Noble rot causes major reprogramming of grape berry metabolism by activating stress responses and ripening processes, including pathways that are inactive or with limited flux in white-skinned berries.
Abstract
Noble rot results from exceptional infections of ripe grape (Vitis vinifera) berries by Botrytis cinerea. Unlike bunch rot, noble rot promotes favorable changes in grape berries and the accumulation of secondary metabolites that enhance wine grape composition. Noble rot-infected berries of cv Sémillon, a white-skinned variety, were collected over 3 years from a commercial vineyard at the same time that fruit were harvested for botrytized wine production. Using an integrated transcriptomics and metabolomics approach, we demonstrate that noble rot alters the metabolism of cv Sémillon berries by inducing biotic and abiotic stress responses as well as ripening processes. During noble rot, B. cinerea induced the expression of key regulators of ripening-associated pathways, some of which are distinctive to the normal ripening of red-skinned cultivars. Enhancement of phenylpropanoid metabolism, characterized by a restricted flux in white-skinned berries, was a common outcome of noble rot and red-skinned berry ripening. Transcript and metabolite analyses together with enzymatic assays determined that the biosynthesis of anthocyanins is a consistent hallmark of noble rot in cv Sémillon berries. The biosynthesis of terpenes and fatty acid aroma precursors also increased during noble rot. We finally characterized the impact of noble rot in botrytized wines. Altogether, the results of this work demonstrated that noble rot causes a major reprogramming of berry development and metabolism. This desirable interaction between a fruit and a fungus stimulates pathways otherwise inactive in white-skinned berries, leading to a greater accumulation of compounds involved in the unique flavor and aroma of botrytized wines.
Noble rot results from atypical Botrytis cinerea infections of ripe or overripe grape (Vitis vinifera) berries that occur under particular environmental conditions (Magyar, 2011; Vannini and Chilosi, 2013). White-skinned grape berries affected by noble rot are used to produce some of the highest quality dessert wines in the world, known as botrytized wines. Botrytized wines have been made since the 16th century, primarily in the Tokaj (Hungary), Rheingau (Germany), and Sauternais (France) regions. Their production has extended more recently to Italy, Australia, New Zealand, South Africa, and California (Magyar, 2011). These naturally sweet wines have distinctive flavor and aroma profiles, which include pineapple, apricot, pear, raisin, clove, and honey features combined with roti (i.e. roasted) characters (Magyar, 2011; Teissedre and Donèche, 2013).
B. cinerea is a necrotrophic fungal pathogen with a broad host range (Choquer et al., 2007). Under specific climatic (i.e. moist nights, foggy mornings, and dry days) and edaphic (i.e. low-nutrient and well-drained soils) conditions, B. cinerea infections may occur slowly, causing noble rot (Ribéreau-Gayon et al., 1980). This preferred type of infection promotes the accumulation of aroma and flavor compounds as well as the concentration of sugars (Vannini and Chilosi, 2013). In contrast, heavy rainfalls and high humidity conditions favor a more aggressive type of B. cinerea colonization of grape berries known as bunch rot (Gubler et al., 2013). As opposed to noble rot, bunch rot negatively impacts wine production by interfering with the fermentation process and altering the sensory properties of the resulting product (Hornsey, 2007; Morales-Valle et al., 2011). The molecular and biochemical processes that lead to either noble rot or bunch rot during B. cinerea-grape berry interactions have not been elucidated yet. It has been proposed that the combinatory effects of the environment, cultural practices, intrinsic characteristics of the grape cultivar, and the berry microbiome influence the type of rot developed (Ribéreau-Gayon et al., 1980; Sipiczki, 2006).
Noble rot causes two distinctive phenotypic alterations in the white-skinned berries that do not occur during bunch rot: (1) color change of the skin to pink and subsequently to purple-brown; and (2) dehydration at the final stages of infection caused by cracks in the cuticle layers. The latter explains the high sugar concentration in fully rotted and shriveled (i.e. pourri roti) berries (Magyar, 2006). Physicochemical changes induced by noble rot in grape berries include higher levels of sugar alcohols (i.e. glycerol, arabitol, mannitol, sorbitol, and inositol), enhanced sugar content, reduced Glc-to-Fru ratio, higher concentration of malic acid, and lower concentration of tartaric acid when compared with uninfected berries (Ribéreau-Gayon et al., 2006). Mucic acid and gluconic acid, two organic acids that result from B. cinerea catabolism of plant sugars, also accumulate in noble-rotted berries. Infected berries usually have high concentrations of free GalA, Rha, Gal, Man, Ara, and Xyl, which may result from the B. cinerea-induced breakdown of pectin and cross-linking glycans present in the fruit cell walls (Magyar, 2011). Greater abundance of total polyphenolic compounds has also been detected in berries affected by noble rot (Magyar, 2006). However, the effects of noble rot in the chemical composition of the berries vary depending on the grape cultivar, terroir, and degree of B. cinerea infection. To date, only targeted compositional analyses have been performed in noble-rotted berries. Comprehensive metabolomic approaches will help to further understand the influence of noble rot in grape berry metabolism.
Due to their high predisposition to noble rot, grapevine cultivars commonly used for botrytized wines include cv Sémillon, cv Riesling, cv Sauvignon Blanc, cv Muscadel, and cv Chenin Blanc (Magyar, 2011). The compact cluster architecture and berry cuticle with lower mechanical resistance of these white-skinned varieties seem to facilitate B. cinerea penetration (Vail and Marois, 1991; Kretschmer et al., 2007). During ripening, grape berries undergo a series of complex biochemical and physiological processes that enhance their quality for wine production but also their vulnerability to pathogen infection (Pezet et al., 2003; Deytieux-Belleau et al., 2009). Some ripening processes that may promote B. cinerea infections of grape berries are cell wall and cuticle modifications that lead to fruit softening and microfractures on the berry surface, modulation of the fruit’s synthesis and perception of plant hormones, changes in the levels of organic acids and sugars, and the loss of preformed defenses (Miedes and Lorences, 2007; Cantu et al., 2008, 2009; Centeno et al., 2011; Blanco-Ulate et al., 2013b; Prusky et al., 2013). Moreover, B. cinerea infections can accelerate some of these ripening processes in unripe fruit (Cantu et al., 2009; Blanco-Ulate et al., 2013b; Agudelo-Romero et al., 2015).
In this study, we integrated transcriptomics, metabolomics, and enzyme activity assays to characterize the impact of noble rot on the development and metabolism of white-skinned grape berries under field conditions. Berries at distinct stages of noble rot were sampled during multiple years from a commercial vineyard. We identified grape molecular pathways that are triggered during distinct stages of noble rot. Some of these pathways correspond to plant responses to biotic and abiotic stresses, while others are involved in the generation of aroma and flavor metabolites. Particular metabolic changes induced by noble rot were also similar to events that occur during the normal development of red-skinned berries, including the induction of pathways of secondary metabolism that have limited flux in white-skinned cultivars. We detected in noble-rotted berries an enhanced expression of transcription factors that are known to regulate secondary metabolic pathways during berry ripening. We validated the relevance of these pathways during noble rot using targeted gene expression and enzymatic analyses in B. cinerea-infected berries harvested during an additional 2 years. Finally, we profiled the metabolites of commercial botrytized wines produced from the same vineyard where the berries were collected and corroborated that key compounds that result from noble rot are carried over to the wines.
RESULTS
Sampling of Naturally Occurring B. cinerea Infections
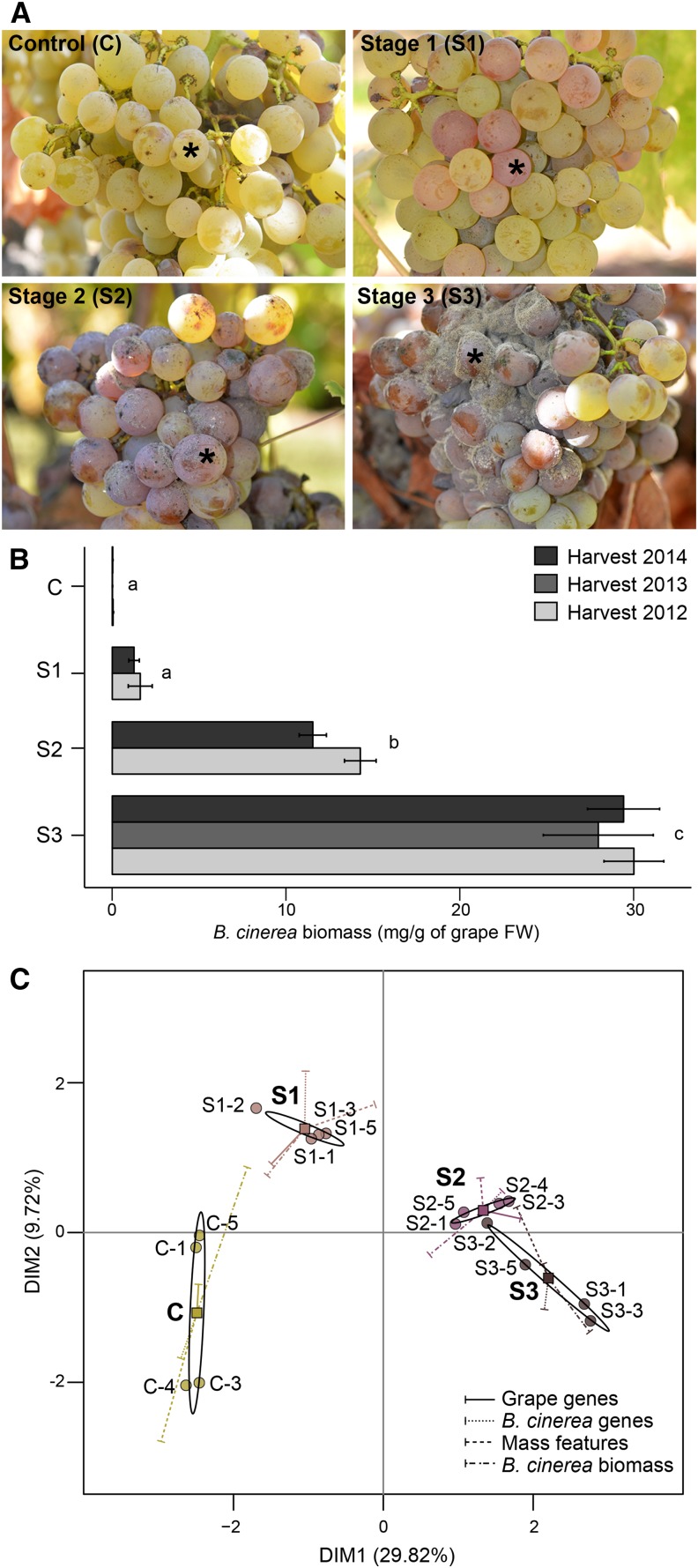
Ripe grape berries (cv Sémillon; soluble solids, 23.9% ± 1%, w/v) displaying noble rot symptoms were harvested in 2012, 2013, and 2014 from a commercial vineyard in Napa, California. Samples were collected on the same day as grapes that were harvested for botrytized wine production. B. cinerea infections occurred naturally in the field and were categorized in three stages of noble rot development. The initial stage of infection (stage 1; Fig. 1) was characterized by a color change of the berries from yellow to pink. As the infection progressed, the berry skin became softer and dark pink (stage 2; Fig. 1). Berries at stage 3 (i.e. pourry plein) were fully rotten but not dry. Stage 3 berries presented cracked skins with evident mycelia growth and sporulation and were purple-brown (Fig. 1). Berries that did not show any symptoms of infection were harvested simultaneously and used as controls (Fig. 1). Measurements of B. cinerea biomass using the monoclonal antibody BC12.CA4 (Meyer and Dewey, 2000) in the collected samples confirmed that the stages correspond to distinct degrees of fungal infection and presented very similar patterns during the three independent harvesting seasons (Fig. 1). Two-way factorial ANOVA demonstrated a significant effect of infection stage on B. cinerea biomass (P < 2.20 × 10−16), while the year of harvest (P = 0.23) and the infection stage × year interaction (P = 0.59) were not significant.
Figure 1.
Stages of noble rot in ripe cv Sémillon berries. A, Representative grape clusters with asymptomatic berries (control [C]) and berries at stages 1 (S1), 2 (S2), and 3 (S3) of noble rot. Asterisks indicate berries with characteristic phenotype of each noble rot stage. B, Accumulation of B. cinerea biomass in control and noble-rotted berries from three independent harvests. The harvest of 2013 includes control and stage 3 berries only. Letters correspond to significant differences between stages (P ≤ 0.01). FW, Fresh weight. C, MFA of infected and control berries using four types of quantitative variables (represented by vectors): transcript abundance of 24,373 grape genes and 15,550 B. cinerea genes, accumulation of 2,084 mass features, and B. cinerea biomass measurements. Samples used in this analysis were harvested in 2012. Each point represents a biological replicate for a given stage of noble rot. Ellipses define confidence areas (95%) for each stage, while squares represent their corresponding centers of gravity. DIM, Dimension.
Sugars, sugar alcohols, and organic acids were quantified as reported previously (Magyar, 2011; Teissedre and Donèche, 2013). As expected, glycerol, mannitol, and sorbitol levels increased significantly (P ≤ 0.01) in the infected samples (Supplemental Table S1). Although Glc and Fru accumulated as a result of noble rot, only a slight reduction in the Glc-Fru ratio was observed at stages 2 and 3 of infection (Supplemental Table S1).
Integration of Transcriptomics and Metabolomics to Study B. cinerea Infections of Grape Berries
RNA sequencing (RNAseq) and metabolomic analyses were performed with samples harvested in 2012. mRNA was isolated from four biological replicates per infection stage (i.e. stages 1–3) and the control, and sequenced using an Illumina HiSeq2000 platform. Supplemental Table S2 provides a summary of the parsed reads from all samples as well as the number of reads that mapped onto the predicted transcriptomes of B. cinerea (strains B05.10 and BcDW1) and grape (cv PN40024). The expression of 15,550 B. cinerea genes (95.42% of total B. cinerea genes) and 24,373 grape genes (81.32% of total grape genes) was detected across all stages of noble rot and control samples. The relative abundance of grape to fungal transcripts decreased at later stages of infection, reflecting the increase in fungal biomass with the progression of noble rot; however, most grape genes (71.34% of total genes) were still detectable at advanced stages of infection (Supplemental Table S2). B. cinerea genes were functionally annotated based on similarity to ascomycete peptides in GenBank, and Gene Ontology (GO) terms were assigned using Blast2GO (Conesa et al., 2005). The VitisNet functional annotations were used to assign grape genes to functional categories and subcategories (Grimplet et al., 2009).
The detection and quantification of metabolites in noble-rotted (stages 1, 2, and 3) and control berries were carried out by ultrahigh pressure liquid chromatography quadrupole time-of-flight mass spectrometry using five biological replicates and six technical replications. A total of 2,163 non-redundant mass features were identified among all samples, comprising products of both plant and fungal metabolism (Supplemental Table S3). The possible identities of the mass features were determined by comparisons with the METLIN database (less than 30 ppm cutoff; Smith et al., 2005). Based on statistical testing (i.e. multiple factor and cluster analyses), a set of 688 metabolites with significant accumulation patterns were manually curated by evaluating their retention times and by matching their tandem mass spectrometry (MS/MS) spectra to the Tandem Mass Spectrum Database (for details, see “Materials and Methods”). The curated metabolites were then classified into compound classes and subclasses using the chemical taxonomy of the Human Metabolome Database (Wishart et al., 2013).
RNAseq and metabolomics data were integrated into a multiple factor analysis (MFA) to determine if the distinct stages of noble rot could be discerned based on the abundance of (1) B. cinerea transcripts, (2) grape transcripts, and (3) mass features. Figure 1 displays the results of the MFA, which included measurements of B. cinerea biomass as an additional class of quantitative variables. The MFA revealed that the noble rot stages were significantly different (95% confidence) from each other at both transcriptome and metabolome levels. The first dimension (DIM1; 29.82%) distinguished the heavily infected samples (i.e. stages 2 and 3) from samples at stage 1 and the asymptomatic controls. The second dimension (DIM2; 9.72%) further separated the samples belonging to stage 1 and the control (Fig. 1). Eigenvalues and percentages of explained variance for the 15 dimensions of the MFA are provided in Supplemental Table S4.
Quantitative variables (i.e. gene transcripts and mass feature abundances) that significantly correlated (−0.7 ≥ r ≥ 0.7, P ≤ 0.01) with the first two dimensions of the MFA were analyzed to provide insights into the molecular pathways responsible for the development of noble rot (Supplemental Table S5). Enrichment analyses (P ≤ 0.05) of GO terms using B. cinerea genes with significant correlations identified functions employed by the pathogen during berry infection (Supplemental Fig. S1). At initial stages of noble rot (i.e. positive DIM2), B. cinerea expressed functions related to cell growth and host tissue colonization. These included the expression of genes involved in plant cell wall degradation (e.g. endo-β-1,4-glucanases; Blanco-Ulate et al., 2014) and the metabolism of reactive oxygen species (e.g. superoxide dismutase; Supplemental Table S5). Another function that was induced early during B. cinerea infections was the biosynthesis of unsaturated fatty acids. At stages 2 and 3 of noble rot (i.e. positive DIM1), the transport of organic molecules (e.g. carbohydrates) was a prominent feature. This is in agreement with the necrotrophic lifestyle of B. cinerea once established in the berry tissues. Potential virulence factors were identified among the B. cinerea genes with the highest significant correlations (r ≥ 0.9, P ≤ 1 × 10−6) to the positive sides of DIM1 and DIM2 (Supplemental Table S5; Amselem et al., 2011; Giesbert et al., 2012; Yang et al., 2013; Ambrose et al., 2015). These comprised two putative salicylate hydroxylases (BC1T_14568 and BC1T_15387), a protein-Tyr phosphatase (BC1T_07726), a Ser/Thr protein phosphatase type 2A (BC1T_05741), and a nonribosomal peptide synthetase (BC1T_01317).
