Abstract
The exploration of plant behavior on a cellular scale in a minimal invasive manner is key to understanding plant adaptations to their environment. Plant hormones regulate multiple aspects of growth and development and mediate environmental responses to ensure a successful life cycle. To monitor the dynamics of plant hormone actions in intact tissue, we need qualitative and quantitative tools with high temporal and spatial resolution. Here, we describe a set of biological instruments (reporters) for the analysis of the distribution and signaling of various plant hormones. Furthermore, we provide examples of their utility for gaining novel insights into plant hormone action with a deeper focus on the drought hormone abscisic acid.
Keywords: abscisic acid, fluorescent reporter, in vivo visualization, plant hormone
Introduction
Plant hormones are mobile chemical compounds that mediate growth and developmental processes on a global and also on a cellular scale to enable plants a successful life cycle under diverse environmental conditions. Chemical compounds designated as plant hormones are auxins (IAA), abscisic acid (ABA), brassinosteroids (BR), cytokinins (CK), ethylene (ET), gibberellins (GA), jasmonates (JA), karrikins (KAR), salicylic acid (SA), and strigolactones (SL). Plant hormones are involved in the promotion (ET, GA, KAR, SL) or inhibition (ABA) of seed germination, the inhibition of root growth (ABA, IAA, ET, JA), apical dominance (IAA, CK, SL), flower and fruit development (IAA, ET, GA, JA), delay (CK) or promotion (ET) of senescence, abiotic (ABA, BR) and biotic (BR, JA, SA, ABA) interactions, gravitropic responses (IAA, BR), and cell division and/or expansion (IAA, BR, CK, GA). This short summary does not present the whole picture of plant hormone functions. For more recent and specific overviews of plant hormones, we refer the reader to other publications [1–16]. Plant hormone signaling pathways communicate with each other on several levels. These include transcriptional responses, interactions of core transcription factors that are regulated by plant hormones, and regulation of hormone metabolism and transport [17–21].
Plant hormones are chemically diverse, and biosynthesis, degradation, or inactivation often involve multiple cellular compartments. For example, precursors or building blocks for the synthesis of IAA, ABA, GA, JA, and SL originate from plastids [11, 22–24]. SA and the most biologically active CKs are fully synthesized in plastids [25, 26]. An overview of plant hormone metabolic pathways and genes involved in these processes is provided from the RIKEN Plant Hormone Research Network (http://hormones.psc.riken.jp/pathway_hormones.shtml).
The distribution of plant hormones is not only dependent on the expression of the respective biosynthetic and metabolizing genes but also on the expression and activity of plant hormone transporters. Plant hormone transport is facilitated through various transport proteins. However, hormone transporters often belong to the ATP-binding cassette (ABC)-type transporter or Nitrate Transporter 1/Peptide Transporter Family (NPF) [27–29]. The hormone best understood in terms of its transport is IAA [30]. IAA uptake is facilitated through proton-coupled transporters of the AUX1/LAX family and through NRT1.1/ NPF6.3 [31, 32]. Auxin efflux is regulated by PIN-type and ABC-type (ABCB1, 4, 19) transporters (summarized in [30]). ABA transport is also relatively well understood [33]. ABA is imported by ABCG40 and AIT/NPF-type transporters (AIT1/NPF4.6, AIT3/NPF4.1) [34, 35] and exported by ABCG25 and DTX50 [36, 37]. However, just recently it became evident that NPF4.1/AIT3 not only transports ABA, but also GA and JA-Ile [29, 35]. Other recent reports identified ABC-type transporters for CK (ABCG14) [38, 39] and SL (PhPDR1) [40]. In the case of brassinosteroids, it is believed that they are not transported over long distances, and rather affect long-distance signaling through modulation of IAA transport [41].
There are certain similarities in hormone perception and signal transduction mechanisms. The mechanism of IAA, GA, and JA signal transduction is based on the hormone-dependent SKP1, Cullin, F-box (SCF)-complex ubiquitin ligase (E3)-mediated ubiquitination and 26S proteasomal degradation of transcriptional repressors [42, 43]. There are indications that KAR, SA, and SL also might signal through such mechanisms [13, 14, 44]. Core components of the ABA and ET signaling pathway are also subject to proteasomal degradation [42]. Other hormones signal through phosphorylation/ de-phosphorylation cascades (ABA, BR, CK, ET) [8, 43].
The coordination of plant hormone metabolism and transport and the regulation of core signaling component expression are crucial for maintaining tissue and cell-type specific hormone concentrations and signaling efficiency to facilitate adequate growth and developmental responses. Here, we describe recent advances in monitoring plant hormone distribution and signaling, and present an in-depth discussion of ABA.
Methods for the quantification and visualization of plant hormone distribution and signaling
Several methods can be used to detect and quantify plant hormones as well as plant hormone signaling. These methods encompass classical assays that require hormone extraction from ground tissue as well as the use of genetically encoded hormone reporters that enable in vivo hormone analyses. In the following, we will discuss current methods to analyze plant hormones and finish with a detailed discussion on methods to visualize ABA distribution and signaling.
Classical methods for the quantification of plant hormones
Quantitative measurements of plant hormones are often performed by liquid- and gas-chromatography coupled to mass spectrometry [45–47]. Other techniques are biological assays or immunochemical methods such as enzyme-linked immunosorbent assays (ELISA) (summarized in [48, 49]). Although such methods have a high sensitivity for detecting hormones, their ability to resolve spatial and cell-type specific differences in hormone concentration is relatively low [45, 50]. Also, such methods most likely require hormone extraction from ground tissue. Therefore, these methods are not preferred for in vivo detection and measurements of plant hormones.
Fluorescently labeled plant hormones
Recently, it has become possible to fluorescently label IAA, BL, GA, and JA [51–55]. Except of NBD-IAA [55], such compounds appeared to be biologically active. Uptake experiments of externally applied fluorescently labeled hormones revealed that NBD-IAA accumulated in the endoplasmatic reticulum (ER) [55]. GA-FL accumulated in the root endodermis and this distribution pattern was affected by ET signaling [53]. The fluorescent BR analog AFCS was used to visualize BR receptor-ligand complexes in plants and its cellular uptake followed an endosomal trafficking route to the vacuole [51]. Fluorescently labeled plant hormones are ideal tools for the investigations of hormone uptake and transport. However, these compounds can only be used for exogenous treatments and their cellular distribution reflects only the distribution of actively transported hormone pools.
Expression-based hormone reporters
Expression-based hormone reporters are natural occurring hormone-responsive promoters or synthetic promoters containing repeats of hormone-responsive elements that are enhanced by a minimal p35S promoter and fused to a reporter gene (Table 1A) [56]. It should be clarified that such constructs report hormone signaling events rather than hormone concentration, because the reporter gene expression requires the whole hormone signaling cascade. The most prominent example of an expression-based reporter is the synthetic IAA-responsive pDR5 promoter [60], which has now been improved in terms of hormone sensitivity (pDR5v2 [62]). In addition, expression-based reporters have also been generated for ABA, CK, ET, JA, and SA (Table 1A and references therein). To date, natural hormone-responsive promoters/genes can be identified, for example, through search in publicly available gene expression datasets using the Genevestigator gene search tool [69]. However, synthetic hormone-responsive promoters might be more specific for a certain plant hormone or hormone-activated transcription factor (gene family).
Table 1.
List of hormone-responsive reporters categorized by (A) expression-based reporters, (B) protein degradation-based reporters, and (C) FRET-based reporters with indicated
| Hormone | Promoter | Reference |
|---|---|---|
| (A) Expression-based reporters | ||
| Abscisic acid | pRD29A/B | [57] |
| Abscisic acid | pAtHB6 | [58] |
| Abscisic acid | pRAB18 | [59] |
| Auxin | pDR5 | [60] |
| Auxin | pBA | [61] |
| Auxin | pDR5v2 | [62] |
| Cytokinins | pARR5 | [63] |
| Cytokinins | pTCS | [64] |
| Cytokinins | pTCSn | [65] |
| Ethylene | pobs1 | [66] |
| Jasmonates | p4D-47 | [67] |
| Salicylic acid | p4xWT-46 | [68] |
| Hormone | Reporter | Reference |
| (B) Protein degradation-based reporters | ||
| Auxin | DII-Venus | [75] |
| Auxin | L2min17-Luc | [76] |
| Auxin | R2D2 | [62] |
| Gibberellins | GFP-RGA | [77] |
| Jasmonates | JAI3-GFP | [78] |
| Jasmonates | Jas9-Venus | [79] |
| Hormone | Reporter | Reference |
| (C) FRET-based reporters | ||
| Abscisic acid | ABACUS | [82] |
| Abscisic acid | ABAleon | [83] |
Reporter genes used for fusion to the hormone-responsive promoter are β-glucuronidase (GUS), luciferases (Luc), or fluorescent proteins (FP). A comparison of these reporter genes for expression studies in plants is provided in [70]. Although the use of the reporter gene depends on experimental needs, GUS assays require histochemical or fluorometric methods [71] that do not allow for direct in vivo investigations. On the other hand, Luc requires luciferin as substrate [70] and FPs require a certain maturation time before becoming fluorescent [72, 73]. In comparison, GUS and FPs are preferred at cellular resolution, while Luc is preferred at tissue and whole plant resolution [70].
