Programmed cell death occurring as an integral part of plant development is characterized by the transcriptional activation of a distinct core of conserved cell death-associated genes.
Abstract
A plethora of diverse programmed cell death (PCD) processes has been described in living organisms. In animals and plants, different forms of PCD play crucial roles in development, immunity, and responses to the environment. While the molecular control of some animal PCD forms such as apoptosis is known in great detail, we still know comparatively little about the regulation of the diverse types of plant PCD. In part, this deficiency in molecular understanding is caused by the lack of reliable reporters to detect PCD processes. Here, we addressed this issue by using a combination of bioinformatics approaches to identify commonly regulated genes during diverse plant PCD processes in Arabidopsis (Arabidopsis thaliana). Our results indicate that the transcriptional signatures of developmentally controlled cell death are largely distinct from the ones associated with environmentally induced cell death. Moreover, different cases of developmental PCD share a set of cell death-associated genes. Most of these genes are evolutionary conserved within the green plant lineage, arguing for an evolutionary conserved core machinery of developmental PCD. Based on this information, we established an array of specific promoter-reporter lines for developmental PCD in Arabidopsis. These PCD indicators represent a powerful resource that can be used in addition to established morphological and biochemical methods to detect and analyze PCD processes in vivo and in planta.
Programmed cell death (PCD) is a fundamental process of life. Already present in clonal colonies of prokaryotes (Bayles, 2014), PCD has evolved to become an essential mechanism in multicellular eukaryotes (Wang and Bayles, 2013). Many different forms of PCD have been recognized, but a unifying definition characterizes PCD as genetically encoded, actively controlled cellular suicide.
In animals and plants, PCD is involved in many aspects of development, sculpting structures or deleting unwanted tissues (Fuchs and Steller, 2011; Van Hautegem et al., 2015). Over the last two decades, intensive investigations have revealed mechanisms controlling different forms of animal PCD; the most prominent among them is apoptotic PCD (Green, 2011). In comparison, there is still little knowledge of the molecular networks controlling PCD in plants, despite its abundance and its importance for plant life: plant PCD occurs as an integral part of development (dPCD) as well as of the plant’s reactions to biotic and abiotic environmental challenges (ePCD; Lam, 2004). Concerning dPCD, a distinction can be made between (1) differentiation-induced PCD that occurs as final differentiation step in specific cell types, for instance, in xylem tracheary elements, the root cap, or the anther tapetum layer (Plackett et al., 2011; Bollhöner et al., 2012; Fendrych et al., 2014), and (2) age-induced PCD as the last step of organ senescence that occurs in all tissues of an organ or even the entire plant at the end of its life cycle (Thomas, 2013). Regarding ePCD, one of the most studied PCD processes occurs during the hypersensitive response (HR), a localized plant response upon pathogen recognition (Coll et al., 2011; Wu et al., 2014). Also abiotic stresses such as heat, UV radiation, or salt stress can lead to cell death displaying certain hallmarks of PCD (Chen et al., 2009; Qi et al., 2011; Nawkar et al., 2013; Petrov et al., 2015).
It is still unclear whether different PCD types in plants share common regulatory mechanisms or if they are controlled by distinct pathways. Due to the scarcity of molecular information, most comparative analyses have been based on morphological and biochemical characteristics. Vacuolar cell death, defined by accumulation of autophagosomes, vacuolar collapse, and corpse degradation, has been opposed to necrotic cell death, with swelling of mitochondria, protoplast shrinkage and unprocessed cell corpses (van Doorn et al., 2011). However, some types of PCD, including HR cell death, pollen self-incompatibility, or endosperm cell death, do not fall into either of these proposed classes (van Doorn et al., 2011).
Here, we exploited publicly available genome-wide transcriptome data that were associated with different forms of cell death in the model plant Arabidopsis (Arabidopsis thaliana), with the aim to comparatively characterize plant PCD types. We identified distinct sets of differentially regulated genes in several developmental and environmental situations known to provoke plant cell death, suggesting that dPCD and ePCD processes are characterized by separate regulatory pathways. Focusing on dPCD, we identified a conserved core of transcriptionally controlled dPCD-associated genes. Based on this information, we created and analyzed an array of promoter-reporter lines that are expressed in cells preparing for different types of dPCD. The presented data will be a powerful tool to complement morphological analysis when attempting PCD discovery, recognition, and analysis of dPCD types in plants.
RESULTS
Meta-Analysis of Available ATH1 Data Sets Reveals Distinct Gene Expression Patterns Characterizing dPCD and ePCD
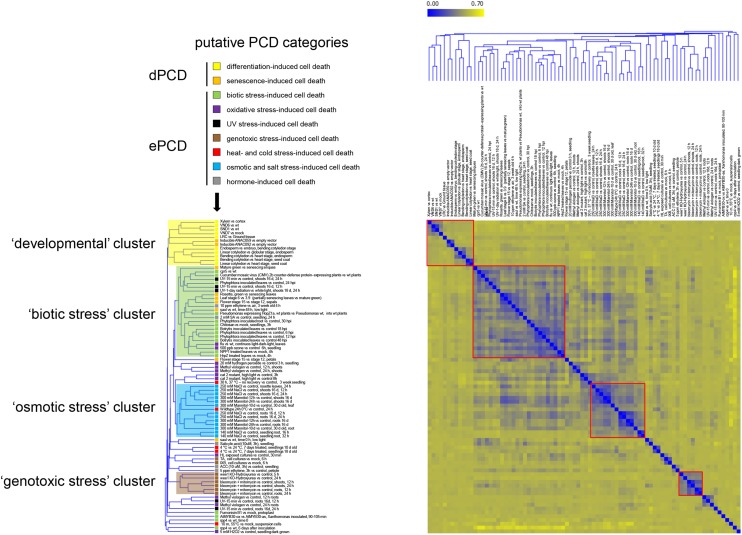
To get a view on similarities and differences in the gene expression profiles of different PCD types, we carried out a meta-analysis of Arabidopsis Affymetrix GeneChip Genome Array (ATH1) data sets. Based on their accompanying experimental descriptions, we selected a total of 59 ATH1 data sets associated with a range of generally accepted or hypothetical PCD contexts. For simplicity, we will refer to all of these contexts as PCD, though for some of them, the actively controlled nature of the cell death has not been unambiguously shown. From this compendium, we extracted 82 conditions, contrasting different cell death situations with their corresponding non-PCD controls (Table I; Supplemental Tables S1 and S2). We assigned these contrasts to nine categories based on their experimental context. The dPCD category differentiation-induced cell death contains experiments describing specific cell types undergoing cell death as part of their differentiation program, while the senescence-induced cell death category comprises data sets produced from entire organs during late stages of senescence. In the ePCD categories, data sets produced from pathogen assays (biotic stress), from plants experiencing oxidative stress, and from plants exposed to UV irradiation, genotoxic compounds, high or low temperatures, and osmotic and salt stresses were included. Finally, data sets from hormone treatments leading to cell death complete the list of putative PCD categories (Fig. 1).
Table I. Overview of the number of conditions profiled per PCD subcategory in the ATH1 compendium.
| PCD Category | PCD Subcategory | Tissue, Organ, and Stress Type | No. of Conditions |
|---|---|---|---|
| Tracheary elements | 4 | ||
| LRC | 1 | ||
| Differentiation-induced | Endosperm | 3 | |
| Seed coat | 2 | ||
| Developmental (dPCD) | Leaves | 4 | |
| Senescence-induced | Petals | 1 | |
| Sepals | 1 | ||
| Siliques | 1 | ||
| Mutant seedlings | 2 | ||
| Biotic stress-induced | Fungal elicitor | 12 | |
| Bacterial elicitor | 3 | ||
| Viral protein | 1 | ||
| Environmental (ePCD) | Abiotic stress-induced | Oxidative stress | 11 |
| UV stress | 5 | ||
| Genotoxic stress | 8 | ||
| Heat stress | 2 | ||
| Cold stress | 3 | ||
| Osmotic stress | 6 | ||
| Salt stress | 7 | ||
| Hormone treatment | Ethylene | 3 | |
| SA | 2 | ||
| Total | 82 |
Figure 1.
PCD-associated ATH1 transcriptome data sets group in distinct clusters. HCA showing the clustering of 82 putative dPCD and ePCD conditions based on the log-fold expression values of differentially regulated genes and indicating their affiliation to different putative PCD categories (arrow). Four clusters are highlighted indicating the relatedness of data sets falling in the developmental, the biotic stress, the osmotic stress, and the genotoxic stress clusters. The color coding from blue to yellow indicates an increase in the Pearson’s correlation distance.
To define the relatedness of the ATH1 data sets, independent of predefined PCD categories, we performed a hierarchical clustering analysis (HCA) based on the expression profiles of all genes that are differentially expressed in at least one condition. Although the overall similarity of the entire compendium is low, it was found to contain several functionally coherent clusters (Fig. 1). At a Pearson’s correlation distance threshold of 0.4, three clusters of more than five conditions could be defined. The biotic stress cluster mainly contains pathogen-related data sets but also contains some senescence, UV stress, and oxidative stress conditions. The osmotic stress cluster contains salt stress and osmotic stress conditions, and a third cluster indicates the tight relationship of most genotoxic stress conditions. At a more relaxed correlation distance threshold, a fourth sizeable cluster emerges. This cluster, although containing more diverse expression patterns than the other three, is also functionally coherent, encompassing all differentiation-induced dPCD conditions along with two senescence-related conditions (Fig. 1, developmental cluster).
