A major QTL encodes a small protein with an Arabidopsis homolog and promotes the formation of functional ovules, thus yielding more seeds per silique in Brassica napus.
Abstract
Number of seeds per silique (NSS) is an important determinant of seed yield potential in Brassicaceae crops, and it is controlled by naturally occurring quantitative trait loci. We previously mapped a major quantitative trait locus, qSS.C9, on the C9 chromosome that controls NSS in Brassica napus. To gain a better understanding of how qSS.C9 controls NSS in B. napus, we isolated this locus through a map-based cloning strategy. qSS.C9 encodes a predicted small protein with 119 amino acids, designated as BnaC9.SMG7b, that shows homology with the Ever ShorterTelomere1 tertratricopeptide repeats and Ever Shorter Telomere central domains of Arabidopsis (Arabidopsis thaliana) SUPPRESSOR WITH MORPHOGENETIC EFFECTS ON GENITALIA7 (SMG7). BnaC9.SMG7b plays a role in regulating the formation of functional female gametophyte, thus determining the formation of functional megaspores and then mature ovules. Natural loss or artificial knockdown of BnaC9.SMG7b significantly reduces the number of functional ovules per silique and thus, results in decreased seed number, indicating that qSS.C9 is a positive regulator of NSS in B. napus. Sequence and function analyses show that BnaC9.SMG7b experiences a subfunctionalization process that causes loss of function in nonsense-mediated mRNA decay, such as in Arabidopsis SMG7. Haplotype analysis in 84 accessions showed that the favorable BnaC9.SMG7b alleles are prevalent in modern B. napus germplasms, suggesting that this locus has been a major selection target of B. napus improvement. Our results represent the first step toward unraveling the molecular mechanism that controls the natural variation of NSS in B. napus.
Elite Brassica spp. (majorly Brassica napus) with low erucic and glucosinolate content is the third leading source of both vegetable oil and oil extraction meal worldwide (Rondanini et al., 2012). Yield enhancement has been one of the most important goals of B. napus production and genetic improvement. As a complex quantitative trait, the seed yield of a B. napus plant is comprehensively determined by three components: number of seeds per silique (NSS), number of siliques per plant, and seed weight, all of which are typical quantitative traits (Clarke and Simpson, 1978). Although there is rich variation for each of the three components (Zhang et al., 2012a), it is critical to balance the three traits during the breeding program, because significant negative correlations among them have been extensively observed (Shi et al., 2009; Zhang et al., 2011). Thus, it is highly desirable to identify the genes responsible for each component and subsequently, understand the genetic mechanisms underlying the natural variations. Toward this goal, a large number of quantitative trait loci (QTLs) associated with these yield components in B. napus has been identified in different mapping populations (Quijada et al., 2006; Udall et al., 2006; Chen et al., 2007, 2011; Li et al., 2007; Radoev et al., 2008; Shi et al., 2009; Basunanda et al., 2010; Fan et al., 2010; Zhang et al., 2011, 2012b; Yang et al., 2012; Cai et al., 2014; Qi et al., 2014). Among these QTLs, 24 are reported to be associated with NSS, and they are distributed on the majority of B. napus chromosomes (A1, A2, A5, A7–A9, C1–C4, C6, C7, and C9), explaining phenotypic variances ranging from 0.78% to 57.77% of each mapping population (Radoev et al., 2008; Shi et al., 2009; Chen et al., 2011; Zhang et al., 2011, 2012b; Qi et al., 2014). However, no yield-related QTLs have been cloned thus far because of the complexity of the genome of allotetraploid B. napus and the relatively limited genomic sequence information before the release of B. napus.
In spermatophytes, ovules deriving within the carpel’s ovary provide the structural support or enclosure for female gametophytes (FGs) and develop into seeds after fertilization (Shi and Yang, 2011). Thus, to some degree, ovule numbers determine NSS and thereby, contribute to the final seed yield in crop plants (Galbiati et al., 2013). From an agronomic perspective, the number of ovules can be attributed to two components: the number of ovule primordia and the developmental rate from the ovule primordium initiation to a functional ovule (Li et al., 2014). It has been suggested that transcription factors and auxin synthesis play important roles in ovule primordial formation (Reyes-Olalde et al., 2013), whereas after the birth of the primordia, meiosis, a key event during ovule development, largely determines the formation of a functional ovule. Meiosis gives rise to a functional megaspore that further develops into an FG, which plays a critical role in every step of the seed formation from the direction of the pollen tube (Higashiyama, 2002) and the completion of double fertilization (Lord and Russell, 2002) to the initiation and accomplishment of seed development (Chaudhury and Berger, 2001; Chaudhury et al., 2001). Two key processes are completed during meiosis: one is the chromosome exchange between parents that generates genetic variations in the daughter generations, and one is the two rounds of cell division that produce four haploid gametes. The molecular mechanisms involved in meiotic recombination have been widely investigated (Jones et al., 2003; Hamant et al., 2006; Liu and Makaroff, 2006; Mercier and Grelon, 2008); however, relatively little is known about the control of cell cycle transition underlying meiotic division in plants (Cromer et al., 2012; Wijnker and Schnittger, 2013; Zhao et al., 2014).
Although meiotic cell cycle progression is known to rely on the quantitative and qualitative aspects of cyclin-dependent kinase (CDK)-cyclin complexes in Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila spp., and mammals, which of the Arabidopsis (Arabidopsis thaliana) cyclins constitutes the core CDK complex with CDKA;1 or possibly, other CDKs remains unclear (Cromer et al., 2012; Harashima et al., 2013). Emerging findings have characterized a regulatory network including the interactions among SUPPRESSOR WITH MORPHOGENETIC EFFECTS ON GENITALIA7 (SMG7), THREE DIVISION MUTANT1 (TDM1), and TARDY ASYNCHRONOUS MEIOSIS (TAM) in Arabidopsis, which modulates the cyclin-CDK activity to drive the progression from anaphase II to meiotic exit (Bulankova et al., 2010). Among these members, SMG7 proteins, as defined by the N-terminal Ever Shorter Telomere1 (EST1) domain, were initially identified as participating in nonsense-mediated mRNA decay (NMD); however, in Arabidopsis, an unusual cell cycle arrest in anaphase II was observed in a hypomorphic allele of smg7, indicating its pivotal function in meiosis exit (Riehs et al., 2008).
Using a double-haploid population derived from the F1 cross between two accessions (Y106 and HZ396) differing significantly in NSS (27.3 ± 3.7 and 11.0 ± 2.6, respectively), we previously mapped a major QTL (qSS.C9) controlling NSS on the B. napus C9 chromosome (Zhang et al., 2011, 2012b). In this study, we report the fine mapping, map-based cloning, and initial characterization of this gene. We also showed that qSS.C9 has experienced positive selection during B. napus improvement. Our work sheds light on the molecular mechanism of natural variations of NSS and suggests a significant implication for B. napus yield improvement.
RESULTS
Phenotypic Characterization of Near-Isogenic Line(HZ396) and Near-Isogenic Line(Y106)
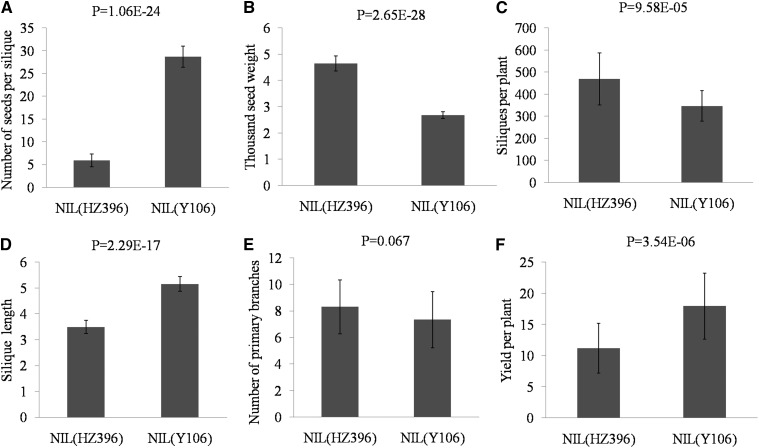
To investigate the effect of qSS.C9 on the grain yield of B. napus, we previously established a pair of near-isogenic lines (NILs), designated as NIL(HZ396) and NIL(Y106), and a heterozygote by selfing a BC3F1 plant heterozygous for qSS.C9, which was obtained by backcrossing a low-NSS parent HZ396 to a high-NSS parent Y106 three times (Zhang et al., 2012b). A molecular marker assay showed that NIL(Y106) carried a small region between the markers SRC9-298 and SRC9-397 from Y106, while containing approximately 97.2% genetic background of HZ396. To further evaluate the effects of qSS.C9 on single-plant yield, we further compared the performances of three yield components between NILs in the BC5F3 families. The results showed that NIL(Y106) displayed an extremely significant increase in NSS over NIL(HZ396) but substantial decreases in thousand-seed weight (TSW) and number of siliques per plant (Fig. 1, A–C). In addition, the silique length was significantly reduced in NIL(HZ396), but comparable numbers of primary branches were found between NILs (Fig. 1, D and E). Collectively, NIL(Y106) outyielded NIL(HZ396) by 6.77 g of seeds (58.7%) per plant (Fig. 1F). Thus, our results indicate that qSS.C9 is an elite allele for B. napus yield potential through the increase of NSS.
Figure 1.
Yield-related traits of NIL(HZ396) and NIL(Y106). A, Silique length. B, NSS. C, TSW. D, Number of primary branches. E, Silique per plant. F, Yield per plant. All phenotype data in A to F were measured from plants grown under normal cultivation conditions. Data are given as means ± sem (n = 30). A two-tailed Student’s t test was used to generate the P values.