Expression-based hormone-responsive reporters are not only suitable for the analyses of hormone signaling but are also powerful tools for genetic screens [57, 61, 74].
Protein degradation-based hormone reporters
Protein degradation-based hormone reporters utilize the hormone-specific SCF-complex-mediated proteasomal degradation of transcriptional repressors or repressor domains that are fused to FP or Luc (Table 1B, summarized in [56]). Such reporters have been developed for IAA, GA, and JA (Table 1B and references therein). Protein degradation-based hormone reporters can be subdivided into classical reporters that have been initially used to study GA (GFP-RGA [77]) or JA (JAI3-GFP [78]) signaling, and more advanced synthetic reporters for IAA (DII-Venus [75]) and JA (Jas9-Venus [79]) that opened the field for protein degradation-based hormone reporter analyses.
Protein degradation-based hormone reporters have the potential to semi-quantitatively report hormone concentrations because they most likely directly bind to the respective hormone receptor in a hormone concentration-dependent manner. Note that the abundance of the reporter not only depends on the efficiency of the hormone-specific SCF-complex but also on the efficiency of the expression cassette (promoter and terminator) that drives the reporter expression in a certain tissue or cell-type. While, the kinetics of hormone reporter degradation provide an approximate measure for the respective hormone concentration in a certain tissue or cell-type [75, 79], their ability to report hormone concentration reduction depends on the timescale of the respective hormone response and on the rate of de novo reporter synthesis.
For quantitative measurements, ratiometric reporters are preferred because they can be normalized for expression levels and because they are more suitable for prolonged microscopic analyses [80]. Approaches for normalizing the protein degradation-based IAA reporter DII-Venus [75] led to the development of R2D2 [62] and L2min17-Luc [76]. For ratiometric analyses, R2D2 combines the IAA-degradable DII domain of IAA28 [75] fused to n3xVenus (DII-n3xVenus) with the non-degradable mDII fused to ntdTomato (mDII-ntdTomato) [62]. To minimize effects of differential expression levels, both constructs were expressed under control of the pRPS5A promoter and located together on a single T-DNA [62]. L2min17-Luc is a protein consisting of Renilla and Firefly Luc linked by the A2 peptide and a 13 amino acid peptide of the IAA-degradable IAA17 [76]. Upon expression, the A2 peptide is cleaved, resulting in stoichiometric amounts of the non-degradable Renilla Luc and the IAA-degradable min17-Firelfy Luc [76]. Both Luc signals are then ratiometrically quantified.
Protein degradation-based hormone reporters enabled the mapping of IAA and JA distribution and signaling in certain tissues and cell-types and investigations of changes in hormone distribution and signaling in response to gravitropic changes and wounding [62, 75, 79, 81].
FRET-based hormone reporters
FRET-based hormone reporters have been developed so far only for ABA [82, 83]. They consist of a fluorescent protein FRET-pair, usually improved variants of CFP and YFP that are linked together by a hormone-responsive sensory module. Upon hormone binding, the sensory module changes its structural conformation, thereby affecting the distance and orientation of the FRET-pair and their FRET efficiency [56, 84]. The FRET-based ABA reporters bind directly to ABA and therefore instantaneously visualize ABA concentration changes rather than ABA signaling [82, 83]. Note that the amount of bound hormone is not quantified through FRET efficiency measurements, but through simple YFP/CFP emission ratio measurements, enabling an internal normalization. In the discussion that follows, the FRET-based ABA reporters ABACUS [82] and ABAleon [83] will be compared and discussed in greater detail.
ABACUS and ABAleon are FRET-based reporters for ABA
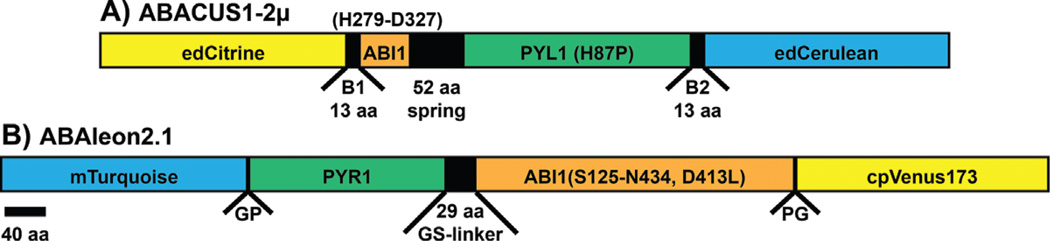
Both ABA reporters ABACUS [82] and ABAleon [83] use FRET (“energy transfer from an excited donor fluorescent protein – CFP variant – to an acceptor fluorescent protein – YFP variant”) [85, 86] as a measure for the ABA concentration. Based on the mechanism of ABA perception and signaling and on structural analyses [87–90], their sensory modules were designed of an ABA receptor (PYR1 or PYL1) that is fused via a linker to a domain of the protein phosphatase type 2C (PP2C) ABI1. The sensory module is then linked to and sandwiched by the fluorescent protein FRET-pair (Fig. 1). Although both FRET-based ABA reporters use the same principle, their features in terms of used FRET-cassette, sensory module, and linkers differ quite strikingly (Fig. 1). Both ABA reporters appear to be flipped relative to the other in terms of the orientation of the FRET-pair and the orientation of the sensory module.
Figure 1.
Schematic presentation of the FRET-based ABA reporters ABACUS1-2µ (A) and ABAleon2.1 (B). As FRET donor and acceptor pair, ABACUS1-2µ uses the enhanced dimerization CFP- and YFP-variants edCerulean and edCitrine. ABAleon2.1 encompasses mTurquoise and Venus circularly permutated at amino acid 173 (cpVenus173) as FRET-pair. The N- and C-terminal orientation of the respective fluorescent protein is interchanged in both reporters. The sensory modules consist of a full length ABA receptor protein (PYL1 with the H87P mutation in ABACUS1-2µ and PYR1 in ABAleon2.1) fused to a fragment of the type 2C protein phosphatase (PP2C) ABI1. ABACUS1-2µ harbors 49 amino acids of the ABI1 phosphatase domain (amino acids H279-D327), while ABAleon2.1 contains the complete phosphatase domain (amino acids S125-N434), but with a D413L mutation that abolishes phosphatase activity. The orientation between PYR1/PYL1 and the ABI1 fragment is interchanged in both reporters. PYR1/PYL1 and ABI1 are linked through a 52 amino acid spring linker in ABACUS1-2 µ, and through a flexible 29 amino acid glycine-serine-(GS)-linker in ABAleon2.1. ABACUS1-2µ was generated using the gateway-cloning strategy, therefore the sensory module is linked to the fluorescent protein pair via 13 amino acid attB1 and attB2 sites. ABAleon2.1 was generated using classical cloning and linked to the fluorescent proteins through GP and PG linkers encoded in the Apal and XmaI restriction sites. Informations about both reporters are derived from [82, 83].
The differences in both ABA reporters are also reflected in their biochemical properties (Table 2). While ABACUS versions exhibit a low ABA affinity and high dynamic range, ABAleon versions bind ABA with high affinity but respond only with a low dynamic range (Table 2) [82, 83]. Still, compared to the advanced FRET-based calcium reporter yellow cameleon YC3.6 that has a dynamic range of 5.6 [91], the first generation FRET-based ABA reporters exhibit a relatively low dynamic range. The design of ABA reporters with higher dynamic range should be considered for future reporter engineering. It also needs to be mentioned that upon ABA binding, ABACUS exhibits a higher energy transfer to the YFP variant, while ABAleon exhibits a lower energy transfer [82, 83].
Table 2.