Grape genes whose transcript accumulation significantly correlated to each of the main MFA dimensions (Supplemental Table S5) were evaluated to determine plant cellular and metabolic processes altered in response to noble rot (Supplemental Fig. S1). Enriched biological functions (P ≤ 0.01) associated with stages 1 and 2 (i.e. positive DIM2) included terpene metabolism, anthocyanin biosynthesis, and fatty acid metabolism. Stages 2 and 3 (i.e. positive DIM1) presented significant enrichments in a variety of functions, such as shikimate metabolism, phenylpropanoid metabolism, alkaloid metabolism, Arg and Pro metabolism, monosaccharide and polysaccharide metabolism, ethylene and jasmonic acid (JA) signaling, and responses to abiotic stress (e.g. oxidative stress and wounding).
Compound classes associated with noble rot were identified using metabolite variables with significant correlations to the major dimensions (Supplemental Table S5). The abundance of only 18 metabolites correlated significantly with stages 1 and 2 (i.e. positive DIM2), most of which belonged to compound classes with relevant functions in response to plant stress as well as in grape aroma and flavor composition, such as benzene derivatives, cinnamic acid derivatives, and flavonoids (Supplemental Fig. S1). The 133 metabolites that showed greater accumulation in heavily noble-rotted samples (i.e. positive DIM1) included fatty acids, prenol lipids, benzene derivatives, cinnamic acid derivatives, and peptides (Supplemental Fig. S1).
The MFA results provided an initial survey of relevant fungal and plant functions that occur throughout the progression of noble rot. Due to the distinctive and prominent effects of noble rot on grape berry metabolism, we focused our subsequent analyses on characterizing changes in grape transcript levels and metabolite abundances at specific stages of infection.
Grape Berry Responses to Noble Rot
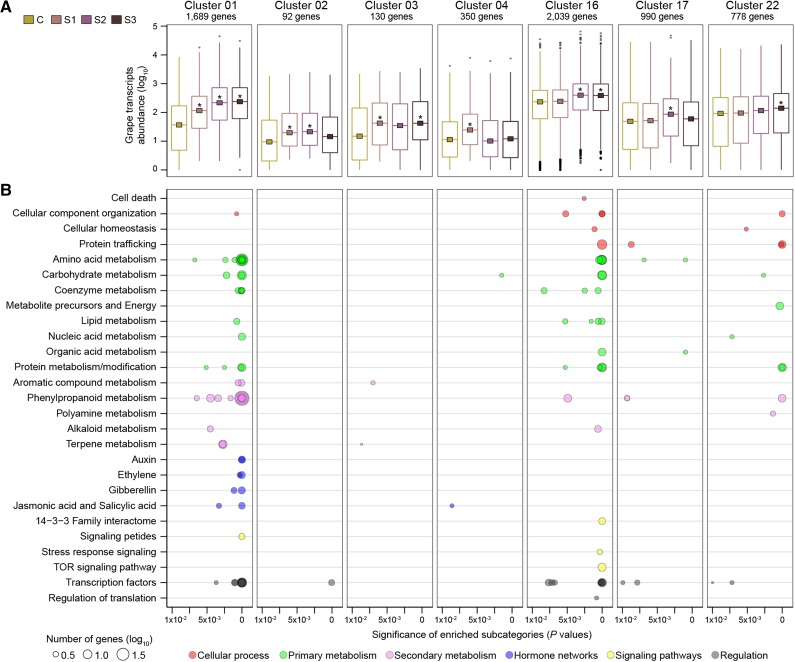
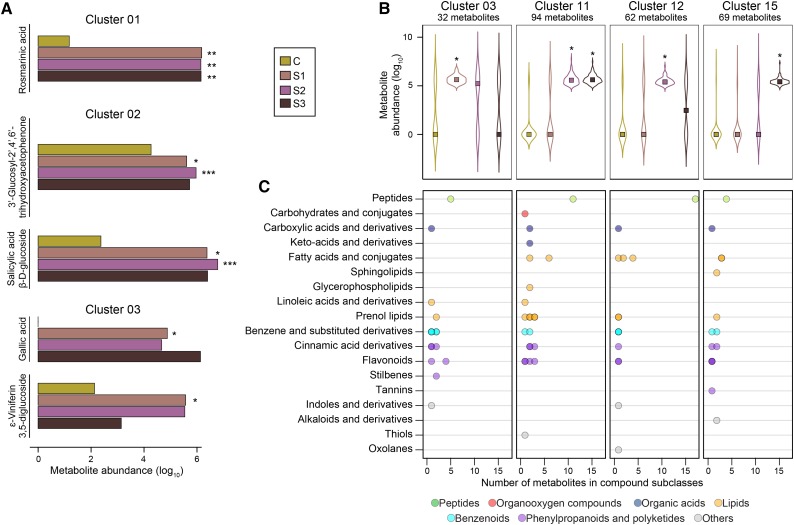
Grape genes that have significant differential expression (DE genes; adjusted P ≤ 0.05) at distinct stages of noble rot were determined by comparisons between samples from each stage and the asymptomatic control using DESeq2 (Love et al., 2014). The DE genes were grouped in 23 clusters according to their patterns of expression across all comparisons (Supplemental Table S6). Among these, seven clusters included only up-regulated genes in response to noble rot (Fig. 2). Metabolite data were processed following a similar strategy. To detect compounds with significant changes in abundance, statistical tests between samples at each infection stage and the asymptomatic controls were performed for each metabolite. Metabolites with statistically significant changes (adjusted P ≤ 0.05) were grouped in 16 distinct clusters based on their patterns of accumulation across the stages of noble rot (Supplemental Table S7). Six clusters consisted of metabolites with increased abundance due to B. cinerea infections (Fig. 3).
Figure 2.
Clusters of significant up-regulated grape genes in response to noble rot. A, Patterns of transcript accumulation in the gene clusters. Squares correspond to the median counts of the genes in control (C) and each of the noble rot stages: 1 (S1), 2 (S2), and 3 (S3). Asterisks indicate significant (adjusted P ≤ 0.05) fold changes when comparing each stage with the control. B, Enriched functional subcategories (P ≤ 0.01) in the clusters. The sizes of the circles reflect the number of genes (log10) for each functional subcategory. The complete data set is available in Supplemental Table S6.
Figure 3.
Clusters of metabolites with significant enhanced accumulation in noble-rotted berries. A, Bar plots show the abundance of some metabolites belonging to clusters 01, 02, and 03. B, Patterns of metabolite accumulation in the clusters that included more than 30 metabolites with significantly higher abundance at any of the noble rot stages compared with the control. Squares correspond to the median abundance of the metabolites in each noble rot stage (S1, stage 1; S2, stage 2; and S3, stage 3) and the control (C). Asterisks indicate significant fold changes (*, adjusted P ≤ 0.05; **, adjusted P ≤ 0.01; and ***, adjusted P ≤ 0.001) when comparing each stage with the control. C, Number of metabolites in compound classes and subclasses. Metabolites with unknown identities were not included. The complete data set is available in Supplemental Table S7.
Biological processes activated as a result of noble rot were determined by enrichment analyses (P ≤ 0.01) of functional subcategories in the clusters of up-regulated genes (Fig. 2; Supplemental Table S8) and by identifying compound classes in the clusters with metabolites that significantly accumulated in infected samples (Fig. 3). Distinct families of transcription factors were induced as a function of B. cinerea infection (Supplemental Table S8). Among these, APETALA2/ETHYLENE-RESPONSIVE FACTOR (AP2-ERF), which play important roles in plant-pathogen interactions and fruit ripening (Gutterson and Reuber, 2004; Licausi et al., 2013), were up-regulated at all stages of noble rot (gene cluster 01). The expression of NON APICAL MERISTEM/ARABIDOPSIS TRANSCRIPTION FACTOR/CUP-SHAPED COTYLEDON (NAC) transcription factors, which regulate key plant developmental processes such as fruit ripening and senescence (Kou et al., 2014; Podzimska-Sroka et al., 2015), increased throughout the progression of noble rot (gene clusters 01, 02, and 16). The expression of transcription factors associated with responses to multiple stresses, such as heat shock factors (gene clusters 01 and 16; Scharf et al., 2012), was also enhanced by infection.
Induction of hormone pathways, specifically ethylene, auxin, GA, and JA, was a common berry response at all stages of noble rot (gene cluster 01; Fig. 2). The induction of GA biosynthesis was supported by the enhanced accumulation of two GAs at stage 2 of noble rot (metabolite cluster 11). Similarly, the higher abundance of a JA and derivatives at the last stages of noble rot (metabolite clusters 03 and 11) confirmed the activation of JA biosynthesis and signaling pathways, including the up-regulation of jasmonate-ZIM domain (JAZ) proteins (gene cluster 01). Salicylic acid (SA) and derivatives, such as SA β-d-glucoside (e.g. a storage form of SA), accumulated at most stages of noble rot (metabolite clusters 02, 03, 12, and 15). A role of SA during noble rot is suggested by the up-regulation at stage 1 (gene cluster 04) of an SA-responsive gene with a putative role in defense responses, a PATHOGENESIS-RELATED PROTEIN1 precursor (VIT_03s0088g00710). Hypothetically, B. cinerea may interfere with SA signaling by expressing the two fungal salicylate hydroxylases mentioned before, which could degrade the hormone (Ambrose et al., 2015). Significant accumulation of abscisic acid (ABA) was detected at the last stages of infection (metabolite clusters 11 and 16), while the expression of ABA biosynthetic and ABA-responsive genes was differentially regulated at various stages of noble rot (Supplemental Table S6).
B. cinerea infections caused transcriptional alterations of particular primary metabolic pathways (Fig. 2; Supplemental Table S8). Several processes involved in the metabolism of amino acids and carbohydrates were triggered by noble rot (Fig. 2). The activation of pathways related to protein metabolism and modification, in particular proteolysis (gene clusters 01, 16, and 22; Fig. 2), may explain the large number of peptides that accumulated in the noble-rotted berries (metabolite clusters 02, 03, 04, 11, 12, and 15; Fig. 3). The observed induction of fungal proteolytic enzymes (e.g. trypsin-like proteases, Cys proteases, and Ser proteases; Supplemental Table S5) may also contribute to the release of peptides. Another specific feature of noble rot was the enhanced expression of fatty acid and glycerolipid biosynthetic genes (gene clusters 01 and 16), which can partially explain the accumulation of fatty acids, hydroxy fatty acids, glycerophospholipids, and glycosphingolipids mostly at stages 2 and 3 (metabolite clusters 01, 11, 12, and 15). The up-regulation of unsaturated fatty acid biosynthesis by B. cinerea at stages 1 and 2 (mentioned above) also could have favored the greater accumulation of lipids in the infected berries.
The secondary metabolism of grape berries was highly induced as noble rot progressed (Fig. 2; Supplemental Table S8), especially the main and peripheral phenylpropanoid pathways (gene clusters 01, 16, 17, and 22). Early products of the phenylpropanoid metabolism (i.e. cinnamic acid derivatives) accumulated in noble-rotted berries (metabolite clusters 01, 03, 11, 12, and 15). Rosmarinic acid, a cinnamic acid derivative with antioxidant and aromatic properties (Petersen, 2013), showed a significant accumulation relative to the control at all stages of noble rot (metabolite cluster 01; Fig. 3). The presence of rosmarinic acid in the noble-rotted berries was validated by 1H-NMR (Supplemental Table S9). The biosynthesis of stilbenes, known to be phytoalexins and strong antioxidants (Cassidy et al., 2000; Chong et al., 2009), was also induced in response to B. cinerea infection (gene cluster 01). Two glycosides of a resveratrol dimer (i.e. viniferin diglucosides) significantly accumulated at stage 1 (metabolite cluster 03; Fig. 3). The biosynthesis of flavonoids was also triggered in response to B. cinerea infection (gene clusters 01 and 16), which explained the significant accumulation of several flavonoid glycosides and flavanones in noble-rotted berries (metabolite clusters 04, 11, and 15). Remarkably, the biosynthesis of anthocyanins, which is normally impaired in white-skinned grape berries, was activated by noble rot (gene clusters 01 and 17). Significant accumulation of cyanidin-3-rutinoside and delphinidin-3-rutinoside (confirmed by 1H-NMR; Supplemental Table S9) and of cyanidin-3-gentiobioside and delphinidin-3-gentiobioside was detected at stages 1 and 2 (metabolite clusters 02, 03, and 12).
Other aromatic compounds, including acetophenones, benzoic acid derivatives, methoxyphenols, and phenolic glycosides, showed increased abundance at all stages of noble rot (metabolite clusters 01, 02, 03, 11, 12, and 15). These metabolites play biological roles as antioxidants and antimicrobial compounds, whereas from an enological perspective they are known to modify the aroma profiles of grape berries and their derived wines (Cheynier et al., 2013; Teixeira et al., 2013; Robinson et al., 2014). Among these, 3′-glucosyl-2′,4′,6′-trihydroxyacetophenone accumulated at stages 1 and 2 (Fig. 3). Gallic acid, a precursor of tannin biosynthesis (Yilmaz and Toledo, 2004), also had increased levels as a consequence of noble rot (Fig. 3; Supplemental Tables S8 and S9). Genes associated with the biosynthesis of these phenolic derivatives showed up-regulation mostly at the first stage of noble rot (gene cluster 03; Fig. 2; Supplemental Table S8).
Secondary metabolic pathways involved in terpene metabolism were also up-regulated by noble rot (Fig. 2; Supplemental Table S8). Besides their biological functions in plant defense and interaction with other organisms, terpenes can markedly influence grape and wine aroma (Hjelmeland and Ebeler, 2014). Increased accumulation of mevalonic acid, a key precursor in terpene biosynthesis, was detected in the noble-rotted berries (metabolite clusters 11 and 15, further validated by 1H-NMR; Supplemental Table S9). Monoterpenes, triterpenes, sesquiterpenes, and terpene glycosides accumulated at all stages of noble rot (metabolite clusters 03, 11, 12, and 15). While this may have resulted from the activation of plant terpene biosynthetic pathways, the fungal origin of these metabolites cannot be ruled out (Collado et al., 2007).
Intersection between Noble Rot and Ripening
A phenotypic hallmark of noble rot is the color change of the grape berries from yellow to pink to purple-brown (Fig. 1). The pink coloration that develops at the first stage of infection resembles the same process that healthy berries from red-skinned varieties undergo at véraison. The results from the analysis of responses to noble rot provided evidence that certain transcriptional and metabolic changes (e.g. biosynthesis of anthocyanins) that occurred in the infected white-skinned berries are similar to those that occur during normal ripening of red-skinned grape berries. Therefore, a meta-analysis of publicly available data sets was carried out to identify alterations in developmental and metabolic pathways that are shared between B. cinerea infections and healthy ripening of grape berries. Raw RNAseq data of red-skinned grape berries at two developmental stages, pre-véraison (i.e. touch stage) and véraison (i.e. soft stage), belonging to five different grape cultivars (Sangiovese, Barbera, Negroamaro, Refosco, and Primitivo) were obtained from Palumbo et al. (2014). The data from these samples were processed following the same bioinformatics pipeline used for the noble-rotted samples. DESeq2 was used to determine DE genes between the two developmental stages of each grape variety. A total of 6,847 DE genes that showed a common trend across all red-skinned varieties were defined as core ripening genes (3,183 up-regulated and 3,664 down-regulated genes; Supplemental Table S10).
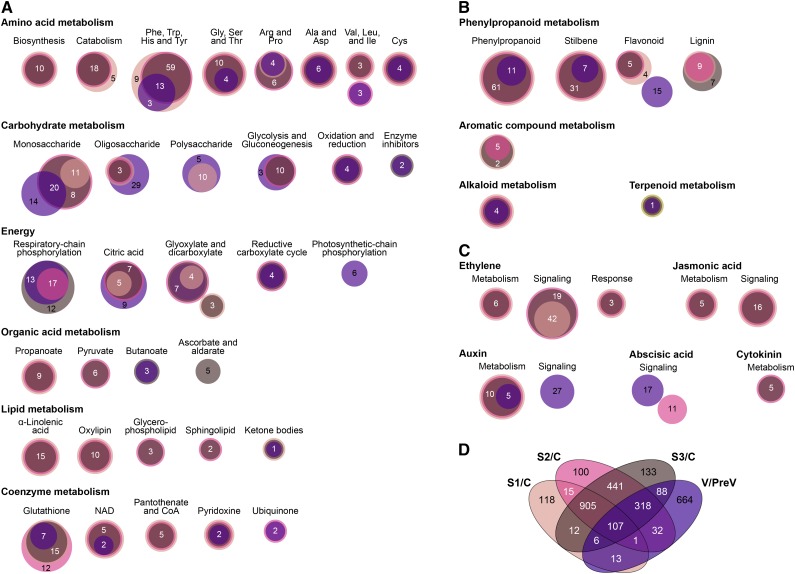
Noble rot-responsive genes (i.e. DE genes at the noble rot stages) were compared with the core ripening genes to identify changes in gene expression shared between noble rot and ripening. A total of 1,382 grape genes were commonly up-regulated in noble-rotted and ripening berries, while 1,249 grape genes were commonly down-regulated (Supplemental Table S11). Key biological processes that were concomitantly activated or suppressed as a consequence of noble rot and normal ripening were identified by enrichment analyses (P < 0.01) using the lists of common and unique DE genes (Fig. 4; Supplemental Table S12).