Map-Based Cloning of qSS.C9
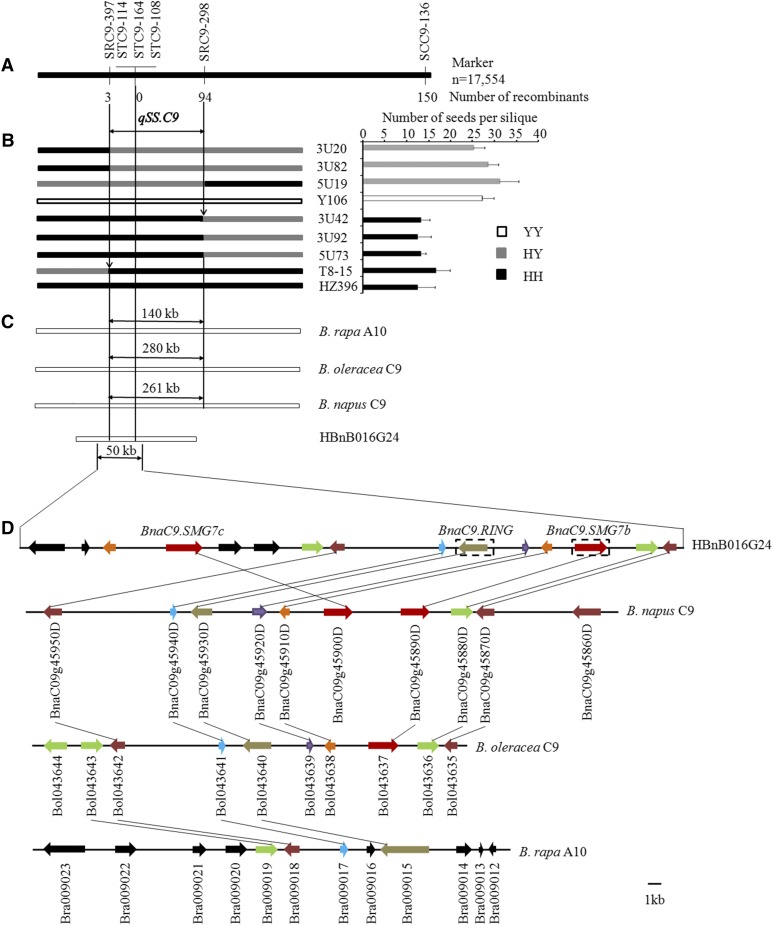
qSS.C9 was previously mapped to a genomic region between the markers SRC9-22 and SCC9-005 on the B. napus C9 chromosome, which could delimit a syntenic interval of 1,005 kb on the Brassica rapa A10 chromosome (Zhang et al., 2012b), 911 kb on the Brassica oleracea C9 chromosome, and 800 kb on the B. napus C9 chromosome. To fine map and clone qSS.C9, we developed three unique markers (SCC9-136, SRC9-298, and SRC9-397) from the candidate region (Fig. 2A); however, SRC9-298 and SRC9-397 remained cosegregated with qSS.C9 in the population used previously (Zhang et al., 2012b). To further narrow down the candidate region of qSS.C9, we field planted a BC4F2 population consisting of 17,554 individuals. From this population, 97 recombinants between the markers SRC9-298 and SRC9-397 were identified, and their genotypes at the qSS.C9 locus were deduced by progeny testing (Fig. 2, A and B; Supplemental Table S1). Finally, the candidate region of qSS.C9 was restricted between SRC9-397 and SRC9-298 (Fig. 2, A and B), corresponding to a DNA fragment of 140, 280, or 262 kb on the B. rapa A10 chromosome, the B. oleracea C9 chromosome, or the B. napus C9 chromosome, respectively (Fig. 2C).
Figure 2.
Map-based cloning of qSS.C9. A, Fine mapping of the qSS.C9 region with 17,554 BC4F2 plants. B, Genotypes and phenotypes of the recombinants. The NSS phenotype of each recombinant was determined by progeny testing (Supplemental Table S1). C, Physical maps of the qSS.C9 candidate region in different reference genomes and the target BAC clone HBnB016G24. D, Gene distribution in the partial candidate region of HBnB016G24 and the reference genomes. Annotated or predicted genes are indicated by rectangles with arrows. Lines connect homologous genes from the region showing perfect microsynteny between different genomes or HBnB016G24. Other homologous (or duplicated) genes are marked in the same color. The two genes denoted by dashed-line boxes were deleted in HZ396. Bar = 1 kb.
Because the B. napus C9 chromosome was evolutionarily derived from the C9 chromosome of B. oleracea, we used the reference genome of B. oleracea to annotate the qSS.C9 candidate region before the release of the B. napus reference genome. According to the B. napus database (http://brassicadb.org/brad/index.php), there are 45 predicted genes in the 280-kb candidate region of B. oleracea. We scanned the predicted proteins and concluded that 16 of them are unlikely to be associated with the trait of NSS according to the available function annotations. For each of the remaining 29 genes, we comparatively sequenced the genomic fragment covering the promoter region and the complete coding region from both homozygous NILs. The results showed that two predicted genes in NIL(Y106) that were individually homologous to Bol043637 and Bol043640 were deleted in NIL(HZ396), whereas no reproducible DNA polymorphism was observed for the other genes. Bol043637 encodes a small protein homologous to the Arabidopsis SMG7 gene that carries the EST1 domain. Previous research revealed that the disruption of AtSMG7 can cause embryonic lethality in Arabidopsis (Riehs et al., 2008; Bulankova et al., 2010). Bol043640 encodes a protein with unknown function homologous to the Arabidopsis Real Interesting New Gene (RING)/U-box superfamily protein. Moreover, we developed one (STC9-114) and two polymorphic markers (STC9-108 and STC9-164) based on the genomic sequences of Bol043640 and Bol043637, respectively, all of which were cosegregated with qSS.C9 in the recombination events described above (Fig. 2A). To characterize the genomic sequence around the candidate genes in B. napus, we further screened a B. napus bacterial artificial chromosome (BAC) clone library constructed from high-NSS accessions (195-14A) by using SRC9-397 and STC9-108 and successfully identified a BAC clone (HBnB016G24) with a 210-kb insert. From this clone, approximately a 50-kb continuous sequence covering the homologs of Bol043637 and Bol043640 was obtained, which shows overall good microcollinearity with the corresponding region of B. oleracea C9 chromosome, while displaying obvious structural variations from the B. napus C9 or B. rapa A10 chromosome (Fig. 2D). Interestingly, a local fragment duplication event was observed in HBnB016G24 relative to B. oleracea (Fig. 2D), which resulted in two homologous copies of SMG7 (hereafter designated as BnaC9.SMG7b and BnaC9.SMG7c). By using the BAC insert sequence as a reference, we obtained the corresponding sequences from both homozygous NILs by PCR amplification. Sequence comparison showed that the fragments from both NILs were highly identical to the fragment from the BAC insert, whereas a 6.1-kb fragment harboring BnaC9.SMG7b and a 5.8-kb fragment harboring BnaC9.RING (a homolog of Bol043640) that existed in NIL(Y106) were deleted in NIL(HZ396) (Fig. 2D). These facts strongly suggest that BnaC9.SMG7b or BnaC9.RING, especially the former, was likely the target of qSS.C9.
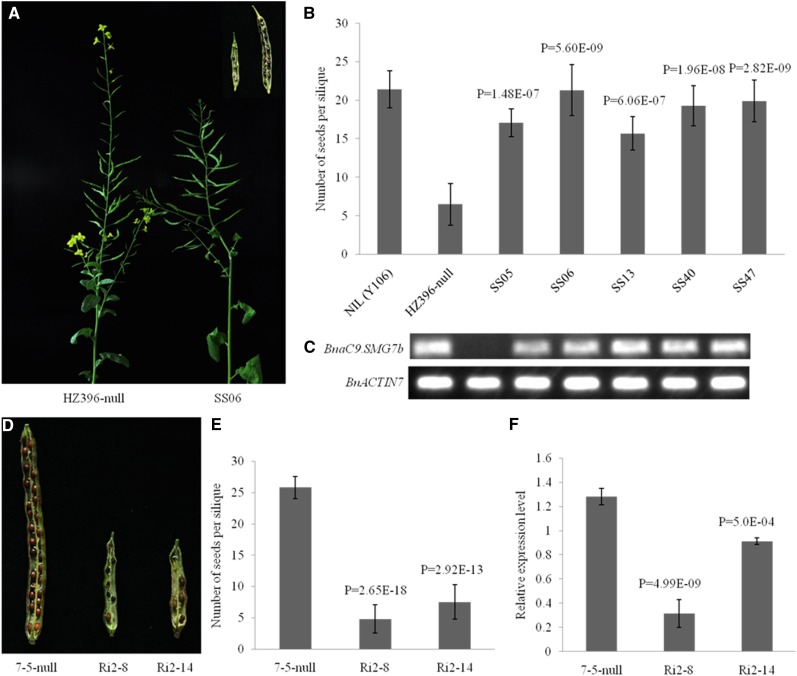
To validate this prediction, we cloned the 5.3-kb genomic fragments from NIL(Y106) that covered the predicted BnaC9.SMG7b into the binary vector pCAMBIA3300 and introduced the construct into HZ396 by Agrobacterium spp.-mediated transformation. In the resulting T0 generation, we observed that all five transgene-positive plants carrying BnaC9.SMG7b had higher NSSs than the transgene-negative plants (Fig. 3, A–C). Further analysis of phenotypes and genotypes confirmed that the transgenic events were cosegregated with the higher NSS in the T1 progeny (Supplemental Table S2). In addition, to suppress the expression level of BnaC9.SMG7b, we transformed an RNA interference (RNAi) construct for BnaC9.SMG7b into an inbred line (7-5), which has the same sequence as Y106 in BnaC9.SMG7b and typically displays a high NSS (28.1 ± 0.6). Compared with the transgene-negative plants, the T0-positive plants had significantly reduced NSS (P < 0.01; Fig. 3, D–F), which is consistent with the lowered expression level of BnaC9.SMG7b in the transgene-positive plants. Furthermore, correlation analysis indicated that the expression level of BnaC9.SMG7b in the T1 families was positively correlated with NSS (Supplemental Table S3). Meanwhile, we transformed a complementation construct containing the predicted BnaC9.RING into HZ396; however, no significant difference in NSS was observed between the T0 transgene-negative and transgene-positive plants (Supplemental Fig. S1). Taken together, these results confirm that BnaC9.SMG7b gene is the target gene for qSS.C9.
Figure 3.
