Abstract
High-throughput mapping of retroviral vector integration sites (RIS) has become an invaluable tool to evaluate novel gene therapy vectors and to track clonal contribution in preclinical and clinical studies. Beard et al. (Methods Mol Biol 2014;1185:321–344) described an improved protocol developed for efficient capture, sequencing, and analysis of RIS that preserves gene-modified clonal contribution information. Here we describe adaptations to the previously published modified genomic sequencing PCR (MGS-PCR) protocol using the Illumina MiSeq paired-end sequencing platform. Lentiviral, gammaretroviral, and foamy virus vector integrations were analyzed. MGS-PCR using the MiSeq platform allows for the use of merged paired-end reads, which allows for efficient localization of RIS to published genomes.
Introduction
Next-generation sequencing (NGS) has been instrumental for retroviral vector integration site (RIS) analysis for gene therapy studies. Mapping RIS is crucial for the development of improved vectors and to study clonal contribution in both preclinical and clinical studies. We set out to adapt the modified genomic sequencing PCR (MGS-PCR) protocol described by Beard et al. for the Illumina MiSeq paired-end sequencing platform.1 MGS-PCR allows for the rapid identification of RIS without restriction enzyme bias. The MGS-PCR method utilizes shearing that allows users to analyze integration sites by the number of unique span or shear sites for an integration, in addition to the read frequency, thereby improving the accuracy of clonal identification and reducing potential PCR bias. Coupling MGS-PCR with the Illumina MiSeq platform has several advantages, including the widespread use of the MiSeq platform, cost-effective sequencing, low error rate, and the ability of paired-end sequencing to improve sequence quality by sequencing both ends of the DNA fragments.2 MiSeq paired-end sequencing provides longer sequence reads when merged, which improves the alignment of sequences to the reference genome to identify RIS. Here we describe modifications to the MGS-PCR protocol using the Illumina MiSeq paired-end sequencing platform for lentiviral, gammaretroviral, and foamy virus vectors.
Materials and Methods
Reagents
The following reagents were used for MGS-PCR. Agencourt AMPure XP Beads (102492-756), Formamide (97062-006), Triton X-100 (AAA16046-AE), and Phusion High-Fidelity PCR Master Mix with HF Buffer (101641-016) were all purchased from VWR (Radnor, PA). Fast-link DNA Ligation Kit (LK0750H), Dynabeads M-280 Streptavidin (11205D), and BSA (20 mg/mL) (BP675-1) were all purchased from Fisher (Waltham, MA). MinElute PCR Purification Kit (28004) and QIAquick Gel Extraction Kit (28704) were purchased from Qiagen (Valencia, CA). DNATerminator End Repair Kit was purchased from Lucigen Corp. (Middleton, WI).
Modified MGS-PCR sequencing
Samples were prepared as previously described.1 Briefly, 3 μg of genomic DNA was sheared using either a fluidic HydroShear device (Digilab Inc., Marlborough, MA) using the standard shearing assembly and set to speed code 3 for 20 cycles to obtain 1,500 bp fragments, or an M220 Focused-ultrasonicator (Covaris, Woburn, MA) using AFA snap cap microTUBEs, and a 1,500 bp fragmentation setting. The fragments were blunt ended using the DNATerminator End Repair Kit (Lucigen Corp.), and the linker cassette described previously was ligated to both ends of the genomic fragment using the Fast-link DNA Ligation Kit (LK0750H; Fisher). Before exponential PCR, samples were captured with AMPure XP Beads (102492-756; VWR) to exclude small DNA fragments. All PCR were conducted using a 2720 Thermo Cycler (Applied Biosystems, Waltham, MA). Thirty rounds of exponential PCR were conducted using a biotin-conjugated LTR-specific primer and a linker-specific primer.
LTR-containing amplicons were enriched with Dynabeads M-280 Streptavidin (11205D; Fisher), and purified using MinElute PCR Purification Kit (28004; Qiagen). Amplicons were then amplified for 30 more rounds of nested PCR with LTR-specific and linker-specific Illumina universal forward and reverse primers. Primers contained the D-series Illumina indices (Table 1) allowing for multiple samples to be pooled and sequenced in a single run. Samples were electrophoresed on 2% agarose gels, and 400–800 bp amplicons were recovered using the QIAquick Gel Extraction Kit (28704; Qiagen).