Figure 4.
Main molecular pathways commonly activated in response to noble rot and healthy ripening. A to C, Euler diagrams represent commonly and uniquely up-regulated (adjusted P ≤ 0.05) genes between B. cinerea infections of cv Sémillon grape berries and the ripening of red-skinned grape berries, which are enriched (P ≤ 0.01) in functional subcategories of biological importance (Supplemental Table S12). Circles are scaled to the log10 of the number of genes included in each comparison. D, Venn diagram displaying common and unique up-regulated (adjusted P ≤ 0.05) genes when comparing responses to noble rot in cv Sémillon berries and the normal ripening of red-skinned berries; only genes enriched (P ≤ 0.01) in grape biological processes are included. C, Control; PreV, pre-véraison; S1, stage 1; S2, stage 2; S3, stage 3; V; véraison.
High expression of genes involved in the biosynthesis of amino acids, particularly aromatic amino acids, was shared by B. cinerea infection and normal development of red-skinned grape berries (Fig. 4). The induction of glutathione biosynthetic genes was another common response. Glutathione is a strong antioxidant with key roles in wine aroma, particularly when it is conjugated to fatty acid derivatives (Kobayashi et al., 2011). Other primary metabolic pathways showed a similar activation in noble-rotted cv Sémillon berries and red-skinned berries (Fig. 4). These included monosaccharide and oligosaccharide metabolism, glycolysis and gluconeogenesis, citric acid metabolism, and glyoxylate and dicarboxylate metabolism.
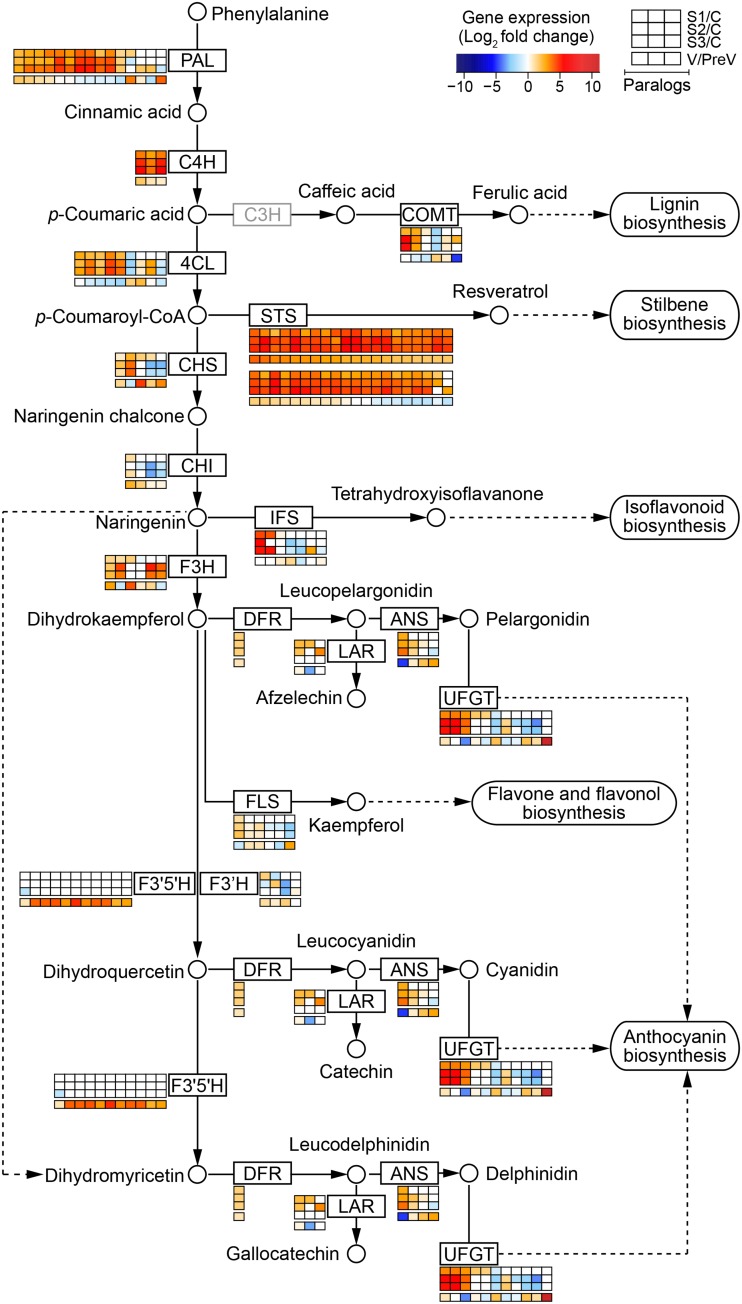
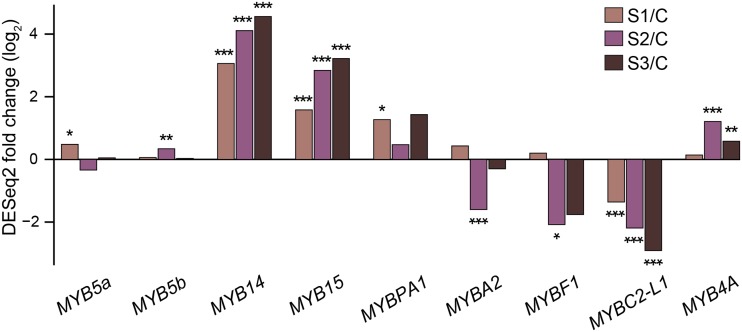
A prominent and common outcome of noble rot and ripening was the enhancement of phenylpropanoid metabolism (Fig. 4). Figure 5 depicts the central phenylpropanoid and flavonoid pathways, in which specific genes were among the up-regulated noble rot-responsive genes and core ripening genes. The concurrent activation of 100 genes in the central and peripheral phenylpropanoid pathways (Supplemental Table S11) suggests that similar transcriptional regulators could be functioning during B. cinerea infections of white-skinned berries and véraison of red-skinned berries. The R2R3-MYB family of transcription factors has been associated with the control of the phenylpropanoid pathways during berry development (Deluc et al., 2006, 2008; Bogs et al., 2007; Walker et al., 2007; Matus et al., 2008; Höll et al., 2013; Cavallini et al., 2015). The VvMYB5a (VIT_08s0007g07230) and VvMYB5b (VIT_06s0004g00570) transcription factors, which are known master regulators of the central phenylpropanoid and flavonoid pathways in grape berries, were significantly up-regulated at different stages of noble rot (Fig. 6). The activators of stilbene biosynthesis VvMYB14 (VIT_07s0005g03340) and VvMYB15 (VIT_05s0049g01020) are known to be induced during berry ripening and were highly up-regulated as noble rot progressed. The expression of a transcription factor that regulates the biosynthesis of proanthocyanidins during grape berry development, VvMYBPA1 (VIT_15s0046g00170), was significantly increased at stage 1. Two suppressors of phenylpropanoid pathways, VvMYBC2-L1 (VIT_01s0011g04760) and VvMYB4a (VIT_03s0038g02310), showed differential gene expression as a consequence of noble rot. In contrast, the transcriptional activators VvMYBA2 (VIT_02s0033g00390) and VvMYBF1 (VIT_07s0005g01210) were down-regulated at stage 2. VvMYBA2 is a regulator of a key anthocyanin biosynthetic gene, whereas VvMYBF1 controls the first step of flavonol biosynthesis (Czemmel et al., 2009).
Figure 5.
Activation of the phenylpropanoid metabolism as a common response between noble rot and healthy ripening. A representation of the central phenylpropanoid and flavonoid pathways based on Kyoto Encyclopedia of Genes and Genomes pathways (www.genome.jp/kegg/pathway.html) is provided. Dashed lines indicate that some steps have been omitted. The gray box indicates an enzyme that has not been completely characterized and, therefore, is not found in the grape gene annotations. Significant fold changes (log2) in expression of noble rot-responsive genes (Supplemental Table S6) and ripening genes from five red-skinned cultivars (median value; Supplemental Table S10) are depicted next to each of the biosynthetic enzymes that they encode. The colors in the heat maps represent the intensity of the expression changes. C, Control; PreV, pre-véraison; S1, stage 1; S2, stage 2; S3, stage 3; V; véraison. Enzymes, from the top, are as follows: PAL, Phe ammonia lyase; C4H, trans-cinnamate 4-monooxygenase; C3H, p-coumarate 3-hydroxylase; COMT, caffeic acid 3-O-methyltransferase; 4CL, 4-coumaroyl-CoA ligase; CHS, chalcone synthase; STS, stilbene synthase; CHI, chalcone isomerase; IFS, isoflavonoid synthase; F3H, flavonone 3-hydroxylase; DFR, dihydroflavanol 4-reductase; LAR, leucoanthocyanidin reductase; ANS, anthocyanidin synthase; UFGT, UDP-Glc:flavonoid 3-O-glucosyltransferase; FLS, flavonol synthase; F3′5′H, flavonoid 3′,5′-hydroxylase; F3′H, flavonoid 3′-monooxygenase.
Figure 6.
Noble rot induction of transcription factors that control the phenylpropanoid metabolism. The bar plot depicts the expression fold changes (log2) of MYB transcription factors between each of the three stages of noble rot (S1, stage 1; S2, stage 2; and S3, stage 3) and the asymptomatic control (C). VvMYBC2-L1 and VvMYB4a are known to encode repressors of phenylpropanoid pathways. Asterisks indicate significant fold changes (*, adjusted P ≤ 0.05; **, adjusted P ≤ 0.01; and ***, adjusted P ≤ 0.001).
Boost in Phenylpropanoid Metabolism Is a Hallmark of Noble Rot
The transcriptomic and metabolomic analyses described above were performed with replicated grape samples harvested in 2012. To confirm that the induction of the phenylpropanoid metabolism was a consistent response to noble rot and not a confounding effect of a particular harvest, noble-rotted and control berries collected from the same vineyard in 2013 and 2014 were analyzed. Temperature and relative humidity during the growing seasons of 2012 and 2014 were more suitable for the development of noble rot than during the season of 2013 (i.e. rains occurred earlier and most of the B. cinerea infections took place 1 month earlier than in 2012 and 2014). Due to differences in the progression of noble rot, only berries at stage 3 and the asymptomatic control from the harvest in 2013 were included in this study.
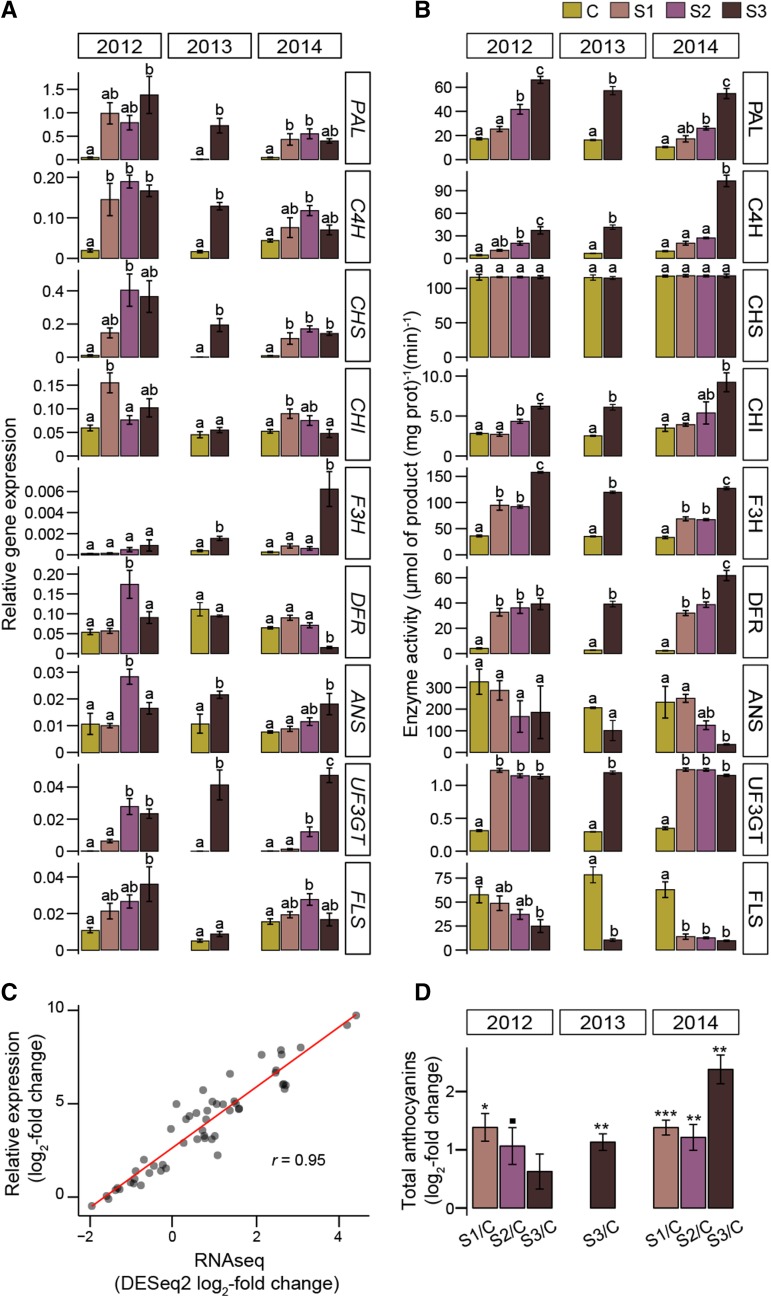
The transcriptional induction of 18 key noble rot-responsive genes in the central phenylpropanoid and flavonoid pathways was confirmed by quantitative reverse transcription (qRT)-PCR (Supplemental Table S13). As shown in Figure 7, most genes presented similar patterns of expression as a result of noble rot across the 3-year evaluation; however, differences in the intensity of the up-regulations were detected. For example, the expression of two PAL-encoding genes (VIT_11s0052g01090 and VIT_08s0040g01710) was higher in 2012 than in subsequent harvests. The qRT-PCR results from samples harvested in 2012 validated the RNAseq expression data with a strong significant correlation (r = 0.95, P < 2.2 × 10−16; Fig. 7).
Figure 7.
Induction of key steps in the central phenylpropanoid and flavonoid pathways during noble rot. A, Bar plots represent the relative gene expression of a paralog involved in the biosynthesis of phenylpropanoids and flavonoids measured by qRT-PCR. B, Activity of their corresponding biosynthetic enzymes. C, Scatterplot of the correlation between the fold changes (log2) in expression obtained by processing the RNAseq data with DESeq2 and the fold changes (log2) in relative expression measured by qRT-PCR. A linear trend is shown. The relative expression data were linearized using VvACTIN as a reference gene. D, Log2 fold change in total anthocyanins when comparing B. cinerea-infected samples against the median value of anthocyanin levels in the control samples of a given harvest. Noble-rotted berries used in these analyses were harvested in 2012, 2013, and 2014. Symbols indicate significant fold changes (■, P < 0.1; *, P < 0.05; **, P < 0.01; and ***, P < 0.001). C, Control; S1, stage 1; S2, stage 2; S3, stage 3.
To demonstrate that the boost in phenylpropanoid metabolism occurred not only at the transcriptional level, the activity of key enzymes of these pathways was measured in the noble-rotted and control samples collected in 2012, 2013, and 2014 as described in “Materials and Methods” (Fig. 7). In most cases, the enzyme activities reflected the patterns of gene expression. A few differences were observed: (1) a constant CHS activity and (2) increased dihydroflavonol 4-reductase activity in noble-rotted berries when compared with the control.
To verify the activation of the anthocyanin biosynthetic pathway, total amounts of anthocyanins were measured in the noble-rotted and control berries. Anthocyanin levels were significantly higher in the infected samples than in the control across all harvests (Fig. 7). Variation in pigment accumulation was expected among years because anthocyanin biosynthesis and oxidation are known to be influenced by a variety of environmental factors, such as temperature, sunlight exposure, and differences in the progression of noble rot (Ortega‐Regules et al., 2006; Tarara et al., 2008). Specific anthocyanin classes that could explain the change in coloration of the berries during noble rot include the cyanidin and delphinidin glycosides mentioned previously (Supplemental Tables S3 and S9).
Metabolic Changes in Noble-Rotted Berries Reflect Differences in the Chemical Composition of Botrytized Wines
Botrytized wines are produced using berries at different stages of infection, ranging from stage 1 to stage 3 of noble rot or even later stages (i.e. pourri roti). Metabolic profiles of botrytized wines were analyzed to determine if compound classes associated with noble rot in the vineyard were detectable in the resulting wines. These corresponded to seven distinct vintages of commercial botrytized wines (i.e. BOT1 wines) produced with naturally infected berries (cv Sémillon) harvested from the same vineyard blocks where the samples of this study were collected. The BOT1 wines were blended with late-harvest wine (e.g. cv Sauvignon Blanc varietal, non-botrytized) during their production. Additional wine samples were included in the analysis to be able to distinguish specific repertoires of metabolites associated with botrytized wines: (1) five botrytized wines produced by a different winery using cv Sémillon grape berries grown in a different location (i.e. BOT2 wines); (2) two wines from non-botrytized cv Sémillon berries; and (3) two wines from non-botrytized cv Sauvignon Blanc berries. Measurements of a specific B. cinerea antigen confirmed that the non-botrytized wines were produced from grape berries without detectable B. cinerea (for details, see “Materials and Methods”).