Functional analysis of BnaC9.SMG7b. A, Plant and silique phenotypes of the low-NSS parental HZ396-null and BnaC9.SMG7b-transgenic T0 lines (SS06). B, Comparison of NSS among NIL(Y106), HZ396-null, and BnaC9.SMG7b-transgenic T0 lines. The statistical significance was at P < 0.01 based on a two-tailed Student’s t test. Error bars represent the sds. C, Expression of BnaC9.SMG7b in the transgenic B. napus lines as revealed by reverse transcription (RT)-PCR. D, Silique phenotypes of 7-5-null and 7-5 RNAi-transformed T0 plants. E, Comparison of NSSs between 7-5-null and 7-5 RNAi-transformed T0 plants. F, Expression of BnaC9.SMG7b in flower buds was analyzed by qPCR. BnACTIN7 was used as a control. A two-tailed Student’s t test between the transgenic plants and the negative control was used to generate the P values in E and F. Error bars represent sds from three independent RNA samples.
qSS.C9 Encodes a Small Protein Showing Homology to AtSMG7
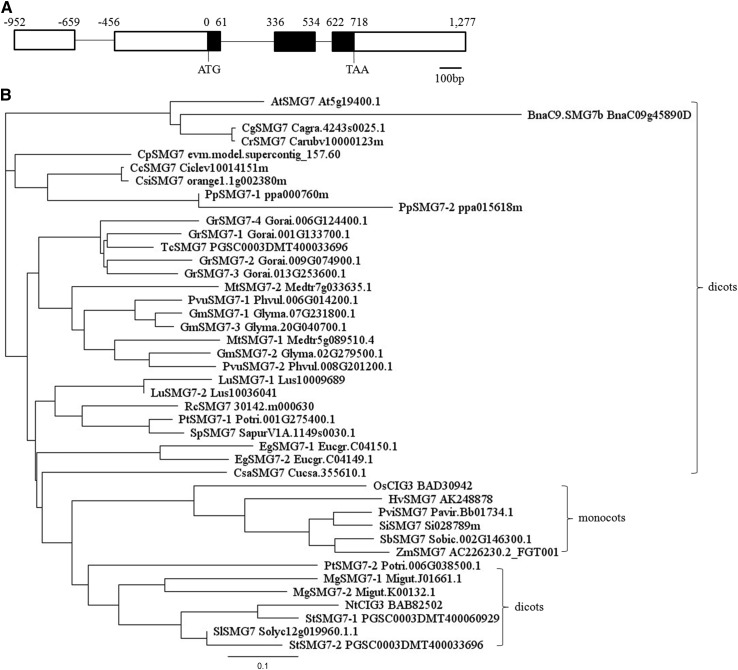
The full-length complementary DNA (cDNA) of BnaC9.SMG7b is 1,666-bp in length, which is composed of four exons and consists of a 750-bp 5′ untranslated region, a 357-bp open reading frame, and a 559-bp 3′ untranslated region (Fig. 4A). Surprisingly, the predicted BnaC9.SMG7b only encodes a small protein of 119 amino acids and contains a conserved EST1-central domain (CD), as indicated by PROSITE analysis (http://prosite.expasy.org/). However, relative to its Arabidopsis homolog SMG7 (1,059 amino acids in length), the EST1-CD was greatly reduced in length, and the N-terminal EST1-tertratricopeptide repeat domain (TPR) and the region following EST1-CD were nearly completely missed (Supplemental Fig. S2). To understand the possible evolutionary process of BnaC9.SMG7b, we searched the protein sequences showing homology to SMG7 from the Phytozome database (www.phytozome.net) and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/); 42 SMG7 homologs were collected from 6 monocot species and 22 dicot species, and they encode proteins varying from 55 to 1,059 amino acids in length. The SMG7 homologs can be divided into three main branches on the phylogenetic tree (Fig. 4B). Although BnaC9.SMG7b falls in the same branch as most of the dicot SMG7 homologs, it has the most distant relationship with any of the SMG7 homologs.
Figure 4.
Gene structure and phylogenetic analysis of BnaC9.SMG7b. A, BnaC9.SMG7b gene structure. Black lines represent introns. Black bars represent exons, and white bars represent 5′ and 3′ untranslated regions. Bar = 100 bp. B, Phylogenetic tree of the plant SMG7 protein. The bootstrap neighbor-joining phylogenetic tree was constructed using Geneious 4.8.3. The lengths of the branches refer to the amino acid variation rates. Cc, Citrus clementina; Cg, Capsella grandiflora; Cp, Carica papaya, Cr, Capsella rubella; Csa, Cucumis sativus; Csi, Citrus sinensis; Eg, Eucalyptus grandis; Gm, Glycine max; Gr, Gossypium raimondii; Hv, Hordeum vulgare; Lu, Linum usitatissimum; Mg, Mimulus guttatus; Mt, Medicago truncatula; Nt, Nicotina tabacum; Os, Oryza sativa; Pp, Prunus persica; Pt, Populus trichocarpa; Pui, Panicum virgatum; Pvu, Phaseolus vulgaris; Rc, Ricinus communis; Sb, Sorghum bicolor; Si, Setaria italica; Sl, Solanum lycopersicum; Sp, Salix purpurea; St, Solanum tuberosum; Tc, Theobroma cacao; Zm, Zea mays.
To understand the origination and evolution of BnaC9.SMG7b in greater detail, we performed BLAST analysis using the protein sequence of BnaC9.SMG7b as a query. Aside from BnaC9.SMG7b, 12 homologs of BnaC9.SMG7b were identified from four species of Brassicaceae, including two from B. rapa, three from B. oleracea, five from B. napus, and two from Arabidopsis. Neighbor-joining cluster analysis using coding sequences grouped all BnaC9.SMG7b homologs into two groups: BnaC9.SMG7b and its 10 homologs from Brassicaceae were classified into one large group with AtSMG7 (group I), whereas AtSMG7L was individually located in the other group (group II; Supplemental Fig. S3), suggesting that AtSMG7L might have evolved after the divergence between Arabidopsis and B. napus. In clades 1 and 3 from group I, each of the B. napus BnaC9.SMG7b homologs corresponds to a highly similar copy from the corresponding B. rapa or B. oleracea chromosome, and all of the homologs derived from AtSMG7 (At5g19400) are located in the ancestral karyotype R block, indicating that these copies were directly inherited from their diploid ancestors and are the products of whole-genome triplication. However, BnaC9.SMG7b was localized in clade 2, where another two homologous copies (BnaC9.SMG7c and BolC9.SMG7b) were identified only from the C9 chromosome of B. napus or B. oleracea. Moreover, collinear comparison revealed that both BnaC9.SMG7b and BnaC9.SMG7c originated from BolC9.SMG7b, and the flanking regions of all of these three homologs belong to the R block (Supplemental Fig. S4). Thus, the above results suggest that the location of the three BnaC9.SMG7b homologs from clade 2 in the R block might be because of a fragment insertion in the C9 chromosome BnaC9.SMG7b.
To determine whether the duplication status of BnaC9.SMG7b and BnaC9.SMG7c is universal in natural B. napus, B. rapa, and B. oleracea accessions, we analyzed 295 B. napus accessions (including HZ396 and Y106), 22 B. rapa accessions, and 50 B. oleracea accessions by the codominant marker (STC9-164) to specifically distinguish between BnaC9.SMG7b and BnaC9.SMG7c. The results showed that, except for eight accessions that did not harbor BnaC9.SMG7b and nine accessions that did not harbor BnaC9.SMG7c, all other B. napus accessions contained both of these highly homologous copies. Meanwhile, none of the B. rapa accessions harbored either of the two highly homologous copies, and all B. oleracea accessions contained the BnaC9.SMG7b homologous copy BolC9.SMG7b. Collectively, these facts show that BnaC9.SMG7b is derived from a gene duplication event of BolC9.SMG7b, which is a B. oleracea genome-specific gene emerging after the divergence of B. rapa and B. olearacea.
Expression Pattern and Subcellular Localization of BnaC9.SMG7b
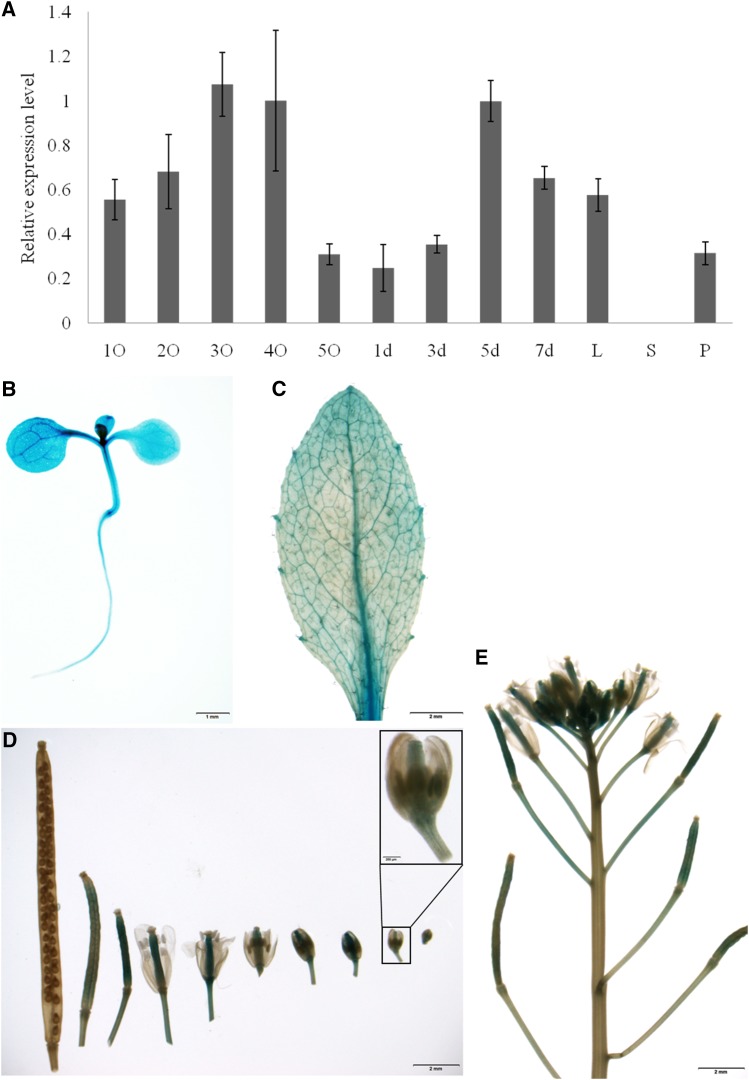
To investigate the expression profile of BnaC9.SMG7b, quantitative real-time PCR (qPCR) analysis was performed using the total RNA extracted from various organs of NIL(Y106). The results indicated that relatively low expression of BnaC9.SMG7b was observed in young leaves and pedicels, with extremely low transcript accumulation in stamens; in contrast, overall high but variable expression levels were detected in developing pistils before and after anthesis (Fig. 5A). To examine BnaC9.SMG7b activity in greater detail, a BnaC9.SMG7b promoter-GUS fusion construct was transformed into Arabidopsis wild-type plants. The expression of GUS in the transgenic plants was detected mainly in the vascular tissue of various organs, including cotyledons, rosette leaves, roots, young pedicels, and pistils, before and after pollination but not in stamens, petals, stems, and mature siliques (Fig. 5, B–E). Consistent with the qPCR analysis, GUS activity was gradually enhanced in the pistils before anthesis and decreased after anthesis. Although a previous study has indicated that AtSMG7 regulates both male and female meiosis cell cycles in Arabidopsis (Riehs et al., 2008), the distinct expression pattern of BnaC9.SMG7b in the stamen and pistil suggests that BnaC9.SMG7b might play an exclusive role in megasporogenesis but not in microsporogenesis in B. napus. This speculation is supported by the fact that the stamen development and pollen viability were not affected in NIL(HZ396) (Supplemental Fig. S5).
Figure 5.
Expression patterns of BnaC9.SMG7b. A, qPCR analysis of BnaC9.SMG7b. 1O to 5O indicate ovaries from NIL(Y106) with <1, 1 to 2, 2 to 3, 3 to 4, and 4 to 5 mm before pollination, respectively; 1 to 7 d indicate ovaries from NIL(Y106) grown for 1, 3, 5, and 7 d after pollination, respectively. BnACTIN7 expression was used as a control. Error bars represent ses from three independent RNA samples. L, Leaves; P, pedicels; S, stamens. B to E, Representative histochemical analysis of GUS expression in tissues under the control of the BnaC.SMG7b promoter in the transgenic Arabidopsis T2 plants. B, Seedling 7 d after germination. C, Rosette leaf. D and E, Inflorescence and mature silique. At least five independent transformants were subjected to histochemical GUS assays. Bars = 1 mm (B) and 2 mm (C–E).