We compared the gene expression profiles of the conditions that fell in these four clusters and identified commonly regulated genes within the clusters. In the developmental cluster, we found SERINE CARBOXYPEPTIDASE-LIKE48 (SCPL48), the aspartic protease PASPA3, BIFUNCTIONAL NUCLEASE1 (BFN1), RIBONUCLEASE3 (RNS3), CALCIUM-DEPENDENT NUCLEASE1 (CAN1), and a DOMAIN OF UNKNOWN FUNCTION679 MEMBRANE PROTEIN2 (DMP2) of unknown function up-regulated in at least 10 out of 12 conditions (Supplemental Table S3, developmental cluster). Additionally, 19 genes were found to be commonly up-regulated in at least eight out of 12 conditions, including genes of families previously implicated in different PCD processes, e.g., VACUOLAR PROCESSING ENZYMES (Hara-Nishimura and Hatsugai, 2011). The biotic cluster exhibits up-regulation of genes involved in salicylic acid (SA) and Ca2+ signaling: the SA-induced genes ENHANCED DISEASE SUSCEPTIBILITY5, PHYTOALEXIN DEFICIENT3, and WRKY DNA-BINDING PROTEIN75; the calcium-binding protein-encoding gene IQ-MOTIF PROTEIN1; and AUTOINHIBITED CA2+ ATPASE12 (Supplemental Table S3, biotic cluster). These results reflect the importance of calcium and SA signaling in the HR (Ma and Berkowitz, 2007; Mur et al., 2008). The integration of biotic stress conditions as well as senescence conditions in the biotic cluster suggests the activation of conserved processes during biotic stress and senescence conditions. In the osmotic cluster, 12 genes were up-regulated in all 14 conditions of mannitol, salt, and cold stress treatments (Supplemental Table S3, osmotic cluster), including SENESCENCE-ASSOCIATED GENE113 and several LATE EMBRYOGENESIS ABUNDANT genes, which are known to be involved in cellular protection and stress tolerance (Olvera-Carrillo et al., 2010; Candat et al., 2014). The genotoxic cluster comprised DNA repair genes such as BREAST CANCER SUSCEPTIBILITY1, RAD51 (At5g20850), and two of its paralogs, RAD17 and RAD21 (Trapp et al., 2011). Furthermore, nucleotide metabolism genes such as TSO MEANING UGLY IN CHINESE2 (TSO2, AT3G27060) and THYMIDINE KINASE1A (Roa et al., 2009) and several plant-specific SIAMESE (SIM)/SIAMESE-RELATED (SMR) CYCLIN-DEPENDENT KINASE (CDK) inhibitors (Yi et al., 2014) were commonly up-regulated in this cluster (Supplemental Table S3, genotoxic cluster).
In contrast to the observed correlation within each of the four clusters, there was little similarity between the gene expression profiles across the clusters. These results indicate that distinct gene expression patterns characterize different forms of PCD, in particular differentiation-induced dPCD and ePCD types. However, which of the differentially expressed genes are effectively involved in PCD regulation and which ones are elicited as part of processes other than PCD remains to be investigated case by case.
Most dPCD-Regulated Genes Are Not Up-Regulated in ePCD Situations
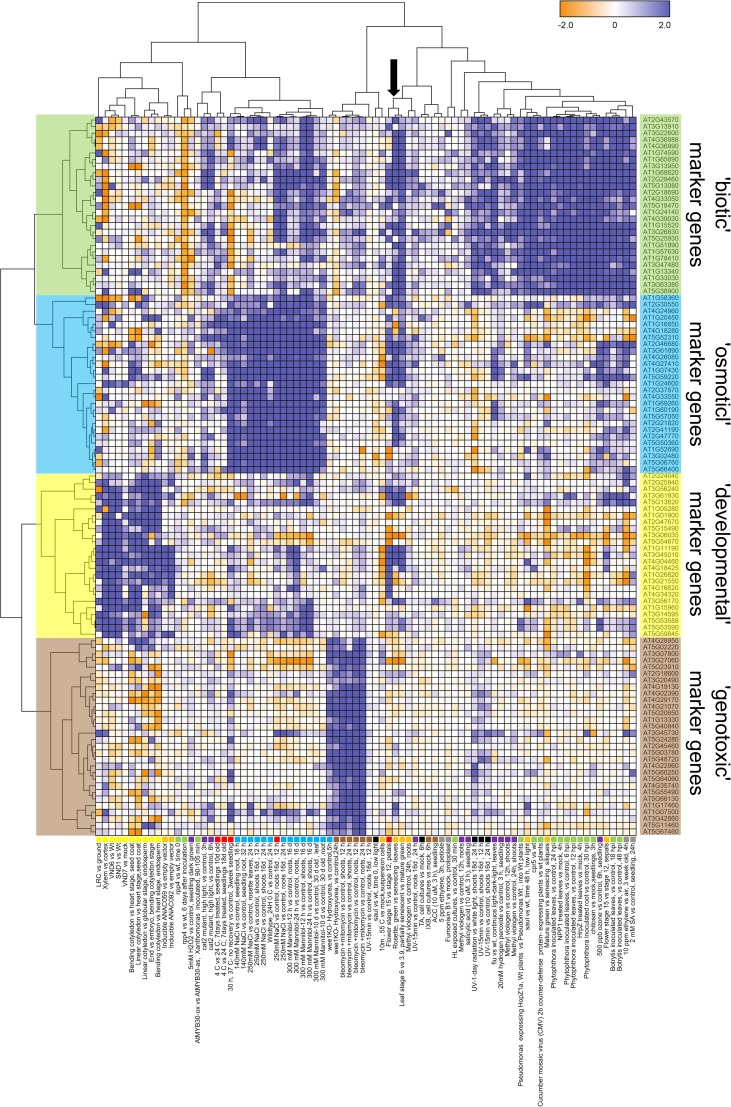
To test the hypothesis of distinct gene regulation occurring in differentiation-induced dPCD and ePCD conditions, we applied two-dimensional clustering to the expression profiles of a curated gene set, which contains the genes that are most commonly up-regulated in the four clusters described above, as identified using custom R scripts (see “Materials and Methods” and Supplemental Table S3). The resulting gene clades mirror the original four clusters, and it again appears there is little common regulation of these genes across clusters (Fig. 2).
Figure 2.
Commonly up-regulated genes within PCD clusters are largely distinct between clusters. Two-dimensional clustering of the gene-condition matrix plotting the expression profiles of the most commonly regulated genes of the four clusters highlighted in Figure 1 over all conditions. The separate blocks of dark blue fields indicate that the regulation of commonly expressed marker genes is largely distinct for each cluster. The arrow indicates a cluster of senescence-related data sets that show up-regulation of biotic, osmotic, and developmental marker genes.
The genotoxic cluster appears most distinct; only few genotoxic marker genes were up-regulated in the other conditions. One example is the CDK-inhibitor SMR5 (At1g07500) that is up-regulated in several salt and osmotic stress conditions (Fig. 2). Gene expression profiles in the osmotic stress and biotic stress clusters have larger overlaps; many biotic marker genes are up-regulated during mannitol and salt treatments, and vice versa some osmotic marker genes are up-regulated as a consequence of inoculation with the necrotrophic pathogen Botrytis cinerea (Fig. 2). The up-regulation of developmental marker genes is largely confined to the differentiation-induced dPCD data sets. Some genes, however, are also up-regulated in osmotic and salt stress conditions, suggesting a certain degree of common gene regulation (see the lower tier of developmental marker genes in Fig. 2). Interestingly, conditions of organ senescence lead to up-regulation of several biotic, osmotic, and developmental marker genes (Fig. 2, arrow), suggesting that plant senescence activates a combination of pathways. Most developmental marker genes, however, are almost exclusively up-regulated in differentiation-induced dPCD situations, suggesting that the transcriptional regulation differs substantially between these and ePCD contexts.
Supervised Classification of PCD Samples Based on Their Gene Expression Profiles Is Possible for Some PCD Types But Not for Others
Prompted by the results of the unsupervised clustering approaches in distinguishing PCD types (Figs. 1 and 2), we attempted to classify the different putative PCD categories (Fig. 1) using supervised classification algorithms, based on their ATH1 expression profiles and the putative PCD class labels assigned to them from the experimental descriptions (see “Materials and Methods”). The aim of building such classifiers is to assess the feasibility of predicting the PCD type of an unlabeled experimental sample based on its gene expression profile.
We first built Support Vector Machine (SVM; Cortes and Vapnik, 1995) and Random Forest (RF; Breiman, 2001) classifiers distinguishing ePCD- from dPCD-related conditions, based on the expression profiles of all genes. A moderate classification performance was obtained on the full data set of ePCD and dPCD conditions (Supplemental Table S4). The performance increased markedly when excluding minority subclasses, i.e., senescence (for dPCD) and/or temperature stress, UV stress, oxidative stress, and hormone treatments (for ePCD). Using the curated set of putative PCD indicators for the four major PCD clusters described above (Fig. 2; Supplemental Table S3 instead of all genes as classification features did not generally lead to improved classification performance (Supplemental Table S4). These results indicate that a clear molecular distinction of ePCD versus dPCD is hampered by expression similarities between certain subtypes of ePCD and dPCD. In particular, the expression profile similarities between senescence-induced dPCD and various ePCD conditions (see Fig. 2) appear to have a negative impact on the dPCD/ePCD classification performance (Supplemental Table S4). To investigate which PCD subtypes suffer the most from expression similarities with other subtypes, we attempted to classify particular PCD subtypes against all other types (Supplemental Table S4). Whereas the maximum classification performance is high for differentiation-induced dPCD, genotoxic cell death, and osmotic cell death, the performance values for other PCD subtypes are moderate to low, reflecting a lack of adequately distinctive expression signatures to separate these poorly performing PCD subtypes from some of the other PCD types grouped together as the alternative label set. Taken together, with the ATH1 data sets that are publically available at this point, unconditionally distinctive sets of marker genes are hard to find for many PCD subtypes, even when using supervised classification strategies.
Identification of Unique dPCD Indicator Genes
The information that genes are predominantly up-regulated in differentiation-induced dPCD types opened the possibility of testing some of these genes for their aptitude as dPCD reporter genes. We took three complementary approaches to identify individual genes that could potentially be used as dPCD markers.
First, we compiled a list of genes that are significantly up-regulated at least 2-fold in at least 60% of the dPCD data sets in the ATH1 compendium described above. SCPL48 and PASPA3 show the highest frequency of up-regulation in all dPCD data sets (89% and 84%, respectively). TELOMERIC DNA-BINDING PROTEIN1 (At5g13820) is up-regulated in 79% of all dPCD contrasts, and BFN1 is up-regulated in 74% of the contrasts. Additional commonly up-regulated genes in more than 60% of all 19 dPCD contrasts include RNS3, CAN1, THIOREDOXIN H-TYPE5, and three genes of unknown function (Supplemental Table S3, dPCD contrasts).
Second, we used the Genevestigator Condition Search and Similarity Search tools (Hruz et al., 2008) to identify genes that are commonly coregulated with BFN1, PASPA3, METACASPASE9 (MC9), and CYSTEINE ENDOPEPTIDASE (CEP1), four genes that have been associated with or functionally implicated in dPCD in several Arabidopsis cell types (Farage-Barhom et al., 2008; Helm et al., 2008; Ohashi-Ito et al., 2010; Bollhöner et al., 2013; Fendrych et al., 2014; Zhang et al., 2014). Seven genes were found to be commonly coregulated with these four target genes (Table II; Supplemental Table S5). Reiterating the analysis with these seven genes, we obtained a list of 154 genes that are coregulated with at least two out of the seven genes (Supplemental Table S5). Of these genes, four are coregulated with all target genes: BFN1 and MC9, as well as RNS3 and DMP4 (At4g18425, a paralog of DMP2). An unknown gene that we dubbed EXITUS1 (EXI1; At2g14095) and the transcription factor ANAC083 (for NO APICAL MERISTEM; ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR; CUP-SHAPED COTYLEDON [NAC] DOMAIN CONTAINING PROTEIN83) are coregulated with at least six of the seven target genes.
