Abstract
Chemotaxis is important for Helicobacter pylori to colonize the stomach. Like other bacteria, H. pylori uses chemoreceptors and conserved chemotaxis proteins to phosphorylate the flagellar rotational response regulator, CheY, and modulate the flagellar rotational direction. Phosphorylated CheY is returned to its non-phosphorylated state by phosphatases such as CheZ. In previously studied cases, chemotaxis phosphatases localize to the cellular poles by interactions with either the CheA chemotaxis kinase or flagellar motor proteins. We report here that the H. pylori CheZ, CheZHP, localizes to the poles independently of the flagellar motor, CheA, and all typical chemotaxis proteins. Instead, CheZHP localization depends on the chemotaxis regulatory protein ChePep and reciprocally, ChePep requires CheZHP for its polar localization. We furthermore show that these proteins interact directly. Functional domain mapping of CheZHP determined the polar localization motif lies within the central domain of the protein, and that the protein has regions outside of the active site that participate in chemotaxis. Our results suggest that CheZHP and ChePep form a distinct complex. These results therefore suggest the intriguing idea that some phosphatases localize independently of the other chemotaxis and motility proteins, possibly to confer unique regulation on these proteins’ activities.
Introduction
Chemotaxis is the ability to sense external environmental cues and respond by moving toward beneficial situations and away from harmful ones. Bacteria utilize chemotaxis to colonize a variety of habitats, including the mammalian host (Josenhans and Suerbaum, 2002; Miller et al., 2009). Bacterial chemotaxis depends on a set of signal transduction proteins comprised of chemoreceptors and chemotaxis signal transduction proteins (Wadhams and Armitage, 2004). These proteins typically localize to the bacterial pole in a supermolecular cluster with extensive interactions between chemoreceptors and chemotaxis signaling proteins (Sourjik and Armitage, 2010). These interactions serve to amplify small signals, allow integration of multiple chemoreceptors, and provide protein-protein regulatory contacts. To date, all chemotactic microbes examined display clusters of chemotaxis proteins, supporting the importance of this organization (Briegel et al., 2009).
Helicobacter pylori is a motile human gastric pathogen that relies on chemotaxis to colonize mammalian stomachs (Foynes et al., 2000; Terry et al., 2005; Lertsethtakarn et al., 2011). H. pylori infection results in ulcers and gastric cancer, and affects millions worldwide (Polk and Peek, 2010; Salama et al., 2013). H. pylori swims utilizing a cluster of 3–7 flagella localized to one pole. The chemotaxis signal transduction system of H. pylori likely localizes to the flagellar pole, based on studies with many bacteria including the related one, Helicobacter hepaticus (Briegel et al., 2009). The composition of the H. pylori chemotaxis signal transduction system is comparable to that of the model bacterium Escherichia coli (Lertsethtakarn et al., 2011). Both organisms utilize specific chemoreceptors to sense their environments. H. pylori has four chemo-receptors—TlpA, TlpB, TlpC and TlpD—that have been reported to sense arginine, bicarbonate (Cerda et al., 2003), pH (Croxen et al., 2006; Sweeney et al., 2012), the quorum sensing molecule, autoinducer-2 (AI-2) (Rader et al., 2011) and energy (Schweinitzer et al., 2008). The receptors transmit ligand-binding information via coupling proteins, CheW or CheV, to the CheA histidine kinase, which is sometimes called CheAY in H. pylori because it has a receiver (REC) domain fused at the C-terminus. H. pylori possesses more coupling proteins than E. coli: one CheW and three CheV proteins. CheV proteins are chimeras of CheW and a phosphorylatable REC domain (Pittman et al., 2001; Lowenthal, Simon, et al., 2009; Alexander et al., 2010). CheAY phosphorylates the response regulator, CheY, which consists of a REC domain (Jiménez-Pearson et al., 2005; Lertsethtakarn and Ottemann, 2010). Phosphorylated CheY (CheY-P) interacts with the flagellar motor to cause clockwise flagellar rotation and bacterial reversals, as opposed to straight swimming when CheY is non-phosphorylated. CheY-P is returned to the non-phosphorylated state by both its own auto-dephosphorylation, as well as the action of phosphatases. In H. pylori, the only known phosphatase is CheZ, called CheZHP in this system.
In addition to the proteins mentioned above, H. pylori possesses a chemotaxis protein called ChePep, which is found only in the Epsilon proteobacteria (Howitt et al., 2011). ChePep was previously annotated as a hypothetical poly E-rich protein, and like most of the H. pylori chemotaxis genes, is encoded in an operon without other chemotaxis genes. ChePep is critical for efficient chemotaxis (Howitt et al., 2011). ChePep deletion mutants migrate poorly through soft agar, displaying a ~ 25% reduction in soft agar colony diameter (Howitt et al., 2011). Additionally, they display about 11-times greater reversals than wild type, a phenotype that is dependent on CheY. This finding suggests that ChePep is required for the efficient dephosphorylation of CheY-P, similar to CheZHP (Howitt et al., 2011). ChePep has a preponderance of glutamic acids and an N-terminal REC domain of unknown function, but otherwise contains no recognizable domains. It localizes to the bacterial pole by an as-yet unknown mechanism (Howitt et al., 2011). Specifically, ChePep is found at only the flagellar pole in short, recently divided cells, and then is seen at both poles as the cells elongate before division (Howitt et al., 2011).
The return of CheY-P to its non-phosphorylated state is a critical aspect of chemotaxis because this action allows the system to reset and the bacteria to respond to new signals. Across different microbes, CheY-P dephosphorylation is promoted by either specific phosphatases or by alternate CheA kinase targets (Wadhams and Armitage, 2004; Silversmith, 2010). The first identified and best-studied chemotaxis phosphatase is CheZ, although there are several other types of phosphatases, including CheC, FliY, and CheX that all share mechanisms similar to that of CheZ (Hess et al., 1988; Zhao et al., 2002; Silversmith, 2010). Regardless of the exact phosphatase, phosphatase activity is generally restricted to one cellular location to prevent formation of a CheY-P gradient throughout the cell (Rao et al., 2005; Lipkow, 2006). This localization, in turn, allows all flagellar complexes—spread throughout the cell in the peritrichously flagellated E. coli—to receive the same signal and therefore rotate all motors in the same direction. In support of this idea, CheZ and CheC are localized to the chemotaxis signaling complex while another phosphatase, FliY, localizes to the flagellar motor (Rao et al., 2005). E. coli CheZ localizes to the chemotaxis cluster via an interaction with both CheA and a variant of CheA called CheA short (CheAs) that results from internal translation initiation (Wang and Matsumura, 1996; Cantwell et al., 2003; Kentner and Sourjik, 2009). This form of CheA lacks the first 97 amino acids, a truncation that enhances a weak full-length CheA-CheZ interaction. FliY, on the other hand, appears to localize via interactions with the FliM and FliN components of the flagellar motor (Szurmant et al., 2003). These studies thus show that phosphatases localize at either the input of the chemotaxis system (the chemotaxis signaling complex), or to the output at the flagellar motor.
H. pylori relies on a CheZHP for CheY-P dephosphorylation, but CheZ proteins share only small regions of homology when compared across different bacterial families, making it difficult to identify them and define functional regions (Terry et al., 2006; Wuichet et al., 2007; Lertsethtakarn and Ottemann, 2010). The main region of homology between different CheZ proteins is located in the C-terminal half. This region contains the active site aspartate and glutamine (D143/189 and Q147/193 in E. coli CheZ and CheZHP, respectively) and a CheY-P binding peptide within the last 12 amino acids (Fig. 1A). Lertsethtakarn and Ottemann used purified CheZHP in several in vitro assays to determine that the protein has phosphatase activity that was dependent on D189, Q193, and the C-terminal 12 amino acids, suggesting it used the same mechanism as E. coli CheZ (Lertsethtakarn and Ottemann, 2010). CheZHP was able to dephosphorylate CheY-P as E. coli CheZ does, but additionally dephosphorylated the REC-domain proteins CheV2 and CheAY (Lertsethtakarn and Ottemann, 2010). While the C-terminal half of CheZHP is moderately conserved, the N-terminal 140 amino acids of CheZHP shows low amino acid homology with E. coli CheZ (Terry et al., 2006; Lertsethtakarn and Ottemann, 2010), suggesting that the functions of this region may not be conserved. Of note, this region is longer by approximately 30 amino acids as compared to E. coli’s, and bears multiple repeats of lysine and glutamic acid, making the protein very charged (Fig. 1A). In E. coli CheZ, increased phosphatase activity gain of function mutations all map to the N-terminal region (Sanna and Simon, 1996). Located within this N-terminal region, specifically amino acids 70–133, is the portion of E. coli CheZ that is required for interaction with CheAs and subcellular localization (Cantwell et al., 2003).
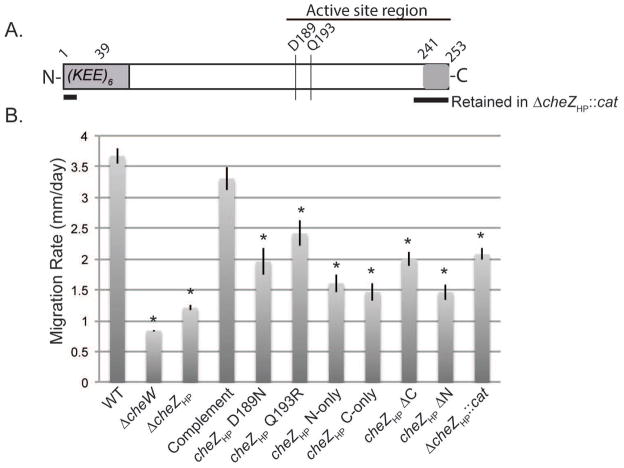
Figure 1.
(A) Schematic of CheZHP protein. The active site region is indicated by a horizontal line above. The N terminal region (1–39) contains six copies of a highly charged amino acid se- quence (KEE). The active site residues are indicated by vertical lines (D189 and Q193R). The C terminal region (241–253) binds CheY-P. The portions retained in the original ΔcheZHP::cat allele are shown with thick horizontal lines below the CheZHP schematic. (B) Soft agar migration rates of H. pylori G27 wild type (WT), ΔcheW, and cheZHP isogenic mutants. Strains were stabbed into Brucella broth-FBS soft agar, and the diameter of the expanded colony measured after 4–5 days. The data represents the average of at least two biological replicates with at least three technical replicates. Error bars show standard error. * indicates significantly different from WT (P value <0.05) using Student’s t test.