Table 1.
MGS-PCR primers and indices
| A. Exponential PCR primers | |
|---|---|
| Linker | GACCCGGGAGATCTGAATTC |
| Biotin-Lenti | 5′Biosg/AGCTTGCCTTGAGTGCTTCA |
| Biotin-Foamy | 5′Biosg/ACCGACTTGATTCGAGAACC |
| Biotin-Gamma | 5′Biosg/CAGTTCGCTTCTCGCTTCTG |
| B. i5 and i7 Illumina primer library indices | |||||||
|---|---|---|---|---|---|---|---|
| i5 index | Sequence | i5 index | Sequence | i7 index | Sequence | i7 index | Sequence |
| D501 | TATAGCCT | D505 | AGGCGAAG | D701 | ATTACTCG | D709 | CGGCTATG |
| D502 | ATAGAGGC | D506 | TAATCTTA | D702 | TCCGGAGA | D710 | TCCGCGAA |
| D503 | CCTATCCT | D507 | CAGGACGT | D703 | CGCTCATT | D711 | TCTCGCGC |
| D504 | GGCTCTGA | D508 | GTACTGAC | D704 | GAGATTCC | D712 | AGCGATAG |
| C. Nested Illumina MiSeq PCR primers | |
|---|---|
| Foamy2 Nested 504 (F.Stem/i504/F.Adapter/Primer) | AATGATACGGCGACCACCGAGATCTACAC/GGCTCTGA/ACACTCTTTCCCTACACGACGCTCTTCCGATCT/GCTAAGGGAGACATCTAGTG |
| Foamy2 Nested 507 (F.Stem/i507/F.Adapter/Primer) | AATGATACGGCGACCACCGAGATCTACAC/CAGGACGT/ACACTCTTTCCCTACACGACGCTCTTCCGATCT/GCTAAGGGAGACATCTAGTG |
| Lenti2 Nested 505 (F.Stem/i505/F.Adapter/Primer) | AATGATACGGCGACCACCGAGATCTACAC/AGGCGAAG/ACACTCTTTCCCTACACGACGCTCTTCCGATCT/AGTAGTGTGTGCCCGTCTGT |
| Lenti2 Nested 508 (F.Stem/i508/F.Adapter/Primer) | AATGATACGGCGACCACCGAGATCTACAC/GTACTGAC/ACACTCTTTCCCTACACGACGCTCTTCCGATCT/AGTAGTGTGTGCCCGTCTGT |
| GammaLTR1 Nested506 (F.Stem/i506/F.Adapter/Primer) | AATGATACGGCGACCACCGAGATCTACAC/TAATCTTA/ACACTCTTTCCCTACACGACGCTCTTCCGATCT/GTGGTCTCGCTGTTCCTTGG |
| Gamma LTR6 Nested 502 (F.Stem/i502/F.Adapter/Primer) | AATGATACGGCGACCACCGAGATCTACAC/ATAGAGGC/ACACTCTTTCCCTACACGACGCTCTTCCGATCT/TCTGCTCCCCGAGCTCAATA |
| GammaLTR1501 (F.Stem/i501/F.Adapter/Primer) | AATGATACGGCGACCACCGAGATCTACAC/TATAGCCT/ACACTCTTTCCCTACACGACGCTCTTCCGATCT/GTGGTCTCGCTGTTCCTTGG |
| Linker Primer2 701 (R.Stem/i701/R.Adapter/Primer) | CAAGCAGAAGACGGCATACGAGAT/ATTACTCG/GTGACTGGAGTTCAGACGTGTGCTCTTCCGATC/GATCTGAATTCAGTGGCACAG |
| Linker Primer2 702 (R.Stem/i702/R.Adapter/Primer) | CAAGCAGAAGACGGCATACGAGAT/TCCGGAGA/GTGACTGGAGTTCAGACGTGTGCTCTTCCGATC/GATCTGAATTCAGTGGCACAG |
| Linker Primer2 703 (R.Stem/i703/R.Adapter/Primer) | CAAGCAGAAGACGGCATACGAGAT/CGCTCATT/GTGACTGGAGTTCAGACGTGTGCTCTTCCGATC/GATCTGAATTCAGTGGCACAG |
| Linker Primer2 704 (R.Stem/i704/R.Adapter/Primer) | CAAGCAGAAGACGGCATACGAGAT/GAGATTCC/GTGACTGGAGTTCAGACGTGTGCTCTTCCGATC/GATCTGAATTCAGTGGCACAG |
| Linker Primer2 705 (R.Stem/i705/R.Adapter/Primer) | CAAGCAGAAGACGGCATACGAGAT/ATTCAGAA/GTGACTGGAGTTCAGACGTGTGCTCTTCCGATC/GATCTGAATTCAGTGGCACAG |
| Linker Primer2 706 (R.Stem/i706/R.Adapter/Primer) | CAAGCAGAAGACGGCATACGAGAT/GAATTCGT/GTGACTGGAGTTCAGACGTGTGCTCTTCCGATC/GATCTGAATTCAGTGGCACAG |
