A Pooideae orphan protein that enhances plant resistance against a toxigenic fungus physically interacts with a key signaling protein.
Abstract
All genomes encode taxonomically restricted orphan genes, and the vast majority are of unknown function. There is growing evidence that such genes play an important role in the environmental adaptation of taxa. We report the functional characterization of an orphan gene (Triticum aestivum Fusarium Resistance Orphan Gene [TaFROG]) as a component of resistance to the globally important wheat (T. aestivum) disease, Fusarium head blight. TaFROG is taxonomically restricted to the grass subfamily Pooideae. Gene expression studies showed that it is a component of the early wheat response to the mycotoxin deoxynivalenol (DON), which is a virulence factor produced by the causal fungal agent of Fusarium head blight, Fusarium graminearum. The temporal induction of TaFROG by F. graminearum in wheat spikelets correlated with the activation of the defense Triticum aestivum Pathogenesis-Related-1 (TaPR1) gene. But unlike TaPR1, TaFROG induction by F. graminearum was toxin dependent, as determined via comparative analysis of the effects of wild-type fungus and a DON minus mutant derivative. Using virus-induced gene silencing and overexpressing transgenic wheat lines, we present evidence that TaFROG contributes to host resistance to both DON and F. graminearum. TaFROG is an intrinsically disordered protein, and it localized to the nucleus. A wheat alpha subunit of the Sucrose Non-Fermenting1-Related Kinase1 was identified as a TaFROG-interacting protein based on a yeast two-hybrid study. In planta bimolecular fluorescence complementation assays confirmed the interaction. Thus, we conclude that TaFROG encodes a new Sucrose Non-Fermenting1-Related Kinase1-interacting protein and enhances biotic stress resistance.
It is estimated that wheat (Triticum aestivum) yields must at least double by 2050 to meet growing future demands. We lose billions of dollars’ worth of wheat grain annually due to diseases caused by fungi (Dean et al., 2012). Some fungal diseases also threaten health because the causal organisms contaminate grain with mycotoxins that are harmful to humans and animals (Arunachalam and Doohan, 2013). Breeding for disease resistance is the most environmentally sustainable method of disease control. Resistance to disease can be quantitative nonspecific or qualitative, pathogen race specific. We must expand our portfolio of wheat resistance genes selected for within-breeding programs if we want to deploy sustainable resistance to a broad spectrum of pathogens. This will be most easily achieved if we obtain better knowledge of the mechanisms and the transcriptional patterns important to disease resistance within wheat (Bischof et al., 2011).
Many of the plant genes activated in response to pathogens are of unknown function. This includes taxonomically restricted orphan genes that lack homologs in other plant families (Arendsee et al., 2014). A role for orphan genes in lineage-specific adaptations is supported by a growing number of examples from a spectrum of diverse organisms, including plants (Tautz and Domazet-Lošo, 2011; Arendsee et al., 2014). Orphans were enriched in differentially expressed gene sets from Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) following treatments with hormones, abiotic, or biotic stressors (Guo et al., 2007; Donoghue et al., 2011). Arabidopsis mutant screens identified several plant orphan genes that alter abiotic stress resistance (Luhua et al., 2013). Examples detailing the functional role of orphan genes in plant disease responses are rare. The Brassicaceae-specific Enhancer of Vascular Wilt Resistance1 orphan gene provides resistance to vascular wilt pathogens (Yadeta et al., 2014), whereas the rice orphan gene Oryza sativa Defense-Responsive Gene10 has a negative effect on plant resistance to bacterial blight disease (Xiao et al., 2009).
Here, we characterize a wheat orphan gene that enhances resistance to Fusarium head blight (FHB) disease, and thus was designated Triticum aestivum Fusarium Resistance Orphan Gene (TaFROG). This gene was originally identified as being responsive to a mycotoxic virulence factor, deoxynivalenol (DON), which is produced by the causal fungal agent of FHB, Fusarium graminearum. This fungus produces DON to facilitate disease spread within wheat head tissue (Bai et al., 2002). The disease results in yield loss and contamination of grain with DON that is harmful to human and animal health (Rocha et al., 2005). Gene expression studies evaluated the effect of DON synthesis on the responsiveness of this gene to F. graminearum. Gene silencing and overexpression studies assessed the contribution of this gene to DON tolerance and FHB resistance. Microscopic analysis of transgenic plants was used to determine the subcellular location of TaFROG. A yeast two-hybrid screen against a wheat complementary DNA (cDNA) library identified a wheat alpha subunit of the Sucrose Non-Fermenting1-Related Kinase1 (TaSnRK1α) as a TaFROG-interacting protein, and the interaction was confirmed in planta using bimolecular fluorescence complementation. On the basis of this study, we have identified a wheat orphan gene that enhances disease resistance and determined that it interacts with a central stress regulator, TaSnRK1α.
RESULTS
TaFROG Is an Orphan Gene, Taxonomically Restricted to the Pooideae
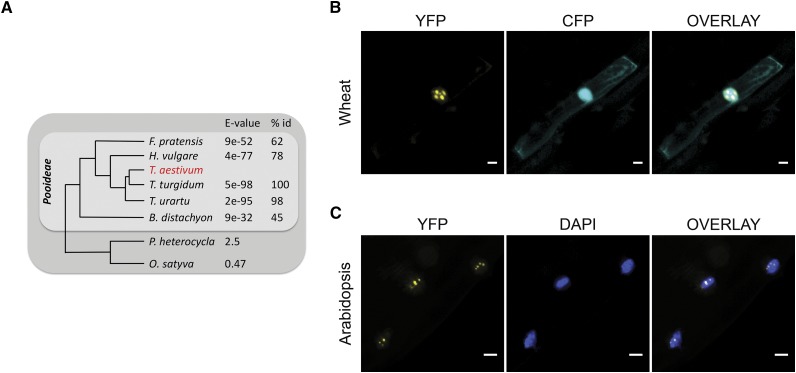
Previous studies within our laboratory identified a unique wheat transcript, responsive to F. graminearum and DON. Using RACE-PCR, we cloned the full-length open reading frame of this gene (TaFROG) from bread wheat ‘CM82036.’ The deduced coding sequence shares 100%, 93%, and 85% homology with sequences on chromosomes 4A, 4D, and 4B, respectively, of the recently published wheat cv Chinese Spring genome (International Wheat Genome Sequencing Consortium, 2014). TaFROG homologs were found within the Triticum spp. genus (E < 10−83). They were restricted to the Pooideae subfamily of Poaceae (E < 10−4; Fig. 1A). TaFROG gene variants were not identified outside this subfamily, either in transcript databases or in any other sequenced monocotyledon genome. TaFROG shared no significant homology with any characterized gene or protein. The protein did not contain any characterized domain. Thus, we conclude that TaFROG is a unique orphan gene: orphan genes are strictly defined as having a narrow phylogenetic distribution and not encoding previously identified protein domains (Khalturin et al., 2009).
Figure 1.
Phylogenetic analysis and subcellular localization of TaFROG. A, Phylogenetic tree showing grass species that contain homologs of TaFROG. BLAST E-value and the percentage of protein sequence identity (% id) calculated using TaFROG from the chromosome 4A are presented. B and C, Microscopic analyses of TaFROG within the nucleus of wheat and Arabidopsis cells, respectively. Wheat seedling leaves were biolistically cotransformed with vectors harboring either CFP or TaFROG-YFP. Stable plant roots overexpressing TaFROG-YFP were analyzed for Arabidopsis. Cells were observed by confocal microscopy for wheat leaves or by light sheet fluorescence microscopy for Arabidopsis roots. YFP and either CFP or 4′,6- diamidino-phenylindole (DAPI) images are shown both separately and as an overlay. Scale bar = 10 µm.
FROG Represents a New Family of Intrinsically Disordered Nuclear Proteins
The FROG family are small basic proteins of between 121 and 142 amino acids (Supplemental Fig. S1) and represent new intrinsically disordered proteins (IDPs) with a highly negative folding index profile determined using the FoldIndex prediction tool (Prilusky et al., 2005; Supplemental Fig. S1). Between 62% and 72% of protein residues are predicted to be disordered, and these lie within four conserved regions (Supplemental Fig. S1). FROG encodes two conserved motifs: a nuclear export signal and a nuclear localization sequence, as shown in Supplemental Figure S1, suggesting FROG can localize in the nucleus. We fused wheat FROG (TaFROG, cloned from chromosome 4A of cv CM82036) to yellow fluorescent protein (YFP) and transiently or stably expressed TaFROG-YFP in wheat leaves or Arabidopsis plants, respectively. In wheat epidermal cells, confocal microscopy analyses showed that TaFROG-YFP was restricted to the nucleus within distinct nuclear bodies (Fig. 1B), unlike coexpressed Cyan Fluorescent Protein (CFP), which was found in both the cytosol and the nucleus. In stable transgenic Arabidopsis lines, light sheet fluorescence microscopy detected TaFROG-YFP in bodies localized within the DAPI-stained nuclei (Fig. 1C). The integrity of Arabidopsis TaFROG-YFP fusion was confirmed via western-blot analysis (Supplemental Fig. S2).