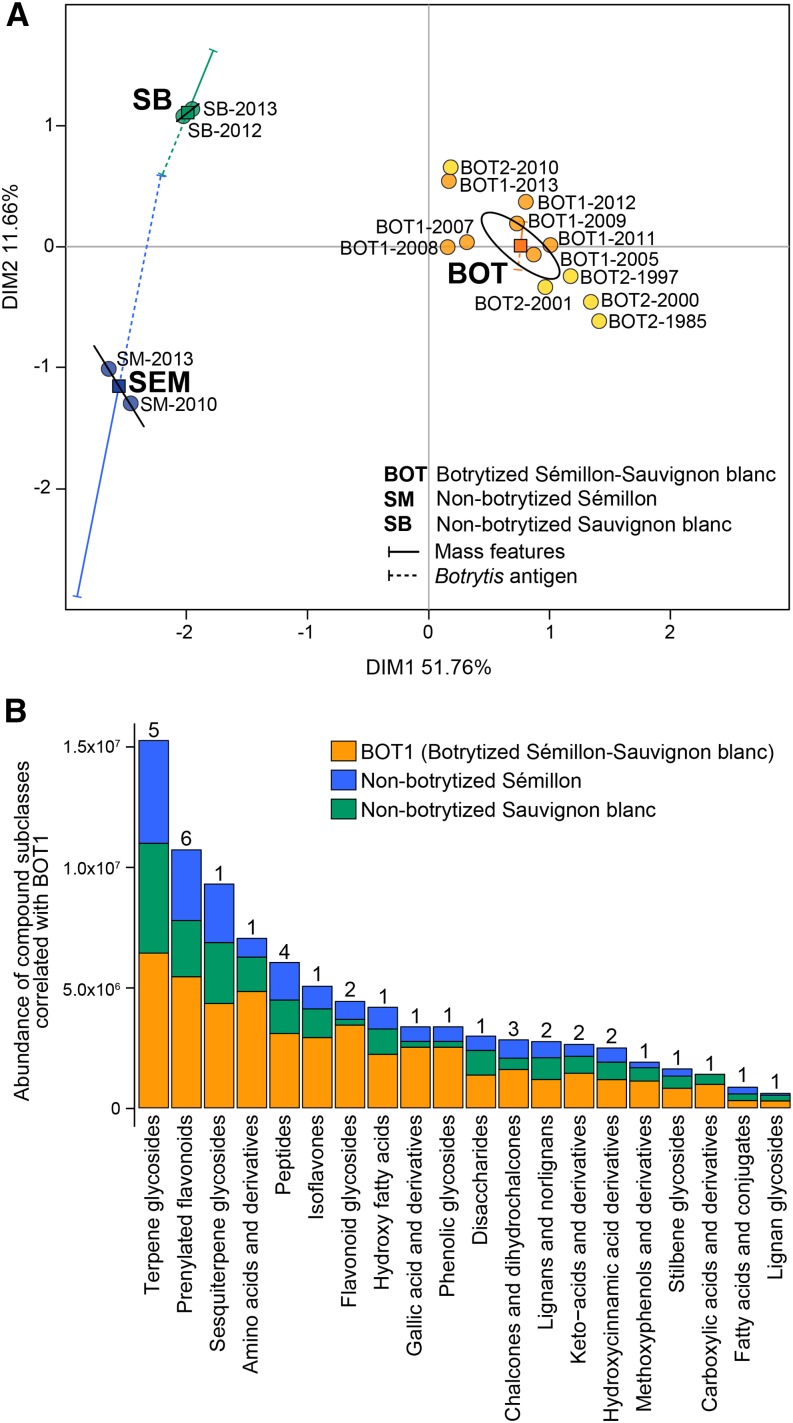
The ultrahigh pressure liquid chromatography quadrupole time-of-flight mass spectrometry approach detected a total of 299 mass features in the wines analyzed (Supplemental Table S14). The abundance of the detected metabolites and the measurements of the level of B. cinerea antigen were used to construct an MFA (Fig. 8). The first dimension (51.76%) of the MFA separated the botrytized wines from the non-botrytized wines, while the second dimension (11.66%) distinguished wine varietals (cv Sémillon and cv Sauvignon Blanc). BOT1 and BOT2 likely landed in the middle of DIM2, since they were a blend of both varietals. Supplemental Table S15 provides the details for the other dimensions calculated.
Figure 8.
Metabolite profiles of botrytized wines. A, MFA of botrytized and control (i.e. non-botrytized) wines using two types of quantitative variables (represented by vectors): accumulation of 299 mass features and the measurements of the B. cinerea antigen. Each point represents a different wine sample. The ellipse defines the confidence area (95%). Squares represent the corresponding centers of gravity for each wine class. DIM, Dimension. BOT1 and BOT2 are botrytized wines. B, Abundance of metabolites that significantly correlated (r ≥ 0.70, P ≤ 0.01) with BOT1 wines when compared with non-botrytized wines (Supplemental Table S16). Metabolites with unknown identities were not included. The number of metabolites per compound class is given on top of each bar.
A correlation analysis was performed to detect metabolites that were significantly more abundant (r ≥ 0.70, P ≤ 0.01) in BOT1 wines compared with non-botrytized wines. In total, 46 metabolites classified in 20 compound subclasses accumulated preferentially in the wines produced from noble-rotted berries (Fig. 8; Supplemental Table S16). Terpene glycosides were the most abundant class in the botrytized wines and appear to be key in defining their aroma and flavor profiles (Genovese et al., 2007). Products of the phenylpropanoid metabolism were expected to be present in the botrytized wines, as they were associated with noble rot development in grape berries. Among these, six prenylated flavonoids, two flavonoid glycosides, three chalcone derivatives, two hydroxycinnamic acid derivatives, and one stilbene glycoside were putatively identified and had enhanced levels in the botrytized wines (Supplemental Table S16). Other aromatic compounds that seem to be relevant during berry responses to noble rot, such as gallic acid derivatives and phenolic glycosides, were also detected in the botrytized wines.
DISCUSSION
Developmental Plasticity of White-Skinned Grape Berries in Response to Noble Rot
In this study, we demonstrate that noble rot causes not only the activation of stress responses but also a reprogramming in the development of white-skinned berries, which results in the activation of secondary metabolic pathways that are normally associated with the ripening of red-skinned cultivars. During noble rot, B. cinerea induced the expression of transcriptional regulators that function in the healthy development (i.e. prior to or during véraison) of red-skinned berries, including five R2R3-MYBs that control the phenylpropanoid metabolism (Deluc et al., 2006, 2008; Bogs et al., 2007; Walker et al., 2007; Matus et al., 2008; Höll et al., 2013). Candidate master regulators of véraison were also up-regulated by noble rot, such as VvNAC33 (VIT_19s0027g00230), VvNAC60 (VIT_08s0007g07670), a zinc-finger transcription factor (VIT_08s0040g01950), and a MYB transcription factor (VIT_07s0005g02730; Palumbo et al., 2014). Noble rot triggered the expression of 55 AP2/ERFs, most of which have been associated with the ripening of healthy red-skinned berries from cv Corvina, cv Cabernet Sauvignon, and cv Trincadeira (Deluc et al., 2007; Licausi et al., 2010; Fortes et al., 2011; Cramer et al., 2014). The expression of other potential berry-ripening and plant senescence regulators, such as the putative ortholog of SlNAC-NONRIPENING (VIT_19s0014g03300) and all WRKY53 genes annotated in the grape genome (VIT_02s0025g01280, VIT_15s0046g01140, and VIT_16s0050g02510), was also induced in noble-rotted berries. Besides their roles in berry development, some of the transcription factors activated during noble rot (e.g. R2R3-MYBs and AP2-ERFs) could regulate responses to biotic and abiotic stresses (Chen et al., 2012; Mizoi et al., 2012; Nakashima et al., 2012).
B. cinerea was recently reported to induce in grape berries the expression of transcription factors associated with ripening and defense responses, mainly ERFs and WRKY33-related genes (Agudelo-Romero et al., 2015). To detect berry responses that could differentiate noble rot from bunch rot, we compared our transcriptomic data on noble rot of cv Sémillon berries with the microarray data from a field study on bunch rot of cv Trincadeira berries at two developmental stages (Agudelo-Romero et al., 2015). This comparative analysis provides a preliminary view of the transcriptional signatures of noble rot and bunch rot. Additional factors, such as discrepancies in the ripening stage of the berries and in the cultivars, may confound the different transcriptional profiles detected in the two studies. Only 11.9% of the 12,575 noble rot-responsive genes showed similar expression patterns as a result of bunch rot. Among the key noble rot-responsive transcription factors, differential expression of three of the R2R3-MYB activators (VvMYB5a, VvMYB5b, and VvMYBPA1) was not detected during bunch rot. Out of the 22 AP2/ERFs with increased gene expression during bunch rot, only 14 genes were commonly up-regulated as a result of noble rot and bunch rot. Notably, a Hypoxia Responsive ERF gene (VIT_07s0005g00820) and a Related to AP2 gene (VIT_05s0077g01860), homologs of the tomato (Solanum lycopersicum) ripening regulators SlERF2 and SlERF6, respectively, were induced by noble rot but not by bunch rot (Pirrello et al., 2006; Lee et al., 2012; Cramer et al., 2014). Although these observations suggest that distinct transcriptional regulators may coordinate part of the berry responses to each type of infection, a more specific experimental design (e.g. the same grape cultivar, berry developmental stage, and transcriptomics technology) is needed to unequivocally conduct these comparisons.
Metabolic Plasticity in White-Skinned Grape Berries Affected with Noble Rot Results in Distinctive Wine Grape Flavor and Aroma
Noble rot induced metabolic alterations favoring the synthesis of a variety of plant compounds (e.g. glycosylated and glutathionylated fatty acid derivatives) that contribute to wine flavor and aroma (Hjelmeland and Ebeler, 2014; Robinson et al., 2014). Grapes showing noble rot symptoms were enriched in prenol lipids (i.e. terpenes), which were carried over into the botrytized wines (Genovese et al., 2007). Besides their contribution to grape and wine aroma, terpenes are involved in plant development (e.g. GAs) and responses to environmental stimuli (Cheng et al., 2007). We determined that the expression of eight terpene biosynthetic genes increased during noble rot (Table I), which corresponded to the only annotated farnesyl-diphosphate synthase (VvFPPS) and distinct classes of terpene synthases (TPSs; Martin et al., 2010), including the functionally characterized β-caryophyllene synthase (VvTPS13), α-humulene synthase (VvTPS11), and β-ocimene synthase (VvTPS34). When mining the microarray data from bunch rot (Agudelo-Romero et al., 2015), we did not observe an increase of expression of VvFPPS or any of the 69 candidate grape TPSs (Martin et al., 2010). Notably, these genes were either not differentially expressed or down-regulated in the bunch-rotted berries (Agudelo-Romero et al., 2015). Moreover, the levels of grape terpenes have been reported to decrease during typical B. cinerea infections (Shimizu et al., 1982; Agudelo-Romero et al., 2015), as B. cinerea is able to degrade and transform plant-derived terpenes (Collado et al., 2007). These observations indicate that the accumulation of terpenes may be a specific feature of noble rot.
Table I. Key genes from secondary metabolic pathways that are induced by noble rot in ripe cv Sémillon berries.
Functional annotations and gene accessions are provided, in addition to their membership in expression clusters (Fig. 2; Supplemental Table S6).
| Functional Annotation | Gene Accession | Response to Noble Rot |
|---|---|---|
| Gallic acid biosynthesis | ||
| Shikimate dehydrogenase | VIT_14s0030g00650 | Cluster 01 |
| Central phenylpropanoid biosynthesis | ||
| Phe ammonia lyase (VvPAL1) | VIT_08s0040g01710 | Cluster 01 |
| Phe ammonia lyase (VvPAL2) | VIT_16s0039g01300 | Cluster 01 |
| trans-Cinnamate 4-monooxygenase | VIT_06s0004g08150 | Cluster 01 |
| trans-Cinnamate 4-monooxygenase | VIT_11s0065g00350 | Cluster 01 |
| trans-Cinnamate 4-monooxygenase | VIT_11s0078g00290 | Cluster 01 |
| 4-Coumarate-CoA ligase | VIT_02s0025g03660 | Cluster 01 |
| 4-Coumarate-CoA ligase | VIT_11s0052g01090 | Cluster 01 |
| 4-Coumarate-CoA ligase | VIT_16s0050g00390 | Cluster 16 |
| Flavonoid biosynthesis | ||
| Chalcone synthase (VvCHS1) | VIT_14s0068g00930 | Cluster 08 |
| Chalcone synthase (VvCHS3) | VIT_05s0136g00260 | Cluster 04 |
| Chalcone isomerase | VIT_13s0067g03820 | Cluster 03 |
| Flavonone 3-hydroxylase | VIT_02s0025g02960 | Cluster 01 |
| Flavonone 3-hydroxylase | VIT_02s0025g02970 | Cluster 16 |
| Flavonone 3-hydroxylase | VIT_04s0023g03370 | Cluster 01 |
| Flavonone 3-hydroxylase | VIT_15s0048g02430 | Cluster 16 |
| Flavonone 3-hydroxylase | VIT_16s0098g00860 | Cluster 04 |
| Dihydroflavonol 4-reductase | VIT_18s0001g12800 | Cluster 01 |
| Anthocyanin biosynthesis and transport | ||
| UDP-Glc:flavonoid 3-O-glucosyltransferase | VIT_03s0017g02000 | Cluster 01 |
| UDP-Glc:flavonoid 3-O-glucosyltransferase | VIT_03s0017g02110 | Cluster 01 |
| UDP-Glc:flavonoid 3-O-glucosyltransferase | VIT_03s0017g02120 | Cluster 01 |
| UDP-Glc:flavonoid 3-O-glucosyltransferase | VIT_12s0034g00030 | Cluster 04 |
| UDP-Glc:flavonoid 3-O-glucosyltransferase | VIT_12s0055g00290 | Cluster 04 |
| Glutathione S-transferase (VvGST1) | VIT_19s0093g00320 | Cluster 01 |
| Terpene biosynthesis | ||
| Farnesyl-diphosphate synthase (VvFPPS) | VIT_19s0015g01010 | Cluster 03 |
| Terpene synthase class A (VvTPS13) | VIT_18s0001g05230 | Cluster 01 |
| Terpene synthase class A (VvTPS04) | VIT_18s0001g04120 | Cluster 04 |
| Terpene synthase class A (VvTPS11) | VIT_18s0001g04870 | Cluster 04 |
| Terpene synthase class A (VvTPS26) | VIT_19s0014g04810 | Cluster 03 |
| Terpene synthase class B (VvTPS32) | VIT_12s0059g02710 | Cluster 01 |
| Terpene synthase class B (VvTPS34) | VIT_12s0134g00020 | Cluster 01 |
| Terpene synthase class C (VvTPS68) | VIT_07s0151g01070 | Cluster 04 |
| Terpene synthase class G (VvTPS51) | VIT_12s0134g00090 | Cluster 17 |
| Oxylipin biosynthesis | ||
| Lipoxygenase (VvLOXB) | VIT_14s0128g00790 | Cluster 01 |
| Lipoxygenase (VvLOXC) | VIT_14s0128g00780 | Cluster 01 |
| Lipoxygenase (VvLOXO) | VIT_09s0002g01080 | Cluster 01 |
| Lipoxygenase (VvLOXP) | VIT_01s0010g02750 | Cluster 01 |
| Lipoxygenase (VvLOXK) | VIT_13s0064g01490 | Cluster 16 |
| Volatile thiol biosynthesis | ||
| Glutathione S-transferase (VvGST3) | VIT_12s0028g00920 | Cluster 01 |
| Glutathione S-transferase (VvGST4) | VIT_04s0079g00690 | Cluster 04 |
| γ-Glutamyltranspeptidase (VvGGT) | VIT_11s0016g02830 | Cluster 01 |
| Carboxypeptidase | VIT_00s0285g00070 | Cluster 04 |
| Carboxypeptidase | VIT_08s0032g00670 | Cluster 03 |
A number of important wine flavor volatiles are produced from the lipoxygenase (LOX) pathway during berry development (Schwab et al., 2008). LOXs catalyze the dioxygenation of unsaturated fatty acids as one of the initial steps in the synthesis of a wide range of oxylipins (Liavonchanka and Feussner, 2006). As a consequence of noble rot, VvLOXB, VvLOXC, VvLOXO, VvLOXK, and VvLOXP were up-regulated (Table I). Jasmonates are C12 oxylipins with important roles in hormone signaling and defense responses against fungal pathogens (Wasternack, 2007). Therefore, the induction of several LOXs and the activation of the jasmonate signaling network appear to be common outcomes of B. cinerea infections in grape berries and are not specific to noble rot or berry ripening (Podolyan et al., 2010; Agudelo-Romero et al., 2015).