Linker- and biotin-tagged LTR-specific primers are reported for lentiviral, gammaretroviral, and foamy viral vectors. Nested Illumina MiSeq–adapted primers are reported for lentiviral, gammaretroviral, and foamy virus vectors. The i5 and i7 indices library is also given with the corresponding sequences.
Exponential and nested PCR primers
All PCR primers for lentiviral, gammaretroviral, and foamy virus vectors and linkers were synthesized using Integrated DNA Technologies (IDT, Coralville, IA). Exponential LTR-specific primers were conjugated with biotin. Nested Illumina universal primers were ordered with HPLC purification and made using the following templates containing the Illumina adapters, library indices (i5, i7), Illumina stems, and LTR/linker-specific sequences. For the complete list of exponential and nested PCR primers with indices, see Table 1.
MiSeq paired-end amplicon sequencing
Gel-isolated amplicons were analyzed at the Genomic Sequencing and Analysis Facility, University Texas at Austin, and sequenced on the MiSeq PE 2x300 platform. Sequences were de-multiplexed and sorted postsequencing using the i5 and i7 indices from the Illumina library by University Texas at Austin, and returned in individual FASTQ files corresponding to the appropriate vectors. Error rates for Illumina MiSeq have been previously described.2
Merging paired-end reads
To merge the forward and reverse reads into a single sequence read, PEAR—a fast and accurate Illumina Paired-end reAd mergeR—was used on a Linux operating system using the standard settings.3 PHRED: 33; Using empirical frequencies: YES; Statistical method: OES; Maximum assembly length: 999999; Minimum assembly length: 50; p-value: 0.01; Quality score threshold (trimming): 0; Minimum read size after trimming: 1; Maximal ratio of uncalled bases: 1; Minimum overlap: 10; Scoring method: Scaled score; Threads: 1.
Bioinformatics for vector integration site analysis
Merged paired sequence reads were processed using the Vector Integration Site Analysis software (VISA).4 VISA recognizes and removes the LTR and linker sequences from the merged paired sequence read. VISA then aligns the trimmed query sequence to the genome and reports the chromosomal location of integration (i.e., Ch11;134,408,796–134,408,978:), and also reports the proximity of RefSeq genes and transcription start sites near identified integrations.
Results and Discussion
There have been many recent gene therapy successes, and gene-modified cells have been used in hundreds of patients.5–7 However, vector-mediated genotoxicity is still a major concern and careful tracking of RIS has been mandated by the U.S. Food and Drug Administration. Furthermore, the use of retroviral vectors in forward mutagenesis screens requires a rapid high-throughput methodology to recover candidate genes for validation and clinical translation.