TaFROG Is a Mycotoxin-Responsive Orphan Gene
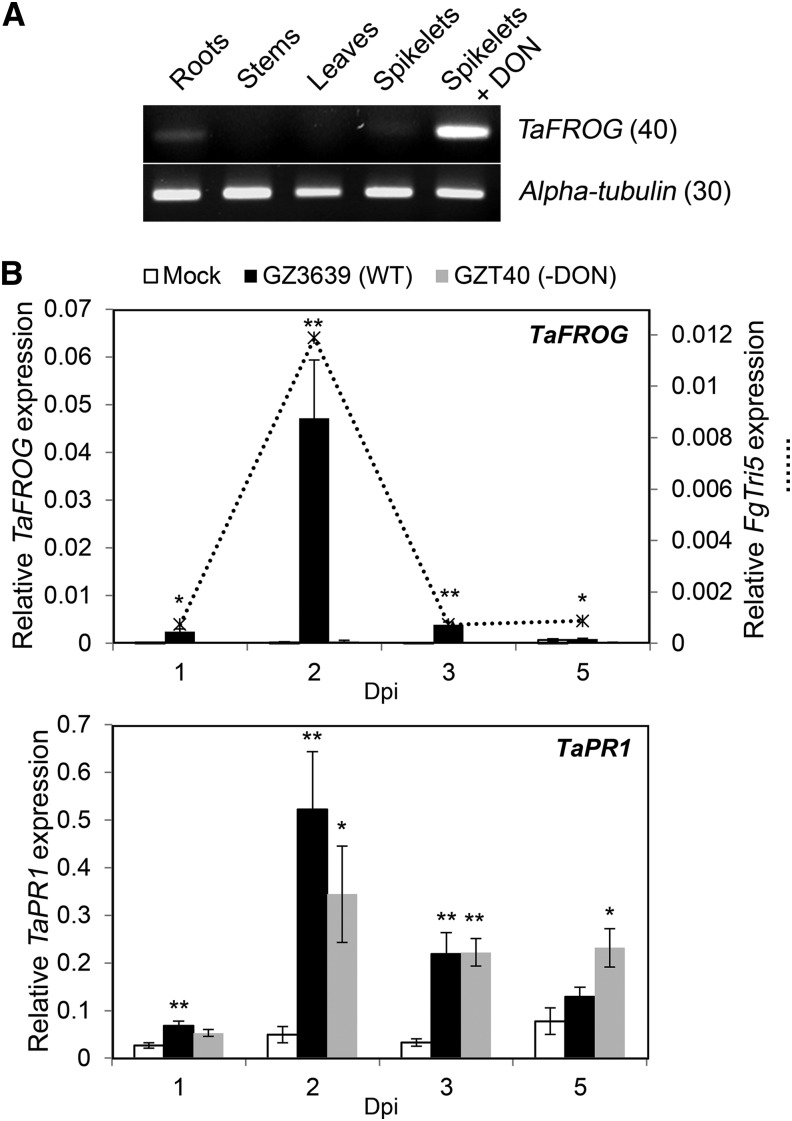
The tissue specificity and DON responsiveness of TaFROG (chromosome 4A homeolog) were analyzed using semiquantitative reverse transcription (sqRT)-PCR and real-time quantitative reverse transcriptase (qRT)-PCR analyses. In mock-treated wheat tissues, the basal expression of TaFROG was below or near detectable limits, in contrast to the high level of gene expression in DON-treated tissues (Fig. 2A; Supplemental Fig. S3). Thus, TaFROG transcription is induced by the toxigenic F. graminearum virulence factor DON. To determine if the gene was activated by F. graminearum, the level of expression was measured by qRT-PCR in wheat head spikelets at 1 to 5 d postfungal inoculation (dpi). The result showed that the fungus induces TaFROG as early as 1 dpi, induction peaking at 2 dpi (Fig. 2B). We also studied Fusarium graminearum Trichodiene synthase5 (FgTri5) transcription in the same tissue, which is a key gene in the fungal DON biosynthetic pathway, and its expression positively correlates with the production of DON (Gardiner et al., 2009). In F. graminearum-infected wheat spikelets, qRT-PCR showed that the peak FgTri5 expression coincided with the peak in wheat TaFROG expression (2 dpi), both the fungal and wheat gene presenting the same temporal expression profile (Fig. 2B). Based on the fact that TaFROG was responsive to both DON and its producer, we hypothesized that toxin biosynthesis might be a requirement for the fungal activation of this gene. Comparative analysis of the response of TaFROG to both wild-type fungus and its less virulent non-DON-producing mutant derivative (GZT40) enabled us to test this hypothesis. Both the mutant and the wild type elicited a similar defense response, as represented by the defense Pathogenesis-Related Gene1 marker gene Triticum aestivum Pathogenesis-Related-1 (TaPR1; Fig. 2B), previously described to be induced as an early response to F. graminearum infection (Pritsch et al., 2000). But, unlike wild-type strain GZ3639, mutant GZT40 did not induce TaFROG expression in wheat spikelets (Fig. 2B). Thus, we conclude that TaFROG is a component of the early host response to F. graminearum, elicited in response to DON. The role of TaFROG in the mycotoxin response is not restricted to the chromosome 4A homeolog. The chromosome 4B, 4D homeologs and the barley (Hordeum vulgare) TaFROG homolog share the same DON-dependent expression pattern in tissues treated with DON or heads infected with F. graminearum (Supplemental Fig. S3; Boddu et al., 2007; Gardiner et al., 2010).
Figure 2.
TaFROG transcript levels in wheat tissues after treatment with DON or F. graminearum. A, Tissues at different wheat development stages and flowering spikelets treated with DON. After 24 h, TaFROG gene expression was assessed via sqRT-PCR (number in parenthesis = number of PCR cycles). B, TaFROG, FgTri5 (top graph) and TaPR1 (bottom graph) gene expression in wheat heads treated with F. graminearum was assessed via qRT-PCR. The wheat alpha-tubulin and F. graminearum FgActin genes were used as internal reference to calculate the relative expression of FgTri5 and TaFROG genes respectively, using the equation 2-(Ct target gene – Ct housekeeping gene). Wheat spikelets were treated with wild-type (WT) F. graminearum GZ3639, its DON-minus mutant derivative GZT40, or mock. The tissues were harvested at various days postinoculation as indicated. Results represent the mean (two biological replicates and four technical replicates per treatment from a pooled biological sample) and error bars indicate ±sem (n = 8). Asterisks show significant differences between treatments and mock (Kruskal-Wallis test; *, P < 0.05; and **, P < 0.01).
TaFROG Contributed to DON Tolerance
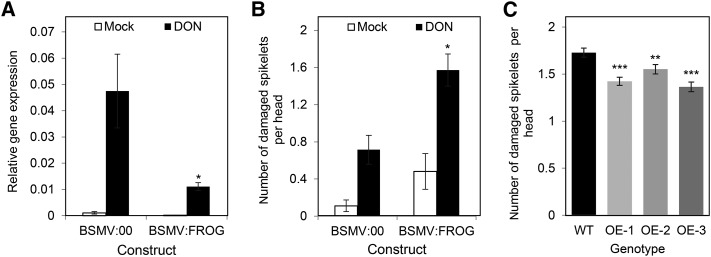
Virus-induced gene silencing (VIGS) was used to determine if reducing TaFROG transcript levels altered the phenotypic response to DON in a toxin-resistant wheat genotype. qRT-PCR confirmed that VIGS worked efficiently. DON treatment of central head spikelets induced TaFROG, but in gene-silenced plants (barley stripe mosaic virus [BSMV]:FROG plants), the DON induction of TaFROG was reduced by 77%, as compared with the effect of DON on plants treated with the mock virus (BSMV:00; Fig. 3A). In the absence of DON, minimal TaFROG expression was observed, both in BSMV:FROG- and BSMV:00-treated plants (Fig. 3A). At a phenotypic level, assessment of heads at 14 d post-toxin treatment showed that BSMV:FROG plants were more sensitive to DON-induced damage than the BSMV:00 plants (Fig. 3B). Silencing of TaFROG resulted in a 2.2-fold increase in the number of DON-damaged spikelets (in BSMV:FROG versus BSMV:00 plants). Note that we obtained similar results when we conducted an independent experiment using a different BSMV construct targeting a larger fragment of the TaFROG gene (Supplemental Figs. S4 and S5).
Figure 3.
Effect of TaFROG silencing and overexpression on DON tolerance. A and B, In the VIGS experiment, gene silencing of TaFROG in wheat spikelets was quantified by qRT-PCR analysis (A) and the phenotypic response to DON (B) was assessed at 14 d post-toxin treatment. For the qRT-PCR analysis, the wheat alpha-tubulin gene was used as an internal reference to calculate the relative expression of the TaFROG gene using the equation 2-(Ct target gene – Ct housekeeping gene). VIGS constructs used were empty vector BSMV:00 or the construct BSMV:FROG that targets the TaFROG gene for silencing. Constructs were applied to the flag leaf, and the central flowering spikelets were treated with either 16.87 mm DON or Tween 20 (mock). C, The phenotypic response of TaFROG overexpressor lines (OE-1, OE-2, and OE-3) and control plants (wild type [WT]) to DON treatment (as above) at 21 d post-toxin treatment. Asterisks show significant differences between DON-treated (A and B) BSMV:00 and BSMV:FROG constructs or (C) overexpressor lines and the wild type (Mann-Whitney U test; *, P < 0.05; **, P < 0.01; and ***, P < 0.001). Error bars indicate ±sem (A and B: n = 20–36; C: n = 60).
The results of the VIGS experiments suggested a direct role for TaFROG in DON tolerance. To confirm this, we generated transgenic wheat (‘Fielder’) lines overexpressing TaFROG and tested their phenotypic response to DON and F. graminearum. Transgenic lines OE-1, OE-2, and OE-3 were selected for F. graminearum studies on the basis that they exhibited a 218-, 445-, and 80-fold increase, respectively, in TaFROG gene expression, as compared with wild-type plants (Supplemental Fig. S6). We evaluated the phenotypic effect of DON on spikelets 21 d post-toxin treatment. Figure 3C illustrates that DON-induced damage of spikelets was significantly reduced for the three overexpressor lines, as compared with the wild-type plants, thus confirming the role of TaFROG in DON tolerance.