C6 oxylipins (e.g. C6 aldehydes, alcohols, and esters) are important determinants of grape berry aroma (Baumes, 2009) and are considered to be the precursors of volatile thiols that define key aromas in several white wine varietals, Bordeaux reds, and botrytized wines (Bouchilloux et al., 1998; Sarrazin et al., 2007; Bailly et al., 2009; Lund et al., 2009). High contents of 3-sulfanylhexanol, 3-sulfanylpentan-1-ol, and 3-sulfanylheptan-1-ol provide most of the fruit and citrus aromas in botrytized wines (Sarrazin et al., 2007; Thibon et al., 2009). The proposed pathway for the generation of volatile thiols starts with the conjugation of a C6 oxylipin, such as (E)-2-hexenal, to glutathione by glutathione S-transferases (GSTs; Kobayashi et al., 2011). Glutathione conjugations are part of the detoxification pathways that occur during normal grape berry development and as a response to multiple stresses (Dixon et al., 2010; Zamboni et al., 2010). We detected the up-regulation of 50 GSTs during all the stages of noble rot, including two specific GSTs (VvGST3 and VvGST4) that have been demonstrated to be involved in the generation of volatile thiols (Table I; Kobayashi et al., 2011). Only seven GST-encoding genes were significantly up-regulated due to bunch rot (Agudelo-Romero et al., 2015). The biosynthesis of volatile thiols proceeds with the breakdown of the glutathione S-conjugates by γ-glutamyltranspeptidases and carboxypeptidases to generate Cys S-conjugates. We determined that a putative grape γ-glutamyltranspeptidase gene and two candidate carboxypeptidases were among the up-regulated noble rot-responsive genes, and none of these had differential expression during bunch rot (Table I; Agudelo-Romero et al., 2015). The Cys S-conjugates are then cleaved during alcoholic fermentation to produce a wide variety of volatile thiols (Thibon et al., 2010). A 3-sulfanylpentan-1-ol Cys S-conjugate drastically accumulated in berries at stages 2 and 3 of noble rot while being absent in healthy berries, which provides further evidence that the biosynthesis of volatile thiol precursors is highly induced during noble rot of cv Sémillon berries.
Noble Rot Induction of Phenylpropanoid Metabolism: Intersection between Plant Stress Responses and Ripening-Associated Events
Noble rot stimulated the production of phenolic compounds that derive from the shikimate and phenylpropanoid metabolism. This could explain the higher levels of polyphenols that are present in white botrytized wines when compared with non-botrytized white wines (Nikfardjam et al., 2006). Phenolic compounds play important roles in the flavor and aroma, color, physical mouth feel, and health-promoting properties of wine (Gawel, 1998; Boulton, 2001; Waterhouse, 2002). In the context of plant-pathogen interactions, they have antimicrobial functions and protect the berry tissues from oxidation (Cheynier et al., 2013; Teixeira et al., 2013). We confirmed the induction of the shikimate metabolism by the enhanced expression of enzymes involved in the flux toward gallic acid biosynthesis (e.g. a shikimate dehydrogenase; Table I) and by the accumulation of gallic acid and derivatives in both noble-rotted berries and botrytized wines.
A strong induction of the initial steps in the phenylpropanoid metabolism was found in noble-rotted berries. PALs catalyze the nonoxidative deamination of Phe to trans-cinnamic acid. PAL gene expression and activity have been reported to be absent or reduced in white-skinned berries when compared with red-skinned berries (Boss et al., 1996). During noble rot, we observed the up-regulation of 15 PAL-encoding genes, most of which are also induced during véraison and bunch rot of red-skinned berries (Palumbo et al., 2014; Agudelo-Romero et al., 2015). The expression of two of these genes, VvPAL1 and VvPAL2, has been reported to increase during ripening of the white-skinned cv Chardonnay berries (Table I; Guillaumie et al., 2011); however, their up-regulation is not as pronounced as in noble-rotted grape berries: 1-fold change (log2) in cv Chardonnay berries compared with more than 3-fold change (log2) in B. cinerea-infected cv Sémillon berries. The C4Hs and 4CLs are indispensable for all subsequent steps of the central and peripheral phenylpropanoid pathways (Weisshaar and Jenkins, 1998; Winkel-Shirley, 1999). The up-regulation of three C4H-encoding genes and eight 4CL-encoding genes was validated by their enhanced enzymatic activities in noble-rotted berries collected during the 2012, 2013, and 2014 harvests (Table I). The expression of these biosynthetic genes also increased during bunch rot and véraison of red-skinned cultivars (Palumbo et al., 2014; Agudelo-Romero et al., 2015). Activation of the phenylpropanoid metabolism is known to occur in response to oxidative stress in plant tissues (Grace and Logan, 2000; Vranová et al., 2002; Cheynier et al., 2013). Therefore, the accumulation of reactive oxygen species at véraison or as a consequence of B. cinerea infections may explain the common induction of the central phenylpropanoid pathway (Torres et al., 2006; Pilati et al., 2014).
Our data showed that the activation of the pathways leading to lignin and stilbene biosynthesis is a common response of grape berries to noble rot and bunch rot (Jeandet et al., 1995; Agudelo-Romero et al., 2015). Several genes encoding putative lignin biosynthetic enzymes were up-regulated in noble-rotted berries, including five caffeic acid 3-O-methyltransferases, seven cinnamyl alcohol dehydrogenases, six cinnamoyl-CoA reductases, and 23 laccases. The up-regulation of 40 genes coding for stilbene synthases together with the accumulation of stilbene glycosides in infected berries and botrytized wine verified the induction of stilbene biosynthesis during noble rot. Some stilbene dimers, in particular ε-viniferin glycosides, have been reported to be more effective phytoalexins and to have stronger health-promoting effects than the monomeric stilbenoids (Schouten et al., 2002; Zghonda et al., 2012). The concomitant expression of the stilbene synthases directly correlated with the highly significant up-regulation of the VvMYB14 and VvMYB15 transcription factors, which are the master regulators of the stilbene pathway (Höll et al., 2013). Although stilbenes are generally induced in berries under biotic and abiotic stress, they also accumulate after véraison in healthy red-skinned and white-skinned berries (Gatto et al., 2008; Höll et al., 2013).
Key steps in flavonoid biosynthesis were triggered by noble rot. Consistently over the 3 years of sampling, a CHI, five F3Hs, and a DFR were induced at the transcriptional level and displayed higher enzymatic activities in noble-rotted berries (Table I). During bunch rot, CHI-encoding genes did not show differential expression, whereas DFR-encoding genes were either not differentially regulated or down-regulated (Agudelo-Romero et al., 2015). The induction of the main flavonoid biosynthetic genes in noble-rotted berries can be explained by the significant up-regulation of the transcriptional activators VvMYB5a and VvMYB5b and the down-regulation of the transcriptional inhibitor VvMYB2-L1, all of which have been shown to control this pathway (Deluc et al., 2006, 2008; Cavallini et al., 2015). On the other hand, the production of flavones and flavonols was suppressed during noble rot. The reduced levels of these flavonoids in noble-rotted berries were congruent with the significant down-regulation of the transcriptional regulator VvMYBF1 (Fig. 6; Czemmel et al., 2009) and the FLS gene expression and activity.
Interestingly, noble rot induced the synthesis and accumulation of anthocyanins in cv Sémillon berries (Figs. 5 and 7). To our knowledge, this is the first report of anthocyanin accumulation in berries from a white-skinned grape cultivar as a result of pathogen infection. White-skinned berries arose from multiallelic mutations in the VvMYBA1 and VvMYBA2 genes, which regulate the gene expression of a UDP-Glc:flavonoid 3-O-glucosyltransferase (VvUF3GT; Kobayashi et al., 2004; Lijavetzky et al., 2006; This et al., 2007; Walker et al., 2007; Shimazaki et al., 2011). UF3GTs catalyze the 3-O-glycosylation of anthocyanidins (Kobayashi et al., 2004). The expression of the VvUF3GT gene and the resultant enzymatic activity are absent in white-skinned berries during normal development (Boss et al., 1996; Kobayashi et al., 2001). Although the grapevine genome contains at least 22 putative UF3GTs (Grimplet et al., 2009), only VvUF3GT (VIT_16s0039g02230) has been widely studied. Even though this UF3GT was not significantly up-regulated, the expression of six putative UF3GTs increased in response to noble rot (Table I) and during ripening in red-skinned berries (Ali et al., 2011; Palumbo et al., 2014). The up-regulation of these genes resulted in enhanced UF3GT activity in the noble-rotted berries across all three growing seasons. We did not detect the expression of VvMYBA1 in the noble-rotted berries and only found a significant down-regulation of VvMYBA2 during late stages of noble rot. The expression of VvMYBA2 is not expected to influence anthocyanin biosynthesis in cv Sémillon berries, because this cultivar carries a mutated allele of this gene, which encodes a nonfunctional protein (Walker et al., 2007). Therefore, this information suggests that transcriptional regulators independent or downstream of VvMYBA1/2 control the expression of the additional UF3GT genes and that a signaling molecule derived from the B. cinerea-grape berry interaction may activate this pathway.
The up-regulation of 35 genes with potential functions in the regulation of anthocyanin content and composition provided further evidence of the activation of anthocyanin metabolism in the noble-rotted berries (Costantini et al., 2015). These included genes encoding several transcription regulators (e.g. VIT_02s0033g00450, VIT_09s0018g00240, VIT_10s0116g01200, and VIT_12s0028g03270), hormone-responsive proteins (e.g. VIT_03s0180g00320, VIT_04s0023g03600, VIT_12s0057g00420, and VIT_17s0000g07560), and enzymes involved in anthocyanin modifications (e.g. VIT_18s0041g00710, VIT_18s0041g00740, and VIT_18s0041g00840). Two GSTs (VvGST1 and VvGST4), shown to transport anthocyanins from the cytosol to the vacuole (Conn et al., 2008), also had higher expression in the noble-rotted berries (Table I). These observations confirmed that the induction of anthocyanin metabolic pathways is a hallmark of noble rot in cv Sémillon berries.
B. cinerea Infection Strategies during Noble Rot
Future research focused on the understanding of the virulence factors and the signaling molecules produced by B. cinerea to promote susceptibility in grape berries still needs to be pursued. These signals may include plant hormones, small RNAs, reactive oxygen species, and/or pathogen- and damage-associated molecular patterns generated during B. cinerea infection (Melotto et al., 1994; Chagué et al., 2002; Siewers et al., 2004; Sharon et al., 2007; Segmüller et al., 2008; Weiberg et al., 2013).
We report here that several plant hormone networks were altered during noble rot of white-skinned berries, including ethylene, auxin, JA, GA, SA, and ABA pathways. ABA and ethylene are known modulators of both berry ripening and plant responses to pathogen attack (Chervin et al., 2004; van Loon et al., 2006; Ton et al., 2009; Wheeler et al., 2009; Böttcher et al., 2013; Nicolas et al., 2014). B. cinerea is able to produce ethylene via the keto-γ-methylthiobutyric acid pathway (Chagué et al., 2002) and ABA from farnesyl diphosphate (Siewers et al., 2006). However, we did not find strong transcriptional evidence that the pathogen was producing these two hormones (i.e. no significant up-regulation of biosynthetic genes) during noble rot of cv Sémillon berries. Instead, the data suggested that B. cinerea infection caused an imbalance in the berry synthesis and perception of hormones, which in turn activated several ripening-associated pathways and facilitated the colonization of the host tissues. The defense mechanisms established by the plant, which could have slowed down the infection, were inadequate to contain the spread of B. cinerea mycelia in the berry. B. cinerea may also rely on virulence factors, such as fungal protein phosphatases (Amselem et al., 2011; Giesbert et al., 2012; Yang et al., 2013), to alter plant signaling cascades (i.e. mitogen-activated protein kinase cascades) that are crucial for berry development and responses to environmental cues. An experimental setup that allows a direct comparison between noble rot and bunch rot will be necessary to define and further characterize the specific molecular mechanisms employed by B. cinerea in each type of infection.
Overall, our study shows that the developmental and metabolic plasticity displayed by white-skinned grape berries during noble rot result from two opposing effects: (1) the induction of ripening-associated pathways by B. cinerea to promote susceptibility; and (2) the activation of plant responses to cope against multiple stresses that result from interaction with the pathogen.
MATERIALS AND METHODS
Biological Material
Ripe grape (Vitis vinifera ‘Sémillon’) berries with symptoms of noble rot, caused by Botrytis cinerea strain BcDW1 (Blanco-Ulate et al., 2013a) and other B. cinerea strains present in the field, were harvested from the Dolce Winery vineyard in Oakville, California, on November 6, 2012, October 13, 2013, and November 6, 2014. These dates were chosen to coincide with the days of harvest selected by the Dolce winemaker for the production of botrytized wines. All sample collections were performed between 8 and 10 am. The weather conditions at the time of collection were as follows: 17.2°C ± 3.4°C with a relative humidity of 68.3% ± 12.1% for the harvest of 2012, 10.6°C ± 5.1°C with a relative humidity of 83% ± 14.7% for the harvest of 2013, and 14.4°C ± 3.1°C with a relative humidity of 78% ± 15% for the harvest of 2014.
Three distinct stages of noble rot (stages 1–3) were determined based on visual assessment of the severity of the symptoms and confirmed by measurements of fungal biomass (described below). At the same time, ripe berries without visible B. cinerea infection were harvested as asymptomatic controls. During the three independent harvests, samples were randomly collected from vines in the same rows of the vineyard (Supplemental Fig. S2). Individual berries were collected from clusters at different positions in the vine to avoid confounding effects, such as differences in cluster sun exposure. Five biological replicates consisting of independent pools of 20 berries from at least five different vines were obtained for each stage of infection and asymptomatic control. Berries of each biological replicate were deseeded, frozen, and ground (skin and pulp) to powder in liquid nitrogen. Soluble solid measurements on the homogenized biological replicates were performed.
Commercial control and botrytized wines were used for metabolomic analyses. These correspond to seven vintages (2005, 2007, 2008, 2009, 2011, 2012, and 2013) of botrytized wines produced with berries from the Dolce Winery vineyard, five vintages (1985, 1997, 2000, 2001, and 2010) of botrytized wines produced with cv Sémillon berries by another winery in Napa Valley, California, two vintages (2010 and 2013 produced in Australia) of non-botrytized cv Sémillon wine, and two vintages (2012 produced in California and 2013 produced in New Zealand) of non-botrytized cv Sauvignon Blanc wine.
B. cinerea Biomass Measurements
The amount of fungal biomass present in the noble-rotted grape berries and botrytized wines was measured with the QuickStix Kit for B. cinerea (EnviroLogix), which utilizes the monoclonal antibody BC12.CA4 (Meyer and Dewey, 2000). Fungal biomass quantification in grape berries was performed with 1 g of ground tissue from each biological replicate suspended in the kit buffer, 1:5 (w/v) for the control samples, 1:20 (w/v) for the samples at stage 1, 1:250 (w/v) for the samples at stage 2, and 1:500 (w/v) for the samples at stage 3. The presence of the BC12.CA4 antigen in the wines was measured using dilutions of 1:10 (v/v) for the control wines and 1:800 (v/v) for the botrytized wines. The antibody cross-reactive material was quantified in 500 μL of the tissue suspension or wine dilution, and the intensity of the test line was determined using the QuickStix reader (EnviroLogix). The material that cross-reacted with the antibody was measured in 500 μL of the tissue suspension or wine dilution. Two standard curves were obtained using known amounts of dry B. cinerea mycelia spiked into tissue extracts from control berries (y = 2.36x – 7.10; r2 = 0.96) and into control wines (y = 2.09x − 28.39; r2 = 0.98). The signal intensity measured by the QuickStix reader was then converted into fungal biomass (μg g−1 fresh weight of berry or μg mL−1 wine) based on the standard curves.
Analyses of Sugars and Sugar Alcohols
Samples were prepared by diluting 1 g of the ground tissue per biological replicate into 2 mL of deionized water (1:2, w/v). Sugars and sugar alcohols were analyzed in an Agilent Technologies 1100 HPLC System coupled to a Hewlett-Packard 1047A Refractive Index Detector. Three technical replications of each sample were loaded into an Aminex HPX-87C calcium form carbohydrate analysis column (300 mm × 7.8 mm; Bio-Rad Laboratories) and run using the following method: water as mobile phase, 0.6 mL min−1 (isocratic) flow rate, 80°C column temperature, 20-µL sample injection volume, and 25-min run time. Standards for Glc, Fru, glycerol, mannitol, and sorbitol (Sigma-Aldrich) were run in parallel with the samples and used to obtain standard curves. Data acquisition and analyses were performed with the ChemStation for LC 3D, Rev. A.10.02 software (Agilent Technologies).
RNA Extraction
Five biological replicates representing the three stages of noble rot and the control from the three harvesting seasons (2012, 2013, and 2014) were used for RNA extraction. Total RNA was obtained from 2 g of ground tissue (skin and pulp) using the protocol described by Blanco-Ulate et al. (2013b). The concentration and purity of the RNA were measured using the NanoDrop 2000c Spectrophotometer (Thermo Scientific). RNA integrity was confirmed by agarose gel electrophoresis.
mRNA Sequencing and Bioinformatics Pipeline
Sequencing of mRNA was performed with four biological replicates of each noble rot stage and control from the 2012 harvest. Complementary DNA (cDNA) libraries were prepared from 4 μg of total RNA using the Illumina TruSeq RNA Sample preparation kit version 2 with the low-throughput protocol (Illumina). Each cDNA library was barcoded individually and analyzed for quantity and quality with the High Sensitivity DNA Analysis Kit in the Agilent 2100 Bioanalyzer (Agilent Technologies). cDNA libraries were combined in equal amounts for sequencing (single end, 50 bp) at the Expression Analysis Core Facility (University of California, Davis) in an Illumina HiSeq2000 sequencer.