Due to the poor conservation of CheZHP in general, we questioned whether CheZHP would localize to the chemotaxis signaling cluster. To place CheZHP cellular localization in the context of H. pylori chemotaxis system, we also determined the cellular localization of H. pylori core chemotaxis proteins. We found that CheZHP localizes to the bacterial pole as do other H. pylori chemotaxis signaling proteins, but surprisingly, CheZHP polar localization is independent of known chemotaxis and flagellar-related proteins. Instead, CheZHP polar localization depends on the chemotaxis regulatory protein, ChePep (Howitt et al., 2011). ChePep localization furthermore depends on CheZHP. Functional domain mapping of CheZHP determined the polar localization motif lies within the central domain of the protein, and that the protein has regions outside of the active site that contribute to chemotactic function. This unexpected localization pattern of CheZHP and ChePep suggest that they form a protein complex that is distinct from the chemoreceptors and flagellar motor, suggesting that H. pylori localizes its phosphatase not at the input or output of chemotaxis as do other known phosphatases, but at a third location.
Results
CheZHP controls swimming reversals and possesses multiple functional regions
In vitro, CheZHP has been demonstrated to dephosphorylate CheY-P, as expected for a CheY phosphatase (Lertsethtakarn and Ottemann, 2010). Its reported in vivo behavior, however, did not match this activity: cheZHP mutants were shown to be straight swimming-biased by Terry et al., instead of the predicted hyper-reversal behavior associated with elevated CheY-P (Terry et al., 2006). The cheZHP allele used by Terry et al. was a partial deletion mutant that replaced the majority of cheZHP with a cat gene, but retained coding potential for CheZ amino acids 1–13 and 239–253 (Fig. 1A) (Terry et al., 2006). The N-terminal peptide has no known function or homology, but the C-terminal region of CheZ contains a conserved CheY-P binding sequence within the last 12 amino acids (Blat and Eisenbach, 1996; Lertsethtakarn and Ottemann, 2010). We hypothesized that these regions might affect H. pylori chemotactic ability, possibly via interactions with other chemotaxis proteins, so we first set out to create a complete cheZHP deletion, and to analyze possible roles of other regions of CheZHP.
cheZHP mutants were created by replacing the endogenous chromosomal copy of cheZHP with various mutant alleles, creating unmarked mutations that were under wild-type cheZHP transcriptional control. Deletion of the entire cheZHP coding region (ΔcheZHP) resulted in a strain that migrated poorly through Brucella Broth-FBS soft agar, with a similar degree of defect as other fully non-chemotactic strains such as a full deletion of cheW (Fig. 1B). When a wild-type copy of cheZHP was introduced back into the original locus (complement), soft agar migration ability was restored to levels that were equivalent to wild type, supporting that the cheZHP mutation caused the soft agar chemotaxis phenotype (Fig. 1B). The original cheZHP mutant (ΔcheZHP::cat), which retained CheZHP amino acids 1–13 and 239–253, was able to migrate somewhat better in the soft agar assay as compared to the ΔcheZHP strain (Fig. 1B).
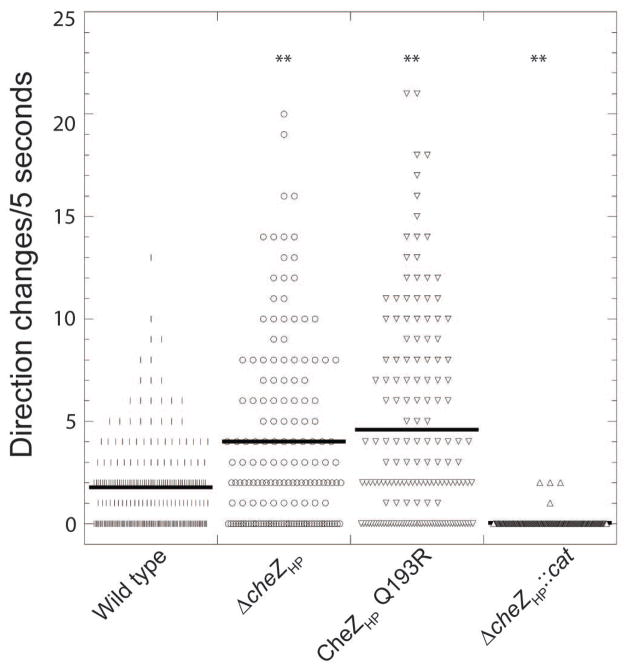
We next examined the swim behavior of the complete cheZHP deletion (ΔcheZHP). We filmed swimming H. pylori and counted the number of direction switches over a five second swim period. Using this method, we found that wild-type H. pylori displayed approximately two direction changes in five seconds (21.6 per minute), while the ΔcheZHP mutant had a statistically significant 2-fold increase in the number of direction changes (4 per five seconds or 48 per minute) (Fig. 2). As described before, strains bearing the ΔcheZHP::cat allele almost never changed direction (Fig. 2). These results suggest several things. First, the original ΔcheZHP::cat allele is not a null allele, while the full deletion (ΔcheZHP) is. Second, loss of cheZHP leads to elevated CheY-P, based on the increase in bacterial reversals. Third, the regions of CheZHP retained in the ΔcheZHP::cat allele retain some ability to function in the chemotaxis pathway. Specifically, they enhance chemotactic migration and promote straight swimming behavior, possibly via an actual function or ability to interact with particular chemotaxis proteins.
Figure 2.
Swimming behavior of cheZHP mutant strains. H. pylori G27 cells in Brucella Broth with FBS (BB10) were filmed using microscopy, and then the number of directional changes in five seconds were counted. The number of examined cells (n) and average directional changes per cell (indicated by black solid lines) are as follow: WT (n = 230, 1.8), ΔcheZHP (n =156, 4.0), and cheZHP Q193R (n = 155, 4.6), cheZHP N-only (n=186, 7.4), ΔcheZHP::cat (n=211, 0.03). At least two biological replicates were used for each strain. ** indicates significantly different from wild type (P value <0.01) using Student’s t test.
We then expanded our analysis to explore additional CheZHP alleles. CheZHP conserves two main regions compared to E. coli CheZ, both of which were shown experimentally to be required for phosphatase activity in vitro (Lertsethtakarn and Ottemann, 2010): the region including the active site residues of D189 and Q193, and the 12 amino acid C-terminal CheY-P binding region (CheZHP 241–253) (Fig. 1A). Mutants that altered the CheZHP active site (D189N or Q193R) created strains that behaved similar to cheZHP null mutants: they displayed hyper-reversal behavior (Fig. 2), and migrated poorly through the soft agar (Fig. 1B), although not as poorly as complete nulls. Combining these findings with previous in vitro work that showed that CheZHP D189N and Q193R lose phosphatase activity (Lertsethtakarn and Ottemann, 2010), suggests that these mutants lose phosphatase activity in vivo, but retain some function, perhaps in interactions with other parts of the chemotaxis signaling pathway that enhance soft agar migration.
To further home in on the regions of CheZHP that promote chemotactic function, we analyzed the soft-agar phenotypes of several additional truncated mutants. These included a variant that removed the 39 amino acids at the N-terminus (CheZHP ΔN39), the last 12 amino acids (CheZ ΔC12), one that retained only the first 39 amino acids (CheZHP N-only), and one that retained only the last 12 amino acids (CheZHP C-only). Interestingly, each of these strains was able to migrate better than a strain with the ΔcheZHP null allele, although all were defective compared to wild type (Fig. 1B). Overall, these results are consistent with the idea that both the N and C terminal regions of CheZHP contribute to H. pylori s overall chemotactic ability.
H. pylori core chemotaxis proteins form a polar cluster
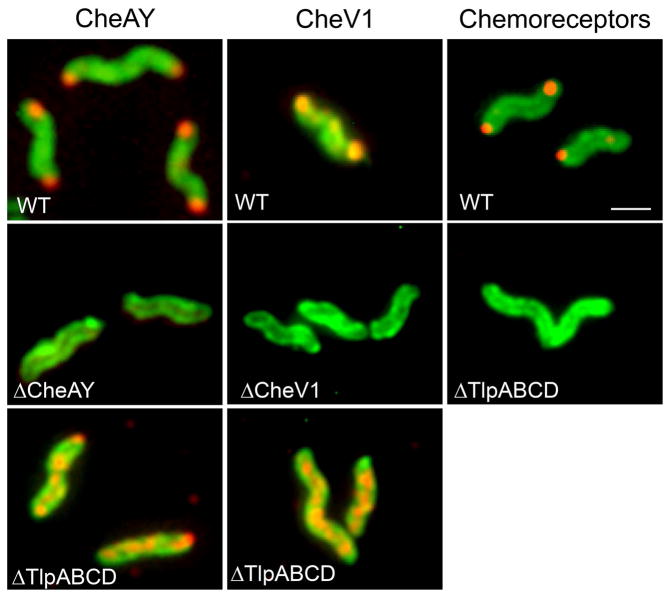
Based on the lack of conservation between CheZHP and E. coli CheZ, we were interested in whether CheZHP would localize to the chemotaxis signaling cluster as does E. coli CheZ (Cantwell et al., 2003). To determine protein location, we used immunofluorescence with anti- bodies specific to each protein. Of note, this approach allowed us to use native proteins expressed at wild-type levels, as opposed to fusion or overexpressed proteins. First, we determined the location of proteins of the H. pylori chemotaxis signaling cluster, by examining the location of two core signaling proteins, CheAY and CheV1. Each of these proteins was polar, with protein detected at either one or both poles (Fig. 3). This distribution is likely due to the age of the cells, with recently-divided cells having proteins at only the flagellated pole and older cells having proteins localized at both poles, as documented previously for the H. pylori chemotaxis protein ChePep (Howitt et al., 2011) as well as E. coli chemotaxis proteins (Ping et al., 2008). The chemoreceptors similarly localized to the poles, although some cytoplasmic staining could also be seen (Fig. 3). Control reactions with H. pylori strains lacking the proteins under study confirmed that each antibody was specific (Fig. 3). We next assessed whether these proteins were part of a supermolecular cluster anchored by the chemoreceptors, by observing localization in a mutant that lacks all of the chemoreceptors (ΔTlpABCD). As predicted for a chemoreceptor-anchored cluster, CheAY and CheV1 lost their polar localization in the absence of the chemoreceptors, and instead distribute throughout the cell in a punctate or clustered pattern (Fig. 3). These results together suggest that H. pylori chemotaxis signaling proteins reside in a polar cluster that is anchored by the chemoreceptor proteins, in an organization similar to that seen in other bacterial species, including close relatives of H. pylori (Briegel et al., 2009).