TaFROG Enhances Wheat Leaf Resistance to F. graminearum
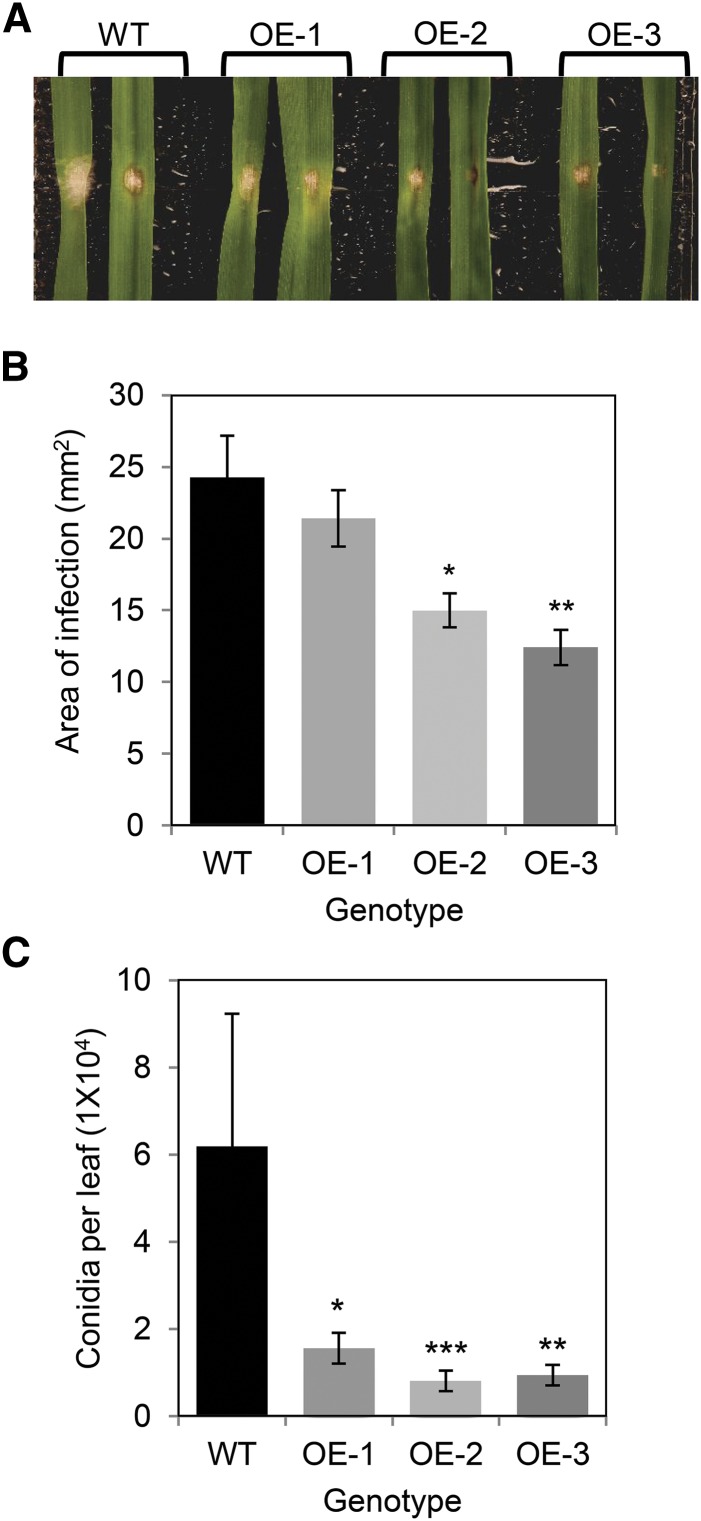
DON facilitates disease spread in wheat heads (Bai et al., 2002) and, based on its contribution to DON tolerance, we hypothesized that TaFROG might enhance Fusarium resistance. Using a detached leaf assay, we evaluated the spread and sporulation of F. graminearum (strain GZ3639) on transgenic lines OE-1, OE-2, and OE-3, relative to wild-type plants. Amending the fungal inoculum with DON (75 µm) was previously reported to increase the reproducibility of results within detached leaf assays (Chen et al., 2006). Thus, we performed two experiments: one using F. graminearum plus DON as the treatment (Supplemental Fig. S7) and the other using only the fungus (Fig. 4). Similar results were obtained for both experiments, disease and sporulation being reduced in the overexpressor as compared with wild-type plants. Thus, the overexpression of TaFROG enhanced wheat resistance to colonization by F. graminearum.
Figure 4.
Effect of TaFROG overexpression on wheat leaf resistance to F. graminearum. TaFROG overexpressor lines (OE-1, OE-2, and OE-3) and control plants (wild type [WT]) were used for phenotypic analysis. Representative leaf symptoms (A), diseased leaf area (B), and conidia production (C) were determined 4 d post-treatment of detached wheat leaves with F. graminearum. Error bars indicate ±sem (B and C: n = 36). Asterisks show significant differences compared with the wild type (B: Tukey's honestly significant difference test; C: Mann-Whitney U test; *, P < 0.05; **, P < 0.01; and ***, P < 0.001).
Overexpression of TaFROG Retards the Spread of FHB Disease Symptoms
We evaluated the effect of TaFROG overexpression on the spread of FHB symptoms from inoculated central spikelets of wheat heads. Results showed that wild-type cv Fielder had an average of 5.1 diseased spikelets at 21 dpi. All three transgenic lines exhibited less disease spread; significant reductions of 23% and 17% were observed for OE-1 and OE-2, respectively, relative to wild-type plants (Fig. 5A; Supplemental Fig. S8). The reduction observed for OE-3 at 21 d post-treatment was not significant; however, like OE1, the disease progression for line OE3 (evaluated as AUDPC calculated using disease scores from 7, 14, and 21 dpi) was significantly lower than on wild-type plants (Fig. 5B).
Figure 5.
Effect of TaFROG overexpression on the susceptibility of wheat to FHB disease. At mid-anthesis, central flowering spikelets from overexpressor lines (OE-1, OE-2, and OE-3) or control plants (wild type [WT]) were point inoculated (A and B) or spray inoculated (C and D) with F. graminearum. Disease was assessed at different days postinoculation, and data presented correspond to the score (A) or percentage (C) of infected spikelets per head at 21 d, or to the area under the disease progress curve (AUDPC; B and D). Error bars indicate ±sem (n = 60). Asterisks show significant differences compared with the wild type (Mann-Whitney U test; *, P < 0.05; **, P < 0.01; and ***, P < 0.001).
The enhanced resistance of the transgenic lines relative to the wild type was confirmed under higher disease pressure, where whole heads were sprayed with the pathogen. All three transgenic lines showed fewer disease symptoms at 10 dpi, as depicted in Supplemental Figure S8. By 21 dpi, 51% and 49% of the spikelets on OE-1 and OE-2 exhibited disease symptoms as compared with 57% on the wild-type plants (Fig. 5C). No reduction in disease was observed for OE-3 at this time point, but like OE-1 and OE-2, the disease progression for line OE3 (evaluated as AUDPC) was significantly lower than on wild-type plants (25%–15% lower, as depicted in Fig. 5D). Overall, the results of both point and spray inoculation experiments demonstrated that overexpression of TaFROG provided quantitative resistance to FHB.
TaFROG, a New SnRK1-Interacting Protein
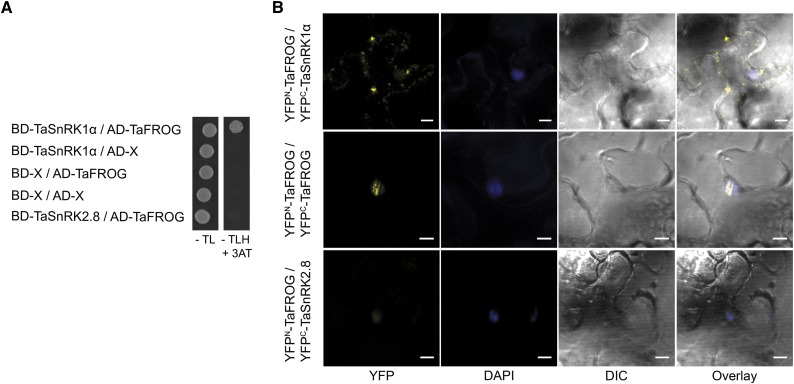
TaFROG is an IDP; the unstructured properties of IDPs allow them to bind multiple protein partners and form an important component of the cellular signaling machinery (Wright and Dyson, 2015). A yeast two-hybrid screen was conducted to determine if TaFROG could interact with components of the plant cell signaling machinery. Using TaFROG as bait, we screened a cDNA expression library generated from DON-treated wheat tissue and thus identified putative interacting proteins, including NAC transcription factors, a histone-binding protein, and the kinase TaSnRK1α. Among the putative interacting enzymes, TaSnRK1α was selected for further analysis. TaSnRK1α is a wheat alpha subunit of an SNF1-related kinase 1, a central regulator of energy and stress signaling in plants (Halford and Hey, 2009). The protein-protein interaction in yeast (Saccharomyces cerevisiae) was confirmed using a galactose-responsive transcription factor GAL4 (GAL4)-based yeast two-hybrid system (Fig. 6A; Supplemental Fig. S9 for serial dilution results; Supplemental Fig. S9 for vector-swapping results). On the contrary, no growth was observed when we combined TaFROG with another SnRK, TaSnRK2.8, or with the empty vectors. Protein expression from constructs was confirmed by western-blot analysis (Supplemental Fig. S9). Thus, we confirmed that TaFROG is able to interact with TaSnRK1α in yeast.
Figure 6.