Table II. Commonly coexpressed genes of BFN1, MC9, PASPA3, and CEP1.
Coexpression scores as calculated by Genevestigator. AGI, Arabidopsis Genome Initiative gene code.
| AGI | Gene Name | Coexpression Score BFN1 | Coexpression Score MC9 | Coexpression Score PASPA3 | Coexpression Score CEP1 |
|---|---|---|---|---|---|
| AT5G04200 | MC9 | 0.7951 | 1 | 0.6195 | 0.6754 |
| AT4G18550 | DSEL | 0.6771 | 0.649 | 0.7146 | 0.6622 |
| AT4G18425 | DMP4 | 0.8552 | 0.8989 | 0.7395 | 0.7205 |
| AT4G04460 | PASPA3 | 0.6586 | 0.668 | 1 | 0.6105 |
| AT1G11190 | BFN1 | 1 | 0.882 | 0.7808 | 0.7112 |
| AT2G14095 | EXI1 | 0.837 | 0.7802 | 0.7406 | 0.6818 |
| AT1G26820 | RNS3 | 0.7672 | 0.9078 | 0.6587 | 0.6031 |
Third, we constructed a list of genes potentially involved in dPCD by comparing the gene expression profiles of two root tissues that are known to execute dPCD as a final differentiation step, the root cap (Fendrych et al., 2014), and the xylem tracheary elements (Bollhöner et al., 2012) with expression profiles of other tissues. Using the Visual Lateral Root Transcriptome Compendium (VLRTC; Parizot et al., 2010) based on a gene expression atlas of the Arabidopsis root (Brady et al., 2007), we found 95 genes commonly up-regulated more than 2-fold in xylem and lateral root cap (LRC) compared with root tissues not undergoing PCD (Supplemental Table S6). Eight of these genes are among the 154 genes identified by Genevestigator as coexpressed with at least two out of seven genes in the target gene set, significantly more than expected by chance (P = 1.5267e-06, hypergeometric test). Next to BFN1, MC9, PASPA3, SCPL48, and RNS3, a fatty acid desaturase family gene (At1g06090), the transcription factor ANAC046 (At3g04060), and SCPL20 are commonly up-regulated, suggesting that these genes might be involved in dPCD processes in the xylem and the LRC.
Although the data sets used in the ATH1 meta-analysis and VLRTC approaches overlap to some extent with each other (the root cap data sets in VLRTC and the meta-analysis are the same) and with the Genevestigator data, the different screening methodologies used led to the identification of candidate reporter gene sets that are only partially overlapping. By virtue of being commonly up-regulated in different differentiation-induced dPCD contexts, these genes can be considered potential dPCD reporters. To test the aptitude of these genes in this respect, we picked a set of 10 genes for in-depth characterization of their expression patterns: CEP1, PASPA3, BFN1, MC9, ANAC046, CAN1, RNS3, SCPL48, EXI1, and DMP4.
dPCD Reporters Are a Powerful Resource to Detect Putative dPCD Processes in Planta
The putative 5′-regulatory regions (promoters) of the eight candidate dPCD reporter genes were cloned and fused to a Gal4 DNA binding domain fused to the transcriptional activator domain of the herpes simplex virus VP16 protein (GAL4-VP16) transcriptional activator, combined with a GAL4-activated upstream activation sequence (UAS) driving a nuclear-localized histone 2A-GFP (>>H2A-GFP) reporter gene. These lines can be used in a versatile manner: as marker lines to detect and analyze PCD processes in planta, as driver lines to control the transcription of transgenes in a PCD-specific spatial and temporal pattern, and as tools to sort GFP-tagged protoplasts or nuclei for tissue-specific -omics analyses.
In a first round, over a dozen independent lines per promoter-reporter construct were investigated in T2, and lines with a single transfer DNA insertion locus and a representative GFP expression pattern were selected for in-depth analysis in T3. As the pEXI1 >>H2A-GFP, the pANAC046 >>H2A-GFP and the pCAN1 >>H2A-GFP constructs conferred weak or inconsistent GFP signals, these lines were not included for further analysis. From CEP1, PASPA3, MC9, and BFN1, which have been previously reported as PCD-associated (Farage-Barhom et al., 2008; Helm et al., 2008; Bollhöner et al., 2013; Fendrych et al., 2014), we chose to display the expression patterns conferred by pCEP1 and pPASPA3 as a reference for the expression pattern of the remaining genes.
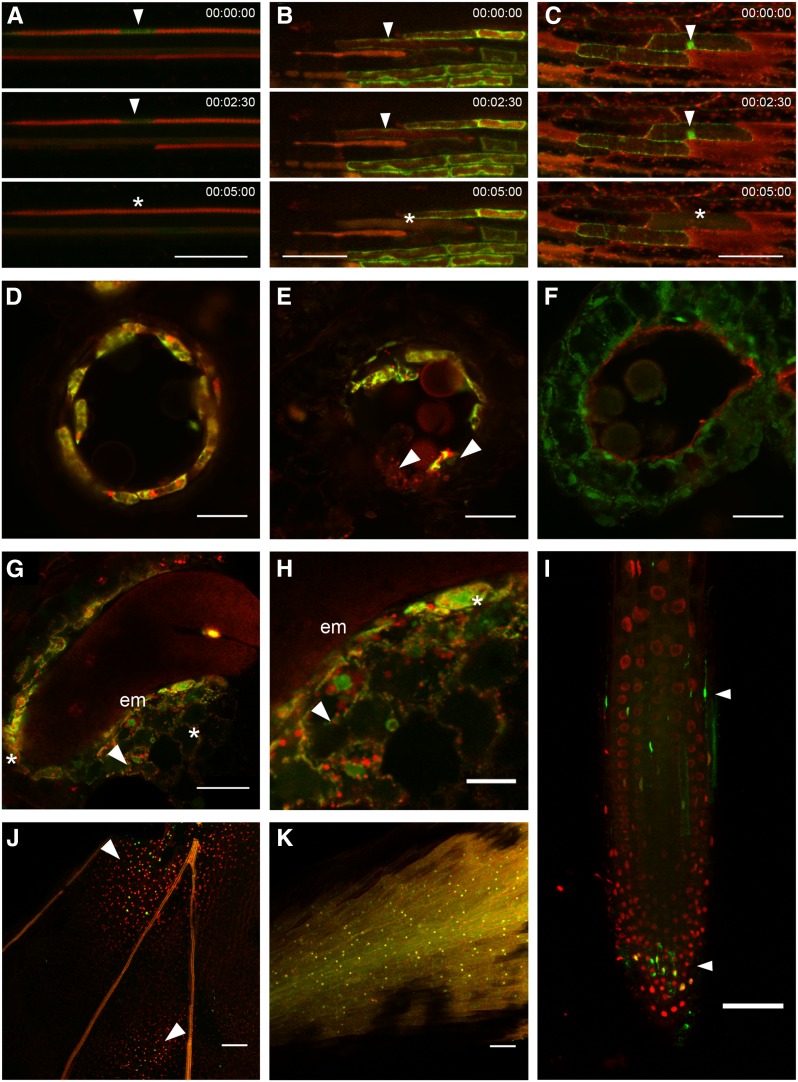
We focused our expression analysis on Arabidopsis tissues or cell types known to undergo differentiation-induced dPCD: the tapetum layer in the developing anther, the protoxylem cells in the growing root, and the cells of the LRC. Cells are also dying in the central endosperm and in senescing petals, but much less is known about the nature of the cell death in these tissues (for review, see Van Hautegem et al., 2015). We exploited a tonoplast integrity marker (ToIM; Fendrych et al., 2014) to investigate vacuolar collapse, a hallmark of vacuolar PCD (van Doorn et al., 2011). In all tissues or cell types, the ToIM expression controlled by the pPASPA3 promoter shows that vacuolar collapse precedes cell death in PASPA3-expressing cells (Fig. 3). In petals and the root cap, we additionally performed whole-mount terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assays, indicating that DNA fragmentation occurs in these tissues in the stages investigated for the expression pattern of the promoter-reporter constructs (Fig. 3).
Figure 3.
Cell death processes occur in different developmental contexts. A ToIM combines expression of a free GFP accumulating in the cytoplasm and the nucleus (green) and of a vacuolar-localized tagRFP (a monomeric derivative of a red fluorescent protein from Entacmaea quadricolor; red). Vacuolar rupture is indicated by the loss of compartmentalization and the merging of the two fluorescent signals. Note that in some dPCD cases, cytoplasmic acidification dampens the GFP signal, making the tagRFP signal more prominent. The ToIM is expressed under the control of the pPASPA3 promoter. A, Time lapse imaging of dPCD in a protoxylem element. The arrowheads indicate the cytoplasm around the cell’s nucleus, which is invaded by tagRFP upon vacuolar rupture (asterisk). B, Time lapse imaging of dPCD in a root cap cell. The arrowheads indicate the cell with intact vacuole, while the asterisk marks the cell once vacuolar rupture has occurred. C, Time lapse imaging of dPCD in petal cells at the base of a petal. The arrowheads indicate the cell with intact vacuole, while the asterisk marks the cell once vacuolar rupture has occurred. D to F, Vibratome sections through developing anthers around the time point of tapetum dPCD. D shows a locule lined by pPASPA3::ToIM-expressing, viable tapetum cells. E shows a locule in which dPCD is ongoing; the arrowheads point at partly degenerated cells. F shows a locule after tapetum dPCD in which degraded remains of tapetum cells line the inside of the locule. G and H, Vibratome sections through a seed in the walking stick state of embryo (em) development. H is a detail of G. Arrowheads point at ToIM-expressing but intact endosperm cells, while the asterisks indicate cells in the process of degeneration. I to K, TUNEL of whole-mount petals and root tips. 4′,6-diamidino-2-phenylindole (DAPI) staining is shown in red, and TUNEL signal is shown in green. I, Arrowheads indicate dying or dead TUNEL-positive root cap cells. J, Arrowheads indicate two fields of TUNEL-positive petal cells. K, TUNEL-positive control treated with DNase to induce tissue-wide DNA fragmentation, showing the overlap of TUNEL and DAPI signals. In A to C, time is indicated in minutes. Bars = 50 µm (A–C, G, and I–K) and 20 µm (D–F and H).