Quality trimming and filtering of the Illumina reads was done with Sickle version 1.21 (https://github.com/ucdavis-bioinformatics/sickle) with a threshold of 20 (quality score > 20) and Scythe version 0.991 (https://github.com/ucdavis-bioinformatics/scythe) with a prior of 0.4. A nonredundant B. cinerea mapping reference was obtained by combining B05.10 transcripts (http://www.broadinstitute.org/annotation/genome/botrytis_cinerea) with BcDW1 (http://www.ncbi.nlm.nih.gov/nuccore/AORW00000000.1) transcript using CD-HIT (version 4.5.4). B. cinerea transcripts were combined with the predicted transcriptome of cv PN40024 (http://www.genoscope.cns.fr/externe/Download/Projets/Projet_ML/data/12X/annotation/) and used as a reference for mapping the quality-trimmed reads with Bowtie2 version 2.1.0 (Langmead and Salzberg, 2012). The mapping parameters were as follow: -q–end-to-end–sensitive–no-unal-p20. The resultant Sequence Alignment/Map files were parsed to obtain read counts per grape gene with the python script sam2counts.py version 0.91 (https://github.com/vsbuffalo/sam2counts). The Bioconductor package DESeq2 (Love et al., 2014) was used to normalize the read counts of B. cinerea and grape genes across the biological replicates of B. cinerea-infected and control samples. Differential expression analyses of the grape genes across stages of noble rot were tested with DESeq2.
The DESeq2-normalized counts of B. cinerea and grape transcripts for each of the samples and their correspondent mass feature abundances (see below) and biomass measurements (see above) were used as quantitative variables for an MFA performed in R with the FactoMineR package (http://factominer.free.fr). Only quantitative variables that were different from zero in at least one of the samples were used for the MFA, which included (1) 24,373 grape genes, (2) 15,550 B. cinerea genes, and (3) 2,084 mass features.
The functional annotation of B. cinerea genes was performed using BLASTP (National Center for Biotechnology Information, nonredundant, Asomycota database; e-value < 10−3, -v 10, -b 10, -F F) followed by GO annotation using Blast2GO (Conesa et al., 2005). GO enrichments among the B. cinerea genes were performed using the Bioconductor package topGO (http://www.bioconductor.org/packages/release/bioc/html/topGO.html), and enriched GO terms (P ≤ 0.05) were visualized using the Reduce + Visualize Gene Ontology Web server (http://revigo.irb.hr; Supek et al., 2011). Grape genes were classified into functional categories and subcategories using the annotations provided in VitisNet (https://www.sdstate.edu/ps/research/vitis/pathways.cfm; Grimplet et al., 2009). Enrichment analyses of grape biological functions were computed in R using a hypergeometric test, and a cutoff of P ≤ 0.01 was set to determine statistical significance.
For the comparative analysis between the responses of cv Sémillon berries to noble rot and the processes that occur during ripening of red-skinned berries, raw Illumina reads from red-skinned grape berries (cv Sangiovese, cv Barbera, cv Negroamaro, cv Refosco, and cv Primitivo) at three developmental stages were downloaded from the Gene Expression Omnibus (accession no. GSE62744). Data were processed, mapped, normalized, and analyzed for differential expression following the same methods described above.
UltraHigh Pressure Liquid Chromatography-MS/MS and Classification of Metabolites into Compound Classes
Five biological replicates of noble-rotted and control samples from the harvest of 2012 were used for metabolite profiling. The samples were prepared by diluting 1 g of the ground tissue per biological replicate into 2 mL of deionized water (1:2, w/v). Two independent dilutions were performed for each biological replication. Three technical replications (i.e. machine replications) of each dilution were analyzed using the Agilent 1290 ultra-high-pressure liquid chromatograph coupled to an Agilent 6530 quadrupole time-of-flight mass spectrometer as described by Collins et al. (2014).
Initial processing of the metabolomics data was performed with the Agilent MassHunter Qualitative Analysis software version 6.00. The Molecular Feature Extractor (MFE) tool was used to determine the presence of nonredundant mass features by screening potential peaks for the presence of a C13 isotope and to detect any adduct ions that might be present. MFE was run using default settings with the peak filter set to include only peaks larger than 1,000 counts or approximately 3 times the electronic noise. Mass features found using MFE were further processed with the Agilent Mass Profiler Professional software version 12.1. The default retention time and mass windows settings of 0.1 min and 5 ppm, respectively, were used to align the mass features across the data set with a minimum absolute abundance of 5,000 counts. Mean retention times and mass sizes of each feature are reported in Supplemental Tables S3 and S14. Feature signal intensities were scaled using Z-transformation as implemented in the Mass Profiler Professional software.
After data processing, the resulting abundance of each mass feature was averaged to obtain a value for each of the two independent dilutions performed per sample. If a mass feature was detected in only one out of the three machine replications, the abundance was set to zero, as it is likely to represent a software miscall or noise. The mass feature abundance of both independent dilutions was then averaged to obtain a value per biological replication. If the mass feature abundance in one of the dilutions was zero (i.e. no detection or detection in only one machine replication), we reported the abundance measured in the other dilution. Differential accumulation of metabolites at the noble rot stages S1 to S3 was determined by Student’s t test against the nonsymptomatic control samples. P values were corrected using the Benjamini-Hochberg method implemented in the R stats package. A cutoff of adjusted P ≤ 0.05 was applied.
Possible compound identities were obtained based on mass matches to the METLIN metabolite database (https://metlin.scripps.edu; Smith et al., 2005) using a 30-ppm mass window. As MFE provides the neutral mass, the database was only searched for the neutral charge. Based on statistical significance (i.e. in the MFA and the Student’s t test), a set of 688 metabolites (642 compounds from control and noble-rotted berries and 46 from wines) was manually curated by evaluating their retention time and MS/MS spectra. Polarity of the potential matches was compared with the polarity of authentic standards of a range of polarities whose retention times in this chromatographic system had been established previously. MS/MS fragmentation data were collected in a data-dependent automated mode after ultrahigh pressure liquid chromatography. The data obtained were qualitatively compared with available spectra in METLIN and the Tandem Mass Spectrum Database (http://www.tmsdatabase.org). The manually curated metabolites were then assigned to classes and subclasses based on the chemical taxonomy established in the Human Metabolome Database version 3.0 (http://www.hmdb.ca; Wishart et al., 2013).
NMR Analyses
Three biological replicates of noble-rotted and control samples from the harvests of 2012 to 2014 were used to confirm the identities of seven metabolites using an NMR approach (Supplemental Table S9). Metabolites were chosen for NMR analysis based on three criteria: (1) they had significant accumulation (adjusted P ≤ 0.05) in most of the noble rot stages; (2) they represented compounds with biological significance (e.g. products of the phenylpropanoid pathway and plant hormones); and (3) most presented an unambiguous compound identity based on mass, retention time, and MS/MS spectra. The chemical extraction was done in 50 mg of freeze-dried grape tissues using methanol:chloroform:water (2:2:1) following by sonication for 1 h. After centrifugation at 11,000 rpm for 20 min at 4°C, the upper hydroalcoholic phase was carefully separated and dried under a Speed-Vac for 24 h. To evaluate the reproducibility of sample preparation, three portions of one of the samples were used to perform three extractions. For the 1H-NMR quantification, the dry residue of the hydroalcoholic phase was dissolved in 0.75 mL of deuterium oxide phosphate buffer (150 mm, pH 7) containing 3 mm 4,4-trimethyl-4-silyl propanoic acid (TSP) as an internal standard.
All 1H-NMR spectra were recorded at 298 K on a Bruker AVIII HD 500 NMR spectrometer (500.13 MHz for 1H) equipped with a 5-mm BroadBand Observe cryogenic probe (Bruker Biospin). 1H spectra were referenced to TSP signal (δ = 0 ppm), whereas 13C spectra were referenced to CH-1 resonance of α-d-Glc (δ = 93.1 ppm). A standard one-dimensional pulse sequence noesypr1d (recycle delay-90°-t1-90°-tm-90°-acquisition) was used to obtain metabolic profiles of the grape berry extracts with the 90° pulse length of about 11 µs and t1 of 3 µs. Water suppression was achieved with a weak irradiation during the recycle delay (2 s) and mixing time (100 ms). Sixty-four transients were collected into 32,768 data points for each spectrum with a spectral width of 12 kHz. An exponential window function with line-broadening factor of 0.5 Hz was applied to free induction decays prior to Fourier transformation.
For resonance assignment purposes, 1H-1H-correlation spectroscopy (COSY; cosygpqfpr), 1H-1H-total correlation spectroscopy (TOCSY; mlevphpr), and 1H-13C-heteronuclear single quantum coherence (HSQC; hsqcetgp) two-dimensional NMR spectra were acquired from the Human Metabolome Database (Wishart et al., 2013) and from previous literature (Nakayama et al., 1999; Seeram et al., 2002; Du et al., 2008; Abedini et al., 2013), in which complex mixtures were used to specifically and selectively identify the chemical shifts. In COSY and TOCSY experiments, 48 transients were collected into 2,048 data points for each of 256 increments with the spectral width of 10 ppm for both dimensions. Phase-insensitive mode was used with gradient selection for the COSY experiments, whereas the well-known MLEV-17 was employed as the spin-lock scheme in the sensitive phase. The TOCSY experiment (time-proportional phase incrementation) had a mixing time of 100 ms. 1H-13C-HSQC NMR spectra were determined using the gradient-selected sequences with 200 transients and 2,048 data points for each of 128 increments. The spectral widths were 26,410 Hz for 1H and 26,410 Hz for 13C in HSQC experiments. The data were Fourier transformed into a 4 × 2k matrix with appropriate apodization functions. 1H-NMR spectra were manually corrected for phase and baseline distortions using TOPSPIN version 3.2 (Bruker Biospin), and the spectral region of δ = 0.5 to 9.5 was uniformly integrated into 3,166 buckets with width of 0.003 ppm (1.8 Hz) using the Chenomx software version 3.8.3 (Bruker Biospin). The region δ = 4.67 to 5.15 was discarded to eliminate the effects of imperfect water presaturation. The spectral areas of all buckets were normalized to the weight of extracts employed for measurements. The intensities of 30 selected 1H resonances due hydroalcoholic metabolites were measured with respect to the intensity of TSP signal used as an internal standard with a concentration of 3 mm.
qRT-PCR
cDNA was prepared from total RNA using Moloney murine leukemia virus reverse transcriptase (Promega). qRT-PCR was performed on a StepOnePlus PCR System using Fast SYBR Green Master Mix (Applied Biosystems). The qRT-PCR conditions used to amplify all genes were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 3 s and 60°C for 30 s. Based on their previously reported stability in grape berries (Licausi et al., 2010), VvACTIN (VIT_04s0044g00580) and VvUBIQUITIN (VIT_16s0098g01190) were initially chosen as reference genes to linearize the transcript levels for all genes of interest as described in Chen and Dubcovsky (2012). The linearized values correspond to the relative gene expression within a given sample and are comparable across genes. Three to five biological replicates of the B. cinerea-infected and control grape berries were used to obtain the relative gene expression data. Supplemental Table S12 provides the sequences of the primers designed for this study. The efficiency of the primer sets was calculated using 4-fold cDNA dilutions (1:1, 1:4, 1:16, 1:64, and 1:256) in duplicate, while their specificity was checked by analyzing the dissociation curves at temperatures ranging from 60°C to 95°C.
The relative expression of 18 genes in the samples of 2012 was used to validate the RNAseq data. The results linearized with VvACTIN provided slightly better correlation than those linearized with VvUBIQUITIN (Supplemental Fig. S3). In addition, VvACTIN had higher expression levels in the noble-rotted and control berries than VvUBIQUITIN, which makes it more suitable for qRT-PCR experiments. Therefore, VvACTIN was selected as the reference gene for the subsequent expression analyses using the samples of 2013 and 2014.
Enzymatic Assays
The extraction procedures for the enzymatic analyses were performed as described by Li et al. (2013) with some modifications. For PAL (EC 4.1.3.5) and CHI (EC 5.5.1.6), the extraction was done in 0.5 g of frozen tissue. Tissues were homogenized at 4°C in 1 mL of 100 mm Tris-HCl buffer (pH 8.8) containing 14 mm β-mercaptoethanol, 5 mm dithiothreitol (DTT), 1% (w/v) bovine serum albumin (BSA), and 5% (w/v) polyvinylpolypyrrolidone (PVPP). For C4H (EC 1.14.13.11) and DFR (EC 1.1.1.219), the same extraction buffer was used as for PAL but with pH 7.5. For CHS (EC 2.3.1.74) and F3H (EC 1.14.11.9), 0.5 g of frozen tissues was homogenized at 4°C with 5% PVPP in 1 mL of 100 mm sodium phosphate buffer (pH 6.8) containing 14 mm β-mercaptoethanol, 5 mm DTT, 40 mm sodium ascorbate, 3 mm EDTA, and 2% BSA at 0°C to 4°C. The homogenate was centrifuged at 12,000g for 20 min at 4°C. The supernatant was precipitated with ammonium sulfate (70% [w/v] saturation), kept on ice for 1 h, and centrifuged at 16,000g for 10 min at 4°C. The resulting precipitate was dissolved in 1 mL of 100 mm sodium phosphate buffer (pH 6.8) containing 8 mm DTT, 40 mm sodium ascorbate, and 1% (w/v) BSA. For FLS (EC 1.14.11.23) and ANS (EC 1.14.11.19), 0.5 g of frozen tissues was homogenized with 5% (w/v) PVPP and 1 mL of 100 mm Tris-HCl buffer (pH 8) containing 14 mm β-mercaptoethanol, 5 mm DTT, 5 mm EDTA, 15 mm MgCl2, and 2% (w/v) BSA. After centrifugation, the enzyme extract was purified with ammonium sulfate as described above. All the above enzyme extracts were desalted by passage through PD10 columns and then used for enzyme analysis immediately.
Enzyme quantifications were performed spectrophotometrically using a plate reader with temperature control (Biotek PowerWave HT; Biotek Instruments). PAL activity was assayed in a mixture (250 µL) containing 100 mm Tris-HCl buffer, pH 8, and enzyme extract. The reaction was initiated by the addition of 150 µL of 200 mg mL−1 l-Phe (final concentration, 6 mg mL−1), and the production of cinnamic acid was measured over 10 min with variation of the A290. C4H activity was assayed in a mixture (250 µL) containing 100 mm phosphate buffer (pH 7.5), 1 mm DTT, 1 mm NADPH, and 100 µL of enzyme extract. The reaction was initiated by the addition of 10 mm trans-cinnamic acid (final concentration, 1 mm), and the variation of the A290 was recorded during 10 min. F3H activity was assayed in two independent reactions following the conversion of naringenin to dehydrokaempferol and the conversion of eridictyol to dihydroquercetin. The reaction mixture (250 µL) contained 100 mm Tris-HCl buffer (pH 7.5), 0.5 mm DTT, 0.25 mm 2-oxoglutarate, 0.05 mm ferrous sulfate, 5 mm sodium ascorbate, and 0.5 mm of the substrate (naringenin or eridictyol). The addition of the substrate initiated the reaction, and the variation of the A280 was followed over 10 min. Results obtained from each substrate were combined and plotted as F3H activity. CHI activity was assayed as follows: 2′,4,4′,6′-tetrahydroxychalcone was synthesized from naringenin by treatment with 50% KOH followed by acidification and recrystallization from aqueous ethanol. Enzyme extract (100 µL) was added to 50 mm Tris-HC1, pH 7.4, containing BSA (final concentration, 7.5 mg mL−1) and 50 mm KCN to give a final volume of 250 µL mL−1. The reaction was initiated by adding 5 µL of 1 mg mL−1 tetrahydroxychalcone in 2-ethoxyethanol, and changes in A381 were recorded with the cell holder maintained at 30°C. To allow for spontaneous isomerization of the chalcone, the reference cell contained the assay mixture without enzyme. The rate of disappearance of the chalcone (change in A381), in the presence of enzyme, was used to estimate CHI activity. DFR activity was measured according to Xie et al. (2004) with some modifications. The reaction mixture contained 25 mm Tris-HCl (pH 7), 4 mm NADPH, 100 mm taxifolin (dihydroquercetin), and enzyme extract. The reaction was initiated by the addition of NADPH at 25°C, followed by measuring the rate of NADPH oxidation at 340 nm. The enzyme activity was calculated using the extinction coefficient of NADPH, 6.22 mm−1 cm−1.
CHS and UFGT activities were assayed as described by Ju et al. (1995) and Ghasemzadeh et al. (2012), respectively. Briefly, the reaction mixture for UFGT activity contained 100 µL of enzyme extract, 1 mm UDP-Glc dissolved in 20 µL of a 330 µm solution of quercetin in methanol, and additional extraction buffer to produce a 250-µL total volume. The reaction started by the addition of the quercetin solution and changes in A290 at 30°C were recorded during 10 min. Samples were incubated for 15 min at 30°C. The CHS assay was performed with 20 µL of enzyme extract mixed with 250 mL of 50 mm Tris-HCI buffer, pH 7.6, containing 10 mm KCN. The enzyme reaction was allowed to proceed for 1 min at 30°C after adding 10 mg of chalcone (prepared as described for CHI activity) to 10 µL of ethylene glycol monomethylether. The activity was determined by measuring the A370.