Interaction of TaFROG with TaSnRK1α. A, Yeast two-hybrid assay using the yeast cotransformed with TaSnRK1α and TaFROG cloned in the Gal4 bait/prey vectors. Yeast was grown for 5 d on Trp/Leu drop-out medium (−TL) or on selective Trp/Leu/His drop-out medium (−TLH) plus 3-amino-1,2,4-triazole (3-AT). B, In planta protein-protein interaction visualized by the bimolecular fluorescence complementation (BiFC) assay. Confocal microscopy images of representative N. benthamiana epidermal leaf cells expressing proteins fused to the N- or C-terminal part of the YFP as indicated. YFP, DAPI fluorescence, and differential interference contrast (DIC) images are shown both separated and as an overlay. Scale bar indicates 10 µm. In A and B, TaSnRK2.8 was used as a negative control. BD, Binding domain; AD, activating domain.
We further investigated the interaction between TaFROG and TaSNRK1α in planta using the BiFC system (Gehl et al., 2009). TaFROG and TaSNRK1α genes were fused at their N terminus with either the N- or C-terminal part of YFP. The resulting constructs were transiently coexpressed in Nicotiana benthamiana leaves. We observed a strong YFP signal, predominantly in punctate cytoplasmic bodies, when YFPN-TaFROG was combined with YFPC-TaSNRK1α (Fig. 6B). A smaller number of larger cytoplasmic bodies with an irregular shape was also detected, potentially corresponding to protein aggregates. Similar results were obtained with different combinations of TaSNRK1α and TaFROG (TaFROG-YFPC/TaSnRK1α-YFPN or YFPC-TaFROG/TaSnRK1α-YFPN), the only difference being that we occasionally detected fluorescent punctate bodies in the nucleus of cells (see YFPC-TaFROG/TaSnRK1α-YFPN, Supplemental Fig. S10). The ability of TaFROG to homodimerize was also evaluated as part of the BiFC experiment. The combination of YFPN-TaFROG/YFPC-TaFROG gave a strong YFP signal, restricted within nuclear bodies (Fig. 6B). As a negative control, the combination YFPN-TaFROG/ YFPC-TaSnRK2.8 gave no clear fluorescence (Fig. 6B). Protein expression from constructs was confirmed by western-blot analysis (Supplemental Fig. S10). Collectively, these BiFC data indicated that TaFROG can homodimerize in the nucleus, and that it can interact with TaSnRK1α in planta, predominantly in cytosolic bodies and potentially (rarely) in nuclear bodies.
DISCUSSION
FHB is one of the most devastating diseases of small-grain cereals. Resistance to FHB in wheat is quantitative, nonspecies specific, and nonrace specific. Many quantitative trait loci (QTL) are confirmed to contribute to FHB resistance, but the causative genes underpinning them remain to be determined. The overexpression of classical plant defense genes was shown to enhance FHB resistance in wheat; examples include β-1,3-glucanase, the defensin protein α-1-purothionin (Mackintosh et al., 2007), and the key regulator of the salicylic defense signaling pathway Nonexpressor of Pathogenesis-Related Genes1 (NPR1; Makandar et al., 2006). To our knowledge, TaFROG is the first wheat orphan gene to be functionally characterized and demonstrated to positively enhance biotic stress resistance, specifically FHB resistance. TaFROG and its homeologs map to group 4 chromosomes, but with the wheat genome data available at present, there is no evidence that TaFROG colocates with FHB QTL previously identified on these chromosomes (Buerstmayr et al., 2009).
Expression of TaFROG is induced in response to the F. graminearum virulence factor DON. DON inhibits eukaryotic protein synthesis (Arunachalam and Doohan, 2013), leading to cell death (Desmond et al., 2008), but lower concentrations can also inhibit plant-programmed cell death (Audenaert et al., 2014). Fusarium species are considered hemibiotrophic, and Audenaert et al. (2014) postulated that low levels of DON could facilitate the initial biotrophism, and the subsequent increase in DON production might facilitate the switch to necrotrophism. Wheat may defend itself against the necrotrophic phase by counteracting oxidative stress and inducing cell death (Walter et al., 2010), a theory supported by the positive association between the activity of genes involved in the alleviation of oxidative stress and the inheritance of DON/FHB resistance QTL Fhb1 (Walter et al., 2008). QTL Fhb1 is also associated with the detoxification of DON to DON-3-O-glucoside (Lemmens et al., 2005).
The DON-responsiveness of TaFROG, combined with the effects of gene silencing and overexpression on DON/FHB resistance, led us to deduce that the enhanced FHB resistance afforded by TaFROG is likely a consequence of enhanced DON resistance. Fungal toxin productivity varies from strain to strain, and there may be a dose-dependent effect on the TaFROG-associated resistance response. TaFROG is an IDP, and there is growing evidence that these proteins control protein signaling complex assembly in cytosolic and nuclear bodies to regulate cellular signaling pathways (Wright and Dyson, 2015). This information, coupled with the lack of protein domains, led us to speculate that TaFROG is not involved directly in any DON detoxification process, but might be a component of the signaling response to DON, perhaps upstream of DON and FHB resistance responses. The fact that TaFROG is induced as an early response to DON and its interaction with the plant cell signaling machinery suggests that this orphan protein might be part of the signaling events established to promote plant resistance to the fungus and the toxin. We confirmed that TaFROG can physically interact with TaSnRK1α, both in yeast and in planta. TaSnRK1α is a wheat alpha subunit of an SNF1-related kinase 1, which is analogous to the yeast SNF1 and the mammalian AMP-activated protein kinase (AMPK). SnRK1 proteins are central regulators of plant sugar and stress signaling (Halford and Hey, 2009). It may be that TaFROG is a new component of the SnRK1 signaling pathway. Interestingly, human macrophages show a change in the phosphorylation state of AMPK in response to DON, indicating that the associated signaling pathway is activated by the mycotoxin (Pan et al., 2013). There is growing evidence that the interaction of the SnRK1/SNF1 with unique proteins, including orphan proteins, regulates cellular adaptability. For example, the yeast orphan protein Mating Depressing Factor1 (MDF1) physically interacts with SNF1, thereby conferring a selective advantage on yeast grown in rich medium by virtue of their rapid consumption of Glc in early generational stages (Li et al., 2014).
To date, there is no evidence that SnRK1 plays a role in plant defense responses against a fungal toxin that act as virulence factors. However, there are studies supporting the role of SnRK1 in plant defense against viruses, fungus, bacteria, and herbivores. For example, tobacco plants overexpressing SnRK1 were more resistant to geminivirus infection (Hao et al., 2003); an SnRK1 null mutant was more susceptible to Magnaporthe oryzae (Kim et al., 2015); SnRK1-silenced plants were impaired in the hypersensitive response induced by the Xanthomonas campestris pv. vesicatoria effector AvrBs1 (Szczesny et al., 2010); GAL83, the β-subunit of the SnRK1 complex, was shown to have an important role in carbon resource allocation to roots, enhancing tolerance to herbivory (Schwachtje et al., 2006). Thus it is conceivable that the complex TaSnRK1α/TaFROG plays a role in the wheat defense response against DON and F. graminearum.
TaFROG is a nuclear protein, restricted within nuclear bodies. BiFC analysis showed that TaFROG can homodimerize within nuclear bodies, and that TaFROG/TaSnRK1α complexes were primarily localized in small cytosolic bodies (very rarely within nuclear bodies). Although further study is required to validate the subcellular location of the complexes, SnRK1 proteins have been previously reported to localize in both the cytoplasm and the nucleus, in agreement with the dual functions of SnRK1 as a regulator of cytosolic enzyme activity and gene expression (Halford and Hey, 2009; Cho et al., 2012). Moreover, SnRK1 can localize in nuclear bodies when interacting with the Domain of Unknown Function581 proteins (Nietzsche et al., 2014) or in cytosolic small fluorescent structures, reflecting both Golgi bodies (O’Brien et al., 2015) or bodies from unknown origins stimulated from mechanical wounding (Williams et al., 2014). Similarly, upon oxidative, metabolic, or drug-induced stress, the AMPK localize in cytosolic bodies (Mahboubi et al., 2015).
CONCLUSION
In the current study, we present the functional characterization of TaFROG as a wheat orphan gene validated to contribute to disease resistance in wheat, providing a new source of resistance genes for crop-breeding programs. Future work will determine the biochemical means by which TaFROG enhances plant disease resistance, including a study to determine if this orphan protein affects the SnRK1 signaling pathway. Because TaFROG is an intrinsically disordered protein localized in cytosolic bodies when interacting with TaSnRK1α, we cautiously speculate that this localization to specific subcellular bodies may affect downstream signaling pathways. Future experiments will determine the nature of these bodies, clarify the subcellular localization of the complex TaFROG/TaSnRK1α using alternative approaches, and determine the effect of this interaction on SnRK1 signaling.
MATERIALS AND METHODS
Plant and Fungal Material
Arabidopsis (Arabidopsis thaliana) accession Columbia-0 and wheat (Triticum aestivum) cultivars ‘Fielder’ and ‘CM82036’ were used in this study. Wheat ‘Fielder’ is susceptible to FHB disease (Badea et al., 2013), whereas cv CM82036 is resistant to FHB and DON, a trait that is associated with QTL located on chromosomes 3B and 5A (Buerstmayr et al., 2003). The wheat ‘CM82036’ was used for gene expression and virus-induced gene-silencing studies, whereas cv Fielder and its transgenic derivatives were used for the disease assessment studies. Wild-type Fusarium graminearum strain GZ3639 and its DON-minus mutant derivative GZT40 were used for pathogenicity studies (Proctor et al., 1995; Bai et al., 2002). Asexual conidial inoculum (macroconidia) was produced in Mung bean broth (Bai and Shaner, 1996) and was harvested, washed, and adjusted to 106 conidia mL−1, as previously described (Brennan et al., 2005).