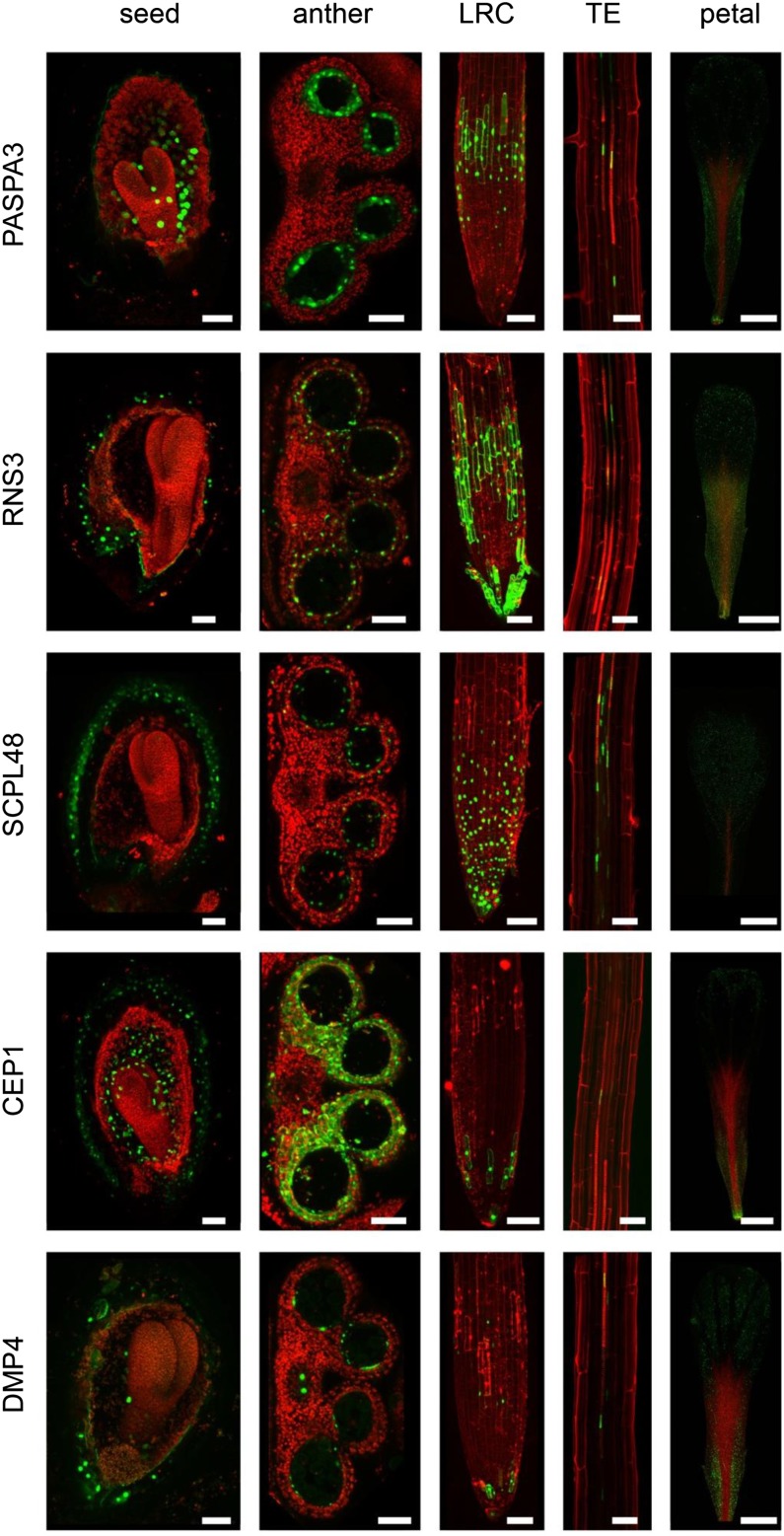
The promoters of RNS3, PASPA3, and DMP4 conferred largely similar expression patterns in the degenerating endosperm from torpedo stage onwards, in the anther tapetum layer before tapetum cell death, in differentiating LRC cells and tracheary elements, and in senescing petals (Fig. 4; Supplemental Figs. S1–S3). In accordance with the ATH1-derived expression data, the pRNS3 promoter conferred the strongest GFP expression, while pDMP4 >>H2A-GFP produced weaker GFP signals. Note that very high expression levels led to a failure to contain the H2A-GFP protein in the nucleus. Similar to pPASPA3, pRNS3 is activated many hours before PCD in the LRC, leading to a broader expression pattern compared with the one conferred by pDMP4, which only activated H2A-GFP expression shortly before PCD, leading to a narrower expression pattern in the LRC (Fig. 4). In developing petals of pRNS3, pPASPA3, and pDMP4 reporter lines, expression was first restricted to the tracheary elements, while expression spread throughout the entire organ during petal senescence (Supplemental Fig. S1). During anther development, pPASPA3 activation was confined to the differentiating tapetum layer, while both pRNS3 and pDMP4 were active in the outer layers of the anthers in later stages of flower development (Supplemental Fig. S2). In developing seeds, both pDMP4 and pPASPA3 exclusively conferred expression in differentiating endosperm from the torpedo stage onwards, while pRNS3 >>H2A-GFP signals were also detected in the differentiating seed coat of later seed stages (Supplemental Fig. S3).
Figure 4.
Selected promoter-reporter lines highlighting cells preparing for dPCD. PASPA3, RNS3, SCPL48, CEP1, and DMP4 expression patterns in developing seeds, developing anthers, the root cap, the xylem, and senescing petals (columns from left to right). pPASPA3 >>H2A-GFP is expressed in the embryo-surrounding region of the endosperm from torpedo stage onwards in the tapetum layer of the anther, in the LRC and the xylem, and in mature petals nearing floral organ senescence. pRNS3 >>H2A-GFP shows a very similar pattern. The SCPL48 promoter confers a broader spatial and temporal expression pattern and is not only restricted to cells preparing for dPCD. pCEP1 >>H2A-GFP shows GFP expression in the endosperm and seed coat of developing seeds, the tapetum and its surrounding anther tissues, cells from the lowest tier of the LRC, differentiating xylem vessels, and the aging petals. pDMP4 >>H2A-GFP is again more similar in expression to pPASPA3 and pRNS3. TE, Tracheary elements. Bar = 50 µm.
Compared with pRNS3, pPASPA3, and pDMP4, the pSCPL48 promoter conferred a broader spatial and temporal expression pattern; it was, for instance, already expressed in petals at anthesis (Supplemental Fig. S1) and in the entire LRC, and not only confined to tracheary elements, but also expressed in their neighboring cells (Fig. 4). In developing anthers, pSCPL48 activity was not confined to the tapetum but spread to the outer anther layers (Supplemental Fig. S3). During seed development, pSCPL48 was not activated in the endosperm but strongly up-regulated in the different layers of the differentiating seed coat (Supplemental Fig. S3).
Finally, the pCEP1 >>H2A-GFP expression pattern was confined to the dying LRC cells close to the root tip (Fig. 4), in accordance with earlier reports (Helm et al., 2008). Additionally, pCEP1 >>H2A-GFP conveys a strong expression in epidermal cells in the root hair zone, though these are not known to undergo cell death (data not shown). Interestingly, the close CEP1 homolog CEP2 is highly expressed in LRC cells in the root transition zone (Hierl et al., 2014) and might take over CEP1 functions here. In the developing seed, pCEP1 is active in the embryonic suspensor during early embryo development (data not shown) and is present in later stages both in the seed coat and the differentiating endosperm (Supplemental Fig. S3).
In summary, most promoter-reporter lines are specifically expressed in differentiating cells known to undergo dPCD or are associated with cellular degradation events that are thus far not well defined. Not all T promoter-reporter lines are present in every dPCD process, and not all gene expression patterns are restricted to cells preparing for dPCD, but the combination of these marker lines provides a powerful tool to identify and analyze putative dPCD processes in a developmental context in vivo and in planta.