Processing bodies (P bodies) are small cytoplasmic foci that contain all NMD factors, including SMG7 in eukaryotes (Eulalio et al., 2007; Riehs, 2009; Kulkarni et al., 2010; Mérai et al., 2013). To investigate the subcellular localization of BnaC9.SMG7b, we fused the coding region of BnaC9.SMG7b with the coding region of an enhanced GFP driven by a Cauliflower mosaic virus 35S promoter. This chimeric plasmid and a P bodies marker (Arabidopsis DECAPPING COMPLEX PROTEIN1 [AtDCP1]; Mérai et al., 2013) were cotransformed into Arabidopsis protoplasts. The results showed that the BnaC9.SMG7b-GFP fusion protein was colocalized with the marker in the P bodies (Fig. 6), suggesting that BnaC9.SMG7b should retain some conserved functions of SMG7.
Figure 6.
Subcellular localization of BnaC9.SMG7b. Colocalization of BnaC9.SMG7b-GFP (A) and AtDCP1-RFP (B) in the P bodies. The bright-field image (C) and merged image (D) are also shown. Bars = 10 μm.
BnaC9.SMG7b Promotes the Meiotic Cell Cycle
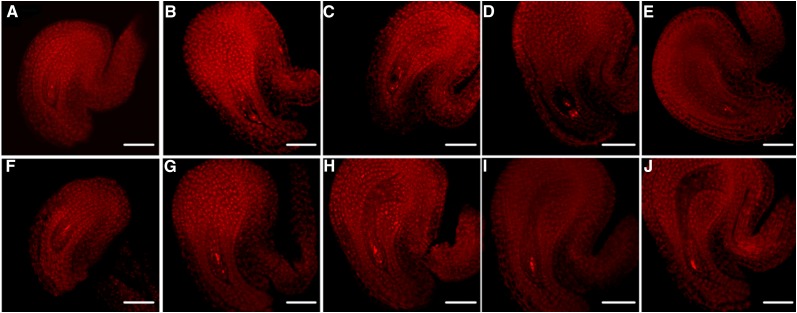
There were no significant differences in ovule number between NIL(HZ396) and NIL(Y106) before anthesis (Supplemental Fig. S6), suggesting that the decrease of NSS in NIL(HZ396) should not result from fewer ovule primordia per silique but from defects in late ovule or early seed development. Further observation of the pollen germination and pollen tube behavior showed that most pollen tubes could grow into the style normally, but some of them failed to enter the embryo sacs successfully in NIL(HZ396) (Supplemental Fig. S7). Considering the fact that the FG plays a central role in pollen tube guidance during fertilization (Ray et al., 1997), these results indicate that some ovules in NIL(HZ396) possibly failed to form the normal FGs or even the functional megaspores. Therefore, we first compared the morphology of FG at the stage of FG5 (Christensen et al., 1997) between NILs by confocal laser-scanning microscopy. As expected, we found that normal FGs appeared in most of the ovules (96.5%) in NIL(Y106) but in only 34.4% of ovules in NIL(HZ396) (Supplemental Table S4). We also compared the initial ovule numbers per silique of both NILs with the corresponding NSSs (Supplemental Table S5). The ratio (65.6%) of abnormal FGs was highly close to the proportion (69.8%) of abortion ovules per silique in NIL(HZ396), indicating that the defective FGs should be responsible for the reduction of NSSs in NIL(HZ396). Moreover, morphologically abnormal FGs (or no FGs) can be observed from the very beginning stage (FG1) to the middle-late stage (FG5; Christensen et al., 1997) in the ovules of NIL(HZ396) (Fig. 7; Supplemental Table S4).
Figure 7.
Confocal laser-scanning microscopy analysis of ovule development in NIL(Y106) and NIL(HZ396). Normal FGs in NIL(Y106) and NIL(HZ396) (A–E) and abnormal FGs in NIL(HZ396) (F–J). A and F, FG1; B and G, FG2; C and H, FG3; D and I, FG4; and E and J, FG5. Bars = 50 μm.
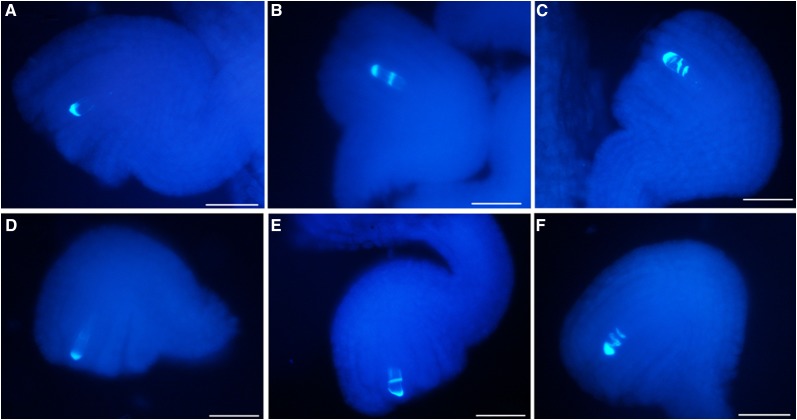
Because the FG (FG1) develops directly from the functional megaspore, we further examined whether the arrested FGs in NIL(HZ396) result from abnormal functional megaspore. Above all, we investigated the megasporogenesis between two NILs by staining callose in the cell plates of megasporocyte undergoing meiosis in ovules with aniline blue (Rodkiewicz, 1970). The dynamic callose pattern as indicated by aniline blue can be used to monitor the female meiotic division progression in Arabidopsis (Qin et al., 2014). Comparable frequencies of aniline blue-stained callose bands were observed between NIL(HZ396) and NIL(Y106) throughout all meiosis stages (Fig. 8; Supplemental Table S6), indicating that meiosis in the majority of the megasporocytes of both NILs can progress at least to anaphase II. However, because of the technique limitation in observing the very short period from anaphase II to the formation of a functional megaspore in B. napus, it remains to be clarified exactly when the developmental abnormity occurs in some ovules of NIL(HZ396). Nevertheless, our results clearly showed that BnaC9.SMG7b plays an important role in determining the differentiation of a haploid gametophyte, but the role is specifically restricted to ovules, because we observed identical and normal pollen development in both NILs (Supplemental Fig. S5).
Figure 8.
Typical meiotic divisions of megasporogenesis in NIL(Y106) (A–C) and NIL(HZ396) (D–F). A and D, Callose staining in the transverse cell plate of dyads. B and E, Two callose bands in triads. C and F, Three callose bands in tetrads. Bars = 50 μm.
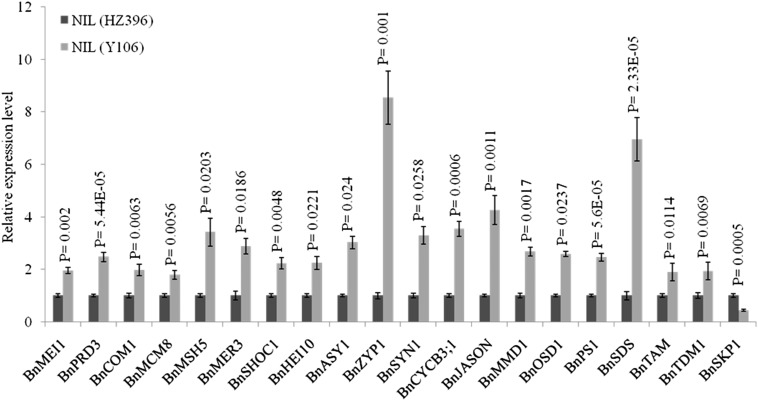
The Arabidopsis SMG7 controls the female fertility and then NSS by regulating the cell cycle progression out of meiosis II (Riehs et al., 2008). To understand whether BnaC9.SMG7b may function in a similar manner, ovaries with a length <1 mm from NIL(HZ396) and NIL(Y106) were collected to analyze the changes in meiotic gene expression by RNA sequencing (RNA-seq). First, to confirm that these differentially expressed genes (DEGs) from the RNA-seq and bioinformatics analyses were essentially differentially expressed, in total, 10 genes were selected randomly from the DEGs for qPCR assays. For all of the 10 genes, qPCR analysis revealed the same expression trends as the RNA-seq data, which confirmed the reliability of RNA-seq data (Supplemental Fig. S8; Supplemental Table S7). Second, in total, 79 putative meiotic gene families were identified from the RNA-seq data according to the previous results (Wijnker and Schnittger, 2013; Mercier et al., 2015). Among them, 20 gene families showed down-regulated expression in NIL(HZ396), and only one (Brassica napus S-phase-kinase-associated protein1 [BnSKP1]) showed up-regulated expression in NIL(HZ396) (Supplemental Table S8). The 21 meiotic genes were further confirmed by qPCR assay (Fig. 9). According to the function annotations of these differentially expressed meiotic gene families, they can be divided into two groups. The first group includes 11 members that are involved in some key meiotic events, such as entry into meiosis, sister chromatid cohesion, double-strand break repair, cross over and noncross over outcome, and assembly of synaptonemal complex; the second group contains nine gene families that are involved in cell cycle control (Fig. 9). Especially, except for BnSKP1, which is a negative regulator of cell cycle, the Arabidopsis orthologs of all of the other eight members (Brassica napus B-TYPE CYCLIN3.1 [BnCYCB3;1], Brassica napus PARALLEL SPINDLE1 [BnJASON], Brassica napus MALE MEIOCYTE DEATH1 [BnMMD1], Brassica napus OMISSION OF SECOND DIVISION1 [BnOSD1], Brassica napus PARALLEL SPINDLE1 [BnPS1], Brassica napus SOLO DANCERS [BnSDS], BnaTAM, and BnaTDM1) from the second group were reported to positively regulate the cell cycle (Wijnker and Schnittger, 2013; Mercier et al., 2015). Taken together, our results show that the function of BnaC9.SMG7b in FG differentiation likely depends on the positive regulation of female meiotic cell cycle-related genes.
Figure 9.
Expression level of meiotic genes in NIL(HZ396) and NIL(Y106). Total RNA was extracted from ovaries during meiotic stages with length <1 mm before pollination. A two-tailed Student’s t test was used to generate the P values. Error bars represent sds from three independent RNA samples.