For FLS and ANS activities, the methods of Wellmann et al. (2002) and Pang et al. (2007), respectively, were used. The assay mixture for FSL activity (total volume, 300 µL) contained 100 µm dihydroquercetin as a substrate, 83 µm 2-oxoglutarate, 42 µm ammonium iron (II) sulfate, 2.5 mm sodium ascorbate, and 2 mg mL−1 bovine catalase. FLS activity was calculated by the disappearance of the dihydroquercentin at 330 nm during 10 min. ANS activity was determined for the production of cyanidin from leucocyanidin (previously synthetized from eridictyol and NADPH) in a final volume of 250 µL containing 20 mm potassium phosphate buffer (pH 7), 200 mm NaCl, 10 mm maltose, 5 mm DTT, 4 mm sodium ascorbate, 1 mm 2-oxo-Glu, 0.4 mm FeSO4, and enzyme extract. Changes in the A330 at 30°C were recorded during 10 min. All the enzymatic activities were expressed as a function of the protein concentration of the extracts, which were assayed and calculated following the method proposed by Bradford (1976).
Quantification of Total Anthocyanin
Samples for total anthocyanin measurements were prepared using 100 μg of ground tissue (skin and pulp) suspended in 350 µL of methanol 1% (v/v) HCl. The samples were homogenized using 3-mm stainless-steel beads in a TissueLyser II (Qiagen) for 30 s with a frequency of 30 s−1. The homogenized samples were centrifuged at 10,000 rpm for 10 min, and then the supernatant was recovered. The sample extracts were incubated at room temperature for 10 min and then measured spectrophotometrically at 520 nm using a plate reader (Biotek PowerWave HT; Biotek Instruments). To calculate the total anthocyanins (μg g−1 fresh weight of berry) present in the samples, a standard curve was prepared with known amounts of malvidin-3-O-glucoside chloride (Sigma-Aldrich) diluted in methanol 1% (v/v) HCl. Malvidin-3-O-glucoside is the major anthocyanin in grape berries; therefore, the results are expressed as malvidin equivalents for comparative purposes.
The Illumina reads were deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus under accession number GSE67932 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE67932).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Summary of quantitative variables from the multiple factor analysis from Figure 2.
Supplemental Figure S2. Map of the vineyard where the noble-rotted and control samples were collected over 3 years.
Supplemental Figure S3. Scatterplots showing the correlation between the fold changes in expression obtained by RNAseq data and the fold changes measured by qRT-PCR.
Supplemental Table S1. Sugar and sugar alcohol levels in B. cinerea-infected and control grape berries.
Supplemental Table S2. Summary of parsed and mapped mRNA reads from noble-rotted samples and controls.
Supplemental Table S3. Scaled peak areas of detected mass features in noble-rotted and control samples, per biological replicate and technical replication.
Supplemental Table S4. Eigenvalues and percentage of explained variance from the multiple factor analysis of samples at three stages of noble rot and controls.
Supplemental Table S5. Quantitative variables significantly correlated with the main dimensions of the multiple factor analysis of samples at distinct noble rot stages.
Supplemental Table S6. DESeq2 and cluster analyses of grape gene expression as noble rot progressed.
Supplemental Table S7. Cluster analyses based on comparisons of metabolite abundance between samples of each noble rot stage and the asymptomatic controls.
Supplemental Table S8. Enriched grape biological processes unique to gene clusters with enhanced differential expression due to B. cinerea infection.
Supplemental Table S9. 1H-NMR spectral data of key metabolites present in noble-rotted cv Sémillon berries.
Supplemental Table S10. Core ripening genes and their functional annotations based on VitisNet.
Supplemental Table S11. List of commonly up- and down-regulated DE grape genes between the three stages of noble rot in cv Sémillon berries and the normal ripening of red-skinned berries.
Supplemental Table S12. Enriched grape biological processes among the common and unique DE genes when comparing the three stages of noble rot in cv Sémillon berries and the normal ripening of red-skinned berries.
Supplemental Table S13. Relative expression of 18 genes involved in the central phenylpropanoid and flavonoid pathways in noble-rotted and control berries harvested during the years of 2012, 2013, and 2014.
Supplemental Table S14. Scaled peak areas of detected mass features in botrytized and control wines, per vintage and technical replication.
Supplemental Table S15. Eigenvalues and percentage of explained variance from the multiple factor analysis of botrytized (BOT1 and BOT2) and non-botrytized wines (cv Sémillon and cv Sauvignon Blanc).
Supplemental Table S16. Metabolites significantly correlated with the botrytized wine BOT1 when compared with two classes of non-botrytized wines.
Acknowledgments
We thank Agilent Technologies for its support of instrumentation in the Food Safety and Measurement Facility at the University of California, Davis, and Dr. Jose Enrique Yuste (Centro de Edafología y Biología Aplicada del Segura-Consejo Superior de Investigaciones Científicas, Metabolomics Service) for assistance in the NMR experiments.
Glossary
- RNAseq
RNA sequencing
- MS/MS
tandem mass spectrometry
- MFA
multiple factor analysis
- GO
Gene Ontology
- JA
jasmonic acid
- DE genes
genes with significant differential expression
- SA
salicylic acid
- ABA
abscisic acid
- qRT
quantitative reverse transcription
- cDNA
complementary DNA
- MFE
Molecular Feature Extractor
- TSP
4,4-trimethyl-4-silyl propanoic acid
- DTT
dithiothreitol
- BSA
bovine serum albumin
- PVPP
polyvinylpolypyrrolidone
Footnotes
This work was supported by the College of Agricultural and Environmental Sciences and the Department of Viticulture and Enology, University of California, Davis (startup funds to D.C.).
Articles can be viewed without a subscription.
References
- Abedini A, Roumy V, Mahieux S, Biabiany M, Standaert-Vitse A, Rivière C, Sahpaz S, Bailleul F, Neut C, Hennebelle T (2013) Rosmarinic acid and its methyl ester as antimicrobial components of the hydromethanolic extract of Hyptis atrorubens Poit. (Lamiaceae). Evid Based Complement Alternat Med 2013: 604536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agudelo-Romero P, Erban A, Rego C, Carbonell-Bejerano P, Nascimento T, Sousa L, Martínez-Zapater JM, Kopka J, Fortes AM (2015) Transcriptome and metabolome reprogramming in Vitis vinifera cv. Trincadeira berries upon infection with Botrytis cinerea. J Exp Bot 66: 1769–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali MB, Howard S, Chen S, Wang Y, Yu O, Kovacs LG, Qiu W (2011) Berry skin development in Norton grape: distinct patterns of transcriptional regulation and flavonoid biosynthesis. BMC Plant Biol 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose KV, Tian Z, Wang Y, Smith J, Zylstra G, Huang B, Belanger FC (2015) Functional characterization of salicylate hydroxylase from the fungal endophyte Epichloë festucae. Sci Rep 5: 10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amselem J, Cuomo CA, van Kan JA, Viaud M, Benito EP, Couloux A, Coutinho PM, de Vries RP, Dyer PS, Fillinger S, et al. (2011) Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet 7: e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly S, Jerkovic V, Meurée A, Timmermans A, Collin S (2009) Fate of key odorants in Sauternes wines through aging. J Agric Food Chem 57: 8557–8563 [DOI] [PubMed] [Google Scholar]
- Baumes R. (2009) Wine aroma precursors. In Wine Chemistry and Biochemistry. Springer, New York, pp 251–274 [Google Scholar]
- Blanco-Ulate B, Allen G, Powell AL, Cantu D (2013a) Draft genome sequence of Botrytis cinerea BcDW1, inoculum for noble rot of grape berries. Genome Announc 1: e00252-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Ulate B, Morales-Cruz A, Amrine KC, Labavitch JM, Powell AL, Cantu D (2014) Genome-wide transcriptional profiling of Botrytis cinerea genes targeting plant cell walls during infections of different hosts. Front Plant Sci 5: 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Ulate B, Vincenti E, Powell AL, Cantu D (2013b) Tomato transcriptome and mutant analyses suggest a role for plant stress hormones in the interaction between fruit and Botrytis cinerea. Front Plant Sci 4: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs J, Jaffé FW, Takos AM, Walker AR, Robinson SP (2007) The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol 143: 1347–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Davies C, Robinson SP (1996) Expression of anthocyanin biosynthesis pathway genes in red and white grapes. Plant Mol Biol 32: 565–569 [DOI] [PubMed] [Google Scholar]
- Böttcher C, Burbidge CA, Boss PK, Davies C (2013) Interactions between ethylene and auxin are crucial to the control of grape (Vitis vinifera L.) berry ripening. BMC Plant Biol 13: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchilloux P, Darriet P, Henry R, Lavigne-Cruège V, Dubourdieu D (1998) Identification of volatile and powerful odorous thiols in Bordeaux red wine varieties. J Agric Food Chem 46: 3095–3099 [Google Scholar]
- Boulton R. (2001) The copigmentation of anthocyanins and its role in the color of red wine: a critical review. Am J Enol Vitic 52: 67–87 [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cantu D, Blanco-Ulate B, Yang L, Labavitch JM, Bennett AB, Powell AL (2009) Ripening-regulated susceptibility of tomato fruit to Botrytis cinerea requires NOR but not RIN or ethylene. Plant Physiol 150: 1434–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu D, Vicente AR, Greve LC, Dewey FM, Bennett AB, Labavitch JM, Powell AL (2008) The intersection between cell wall disassembly, ripening, and fruit susceptibility to Botrytis cinerea. Proc Natl Acad Sci USA 105: 859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy A, Hanley B, Lamuela‐Raventos RM (2000) Isoflavones, lignans and stilbenes: origins, metabolism and potential importance to human health. J Sci Food Agric 80: 1044–1062 [Google Scholar]
- Cavallini E, Matus JT, Finezzo L, Zenoni S, Loyola R, Guzzo F, Schlechter R, Ageorges A, Arce-Johnson P, Tornielli GB (2015) The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol 167: 1448–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centeno DC, Osorio S, Nunes-Nesi A, Bertolo AL, Carneiro RT, Araújo WL, Steinhauser MC, Michalska J, Rohrmann J, Geigenberger P, et al. (2011) Malate plays a crucial role in starch metabolism, ripening, and soluble solid content of tomato fruit and affects postharvest softening. Plant Cell 23: 162–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagué V, Elad Y, Barakat R, Tudzynski P, Sharon A (2002) Ethylene biosynthesis in Botrytis cinerea. FEMS Microbiol Ecol 40: 143–149 [DOI] [PubMed] [Google Scholar]
- Chen A, Dubcovsky J (2012) Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genet 8: e1003134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Song Y, Li S, Zhang L, Zou C, Yu D (2012) The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta 1819: 120–128 [DOI] [PubMed] [Google Scholar]
- Cheng AX, Lou YG, Mao YB, Lu S, Wang LJ, Chen XY (2007) Plant terpenoids: biosynthesis and ecological functions. J Integr Plant Biol 49: 179–186 [Google Scholar]
- Chervin C, El-Kereamy A, Roustan JP, Latché A, Lamon J, Bouzayen M (2004) Ethylene seems required for the berry development and ripening in grape, a non-climacteric fruit. Plant Sci 167: 1301–1305 [Google Scholar]
- Cheynier V, Comte G, Davies KM, Lattanzio V, Martens S (2013) Plant phenolics: recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiol Biochem 72: 1–20 [DOI] [PubMed] [Google Scholar]
- Chong J, Poutaraud A, Hugueney P (2009) Metabolism and roles of stilbenes in plants. Plant Sci 177: 143–155 [Google Scholar]
- Choquer M, Fournier E, Kunz C, Levis C, Pradier JM, Simon A, Viaud M (2007) Botrytis cinerea virulence factors: new insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol Lett 277: 1–10 [DOI] [PubMed] [Google Scholar]
- Collado IG, Sánchez AJM, Hanson JR (2007) Fungal terpene metabolites: biosynthetic relationships and the control of the phytopathogenic fungus Botrytis cinerea. Nat Prod Rep 24: 674–686 [DOI] [PubMed] [Google Scholar]
- Collins TS, Zweigenbaum J, Ebeler SE (2014) Profiling of nonvolatiles in whiskeys using ultra high pressure liquid chromatography quadrupole time-of-flight mass spectrometry (UHPLC-QTOF MS). Food Chem 163: 186–196 [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676 [DOI] [PubMed] [Google Scholar]
- Conn S, Curtin C, Bézier A, Franco C, Zhang W (2008) Purification, molecular cloning, and characterization of glutathione S-transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. J Exp Bot 59: 3621–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini L, Malacarne G, Lorenzi S, Troggio M, Mattivi F, Moser C, Grando MS (2015) New candidate genes for the fine regulation of the colour of grapes. J Exp Bot 66: 4427–4440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer GR, Ghan R, Schlauch KA, Tillett RL, Heymann H, Ferrarini A, Delledonne M, Zenoni S, Fasoli M, Pezzotti M (2014) Transcriptomic analysis of the late stages of grapevine (Vitis vinifera cv. Cabernet Sauvignon) berry ripening reveals significant induction of ethylene signaling and flavor pathways in the skin. BMC Plant Biol 14: 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czemmel S, Stracke R, Weisshaar B, Cordon N, Harris NN, Walker AR, Robinson SP, Bogs J (2009) The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol 151: 1513–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L, Barrieu F, Marchive C, Lauvergeat V, Decendit A, Richard T, Carde JP, Mérillon JM, Hamdi S (2006) Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol 140: 499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L, Bogs J, Walker AR, Ferrier T, Decendit A, Merillon JM, Robinson SP, Barrieu F (2008) The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol 147: 2041–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc LG, Grimplet J, Wheatley MD, Tillett RL, Quilici DR, Osborne C, Schooley DA, Schlauch KA, Cushman JC, Cramer GR (2007) Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development. BMC Genomics 8: 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deytieux-Belleau C, Geny L, Roudet J, Mayet V, Donèche B, Fermaud M (2009) Grape berry skin features related to ontogenic resistance to Botrytis cinerea. Eur J Plant Pathol 125: 551–563 [Google Scholar]
- Dixon DP, Skipsey M, Edwards R (2010) Roles for glutathione transferases in plant secondary metabolism. Phytochemistry 71: 338–350 [DOI] [PubMed] [Google Scholar]
- Du Q, Zheng J, Xu Y (2008) Composition of anthocyanins in mulberry and their antioxidant activity. J Food Compos Anal 21: 390–395 [Google Scholar]
- Fortes AM, Agudelo-Romero P, Silva MS, Ali K, Sousa L, Maltese F, Choi YH, Grimplet J, Martinez-Zapater JM, Verpoorte R, et al. (2011) Transcript and metabolite analysis in Trincadeira cultivar reveals novel information regarding the dynamics of grape ripening. BMC Plant Biol 11: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto P, Vrhovsek U, Muth J, Segala C, Romualdi C, Fontana P, Pruefer D, Stefanini M, Moser C, Mattivi F, et al. (2008) Ripening and genotype control stilbene accumulation in healthy grapes. J Agric Food Chem 56: 11773–11785 [DOI] [PubMed] [Google Scholar]
- Gawel R. (1998) Red wine astringency: a review. Aust J Grape Wine Res 4: 74–95 [Google Scholar]
- Genovese A, Gambuti A, Piombino P, Moio L (2007) Sensory properties and aroma compounds of sweet Fiano wine. Food Chem 103: 1228–1236 [Google Scholar]
- Ghasemzadeh A, Jaafar HZ, Karimi E (2012) Involvement of salicylic acid on antioxidant and anticancer properties, anthocyanin production and chalcone synthase activity in ginger (Zingiber officinale Roscoe) varieties. Int J Mol Sci 13: 14828–14844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesbert S, Schumacher J, Kupas V, Espino J, Segmüller N, Haeuser-Hahn I, Schreier PH, Tudzynski P (2012) Identification of pathogenesis-associated genes by T-DNA-mediated insertional mutagenesis in Botrytis cinerea: a type 2A phosphoprotein phosphatase and an SPT3 transcription factor have significant impact on virulence. Mol Plant Microbe Interact 25: 481–495 [DOI] [PubMed] [Google Scholar]
- Grace SC, Logan BA (2000) Energy dissipation and radical scavenging by the plant phenylpropanoid pathway. Philos Trans R Soc Lond B Biol Sci 355: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimplet J, Cramer GR, Dickerson JA, Mathiason K, Van Hemert J, Fennell AY (2009) VitisNet: “omics” integration through grapevine molecular networks. PLoS One 4: e8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler WD, Hashim JM, Smilanick JL, Leavitt GM (2013) Gray mold (Botrytis cinerea). In LJ Bettiga, ed, Grape Pest Management, Ed 3 University of California, Agriculture and Natural Resources, Richmond, CA, p 133–135 [Google Scholar]