Biochemical properties of FRET-based ABA reporters
| ABA reporter | ABA receptor |
K’d [µM] |
In vitro DR |
|---|---|---|---|
| ABACUS1-2 µ | PYL1 H87P | 2 | 1.6 |
| ABACUS1-80 µ | PYL1 | 80 | 2.6 |
| ABAleon2.1 | PYR1 | 0.1 | 1.1 |
| ABAleon2.15 | PYR1 H115A | 0.5 | 1.1 |
Indicated are the reporter names, the ABA receptor incorporated in the sensory module, their apparent ABA affinity (K’d) and in vitro dynamic range (DR) calculated from ABA-bound emission ratio/ ABA-unbound emission ratio.
“Dimeric” ABA receptors, including PYR1 and PYL1, exhibit ABA affinities of Kd ~50–100 µM, while “monomeric” ABA receptors bind ABA with Kd ~1 µM (summarized in [92]). Conversion of PYR1 to an enhanced “monomeric” state by introducing the H60P mutation resulted in a Kd for ABA of ~3 µM [92]. Consistently, a homologous mutation in PYL1 H87P led to the conversion of ABACUS1-80µ to the higher affinity ABACUS1-2µ (Table 2) [82]. Introduction of the PYR1 H60P mutation in the ABAleon version ABAleon2.11 resulted in a non-ABA-responsive reporter [83]. “Monomeric” ABA receptors have the potential to interact with PP2Cs in the absence of ABA [87, 88, 93] which could also be the case for the sensory module of ABAleon2.11. ABAleon-type ABA reporters have a K’d for ABA comparable to ABA affinities of PYR1/PYL/RCAR-PP2C complexes that were measured between 18 and 125 nM (Table 2 and summarized in [94]). This implies that the ABAleon sensory module reflects the natural ABA-binding ability of PYR1 in complex with ABI1.
Based on structural and biochemical information, lower affinity ABAleon-type ABA reporters have been generated of which ABAleon2.15 – containing the PYR1 H115A mutation (Table 2) – exhibited a strong stereospecificity for the natural ABA enantiomer (+)-ABA [83]. A homologous amino acid in PYL3 (H139) was also found to be important for ABA stereospecificity [95], suggesting that ABAleon can be utilized to study biochemical properties of ABA receptors.
ABACUS1-2µ and ABAleon affect ABA responses
One feature of both ABA reporters is that they in some way affect ABA signaling. When Arabidopsis plants overexpress ABA receptors, as positive regulators of ABA signaling, they appear more sensitive to ABA [83, 88, 93, 96]. In contrast, over-expression of PP2Cs, as negative regulators of ABA signaling, renders plants less sensitive to ABA [93, 97]. ABACUS1-2µ lines exhibit an increased ABA sensitivity in seed germination and root growth inhibition assays [82]. These data indicate that PYL1H87P encoded in the ABACUS1-2µ sensory module might affect ABA signaling.
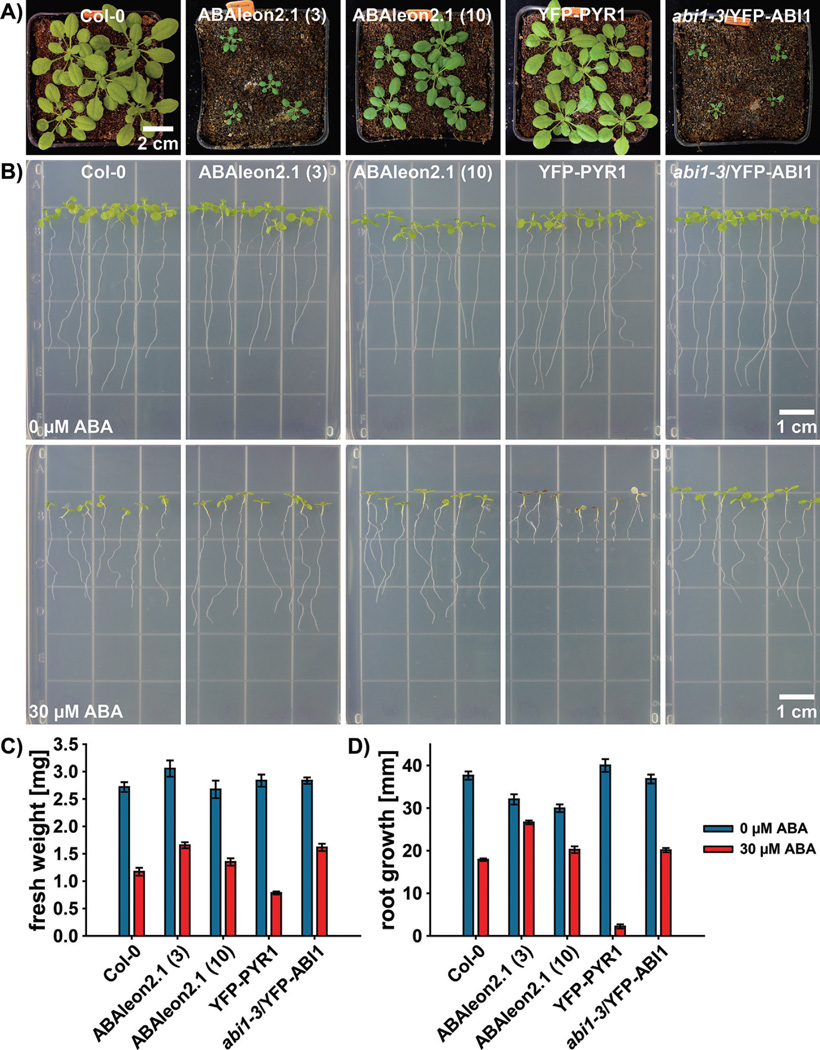
ABAleon2.1 lines were less sensitive to ABA during seed germination and seedling growth [83], and exhibit an overall reduction in growth when compared to Col-0 wild-type plants (Fig. 2A and B; [83]). The reduced growth was particularly evident for the high expressing ABAleon2.1 line 3 compared to the low expressing line 10 when grown in soil at relative air humidity of ~40% (Fig. 2A). Interestingly, growth of ABAleon2.1 line 3 was comparable to a line over-expressing the PP2C ABI1 fused to YFP (abi1-3/YFP-ABI1 [97]) (Fig. 2A). Note that ABAleon2.1 plant growth in soil can be improved when grown at higher relative air humidity (e.g. 70%; Waadt et al., unpublished).
Figure 2.
ABAleon2.1 plants exhibit a growth phenotype and a reduced sensitivity to ABA. A: Twenty-five-day-old plants grown in parallel in a growth chamber at a relative humidity of ~40%. B: Four-day-old seedlings were transferred to 0.5 MS agar plates supplemented with 0 µM ABA (top row) or 30 µM ABA (bottom row) and grown for 5 additional days. C and D: Quantification of growth parameters (C) fresh weight and (D) root growth of seedlings presented in (B) 5 days after transfer to media supplemented with 0 µM ABA (blue bars) or 30 µM ABA (red bars). Data represent means ±SEM of n = 5 experiments with seven seedlings per experiment. Plants were grown and analyzed as described in [83].
Previous analyses revealed that ABAleon2.1 lines were less sensitive to ABA-mediated growth inhibition when 10 µM ABA was applied externally to in vitro grown seedlings [83]. We performed similar (root) growth inhibition analyses and transferred 4-day-old Arabidopsis seedlings to 0.5 MS media supplemented with either 0 µM ABA (control) or 30 µM ABA and grew the seedlings for additional 5 days (Fig. 2B–D). Growth on media containing 30 µM ABA reduced the overall growth, measured as fresh weight, in all investigated lines (Fig. 2C). Compared to Col-0 wild-type and in line with their role in ABA signaling, ABA-induced fresh weight reduction was less pronounced for abi1-3/YFP-ABI1 and increased for YFP-PYR1 lines. ABAleon2.1 lines were also less sensitive to ABA in this respect (Fig. 2C). In 0 µM ABA conditions, ABAleon2.1 lines exhibited a reduced root growth when compared to Col-0 wild-type, consistent with the overall reduced growth in soil (Fig. 2A, B, and D). In the presence of 30 µM ABA, root growth of ABAleon2.1 lines and abi1-3/ YFP-ABI1 was less inhibited compared to Col-0 wild-type (Fig. 2D). YFP-PYR1 roots barely grew in these conditions (Fig. 2B and D).
There are two hypotheses that could explain the growth and ABA-response phenotypes of ABAleon2.1 plants: (i) due to the high ABA affinity, ABAleon2.1 might sequester physiologically relevant ABA pools and therefore suppress ABA responses [83]. In line with this hypothesis, it was found that the degree of ABA signaling suppression by ABAleon2.1 was dependent on its expression levels (Fig. 2D; [83]); (ii) the ABI1 domain encoded in the ABAleon sensory module might affect ABA signaling, even though it carries a phosphatase inactive mutation [83]. In line with this hypothesis, various transgenic lines of low affinity ABAleon versions also exhibited a reduced growth in soil (Waadt et al., unpublished). In the future, it will be important to develop FRET-based ABA reporters with reduced impact on ABA signaling.