BnaC9.SMG7b Is Not Involved in NMD
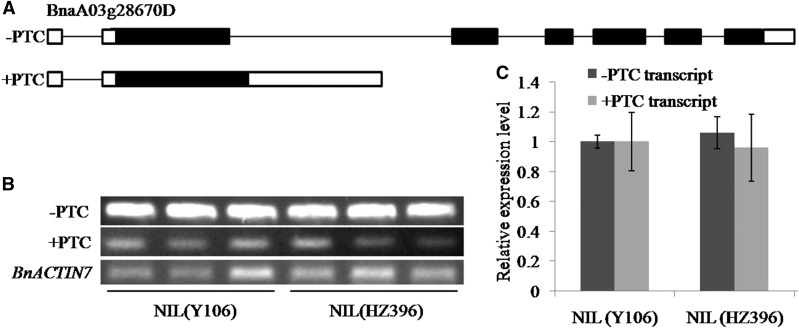
Other than the function in progression out of meiosis II, AtSMG7 is also involved in the NMD, one of the approaches to eliminate the transcripts containing premature termination codon (PTC; Riehs et al., 2008; Riehs-Kearnan et al., 2012). To identify whether BnaC9.SMG7b is also involved in NMD in B. napus, we analyzed the different types of alternative splicing (AS) events between NIL(HZ396) and NIL(Y106) from the RNA-seq data. The results indicate that no significant difference was observed in any type of the AS events (Supplemental Fig. S9). We further investigated the expression levels of two different transcripts of a single-copy gene (BnaA03g28670D) by qPCR analysis and found that the accumulation of PTC-containing transcripts (+PTC) and normal transcripts (−PTC) is not significantly elevated in NIL(HZ396) compared with NIL(Y106) (Fig. 10). Consistently, we did not detect obvious difference in vegetative growth between the two NILs (Supplemental Fig. S10), which is regarded as the major negative effect of NMD deficiency in Arabidopsis (Hori and Watanabe, 2005; Arciga-Reyes et al., 2006; Yoine et al., 2006; Riehs et al., 2008; Riehs-Kearnan et al., 2012). Therefore, these data indicate that BnaC9.SMG7b is not involved in the NMD process.
Figure 10.
NMD efficiency in NIL(Y106) and NIL(HZ396). A, AS of BnaA03g28670D without PTC (−PTC) and with PTC (+PTC) results in a PTC in the second exon. Black lines represent introns. Black bars represent exons, and white bars represent 5′ and 3′ untranslated regions. Expression of the BnaA03g28670D transcripts in NIL(Y106) and NIL(HZ396) as revealed by RT-PCR (B) and qPCR (C). Three independent RNA samples from each NIL were analyzed. BnACTIN7 was used as a control. Error bars represent sds.
Sequence Polymorphisms of BnaC9.SMG7b in B. napus Germplasms
To understand whether the deletion or potential sequence variation in BnaC9.SMG7b is associated with NSS in natural B. napus populations, we sequenced the 3.48-kb BnaC9.SMG7b genomic fragment (covering the 1.25-kb promoter region and the 2.23-kb gene region) in 76 randomly selected lines containing BnaC9.SMG7b and measured the NSSs of these lines and the eight accessions in which BnaC9.SMG7b was deleted as described above (Supplemental Fig. S11; Supplemental Table S9). According to the sequence variations of BnaC9.SMG7b, four haplotypes were identified from these B. napus accessions (Supplemental Fig. S11). The first haplotype (HAP1) characterized by the deletion of BnaC9.SMG7b was observed in NIL(HZ396) and seven other lines. The second haplotype (HAP2) appeared in 69 lines with the same sequence as BnaC9.SMG7b in NIL(Y106). The third haplotype (HAP3) was found in six accessions carrying only one nucleotide change in the promoter region relative to HAP2. The fourth haplotype only existed in one accession, which displayed five single-nucleotide polymorphism variations relative to HAP2, but none of the nucleotide changes affected the BnaC9.SMG7b coding sequence. Further statistical analysis indicated that HAP2 or HAP3 showed a significantly higher value of NSS than HAP1, but no significant difference was observed between HAP2 and HAP3 (HAP4 was not analyzed, because it is an extremely rare allele; Supplemental Table S10). Therefore, the presence/absence of BnaC9.SMG7b might be an important factor that affects the natural variation of NSS in B. napus, whereas the single-nucleotide polymorphism in the promoter region of BnaC9.SMG7b has no effect on NSS variation.
DISCUSSION
Through transgenic complementation and RNAi and microscopy observations, we cloned and primarily characterized BnaC9.SMG7b, which underlies a major QTL controlling NSS in Brassica spp. BnaC9.SMG7b functions during the stage from female meiosis anaphase II to the formation of a functional megaspore and subsequently, determines the generation of functional FGs. The natural deletion or reduction of BnaC9.SMG7b expression will lead to a decreased NSSs, indicating it as a positive regulator of NSS. Our results show that, although BnaC9.SMG7b encodes a small protein homologous to AtSMG7, BnaC9.SMG7b shows clear sequence and functional divergences from AtSMG7. Analysis of the sequence variations of BnaC9.SMG7b among natural populations showed that the elite allele of BnaC9.SMG7b has been nearly fixed in modern B. napus accessions.
As a key step in the lifecycle of sexually reproducing eukaryotes, the completion of meiosis depends on the transition of the meiotic cell cycle (d’Erfurth et al., 2010). CDKA;1 is the only CDK that has been proposed to play a central role in meiosis (Dissmeyer et al., 2007; Bulankova et al., 2010). TAM, as a main cyclin, drives the cell from G2 into the first meiosis by promoting the activities of the core CDK complexes and promotes entry into the second meiosis by antagonizing the inhibitory effects of SMG7 and TDM1. TAM can also form an active complex with CDKA;1 to phosphorylate OSD1, which is another regulator to promote meiotic progression through Anaphase Promoting Complex/Cyclosome inhibition (Cromer et al., 2012). SMG7 down-regulates CDK activity in anaphase I and is required after the first meiotic division for full chromatin decondensation (Bulankova et al., 2010). In anaphase II, SMG7 acts through TDM1 to promote meiotic exit and then the transition to mitotic G1 by down-regulating CDK activity again (Bulankova et al., 2010). Based on the above participants in meiotic cell cycle regulation in Arabidopsis, a molecular model was proposed and complemented: the meiotic progression in Arabidopsis pollen mother cells is driven by cyclin-CDK activity modulated by the regulatory interactions among TAM, SMG7, TDM1, and OSD1 (Bulankova et al., 2010; Cromer et al., 2012). In this study, we showed that the deletion of BnaC9.SMG7b possibly resulted in a similar defect in female meiotic progression regulation, although the cytological detail of the arrested embryo sacs in NIL(HZ396) remains to be clarified. Furthermore, the expression levels of BnCYCB3;1, BnSDS, BnTAM, BnOSD1, and BnTDM1 were significantly decreased during female meiosis in the absence of BnaC9.SMG7b. These data collectively suggest that BnaC9.SMG7b likely also plays a conserved and important role in meiotic cell cycle regulation, similar to SMG7 in Arabidopsis, and the regulatory model is also applicable for female meiosis in B. napus. In addition to these cyclin-CDK activity-related genes, four cell cycle-related genes (JASON, PS1, MMD1, and SKP1) showed different expression levels between NIL(HZ396) and NIL(Y106) by RNA-seq and qPCR analyses (Fig. 9; Supplemental Table S8). JASON is required for metaphase II spindle orientation through positively regulating PS1 expression (d’Erfurth et al., 2008; De Storme and Geelen, 2011). Because the smg7 mutation exhibits delayed chromosome decondensation and aberrant rearrangement of the meiotic spindle in anaphase II (Riehs et al., 2008), SMG7 may interact with JASON and PS1. MMD1 and SKP1 are involved in both chromatin structure and the 26S ubiquitination pathway (Yang et al., 1999, 2003; Reddy et al., 2003; Smalle and Vierstra, 2004; Wang and Yang, 2006; Zhao et al., 2006). Observation of the aniline blue-stained callose bands of NILs indicates that the chromosome organization and progression are normal (Fig. 8). Therefore, the deletion of BnaC9.SMG7b likely leads to abnormal programmed cell death in NIL(HZ396), which induces the abnormal expression of MMD1 and SKP1.
Functional divergence of duplicated genes provides a source of evolutionary novelties in polyploids, including pseudogenization, neofunctionalization, and subfunctionalization (Force et al., 1999; Conant and Wolfe, 2008; Liu and Adams, 2010). Our study showed that an obvious subfunctionalization occurred in BnaC9.SMG7b, because it only assumes the role as a positive regulator of female meiotic exit but discards its function during male meiotic exit and NMD decay compared with SMG7 in Arabidopsis (Riehs et al., 2008; Bulankova et al., 2010; Riehs-Kearnan et al., 2012). There are two possible reasons for this functional divergence of BnaC9.SMG7b. First, the N-terminal EST1-TPR domain of SMG7 contains a phospho-Ser binding domain that is required for the early steps of NMD (Fukuhara et al., 2005; Mérai et al., 2013), whereas this conserved domain is nearly completely deleted in BnaC9.SMG7b (Supplemental Fig. S2). This observation also indicates that the EST1-TPR domain is actually not necessary for the meiotic cycle progression, and AtSMG7 does not antagonize CDK activity by mediating the dephosphorylation of the CDK-cyclin-B substrates during anaphase as suggested previously (Riehs et al., 2008). Second, the promoter sequence of BnaC9.SMG7b has been greatly modified compared with the corresponding sequence in AtSMG7; BnaC9.SMG7b is not expressed in anthers and shows no effect on the development of male reproduction units. Similarly, although BnaC9.SMG7b and BnaC9.SMG7c have highly conserved amino acid sequences (Supplemental Fig. S2) and predicted gene structure and are both located in P bodies (Supplemental Fig. S12), they further experienced a process of subfunctionalization, which was indicated by the clear sequence variation in the promoter regions and the distinct expression and regulation patterns of genes in the reproductive organs (Supplemental Fig. S13). In combination with the normal male fertility in both NIL(HZ396) and NIL(Y106), BnaC9.SMG7b and BnaC9.SMG7c may specifically function in female and male meiotic cell cycle progression, respectively, whereas other homologous BnSMG7 copies may be mainly involved in NMD decay, although they can complement the function of BnaC9.SMG7b to some extent. In polyploid crops, the functional divergence of duplicated genes may be an important source of quantitative trait variation, such as the flowering time regulation in B. napus (Wang et al., 2009).
After polyploidization, a series of common events, including gene loss, chromosome rearrangement, and tandem duplication, occurred continually throughout the evolutionary history of Brassicaceae species (Wang et al., 2011; Cheng et al., 2013; Liu et al., 2014; Chalhoub et al., 2014). From the ancestral precursor to modern BnaC9.SMG7b, SMG7 homologs in B. napus also experienced these evolutionary events. First, whole-genome triplication produced three copies of SMG7 homologs in the Brassica spp. diploid ancestor, and the one located in the medium fractionated subgenome (Wang et al., 2011) was deleted before the divergence of B. rapa and B. oleracea. Subsequently, a new copy of SMG7 was generated through a local duplication of BolC9.SMG7a and transported to the R block of the same C9 chromosome. A sequence divergence occurred in this B. oleracea-specific copy, resulting in the production of BolC9.SMG7b. After the formation of B. napus through wide hybridization between B. rapa and B. oleracea, BolC9.SMG7b further experienced a tandem duplication event and gave rise to BnaC9.SMG7b and BnaC9.SMG7c. Finally, there are six copies of SMG7 in the B. napus genome. Based on these analyses, we proposed a model to describe the possible evolutionary process from AtSMG7 to its homologs in B. napus (Supplemental Fig. S4).