Plant Material for Gene Expression Studies
For root gene expression studies, wheat plants were germinated for 3 d at 20°C on moist Whatman No. 1 filter paper, and then placed in a new petri dish on filter paper soaked with 6 mL of either 67.5 µm DON (Santa Cruz) in 0.02% (v/v) Tween 20 or 0.02% (v/v) Tween 20 (mock). Dishes were sealed with Parafilm and incubated for 24 h at 20°C in the dark before harvesting the roots. There were three biological replicates, each including 12 plants (three petri dishes) per treatment.
For detached leaf gene expression studies, wheat plants were cultivated under contained environment conditions at 20°C to 22°C with a 16-h-light/8-h-dark photoperiod at 300 μmol m−2 s−1 and 70% relative humidity, as previously described (Ansari et al., 2007). The second leaves (growth stage 10; Zadoks et al., 1974) were collected, and 8-cm sections were placed in a square petri dish on moist Whatman No. 1 filter paper soaked with 10 mL 0.67 mm benzimidazole (Sigma-Aldrich), adaxial side facing upward. The ends of the leaf were sandwiched between two slices of 1% agar containing 0.67 mm benzimidazole. Leaves were gently wounded with a glass Pasteur pipette, and the wound site was treated with 10 μL of either 0.02% (v/v) Tween 20 (mock) or this solution containing 337.5 µm DON. Dishes were sealed as above and incubated in a growth chamber at 20°C to 22°C with a 16-h-light/8-h-dark photoperiod at 200 μmol m−2 s−1 and 70% relative humidity. There were three biological replicates, each including 14 leaves (seven petri dishes) per treatment.
For adult plant DON and FHB studies, plants were cultivated as above under contained environment conditions. At mid-anthesis (growth stage 65; Zadoks et al., 1974), two central spikelets of the heads were treated; for the DON experiment, they were treated with 15 μL of either 0.02% (v/v) Tween 20 (mock) or 16.87 mm DON in 0.02% (v/v) Tween 20, while in a separate FHB experiment, the central spikelets were treated with either 20 μL 0.02% (v/v) Tween 20 (mock) or this solution augmented with 2 × 104 conidia of either F. graminearum strain GZ3639 (wild type) or its non-DON-producing mutant derivative GZT40. In both the DON and FHB experiment, the treated heads were covered with plastic bags for 2 d to maintain high humidity. Treated spikelets were harvested at various time points post-treatment, and plant materials were flash frozen in liquid N2 and stored at −70°C prior to RNA extraction. The DON experiment comprised three biological replicates, whereas the FHB experiment comprised two biological replicates. In each biological replicate, RNA was extracted from one pooled sample per treatment (representing a pool of four heads from individual plants).
DNA, RNA Extraction, and cDNA Synthesis
DNA was extracted using the HP plant DNA mini kit (OMEGA) following the manufacturer’s instructions. Wheat head RNA was extracted as described previously (Ansari et al., 2007). RNA from all other plant tissues was extracted using the RNeasy plant kit (Qiagen), according to the manufacturer’s instructions. DNase treatment of RNA was performed using the TURBO DNA-free TM kit (Ambion Inc.), according to the manufacturer’s instructions. The quality, yield, and integrity of the RNA were analyzed by measuring the UV absorbance with a NanoDrop 1000 (Thermo Scientific), and visualized following electrophoresis through an agarose gel. The absence of DNA contamination was confirmed by PCR primers targeting the endogenous gene Triticum aestivum glyceraldehyde-3-phosphate dehydrogenase (primer sets listed in Supplemental Table S1). Reverse transcription of total RNA was performed as described previously (Walter et al., 2008).
Amplification and Sequencing of TaFROG
The mRNA sequence of TaFROG was obtained by three successive rounds of RACE using RNA extracted from DON-treated heads of wheat ‘CM82036’ and the GeneRacer TM kit (Invitrogen). RNA was dephosphorylated, decapped, and reverse transcribed using SuperScript III RT (Invitrogen), according to the manufacturer’s instructions. Gene-specific primers are detailed in Supplemental Table S1. The initial touchdown PCR reaction (50-µL final volume) contained 200 nm forward or reverse gene-specific primers (GSP1 primer), 600 nm 5′ or 3′ GeneRacer primer, 1× high fidelity buffer, 2.5 units of high fidelity platinum Taq polymerase (Invitrogen), 2 mm MgSO4, 200 µm of each deoxynucleotide triphosphate, and 1 µL of cDNA. PCR reaction conditions consisted of an initial denaturation step at 94°C for 2 min followed by five cycles at 94°C for 30 s, 72°C for 1 min; five cycles at 94°C for 30 s, 70°C for 1 min; 25 cycles at 94°C for 30 s, final annealing at 68°C for 30 s; and one cycle at 72°C for 1 min. The two subsequent rounds of nested PCR (50-µL volume) contained 1 μL of a 1:300 dilution of PCR product, 2.5 units of LA Taq, and 1× TaKaRa LA Taq Mg2+ plus buffer (Takara); 200 µm dNTP (Gibco, UK); 200 nm of either the 5′ or 3′ nested GeneRacer primer; and 200 nm of either the forward or reverse nested gene-specific primers (GSP2). PCR reaction conditions consisted of an initial denaturation at 94°C for 1 min followed by 25 cycles of 94°C for 30 s, 65°C for 30 s, and 68°C for 2 min. The final extension was set at 68°C for 10 min. Amplified fragments were cloned into the TOPO TA Cloning kit (Invitrogen) and sequenced.
Bioinformatic Analysis
Using the sequence obtained via RACE-PCR of TaFROG, the open reading frame and the coding sequence (CDS) were deduced using National Center for Biotechnology Information’s (NCBI’s) open reading frame finder (http://wheat-urgi.versailles.inra.fr/). Homologs and homeologs were identified via BLASTn query of the nucleotide sequences in the NCBI GenBank and genome databases of wheat (http://wheat‐urgi.versailles.inra.fr/Seq‐Repository and http://www.cerealsdb.uk.net/cerealgenomics/CerealsDB), barley (Hordeum vulgare; http://pgsb.helmholtz-muenchen.de/plant/barley/fpc/searchjsp/index.jsp), Brachypodium distachyon (http://pgsb.helmholtz-muenchen.de/plant/brachypodium/searchjsp/index.jsp), Triticum urartu and Triticum turgidum (http://dubcovskylab.ucdavis.edu/wheat_blast), bamboo (Phyllostachys heterocycla; http://www.bamboogdb.org/), rice (Oryza sativa; http://rice.plantbiology.msu.edu/), Zea mays, Leersia perrieri, Musa acuminata, Setaria italica, and Sorghum bicolor (http://plants.ensembl.org/index.html). Prediction of the nuclear localization sequence and nuclear export signal was performed with WolF PSORT (http://psort.hgc.jp/), NLStradamus (http://www.moseslab.csb.utoronto.ca/NLStradamus/), and NetNES 1.1 (http://www.cbs.dtu.dk/services/NetNES/). Multiple sequence alignment of TaFROG proteins and homologs was performed using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/) and Boxshade (http://www.ch.embnet.org/software/BOX_form.html) programs.
Generation of Transgenic Wheat and Arabidopsis Plants
Transgenic wheat ‘Fielder’ overexpressing TaFROG was generated as follows. The TaFROG CDS was cloned using the gateway cloning strategy (primer sets listed in Supplemental Table S1) and the pDONR207 vector (Invitrogen); the gene was subsequently cloned into binary vector pSc4ActR1R2 (under the control of the rice actin promoter; McElroy et al., 1990). The recombinant plasmid pEW246-TaFROG was then transformed into Agrobacterium tumefaciens strain AGL-1 and cocultivated with immature wheat embryos at 23°C in the dark for 2 d (Ishida et al., 2015). Following removal of the embryonic axis, subsequent tissue culture of the plant material was performed essentially as described previously (Risacher et al., 2009). DNA isolated from regenerated plantlets was analyzed by PCR to determine the copy number of the neomycin phosphotransferase II selectable marker gene, relative to an internal control (M. Craze, unpublished data). T0 transgenic plants carrying transfer DNA (1 copy in lines OE-1 and OE-2; 1–2 copies in line OE-3) and overexpressing TaFROG (Supplemental Fig. S6) were propagated to the T3 generation: plants were grown under contained environment conditions at 20°C to 22°C with a 16-h light/8-h dark photoperiod at 300 μmol m−2 s−1 70% relative humidity, as previously described (Ansari et al., 2007). Homozygosity was analyzed by testing for the presence/absence of the construct and calculating the segregation ratio in each generation.
For Arabidopsis transformation, the TaFROG coding sequence was fused to YFP (TaFROG-YFP) under the control of the 35S promoter. TaFROG CDS was cloned in pDONR207 (Invitrogen) using a gateway cloning strategy (primer sets listed in Supplemental Table S1), and the gene was subsequently cloned into binary vector pAM-PAT-P35S-YFP (Bernoux et al., 2008). pAM-PAT-P35S-TaFROG-YFP was transformed into Arabidopsis ecotype Columbia-0 via Agrobacterium spp.-mediated transformation using the floral dipping method (Logemann et al., 2006). Transgenic plants were selected in vitro on one-half-strength Murashige and Skoog containing 1% Suc and BASTA (7.5 µg mL−1 glufosinate ammonium), pH 5.7. Homozygous lines were propagated to the T2 generation at 20°C to 22°C with a 16-h light/8-h dark photoperiod at 200 μmol m−2 s−1 and 70% relative humidity. Expression of TaFROG-YFP was confirmed by western-blot analysis (Supplemental Fig. S2).