Applications of ABACUS and ABAleon
ABACUS and ABAleon can be utilized for various aspects of ABA research. As both reporters instantaneously respond to ABA but also to the ABA mimic Pyrabactin [82] (Waadt et al., unpublished), they can be used to screen for other ABA analogs or compounds that suppress ABA binding by PYR1/PYL1 or interfere with the ABA-induced interaction of PYR1 with ABI1. The identification of ABA analogs and the engineering of ABA receptors is important for agrochemical approaches to improve water use efficiency [88, 96, 98].
ABACUS is able to detect ABA uptake into yeast cells after co-expression with the ABA importing transporter AIT1 [82]. Screening cDNA or transporter libraries in heterologous systems provides a powerful approach for the identification of novel ABA transporters [35]. In this respect, ABACUS and ABAleon could be used as another experimental readout to identify and characterize ABA transporters.
In Arabidopsis, ABACUS and ABAleon have been initially used to study ABA uptake. A 15 minutes pulse of ABA application induced an emission ratio increase of ABACUS, which declined after the ABA supply stopped [82]. These data indicated that ABA binds to ABACUS reversibly and that ABA is either quickly metabolized or exported from the cytoplasm. Studies with ABAleon2.1 seedlings revealed that ABA uptake was faster in the root elongation – compared to the root maturation zone. ABA was transported from shoot to root and hypocotyl to shoot, but not, in live cells, from root to shoot under the imposed experimental conditions [83]. The ABA transport rate within the hypocotyl was calculated to be ~16 µm/minute. In Arabidopsis, shoot-to-root ABA transport is consistent with the observations that in response to drought the shoot was the main source for rapid de novo ABA synthesis, which was then required to be translocated to the roots [58, 99].
Rapid FRET-based hormone reporters can be used to uncover time-resolved localized hormone concentration changes in response to diverse stimuli. For example, the time course of tissue and cell specific responses to salinity, osmotic stress and humidity changes have been investigated in initial studies [82, 83]. ABACUS and ABAleon detected salt stress (100–150 mM NaCl) induced [ABA] increases in leaves and guard cells [82, 83]. Interestingly, osmotically stressed (300 mM sorbitol) excised leaves did not accumulate ABA in guard cells [83]. In roots, both salt- and osmotic-stress triggered elevations in [ABA] [83].
When ABAleon2.1 plants were subjected to a drop in relative air humidity guard cell [ABA] elevated within 15 minutes [83]. The mechanism of ABA involvement in low humidity responses is still a matter of debate. Some studies suggest that a humidity drop causes a rapid rise in guard cell [ABA] (e.g. within 1–4 minutes), which would be required if ABA mediates the rapid humidity response. Other studies suggest that the humidity response does not require a rapid rise in guard cell [ABA] [100–104]. Stomatal conductance started decreasing within 5 minutes after a relative humidity drop [103, 105]. Knowledge about the exact time frame for reduced humidity-induced ABA synthesis would be important to evaluate the importance of ABA in reduced humidity-mediated stomatal closure. One model has been discussed in which the basal ABA levels may aid in the low humidity response without a requirement for rapid [ABA] changes in guard cells [103]. It has to be noted that in guard cells, the ABA sensitivity of ABAleon2.1 is quite low. Due to the high ABA affinity of ABAleon2.1 and the high basal [ABA] in guard cells [83], ABAleon2.1 appears often saturated. In the future, direct ABA reporters with an increased dynamic range and reduced ABA affinity (e.g. ABAleon2.15 [83] and ABACUS1-2m [82]) should allow direct analyses of the dynamics of humidity-induced [ABA] changes in guard cells.
Many other biological questions can be investigated with the newly emerging rapid FRET-based ABA reporters. For example, in present models [CO2] changes that control stomatal movements suggest that the ABA and CO2 signaling pathways converge downstream of ABA synthesis and perception [106]. Rapidly responding ABA FRET-based reporters will allow future direct studies into this presently untested hypothesis and question whether [CO2] changes cause rapid changes in [ABA] in guard cells.
Distribution of ABA and ABA signaling in Arabidopsis
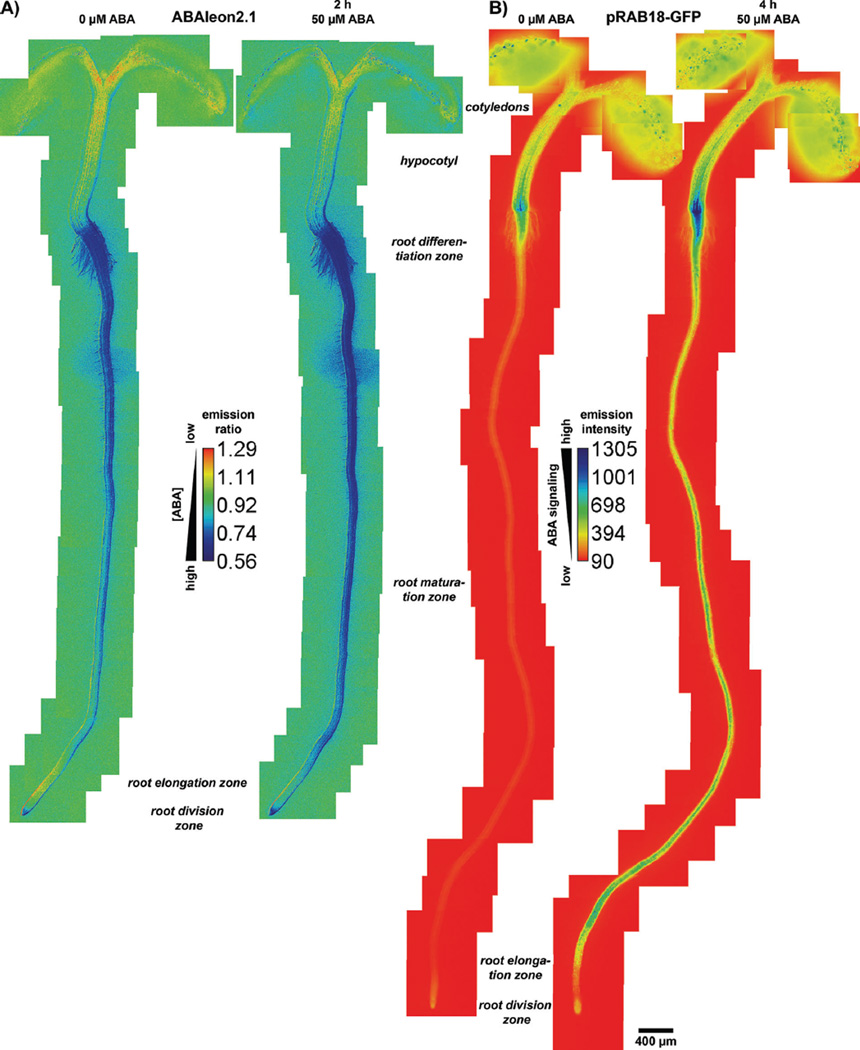
Because the FRET-based ABA reporters ABACUS and ABAleon are ratiometric and normalized, they can be used to visualize the distribution of ABA in intact and live plants. ABA distribution maps on seedlings have been generated using the highly expressing ABAleon2.1 line 3 [83]. These analyses revealed that [ABA] maxima exist in the root-tip, in the hypocotyl–root junction, and in guard cells. Furthermore, an [ABA] gradient appeared to exist in roots. ABAleon2.1 reported high [ABA] in the root base, with decreasing [ABA] toward the root-elongation zone that appeared to be a zone of minimal [ABA]. From there [ABA] increased towards the roottip [83]. These data are consistent with an ABA distribution map generated using the independent ABAleon2.1 line 10 (Fig. 3A). The [ABA] maxima reported by ABAleon2.1 are reflected in the expression pattern of NCED genes involved in ABA biosynthesis [107, 108]. High [ABA] in guard cells is also consistent with their ability to autonomously synthesize ABA [102].
Figure 3.