Four types of haplotypes at the BnaC9.SMG7b locus have been found among natural accessions. HAP1, which represented the absence of BnaC9.SMG7b as revealed in HZ396, greatly reduced NSS compared with any of the other three haplotypes, whereas no significant difference in NSS was observed between HAP2 and HAP3 (Supplemental Table S10). Therefore, the presence of BnaC9.SMG7b in the B. napus genome is an important factor for the increase of NSS in natural accessions. Thus, the natural variations at BnaC9.SMG7b can actually be divided into two groups. One group is represented by HZ396, in which BnaC9.SMG7b is completely deleted. The other group, including accessions possessing HAP2 to HAP4, carries a functional BnaC9.SMG7b allele. Because of the great contribution on NSS, BnaC9.SMG7b might have undergone positive selection during the yield improvement of modern B. napus breeding accessions, and therefore, BnaC9.SMG7b is ubiquitous in most of the germplasms (97.3%) analyzed. However, consistent with previous reports that the trait of NSS is negatively correlated with the trait of TSW (Shi et al., 2009; Zhang et al., 2011), we also observed an extremely low NSS together with a significantly high TSW in HZ396 (Fig. 1). Thus, it is reasonable that several accessions without BnaC9.SMG7b are still retained among modern B. napus breeding accessions, likely as the byproduct of artificial selection of high TSW. Certainly, it would be interesting to determine in the future if the negative correlation between NSS and TSW is caused by BnaC9.SMG7b itself or a tightly linked neighboring member. In addition, even in the presence of BnaC9.SMG7b, a wide range of NSS variations was also observed among inbreeding accessions (Supplemental Table S11), showing that unique QTLs that control NSS variations are present in these lines. In future studies, we expect to identify these QTLs by using populations derived from parental lines with distinct differences in the numbers of both initial ovules and arrested ovules.
MATERIALS AND METHODS
Plant Materials
NILs (BC3F2) segregating at qSS.C9 were successfully constructed using a low-NSS inbred line HZ396 (recurrent parent) and a high-NSS inbred line Y106 (donor parent; Zhang et al., 2012b). Based on the NILs, a BC4F2 population consisting of 17,554 individuals was further developed in 2011 to 2012 to fine map qSS.C9. All of the identified individuals with recombination events at the qSS.C9 locus were further determined by progeny testing in 2012 and 2013. To compare the yield-related traits between NILs, we randomly selected NIL(Y106) and NIL(HZ396) BC5F2 plants homozygous at qSS.C9 to produce the BC5F3 families, and 30 plants from each family were used for trait evaluation. In total, 293 Brassica napus accessions, including 190 materials from an association mapping population (Cai et al., 2014) and 103 inbreeding lines, were used to evaluate the duplication status (BnaC9.SMG7b and BnaC9.SMG7c). Eighty-two of these accessions, listed in Supplemental Table S8, were randomly selected for haplotype analysis; 22 Brassica rapa accessions and 50 Brassica oleracea accessions were used to evaluate the origin and evolution of BnaC9.SMG7b.
Plant Growth Conditions and Treatments
The B. napus plants used for genetic mapping and haplotype analysis were grown in normal growing seasons in the Experimental Station of Huazhong Agricultural University. The planting density was 20 cm between plants within rows and 25 cm between rows. Regular field management was conducted according to local agricultural practices. Arabidopsis (Arabidopsis thaliana) plants and transgene-positive and -negative B. napus plants were all pot grown in a greenhouse at 20°C to 22°C under 40% to 50% humidity and long-day conditions (16-h-light/8-h-dark cycle).
Measurements of NSS and Other Yield-Related Traits
Harvested B. napus was air dried and stored at room temperature before testing. NSS and silique length were estimated as a mean from 10 well-developed siliques sampled from the primary branch in the middle of the harvested individual (Zhang et al., 2011). The seed weight for each plant was estimated by the average of three measurements of 1,000 well-filled seeds (Fan et al., 2010). The number of primary branches was measured as the number of effective primary branches (Shi et al., 2009). The number of siliques per plant was the number of well-filled, normally developed siliques on each harvested individual (Shi et al., 2009). Yield per plant was measured as the average dry weight of seeds of the harvested individuals in a plot (Shi et al., 2009).
BAC Screening, Sequencing, and Annotation
A unique BAC clone library designated as HBnB was constructed, containing 61,440 clones with a mean insert size of 140 kb and 9.2-fold redundant representation of the genome of B. napus. BAC clone plasmid DNA was prepared using the Macherey-Nagel NucleoBondXtra Midi Kit. The target BAC clones were identified through a two-stage PCR screening method with SRC9-397 and STC9-108 (Crooijmans et al., 2000). The target BAC clone was sheared to fragments ranging from 6 to 8 kb and then subcloned into the plasmid PUC118 to obtain a library containing 1,536 subclones. Then, we used PCR screening and end sequencing to construct a clone contig spanning the markers STC9-108 and SRC9-397 and further determined their sequences by Sanger sequencing. The Web site-based software FGENSH (http://www.softberry.com) was applied to predict the putative genes from partial sequences of the BAC clone. The genomic or coding sequences of predicted genes were submitted to the B. napus database (http://brassicadb.org/brad/index.php) and the Genoscope Genome Database (www.genoscope.cns.fr/brassicanapus) for homology search and basic function analysis.
cDNA Preparation and 5′- and 3′-RACE
Total RNA was extracted from various plant tissues using an RNA extraction kit (RNeasy Plant Mini Kit; QIAGEN). The first-strand cDNA was synthesized using 2 μg of RNA and 200 units of M-MLV reverse transcriptase (Promega Kit) in a volume of 25 μL. The 5′- and 3′-RACE reactions were performed using the SMARTer RACE Amplification Kit (Clontech) according to the manufacturer’s instructions.
Constructs and Transformation
To prepare the BnaC9.SMG7b complementation construct, we amplified a 5,306-bp fragment spanning from 1,252 bp upstream of the translation start codon to 1,823 bp downstream of the termination codon of BnaC9.SMG7b from NIL(Y106). The correct fragment, confirmed by sequencing, was then cloned into the binary vector pCAMBIA3300 (Supplemental Fig. S14A; Supplemental Table S12). To prepare the RNAi construct, we cloned a 174-bp PCR fragment from the second exon of BnaC9.SMG7b cDNA into the pFGC5941 vector in both the sense and antisense orientations (Supplemental Fig. S14B; Supplemental Table S12). To prepare the BnaC9.RING complementation construct, we amplified a 6,126-bp fragment spanning from 2,625 bp upstream of the translation start codon to 1,590 bp downstream of the termination codon of BnaC9.RING and inserted the correct fragment into the binary vector pFGC5941 (Supplemental Fig. S14C; Supplemental Table S12). To prepare the ProBnaC9.SMG7b-GUS construct, we amplified the BnaC9.SMG7b promoter region (from −140 to −2,205 bp) from NIL(Y106) and inserted the correct fragment into pMDC162 upstream of the GUS coding sequence (Supplemental Fig. S14D; Supplemental Table S12). These constructs were introduced into the host cells Agrobacterium tumefaciens GV3101. The complementation constructs were transformed into the low-NSS parent HZ396, whereas the RNAi construct was introduced into a high-NSS inbred line (7-5) by Agrobacterium spp.-mediated transformation (Cardoza and Stewart, 2003). The ProBnaC9.SMG7b-GUS construct was introduced into wild-type Arabidopsis (Columbia-0) by floral dipping (Clough and Bent, 1998). The T2 plants were grown for GUS staining.
Expression Analyses
The reverse-transcribed products from various tissues were used as templates for qPCR/RT-PCR assay using the STC9-300 and STC9-333 primers to examine the expressions of BnaC9.SMG7b and BnaC9.SMG7c, respectively (Supplemental Table S12). qPCR was conducted in a total volume of 25 μL containing 2 μL of reverse-transcribed product, 0.2 mm gene-specific primers, and 12.5 μL of Go Taq qPCR Master Mix (Promega Kit) using the ABI 7500 Real-Time PCR System according to the manufacturer’s instructions. The measurements were obtained using the relative quantification method (Livak and Schmittgen, 2001). The BnACTIN7 (AF111812) gene and Arabidopsis ACTIN7 (At5g09810) were used as the internal controls for B. napus and Arabidopsis genes, respectively. All expression level data obtained by qPCR were based on three biological samples and three replicates for each sample.
Histochemical GUS and Callose Staining
Eleven independent T2 lines were subjected for histochemical GUS assays. Various organs of the transgenic plants were incubated at 37°C overnight in 5-bromo-4-chloro-3-indolyl-β-glucuronic acid solution and then cleaned in 75% (v/v) ethanol. The treated tissues were observed on an Olympus IX-70 Microscope equipped with Nomarski Optics. To observe callose formation during meiotic cytokinesis, pistils from NILs were first fixed in formaldehyde-acetic acid for 1 h at room temperature. After washing with distilled, deionized water, the tissues were incubated in sodium hydroxide solution (6 m) for 12 h. The tissues were washed with distilled, deionized water again and then transferred into 0.1% (w/v) aniline blue with 0.14 m K2HPO4 (pH 8.2) for 24 h. Ovules were dissected from the pistils, mounted in antifade mounting medium, and then viewed and photographed on a fluorescent microscope using a UV filter.
Subcellular Localization
The BnaC9.SMG7b and BnaC9.SMG7c coding sequences without the termination codon (TAA) were amplified by PCR using the primer STC9-368 from NIL(Y106). The amplified cDNA fragments were inserted downstream of the Cauliflower mosaic virus 35S promoter through XmaI and BamHI restriction enzyme sites in frame with GFP in the pGY1-EGFP vector. The AtDCP1 (At1g08370) coding sequence without the termination codon (TGA) was amplified by PCR using the primer STC9-391 and inserted into a premade Red Fluorescent Protein (RFP) vector in luminal-binding protein (BiP)-RFP (Peng et al., 2014) through a BglII restriction enzyme site to generate a fluorescent protein marker for the P bodies (Supplemental Table S12). The BnaC9.SMG7b/c-GFP fusion construct and P bodies protein marker (AtDCP1-RFP) were cotransformed into Arabidopsis protoplasts by polyethylene glycol/calcium-mediated transformation (Yoo et al., 2007). Fluorescence signals were detected by confocal laser-scanning microscopy and visualized using a Leica Microsystem LAS AF. All fluorescence experiments were independently repeated at least three times.