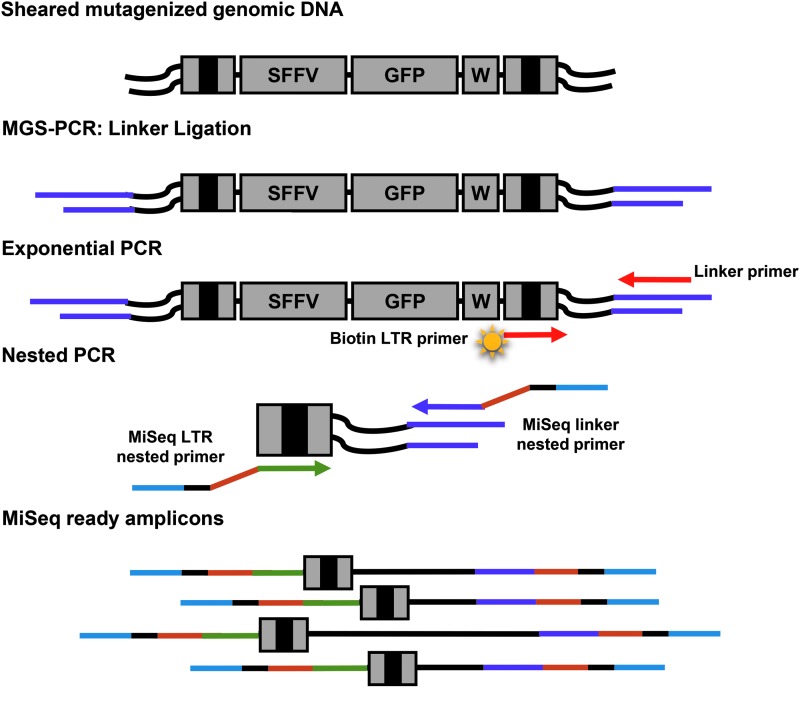
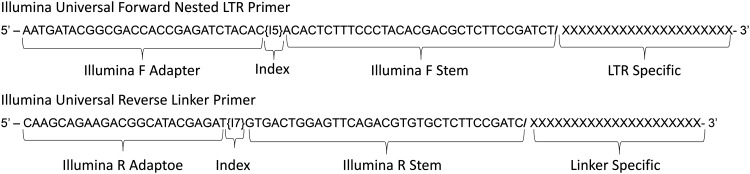
High-throughput mapping of RIS has become an invaluable tool to evaluate oncogenes and novel gene therapy vectors, and to track clonal contribution in preclinical and clinical studies. NGS has greatly improved the sensitivity of these studies. However, NGS platforms continue to evolve and require the adaptation of established protocols for use with these newer platforms. Here we set out to adapt the high-throughput MGS-PCR protocol published by Beard et al.1 for vector RIS analysis with the Illumina MiSeq paired-end sequencing platform (Fig. 1). MGS-PCR utilizes exponential PCR coupled with nested PCR to rapidly identify RIS and allows for analysis of the number of different span or shear lengths to evaluate clonality. The MGS-PCR protocol can be adapted to other NGS platforms as older platforms are discontinued. For adaptation, modified nested PCR primers including the Illumina universal adapters, indices, and stems for MiSeq paired-end sequencing were prepared (Fig. 2).
Figure 1.
Illumina MiSeq MGS-PCR. Genomic DNA is sheared, resulting in smaller genomic fragments that may contain the vector provirus. Fragments are blunt ended and linker cassettes ligated to the ends of the fragment. Thirty rounds of exponential PCR are conducted with a biotin-tagged LTR primer and linker-specific primer. The resulting amplicons undergo selection of biotin-tagged amplicons with streptavidin beads to enrich for fragments containing LTRs. Amplicons are then recovered and 30 rounds of nested PCR performed using an Illumina universal forward LTR-specific primer and an Illumina universal reverse linker-specific primer are performed. The resulting amplicons are gel isolated and ready for sequencing on the MiSeq. Color images available online at www.liebertpub.com/hgtb
Figure 2.
Illumina MiSeq universal primers. The Illumina universal primers (forward and reverse) consist of an adapter, indices, stem, and the user-defined specific primer sequences. In our adaptation for MGS-PCR, the forward primer is designed for the LTR, while the reverse primer is designed for the linker region.