dPCD Indicators Are Not Up-Regulated by Biotic and Abiotic Stresses Causing Cell Death
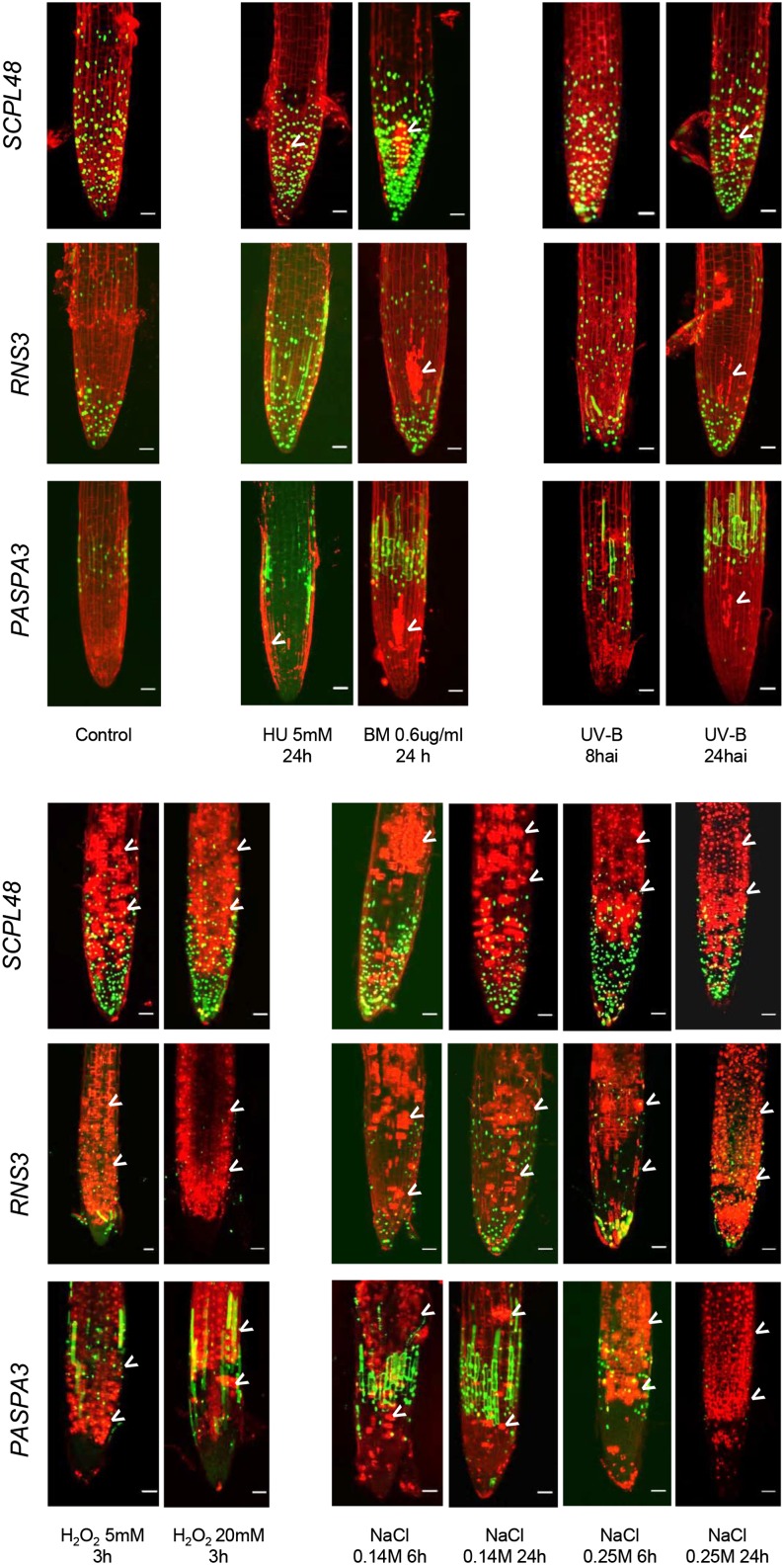
Our meta-analysis indicated that largely nonoverlapping sets of genes are up-regulated in differentiation-induced dPCD and various abiotic stress-induced ePCD types (Fig. 2), indicating that distinct transcriptional programs are activated in these plant cell death types. To further test this hypothesis experimentally, we analyzed dPCD marker expression in roots of Arabidopsis seedlings upon a variety of abiotic stresses. Propidium iodide (PI), which only enters cells with compromised plasma membrane integrity (Truernit and Haseloff, 2008), was used to highlight dead and dying cells. We investigated three marker constructs, pSCPL48, pRNS3, and pPASPA3, which showed a specific expression pattern in the LRC of the control root tips. Upon treatments with hydroxyurea, bleomycin, UV-B irradiation, hydrogen peroxide, and NaCl, increasing numbers of PI-positive cells indicated the occurrence of cell death during the different stress treatments (Fig. 5, arrowheads). While genotoxic and UV stress led to localized cell death of root meristem cells, oxidative and salt stress produced more widespread cell death. In all cases, cell death was neither preceded by ectopic dPCD marker expression at early time points nor accompanied by dPCD marker expression at late time points. However, whether the observed cell death is a result of PCD programs activated by the stress treatments or is caused by direct cellular damage is difficult to ascertain. We performed whole-mount TUNEL and found that hydrogen peroxide treatment leads to TUNEL-positive root cells (Supplemental Fig. S4). All other stress treatments did not lead to clearly TUNEL-positive cells, apart from the dying root cap cells that are TUNEL positive due to stress-independent dPCD (Supplemental Fig. S4). These results confirm that the stresses used to produce the ATH1 data sets meta-analyzed in our study were sufficient to cause cell death, but they leave open whether this cell death is an active PCD or a passive, unregulated form of cell death. Although abiotic stresses have been shown to provoke cell death displaying hallmarks of PCD (Chen et al., 2009; Qi et al., 2011; Nawkar et al., 2013; Petrov et al., 2015), detailed case-by-case investigations have to show if genuine actively controlled, genetically encoded programs are responsible for these types of cell death.
Figure 5.
Abiotic stress treatments cause cell death without the up-regulation of dPCD reporters. Abiotic stress treatments applied to 5-d-old seedlings from dPCD markers SCPL48, RNS3, and PASPA3. Pictures were taken after the indicated time points and treatments at the root tip to show the expression around the LRC and were stained with PI to highlight the cell walls and cells with compromised plasma membrane integrity indicative of cell death (arrowheads). BM, Bleomycin; HU, hydroxyurea.
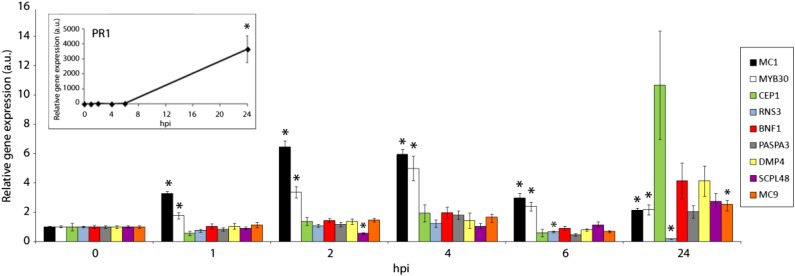
Although there appears to be an overlap between the genes up-regulated during abiotic stress-induced cell death and pathogen-induced ePCD, our meta-analysis suggested that pathogen-related ePCD and differentiation-induced dPCD are regulated largely independently (Fig. 2). To confirm these results experimentally, we performed a quantitative reverse transcription (qRT)-PCR experiment of plants inoculated with an HR-inducing Pseudomonas syringae strain. In contrast to HR marker genes, none of the canonical dPCD marker gene transcripts were significantly up-regulated during HR (Fig. 6; Supplemental Fig. S5). To test if only individual cells express dPCD reporter genes, which might not register on a tissue-wide scale of RNA quantification, we also investigated promoter-reporter lines but did not find any GFP signals in or around HR lesions (data not shown). These results confirm that dPCD marker genes are not transcriptionally regulated during HR-related ePCD.
Figure 6.
dPCD marker genes are not up-regulated during HR PCD. qRT-PCR of Col-0 wild-type plants inoculated with an avirulent HR-inducing P. syringae strain in a time course experiment after infection. Relative expression of the indicated genes both in the inoculated area and in noninoculated tissue was determined by qRT-PCR at the indicated time points. PATHOGENESIS RELATED1 (PR1), MC1, and MYB DOMAIN PROTEIN30 (MYB30) were used as HR marker genes. Expression values were normalized using the SAND family gene as internal standard. Ratios of the expression values for each gene in the inoculated zone with respect to the noninoculated area are presented for each time point. Mean and se of the mean values were calculated from three independent experiments with three replicates. Statistical significance according to a Student’s t test P value of 0.005 is indicated by asterisks. hpi, Hours after inoculation; a.u., arbitrary units.
Core dPCD Marker Genes Are Evolutionary Conserved in Land Plants
The phenomenon of developmentally regulated PCD is most likely evolutionary ancient and occurs also in simple land plants, for instance the moss Physcomitrella patens (Xu et al., 2014). To assess the degree to which the molecular regulation of dPCD might be evolutionary conserved, we investigated the conservation of the dPCD indicator genes identified in Arabidopsis within the plant kingdom as well as between plants and vertebrates. According to the plant comparative genomics platform PLAZA (Proost et al., 2015), RNS3, BFN1, PASPA3, MC9, and SCPL48 are widely conserved in the green plant lineage, while BFN1 appears to be restricted to the land plant lineage (Table III). Using the comparative online tool Phytozome (Goodstein et al., 2012), we identified putative homologs of these dPCD markers in different angiosperm lineages, as well as in the basal angiosperm Amborella trichopoda and in the lower land plants P. patens and Selaginella moellendorffii. In the green alga Chlamydomonas reinhardtii, we identified protein sequences related to SCPL48, PASPA3, and RNS3, sequences with limited blast length for MC9 but no clear homolog for BFN1 (Supplemental Table S7). Outside the plant kingdom, the HomoloGene algorithm (Sayers et al., 2012) indicated conservation of RNS3, SCPL48, and PASPA3 in all eukaryotes, while BFN1 and MC9 appeared not to be conserved between plants and vertebrates (Table III). Interestingly, a putative RNS3 homolog, the RNase T2, has recently been implicated in the control of melanocyte apoptosis via the tumor necrosis factor receptor-associated factor2 pathway in vitiligo patients (Wang et al., 2014). Furthermore, the putative PASPA3 homolog Cathepsin D functions as a proapoptotic gene targeting Bid after release from the lysosome (Appelqvist et al., 2012; Repnik et al., 2014).
Table III. Evolutionary conservation of putative core dPCD markers.
% ID reflects the sequence similarity between the Arabidopsis gene and the best blast hit in vertebrates. All genes belong to PLAZA 3.0 orthologous gene families that encompass the Magnoliophyta (RNS3, BFN1, and MC9) or Viridiplantae (SCPL48 and PASPA3) clade (not shown). AGI, Arabidopsis Genome Initiative gene code; HOM, homologous; Nuclease PA3-like, a predicted protease (GenBank accession number XP_005974529.1); CPVL, carboxypeptidase, vitellogenic-like.
| AGI (Name) | PLAZA 3.0 Homologous Gene Family | PLAZA 3.0 Plant Clade of HOM Family | HomoloGene | Blast Hits in Vertebrates | % ID | Role in Vertebrates |
|---|---|---|---|---|---|---|
| AT1G26820 (RNS3) | HOM03D000496 | Viridiplantae | 31190, conserved in Eukaryota | RNase T2 | 30 | Potentially skin cell apoptosis (Wang et al., 2014) |
| AT1G11190 (BFN1) | HOM03D001490 | Embryophyta | No information | Nuclease PA3-like | 27 | |
| AT5G04200 (MC9) | HOM03D001276 | Viridiplantae | No information | No clear hits | 0 | |
| AT3G45010 (SCPL48) | HOM03D000050 | Viridiplantae | 137548, conserved in Eukaryota | Serine CPVL | 30 | |
| AT1G62290 (PASPA3) | HOM03D000729 | Viridiplantae | 124002, conserved in Eukaryota | Cathepsin D | 50 | Proapoptotic gene (Appelqvist et al., 2012; Repnik et al., 2014) |
These results suggest a high degree of conservation of core dPCD marker genes within the green plant lineage. Whether the proapoptotic roles of PASPA3- and RNS3-related enzymes in mammals is due to functional conservation or due to convergent evolution is difficult to determine. Nevertheless, it is tempting to speculate that similar mechanisms are functional in both animal and plant PCD types.
DISCUSSION
To date, despite the undisputed importance of the diverse forms of plant PCD for development and for environmental interactions (Wu et al., 2014; Petrov et al., 2015; Van Hautegem et al., 2015), still only little is known about the molecular regulation of these processes. During the plant life cycle, PCD is induced at numerous occasions, but it is unclear whether there are common mechanisms involved in controlling different PCD types. Attempts to characterize and relate different plant PCD types have been made, based chiefly on morphological and ultrastructural features of dying cells (van Doorn, 2011; van Doorn et al., 2011). Here, we explored the possibility to characterize different types of plant PCD using molecular information. As transcriptional regulation has been implicated in plant PCD control (Van Hautegem et al., 2015), and as so far only scarce proteomic data in plant PCD contexts exist, this transcriptome meta-analysis is a first step into a systematic molecular characterization of plant PCD processes.