Figure 3.
H. pylori chemoreceptors and core chemotaxis signaling proteins form a polar cluster anchored by the chemoreceptors. The protein examined is indicated across the top, and the strain backgrounds are indicated in white writing within each panel. CheAY, CheV1, and the chemo-receptors are shown in red, detected by immunofluorescence using rabbit polyclonal anti-CheAY, anti-CheV1, or anti-TlpA22 respectively, followed by incubation with anti-rabbit antibodies conjugated with Alexa Fluor® 594 to fluoresce red. H. pylori cells are green, visualized by chicken anti-H. pylori antibodies, followed by anti-chicken antibodies conjugated with Alexa Fluor® 488, to fluoresce green. Multiple bacteria are shown; in some cases these were captured from independent images.
H. pylori CheZ forms a polar cluster that does not depend on the chemotaxis proteins
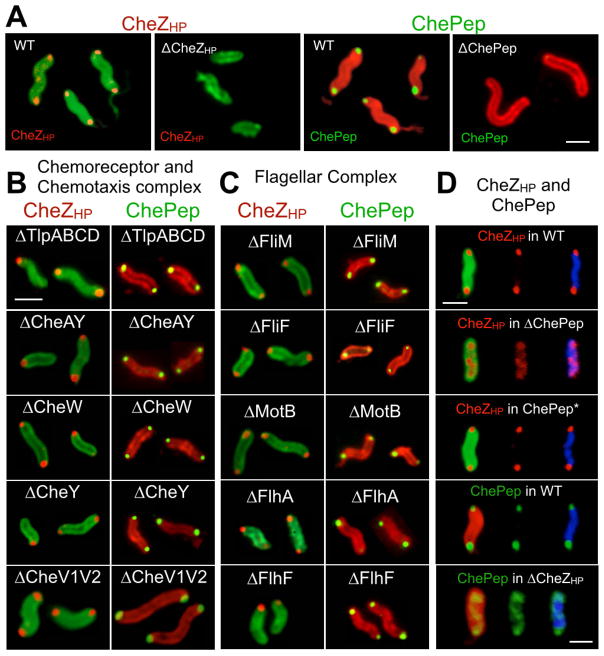
We next examined the localization of CheZHP. As found with the chemoreceptors and core signaling proteins, CheZHP localized to either one or both cellular poles (Fig. 4A and Table 1). Surprisingly, deletion of the chemoreceptors did not alter the polar position of CheZHP (Fig. 4B, Table 1), as it had for CheAY and CheV1 (Fig. 3). We then tested CheZHP localization in mutants lacking each of the chemotaxis signaling proteins. We were particularly interested in CheAY, as the E. coli ortholog of this protein recruits E. coli CheZ to the chemoreceptor complex (Cantwell et al., 2003), although H. pylori does not have a detectable CheA-short form by immunoblotting (data not shown). Again CheZHP remained polar even without CheAY (Fig. 4B, Table 1). We then analyzed CheZHP localization in mutants lacking each additional known chemotaxis signal transduction protein (CheV1, CheV2, CheV3, CheW, CheV1CheV2 double mutant and CheY). CheZHP polar localization was not affected by the removal of any of these chemotaxis proteins (Fig. 4B, Table 1).
Figure 4.
CheZHP and ChePep form a polar cluster that is independent from chemotaxis and flagellar-related proteins. Protein analyzed indicated above each set of relevant panels in a color matching the detection color. Strain background indicated in white writing within each panel. Multiple bacteria are shown for each mutant; in some cases these were captured from independent images. Scale bar represents 1 μm. A. CheZHP (red) was detected using anti-CheZHP antibodies, followed by secondary antibodies conjugated to Alexa Fluor® 594 to fluoresce red. H. pylori cells (green) were visualized chicken anti-H. pylori antibodies, followed by secondary conjugated with Alexa Fluor® 488. B. CheZHP and ChePep localization in chemotaxis signaling mutants. CheZHP (red) was visualized as in Panel A. ChePep (green) was visualized using anti-ChePep antibodies, followed by secondary anti-rabbit antibodies conjugated to Alexa Fluor 488, while whole bacteria were visualized using chicken anti-H. pylori followed by secondary antibodies conjugated to Alexa Fluor 594 to fluoresce red. C. CheZHP and ChePep localization in flagellar mutants. CheZHP and ChePep visualized as in Panel A and B, respectively. D. CheZHP and ChePep are mutually dependent on each other. CheZHP and ChePep visualized as in Panel A and B, respectively, with the addition of cells being visualized by DAPI DNA staining (blue).
Table 1.
CheZHP and ChePep cellular localization in different mutant backgrounds.
| H. pylori strain | CheZHP | ChePep | ||
|---|---|---|---|---|
|
| ||||
| Location (N) | Strains | Location (N) | Strains | |
|
| ||||
| WT | Pole (176) | G27, mG27, G27- MA, SS1 | Pole (187) | G27, G27-MA, SS1, SS2000, 26695, J99 |
| tlpABCD (Δtlp’s) | Pole (82) | mG27 | Pole (117) | mG27 |
| cheAY | Pole (81) | G27, G27-MA, SS1 | Pole (90) | G27-MA, SS1 |
| cheW | Pole (43) | G27, G27-MA, SS1 | Pole (44) | G27-MA, SS1 |
| cheV1 | Pole (38) | G27, G27-MA, SS1 | Pole (30) | G27-MA, SS1 |
| cheV2 | Pole (60) | G27, G27-MA, SS1 | Pole (54) | G27-MA, SS1 |
| cheV1 cheV2 | Pole | G27-MA | Pole | G27-MA |
| cheV3 | Pole (75) | G27, G27-MA, SS1 | Pole (63) | G27-MA |
| cheY | Pole (11) | G27, G27-MA, SS1 | Pole (79) | G27-MA, SS1 |
| ΔcheZHP or | Not detected | G27, G27-MA, | Diffuse (38) | G27-MA |
| ΔcheZHP::KS | (76) | SS1 | ||
| fliG | Pole (72) | G27 | ||
| fliM | Pole (81) | G27 | Pole (27) | G27-MA |
| fliN | Pole (69) | G27 | ||
| fliY | Pole (72) | G27 | ||
| fliF | Pole (58) | G27-MA | Pole (23) | G27-MA |
| motB | Pole (75) | G27 | Pole (54) | G27-MA |
| flhA | Pole (76) | G27 | Pole (76) | G27 |
| flhF | Pole (33) | G27 | Pole (150) | G27 |
| fliA | Pole (77) | G27 | ||
| flhG | Pole (33) | G27 | ||
| hp0062 | Pole (19) | G27 | ||
| chePep | Diffuse (27) | G27-MA, G27, SS1, PMSS1 | Not detected | G27-MA, G27, SS1, PMSS1 |
| cheZHP ΔN39 | Pole (138) | G27 | ||
| cheZHP N-only | ND (70) | G27 | ||
| cheZHP ΔC12 | Pole (20) | G27 | Pole (150) | G27 |
| cheZHP C-only | ND (16) | G27 | ||
N indicates number of individual cells viewed, from > 1 biological replicate; in all cases, CheZHP was observed as indicated. When more than one strain is listed, localization enumeration was done in the first strain and verified in the others..
CheZHP forms a polar cluster that does not depend on flagellar proteins
We expanded our search for proteins that anchor CheZHP to the pole to examine components of the flagellar motor (FliG, FliM, FliN, FliY) (Lowenthal, Hill, et al., 2009), the MS ring (FliF) (Allan et al., 2000), and the motor (MotB) (Ottemann and Lowenthal, 2002). Each of these single mutants retained CheZHP at the pole suggesting none were singly responsible for CheZHP polar localization (Fig. 4C, Table 1). We speculated that perhaps several proteins were sufficient for CheZHP polar localization, so we examined mutants that were missing the flagellar transcriptional regulators and thus lack several flagellar-related proteins. Specifically, we analyzed mutants lacking FlhA and FlhF, which are the master regulators for intermediate and late flagellar biosynthesis genes as well as several non-flagellar genes (Niehus et al., 2004). We also analyzed mutants lacking FliA/ σ28, which regulates several intermediate and late flagellar genes including flagellin (flaA and flaB), flgE1 (hook), along with non-flagellar genes envA, and omp11 (Niehus et al., 2004). None of these mutations resulted in loss of CheZHP from the pole (Fig. 4C, Table 1). Similar results were obtained with the flhG mutant, which encodes a protein that regulates flagellar number and placement (Kazmierczak and Hendrixson, 2013), as well as HP0062, the only protein suggested to interact with CheZHP based solely on a genome-wide two hybrid analysis (Rain et al., 2001) (Table 1). We thus concluded that CheZHP is not anchored at the pole via known chemotaxis or flagellar proteins.
CheZHP and ChePep depend on each other for polar localization
We next turned our attention to ChePep (Howitt et al., 2011). Cells lacking ChePep switch direction frequently (Howitt et al., 2011), as do CheZHP mutants (Fig. 2). ChePep was previously observed to localize to the bacterial pole (Howitt et al., 2011) (Fig. 4A), but the protein components required for its localization were not known. We therefore examined whether ChePep required the chemotaxis signaling or flagella proteins. Similar to what was observed for CheZHP, we found that ChePep localizes to the poles independently of the chemotaxis signaling complex and the flagella (Fig. 4B–C, Table 1). Because ChePep and CheZHP displayed similar localization patterns, we therefore examined whether loss of ChePep would affect CheZHP. We found that CheZHP polar localization was substantially different from wild type in the ΔchePep mutant background (Fig. 4D, Table 1). In particular, we found that a substantial fraction of CheZHP was lost from the pole, and a new population appeared either laterally dispersed or diffuse throughout the cell (Fig. 4C, Table 1, Supplemental movie 1). Control experiments confirmed that loss of either cheZ or chePep did not affect the expression of the other (Fig. 5). Furthermore, CheZHP localization could be restored by complementing ChePep in trans (Fig. 4D and Supplemental movie 1).