Confocal Microscopy of Megagametogenesis
Pistils from NILs were fixed in formaldehyde-acetic acid for 1 h, with the initial 30 min of fixation performed under a house vacuum (approximately 200 torr), and subsequently kept in 70% (v/v) ethanol overnight at 4°C. The tissues were then incubated in different solutions as follows: 2% (w/v) potassium aluminum for 20 min, 4% (w/v) Suc solution with 10 mg L−1 eosin for 10 to 12 h, and 2% (w/v) potassium aluminum for 20 min. Pistils were then dehydrated in a graded ethanol series (20% steps for 30 min each). After dehydration, the tissues were first cleared in a 1:1 mixture of methyl salicylate:ethanol for 1 h and then cleared in 100% methyl salicylate for 24 h. Ovules dissected out and mounted in 30% (v/v) glycerol were observed using a confocal laser-scanning microscope and visualized using a Leica Microsystem LAS AF with an excitation wavelength of 559 nm and detection wavelengths of 603 to 637 nm.
RNA-Seq
Total RNA extracted from ovaries with lengths <1 mm before pollination was collected for transcriptomic analysis. Total RNA was isolated using an RNA extraction kit (RNeasy Plant Mini Kit; QIAGEN). RNA quality was determined using an Agilent Bioanalyser 2100 (Agilent Technologies, Inc.), and the concentration was measured using an ND-2000 (NanoDrop Technologies). RNA-seq was performed by Novogene Bioinformatics Technology Co., Ltd.
Sequence Tag Preprocessing and Mapping
Sequence tag preprocessing was performed according to a previously described protocol with some modifications. From mRNA sequencing, 30,322,517 and 31,937,078 raw reads were obtained from NIL(HZ396) and NIL(Y106), respectively; after dirty raw reads were removed, 24,556,999 (80.99%) and 25,805,093 (80.8%) clean reads remained, respectively. To identify genes corresponding to reads in each library, the clean reads were aligned to the B. napus reference genome (http://www.genoscope.cns.fr/brassicanapus/) using the TopHat2 software with a maximum allowance of two nucleotide mismatches.
Gene Expression Calculation and AS Event Statistics
Gene expression was calculated using the reads per kilobase transcriptome per million mapped reads method with the HTSeq software (union model). AS events were analyzed using the ASprofile software.
Computational and Database Analyses
Homologs of the BnaC9.SMG7b protein sequence were searched using the BLASTP program of the Phytozome database (www.phytozome.net), The Arabidopsis Information Resource database (http://www.arabidopsis.org), the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), the B. napus database (http://brassicadb.org/brad/index.php), and the Genoscope Genome Database (http://www.genoscope.cns.fr/brassicanapus/). The phylogenetic trees for the plant SMG7 homologs and Brassicaceae SMG7 orthologs were constructed using MEGA 4.0 (http://www.megasoftware.net/). The amino acid sequences were aligned using Geneious 4.8.3.
Sequence data from this article have been deposited in the NCBI GenBank database under accession numbers KT947110, BnaC9.SMG7b full length genomic and protein sequence from NIL(Y106); and KT947111, BnaC9.SMG7c full length genomic and protein sequence from NIL(Y106). The transcriptome data for ovary has been deposited in NCBI Sequence Read Archive database (accession number, SRR2820655).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. NSSs in NIL(Y106), HZ396, and BnaC9.RING-transgenic T0 lines.
Supplemental Figure S2. Comparison of the amino acid sequences of Brassicaceae SMG7.
Supplemental Figure S3. Phylogenetic tree of the Brassicaceae SMG7 proteins.
Supplemental Figure S4. Origin and evolution analysis of the SMG7 homologs.
Supplemental Figure S5. Flower phenotype and pollen fertility of NIL(HZ396) and NIL(Y106).
Supplemental Figure S6. Quantitative analysis of ovule numbers in NIL(HZ396) and NIL(Y106).
Supplemental Figure S7. Fluorescence micrographs of pollen tube germination and growth in NIL(HZ396) and NIL(Y106).
Supplemental Figure S8. qPCR confirmation of the DEGs between NIL(HZ396) and NIL(Y106).
Supplemental Figure S9. Statistics of AS events in NIL(HZ396) and NIL(Y106).
Supplemental Figure S10. Ten-week-old NIL(HZ396) and NIL(Y106) plants.
Supplemental Figure S11. Gene structure and natural variations of BnaC9.SMG7b.
Supplemental Figure S12. BnaC9.SMG7c subcellular localization.
Supplemental Figure S13. Expression patterns of BnaC9.SMG7c by RT-PCR.
Supplemental Figure S14. Schematic presentation of transgenic constructs.
Supplemental Table S1. Progeny testing of the seven recombinants occurring in the interval between SRC9-298 and SRC9-397 in the autumn of 2012 in Wuhan, China.
Supplemental Table S2. NSSs in five T1 BnaC9.SMG7b-transgenic B. napus lines.
Supplemental Table S3. NSSs and BnaC9.SMG7b expression level in Ri2-8 and Ri2-14 T1 families of 7-5 RNAi-transformed plants.
Supplemental Table S4. Confocal laser-scanning microscopy observations of NIL(Y106) and NIL(HZ396) ovules.
Supplemental Table S5. Quantitative analysis of initial ovule number and NSSs in NIL(Y106) and NIL(HZ396).
Supplemental Table S6. Meiosis during megasporogenesis in NIL(Y106) and NIL(HZ396).
Supplemental Table S7. Part of DEGs expression quantification in NIL(HZ396) and NIL(Y106) by RNA-seq.
Supplemental Table S8. Meiotic genes expression quantification in NIL(HZ396) and NIL(Y106) by RNA-seq.
Supplemental Table S9. qSS.C9 genotypes, haplotypes, and NSSs in 84 accessions.
Supplemental Table S10. NSSs in three haplotypes.
Supplemental Table S11. Quantitative analyses of initial ovule number and NSSs in 15 B. napus germplasms.
Supplemental Table S12. Summary of primers used in this study.
Supplementary Material
Acknowledgments
We thank Kede Liu (Huazhong Agricultural University) for providing the DNA and NSS data of Genome-wide Association Study lines for haplotype analysis.
Glossary
- AS
alternative splicing
- cDNA
complementary DNA
- NMD
nonsense-mediated mRNA decay
- NSS
number of seeds per silique
- P body
processing body
- PTC
premature termination codon
- qPCR
quantitative real-time PCR
- QTL
quantitative trait locus
- RNAi
RNA interference
- RNA-seq
RNA sequencing
- RT
reverse transcription
- TSW
thousand-seed weight
- P bodies
processing bodies
Footnotes
This work was supported by the National Natural Science Foundation of China (grant no. 31371251), the China Postdoctoral Science Foundation (grant no. 2012T50653), and the National Program on the Development of Basic Research (grant no. 2015CB150202).
References
- Arciga-Reyes L, Wootton L, Kieffer M, Davies B (2006) UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J 47: 480–489 [DOI] [PubMed] [Google Scholar]
- Basunanda P, Radoev M, Ecke W, Friedt W, Becker HC, Snowdon RJ (2010) Comparative mapping of quantitative trait loci involved in heterosis for seedling and yield traits in oilseed rape (Brassica napus L.). Theor Appl Genet 120: 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulankova P, Riehs-Kearnan N, Nowack MK, Schnittger A, Riha K (2010) Meiotic progression in Arabidopsis is governed by complex regulatory interactions between SMG7, TDM1, and the meiosis I-specific cyclin TAM. Plant Cell 22: 3791–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Xiao Y, Yang W, Ye W, Wang B, Younas M, Wu J, Liu K (2014) Association mapping of six yield‑related traits in rapeseed (Brassica napus L.). Theor Appl Genet 127: 85–96 [DOI] [PubMed] [Google Scholar]
- Cardoza V, Stewart CN Jr (2003) Increased Agrobacterium-mediated transformation and rooting efficiencies in canola (Brassica napus L.) from hypocotyl segment explants. Plant Cell Rep 21: 599–604 [DOI] [PubMed] [Google Scholar]
- Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, et al. (2014) Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345: 950–953 [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Berger F (2001) Maternal control of seed development. Semin Cell Dev Biol 12: 381–386 [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Koltunow A, Payne T, Luo M, Tucker MR, Dennis ES, Peacock WJ (2001) Control of early seed development. Annu Rev Cell Dev Biol 17: 677–699 [DOI] [PubMed] [Google Scholar]
- Chen W, Zhang Y, Liu X, Chen B, Tu J, Tingdong F (2007) Detection of QTL for six yield-related traits in oilseed rape (Brassica napus) using DH and immortalized F(2) populations. Theor Appl Genet 115: 849–858 [DOI] [PubMed] [Google Scholar]
- Chen W, Zhang Y, Yao J, Ma C, Tu J, Tingdong F (2011) Quantitative trait loci mapping for two seed yield component traits in an oilseed rape (Brassica napus) cross. Plant Breed 130: 640–646 [Google Scholar]
- Cheng F, Mandáková T, Wu J, Xie Q, Lysak MA, Wang X (2013) Deciphering the diploid ancestral genome of the Mesohexaploid Brassica rapa. Plant Cell 25: 1541–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen CA, King EJ, Jordan JR, Drews GN (1997) Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex Plant Reprod 10: 49–64 [Google Scholar]
- Clarke JM, Simpson GM (1978) Influence of irrigation and seeding rates on yield and yield components of Brassica napus cv. Tower. Can J Plant 58: 731–737 [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH (2008) Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9: 938–950 [DOI] [PubMed] [Google Scholar]
- Cromer L, Heyman J, Touati S, Harashima H, Araou E, Girard C, Horlow C, Wassmann K, Schnittger A, De Veylder L, et al. (2012) OSD1 promotes meiotic progression via APC/C inhibition and forms a regulatory network with TDM and CYCA1;2/TAM. PLoS Genet 8: e1002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooijmans RP, Vrebalov J, Dijkhof RJ, van der Poel JJ, Groenen MA (2000) Two-dimensional screening of the Wageningen chicken BAC library. Mamm Genome 11: 360–363 [DOI] [PubMed] [Google Scholar]