Distribution of (A) [ABA] and (B) ABA signaling. Five-day-old seedlings of (A) ABAleon2.1 line 10 and (B) pRAB18-GFP were imaged before (left panel) or 2 hours (ABAleon2.1) or 4 hours (pRAB18-GFP) after application of 50 µM ABA (right panel). Shown are manually assembled emission ratio (ABAleon2.1 – reports [ABA]) or background subtracted fluorescence emission (pRAB18-GFP – reports ABA signaling) images calibrated to the respective calibration bar. Blue color indicates high [ABA] or signaling and red indicates low [ABA] or signaling. Background colors are according to the respective calibration bars [values are ~1 in (A) and 0 in (B)]. Plants were grown, imaged, and analyzed as described in [83], except that imaging of pRAB18-GFP was conducted using a 71012pH sensitive GFP filter set (CHROMA Technology Corp.).
Differences in hormone concentrations do not necessarily reflect the signaling efficiency of a hormone in certain tissues or cell-types [75, 109]. For example, in Arabidopsis embryos of the heart stage [auxin] was high in the shoot apical meristem detected by the protein degradation-based reporter R2D2. However, the expression-based auxin signaling reporter DR5v2-n3GFP was not detected in the same cells [62]. These data indicate that hormone abundance not necessarily induces hormone signaling.
To compare ABA distribution and ABA signaling, ABAleon2.1 seedlings were investigated together with the expression-based reporter line pRAB18-GFP (Fig. 3). RAB18 is a gene for which expression is induced by ABA [110, 111]. The pRAB18-GFP reporter has been initially established to screen for chemical compounds that affect ABA signaling [59]. Note that in Fig. 3, high [ABA] or signaling is represented in blue color and low [ABA] and signaling in red. Similar to the ABA distribution map generated by ABAleon2.1, pRAB18-GFP reported enhanced ABA signaling in guard cells (seen as blue dots on the cotyledons) and in the hypocotyl–root junction (Fig. 3). Compared to the high [ABA] in the root tip, ABA signaling was only moderate. Striking differences between both reporters were found in the root, where pRAB18-GFP expression was at the detection limit of the microscope and ABAleon2.1 reported comparably high [ABA], except in the root-elongation zone (Fig. 3A and B, left panels). External application of 50 µM ABA resulted in ABA uptake and increased ABA signaling (Fig. 3A and B, right panels). While [ABA] increased in the entire seedling after external ABA application, ABA-induced ABA signaling was more evident in the root and the hypocotyl–root junction. Note that pRAB18-GFP responds slower to externally applied ABA, as expression of the GFP reporter gene requires time for signaling, mRNA transcription, translation, and GFP maturation. The ABA signaling map reported by pRAB18-GFP was consistent with observations using other ABA-responsive expression-based reporters (pRD29B and pAtHB6, [58]). In the future, it will be interesting to re-evaluate the [ABA] map using improved next generation ABA reporters, and to elucidate the role of the hypocotyl–root junction in ABA-mediated responses and the reason for the apparent reduction in the endogenous ABA sensitivity of roots compared to guard cells.
pRAB18-GFP reports changes in ABA signaling in response to abiotic stresses
It is important to know how certain tissues respond to stresses that affect ABA signaling. pRD29A/B-Luc and pAtHB6-Luc reporters have been used to study cold, salt, and osmotic stress responses [57, 58, 112]. These analyses provided evidence for the involvement of ABA in abiotic stress responses and led to the identification of ABA synthesis genes [113, 114]. The pRAB18-GFP reporter has been used for a chemical genetic screening and to study salt stress responses in roots and humidity responses in guard cells [59, 115–117]. As pRAB18-GFP uses a fluorescent protein as reporter gene, this reporter might have a higher potential to resolve tissue and cell-specific differences in ABA signaling.
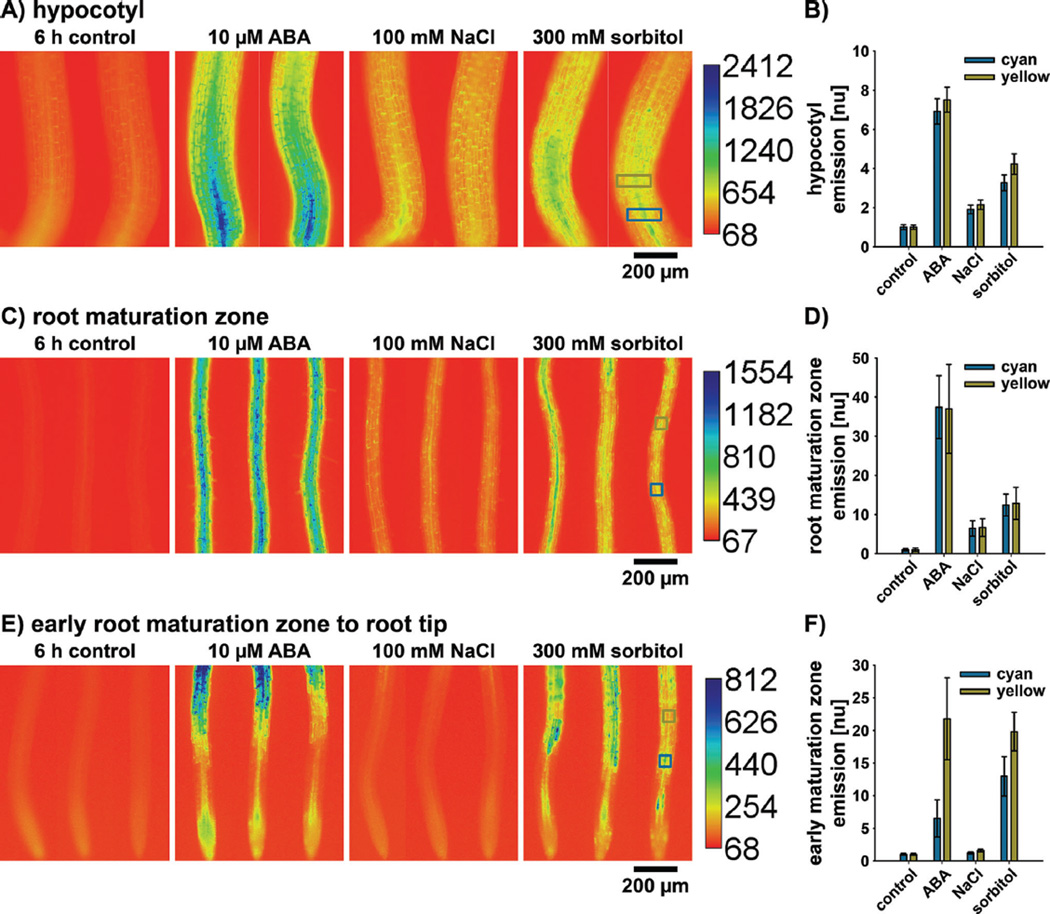
Five-day-old seedlings of the expression-based ABA signaling reporter pRAB18-GFP were incubated for 6 hours in control media and media containing 10 µM ABA, 100 mM NaCl (salt stress), or 300 mM sorbitol (osmotic stress) (Fig. 4). Analyses in hypocotyls (Fig. 4A) and two different zones of the root (Fig. 4B and C) indicated a strong reporter induction in response to 10mM ABA. In the hypocotyl and root-maturation zone, salt and osmotic stress induced pRAB18-GFP expression, but to a lesser extent compared to 10 µM ABA treatment (Fig. 4A – D). Also in these tissues, the osmotic stress response was approximately twofold higher compared to the salt stress response (Fig. 4B and D). The induction of ABA signaling in the root maturation zone in response to salt and osmotic stress was consistent with increased [ABA] measured using ABAleon2.1 [83]. In the early maturation zone, salt stress did not affect pRAB18-GFP expression, while osmotic stress responses were comparable with ABA responses (Fig. 4E and F). Note that in this region salt stress induced [ABA] increases [83], but not ABA signaling (Fig. 4E and F). The root elongation zone, which is located directly below the early maturation zone, appeared to be special in terms of ABA responses. This region exhibited the lowest [ABA] in the root (Fig. 3A), but also a weaker pRAB18-GFP induction in response to ABA and sorbitol when compared to the early maturation zone (Figs. 3 and 4E).
Figure 4.
ABA signaling in response to salt and osmotic stress. Fluorescence emission of the expression-based ABA signaling reporter pRAB18-GFP in (A and B) the hypocotyl, (C and D) the root maturation zone, and (E and F) the early maturation zone, elongation zone, and division zone in response to 6 hours control, 10 µM ABA, 100mM NaCl (salt stress), and 300 µM sorbitol (osmotic stress) treatments. Images were calibrated according to the adjacent calibration bar. B, D, and F: Background subtracted fluorescence emission values measured from two color-coded regions depicted in the right images of (A, C, and E) and normalized to the control. Data represent means ± SEM of n = 8 seedlings. The indicated color-coded regions are representative for regions analyzed in all acquired images with identical positions on the Y-axis.