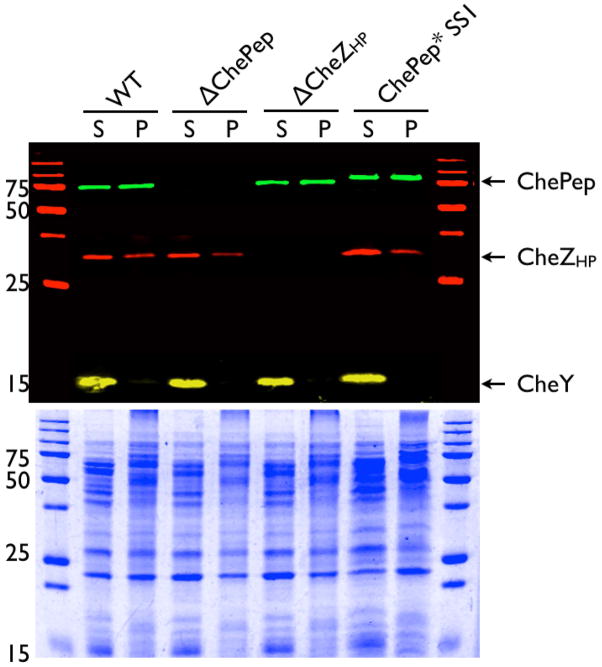
Figure 5. ChePep and CheZHP are expressed independently of each other.
Western blot analysis of 8–16% gradient gels of ChePep, CheZHP and CheY association with triton-insoluble (Pellet, P) and soluble fractions (S). The bottom panel shows coomassie stained identical samples. Molecular weight in kilodaltons indicated at the left of each panel. The predicted molecular weight of ChePep is 56 kilodaltons, but it migrates slower in SDS-PAGE presumably due to its high charge.
We next examined whether ChePep localization would depend on CheZHP. In mutants lacking CheZHP, ChePep no longer tightly localized to the poles and was found along the length of the bacteria organized in what appears to be a helical conformation (Fig. 4D, Table 1, Supplemental Movie 1). Together, these findings indicate that CheZHP and ChePep depend on each other for their polar localization and form a novel chemotaxis protein cluster distinct from the flagellar or chemotaxis signaling complexes.
The CheZHP localization region maps to amino acids 40-229
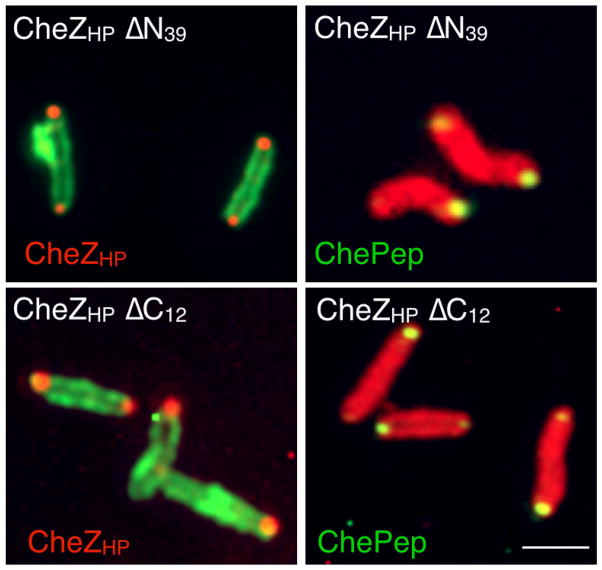
To gain additional insight into the localization requirements of CheZHP and ChePep, we analyzed the CheZHP truncated mutants that lacked either the first 39 amino acids (CheZHP ΔN39) or the last 12 amino acids CheZHP ΔC12). Both of these truncated variants produced protein that was detected by our anti-CheZHP polyclonal antibody (Fig. 6). In contrast the small CheZHP pieces of CheZHP N-only or CheZHP C-only were not detected by our antibodies, so were not analyzed further. Immunofluorescence analysis of whole H. pylori showed that both of these CheZHP ΔN39 and CheZHP ΔC12 localized to the pole in a manner that was indistinguishable from that of full-length CheZHP, suggesting they retain folding requirements for this function (Fig. 6, Table 1). Additionally, ChePep localization was not affected by deletion of either the CheZHP N or C terminus (Fig. 6, Table 1). These results suggest that region responsible for polar localization of CheZHP maps to the middle of CheZHP, corresponding to amino acids 40–241.
Figure 6.
CheZHP N and C termini are dispensable for polar localization of CheZHP (left panels, red) and ChePep (right panels, green). CheZHP and ChePep were detected by immunofluorescence as described in Fig. 4. Protein analyzed indicated in each set of relevant panels in a color matching the detection color. Strain background indicated in white writing within each panel. Multiple bacteria are shown; in some cases these were captured from independent images.
CheZHP and ChePep interact directly
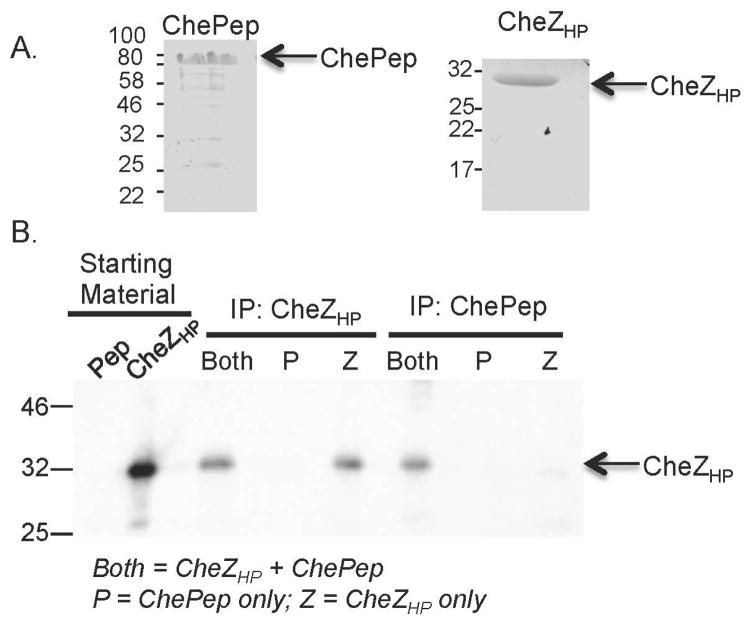
Our results suggest that CheZHP and ChePep form a distinct chemotaxis-regulatory complex, so we next examined whether they interact directly. We used co-immunoprecipitation with purified proteins (Fig. 7A), and found that CheZHP was co-immunoprecipitated with ChePep, suggesting these proteins interact directly (Fig.7B), and ChePep was similarly co-immunoprecipitated with CheZHP (data not shown). Neither CheZHP nor ChePep have predicted transmembrane domains, but both were not detergent soluble as most cytoplasmic proteins, e.g. CheY, are (Fig. 5). Solubility did not change in the presence or absence of either protein (Fig. 5). All together, these findings suggest that CheZHP and ChePep and are poorly soluble.
Figure 7.
CheZHP and ChePep interact directly. A. Coomassie-stained SDS-PAGE gel of purified ChePep (left) and CheZHP (right) proteins. Molecular weight in kilodaltons indicated at the left of each panel. B. Co-immunoprecipitation of CheZHP and ChePep, analyzed by western blotting of 10% SDS-PAGE gels with anti-CheZHP. From left to right: (1) Pep: the ChePep starting material (2) CheZHP: the CheZHP starting materials; (3–5) Immunoprecipitation (IP) with anti-CheZHP, incubated with a mixture of ChePep+CheZHP (both), ChePep (P) or CheZHP (Z); (6–8) IP with anti-ChePep, incubated with each set of proteins as in (3–5). The positions of ChePep and CheZHP are indicated on the right.
Discussion
In this manuscript, we report that the H. pylori CheZ phosphatase (CheZHP) localizes to the bacterial pole as do other phosphatases, but its localization relies on unique interactions. The other H. pylori chemotaxis proteins are also polar, as shown previously for ChePep (Howitt et al., 2011). Unexpectedly, CheZHP localization does not depend on the chemoreceptors or CheA, as would be expected from the E. coli paradigm, or on any flagellar proteins, as one would pre-dict from other chemotaxis phosphatases (Rao et al., 2005). Instead, CheZHP localization depends on the ChePep chemotaxis protein (Howitt et al., 2011) and conversely ChePep localization depends on CheZHP. This finding raises the intriguing possibility that some phosphatases, including CheZHP and ChePep, exist in a complex that is distinct from the core chemotaxis signaling and flagellar complexes. We also show that CheZHP behaves as a phosphatase in vivo, based on the reversal-biased behavior of a cheZHP null mutant. This outcome agrees with previous biochemical analysis (Lertsethtakarn and Ottemann, 2010). Somewhat surprisingly, we found that CheZHP regions outside of the known phosphatase active site and CheY-P binding regions play a role in chemotactic soft agar migration. This finding suggests that these regions retain some function that is not strictly related to phosphatase activity.