One aim of our study was to investigate whether existing transcriptome data might be useful for a molecular categorization of plant PCD types. By comparing transcriptome profiles of different developmental stages and environmental stresses leading to cell death, we expected to find similarities and differences that would allow relating different PCD types based on the degree of common gene regulation. Such information could be used to complement PCD characterization based on morphological and biochemical hallmarks (van Doorn, 2011; van Doorn et al., 2011). Our approach of exploiting publicly available ATH1 data sets by means of several bioinformatics approaches was successful in identifying unique dPCD indicator genes. Promoter-reporter constructs of these genes marked cells preparing for cell death in well-defined PCD settings, e.g., the xylem (Bollhöner et al., 2012), the root cap (Fendrych et al., 2014), or the tapetum (Plackett et al., 2011), but also highlighted cell types in which so far only scarce genetic evidence exists for the occurrence of PCD, e.g., the seed coat (Haughn and Chaudhury, 2005) or the endosperm (Waters et al., 2013) in developing seeds. These results suggest that a conserved core of PCD-associated genes is commonly regulated in diverse dPCD contexts, and our findings will give impulses to investigate developmentally regulated PCD processes in more detail.
Among the genes that we found to be transcriptionally regulated during differentiation-induced dPCD were several genes encoding nucleases, including CAN1, BFN1, and RNS3. BFN1 is a well-known leaf senescence reporter, which has also been shown to function in chromatin breakdown during root cap PCD in Arabidopsis and tracheary element PCD in Zinnia elegans (Ito and Fukuda, 2002; Fendrych et al., 2014). CAN1 is a staphylococcal-like plasma membrane-bound nuclease whose expression has been associated with PCD events before (Leśniewicz et al., 2012), but its exact role remains unclear. RNS3 belongs to the evolutionary conserved family of T2 endoribonucleases that cleave single-stranded RNA. T2 endoribonucleases have been suggested to perform a variety of functions, including scavenging of nucleic acids, degradation of self-RNA, modulating host immune responses, and serving as cellular cytotoxins (Luhtala and Parker, 2010). In plants, T2 ribonucleases are induced during phosphate starvation and have been hypothesized to function in providing phosphates from nucleic acids (Taylor et al., 1993; Bariola et al., 1994). Our results show RNS3 up-regulation during leaf and floral organ senescence, correlating senescence-induced cell death with differentiation-induced dPCD.
Next to nucleases, also protein hydrolases including PASPA3, MC9, SCPL48, and CEP1 are among the core of dPCD-associated proteins. For most of these proteases, little data exist regarding their actual function and substrates. The degradome of MC9 has been investigated in detail, though most substrates identified rather suggested functions other than PCD (Tsiatsiani et al., 2013). Nevertheless, MC9 has been shown to be a part of a proteolytic cascade effecting postmortem cell clearance of tracheary elements in Arabidopsis (Bollhöner et al., 2013). Recently, MC9 activity was found to be important to mediate oxidative stress-dependent cell death via the cleavage of GRIM REAPER (Wrzaczek et al., 2015).
Next to hydrolytic enzymes, genes encoding several proteins of unknown functions such as the plasma membrane-localized DMP4 were up-regulated during several dPCD processes. DMPs represent a unique family of plant-specific plasma membrane proteins of unknown function that have been recently identified in a screen for senescence-associated genes in Arabidopsis (Kasaras and Kunze, 2010). Of the 10 Arabidopsis DMP paralogs, DMP4 is coregulated with the core of dPCD marker genes and up-regulated in several dPCD conditions. Additionally, expression of DMP4 has been described in abscission zones of floral organs (Kasaras and Kunze, 2010), although involvement of PCD in this abscission process has not been investigated. The molecular function of DMP proteins still needs to be determined, but misexpression of Arabidopsis DMP1 led to an aberrant endoplasmic reticulum and in some cases to the death of transfected cells (Kasaras et al., 2012).
Despite the fact that different PCD types appear to exhibit different gene expression profiles, an adequate supervised classification of PCD types based on the available transcriptome data proved to be possible only for some PCD subtypes, possibly due to the nature and quantity of the available transcriptome data sets. Most data sets analyzed were not explicitly designed to characterize gene expression changes associated with cell death processes but rather to identify regulators of processes that precede or even might counteract cell death. What is more, deducing from the experimental metadata which particular PCD subtype, if any, is represented in a given data set is not always straightforward.
To reliably classify different types of plant PCD, a more thorough understanding of their molecular regulation will be necessary. A means to this end will be the generation of specific transcriptome profiles of precisely described PCD systems. For differentiation-induced dPCD, this is a challenging task, as only single cells, or small groups of cells, are undergoing cell death at a time. Techniques of isolating these cells or their nuclei for transcriptome analysis by fluorescent-associated cell sorting or isolation of nuclei tagged in specific cell types (Deal and Henikoff, 2011) will be instrumental to obtain meaningful data sets. The dPCD promoter-reporter constructs presented in this study will facilitate these approaches. At least for closely related PCD types, for instance, different forms of differentiation-induced dPCD, such a comparative approach will become valuable to reveal unique PCD markers and putative core PCD regulators. This approach, accompanied by thorough morphological, molecular genetics and cell biological analyses, will open the way to a more comprehensive understanding of PCD as a fundamental cellular process in plants.
CONCLUSION
Despite the progress achieved over the last decade by a relatively small research community dedicated to plant PCD, the molecular regulation of PCD largely remains a terra incognita. To fill the white spots on the map, and to relate the findings made in different plant PCD systems, we need to understand more of the molecular principles that govern plant cell death. Here, we made a step toward a comprehensive understanding of plant PCD by integrating genome-wide transcriptome profiles of different established as well as less well-known cell death systems. With the recognized expression patterns and dPCD reporter lines, we can now progress to a more specific mode of analysis. Of course, transcriptional regulation is only a fraction of the molecular control that leads to an ordered and timely termination of vital processes of a cell undergoing PCD. A challenge for the next decade will be to define the molecular modes of function of putative PCD regulators and the posttranslational modifications that lead to the rapid execution of cell death observed in many systems.
MATERIALS AND METHODS
Meta-Analysis Data Retrieval
We retrieved Affymetrix ATH1 CEL files for the various transcriptome data sets from ArrayExpress (http://www.ebi.ac.uk/arrayexpress/), Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), Riken Expression Array Database (http://read.gsc.riken.go.jp/), or other third-party data providers. Multiple rounds of preanalysis processing steps (data curation and filtering to remove conditions with little or no differential gene expression) were performed to retain an optimal selection of expression data sets.
Meta-Analysis Detection of Differential Expression
The microarray data were preprocessed with the Robust Multiarray Average procedure, as implemented in BioConductor (Irizarry et al., 2003; Gentleman et al., 2004). An up-to-date Chip Defnition File based on the latest version of the Arabidopsis genome annotation by The Arabidopsis Information Resource was retrieved from BrainArray (http://brainarray.mbni.med.umich.edu) to define probe-gene relations. A filtering of differentially expressed genes was performed using the R/Bioconductor software package Limma (Ritchie et al., 2015) to retain only those genes with an adjusted P ≤ 0.05 and absolute log2 fold change > 1.
HCA
The expression profiles of the ATH1 genes showing differential expression in at least one PCD condition were hierarchically clustered with the Orange Canvas software (http://orange.biolab.si/) using Pearson’s correlation distance as the distance measure and the average linkage clustering option. To identify the most commonly up-regulated genes in particular PCD clusters, we used an R script that, given a cluster of interest, ranks genes according to the number of conditions in the cluster in which they are significantly up-regulated (P < 0.05) at least 2-fold. For each cluster, the resulting ranked gene list was truncated at a specific number of observed up-regulations to obtain lists for all clusters of 25 to 30 genes each (Supplemental Table S3). A similar analysis was done to identify genes that are commonly up-regulated across all conditions labeled as dPCD. A gene was considered to be commonly up-regulated in dPCD when it was designated as significantly up-regulated (P < 0.05) at least 2-fold in 60% of the dPCD-labeled conditions.
Supervised Classification Analyses
SVM (Cortes and Vapnik, 1995) and RF (Breiman, 2001) analyses were performed using the Orange toolbox (Demšar et al., 2013) by writing Python scripts accessing the Orange API. In each analysis, an automated exhaustive search of the algorithm parameter space was performed to optimize the parameter settings. These settings are reported per analysis in Supplemental Table S4. Comparison of the classification performance across analyses and algorithms was done by means of the Matthews Correlation Coefficient (MCC) as reported after 5- or 10-fold cross validation (5-fold cross validation was used when the number of contrasts in one of the classes was <10). The MCC is a balanced measure of binary classification performance that is particularly useful if the classes are of different sizes. MCC scores range from 1 for perfect classifiers to –1 when there is a total disagreement between the predicted and observed class labels, with a score of 0 indicating that the classifier does not perform better than random.
For the analyses on balanced dPCD- and ePCD-labeled data, 19 (or 10) ePCD experiments were randomly sampled without replacement out of the relevant ePCD subset and added to the 19 (or 10) dPCD experiments, after which SVM and RF classifiers were learned. This random selection was performed 100 times, and the average MCC score is reported in Supplemental Table S4.
Genevestigator Coexpression Tool Search
The query genes were screened with the Conditions Search and Similarity Search tools of Genevestigator. To find the relevant conditions that induce the expression of the query gene, all ATH1 microarrays were given as input into the Conditions search (Perturbation tool) and filtered by selecting microarrays showing a log-fold change of the query gene greater than or equal to 2 and a P value greater than or equal to 0.01. The resulting microarrays were saved in a new list and fed in the Coexpression tool to find the top 200 positively correlated genes in the Perturbation option. To identify commonly coregulated genes between different genes, coregulated genes with a Genevestigator score (Pearson’s correlation coefficient) greater than 0.6 were selected for each gene. The resulting gene lists were fed into the online Venn Diagram tool program provided by the VIB-Ghent University Bioinformatics and Systems Biology laboratory at http://bioinformatics.psb.ugent.be/cgi-bin/liste/Venn/calculate_venn.htpl to identify commonly regulated genes.
Identification of Genes Coregulated in Maturing Xylem and LRC
The VLRTC method (Parizot et al., 2010) was used to reanalyze the data from Brady et al. (2007) as described in Fendrych et al. (2014). The candidate genes were first thresholded for their expression in the LRC and in the maturing tracheary elements as follows:
 |
And these genes were further ranked according to:
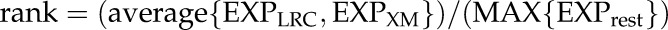 |
MAX refers to the maximum expression value; EXP to normalized expression values, rest to {Stele (wol), Stele(J2501), Protophloem(S32), Phloem + Companion Cells(APL), Phloem Companion Cells (SUC2), Develloping Xylem (S4), Pericycle (J2661), Pericycle Phloem Pole (S17), Pericycle Xylem Pole (JO121), Primordia (rm1000), Ground Tissues (J0571), Endodermis (scr5), CORTEX, Epidermis Atrichoblast (gl2), and Epidermis Trichoblast (COBL9)}; and XM to xylem maturing.