In summary, the pRAB18-GFP reporter represents a useful tool to study ABA signaling in response to abiotic stresses and reveals differences in signaling intensity and distribution between salt- and osmotically stressed seedlings.
Conclusions and outlook
Plant hormones mediate various growth and developmental processes dependent on the specific natural environment and growth stage of a plant. Recent advances in fluorescent labeling of plant hormones and the development of genetically encoded reporters have opened new possibilities for researchers to study hormone uptake, distribution, signaling patterns, and hormone-mediated protein degradation in living intact plants. However, such tools remain to be established for each hormone (see Table 1). The first generation of FRET-based ABA reporters ABACUS and ABAleon affect ABA signaling, but also allow insights into the cellular dynamics and movement of ABA through the plant in real time. Future reporter engineering has the potential for many improvements, including their dynamic range and spectral characteristics. Design of reporters with non-overlapping fluorescence emission would enable the monitoring of various aspects of hormone action or multiple hormones at the same time. Using such reporters, future studies could include investigations of hormone transport, environmental responses, but also the analysis of hormone crosstalk/interdependence. In summary, the use of genetically encoded reporters will advance our understanding about plant hormones and lead to deeper knowledge of how plants grow, develop, and how they face their daily challenges with the environment.
Acknowledgments
We thank Alexander M. Jones for critical reading of the ABACUS and ABAleon comparison and Melissa Lee and Elly Poretsky (UCSD) for technical support. This work was supported by grants from the National Institutes of Health (GM060396-ES010337) and the National Science Foundation (MCB1414339) to JIS, and a fellowship from the Postdoctoral Research Abroad Program sponsored by the Ministry of Science and Technology of Taiwan (104-2917-I-564-008) to P-KH.
Abbreviations
- ABA
abscisic acid
- ABC
ATP-binding cassette
- ABI1
ABA insensitive 1
- AFCS
alexafluor 647-castasterone
- AIT
ABA-importing transporter
- ARR5
Arabidopsis response regulator 5
- AtHB6
Arabidopsis thaliana homeobox protein 6
- AUX1
auxin resistant 1
- BR
brassinosteroid
- CFP
cyan fluorescent protein
- CK
cytokinin
- DTX50
detoxification efflux carrier 50
- ELISA
enzyme-linked immunosorbent assay
- ET
ethylene
- FP
fluorescent protein
- FRET
Förster resonance energy transfer
- GA
gibberellin
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- IAA
auxin
- IAA17
IAA inducible 17
- IAA28
IAA inducible 28
- JA
jasmonate
- JAI3
jasmonate insensitive 3
- KAR
karrikin
- K’d
apparent affinity
- LAX
like AUX1
- LUC
luciferase
- NCED
9-cis-epoxycarotenoid dioxygenase
- NPF
nitrate transporter 1/peptide transporter family
- obs1
optimal binding sequence for TEIL
- p35S
cauliflower mosaic virus 35S promoter
- PDR1
pleiotropic drug resistance 1
- PP2C
protein phosphatase type 2C
- PYL1
pyrabactin resistance 1-like
- PYR1
pyrabactin resistance 1
- RAB18
responsive to ABA 18
- RCAR
regulatory component of ABA receptor
- RD29A/B
responsive to dessication 29 A/B
- RGA
repressor of GA
- RPS5A
ribosomal protein 5A
- SA
salicylic acid
- SCF
SKP1, cullin, F-box-complex
- SKP1
S-phase kinase associated protein 1
- SL
strigolactone
- TCS
two component signaling sensor
- T-DNA
transfer-DNA
- TEIL
tobacco EIN3-like
- YC3.6
yellow cameleon 3.6
- YFP
yellow fluorescent protein
Footnotes
The authors have declared no conflicts of interest.
References
- 1.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 2.Finkelstein R. Abscisic acid synthesis and response. Arabidopsis Book. 2013;11:e0166. doi: 10.1199/tab.0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peer WA. From perception to attenuation: auxin signalling and responses. Curr Opin Plant Biol. 2013;16:561–568. doi: 10.1016/j.pbi.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Wang R, Estelle M. Diversity and specificity: auxin perception and signaling through the TIR1/AFB pathway. Curr Opin Plant Biol. 2014;21:51–58. doi: 10.1016/j.pbi.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim TW, Wang ZY. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- 6.Kieber JJ, Schaller GE. Cytokinins. Arabidopsis Book. 2014;12:e0168. doi: 10.1199/tab.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaller GE, Street IH, Kieber JJ. Cytokinin and the cell cycle. Curr Opin Plant Biol. 2014;21:7–15. doi: 10.1016/j.pbi.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Merchante C, Alonso JM, Stepanova AN. Ethylene signaling: simple ligand, complex regulation. Curr Opin Plant Biol. 2013;16:554–560. doi: 10.1016/j.pbi.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Davière JM, Achard P. Gibberellin signaling in plants. Development. 2013;140:1147–1151. doi: 10.1242/dev.087650. [DOI] [PubMed] [Google Scholar]
- 10.Xu H, Liu Q, Yao T, Fu X. Shedding light on integrative GA signaling. Curr Opin Plant Biol. 2014;21:89–95. doi: 10.1016/j.pbi.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 11.Browse J. Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol. 2009;60:183–205. doi: 10.1146/annurev.arplant.043008.092007. [DOI] [PubMed] [Google Scholar]
- 12.Fonseca S, Chico JM, Solano R. The jasmonate pathway: the ligand, the receptor and the core signalling module. Curr Opin Plant Biol. 2009;12:539–547. doi: 10.1016/j.pbi.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 13.Waters MT, Scaffidi A, Flematti GR, Smith SM. The origins and mechanisms of karrikin signalling. Curr Opin Plant Biol. 2013;16:667–673. doi: 10.1016/j.pbi.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Kumar D. Salicylic acid signaling in disease resistance. Plant Sci. 2014;228:127–134. doi: 10.1016/j.plantsci.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 15.Yan S, Dong X. Perception of the plant immune signal salicylic acid. Curr Opin Plant Biol. 2014;20:64–68. doi: 10.1016/j.pbi.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett T, Leyser O. Strigolactone signalling: standing on the shoulders of DWARFs. Curr Opin Plant Biol. 2014;22:7–13. doi: 10.1016/j.pbi.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 18.Muday GK, Rahman A, Binder BM. Auxin and ethylene: collaborators or competitors? Trends Plant Sci. 2012;17:181–195. doi: 10.1016/j.tplants.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Vanstraelen M, Benková E. Hormonal interactions in the regulation of plant development. Annu Rev Cell Dev Biol. 2012;28:463–487. doi: 10.1146/annurev-cellbio-101011-155741. [DOI] [PubMed] [Google Scholar]
- 20.Song S, Qi T, Wasternack C, Xie D. Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr Opin Plant Biol. 2014;21:112–119. doi: 10.1016/j.pbi.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Wang W, Bai MY, Wang ZY. The brassinosteroid signaling network - a paradigm of signal integration. Curr Opin Plant Biol. 2014;21:147–153. doi: 10.1016/j.pbi.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamaguchi S, Kamiya Y. Gibberellin biosynthesis: its regulation by endogenous and environmental signals. Plant Cell Physiol. 2000;41:251–257. doi: 10.1093/pcp/41.3.251. [DOI] [PubMed] [Google Scholar]
- 23.Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- 24.Yue J, Hu X, Huang J. Origin of plant auxin biosynthesis. Trends Plant Sci. 2014;19:764–770. doi: 10.1016/j.tplants.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Dempsey DA, Vlot AC, Wildermuth MC, Klessig DF. Salicylic acid biosynthesis and metabolism. Arabidopsis Book. 2011;9:e0156. doi: 10.1199/tab.0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ha S, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, et al. Cytokinins: metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 2012;17:172–179. doi: 10.1016/j.tplants.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Saito H, Oikawa T, Hamamoto S, Ishimaru Y, et al. The jasmonate-responsive GTR1 transporter is required for gibberellin-mediated stamen development in Arabidopsis. Nat Commun. 2015;6:6095. doi: 10.1038/ncomms7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Léran S, Varala K, Boyer JC, Chiurazzi M, et al. A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 2014;19:5–9. doi: 10.1016/j.tplants.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 29.Chiba Y, Shimizu T, Miyakawa S, Kanno Y, et al. Identification of Arabidopsis thaliana NRT1/PTR FAMILY (NPF) proteins capable of transporting plant hormones. J Plant Res. 2015;128:679–686. doi: 10.1007/s10265-015-0710-2. [DOI] [PubMed] [Google Scholar]