CheZHP localization depends on ChePep, a protein that functions in the chemotaxis pathway and is found only in Epsilon Protebacteria. ChePep and CheZHP are similar in many ways: as we show here, both localize to the pole, and null mutants of either show hyper-reversal phenotypes (Howitt et al., 2011). Both are highly negatively charged with acidic isoelectric points of 4.29 and 4.63, respectively. The fact that both ChePep and CheZHP mutants display hyper reversals suggests that loss of either protein creates elevated CheY-P. One possible explanation for this phenotype is that loss of a ChePep-CheZHP interactions results in less active CheZHP. Our data showing that CheZHP and ChePep interact directly supports this idea. Protein-protein interactions are known to activate E. coli CheZ; specifically, interactions with CheA-short activate CheZ 2.5-fold (Wang and Matsumura, 1996; Cantwell and Manson, 2009). ChePep contains a REC domain (Howitt et al., 2011) —a type of domain that normally interacts with CheZ. Thus one possibility is that ChePep uses its REC domain to bind CheZHP and enhances its activity. Another possibility is that without ChePep, CheZHP is mislocalized and chemotaxis is inefficient in this situation, as discussed below. A third possibility is that both CheZHP and ChePep have phosphatase activity. Preliminary in vitro experiments, however, did not detect any phosphatase activity associated with ChePep (data not shown). A discrete CheZHP localization was expected, given that other chemotaxis phosphatases localize to specific cellular sites (Rao et al., 2005; Lipkow, 2006). Computer models suggest that phosphatase localization prevents formation of a CheY-P gradient throughout the cell, which in turn allows all flagellar complexes to receive the same signal, rotate their motors in cooperation, and confer efficient cell migration (Rao et al., 2005; Lipkow, 2006). Unexpectedly, CheZHP localization did not depend on the chemoreceptors, CheAY, other chemotaxis proteins, or flagellar proteins. Thus the localization mechanism of CheZHP differs from that of E. coli CheZ, which depends on CheA and the chemoreceptors (Cantwell et al., 2003), and FliY, which depends on other flagellar motor proteins (Szurmant et al., 2003). Instead, its localization depends on the chemotaxis protein ChePep. Conversely, ChePep localization depends on CheZHP. Thus it appears that each of these proteins enhances the polar localization of the other, by an as-yet-unknown mechanism.
An additional finding reported here is that H. pylori chemoreceptors form a polar cluster that includes the core signaling proteins CheAY and CheV1, and presumably others. When the chemoreceptors are absent, CheAY and CheV1 are no longer polar (Fig. 3). Instead, they appear in the cytoplasm in manner that is clearly non-polar , but does retain some punctate aspects for as yet unknown reasons. This finding is not surprising, given that all bacteria analyzed have a polar chemoreceptor supermolecular cluster (Briegel et al., 2009). H. pylori had not been specifically analyzed, although the related microbe Helicobacter hepaticus had been. In H. hepaticus, the chemoreceptor cluster forms at the pole that also contains the flagella (Briegel et al., 2009). The core chemotaxis signaling cluster of H. pylori also appears to form at the flagellar pole in recently-divided cells, and then forms at the second pole prior to cell division (Howitt et al., 2011). We additionally observed minor cytoplasmic or lateral chemoreceptor distribution, as has been observed in E. coli, suggesting that both polar and lateral clusters might occur (Maddock and Shapiro, 1993; Greenfield et al., 2009).
The finding that CheZHP and ChePep localize independently of the two other motility related complexes—the core chemotaxis complex and flagellar basal body— suggests that they may form a third chemotaxis complex. The reason behind this distinct localization is not yet known. However, it should be pointed out that both E. coli and B. subtilis have polar chemotaxis proteins and peritrichous flagella, while H. pylori has chemotaxis and flagella at one pole. No other phosphatases from polarly flagellated bacteria have been analyzed. One possibility is that the CheZHP-ChePep complex is under a distinct regulatory control that is afforded by its separation from the other motility-related complexes. ChePep is found only in the Epsilon proteobacteria, suggesting this Class of bacteria may have evolved unique regulatory mechanisms.
The characterization of various cheZHP mutants uncovered that portions of CheZHP without any known phosphatase activity modulate chemotaxis. Strains completely lacking cheZHP had reduced migration in the soft agar assay and displayed hyper-reversal swimming behavior. This swimming behavior suggests high CheY-P in the cell, consistent with the in vitro CheZHP phosphatase activity (Lertsethtakarn and Ottemann, 2010). The soft-agar migration phenotype is also consistent with the cells possessing a hyper-reversal-bias, as tumble-bias mutants perform slightly better than swim-bias mutants in this assay (Wolfe and Berg, 1989), as we observed comparing ΔcheZHP to ΔcheW (Fig. 1). Several CheZHP mutants that lose phosphatase activity in vitro (CheZHP D189N, Q193R, and lacking the 12 C-terminal amino acids (Lertsethtakarn and Ottemann, 2010)) seemed to retain some in vivo function, as evidenced by intermediate soft-agar migration rates. One possible explanation for this phenotype is that these CheZHP variants retain the weak ability to bind CheY-P. In this case, they might sequester some CheY-P away from the flagellar motor to allow slightly more normal switching between reversals and forward swimming. Similarly, Sanna and Simon (Sanna and Simon, 1996) reported that very high or very low levels of E. coli CheZ caused loss of soft agar migration, highlighting the idea that there is a range of CheY-P levels that supports normal soft agar movement. H. pylori intrinsic CheY dephosphorylation is quite fast–0.28 s−1–a rate that is 8X faster than that of E. coli CheY (Lertsethtakarn and Ottemann, 2010). Thus in H. pylori, CheY may more readily dephosphorylate on its own and allow modest chemotaxis even without a phosphatase. Furthermore, there is precedence for the idea that there are multiple CheY-P binding regions in CheZ from work with E. coli CheZ; three regions of E. coli CheZ bind CheY-P—residues 67–71, 136–151, and the C-terminal 12 amino acids (Zhao et al., 2002). Our finding that the strain bearing cheZHP Q193R had a similar directional change bias as the ΔcheZHP mutant, does not support this model however because there appears to be high CheY-P in this strain. An alternative idea is that the various CheZHP variants retain the ability to interact with some components the chemotaxis signaling pathway. Indeed, there is evidence that CheZHP has interactions with other parts of the chemotaxis pathway. Specifically, CheZHP was discovered based on the finding that cheZHP mutants were able suppress loss of the CheW coupling protein (Terry et al., 2006), and CheZHP has phosphatase activity towards phosphoryl CheAY and CheV2 in addition to CheY (Lertsethtakarn and Ottemann, 2010). While we do not yet know the mechanism behind the ability of cheZHP mutants to suppress loss of cheW, these results suggest there are as-yet poorly understood connections in the H. pylori chemotaxis pathway. One other consideration is that the soft agar assay monitors accumulation of chemotaxis ability over a period of days, whereas monitoring of the swimming behavior spans only seconds, so there are differences in strain behavior in this assay. Specifically, Lowenthal et al. found that H. pylori strains can have few reversals in the swimming assay, but gain the ability to reverse in the soft agar (Lowenthal, Simon, et al., 2009).
In summary, we report that CheZHP and ChePep localize to the pole to a complex that is distinct from the chemoreceptor-signaling and flagellar complexes. CheZHP and ChePep promote each other’s polar localization, and interact directly. Our findings raise the possibility that CheZHP-ChePep form a complex that localizes separately from the other motility-related complexes for a specific purpose. We noted that the polar localization of CheZHP-ChePep did not completely disappear in the absence of one, suggesting that there might be other proteins that participate in the CheZHP and ChePep complex. We also report that CheZHP may have additional functions or interactions in the chemotaxis pathway, beyond its phosphatase activity, based on the partial chemotaxis behavior of several cheZHP mutants. Together, these findings suggest that while CheZHP has conserved CheZ phosphatase function and mechanism, it has diverged significantly in other regards.
Experimental Procedures
Bacterial strains and growth conditions
All H. pylori strains are listed in Table 2, and plasmids listed in Table 3. H. pylori strain G27 or its variants G27-MA and mG27 were used for all localization experiments. These strains are all highly related, were derived from the same parent, and behave the same for chemotaxis and motility. Strains SS1, SS2000, 26695, and J99 were used to confirm protein localization in some cases. H. pylori was grown under microaerobic condition at 37°C in an incubator with a gas mixture of 5–10% O2, 10% CO2, and 80–85% N2, on Columbia horse blood agar (CHBA) with 5% difibrinated horse blood, or in brucella broth with 10% v/v heat inactivated fetal bovine serum (FBS) (BB10). Kanamycin was used at 15 μg/ml, and chloramphenicol was used at 10 μg/ml for selection of mutants.
Table 2.