VIGS
VIGS were performed as described previously (Walter et al., 2015), with some modification. The BSMV-derived VIGS vectors used in this study consisted of the wild-type BSMV ND18 α, β, γ tripartite genome (Holzberg et al., 2002; Scofield et al., 2005). Two different gene fragments were used for VIGS of TaFROG (Supplemental Fig. S4). These were amplified from the CDS of TaFROG (primer sets listed in Supplemental Table S1) via PCR. VIGS target sequences were chosen to preferentially silence TaFROG (chromosome 4A; 100% homology) but could also potentially target the two other homeologs (the two VIGS fragments sharing 92% homology to the variants on chromosomes 4B and 4D, based on cv Chinese spring sequences). PCR amplicons of the silencing fragments were cloned into NotI/PacI-digested γ RNA vector pSL038-1 (Scofield et al., 2005). A BSMV γ RNA construct containing a 1,85-bp fragment of the barley phytoene desaturase (PDS) gene served as a positive control for VIGS and has been previously described (Scofield et al., 2005).
Vectors containing the BSMV α and γ genomes (BSMV:00), the γ RNA genome encoding silencing fragments of FROG (FROG or FROG2), or PDS were linearized with MluI. The vector carrying the BSMV β genome was linearized using SpeI. Capped in vitro transcripts were prepared from the linearized plasmids using the mMessage mMachine T7 in vitro transcription kit (AM1344; Ambion) following the manufacturer’s protocol. Flag leaves of wheat ‘CM82036’ at growth stage 47 (just before the emergence of the first wheat head [Zadoks et al., 1974]) were rub inoculated with BSMV constructs following the protocol described by Scofield et al. (2005) Rub inoculations were done with 1:1:1 ratio mixtures of the in vitro transcripts from BSMV α and β and γ RNA (BSMV:00), BSMV:PDS, BSMV:FROG, or BSMV:FROG2. At mid-anthesis (growth stage 65), two florets of two central spikelets of heads of BSMV-infected tillers were treated with 15 μL of either 0.02% (v/v) Tween 20 (mock) or 16.87 mm DON in 0.02% (v/v) Tween 20. Treated heads were covered with plastic bags for 2 d to maintain high humidity. One spikelet above the treated spikelets was sampled 24 h after DON treatment, flash frozen in liquid N2, and stored at −70°C prior to RNA extraction for gene expression studies. The number of damaged (discolored and necrotic) spikelets (including treated spikelets) was assessed 14 d after DON treatment. In total, there were two VIGS experiments conducted, each including two biological replicates. Experiments 1 and 2 used BSMV:FROG and BSMV:FROG2, respectively, for gene silencing (Supplemental Fig. S5). Each biological replicate included positive and negative controls. For BSMV:FROG, each biological replicate included 10 to 18 heads (five to nine plants) per treatment combination. For BSMV:FROG2, each biological replicate included 10-15 heads (15 plants) per treatment combination.
Quantitiative and Semiquantitative Reverse Transcriptase PCR Analysis
Reverse transcription of total RNA was performed as described previously (Walter et al., 2008). qRT-PCR analysis was conducted using the Stratagene Mx3000TM Real-Time PCR. Each reaction contained 1.25 μL of a 1:5 (v/v) dilution of cDNA, 0.2 μm of each of the primers, and 1× SYBR Premix Ex Taq (Tli RNase H plus, RR420A; Takara) in a total reaction volume of 12.5 μL. PCR conditions were as follows: 1 cycle of 1 min at 95°C; 40 cycles of 5 s at 95°C and 20 s at 60°C; and a final cycle of 1 min at 95°C, 30 s, at 55°C, and 30 s at 95°C for the dissociation curve. All qRT-PCR analyses were conducted in duplicate (two cDNA generated from independent reverse transcriptions). The threshold cycle (Ct) values obtained by qRT-PCR were used to calculate the relative gene expression using the Equation 2-(Ct target gene – Ct housekeeping gene) as described previously (Livak and Schmittgen, 2001). To measure the fold change, an efficiency-corrected calculation model was used using the formula ([Etarget]∆Ct target [control – sample)]/(Ehousekeeping)∆Ct housekeeping (control – sample); (Souazé et al., 1996).
sqRT-PCR was conducted to quantify TaFROG transcript levels in different wheat tissues after calibration of the cDNA template with the wheat alpha-tubulin. PCR conditions were as follows: 1 cycle of 5 min at 95°C; 30 or 40 cycles of 45 s at 95°C, 45 s at 60°C, 45 s at 72°C; and a final cycle of 5 min at 72°C. PCR products were visualized after electrophoresis through a 1.5% agarose gel.
Primers
Primer sets used are listed in Supplemental Table S1. TaFROG, TaFROG-B, and TaFROG-D primers were designed to be specific to the chromosome 4A, 4B, and 4D variants, respectively. Unless otherwise stated, primers used for gene expression studies were specific to the chromosome 4A variant. TaFROG primers used for validation of gene silencing were designed so as to not overlap with the gene fragments used for the VIGS. The wheat alpha-tubulin (GenBank [GB] No.: U76558.1; Xiang et al., 2011) was used as a housekeeping gene for TaFROG gene expression analysis and verified to be not differentially expressed either in the microarray expression data available in the PLEXdb (http://www.plexdb.org/) or in the conditions used in our analysis. For F. graminearum, the FgActin gene (FGSG locus ID: 07335.3; Brown et al., 2011) was used as a reference to calculate the relative expression of the FgTri5 gene (FGSG locus ID: 03537.3). The specificity of all primers used was verified by analysis of the dissociation curve, sequence, and electrophoretic profile of PCR products.
Detached Leaf Disease Assessment Study
Detached leaf disease experiments were performed as described previously (Browne and Cooke, 2004), with some modifications. An 8-cm section was cut from the second leaf of three-leaf-stage plants (growth stage 13; Zadoks et al., 1974). Leaf sections were placed with the adaxial side facing upward on the surface of a petri dish (90-mm diameter). The cut ends were placed between a sandwich of 1% plant agar, pH 5.7 (Duchefa) containing 0.5 mm benzimidazole (agar was removed from the center of the plate to prevent excessive fungal growth at the point of leaf inoculation). The center of each leaf section was punctured and treated with a 4-μL droplet of 0.02% (v/v) Tween 20 (mock) or this solution augmented with either 106 conidia mL−1 F. graminearum strain GZ3639 or 106 conidia mL−1 of strain GZ3639 plus 75 µm DON. Plates were incubated at 20°C under a 16-h light/8-h dark cycle. Leaf sections were analyzed 4 d postinoculation. Disease leaf area was estimated using FIJI software of photographed leaf sections (Schindelin et al., 2012). Leaf sections were dipped in water and vigorously vortexed, and macroconidia production was counted using a hemocytometer (Hycor Biomedical). There were three biological replicates, each including six plates per treatment, and each plate including two leaf sections per wheat genotype.
FHB Assessment and DON Studies
Wheat ‘Fielder’ and transgenic lines overexpressing TaFROG were grown and, for the point inoculation experiment, heads were treated as described above (VIGS) with 10 μL of DON or Tween 20 (mock; DON experiment) or 20 μL of 106 conidia mL−1 of F. graminearum strain GZ3639 or Tween 20 (mock; FHB experiment). In the case of the spray inoculation experiment, heads were sprayed with a total of 2 mL inoculum of 2 × 105 conidia mL−1 of F. graminearum using a hand-held sprayer. Control plants were sprayed with Tween 20 (mock). Spray inoculated heads were covered with plastic bags for 4 d to promote infection. For the DON experiment, the number of damaged spikelets was assessed as above (VIGS) 21 d after DON treatment. For the FHB experiments (point and spray inoculation), the level of infection was calculated by visually scoring the number of infected spikelets at 7, 14, and 21 d postinoculation, and data were used to calculate AUDPC (Shaner, 1977). For each experiment, there were three biological replicates, each including at least 20 heads (10 plants) per treatment.
Yeast Two-Hybrid Analysis
Yeast two-hybrid screening was performed by Hybrigenics Services, S.A.S. (http://www.hybrigenics-services.com). In brief, the CDS for TaFROG was cloned into pB29 as an N-terminal fusion to LexA. The construct was used as a bait to screen a random-primed wheat cDNA library produced with RNA extracted from DON-treated heads of wheat ‘CM82036’ and cloned into pP6. A total of 62 million clones were screened using a mating approach, and positive colonies were selected on a medium lacking Trp, Leu, and His. The prey fragments of the positive clones were amplified by PCR and sequenced. Sequences were used to identify the corresponding proteins in the GenBank database (NCBI). For analysis of specific protein-protein interactions, full-length CDS of TaSnRK1α and TaSnRK2.8 (Zhang et al., 2010) was cloned into the vector pDONR207 using the Gateway cloning technology (primer sets listed in Supplemental Table S1) as described previously for TaFROG. They were then recombined into bait and prey vectors derived from pGADT7 and pGBKT7 plasmids (Clontech). Analysis of protein-protein interactions was performed using the Gal4 two-hybrid assay as described previously (Perochon et al., 2010). 3-AT was included as a competitive inhibitor of the imidazole-glycerolphosphate dehydratase protein to increase the stringency and prevent autoactivation.
Subcellular Localization of Fluorescent Fusion Proteins and BiFC
Vectors pAM-PAT-P35S-CFP and pAM-PAT-P35S-TaFROG-YFP (described above) were used to biolistically transform detached wheat seedling leaves for subcellular localization studies. Transformation was performed with 2 to 5 μg of total DNA coated onto 1 μm gold particles using the Bio-Rad PDS-1000/He biolistic particle delivery system and the manufacturers’ instructions (pressure set at 1,100 psi). After bombardment, leaves were placed on moist Whatman filter paper in a petri dish and incubated for 24 h at 21°C in the dark before analysis with a confocal laser scanning microscope (Olympus fluoview FV1000) equipped with a UPLSAPO 40× objective (numerical aperture, 0.95). CFP and YFP excitation was performed at 405 and 515 nm, respectively, and emission detected in the 460 to 500 nm range for CFP and 530 to 630 nm range for YFP. The experiment comprised three biological replicates, each including three leaves.