Plant Material and Growth Conditions
For the root imaging, seedlings were grown vertically 5 d after sowing on one-half-strength Murashige and Skoog (MS) plates (2.15 g L–1 MS salts [Caisson Labs], 0.1 g L–1 MES [Sigma], pH 5.8 [KOH], and 0.8% [w/v] agar [Lab M]) in a 16-h-light/ 8-h-dark photoperiod at 21°C with 70% humidity. For the imaging of anthers, petals, and developing seeds, 5-week-old plants were grown in jiffy pots in a 16-h-light/8-h-dark photoperiod at 21°C and kept under optimal irrigation and nutrient supply conditions throughout the plant life cycle.
Stress Treatments
Three biological replicates of 5-d-old seedlings from each of the marker lines analyzed were transferred from one-half-strength MS plates to one-half-strength MS plates containing 0, 140, and 250 mm NaCl (VWR), 5 and 20 mm hydrogen peroxide (Merck), 5 mm Hydroxyurea (Sigma), and 0.6 ug mL–1 Bleomycin (Duchefa) for the indicated times before confocal imaging. For UV stress, the seedlings were UV-B treated for 15 min, 30 min, 45 min and 1 h with UV-B 313 EL lamps (Q-Lab) at an intensity of 1 W m–2 measured with the Spectrasense 2+ meter coupled to the compatible UV-B sensor (Skye Instruments). The UV-B lamps were in an incubator where the conditions were 18°C and 67% relative humidity.
Cloning and Transgenic Lines Preparation
The proCEP1 and proPASPA3 were obtained as Gateway cloning-compatible amplicons from the systematic analysis of Arabidopsis promoters collection (Benhamed et al., 2008) and were recombined into the pDONRP4P1r vector (Invitrogen). The proCEP1 spans 1,626 bp, and the proPASPA3 spans 1,997 bp upstream of the respective start codon. The proSCPL48, proRNS3, and proDMP-4 were isolated from Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) genomic DNA using gene-specific primers (Supplemental Table S8) and adding BamHI and XhoI restriction sites to clone directionally into pENTRL4-R1, a Gateway-compatible entry vector containing a cassette with a multiple cloning site (https://gateway.psb.ugent.be/). The proSCPL48 spans 2,054 bp (including the first 24 bp after the start codon), proRNS3 spans 1,440 bp, and proDMP4 spans 1,352 bp upstream of the respective start codon. Sequence information about these genes can be found in The Arabidopsis Information Resource under the following accession numbers: BFN1 (Atg11190), CEP1 (At5g50260), PASPA3 (At4g04460), SCPL48 (At3g45010), RNS3 (At1g26820), and DMP-4 (At4g18425). The promoters were assembled in a multisite Gateway reaction using LR clonase II+ (Invitrogen) with the GAL4 coding sequence and the destination vector pB9-H2A-UAS-7m24GW to create activator lines. These lines can be used for transactivation, and at the same time, the nuclei of the cells where the promoter is expressed are marked with GFP. This vector contains a HISTONE 2A-6 (H2A) coding sequence (At5g59870) fused to eGFP and driven by the repetitive UAS promoter. This vector is part of a transactivation driver line-effector line set as described (Karimi et al., 2005).
The expression clones obtained were transformed into Agrobacterium tumefaciens C58C1 (pMP90)-competent cells using electroporation, and these bacteria were used for a modified floral dip method to stably transform Arabidopsis Col-0 plants. One milliliter of Yeast Extract Broth-grown culture was incubated 6 h at 28°C, and 10 mL of Yeast Extract Broth was added and grown overnight at 28°C. Plants were dipped with the overnight culture, adding 40 mL of floral dip medium (10% [w/v] Suc and 0.05% [v/v] Silwet L-77). All analyses were performed with T3 homozygous plants with a single-locus insertion determined by segregation analysis.
Confocal Imaging and Image Processing
Confocal images were acquired using a Zeiss 710 CLSM microscope. Objectives used were Plan-Apochromat 20×/0.8 Dry (most images) and EC Plan-Neofluar 10×/0.30 Dry. GFP was excited with the 488-nm laser line of the argon laser, and the emission was detected between 495 and 545 nm. Propidium iodide (PI, Sigma) was excited by 561 nm and detected between 580 and 680 nm. PI was dissolved in one-tenth-strength MS (0.43 g L–1 MS salts and 4 μg mL–1 PI).
Siliques and anthers from different developmental stages were fixed for 2 h at room temperature in a 3.7% (w/v) paraformaldehyde solution dissolved in 50 mm PIPES, 5 mm EGTA, and 1 mm MgSO4 buffer, embedded in 5% (w/v) agarose blocks, and sectioned using a vibratome (Campden Instruments). The samples from developing seeds were dissected in a binocular microscope to remove the valves before fixation. The samples from senescing petals were mounted in the glass slides using one-tenth-strength MS and 0.01% (v/v) Triton X-100.
Image processing was done using Fiji (Schindelin et al., 2012). Some panels were assembled using the stiching plugin.
TUNEL Assay
For the TUNEL, seedlings were fixed for 1 h in 4% (v/v) paraformaldehyde in phosphate-buffered saline (PBS), pH 7.4, under vacuum at room temperature. After fixation, seedlings were washed five times in PBS and permeabilized for 2 min on ice in a 0.1% (w/v) sodium citrate solution with 0.1% (v/v) Triton X-100. Afterward, seedlings were washed five times in PBS. For the positive control, fixed and permeabilized wild-type seedlings were treated with DNaseI for 15 min at room temperature and washed three times with PBS. For the TUNEL reaction, label solution and enzyme solution were mixed according to the manufacturer’s manual (In Situ Cell Death Detection Kit, Fluorescein, Roche Applied Science), and 50 µL was added to a 1.5-mL microcentrifuge tube together with the seedlings. For the negative control, only label solution was used. All samples were incubated at 37°C in the dark for 1 h. Afterward, the seedlings were washed three times with PBS and mounted with an antifading agent (citifluor, Citifluor Ltd.) containing 1 μg mL–1 DAPI. The same procedure was used for petals. Stress treatments of seedlings were the same as described before.
Pathogen Assays, RNA Extraction, and qRT-PCR Analysis
Arabidopsis Col-0 4-week-old plants were inoculated with a bacterial suspension of Pseudomonas syringae pv tomato AvrRpm1 (5 × 107 colony forming units mL–1). Leaf samples were harvested at the indicated time points both inside the infiltrated zone and in noninoculated areas and ground in liquid nitrogen. Total RNA was isolated using the Nucleospin RNA plant kit (Macherey-Nagel) according to the manufacturer’s recommendations. Reverse transcription was performed using 1.5 µg of total RNA. Real-time quantitative PCR was performed on a Light Cycler 480 II machine (Roche Diagnostics) using Roche reagents. Primers used for qRT-PCR are shown in Supplemental Table S8. Relative expression was calculated as the crossing point difference between each gene and the internal control SAND family gene (At2g28390). Average crossing point difference was calculated from three independent experiments with three replicates and related to the value of each gene at time 0, which is set at 1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Developmental series for petal senescence.
Supplemental Figure S2. Developmental series for tapetum differentiation.
Supplemental Figure S3. Developmental series for seed development.
Supplemental Figure S4. Whole-mount TUNEL of 5- to 6-day-old root tip after different abiotic stresses provoking cell death.
Supplemental Figure S5. dPCD marker genes are not transcriptionally regulated during HR-related ePCD.
Supplemental Table S1. Detailed overview of the ATH1 microarray experiments used for the meta-analysis.
Supplemental Table S2. Overview of the number of up- and down-regulated genes per condition in the experiments used in the meta-analysis.
Supplemental Table S3. Genes commonly regulated in different PCD clusters.
Supplemental Table S4. Performance results of SVM and RF classification of dPCD versus ePCD instances based on the expression profiles of various gene (feature) sets in various experiment subsets.
Supplemental Table S5. Commonly coregulated genes of MC9, RNS3, BFN1, ARABIDOPSIS THALIANA DAD1-LIKE SEEDING ESTABLISHMENT-RELATED LIPASE (DSEL), EXI1, PASPA3, and DMP4.
Supplemental Table S6. Ninety-five commonly regulated genes between the LRC and differentiating tracheary elements, of which eight genes are common with the 154 coregulated dPCD genes (Supplemental Table S5).
Supplemental Table S7. Phytozome blast search for putative homologs of the Arabidopsis dPCD marker genes MC9, BFN1, PASPA3, RNS3, and SCPL48.
Supplemental Table S8. Primers used for promoter cloning and qRT-PCR.
Supplementary Material
Acknowledgments
We thank all members of the PCD research team at the Vlaams Instituut voor Biotechnologie-Plant Systems Biology Department department for critical reading of the article, Annick Bleys for help in revising the cited references, other members of the Vlaams Instituut voor Biotechnologie-Plant Systems Biology Department for sharing fields of expertise, Dr. Marc Heijde and Dr. Toon Cools for the genotoxic stress experiments, and Dr. Pavel Kerchev for the oxidative stress experiments.
Glossary
- PCD
programmed cell death
- HR
hypersensitive response
- HCA
hierarchical clustering analysis
- SA
salicylic acid
- SVM
Support Vector Machine
- RF
Random Forest
- VLRTC
Visual Lateral Root Transcriptome Compendium
- ToIM
tonoplast integrity marker
- PI
propidium iodide
- MCC
Matthews Correlation Coefficient
- LRC
lateral root cap
- MS
Murashige and Skoog
- Col-0
ecotype Columbia
- PBS
phosphate-buffered saline
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick-end labeling
- DAPI
4′,6-diamidino-2-phenylindole
- qRT
quantitative reverse transcription
Footnotes
This work was supported by the Consejo Nacional de Ciencia y Tecnología (postdoctoral fellowship registration nos. 186253 and 203288 to Y.O.-C.) and the French Laboratory of Excellence (project TULIP ANR–10–LABX–41 and ANR–11–IDEX–0002–02).
Articles can be viewed without a subscription.