H. pylori strains used in this study
| H. pylori strains | Strain # | Genotype/Description | Reference/source |
|---|---|---|---|
| G27 | Wild type | (Censini et al., 1996)/From Nina Salama | |
| mG27 | KO625 | G27, mouse-adapted | (Castillo et al., 2008) |
| G27-MA | G27, MDCK cells adapted | (Amieva et al., 2003) | |
| SS1 | Wild type, mouse adapted | (Lee et al., 1997) | |
| SS2000 | Wild type | (Thompson et al., 2004) | |
| PMSS1 | Wild type, Parent of strain SS1, not mouse adapted | (Arnold et al., 2011) | |
| 26695 | Wild type | (Tomb et al., 1997) | |
| J99 | Wild type | (Alm et al., 1999) | |
| G27 ΔcheZHP::KS | KO1269 | G27 ΔcheZHP::aphA3/sacB | This study |
| G27 ΔcheZHP | KO1315 | KO1269 ΔcheZHP (entire coding region deleted) | This study |
| G27 ΔcheZHP::cheZHP | KO1304 | KO1269 ΔcheZHP:: cheZHP (complement) | This study |
| G27 ΔcheZHP::cat | KO1325 | G27 ΔΔcheZHP::cat (retains coding potential for the first 13 and last 12 amino acids) | This study; allele originally published in (Terry et al., 2006) |
| G27 cheZHP Q193R | KO1307 | KO1269 ΔcheZHP::cheZHP Q193R | This study |
| G27 cheZHP D189N | KO1306 | KO1269 ΔcheZHP::cheZHP D189N | This study |
| G27 cheZHP ΔN39 | KO1313 | KO1269 ΔcheZHP:: cheZHP ΔN (deletion of amino acids 1–39) | This study |
| G27 cheZHP N-only | KO1273 | KO1269 ΔcheZHP:: cheZHP 1–39 (retains amino acids 1–39) | This study |
| G27 ΔcheZHP C12 | KO1300 | KO1269 ΔcheZHP:: ΔcheZHP C (deletion of C-terminal 12 amino acids) | This study |
| G27 cheZHP C-only | KO1312 | KO1269 ΔcheZHP:: cheZHP C-only (retains amino acids 241–253) | This study |
| G27 ΔcheW | KO851 | G27 ΔcheW::aphA3 | (Terry et al., 2005) |
| G27 ΔcheAY | KO857 | G27 ΔcheAY::cat (also called ΔcheA::cat) | This study; cheAY allele published in (Terry et al., 2005) |
| G27 ΔcheY | KO1250 | G27 ΔcheY::aphA3-sacB | This study |
| G27 ΔcheV1 | KO1277 | G27 ΔcheV1::cat | This study; cheV1 allele published in (Lowenthal, Simon, et al., 2009) |
| G27 ΔcheV2 | KO1278 | G27 ΔcheV2::cat | This study; cheV2 allele published in (Lowenthal, Simon, et al., 2009) |
| G27 ΔcheV3 | KO1279 | G27 ΔcheV3::cat | This study; cheV3 allele published in (Lowenthal, Simon, et al., 2009) |
| mG27 ΔtlpA | KO1002 | mG27 ΔtlpA | (Rader et al., 2011) |
| mG27 ΔtlpB | KO1004 | mG27 ΔtlpB | (Rader et al., 2011) |
| mG27 ΔtlpC | KO1005 | mG27 ΔtlpC::aphA3 | (Rader et al., 2011) |
| mG27 ΔtlpD | KO1006 | mG27 ΔtlpD::cat | (Rader et al., 2011) |
| mG27 ΔtlpB::kan-sac | KO1003 | mG27 ΔtlpB::aphA3-sacB | (Rader et al., 2011) |
| mG27 ΔtlpA ΔtlpD | KO1009 | KO1002 ΔtlpD::cat | This study |
| mG27 ΔtlpA ΔtlpB ΔtlpD | KO1015 | KO1009 ΔtlpB | This study |
| mG27 ΔtlpA ΔtlpB ΔtlpC ΔtlpD (Δtlps) | KO1021 | KO1015 ΔtlpC::aphA3 | This study |
| G27-MA ΔchePep | G27-MA ΔchePep::cat | (Howitt et al., 2011) | |
| G27-MA ChePep* | G27-MA ΔchePep::cat rdxA::chePep- ahpA3 | (Howitt et al., 2011) | |
| G27-MA ΔcheV1 ΔcheV2 | This study | ||
| H. pylori G27 flhA | KO1284 | G27 flhA::kan | (Rader et al., 2007). Gift of Karen Guillemin. |
| H. pylori G27 ΔflhF | KO1367 | G27 ΔflhF::cat (165 bp deletion) | This study. Allele provided by Nina Salama (Fred Hutchison Cancer Research Center, Seattle WA). |
| H. pylori G27 ΔflhG | KO1328 | G27 ΔflhG::cat (62 bp deletion) | This study. Allele provided by Nina Salama (Fred Hutchison Cancer Research Center, Seattle WA). |
| H. pylori G27 fliA | KO1285 | G27ΔfliA::kan | (Rader et al., 2007). Gift of Karen Guillemin. |
| G27-MA ΔfliF | G27 ΔfliF::cat | This study | |
| G27 ΔfliG | KO1063 | G27 ΔfliG::cat | This study |
| G27 ΔfliM | KO1060 | G27 ΔfliM::cat | (Lowenthal, Hill, et al., 2009) |
| G27 fliN (KO1061) | KO1061 | G27 ΔfliN::cat | (Lowenthal, Hill, et al., 2009) |
| G27 fliY (KO1062) | KO1062 | G27 ΔfliY::cat | (Lowenthal, Hill, et al., 2009) |
| G27 motB | KO489 | G27 motB::aphA3-sacB | (Ottemann and Lowenthal, 2002) |
| G27 Δhp0062 | KO1310 | G27_57/hp0062::cat | This study. Allele pro- vided by Nina Salama (Fred Hutchison Cancer Research Center, Seattle WA). |
Table 3.
Plasmids used in this study
| Plasmid | Characteristic | Reference |
|---|---|---|
| pKT30 | pBluescript::hp0170SS1 (cheZHP) | (Terry et al., 2006) |
| pKO126 | pBluescript::cheYSS1 | (Terry et al., 2005) |
| pKSFII | pBluescript::aphA3-sacB (kan-sac or KS) | (Copass et al., 1997) |
| pKSF3 | pKSFII XhoI::XmnI | This study |
| pBS-FliG | pBluescript::fliGG27 | This study |
| pBS-FliG::cat-mut | pBluescript::fliGG27::cat-mut | This study |
| pKO126i | pKO126::aphA3-sacB | This study |
| pCat-mut | pBluescript::cat-mut (lacking transcriptional terminator) | (Terry et al., 2005) |
Generation of cheZHP mutants
Plasmid pKT30 containing cheZHP and approximately 500 base pairs flanking the coding sequence (Terry et al., 2006) was used as template for iPCR to remove the entire coding region of cheZHP using primers cheZup2 (5’-TGTCGTTCCTTGCCAATTGGTTTT) and cheZdn2 (5’-TTTTGAACCAAGTTGAATTACTCTC). The resulting iPCR product was ligated with an aphA3-sacB cassette (KS), cut from pKSF3 plasmid with SmaI and XmnI. This cassette confers kanamycin resistance and sucrose sensitivity. pKSF3 was generated from pKSFII (Copass et al., 1997)by ligating XmnI linkers into the XhoI site. The ΔcheZHP::KS plasmid was transformed into E. coli DH10B (Durfee et al., 2008)and verified by sequencing. The ΔcheZHP::KS plasmid was then transformed into H. pylori G27 wild type, via natural transformation, and selection for kanamycin resistance, to replace ΔcheZHP with cheZHP::KS cassette. The resulting kanamycin resistant colonies were screened for sucrose sensitivity and verified by sequencing of a PCR product generated from the cheZHP locus. This strain is called G27 ΔcheZHP::KS (Table 2), and was used as a parental strain for subsequent transformations to generate cheZHP mutants.
Plasmids bearing cheZHP deletion mutants were generated by iPCR as described above using primers that are available upon request. The resulting iPCR products were self-ligated and transformed into E. coli DH10B. The plasmids were verified by sequencing and used to transform G27 ΔcheZHP::KS. Kanamycin-sensitive, sucrose-resistant colonies were selected, screened, and verified by PCR and sequencing of cheZHP loci.
Generation of flhF, flhG, hp0062, fliG, cheY, cheV1 cheV2, and fliF mutants
flhF, flhG, and hp0062 mutant alleles marked with cat were obtained as G27 genomic DNA from Dr. Nina Salama (Fred Hutchison Cancer Research Center, Seattle WA). The DNA was used to transform H. pylori G27 wild type to chloramphenicol resistance. Colonies that were chloramphenicol resistant were selected and screened with PCR using primers that flank each locus. Similar approaches were used to move mutant alleles from other strains, and thus create G27 or G27-MA.
To create the fliG mutant, fliG was cloned from G27 genomic DNA by PCR using primers fliGlocusfor (5’-CACGCCTTTAATCAACTATA) and fliGlocusrev (5’-TAAAGCCGATTTTATAGCCA). The resulting PCR product was cloned into EcoRV-cut pBluescript creating vector pBS-FliG. pBS-FliG, was used as template in iPCR, with primers fliGcircfor-2 (5’-GGTAAGCTTGGTTGCCATTTTTA) and fliGcircrev-2 (5’-GATTCCAAACCGGTGAAGAGGA) that deleted most of the gene. This PCR product was gel purified and ligated with a terminator-less cat gene obtained from the vector pCat-mut (Terry et al., 2005) using HincII, to create the vector pBS-FliG::cat-mut. pBS-FliG::cat-mut was used to transform G27 to chloramphenicol resistance.
The cheY allele used here was generated by digesting pKO126 with Bpu1102I, which cuts about ¼ of the way into the open reading frame, followed by creating blunt ends. This product was ligated with the aphA-sacB genes derived from pKSFII cut with SmaI and XhoI, followed by blunt end generation, to create pKO126i. pKO126i was used to transform G27 to kanamycin resistance. The double mutant in cheV1 and cheV2 was constructed using the allelic replacement strategy as previously described (Chalker et al., 2001). First, G27-MA was naturally transformed with a construct replacing cheV1 with a kanamycin resistance cassette (aphA3) using the primers CheV1-1 (5’-CTAGCGAGTTTAGGAAGCAATTG), CheV1-2 (5’-ATGGTTCGCTGGGTTTATCACTATCAGCCATGATTTCCCCTT), CheV1-3 (5’-TTACTAGGATGAATTGTTTTAGTACCCAATGGTAAAACCTTATTGGAGC), and CheV1-4 (5’-GCTCGCACAACACCCGTTCAATC). Mutants were screened for kanamycin resistance and confirmed by PCR. Then G27-MA ΔcheV1::aphA3 mutants were naturally transformed with a construct replacing cheV2 with an erythromycin resistance cassette using the following primers, CheV2-1 (5’-AGCGTTAGTAACAAGCTCTCC), CheV2-2 (5’-TACTGCAATCTGATGCGATTATTGCTAATTTCCCCTAAAGCCCTATC), CheV2-3 (5’-TTCAATAGCTATAAATTATTTAATAAGTAAGAATCGCTACTCATGGACGAATTG) and CheV2-4 (5’-GGTATTTCAAGCGCAAATCTTCATTC). Transformants were screened for both kanamycin and erythromycin resistance and confirmed by sequencing.
Mutants in fliF were generated by replacing fliF with a cat cassette followed by selection of transformants on chloramphenicol. The allelic replacement construct was generated by joining the cat cassette with sequence upstream of fliF with primers fliF-1(5’-TCGCAGAAACTTAGCGCTTGG), fliF-2(5’-ATCCACTTTTCAATCTATATCAAGCAAAGCGGTGATTAAAACC) and downstream fliF-3(5’-CCCAGTTTGTCGCACTGATAAAAGATAAAAGGTTAAAAATGGCAACC), fliF-4(5’-TGGAGATCTCAGCTTTCATTTCA). The resulting G27-MA ΔfliF mutants were confirmed by sequence analysis.
