- 66.Shen M-Y, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–24. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pons C, Talavera D, la Cruz de X, Orozco M, Fernandez-Recio J. Scoring by intermolecular pairwise propensities of exposed residues (SIPPER): a new efficient potential for protein-protein docking. J Chem Inf Model. 2011;51:370–7. doi: 10.1021/ci100353e. [DOI] [PubMed] [Google Scholar]
- 68.Pierce B, Weng Z. A combination of rescoring and refinement significantly improves protein docking performance. Proteins. 2008;72:270–9. doi: 10.1002/prot.21920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andrusier N, Nussinov R, Wolfson HJ. FireDock: fast interaction refinement in molecular docking. Proteins. 2007;69:139–59. doi: 10.1002/prot.21495. [DOI] [PubMed] [Google Scholar]
- 70.Moal IH, Dapkūnas J, Fernandez-Recio J. Inferring the microscopic surface energy of protein-protein interfaces from mutation data. Proteins. 2015;83:640–50. doi: 10.1002/prot.24761. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.