- d’Erfurth I, Cromer L, Jolivet S, Girard C, Horlow C, Sun Y, To JPC, Berchowitz LE, Copenhaver GP, Mercier R (2010) The cyclin-A CYCA1;2/TAM is required for the meiosis I to meiosis II transition and cooperates with OSD1 for the prophase to first meiotic division transition. PLoS Genet 6: e1000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Erfurth I, Jolivet S, Froger N, Catrice O, Novatchkova M, Simon M, Jenczewski E, Mercier R (2008) Mutations in AtPS1 (Arabidopsis thaliana parallel spindle 1) lead to the production of diploid pollen grains. PLoS Genet 4: e1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Storme N, Geelen D (2011) The Arabidopsis mutant jason produces unreduced first division restitution male gametes through a parallel/fused spindle mechanism in meiosis II. Plant Physiol 155: 1403–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissmeyer N, Nowack MK, Pusch S, Stals H, Inzé D, Grini PE, Schnittger A (2007) T-loop phosphorylation of Arabidopsis CDKA;1 is required for its function and can be partially substituted by an aspartate residue. Plant Cell 19: 972–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E (2007) P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Fan C, Cai G, Qin J, Li Q, Yang M, Wu J, Fu T, Liu K, Zhou Y (2010) Mapping of quantitative trait loci and development of allele-specific markers for seed weight in Brassica napus. Theor Appl Genet 121: 1289–1301 [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E (2005) SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell 17: 537–547 [DOI] [PubMed] [Google Scholar]
- Galbiati F, Sinha Roy D, Simonini S, Cucinotta M, Ceccato L, Cuesta C, Simaskova M, Benkova E, Kamiuchi Y, Aida M, et al. (2013) An integrative model of the control of ovule primordia formation. Plant J 76: 446–455 [DOI] [PubMed] [Google Scholar]
- Hamant O, Ma H, Cande WZ (2006) Genetics of meiotic prophase I in plants. Annu Rev Plant Biol 57: 267–302 [DOI] [PubMed] [Google Scholar]
- Harashima H, Dissmeyer N, Schnittger A (2013) Cell cycle control across the eukaryotic kingdom. Trends Cell Biol 23: 345–356 [DOI] [PubMed] [Google Scholar]
- Higashiyama T. (2002) The synergid cell: attractor and acceptor of the pollen tube for double fertilization. J Plant Res 115: 149–160 [DOI] [PubMed] [Google Scholar]
- Hori K, Watanabe Y (2005) UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J 43: 530–540 [DOI] [PubMed] [Google Scholar]
- Jones GH, Armstrong SJ, Caryl AP, Franklin FCH (2003) Meiotic chromosome synapsis and recombination in Arabidopsis thaliana; an integration of cytological and molecular approaches. Chromosome Res 11: 205–215 [DOI] [PubMed] [Google Scholar]
- Kulkarni M, Ozgur S, Stoecklin G (2010) On track with P-bodies. Biochem Soc Trans 38: 242–251 [DOI] [PubMed] [Google Scholar]
- Li YP, Cheng Y, Cai GQ, Fan CC, Zhou YM (2014) Cytological basis and molecular mechanism of variation in number of seeds per pod in Brassica napus. Sci Sin 44: 822–831 [Google Scholar]
- Li YY, Shen JX, Wang TH, Fu TD, Ma CZ (2007) Construction of a linkage map using SRAP, SSR and AFLP markers in Brassica napus L. Sci Agric Sin 40: 1118–1126 [Google Scholar]
- Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin IA, Zhao M, Ma J, Yu J, Huang S, et al. (2014) The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun 5: 3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Adams KL (2010) Dramatic change in function and expression pattern of a gene duplicated by polyploidy created a paternal effect gene in the Brassicaceae. Mol Biol Evol 27: 2817–2828 [DOI] [PubMed] [Google Scholar]
- Liu Z, Makaroff CA (2006) Arabidopsis separase AESP is essential for embryo development and the release of cohesin during meiosis. Plant Cell 18: 1213–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lord EM, Russell SD (2002) The mechanisms of pollination and fertilization in plants. Annu Rev Cell Dev Biol 18: 81–105 [DOI] [PubMed] [Google Scholar]
- Mérai Z, Benkovics AH, Nyikó T, Debreczeny M, Hiripi L, Kerényi Z, Kondorosi É, Silhavy D (2013) The late steps of plant nonsense-mediated mRNA decay. Plant J 73: 50–62 [DOI] [PubMed] [Google Scholar]
- Mercier R, Grelon M (2008) Meiosis in plants: ten years of gene discovery. Cytogenet Genome Res 120: 281–290 [DOI] [PubMed] [Google Scholar]
- Mercier R, Mézard C, Jenczewski E, Macaisne N, Grelon M (2015) The molecular biology of meiosis in plants. Annu Rev Plant Biol 66: 297–327 [DOI] [PubMed] [Google Scholar]
- Peng B, Kong H, Li Y, Wang L, Zhong M, Sun L, Gao G, Zhang Q, Luo L, Wang G, et al. (2014) OsAAP6 functions as an important regulator of grain protein content and nutritional quality in rice. Nat Commun 5: 4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Mao L, Sun C, Pu Y, Fu T, Ma C, Shen J, Tu J, Yi B, Wen J (2014) Interpreting the genetic basis of silique traits in Brassica napus using a joint QTL network. Plant Breed 133: 52–60 [Google Scholar]
- Qin Y, Zhao L, Skaggs MI, Andreuzza S, Tsukamoto T, Panoli A, Wallace KN, Smith S, Siddiqi I, Yang Z, et al. (2014) ACTIN-RELATED PROTEIN6 regulates female meiosis by modulating meiotic gene expression in Arabidopsis. Plant Cell 26: 1612–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quijada PA, Udall JA, Lambert B, Osborn TC (2006) Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.). I. Identification of genomic regions from winter germplasm. Theor Appl Genet 113: 549–561 [DOI] [PubMed] [Google Scholar]
- Radoev M, Becker HC, Ecke W (2008) Genetic analysis of heterosis for yield and yield components in rapeseed (Brassica napus L.) by quantitative trait locus mapping. Genetics 179: 1547–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray SM, Park SS, Ray A (1997) Pollen tube guidance by the female gametophyte. Development 124: 2489–2498 [DOI] [PubMed] [Google Scholar]
- Reddy TV, Kaur J, Agashe B, Sundaresan V, Siddiqi I (2003) The DUET gene is necessary for chromosome organization and progression during male meiosis in Arabidopsis and encodes a PHD finger protein. Development 130: 5975–5987 [DOI] [PubMed] [Google Scholar]
- Reyes-Olalde JI, Zuñiga-Mayo VM, Chávez Montes RA, Marsch-Martínez N, de Folter S (2013) Inside the gynoecium: at the carpel margin. Trends Plant Sci 18: 644–655 [DOI] [PubMed] [Google Scholar]
- Riehs N (2009) Functional characterization of the Arabidopsis SMG7 protein. PhD thesis. Uniwien, Vienna [Google Scholar]
- Riehs N, Akimcheva S, Puizina J, Bulankova P, Idol RA, Siroky J, Schleiffer A, Schweizer D, Shippen DE, Riha K (2008) Arabidopsis SMG7 protein is required for exit from meiosis. J Cell Sci 121: 2208–2216 [DOI] [PubMed] [Google Scholar]
- Riehs-Kearnan N, Gloggnitzer J, Dekrout B, Jonak C, Riha K (2012) Aberrant growth and lethality of Arabidopsis deficient in nonsense-mediated RNA decay factors is caused by autoimmune-like response. Nucleic Acids Res 40: 5615–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodkiewicz B. (1970) Callose in cell walls during megasporogenesis in angiosperms. Planta 93: 39–47 [DOI] [PubMed] [Google Scholar]
- Rondanini DP, Gomez NV, Agosti MB, Miralles DJ (2012) Global trends of rapeseed grain yield stability and rapeseed-to-wheat yield ratio in the last four decades. Eur J Agron 37: 56–65 [Google Scholar]
- Shi DQ, Yang WC (2011) Ovule development in Arabidopsis: progress and challenge. Curr Opin Plant Biol 14: 74–80 [DOI] [PubMed] [Google Scholar]
- Shi J, Li R, Qiu D, Jiang C, Long Y, Morgan C, Bancroft I, Zhao J, Meng J (2009) Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics 182: 851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Udall JA, Quijada PA, Lambert B, Osborn TC (2006) Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.). II. Identification of alleles from unadapted germplasm. Theor Appl Genet 113: 597–609 [DOI] [PubMed] [Google Scholar]
- Wang J, Long Y, Wu B, Liu J, Jiang C, Shi L, Zhao J, King GJ, Meng J (2009) The evolution of Brassica napus FLOWERING LOCUS T paralogues in the context of inverted chromosomal duplication blocks. BMC Evol Biol 9: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, et al. (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43: 1035–1039 [DOI] [PubMed] [Google Scholar]
- Wang Y, Yang M (2006) The ARABIDOPSIS SKP1-LIKE1 (ASK1) protein acts predominately from leptotene to pachytene and represses homologous recombination in male meiosis. Planta 223: 613–617 [DOI] [PubMed] [Google Scholar]
- Wijnker E, Schnittger A (2013) Control of the meiotic cell division program in plants. Plant Reprod 26: 143–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Hu Y, Lodhi M, McCombie WR, Ma H (1999) The Arabidopsis SKP1-LIKE1 gene is essential for male meiosis and may control homologue separation. Proc Natl Acad Sci USA 96: 11416–11421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Shu C, Chen L, Xu J, Wu J, Liu K (2012) Identification of a major QTL for silique length and seed weight in oilseed rape (Brassica napus L.). Theor Appl Genet 125: 285–296 [DOI] [PubMed] [Google Scholar]
- Yang X, Makaroff CA, Ma H (2003) The Arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell 15: 1281–1295 [PMC free article] [PubMed] [Google Scholar]
- Yoine M, Nishii T, Nakamura K (2006) Arabidopsis UPF1 RNA helicase for nonsense-mediated mRNA decay is involved in seed size control and is essential for growth. Plant Cell Physiol 47: 572–580 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang F, Zhao YG, Gu TC, Zhang DX, Liu FL, Guo RX, Fu GP, Zhang XK (2012a) Yield and agronomic traits of winter rapeseed cultivars registered in China from 2001 to 2010. Chin J Oil Crop Sci 34: 239–244 [Google Scholar]
- Zhang L, Li S, Chen L, Yang G (2012b) Identification and mapping of a major dominant quantitative trait locus controlling seeds per silique as a single Mendelian factor in Brassica napus L. Theor Appl Genet 125: 695–705 [DOI] [PubMed] [Google Scholar]
- Zhang L, Yang G, Liu P, Hong D, Li S, He Q (2011) Genetic and correlation analysis of silique-traits in Brassica napus L. by quantitative trait locus mapping. Theor Appl Genet 122: 21–31 [DOI] [PubMed] [Google Scholar]
- Zhao D, Yang X, Quan L, Timofejeva L, Rigel NW, Ma H, Makaroff CA (2006) ASK1, a SKP1 homolog, is required for nuclear reorganization, presynaptic homolog juxtaposition and the proper distribution of cohesin during meiosis in Arabidopsis. Plant Mol Biol 62: 99–110 [DOI] [PubMed] [Google Scholar]
- Zhao L, He J, Cai H, Lin H, Li Y, Liu R, Yang Z, Qin Y (2014) Comparative expression profiling reveals gene functions in female meiosis and gametophyte development in Arabidopsis. Plant J 80: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.