MiSeq MGS-PCR samples
We performed Illumina MiSeq paired-end sequencing on MGS-PCR samples that had expanded clones, or were oligoclonal, or highly polyclonal: (1) lentiviral vector-mutagenized orthotopic primary prostate tumors that had expanded clones, (2) gammaretroviral vector-mutagenized CD105+, Sca-1+-enriched mouse bone marrow that was expanded in vitro and was oligoclonal, and (3) foamy viral vector-mutagenized human CD34+ cells that were highly polyclonal. Our gammaretroviral vectors were derived from the murine leukemia virus (MLV), and here we report novel primer pairs for MLV-derived gammaretroviral vectors for MGS-PCR (Table 1). Three independent shears were performed per sample, and the barcoded amplicons were sequenced in three Illumina MiSeq paired-end runs.
Analysis of MGS-PCR amplicons
Samples were sequenced by utilizing unique primer barcodes (i5, i7 Illumina D Series Indices) allowing for the multiplexing of several samples in a single run. Cross-contamination from multiplexed samples or back-to-back runs has been calculated for the Illumina MiSeq. Back-to-back runs that had very different sample types (e.g., amplicon library on run 1, whole genome shot gun on run 2) but the same barcodes had an observed contamination frequency of <0.01% based upon the detection of amplicon features in the whole genome shotgun library at the University of Texas at Austin sequencing facility. Sequence reads were returned de-multiplexed and sorted by vector type using the unique i5 and i7 D series indices imbedded from nested PCR. The paired forward and reverse reads were then merged using the paired-end read platform (PEAR) resulting in longer, high-quality reads.3 PEAR can be used with high confidence to align and merge sequence reads even when the paired reads cannot be merged, if the amplicons are of a known size. Since we sequenced a range of amplicons (400–800 bp), we chose to analyze only reads that could be merged. Using PEAR, an average of >90% of sequence reads were successfully aligned and merged (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/hgbt). The quality and length of the reads was dramatically improved by using merged paired reads across all data sets as demonstrated by using the FastQC quality control tool for high-throughput sequence data available from Babraham Bioinformatics (Supplementary Fig. S2). Analysis of each DNA base by PHRED score in the FastQ sequencing files for forward and reverse reads alone, were then compared to merged paired sequences. This demonstrated dramatically improved base pair quality for merged paired sequences (Supplementary Fig. S2).
RIS analysis
Two to five million sequence reads were obtained per sample of lentiviral, gammaretroviral, and foamy viral vector-mutagenized samples (Table 2). Overall, MiSeq paired-end sequencing improved the data set quality (PHRED score) and genomic alignment score and resulted in the identification of several hundred unique RIS in each sample (Table 2). We were interested to see if the merged paired-end sequencing was able to alleviate a common mapping problem in which integrations occurring in repetitive regions of the genome cannot reliably be mapped. Our hypothesis was that increasing the query sequence length using merged paired sequencing would reduce the number of reads that could not be mapped. Therefore, we compared analysis of the unmerged sequence files to the merged paired sequence files and assessed the percent of reads that could be mapped in or near repetitive elements compared with those that could not be mapped near these regions. Merged paired sequences significantly reduced the percentage of reads that could not be mapped because of integration in repeat regions of the genome, and improved alignment or blastbit score with the genome, thus identifying more RIS (Supplementary Table S1). This is consistent with the literature that longer merged paired sequences can help resolve the location of an integration that occurs within a repetitive region of the genome.8,9
Table 2.