- 30.Petrásek J, Friml J. Auxin transport routes in plant development. Development. 2009;136:2675–2688. doi: 10.1242/dev.030353. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y, Hammes UZ, Taylor CG, Schachtman DP, et al. High-affinity auxin transport by the AUX1 influx carrier protein. Curr Biol. 2006;16:1123–1127. doi: 10.1016/j.cub.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 32.Krouk G, Lacombe B, Bielach A, Perrine-Walker F, et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell. 2010;18:927–937. doi: 10.1016/j.devcel.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 33.Boursiac Y, Léran S, Corratgé-Faillie C, Gojon A, et al. ABA transport and transporters. Trends Plant Sci. 2013;18:325–333. doi: 10.1016/j.tplants.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 34.Kang J, Hwang JU, Lee M, Kim YY, et al. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc Natl Acad Sci USA. 2010;107:2355–2360. doi: 10.1073/pnas.0909222107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanno Y, Hanada A, Chiba Y, Ichikawa T, et al. Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc Natl Acad Sci USA. 2012;109:9653–9658. doi: 10.1073/pnas.1203567109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuromori T, Miyaji T, Yabuuchi H, Shimizu H, et al. ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc Natl Acad Sci USA. 2010;107:2361–2366. doi: 10.1073/pnas.0912516107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, Zhu H, Pan Y, Yu Y, et al. A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates ABA sensitivity and drought tolerance in Arabidopsis. Mol Plant. 2014;7:1522–1532. doi: 10.1093/mp/ssu063. [DOI] [PubMed] [Google Scholar]
- 38.Ko D, Kang J, Kiba T, Park J, et al. Arabidopsis ABCG14 is essential for the root-to-shoot translocation of cytokinin. Proc Natl Acad Sci USA. 2014;111:7150–7155. doi: 10.1073/pnas.1321519111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang K, Novak O, Wei Z, Gou M, et al. Arabidopsis ABCG14 protein controls the acropetal translocation of root-synthesized cytokinins. Nat Commun. 2014;5:3274. doi: 10.1038/ncomms4274. [DOI] [PubMed] [Google Scholar]
- 40.Kretzschmar T, Kohlen W, Sasse J, Borghi L, et al. A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature. 2012;483:341–344. doi: 10.1038/nature10873. [DOI] [PubMed] [Google Scholar]
- 41.Symons GM, Ross JJ, Jager CE, Reid JB. Brassinosteroid transport. J Exp Bot. 2008;59:17–24. doi: 10.1093/jxb/erm098. [DOI] [PubMed] [Google Scholar]
- 42.Santner A, Estelle M. Recent advances and emerging trends in plant hormone signalling. Nature. 2009;459:1071–1078. doi: 10.1038/nature08122. [DOI] [PubMed] [Google Scholar]
- 43.Shan X, Yan J, Xie D. Comparison of phytohormone signaling mechanisms. Curr Opin Plant Biol. 2012;15:84–91. doi: 10.1016/j.pbi.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 44.Fu ZQ, Dong X. Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol. 2013;64:839–863. doi: 10.1146/annurev-arplant-042811-105606. [DOI] [PubMed] [Google Scholar]
- 45.Müller A, Düchting P, Weiler EW. A multiplex GC-MS/MS technique for the sensitive and quantitative single-run analysis of acidic phytohormones and related compounds, and its application to Arabidopsis thaliana . Planta. 2002;216:44–56. doi: 10.1007/s00425-002-0866-6. [DOI] [PubMed] [Google Scholar]
- 46.Forcat S, Bennett MH, Mansfield JW, Grant MR. A rapid and robust method for simultaneously measuring changes in the phytohormones ABA, JA and SA in plants following biotic and abiotic stress. Plant Methods. 2008;4:16. doi: 10.1186/1746-4811-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kojima M, Kamada-Nobusada T, Komatsu H, Takei K, et al. Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol. 2009;50:1201–1214. doi: 10.1093/pcp/pcp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiler EW. Immunoassay of plant growth regulators. Annu Rev Plant Physiol. 1984;35:85–95. [Google Scholar]
- 49.Davis GC, Hein MB, Neely BC, Sharp CR, et al. Strategies for the determination of plant hormones. Anal Chem. 1985;57:638–648. [Google Scholar]
- 50.Weiler EW, Schnabl H, Hornberg C. Stress-related levels of abscisic acid in guard cell protoplasts of Vicia faba L. Planta. 1982;154:24–28. doi: 10.1007/BF00385492. [DOI] [PubMed] [Google Scholar]
- 51.Irani NG, Di Rubbo S, Mylle E, Van den Begin J, et al. Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat Chem Biol. 2012;8:583–589. doi: 10.1038/nchembio.958. [DOI] [PubMed] [Google Scholar]
- 52.Gao Y, Yu Y, Hu X, Cao Y, et al. Imaging of jasmonic acid binding sites in tissue. Anal Biochem. 2013;440:205–211. doi: 10.1016/j.ab.2013.04.032. [DOI] [PubMed] [Google Scholar]
- 53.Shani E, Weinstain R, Zhang Y, Castillejo C, et al. Gibberellins accumulate in the elongating endodermal cells of Arabidopsis root. Proc Natl Acad Sci USA. 2013;110:4834–4839. doi: 10.1073/pnas.1300436110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sokołowska K, Kizińska J, Szewczuk Z, Banasiak A. Auxin conjugated to fluorescent dyes-a tool for the analysis of auxin transport pathways. Plant Biol (Stuttg) 2014;16:866–877. doi: 10.1111/plb.12144. [DOI] [PubMed] [Google Scholar]
- 55.Hayashi K, Nakamura S, Fukunaga S, Nishimura T, et al. Auxin transport sites are visualized in planta using fluorescent auxin analogs. Proc Natl Acad Sci USA. 2014;111:11557–11562. doi: 10.1073/pnas.1408960111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waadt R. Plant hormones: on-the-spot reporting. Nat Plants. 2015;1:15001. doi: 10.1038/nplants.2015.1. [DOI] [PubMed] [Google Scholar]
- 57.Ishitani M, Xiong L, Stevenson B, Zhu JK. Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell. 1997;9:1935–1949. doi: 10.1105/tpc.9.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Christmann A, Hoffmann T, Teplova I, Grill E, et al. Generation of active pools of abscisic acid revealed by in vivo imaging of water-stressed Arabidopsis. Plant Physiol. 2005;137:209–219. doi: 10.1104/pp.104.053082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim TH, Hauser F, Ha T, Xue S, et al. Chemical genetics reveals negative regulation of abscisic acid signaling by a plant immune response pathway. Curr Biol. 2011;21:990–997. doi: 10.1016/j.cub.2011.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oono Y, Chen QG, Overvoorde PJ, Köhler C, et al. age Mutants of Arabidopsis exhibit altered auxin-regulated gene expression. Plant Cell. 1998;10:1649–1662. doi: 10.1105/tpc.10.10.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liao CY, Smet W, Brunoud G, Yoshida S, et al. Reporters for sensitive and quantitative measurement of auxin response. Nat Methods. 2015;12:207–210. doi: 10.1038/nmeth.3279. 2 p following 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D’Agostino IB, Deruère J, Kieber JJ. Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 2000;124:1706–1717. doi: 10.1104/pp.124.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Müller B, Sheen J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature. 2008;453:1094–1097. doi: 10.1038/nature06943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zürcher E, Tavor-Deslex D, Lituiev D, Enkerli K, et al. A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol. 2013;161:1066–1075. doi: 10.1104/pp.112.211763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kosugi S, Ohashi Y. Cloning and DNA-binding properties of a tobacco Ethylene-Insensitive3 (EIN3) homolog. Nucleic Acids Res. 2000;28:960–967. doi: 10.1093/nar/28.4.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vom Endt D, Soarese Silva M, Kijne JW, Pasquali G, et al. Identification of a bipartite jasmonate-responsive promoter element in the Catharanthus roseus ORCA3 transcription factor gene that interacts specifically with AT-Hook DNA-binding proteins. Plant Physiol. 2007;144:1680–1689. doi: 10.1104/pp.107.096115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rocher A, Dumas C, Cock JM. A W-box is required for full expression of the SA-responsive gene SFR2. Gene. 2005;344:181–192. doi: 10.1016/j.gene.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 69.Hruz T, Laule O, Szabo G, Wessendorp F, et al. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Ruijter NCA, Verhees J, van Leeuwen W, van der Krol AR. Evaluation and comparison of the GUS, LUC and GFP reporter system for gene expression studies in plants. Plant Biol. 2003;5:103–115. [Google Scholar]