References
- Appelqvist H, Johansson AC, Linderoth E, Johansson U, Antonsson B, Steinfeld R, Kågedal K, Öllinger K (2012) Lysosome-mediated apoptosis is associated with cathepsin D-specific processing of Bid at Phe24, Trp48, and Phe183. Ann Clin Lab Sci 42: 231–242 [PubMed] [Google Scholar]
- Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD, Green PJ (1994) The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J 6: 673–685 [DOI] [PubMed] [Google Scholar]
- Bayles KW. (2014) Bacterial programmed cell death: making sense of a paradox. Nat Rev Microbiol 12: 63–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M, Martin-Magniette ML, Taconnat L, Bitton F, Servet C, De Clercq R, De Meyer B, Buysschaert C, Rombauts S, Villarroel R, et al. (2008) Genome-scale Arabidopsis promoter array identifies targets of the histone acetyltransferase GCN5. Plant J 56: 493–504 [DOI] [PubMed] [Google Scholar]
- Bollhöner B, Prestele J, Tuominen H (2012) Xylem cell death: emerging understanding of regulation and function. J Exp Bot 63: 1081–1094 [DOI] [PubMed] [Google Scholar]
- Bollhöner B, Zhang B, Stael S, Denancé N, Overmyer K, Goffner D, Van Breusegem F, Tuominen H (2013) Post mortem function of AtMC9 in xylem vessel elements. New Phytol 200: 498–510 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Breiman L. (2001) Random forests. Mach Learn 45: 5–32 [Google Scholar]
- Candat A, Paszkiewicz G, Neveu M, Gautier R, Logan DC, Avelange-Macherel MH, Macherel D (2014) The ubiquitous distribution of late embryogenesis abundant proteins across cell compartments in Arabidopsis offers tailored protection against abiotic stress. Plant Cell 26: 3148–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wang Y, Li JY, Jiang A, Cheng Y, Zhang W (2009) Mitochondrial proteome during salt stress-induced programmed cell death in rice. Plant Physiol Biochem 47: 407–415 [DOI] [PubMed] [Google Scholar]
- Coll NS, Epple P, Dangl JL (2011) Programmed cell death in the plant immune system. Cell Death Differ 18: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20: 273–297 [Google Scholar]
- Deal RB, Henikoff S (2011) The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat Protoc 6: 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demšar J, Curk T, Erjavec A, Gorup C, Hočevar T, Milutinovič M, Možina M, Polajnar M, Toplak M, Starič A, et al. (2013) Orange: Data mining toolbox in Python. J Mach Learn Res 14: 2349–2353 [Google Scholar]
- Farage-Barhom S, Burd S, Sonego L, Perl-Treves R, Lers A (2008) Expression analysis of the BFN1 nuclease gene promoter during senescence, abscission, and programmed cell death-related processes. J Exp Bot 59: 3247–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M, Van Hautegem T, Van Durme M, Olvera-Carrillo Y, Huysmans M, Karimi M, Lippens S, Guérin CJ, Krebs M, Schumacher K, et al. (2014) Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Curr Biol 24: 931–940 [DOI] [PubMed] [Google Scholar]
- Fuchs Y, Steller H (2011) Programmed cell death in animal development and disease. Cell 147: 742–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR. (2011) Means to an End. Apoptosis and Other Cell Death Mechanisms. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Hara-Nishimura I, Hatsugai N (2011) The role of vacuole in plant cell death. Cell Death Differ 18: 1298–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn G, Chaudhury A (2005) Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci 10: 472–477 [DOI] [PubMed] [Google Scholar]
- Helm M, Schmid M, Hierl G, Terneus K, Tan L, Lottspeich F, Kieliszewski MJ, Gietl C (2008) KDEL-tailed cysteine endopeptidases involved in programmed cell death, intercalation of new cells, and dismantling of extensin scaffolds. Am J Bot 95: 1049–1062 [DOI] [PubMed] [Google Scholar]
- Hierl G, Höwing T, Isono E, Lottspeich F, Gietl C (2014) Ex vivo processing for maturation of Arabidopsis KDEL-tailed cysteine endopeptidase 2 (AtCEP2) pro-enzyme and its storage in endoplasmic reticulum derived organelles. Plant Mol Biol 84: 605–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Ito J, Fukuda H (2002) ZEN1 is a key enzyme in the degradation of nuclear DNA during programmed cell death of tracheary elements. Plant Cell 14: 3201–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P (2005) Modular cloning in plant cells. Trends Plant Sci 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Kasaras A, Kunze R (2010) Expression, localisation and phylogeny of a novel family of plant-specific membrane proteins. Plant Biol 12: 140–152 [DOI] [PubMed] [Google Scholar]
- Kasaras A, Melzer M, Kunze R (2012) Arabidopsis senescence-associated protein DMP1 is involved in membrane remodeling of the ER and tonoplast. BMC Plant Biol 12: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E. (2004) Controlled cell death, plant survival and development. Nat Rev Mol Cell Biol 5: 305–315 [DOI] [PubMed] [Google Scholar]
- Leśniewicz K, Poręba E, Smolarkiewicz M, Wolff N, Stanisławski S, Wojtaszek P (2012) Plant plasma membrane-bound staphylococcal-like DNases as a novel class of eukaryotic nucleases. BMC Plant Biol 12: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhtala N, Parker R (2010) T2 family ribonucleases: ancient enzymes with diverse roles. Trends Biochem Sci 35: 253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Berkowitz GA (2007) The grateful dead: calcium and cell death in plant innate immunity. Cell Microbiol 9: 2571–2585 [DOI] [PubMed] [Google Scholar]
- Mur LAJ, Kenton P, Lloyd AJ, Ougham H, Prats E (2008) The hypersensitive response: The centenary is upon us but how much do we know? J Exp Bot 59: 501–520 [DOI] [PubMed] [Google Scholar]
- Nawkar GM, Maibam P, Park JH, Sahi VP, Lee SY, Kang CH (2013) UV-Induced cell death in plants. Int J Mol Sci 14: 1608–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K, Oda Y, Fukuda H (2010) Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22: 3461–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olvera-Carrillo Y, Campos F, Reyes JL, Garciarrubio A, Covarrubias AA (2010) Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol 154: 373–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parizot B, De Rybel B, Beeckman T (2010) VisuaLRTC: a new view on lateral root initiation by combining specific transcriptome data sets. Plant Physiol 153: 34–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov V, Hille J, Mueller-Roeber B, Gechev TS (2015) ROS-mediated abiotic stress-induced programmed cell death in plants. Front Plant Sci 6: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plackett ARG, Thomas SG, Wilson ZA, Hedden P (2011) Gibberellin control of stamen development: a fertile field. Trends Plant Sci 16: 568–578 [DOI] [PubMed] [Google Scholar]
- Proost S, Van Bel M, Vaneechoutte D, Van de Peer Y, Inzé D, Mueller-Roeber B, Vandepoele K (2015) PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res 43: D974–D981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Wang H, Zou Y, Liu C, Liu Y, Wang Y, Zhang W (2011) Over-expression of mitochondrial heat shock protein 70 suppresses programmed cell death in rice. FEBS Lett 585: 231–239 [DOI] [PubMed] [Google Scholar]
- Repnik U, Hafner Česen M, Turk B (2014) Lysosomal membrane permeabilization in cell death: concepts and challenges. Mitochondrion 19: 49–57 [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roa H, Lang J, Culligan KM, Keller M, Holec S, Cognat V, Montané MH, Houlné G, Chabouté ME (2009) Ribonucleotide reductase regulation in response to genotoxic stress in Arabidopsis. Plant Physiol 151: 461–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. (2012) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 40: D13–D25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CB, Bariola PA, delCardayré SB, Raines RT, Green PJ (1993) RNS2: a senescence-associated RNase of Arabidopsis that diverged from the S-RNases before speciation. Proc Natl Acad Sci USA 90: 5118–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H. (2013) Senescence, ageing and death of the whole plant. New Phytol 197: 696–711 [DOI] [PubMed] [Google Scholar]
- Trapp O, Seeliger K, Puchta H (2011) Homologs of breast cancer genes in plants. Front Plant Sci 2: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Haseloff J (2008) A simple way to identify non-viable cells within living plant tissue using confocal microscopy. Plant Methods 4: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiatsiani L, Timmerman E, De Bock PJ, Vercammen D, Stael S, van de Cotte B, Staes A, Goethals M, Beunens T, Van Damme P, et al. (2013) The Arabidopsis metacaspase9 degradome. Plant Cell 25: 2831–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn WG. (2011) Classes of programmed cell death in plants, compared to those in animals. J Exp Bot 62: 4749–4761 [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Beers EP, Dangl JL, Franklin-Tong VE, Gallois P, Hara-Nishimura I, Jones AM, Kawai-Yamada M, Lam E, Mundy J, et al. (2011) Morphological classification of plant cell deaths. Cell Death Differ 18: 1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hautegem T, Waters AJ, Goodrich J, Nowack MK (2015) Only in dying, life: programmed cell death during plant development. Trends Plant Sci 20: 102–113 [DOI] [PubMed] [Google Scholar]
- Wang J, Bayles KW (2013) Programmed cell death in plants: lessons from bacteria? Trends Plant Sci 18: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Jiang M, Wu J, Ma Y, Li T, Chen Q, Zhang X, Xiang L (2014) Stress-induced RNASET2 overexpression mediates melanocyte apoptosis via the TRAF2 pathway in vitro. Cell Death Dis 5: e1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters A, Creff A, Goodrich J, Ingram G (2013) “What we’ve got here is failure to communicate”: zou mutants and endosperm cell death in seed development. Plant Signal Behav 8: e24368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzaczek M, Vainonen JP, Stael S, Tsiatsiani L, Help-Rinta-Rahko H, Gauthier A, Kaufholdt D, Bollhöner B, Lamminmäki A, Staes A, et al. (2015) GRIM REAPER peptide binds to receptor kinase PRK5 to trigger cell death in Arabidopsis. EMBO J 34: 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Chen H, Curtis C, Fu ZQ (2014) Go in for the kill. Virulence 5: 710–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Ohtani M, Yamaguchi M, Toyooka K, Wakazaki M, Sato M, Kubo M, Nakano Y, Sano R, Hiwatashi Y, et al. (2014) Contribution of NAC transcription factors to plant adaptation to land. Science 343: 1505–1508 [DOI] [PubMed] [Google Scholar]
- Yi D, Alvim Kamei CL, Cools T, Vanderauwera S, Takahashi N, Okushima Y, Eekhout T, Yoshiyama KO, Larkin J, Van den Daele H, et al. (2014) The Arabidopsis SIAMESE-RELATED cyclin-dependent kinase inhibitors SMR5 and SMR7 regulate the DNA damage checkpoint in response to reactive oxygen species. Plant Cell 26: 296–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Liu D, Lv X, Wang Y, Xun Z, Liu Z, Li F, Lu H (2014) The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell 26: 2939–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.