Analysis of lenti, gamma, and foamy RIS by MGS-PCR with MiSeq
| A. Triplicate data for lentiviral vector samples | ||||||
|---|---|---|---|---|---|---|
| Lenti viral vector | SET A | SET B | Set C | |||
| Total reads | 3,636,088 | 3,163,330 | 1,198,876 | |||
| Alignment filters | N | % | N | % | N | % |
| No LTR-Chr junction | 1,906,919 | 52.44 | 1,594,545 | 50.40 | 618,374 | 51.57 |
| Query less than 30 bp | 696,270 | 19.14 | 596,832 | 18.86 | 223,263 | 18.62 |
| No BLAT alignment | 653,463 | 17.97 | 757,407 | 18.73 | 201,551 | 16.81 |
| Repeat filter | 28,350 | 0.78 | 28,260 | 0.89 | 9,322 | 0.77 |
| Low identity alignments | 9,401 | 0.25 | 10,075 | 0.31 | 1,136 | 0.09 |
| Candidate RIS | 341,685 | 9.39 | 340,845 | 10.77 | 145,230 | 12.11 |
| Unique RIS filters | ||||||
| Total RIS | 341,685 | 340,845 | 145,230 | |||
| Unique RIS | 318 | 0.093 | 290 | 0.085 | 134 | 0.092 |
| B. Triplicate data for gammaretroviral vector samples | ||||||
|---|---|---|---|---|---|---|
| Gammaretroviral vector | SET A | SET B | SET C | |||
| Total reads | 2,705,335 | 3,841,112 | 3,558,352 | |||
| Alignment filters | N | % | N | % | N | % |
| No LTR-Chr junction | 1,443,168 | 53.34 | 1,982,913 | 51.62 | 1,853,803 | 52 |
| Query less than 30 bp | 334,576 | 12.37 | 464,337 | 12.08 | 429,431 | 12 |
| No BLAT alignment | 57,900 | 2.14 | 103,277 | 2.68 | 167,423 | 5 |
| Repeat filter | 46,336 | 0.88 | 794,665 | 20.68 | 668,024 | 19 |
| Low identity alignments | 11,722 | 0.43 | 24,885 | 0.64 | 28,183 | 1 |
| Candidate RIS | 287,545 | 10.63 | 472,764 | 12.30 | 411,488 | 12 |
| Unique RIS filters | ||||||
| Total RIS | 287,545 | 472,764 | 411,488 | |||
| Unique RIS | 337 | 0.117 | 483 | 0.10 | 667 | 0 |
| C. Triplicate data for foamy viral vector samples | ||||||
|---|---|---|---|---|---|---|
| Foamy viral vector | SET A | SET B | SET C | |||
| Total reads | 4,069,518 | 5,250,488 | 4,263,921 | |||
| Alignment filters | N | % | N | % | N | % |
| No LTR-Chr junction | 2,431,679 | 59.75 | 3,142,864 | 59.86 | 2,602,694 | 61.04 |
| Query less than 30 bp | 559,789 | 13.75 | 797,934 | 15.20 | 708,551 | 16.62 |
| No BLAT alignment | 701,473 | 17.23 | 829,096 | 15.80 | 640,730 | 15.03 |
| Repeat filter | 37,702 | 0.92 | 46,336 | 0.90 | 29,864 | 0.70 |
| Low identity alignments | 10,181 | 0.25 | 13,682 | 0.26 | 11589 | 0.27 |
| Candidate RIS | 328,694 | 8.07 | 420,576 | 8.01 | 270,493 | 6.34 |
| Unique RIS filters | ||||||
| Total RIS | 328,694 | 420,576 | 270,493 | |||
| Unique RIS | 10,616 | 3.23 | 11,257 | 2.677 | 13,119 | 4.85 |
Three independent shears were performed for each sample, and sequenced on three MiSeq paired-end runs. Merged paired-end sequences were analyzed. The table reports the total number of retroviral integration sites (RIS), the number and percentage of those potential RIS as they pass through alignment and inclusion filters, and the total number of aligned RIS and the total number of unique RIS.
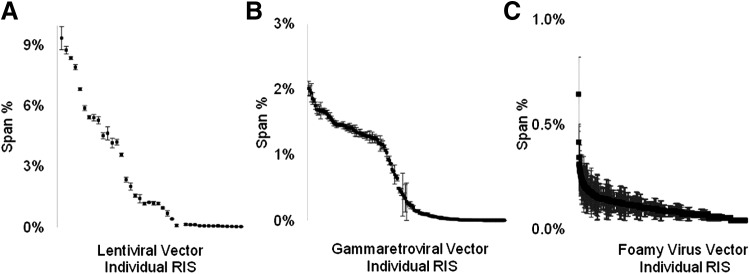
By evaluating the number of span counts for a given RIS, the clonal contribution can be estimated.10 We evaluated the span count frequency for lentiviral samples from a mutagenesis screen where clonal outgrowth was observed (Fig. 3A). As expected, the most frequently observed RIS represented a high percentage (>9%) of all RIS identified. In addition, there was very little variation between the three replicates, further indicating clonal outgrowth in this sample. For the gammaretroviral sample, specific clones were above 2% of all detected RIS by span count analysis (Fig. 3B). For the highly polyclonal foamy viral vector-mutagenized samples, the span count frequency for the most frequently identified RIS was less than 0.4% of all unique integrations (Fig. 3C). In the lentiviral, gammaretroviral, and foamy viral vector data sets, we demonstrated the ability of MGS-PCR to reproducibly capture the representative clonal frequencies within a given sample and detect clonal skewing when present. MGS-PCR when paired with the MiSeq paired-end sequencing was able to reproducibly capture and report clonal contributions within the same vector-modified sample in three independent shears analyzed in different sequencing reactions.