TaFROG fused to the YFP was visualized within roots of three independent stable transgenic Arabidopsis plants (described above) using a light sheet fluorescence microscope (Lightsheet Z.1; Carl Zeiss Microscopy GmbH) equipped with a W Plan-Apochromat 20× objective (numerical aperture 1). In brief, 1-week-old Arabidopsis seedlings grown on one-half-strength Murashige and Skoog 0.8% agar medium were collected and stained by manual vacuum infiltration with a 1 µg mL−1 DAPI (D9542; Sigma-Aldrich) solution in phosphate-buffered saline buffer (incubated for 20 min in the solution). As described by von Wangenheim et al. (2014), seedlings were then embedded within a cylinder of melted 1% agarose type VI (Sigma-Aldrich). DAPI and YFP excitation was performed at 458 and 515 nm, respectively, and emission was detected in the 465 to 514 nm range for DAPI and 525 to 600 nm range for YFP.
For the in planta analysis of protein-protein interactions, the gateway cloning strategy was used to subclone TaFROG, TaSnRK1α and TaSnRK2.8 CDS into the BiFC vectors pDEST-GW VYCE, pDEST-VYCE(R)GW, pDEST- GWVYNE, and pDEST-VYNE(R)GW (Gehl et al., 2009). This generated constructs wherein proteins were fused at either their N or C terminus to the YFP C-terminal (YFPC) or N-terminal fragment (YFPN). Vectors were introduced into A. tumefaciens. A mix of Agrobacterium spp. transformants was prepared: Optical Density at 600 nm = 0.5, 0.5, and 0.1, respectively, for the YFPC construct, YFPN construct, and the P19 silencing construct (http://www.plantsci.cam.ac.uk/research/davidbaulcombe/methods/protocols/pbin61-p19.doc/view). This mix was syringe infiltrated into leaf epidermal cells of 4-week-old Nicotiana benthamiana. Epidermal cells were assayed for fluorescence 2 to 3 d after infiltration. At 30 min prior to observation, cell nuclei were stained by pressure infiltration of the leaves with a DAPI solution at 1 µg mL−1. Image analyses were carried out using a confocal laser scanning microscope (Olympus fluoview FV1000) equipped with the same objective described above. The DAPI excitation was performed at 405 nm and emission detected in the 425 to 475 nm range. Excitation/emission for the YFP was performed as described above.
All microscopy image stacks were saved on the computer as TIFF files and further processed using FIJI software.
Western Blot
Protein extracts from yeast (yeast two-hybrid studies), N. benthamiana leaf cells (BiFC studies), and Arabidopsis leaves (localization studies) were used for western-blot analysis. Yeast (Saccharomyces cerevisiae) cells were grown at 28°C overnight in Trp/Leu drop-out liquid medium and subsequently for 3 to 5 h in yeast peptone dextrose liquid medium. Total protein extracts were prepared from yeast cells according to the yeast protocols handbooks (Clontech). N. benthamiana leaves and Arabidopsis leaves were snap frozen and ground with beads in a Tissuelyser II (Qiagen). Total plant protein extracts were obtained using Protein Extraction buffer (Agrisera) following the manufacturer’s instructions. Proteins were electrophoresed using a NuPAGE system and 10% Bis-Tris PAGE gels (Life Technologies) according to the manufacturer’s instructions. After protein transfer to nitrocellulose membranes using an iBlot gel transfer system (Life Technologies), proteins fused to the Gal4 activating domain or the YFPN were detected via an epitope of the human influenza hemagglutinin (anti-HA) antibody at 1/1,000 dilution (Roche). In the case of the proteins fused to the Gal4 binding domain or YFPC, an antic-Myc antibody at 1/500 dilution (Roche) was used to detect their epitope cMyc. Following electrochemiluminescence assay, emitted signal was imaged with the Fusion-FX (Vilber Lourmat).
Statistical Analysis
All statistical analyses were performed using the SPSS statistic version 20 software. The normality of the data distribution was evaluated with the Shapiro-Wilk test. In the case of the gene expression, conidial production, DON, and FHB data sets, individual treatments were compared using either the Mann-Whitney U or Kruskal-Wallis test. The normally distributed infected leaf area data from detached leaf experiments were assessed by ANOVA incorporating Tukey's honestly significant difference test at the 0.05 level of significance.
TaFROG (GB no. KR611570), TaSnRK1α (GB no. KR611568), and TaSnRK2.8 (GB no. KR611569) were cloned from the wheat cultivar (cv CM82036). TaFROG homeolog sequences were identified within wheat genome contigs (chromosome 4A: contigs_longerthan_200_7074280, chromosome 4B: contigs_longerthan_200_4893246, chromosome 4C: contigs_longerthan_200_2310089). Homologs from T. turgidum and T. urartu were identified within contigs UCW_Tt-k25_contig_48942 and UCW_Tu-k51_contig_30323, respectively. Homologs from Aegilops tauschii (GB no. EMT01121), H. vulgare (GB no. BAJ91423), and Festuca pratensis (GB no. GO795446) were collected from the NCBI GenBank database. The homolog from Brachypodium distachyon (Bradi4g22656.1) was collected from the MIPS Plants database.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Characteristics of FROG protein sequences.
Supplemental Figure S2. Immunoblot analysis with GFP antibody using the total proteins extracted from wild-type or TaFROG-YFP Arabidopsis leaves.
Supplemental Figure S3. Expression of TaFROG homeologs and its barley homolog in different tissues and in response to DON or F. graminearum.
Supplemental Figure S4. Schematic representation of the position of the two VIGS gene fragments (FROG and FROG2) and the qRT-PCR target region within the TaFROG gene.
Supplemental Figure S5. Effect of TaFROG silencing on DON tolerance.
Supplemental Figure S6. Molecular characterization of transgenic wheat ‘Fielder’ overexpressing TaFROG under the control of a rice actin promoter.
Supplemental Figure S7. Effect of TaFROG overexpression on wheat leaf resistance to F. graminearum.
Supplemental Figure S8. Symptoms of FHB on TaFROG overexpression lines.
Supplemental Figure S9. Yeast two-hybrid analysis.
Supplemental Figure S10. Results for positive and negative controls included within the BiFC analysis.
Supplemental Table S1. Primer sets used in this study.
Supplementary Material
Acknowledgments
The authors thank Bredagh Moran, Kirstin Heinrich-Lynch, Brian Fagan, Pierre Grandjean, Melanie Craze, and Ruth Bates for technical assistance; Hermann Buerstmayr (IFA Tulln) for wheat cultivars; Robert Proctor (USDA) for F. graminearum fungi; Chris Power (Carl Zeiss UK Ltd.) and Emmanuel G. Reynaud (UCD) for assistance with light sheet microscopy; and Carl K. Ng (UCD) for assistance with cell biology.
Glossary
- DON
deoxynivalenol
- cDNA
complementary DNA
- DAPI
4′,6- diamidino-phenylindole
- IDP
intrinsically disordered protein
- sqRT
semiquantitative reverse transcription
- qRT
quantitative reverse transcriptase
- dpi
days postfungal inoculation
- VIGS
virus-induced gene silencing
- BSMV
barley stripe mosaic virus
- AUDPC
area under the disease progress curve
- BiFC
bimolecular fluorescence complementation
- QTL
quantitative trait loci
- AMPK
AMP-activated protein kinase
- CDS
coding sequence
- NCBI
National Center for Biotechnology Information
Footnotes
This work was supported by the Science Foundation Ireland (project nos. 10/IN.1/B3028 and 14/1A/2508) and the National Institute of Agricultural Botany Trust. The authors have a patent pending related to this material.
Articles can be viewed without a subscription.