- 71.Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- 72.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 73.Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 74.Chinnusamy V, Stevenson B, Lee BH, Zhu JK. Screening for gene regulation mutants by bioluminescence imaging. Sci STKE. 2002;2002:pl10. doi: 10.1126/stke.2002.140.pl10. [DOI] [PubMed] [Google Scholar]
- 75.Brunoud G, Wells DM, Oliva M, Larrieu A, et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482:103–106. doi: 10.1038/nature10791. [DOI] [PubMed] [Google Scholar]
- 76.Wend S, Dal Bosco C, Kämpf MM, Ren F, et al. A quantitative ratiometric sensor for time-resolved analysis of auxin dynamics. Sci Rep. 2013;3:2052. doi: 10.1038/srep02052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fu X, Harberd NP. Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature. 2003;421:740–743. doi: 10.1038/nature01387. [DOI] [PubMed] [Google Scholar]
- 78.Chini A, Fonseca S, Fernández G, Adie B, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 79.Larrieu A, Champion A, Legrand J, Lavenus J, et al. A fluorescent hormone biosensor reveals the dynamics of jasmonate signalling in plants. Nat Commun. 2015;6:6043. doi: 10.1038/ncomms7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O’Connor N, Silver RB. Ratio imaging: practical considerations for measuring intracellular Ca2+ and pH in living cells. Methods Cell Biol. 2013;114:387–406. doi: 10.1016/B978-0-12-407761-4.00016-6. [DOI] [PubMed] [Google Scholar]
- 81.Band LR, Wells DM, Larrieu A, Sun J, et al. Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proc Natl Acad Sci USA. 2012;109:4668–4673. doi: 10.1073/pnas.1201498109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jones AM, Danielson JA, Manojkumar SN, Lanquar V, et al. Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. Elife. 2014;3:e01741. doi: 10.7554/eLife.01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Waadt R, Hitomi K, Nishimura N, Hitomi C, et al. FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. Elife. 2014;3:e01739. doi: 10.7554/eLife.01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Choi WG, Gilroy S. Plant biologists FRET over stress. Elife. 2014;3:e02763. doi: 10.7554/eLife.02763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 86.Pollok BA, Heim R. Using GFP in FRET-based applications. Trends Cell Biol. 1999;9:57–60. doi: 10.1016/s0962-8924(98)01434-2. [DOI] [PubMed] [Google Scholar]
- 87.Ma Y, Szostkiewicz I, Korte A, Moes D, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 88.Park SY, Fung P, Nishimura N, Jensen DR, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Melcher K, Ng LM, Zhou XE, Soon FF, et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature. 2009;462:602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Miyazono K, Miyakawa T, Sawano Y, Kubota K, et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- 91.Nagai T, Yamada S, Tominaga T, Ichikawa M, et al. Expanded dynamic range of fluorescent indicators for Ca(2+) by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci USA. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dupeux F, Santiago J, Betz K, Twycross J, et al. A thermodynamic switch modulates abscisic acid receptor sensitivity. EMBO J. 2011;30:4171–4184. doi: 10.1038/emboj.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Santiago J, Rodrigues A, Saez A, Rubio S, et al. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009;60:575–588. doi: 10.1111/j.1365-313X.2009.03981.x. [DOI] [PubMed] [Google Scholar]
- 94.Joshi-Saha A, Valon C, Leung J. A brand new START: abscisic acid perception and transduction in the guard cell. Sci Signal. 2011;4:re4. doi: 10.1126/scisignal.2002164. [DOI] [PubMed] [Google Scholar]
- 95.Zhang X, Jiang L, Wang G, Yu L, et al. Structural insights into the abscisic acid stereospecificity by the ABA receptors PYR/PYL/RCAR. PLoS ONE. 2013;8:e67477. doi: 10.1371/journal.pone.0067477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mosquna A, Peterson FC, Park SY, Lozano-Juste J, et al. Potent and selective activation of abscisic acid receptors in vivo by mutational stabilization of their agonist-bound conformation. Proc Natl Acad Sci USA. 2011;108:20838–20843. doi: 10.1073/pnas.1112838108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nishimura N, Sarkeshik A, Nito K, Park SY, et al. PYR/PYL/ RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 2010;61:290–299. doi: 10.1111/j.1365-313X.2009.04054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Park SY, Peterson FC, Mosquna A, Yao J, et al. Agrochemical control of plant water use using engineered abscisic acid receptors. Nature. 2015;520:545–548. doi: 10.1038/nature14123. [DOI] [PubMed] [Google Scholar]
- 99.Ikegami K, Okamoto M, Seo M, Koshiba T. Activation of abscisic acid biosynthesis in the leaves of Arabidopsis thaliana in response to water deficit. J Plant Res. 2009;122:235–243. doi: 10.1007/s10265-008-0201-9. [DOI] [PubMed] [Google Scholar]
- 100.Assmann SM, Snyder JA, Lee Y-RJ. ABA-deficient (aba1) and ABA-insensitive (abi1-1, abi2-1) mutants of Arabidopsis have a wild-type stomatal response to humidity. Plant Cell Environ. 2000;23:387–395. [Google Scholar]
- 101.Xie X, Wang Y, Williamson L, Holroyd GH, et al. The identification of genes involved in the stomatal response to reduced atmospheric relative humidity. Curr Biol. 2006;16:882–887. doi: 10.1016/j.cub.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 102.Bauer H, Ache P, Lautner S, Fromm J, et al. The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Curr Biol. 2013;23:53–57. doi: 10.1016/j.cub.2012.11.022. [DOI] [PubMed] [Google Scholar]
- 103.Merilo E, Laanemets K, Hu H, Xue S, et al. PYR/RCAR receptors contribute to ozone-, reduced air humidity-, darkness-, and CO2-induced stomatal regulation. Plant Physiol. 2013;162:1652–1668. doi: 10.1104/pp.113.220608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Merilo E, Jalakas P, Kollist H, Brosché M. The role of ABA recycling and transporter proteins in rapid stomatal responses to reduced air humidity, elevated CO2, and exogenous ABA. Mol Plant. 2015;8:657–659. doi: 10.1016/j.molp.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 105.Vahisalu T, Kollist H, Wang YF, Nishimura N, et al. SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature. 2008;452:487–491. doi: 10.1038/nature06608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim TH, Böhmer M, Hu H, Nishimura N, et al. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol. 2010;61:561–591. doi: 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tan BC, Joseph LM, Deng WT, Liu L, et al. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003;35:44–56. doi: 10.1046/j.1365-313x.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- 108.Jones AM. A new look at stress: abscisic acid patterns and dynamics at high-resolution. New Phytol. 2015 doi: 10.1111/nph.13552. in press. [DOI] [PubMed] [Google Scholar]
- 109.Vernoux T, Brunoud G, Farcot E, Morin V, et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol. 2011;7:508. doi: 10.1038/msb.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lång V, Palva ET. The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol Biol. 1992;20:951–962. doi: 10.1007/BF00027165. [DOI] [PubMed] [Google Scholar]
- 111.Leonhardt N, Kwak JM, Robert N, Waner D, et al. Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell. 2004;16:596–615. doi: 10.1105/tpc.019000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Christmann A, Weiler EW, Steudle E, Grill E. A hydraulic signal in root-to-shoot signalling of water shortage. Plant J. 2007;52:167–174. doi: 10.1111/j.1365-313X.2007.03234.x. [DOI] [PubMed] [Google Scholar]
- 113.Xiong L, Ishitani M, Lee H, Zhu JK. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell. 2001;13:2063–2083. doi: 10.1105/TPC.010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xiong L, Lee H, Ishitani M, Zhu JK. Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J Biol Chem. 2002;277:8588–8596. doi: 10.1074/jbc.M109275200. [DOI] [PubMed] [Google Scholar]
- 115.Duan L, Dietrich D, Ng CH, Chan PM, et al. Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell. 2013;25:324–341. doi: 10.1105/tpc.112.107227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Geng Y, Wu R, Wee CW, Xie F, et al. A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell. 2013;25:2132–2154. doi: 10.1105/tpc.113.112896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Merilo E, Jalakas P, Laanemets KM, et al. Abscisic acid transport and homeostasis in the context of stomatal regulation. Mol Plant. 2015;8:1321–1333. doi: 10.1016/j.molp.2015.06.006. [DOI] [PubMed] [Google Scholar]