- Guillaumie S, Fouquet R, Kappel C, Camps C, Terrier N, Moncomble D, Dunlevy JD, Davies C, Boss PK, Delrot S (2011) Transcriptional analysis of late ripening stages of grapevine berry. BMC Plant Biol 11: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterson N, Reuber TL (2004) Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr Opin Plant Biol 7: 465–471 [DOI] [PubMed] [Google Scholar]
- Hjelmeland AK, Ebeler SE (2014) Glycosidically bound volatile aroma compounds in grapes and wine: a review. Am J Enol Vitic 66: 1–11 [Google Scholar]
- Höll J, Vannozzi A, Czemmel S, D’Onofrio C, Walker AR, Rausch T, Lucchin M, Boss PK, Dry IB, Bogs J (2013) The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell 25: 4135–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornsey IS. (2007) Botrytis cinerea In The Chemistry and Biology of Winemaking. Royal Society of Chemistry, Cambridge, UK, pp 367–382 [Google Scholar]
- Jeandet P, Bessis R, Sbaghi M, Meunier P (1995) Production of the phytoalexin resveratrol by grapes as a response to Botrytis attack under natural conditions. J Phytopathol 143: 135–139 [Google Scholar]
- Ju Z, Liu C, Yuan Y (1995) Activities of chalcone synthase and UDP-glucose:flavonol-3-O-glycosyltransferase in relation to anthocyanin synthesis in apple. Sci Hortic (Amsterdam) 63: 175–185 [Google Scholar]
- Kobayashi H, Takase H, Suzuki Y, Tanzawa F, Takata R, Fujita K, Kohno M, Mochizuki M, Suzuki S, Konno T (2011) Environmental stress enhances biosynthesis of flavor precursors, S-3-(hexan-1-ol)-glutathione and S-3-(hexan-1-ol)-L-cysteine, in grapevine through glutathione S-transferase activation. J Exp Bot 62: 1325–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Goto-Yamamoto N, Hirochika H (2004) Retrotransposon-induced mutations in grape skin color. Science 304: 982. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Ishimaru M, Ding CK, Yakushiji H, Goto N (2001) Comparison of UDP-glucose:flavonoid 3-O-glucosyltransferase (UFGT) gene sequences between white grapes (Vitis vinifera) and their sports with red skin. Plant Sci 160: 543–550 [DOI] [PubMed] [Google Scholar]
- Kou X, Wang S, Wu M, Guo R, Xue Z, Meng N, Tao X, Chen M, Zhang Y (2014) Molecular characterization and expression analysis of NAC family transcription factors in tomato. Plant Mol Biol Rep 32: 501–516 [Google Scholar]
- Kretschmer M, Kassemeyer HH, Hahn M (2007) Age‐dependent grey mould susceptibility and tissue‐specific defence gene activation of grapevine berry skins after infection by Botrytis cinerea. J Phytopathol 155: 258–263 [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Joung JG, McQuinn R, Chung MY, Fei Z, Tieman D, Klee H, Giovannoni J (2012) Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J 70: 191–204 [DOI] [PubMed] [Google Scholar]
- Li P, Ma F, Cheng L (2013) Primary and secondary metabolism in the sun-exposed peel and the shaded peel of apple fruit. Physiol Plant 148: 9–24 [DOI] [PubMed] [Google Scholar]
- Liavonchanka A, Feussner I (2006) Lipoxygenases: occurrence, functions and catalysis. J Plant Physiol 163: 348–357 [DOI] [PubMed] [Google Scholar]
- Licausi F, Giorgi FM, Zenoni S, Osti F, Pezzotti M, Perata P (2010) Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genomics 11: 719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P (2013) APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol 199: 639–649 [DOI] [PubMed] [Google Scholar]
- Lijavetzky D, Ruiz-García L, Cabezas JA, De Andrés MT, Bravo G, Ibáñez A, Carreño J, Cabello F, Ibáñez J, Martínez-Zapater JM (2006) Molecular genetics of berry colour variation in table grape. Mol Genet Genomics 276: 427–435 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund CM, Thompson MK, Benkwitz F, Wohler MW, Triggs CM, Gardner R, Heymann H, Nicolau L (2009) New Zealand Sauvignon Blanc distinct flavor characteristics: sensory, chemical, and consumer aspects. Am J Enol Vitic 60: 1–12 [Google Scholar]
- Magyar I. (2006) Microbiological Aspects of Winemaking. PhD thesis, Corvinus University of Budapest, Faculty of Food Science, Budapest [Google Scholar]
- Magyar I. (2011) Botrytized wines. Adv Food Nutr Res 63: 147–206 [DOI] [PubMed] [Google Scholar]
- Martin DM, Aubourg S, Schouwey MB, Daviet L, Schalk M, Toub O, Lund ST, Bohlmann J (2010) Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol 10: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus JT, Aquea F, Arce-Johnson P (2008) Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol 8: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto E, Greve LC, Labavitch JM (1994) Cell wall metabolism in ripening fruit: VII. Biologically active pectin oligomers in ripening tomato (Lycopersicon esculentum Mill.) fruits. Plant Physiol 106: 575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer UM, Dewey FM (2000) Efficacy of different immunogens for raising monoclonal antibodies to Botrytis cinerea. Mycol Res 104: 979–987 [Google Scholar]
- Miedes E, Lorences EP (2007) The implication of xyloglucan endotransglucosylase/hydrolase (XTHs) in tomato fruit infection by Penicillium expansum Link. A. J Agric Food Chem 55: 9021–9026 [DOI] [PubMed] [Google Scholar]
- Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819: 86–96 [DOI] [PubMed] [Google Scholar]
- Morales-Valle H, Silva LC, Paterson RR, Venâncio A, Lima N (2011) Effects of the origins of Botrytis cinerea on earthy aromas from grape broth media further inoculated with Penicillium expansum. Food Microbiol 28: 1048–1053 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819: 97–103 [DOI] [PubMed] [Google Scholar]
- Nakayama M, Yamaguchi M, Urashima O, Kan Y, Fukui Y, Yamaguchi Y, Koshioka M (1999) Anthocyanins in the dark purple anthers of Tulipa gesneriana: identification of two novel delphinidin 3-O-(6-O-(acetyl-α-rhamnopyranosyl)-β-glucopyranosides). Biosci Biotechnol Biochem 63: 1509–1511 [DOI] [PubMed] [Google Scholar]
- Nicolas P, Lecourieux D, Kappel C, Cluzet S, Cramer G, Delrot S, Lecourieux F (2014) The basic leucine zipper transcription factor ABSCISIC ACID RESPONSE ELEMENT-BINDING FACTOR2 is an important transcriptional regulator of abscisic acid-dependent grape berry ripening processes. Plant Physiol 164: 365–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikfardjam MP, László G, Dietrich H (2006) Resveratrol-derivatives and antioxidative capacity in wines made from botrytized grapes. Food Chem 96: 74–79 [Google Scholar]
- Ortega‐Regules A, Romero‐Cascales I, López‐Roca JM, Ros‐García JM, Gómez‐Plaza E (2006) Anthocyanin fingerprint of grapes: environmental and genetic variations. J Sci Food Agric 86: 1460–1467 [Google Scholar]
- Palumbo MC, Zenoni S, Fasoli M, Massonnet M, Farina L, Castiglione F, Pezzotti M, Paci P (2014) Integrated network analysis identifies fight-club nodes as a class of hubs encompassing key putative switch genes that induce major transcriptome reprogramming during grapevine development. Plant Cell 26: 4617–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y, Peel GJ, Wright E, Wang Z, Dixon RA (2007) Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula. Plant Physiol 145: 601–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M. (2013) Rosmarinic acid: new aspects. Phytochem Rev 12: 207–227 [Google Scholar]
- Pezet R, Viret O, Perret C, Tabacchi R (2003) Latency of Botrytis cinerea Pers.: Fr. and biochemical studies during growth and ripening of two grape berry cultivars, respectively susceptible and resistant to grey mould. J Phytopathol 151: 208–214 [Google Scholar]
- Pilati S, Brazzale D, Guella G, Milli A, Ruberti C, Biasioli F, Zottini M, Moser C (2014) The onset of grapevine berry ripening is characterized by ROS accumulation and lipoxygenase-mediated membrane peroxidation in the skin. BMC Plant Biol 14: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrello J, Jaimes-Miranda F, Sanchez-Ballesta MT, Tournier B, Khalil-Ahmad Q, Regad F, Latché A, Pech JC, Bouzayen M (2006) Sl-ERF2, a tomato ethylene response factor involved in ethylene response and seed germination. Plant Cell Physiol 47: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Podolyan A, White J, Jordan B, Winefield C (2010) Identification of the lipoxygenase gene family from Vitis vinifera and biochemical characterisation of two 13-lipoxygenases expressed in grape berries of Sauvignon Blanc. Funct Plant Biol 37: 767–784 [Google Scholar]
- Podzimska-Sroka D, O’Shea C, Gregersen PL, Skriver K (2015) NAC transcription factors in senescence: from molecular structure to function in crops. Plants 4: 412–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusky D, Alkan N, Mengiste T, Fluhr R (2013) Quiescent and necrotrophic lifestyle choice during postharvest disease development. Annu Rev Phytopathol 51: 155–176 [DOI] [PubMed] [Google Scholar]
- Ribéreau-Gayon J, Riberau-Gayon P, Seguin G (1980) Botrytis cinerea in enology. In JR Coley-Smith, K Verhoeff, WR Jarvis, eds, The Biology of Botrytis. Academic Press, London, pp 251–274 [Google Scholar]
- Ribéreau-Gayon P, Dubourdieu D, Donèche B, Lonvaud A (2006) Botrytized sweet wines (Sauternes and Tokay). In Pascal RG, ed, Handbook of Enology: The Microbiology of Wine and Vinifications, Ed 2, Vol 1 John Wiley & Sons, West Sussex, UK, pp 449–458 [Google Scholar]
- Robinson AL, Boss PK, Solomon PS, Trengove RD, Heymann H, Ebeler SE (2014) Origins of grape and wine aroma. Part 1. Chemical components and viticultural impacts. Am J Enol Vitic 65: 1–24 [Google Scholar]
- Sarrazin E, Dubourdieu D, Darriet P (2007) Characterization of key-aroma compounds of botrytized wines, influence of grape botrytization. Food Chem 103: 536–545 [Google Scholar]
- Scharf KD, Berberich T, Ebersberger I, Nover L (2012) The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim Biophys Acta 1819: 104–119 [DOI] [PubMed] [Google Scholar]
- Schouten A, Wagemakers L, Stefanato FL, van der Kaaij RM, van Kan JA (2002) Resveratrol acts as a natural profungicide and induces self-intoxication by a specific laccase. Mol Microbiol 43: 883–894 [DOI] [PubMed] [Google Scholar]
- Schwab W, Davidovich-Rikanati R, Lewinsohn E (2008) Biosynthesis of plant-derived flavor compounds. Plant J 54: 712–732 [DOI] [PubMed] [Google Scholar]
- Seeram NP, Schutzki R, Chandra A, Nair MG (2002) Characterization, quantification, and bioactivities of anthocyanins in Cornus species. J Agric Food Chem 50: 2519–2523 [DOI] [PubMed] [Google Scholar]
- Segmüller N, Kokkelink L, Giesbert S, Odinius D, van Kan J, Tudzynski P (2008) NADPH oxidases are involved in differentiation and pathogenicity in Botrytis cinerea. Mol Plant Microbe Interact 21: 808–819 [DOI] [PubMed] [Google Scholar]
- Sharon A, Elad Y, Barakat R, Tudzynski P (2007) Phytohormones in Botrytis-plant interactions. In Y Elad, B Williamson, P Tudzynski, N Delen, eds, Botrytis: Biology, Pathology and Control. Springer, Dordrecht, The Netherlands, pp 163–179 [Google Scholar]
- Shimazaki M, Fujita K, Kobayashi H, Suzuki S (2011) Pink-colored grape berry is the result of short insertion in intron of color regulatory gene. PLoS One 6: e21308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu J, Uehara M, Watanabe M (1982) Transformation of terpenoids in grape must by Botrytis cinerea. Agric Biol Chem 46: 1339–1344 [Google Scholar]
- Siewers V, Kokkelink L, Smedsgaard J, Tudzynski P (2006) Identification of an abscisic acid gene cluster in the grey mold Botrytis cinerea. Appl Environ Microbiol 72: 4619–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siewers V, Smedsgaard J, Tudzynski P (2004) The P450 monooxygenase BcABA1 is essential for abscisic acid biosynthesis in Botrytis cinerea. Appl Environ Microbiol 70: 3868–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M. (2006) Metschnikowia strains isolated from botrytized grapes antagonize fungal and bacterial growth by iron depletion. Appl Environ Microbiol 72: 6716–6724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G (2005) METLIN: a metabolite mass spectral database. Ther Drug Monit 27: 747–751 [DOI] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T (2011) REVIGO summarizes and visualizes long lists of Gene Ontology terms. PLoS One 6: e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarara JM, Lee J, Spayd SE, Scagel CF (2008) Berry temperature and solar radiation alter acylation, proportion, and concentration of anthocyanin in Merlot grapes. Am J Enol Vitic 59: 235–247 [Google Scholar]
- Teissedre PL, Donèche B (2013) Botrytized wines: Sauternes, German wines. In F Mencarelli, P Tonutti, eds, Sweet, Reinforced and Fortified Wines: Grape Biochemistry, Technology and Vinification. Wiley-Blackwell, Chichester, UK, pp 285–299
- Teixeira A, Eiras-Dias J, Castellarin SD, Gerós H (2013) Berry phenolics of grapevine under challenging environments. Int J Mol Sci 14: 18711–18739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibon C, Dubourdieu D, Darriet P, Tominaga T (2009) Impact of noble rot on the aroma precursor of 3-sulfanylhexanol content in Vitis vinifera L. cv Sauvignon Blanc and Sémillon grape juice. Food Chem 114: 1359–1364 [Google Scholar]
- Thibon C, Shinkaruk S, Jourdes M, Bennetau B, Dubourdieu D, Tominaga T (2010) Aromatic potential of botrytized white wine grapes: identification and quantification of new cysteine-S-conjugate flavor precursors. Anal Chim Acta 660: 190–196 [DOI] [PubMed] [Google Scholar]
- This P, Lacombe T, Cadle-Davidson M, Owens CL (2007) Wine grape (Vitis vinifera L.) color associates with allelic variation in the domestication gene VvmybA1. Theor Appl Genet 114: 723–730 [DOI] [PubMed] [Google Scholar]
- Ton J, Flors V, Mauch-Mani B (2009) The multifaceted role of ABA in disease resistance. Trends Plant Sci 14: 310–317 [DOI] [PubMed] [Google Scholar]
- Torres MA, Jones JD, Dangl JL (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol 141: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vail M, Marois J (1991) Grape cluster architecture and the susceptibility of berries to Botrytis cinerea. Phytopathology 81: 188–191 [Google Scholar]
- van Loon LC, Geraats BP, Linthorst HJ (2006) Ethylene as a modulator of disease resistance in plants. Trends Plant Sci 11: 184–191 [DOI] [PubMed] [Google Scholar]
- Vannini A, Chilosi G (2013) Botrytis infection: grey mould and noble rot. In F Mencarelli, P Tonutti, eds, Sweet, Reinforced and Fortified Wines: Grape Biochemistry, Technology and Vinification. Wiley-Blackwell, Chichester, UK, pp 159–169 [Google Scholar]
- Vranová E, Inzé D, Van Breusegem F (2002) Signal transduction during oxidative stress. J Exp Bot 53: 1227–1236 [PubMed] [Google Scholar]
- Walker AR, Lee E, Bogs J, McDavid DA, Thomas MR, Robinson SP (2007) White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J 49: 772–785 [DOI] [PubMed] [Google Scholar]
- Wasternack C. (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot (Lond) 100: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AL. (2002) Wine phenolics. Ann N Y Acad Sci 957: 21–36 [DOI] [PubMed] [Google Scholar]
- Weiberg A, Wang M, Lin FM, Zhao H, Zhang Z, Kaloshian I, Huang HD, Jin H (2013) Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342: 118–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisshaar B, Jenkins GI (1998) Phenylpropanoid biosynthesis and its regulation. Curr Opin Plant Biol 1: 251–257 [DOI] [PubMed] [Google Scholar]
- Wellmann F, Lukačin R, Moriguchi T, Britsch L, Schiltz E, Matern U (2002) Functional expression and mutational analysis of flavonol synthase from Citrus unshiu. Eur J Biochem 269: 4134–4142 [DOI] [PubMed] [Google Scholar]
- Wheeler S, Loveys B, Ford C, Davies C (2009) The relationship between the expression of abscisic acid biosynthesis genes, accumulation of abscisic acid and the promotion of Vitis vinifera L. berry ripening by abscisic acid. Aust J Grape Wine Res 15: 195–204 [Google Scholar]
- Winkel-Shirley B. (1999) Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol Plant 107: 142–149 [Google Scholar]
- Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E (2013) HMDB 3.0: the human metabolome database in 2013. Nucleic Acids Res 41: D801–D807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DY, Jackson LA, Cooper JD, Ferreira D, Paiva NL (2004) Molecular and biochemical analysis of two cDNA clones encoding dihydroflavonol-4-reductase from Medicago truncatula. Plant Physiol 134: 979–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Yu F, Yin Y, Ma Z (2013) Involvement of protein tyrosine phosphatases BcPtpA and BcPtpB in regulation of vegetative development, virulence and multi-stress tolerance in Botrytis cinerea. PLoS One 8: e61307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz Y, Toledo RT (2004) Major flavonoids in grape seeds and skins: antioxidant capacity of catechin, epicatechin, and gallic acid. J Agric Food Chem 52: 255–260 [DOI] [PubMed] [Google Scholar]
- Zamboni A, Di Carli M, Guzzo F, Stocchero M, Zenoni S, Ferrarini A, Tononi P, Toffali K, Desiderio A, Lilley KS, et al. (2010) Identification of putative stage-specific grapevine berry biomarkers and omics data integration into networks. Plant Physiol 154: 1439–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zghonda N, Yoshida S, Ezaki S, Otake Y, Murakami C, Mliki A, Ghorbel A, Miyazaki H (2012) ε-Viniferin is more effective than its monomer resveratrol in improving the functions of vascular endothelial cells and the heart. Biosci Biotechnol Biochem 76: 954–960 [DOI] [PubMed] [Google Scholar]