Generation of mutant lacking all chemoreceptors
Construction of the ΔtlpA, ΔtlpB, ΔtlpC and ΔtlpD individual mutants has been described (Rader et al., 2011). In all cases, the resulting mutations were verified using PCR and sequencing for clean deletions. To create multiple receptor mutants, we started with mG27 ΔtlpA (KO1002) and transformed with genomic DNA bearing ΔtlpD::cat from KO1006, to create ΔtlpA tlpD (KO1009). After verification, this strain was transformed with genomic DNA with tlpB::kan-sac, followed by transformation to sucrose resistance/kanamycin sensitivity using KO1004 (ΔtlpB) chromosome. This created strain KO1015 which was then transformed with KO1005 chromosome (ΔtlpC::kan) to create ΔtlpA ΔtlpB ΔtlpC::kan ΔtlpD::cat (KO1021).
Creation and pre-absorption of antibodies
Antibodies that recognize ChePep, CheY, and all H. pylori chemoreceptors, called TlpA22, have been described previously (Williams et al., 2007; Lowenthal, Hill, et al., 2009; Howitt et al., 2011). Antibodies that recognize CheAY, CheV1, or CheZHP were generated in rabbits using either purified His-CheAY, CheV1, or CheZHP (Lertsethtakarn and Ottemann, 2010). For pre-absorption of these antibodies, strain H. pylori G27 ΔcheZHP::KS, ΔcheZHP, ΔcheAY::cat, ΔcheV1::cat, Δtlp’s, or Δchepep were used. For the preabsorption, each H. pylori strain was grown overnight on three CHBA plates. Cells were resuspended in 2ml 1X PBS (10X: 80g NaCl, 2g KCl,11.5g Na2HPO4·7H2O, 2g KH2 PO4 to 1L adjusted to pH 7.3) collected by centrifugation and resuspended in 1ml PLP (75mM NaPO4, pH 7.4, 2.5mM NaCl, 2% para-formaldehyde in 1X PBS) followed by 10 minutes room temperature incubation to fix the cells. Cells were collected by centrifugation, and washed with 1XPBS three times. To permeabilize the cells, 1ml of permeabilizing buffer (3% BSA, 1% saponin, 0.1% triton X-100, 0.02% sodium azide in PBS) was added and cells were incubated at room temperature for 10 minutes. Permeabilized cells were centrifuged as above to remove supernatant. Cells were resuspended in 700μl of permeabilizing buffer and respective antibody was added at a 1:100 dilution. The mixture was incubated with rotation overnight at 4°C. Cells were removed by centrifugation and the supernatant was collected. To check for complete absorption of the antibody, western analysis was performed.
Immunofluorescence
For immunofluorescence analysis, liquid cultures of the H. pylori strains to be analyzed were grown in BB10 for 6 hours (exponential phase). The culture was visually inspected for motility before slide preparation. 40–65 μl of the culture was placed on a poly-L-lysine coated slides (Ted Pella, Inc), followed by addition of PLP and incubation at room temperature for 10 minutes. These fixed cells were then permeabilized with permeabilizing buffer at room temperature for 10 minutes. Pre-absorbed primary anti-CheZHP, -His-CheAY, -CheV1, or -ChePep were each used at 1:200, anti-TlpA22 was used at 1:1000, and chicken anti-H. pylori (AgriSera AB) was used at 1:500 dilution. The reactions were incubated at room temperature for 30 minutes and the cells were washed with blocking buffer (3% BSA, 0.1% TritonX-100 in 1X PBS) 3 times. Goat anti-rabbit conjugated with Alexa Fluor® 594 (Invitrogen) and goat anti-chicken conjugated with Alexa Fluor® 488 (Invitrogen) were added at 1:300 and 1:500 dilutions, respectively, and incubated in the dark at room temperature for 30 minutes. The samples were washed as above. A drop of Vectashield® with DAPI (Vector Laboratories, Inc.) was added and the samples were sealed with coverslips.
Immunofluorescent cells were viewed using a Nikon ECLIPSE E600 microscope and SPOT software Version 4.7 (Diagnostic instruments, inc.), using a Plan Flour 100X (Nikon) objective. A Texas Red® (Chroma) filter cube was used to view and capture emission from Alexa Fluor® 594 (red) and FITC/GFP (Chroma) filter cube was used to view and capture emission from Alexa Fluor® 488 (green). Images were taken in color for each fluor separately and merged in Adobe® Photoshop® CS2 version 9.0.2 (Adobe®).
Immunoprecipitation, cell fractionation, and immunoblotting
For immunoprecipitation, CheZHP and ChePep were purified as a GST-fusion proteins, as described previously (Lertsethtakarn and Ottemann, 2010; Howitt et al., 2011). For co- immunoprecipitation experiments, the GST-tag was removed using Prescission Protease (GE Healthcare Life Sciences). Anti-CheZHP or anti-ChePep antibodies were conjugated to beads, using Protein A-coupled magnetic Dynabeads (Life Technologies) and crosslinking with BS3. Equimolar ChePep and CheZHP (9 μM each) were mixed, allowed to form complexes for 30–60 minutes at room temperature, diluted to 3μM, and then incubated with the antibody-bound beads as directed by the manufacturers’ protocols. Beads were washed four-times with phosphate buffered saline plus 0.04% Tween-20, before elution with pH 2.8 glycine, following the manufacturers protocols.
To test for expression and differential solubility of CheZHP and ChePep in the different mutant backgrounds, H. pylori cells grown for < 24 hours in microaerobic conditions were harvested directly from blood agar plates into 0.5% Tween-20, 50mM Tris pH 7.4, 200 mM NaCl, 1 mM EDTA, 1M PMSF, vortexed and incubated for 15 minutes on ice. The lysates were then centrifuged at 15,000Xg and the soluble fraction diluted 1:1 in 2X SDS sample buffer. The Tween-insoluble pellets were resuspended in equal volumes of SDS sample buffer, prior to boiling, separation by SDS-PAGE and immunoblotting as described above.
For immunoblots, samples were electrophoreses on either 8–16% or 10% SDS-PAGE gels. After transfer to polyvinylidene difluoride (PVDF) membranes, proteins were detected using either rabbit anti-CheZHP, rabbit anti-ChePep, or rabbit anti-CheY. Fluorescent secondary antibodies were used for the solubility experiments with anti-CheZHP followed by goat anti-rabbit Alexa Fluor 660, anti-ChePep followed by goat anti-rabbit Alex Fluor 800, anti-CheY with both goat anti-rabbit Alex Fluor 660 and 800. All membranes were scanned with a Licor-Odyssey scanner and overlayed to create a single western blot. HRP-conjugated goat-anti rabbit antibodies (Thermo-Fisher) were used for the co-immunoprecipitation experiments.
Soft agar migration assay
cheZHP mutants were inoculated in BB10 containing 0.35% (w/v) of agar (Bacto). Each plate was also inoculated with H. pylori wild type and non-chemotactic ΔcheW to serve as controls. Cultures were incubated as described above for 4–5 days. The diameter of the bacterial colony was measured at the end of incubation period.
Analysis of swimming behavior
H. pylori strains were cultured for six hours with shaking or overnight without shaking in BB10. The swimming behavior of each culture was viewed and recorded using Simple PCI version 5.3.1. (Compix Inc., Imaging Systems) and Hamamatsu Digital Camera C4742-98 on a Nikon ECLIPSE E600 microscope at 100X magnification. At least twenty films were recorded for each culture, from at least two independent biological replicates. Files were randomized to conceal the identity of analyzed strain. For each H. pylori strain, at least 150 cells were tracked for clear directional changes for 5 seconds, using hand-tracing of each swimming bacteria.
Statistical analysis
Two sample Student’s t-test in SYSTAT 13 © (Systat Software, Inc.) was used to perform statistical analyses.