Figure 3.
Evaluation of clonal contribution. The clonal frequency was determined using the span count percentage. All data points represent a single retroviral integration site (RIS), identified in triplicate samples, and are presented with standard error. (A) Lentiviral-mutagenized samples exhibiting clonal expansion, (B) oligoclonal gamma retroviral-mutagenized samples, and (C) polyclonal foamy virus vector-mutagenized samples.
For the highly polyclonal foamy virus vector samples, we expected a lower frequency of overlapping RIS between replicates when using span count analysis. This is in part because there are over 10,000 unique RIS per replicate. Consistent with our hypothesis we observed much more variation in span count frequency for each RIS in the polyclonal foamy viral vector-mutagenized samples. This was evidenced by less than 10% of all unique integrations being recaptured between the three replicates, and low span count frequencies. In contrast, in the lentiviral and gammaretroviral vector samples, we recaptured >60% of all RIS between replicates with minimal variance in gene rank and span count percent (Fig. 3).
In summary, we have adapted the MGS-PCR protocol published by Beard et al.1 with the Illumina MiSeq paired-end sequencing platform for efficient identification of RIS in gene-modified cells. Using this approach we were able to reproducibly achieve high-throughput recovery of high-quality reads for lentiviral, gammaretroviral, and foamy virus vector RIS. The MiSeq adaptation of the MGS-PCR protocol is important for continued development of gene therapy vectors, and analysis of clonality in patients and preclinical studies. In the future, modifications and optimization of the MGS-PCR protocol may further improve upon the technology for tracking of clonal contribution both in preclinical and clinical settings.
Supplementary Material
Acknowledgments
These studies were supported by NIH Grants AI097100, AI102672, and CA173598 and by the Department of Defense Peer Reviewed Cancer Research Program under award number W81XWH-11-1-0576.
Disclaimer
Views and opinions of and endorsements by the authors do not reflect those of the U.S. Army or the Department of Defense.
Author Disclosure
No competing financial interests exist.
References
- 1.Beard BC, Adair JE, Trobridge GD, Kiem HP. High-throughput genomic mapping of vector integration sites in gene therapy studies. Methods Mol Biol 2014;1185:321–344 [DOI] [PubMed] [Google Scholar]
- 2.Quail MA, Smith M, Coupland P, et al. A tale of three next generation sequencing platforms: Comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 2012;13:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014;30:614–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hocum JD, Battrell LR, Maynard R, et al. VISA—Vector Integration Site Analysis server: A web-based server to rapidly identify retroviral integration sites from next-generation sequencing. BMC Bioinform 2015;16:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim Y, Schmidt-Wolf IG. [Gene therapy in Germany: From past to present]. Dtsch Med Wochenschr 2015;140:684–686 [DOI] [PubMed] [Google Scholar]
- 6.Fischer A. Gene therapy: Repair and replace. Nature 2014;510:226–227 [DOI] [PubMed] [Google Scholar]
- 7.Ginn SL, Alexander IE, Edelstein ML, et al. Gene therapy clinical trials worldwide to 2012—an update. J Gene Med 2013;15:65–77 [DOI] [PubMed] [Google Scholar]
- 8.Magoc T, Salzberg SL. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011;27:2957–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ekblom R, Wolf JB. A field guide to whole-genome sequencing, assembly and annotation. Evol Appl 2014;7:1026–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou S, Bonner MA, Wang YD, et al. Quantitative shearing linear amplification polymerase chain reaction: An improved method for quantifying lentiviral vector insertion sites in transplanted hematopoietic cell systems. Hum Gene Ther Methods 2015;26:4–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.