Supplementary Material
Acknowledgments
The authors would like to thank Tate Sessler and Dr. Nina Salama of the Fred Hutchison Cancer Research Center for providing genomic DNA for several mutant alleles; Dr. Karen Guillemin of the University of Oregon for providing several H. pylori mutant strains; Marla Hill for generating the G27 ΔfliG::cat strain; Kieran Collins and Samar Abedrabbo for immunofluorescence technical assistance; and Samar Abedrabbo for critical comments on the manuscript. The described project was supported by Grant Number AI050000 (to K.M.O.) from the National Institutes of Allergy and Infectious Disease (NIAID) at the National Institutes of Health and the University of California Cancer Research Coordinating Committee to K.M.O., and by the Morgridge Faculty Scholar Award and the Stanford Pediatric Research Fund to M.R.A. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Literature cited
- Alexander RP, Lowenthal AC, Harshey RM, Ottemann KM. CheV: CheW-like coupling proteins at the core of the chemotaxis signaling network. Trends Microbiol. 2010;18:494–503. doi: 10.1016/j.tim.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan E, Dorrell N, Foynes S, Anyim M, Wren BW. Mutational analysis of genes encoding the early flagellar components of Helicobacter pylori: evidence for transcriptional regulation of flagellin A biosynthesis. J Bacteriol. 2000;182:5274–5277. doi: 10.1128/jb.182.18.5274-5277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- Amieva MR, Vogelmann R, Covacci A, Tompkins LS, Nelson WJ, Falkow S. Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science. 2003;300:1430–1434. doi: 10.1126/science.1081919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold IC, Lee JY, Amieva MR, Roers A, Flavell RA, Sparwasser T, Müller A. Tolerance rather than immunity protects from Helicobacter pylori-induced gastric prene-oplasia. Gastroenterology. 2011;140:199–209. doi: 10.1053/j.gastro.2010.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat Y, Eisenbach M. Conserved C-terminus of the phosphatase CheZ is a binding domain for the chemotactic response regulator CheY. Biochemistry. 1996;35:5679–5683. doi: 10.1021/bi9530447. [DOI] [PubMed] [Google Scholar]
- Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, et al. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci USA. 2009;106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantwell BJ, Manson MD. Protein domains and residues involved in the CheZ/CheAS interaction. J Bacteriol. 2009;191:5838–5841. doi: 10.1128/JB.00280-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantwell BJ, Draheim RR, Weart RB, Nguyen C, Stewart RC, Manson MD. CheZ phosphatase localizes to chemoreceptor patches via CheA-short. J Bacteriol. 2003;185:2354–2361. doi: 10.1128/JB.185.7.2354-2361.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo AR, Woodruff AJ, Connolly LE, Sause WE, Ottemann KM. Recombination-Based In Vivo Expression Technology Identifies Helicobacter pylori Genes Important for Host Colonization. Infect Immun. 2008;76:5632–5644. doi: 10.1128/IAI.00627-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, et al. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda O, Rivas A, Toledo H. Helicobacter pylori strain ATCC700392 encodes a methyl-accepting chemotaxis receptor protein (MCP) for arginine and sodium bicarbonate. FEMS Microbiol Lett. 2003;224:175–181. doi: 10.1016/S0378-1097(03)00423-3. [DOI] [PubMed] [Google Scholar]
- Chalker AF, Minehart HW, Hughes NJ, Koretke KK, Lonetto MA, Brinkman KK, et al. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J Bacteriol. 2001;183:1259–1268. doi: 10.1128/JB.183.4.1259-1268.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copass M, Grandi G, Rappuoli R. Introduction of unmarked mutations in the Helicobacter pylori vacA gene with a sucrose sensitivity marker. Infect Immun. 1997;65:1949–1952. doi: 10.1128/iai.65.5.1949-1952.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxen MA, Sisson G, Melano R, Hoffman PS. The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J Bacteriol. 2006;188:2656–2665. doi: 10.1128/JB.188.7.2656-2665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T, Nelson R, Baldwin S, Plunkett G, Burland V, Mau B, et al. The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J Bacteriol. 2008;190:2597–2606. doi: 10.1128/JB.01695-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foynes S, Dorrell N, Ward SJ, Stabler RA, McColm AA, Rycroft AN, Wren BW. Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect Immun. 2000;68:2016–2023. doi: 10.1128/iai.68.4.2016-2023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield D, McEvoy AL, Shroff H, Crooks GE, Wingreen NS, Betzig E, Liphardt J. Self-Organization of the Escherichia coli Chemotaxis Network Imaged with Super-Resolution Light Microscopy. PLoS Biol. 2009;7:e1000137. doi: 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess JF, Oosawa K, Kaplan N, Simon MI. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell. 1988;53:79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- Howitt MR, Lee JY, Lertsethtakarn P, Vogelmann R, Joubert LM, Ottemann KM, Amieva MR. ChePep controls Helicobacter pylori Infection of the gastric glands and chemotaxis in the Epsilonproteobacteria. mBio. 2011;2:e00098–11. doi: 10.1128/mBio.00098-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Pearson MA, Delany I, Scarlato V, Beier D. Phosphate flow in the chemotactic response system of Helicobacter pylori. Microbiology (Reading, Engl) 2005;151:3299–3311. doi: 10.1099/mic.0.28217-0. [DOI] [PubMed] [Google Scholar]
- Josenhans C, Suerbaum S. The role of motility as a virulence factor in bacteria. Int J Med Microbiol. 2002;291:605–614. doi: 10.1078/1438-4221-00173. [DOI] [PubMed] [Google Scholar]
- Kazmierczak BI, Hendrixson DR. Spatial and numerical regulation of flagellar biosynthesis in polarly flagellated bacteria. Mol Microbiol. 2013;88:655–663. doi: 10.1111/mmi.12221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentner D, Sourjik V. Dynamic map of protein interactions in the Escherichia coli chemotaxis pathway. 2009;5:238. doi: 10.1038/msb.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, O'Rourke J, De Ungria MC, Robertson B, Daskalopoulos G, Dixon MF. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology. 1997;112:1386–1397. doi: 10.1016/s0016-5085(97)70155-0. [DOI] [PubMed] [Google Scholar]
- Lertsethtakarn P, Ottemann KM. A remote CheZ orthologue retains phosphatase function. Mol Microbiol. 2010;77:225–235. doi: 10.1111/j.1365-2958.2010.07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lertsethtakarn P, Ottemann KM, Hendrixson DR. Motility and Chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol. 2011;65:389–410. doi: 10.1146/annurev-micro-090110-102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkow K. Changing cellular location of CheZ predicted by molecular simulations. PLoS Comput Biol. 2006;2:e39. doi: 10.1371/journal.pcbi.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenthal AC, Hill M, Sycuro LK, Mehmood K, Salama NR, Ottemann KM. Functional analysis of the Helicobacter pylori flagellar switch proteins. J Bacteriol. 2009;191:7147–7156. doi: 10.1128/JB.00749-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenthal AC, Simon C, Fair AS, Mehmood K, Terry K, Anastasia S, Ottemann KM. A fixed-time diffusion analysis method determines that the three cheV genes of Helicobacter pylori differentially affect motility. Microbiology. 2009;155:1181–1191. doi: 10.1099/mic.0.021857-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- Miller LD, Russell MH, Alexandre G. Diversity in bacterial chemotactic responses and niche adaptation. Adv Appl Microbiol. 2009;66:53–75. doi: 10.1016/S0065-2164(08)00803-4. [DOI] [PubMed] [Google Scholar]
- Niehus E, Gressmann H, Ye F, Schlapbach R, Dehio M, Dehio C, et al. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol Microbiol. 2004;52:947–961. doi: 10.1111/j.1365-2958.2004.04006.x. [DOI] [PubMed] [Google Scholar]
- Ottemann KM, Lowenthal AC. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect Immun. 2002;70:1984–1990. doi: 10.1128/IAI.70.4.1984-1990.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping L, Weiner B, Kleckner N. Tsr-GFP accumulates linearly with time at cell poles, and can be used to differentiate 'old' versus “new” poles, in Escherichia coli. Mol Microbiol. 2008;69:1427–1438. doi: 10.1111/j.1365-2958.2008.06372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman MS, Goodwin M, Kelly DJ. Chemotaxis in the human gastric pathogen Helicobacter pylori: different roles for CheW and the three CheV paralogues, and evidence for CheV2 phosphorylation. Microbiology (Reading, Engl) 2001;147:2493–2504. doi: 10.1099/00221287-147-9-2493. [DOI] [PubMed] [Google Scholar]
- Polk DB, Peek RM. Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer. 2010;10:403–414. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rader BA, Campagna SR, Semmelhack MF, Bassler BL, Guillemin K. The Quorum-Sensing Molecule Autoinducer 2 Regulates Motility and Flagellar Morphogenesis in Helicobacter pylori. J Bacteriol. 2007;189:6109–6117. doi: 10.1128/JB.00246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rader BA, Wreden C, Hicks KG, Sweeney EG, Ottemann KM, Guillemin K. Helicobacter pylori perceives the quorum-sensing molecule AI-2 as a chemorepellent via the chemoreceptor TlpB. Microbiology. 2011;157:2445–2455. doi: 10.1099/mic.0.049353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, et al. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- Rao CV, Kirby JR, Arkin AP. Phosphatase localization in bacterial chemotaxis: divergent mechanisms, convergent principles. Phys Biol. 2005;2:148–158. doi: 10.1088/1478-3975/2/3/002. [DOI] [PubMed] [Google Scholar]
- Salama NR, Hartung ML, Müller A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat Rev Microbiol. 2013 doi: 10.1038/nrmicro3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanna MG, Simon MI. In vivo and in vitro characterization of Escherichia coli protein CheZ gain- and loss-of-function mutants. J Bacteriol. 1996;178:6275–6280. doi: 10.1128/jb.178.21.6275-6280.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinitzer T, Mizote T, Ishikawa N, Dudnik A, Inatsu S, Schreiber S, et al. Functional characterization and mutagenesis of the proposed behavioral sensor TlpD of Helicobacter pylori. J Bacteriol. 2008;190:3244–3255. doi: 10.1128/JB.01940-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silversmith RE. Auxiliary phosphatases in two-component signal transduction. Curr Opin Microbiol. 2010;13:177–183. doi: 10.1016/j.mib.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Armitage JP. Spatial organization in bacterial chemotaxis. EMBO J. 2010;29:2724–2733. doi: 10.1038/emboj.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney EG, Henderson JN, Goers J, Wreden C, Hicks KG, Foster JK, et al. Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB. Structure. 2012;20:1177–1188. doi: 10.1016/j.str.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H, Bunn MW, Cannistraro VJ, Ordal GW. Bacillus subtilis hydrolyzes CheY-P at the location of its action, the flagellar switch. J Biol Chem. 2003;278:48611–48616. doi: 10.1074/jbc.M306180200. [DOI] [PubMed] [Google Scholar]
- Terry K, Go AC, Ottemann KM. Proteomic mapping of a suppressor of non-chemotactic cheW mutants reveals that Helicobacter pylori contains a new chemotaxis protein. Mol Microbiol. 2006;61:871–882. doi: 10.1111/j.1365-2958.2006.05283.x. [DOI] [PubMed] [Google Scholar]
- Terry K, Williams SM, Connolly LL, Ottemann KM. Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect Immun. 2005;73:803–811. doi: 10.1128/IAI.73.2.803-811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LJ, Danon SJ, Wilson JE, O'Rourke JL, Salama NR, Falkow S, et al. Chronic Helicobacter pylori infection with Sydney strain 1 and a newly identified mouse-adapted strain (Sydney strain 2000) in C57BL/6 and BALB/c mice. Infect Immun. 2004;72:4668–4679. doi: 10.1128/IAI.72.8.4668-4679.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomb JFJ, White OO, Kerlavage ARA, Clayton RAR, Sutton GGG, Fleischmann RDR, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- Wang H, Matsumura P. Characterization of the CheAS/CheZ complex: a specific interaction resulting in enhanced dephosphorylating activity on CheY-phosphate. Mol Microbiol. 1996;19:695–703. doi: 10.1046/j.1365-2958.1996.393934.x. [DOI] [PubMed] [Google Scholar]
- Williams SM, Chen YT, Andermann TM, Carter JE, McGee DJ, Ottemann KM. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect Immun. 2007;75:3747–3757. doi: 10.1128/IAI.00082-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ, Berg HC. Migration of bacteria in semisolid agar. Proc Natl Acad Sci USA. 1989;86:6973–6977. doi: 10.1073/pnas.86.18.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuichet K, Alexander RP, Zhulin IB. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Meth Enzymol. 2007;422:1–31. doi: 10.1016/S0076-6879(06)22001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Collins EJ, Bourret RB, Silversmith RE. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat Struct Biol. 2002;9:570–575. doi: 10.1038/nsb816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.