References
- Ansari KI, Walter S, Brennan JM, Lemmens M, Kessans S, McGahern A, Egan D, Doohan FM (2007) Retrotransposon and gene activation in wheat in response to mycotoxigenic and non-mycotoxigenic-associated Fusarium stress. Theor Appl Genet 114: 927–937 [DOI] [PubMed] [Google Scholar]
- Arendsee ZW, Li L, Wurtele ES (2014) Coming of age: orphan genes in plants. Trends Plant Sci 19: 698–708 [DOI] [PubMed] [Google Scholar]
- Arunachalam C, Doohan FM (2013) Trichothecene toxicity in eukaryotes: cellular and molecular mechanisms in plants and animals. Toxicol Lett 217: 149–158 [DOI] [PubMed] [Google Scholar]
- Audenaert K, Vanheule A, Höfte M, Haesaert G (2014) Deoxynivalenol: a major player in the multifaceted response of Fusarium to its environment. Toxins (Basel) 6: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea A, Eudes F, Laroche A, Graf R, Doshi K, Amundsen E, Nilsson D, Puchalski B (2013) Antimicrobial peptides expressed in wheat reduce susceptibility to Fusarium head blight and powdery mildew. Can J Plant Sci 93: 199–208 [Google Scholar]
- Bai GH, Desjardins AE, Plattner RD (2002) Deoxynivalenol-nonproducing fusarium graminearum causes initial infection, but does not cause disease spread in wheat spikes. Mycopathologia 153: 91–98 [DOI] [PubMed] [Google Scholar]
- Bai GH, Shaner G (1996) Variation in Fusarium graminearum and cultivar resistance to wheat scab. Plant Dis 80: 975–979 [Google Scholar]
- Bernoux M, Timmers T, Jauneau A, Brière C, de Wit PJ, Marco Y, Deslandes L (2008) RD19, an Arabidopsis cysteine protease required for RRS1-R-mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell 20: 2252–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof M, Eichmann R, Hückelhoven R (2011) Pathogenesis-associated transcriptional patterns in Triticeae. J Plant Physiol 168: 9–19 [DOI] [PubMed] [Google Scholar]
- Boddu J, Cho S, Muehlbauer GJ (2007) Transcriptome analysis of trichothecene-induced gene expression in barley. Mol Plant Microbe Interact 20: 1364–1375 [DOI] [PubMed] [Google Scholar]
- Brennan JM, Egan D, Cooke BM, Doohan FM (2005) Effect of temperature on head blight of wheat caused by Fusarium culmorum and F. graminearum. Plant Pathol 54: 156–160 [Google Scholar]
- Brown NA, Bass C, Baldwin TK, Chen H, Massot F, Carion PW, Urban M, van de Meene AM, Hammond-Kosack KE (2011) Characterisation of the Fusarium graminearum-Wheat Floral Interaction. J Pathogens 2011: 626345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne RA, Cooke BM (2004) Development and evaluation of an in vitro detached leaf assay forc pre-screening resistance to Fusarium head blight in wheat. Eur J Plant Pathol 110: 91–102 [Google Scholar]
- Buerstmayr H, Ban T, Anderson JA (2009) QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: a review. Plant Breed 128: 1–26 [Google Scholar]
- Buerstmayr H, Steiner B, Hartl L, Griesser M, Angerer N, Lengauer D, Miedaner T, Schneider B, Lemmens M (2003) Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. II. Resistance to fungal penetration and spread. Theor Appl Genet 107: 503–508 [DOI] [PubMed] [Google Scholar]
- Chen X, Steed A, Harden C, Nicholson P (2006) Characterization of Arabidopsis thaliana-Fusarium graminearum interactions and identification of variation in resistance among ecotypes. Mol Plant Pathol 7: 391–403 [DOI] [PubMed] [Google Scholar]
- Cho YH, Hong JW, Kim EC, Yoo SD (2012) Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiol 158: 1955–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J. , et al. (2012) The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13: 414–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmond OJ, Manners JM, Stephens AE, Maclean DJ, Schenk PM, Gardiner DM, Munn AL, Kazan K (2008) The Fusarium mycotoxin deoxynivalenol elicits hydrogen peroxide production, programmed cell death and defence responses in wheat. Mol Plant Pathol 9: 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue MT, Keshavaiah C, Swamidatta SH, Spillane C (2011) Evolutionary origins of Brassicaceae specific genes in Arabidopsis thaliana. BMC Evol Biol 11: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner DM, Kazan K, Manners JM (2009) Nutrient profiling reveals potent inducers of trichothecene biosynthesis in Fusarium graminearum. Fungal Genet Biol 46: 604–613 [DOI] [PubMed] [Google Scholar]
- Gardiner SA, Boddu J, Berthiller F, Hametner C, Stupar RM, Adam G, Muehlbauer GJ (2010) Transcriptome analysis of the barley-deoxynivalenol interaction: evidence for a role of glutathione in deoxynivalenol detoxification. Mol Plant Microbe Interact 23: 962–976 [DOI] [PubMed] [Google Scholar]
- Gehl C, Waadt R, Kudla J, Mendel RR, Hänsch R (2009) New GATEWAY vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol Plant 2: 1051–1058 [DOI] [PubMed] [Google Scholar]
- Guo WJ, Li P, Ling J, Ye SP (2007) Significant comparative characteristics between orphan and nonorphan genes in the rice (Oryza sativa L.) genome. Comp Funct Genomics : 21676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford NG, Hey SJ (2009) Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem J 419: 247–259 [DOI] [PubMed] [Google Scholar]
- Hao L, Wang H, Sunter G, Bisaro DM (2003) Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell 15: 1034–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzberg S, Brosio P, Gross C, Pogue GP (2002) Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J 30: 315–327 [DOI] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium (IWGSC) (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345: 1251788. [DOI] [PubMed] [Google Scholar]
- Ishida Y, Tsunashima M, Hiei Y, Komari T (2015) Wheat (Triticum aestivum L.) transformation using immature embryos. Methods Mol Biol 1223: 189–198 [DOI] [PubMed] [Google Scholar]
- Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TC (2009) More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet 25: 404–413 [DOI] [PubMed] [Google Scholar]
- Kim CY, Vo K, An G, Jeon JS (2015) A rice sucrose non-fermenting-1 related protein kinase 1, OSK35, plays an important role in fungal and bacterial disease resistance. J Korean Soc Appl Biol Chem 58: 669-675 [Google Scholar]
- Lemmens M, Scholz U, Berthiller F, Dall’Asta C, Koutnik A, Schuhmacher R, Adam G, Buerstmayr H, Mesterházy A, Krska R. , et al. (2005) The ability to detoxify the mycotoxin deoxynivalenol colocalizes with a major quantitative trait locus for Fusarium head blight resistance in wheat. Mol Plant Microbe Interact 18: 1318–1324 [DOI] [PubMed] [Google Scholar]
- Li D, Yan Z, Lu L, Jiang H, Wang W (2014) Pleiotropy of the de novo-originated gene MDF1. Sci Rep 4: 7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Logemann E, Birkenbihl RP, Ulker B, Somssich IE (2006) An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhua S, Hegie A, Suzuki N, Shulaev E, Luo X, Cenariu D, Ma V, Kao S, Lim J, Gunay MB, et al. (2013) Linking genes of unknown function with abiotic stress responses by high-throughput phenotype screening. Physiol Plant 148: 322–333 [DOI] [PubMed] [Google Scholar]
- Mackintosh CA, Lewis J, Radmer LE, Shin S, Heinen SJ, Smith LA, Wyckoff MN, Dill-Macky R, Evans CK, Kravchenko S, et al. (2007) Overexpression of defense response genes in transgenic wheat enhances resistance to Fusarium head blight. Plant Cell Rep 26: 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahboubi H, Barisé R, Stochaj U (2015) 5′-AMP-activated protein kinase alpha regulates stress granule biogenesis. Biochim Biophys Acta 1853: 1725–1737 [DOI] [PubMed] [Google Scholar]
- Makandar R, Essig JS, Schapaugh MA, Trick HN, Shah J (2006) Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol Plant Microbe Interact 19: 123–129 [DOI] [PubMed] [Google Scholar]
- McElroy D, Zhang W, Cao J, Wu R (1990) Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2: 163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nietzsche M, Schießl I, Börnke F (2014) The complex becomes more complex: protein-protein interactions of SnRK1 with DUF581 family proteins provide a framework for cell- and stimulus type-specific SnRK1 signaling in plants. Front Plant Sci 5: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien M, Kaplan-Levy RN, Quon T, Sappl PG, Smyth DR (2015) PETAL LOSS, a trihelix transcription factor that represses growth in Arabidopsis thaliana, binds the energy-sensing SnRK1 kinase AKIN10. J Exp Bot 66: 2475–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Whitten DA, Wu M, Chan C, Wilkerson CG, Pestka JJ (2013) Global protein phosphorylation dynamics during deoxynivalenol-induced ribotoxic stress response in the macrophage. Toxicol Appl Pharmacol 268: 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perochon A, Dieterle S, Pouzet C, Aldon D, Galaud JP, Ranty B (2010) Interaction of a plant pseudo-response regulator with a calmodulin-like protein. Biochem Biophys Res Commun 398: 747–751 [DOI] [PubMed] [Google Scholar]
- Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL (2005) FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 21: 3435–3438 [DOI] [PubMed] [Google Scholar]
- Pritsch C, Muehlbauer GJ, Bushnell WR, Somers DA, Vance CP (2000) Fungal development and induction of defense response genes during early infection of wheat spikes by Fusarium graminearum. Mol Plant Microbe Interact 13: 159–169 [DOI] [PubMed] [Google Scholar]
- Proctor RH, Hohn TM, McCormick SP (1995) Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol Plant Microbe Interact 8: 593–601 [DOI] [PubMed] [Google Scholar]
- Risacher T, Craze M, Bowden S, Paul W, Barsby T (2009) Highly efficient Agrobacterium-mediated transformation of wheat via in planta inoculation. Methods Mol Biol 478: 115–124 [DOI] [PubMed] [Google Scholar]
- Rocha O, Ansari K, Doohan FM (2005) Effects of trichothecene mycotoxins on eukaryotic cells: a review. Food Addit Contam 22: 369–378 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwachtje J, Minchin PE, Jahnke S, van Dongen JT, Schittko U, Baldwin IT (2006) SNF1-related kinases allow plants to tolerate herbivory by allocating carbon to roots. Proc Natl Acad Sci USA 103: 12935–12940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield SR, Huang L, Brandt AS, Gill BS (2005) Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol 138: 2165–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner G. (1977) The effect of nitrogen fertilization on the expression of slow-mildewing resistance in knox wheat. Phytopathology 67: 1051–1056 [Google Scholar]
- Souazé F, Ntodou-Thomé A, Tran CY, Rostène W, Forgez P (1996) Quantitative RT-PCR: limits and accuracy. Biotechniques 21: 280–285 [DOI] [PubMed] [Google Scholar]
- Szczesny R, Büttner D, Escolar L, Schulze S, Seiferth A, Bonas U (2010) Suppression of the AvrBs1-specific hypersensitive response by the YopJ effector homolog AvrBsT from Xanthomonas depends on a SNF1-related kinase. New Phytol 187: 1058–1074 [DOI] [PubMed] [Google Scholar]
- Tautz D, Domazet-Lošo T (2011) The evolutionary origin of orphan genes. Nat Rev Genet 12: 692–702 [DOI] [PubMed] [Google Scholar]
- von Wangenheim D, Daum G, Lohmann JU, Stelzer EK, Maizel A (2014) Live imaging of Arabidopsis development. Methods Mol Biol 1062: 539–550 [DOI] [PubMed] [Google Scholar]
- Walter S, Brennan JM, Arunachalam C, Ansari KI, Hu X, Khan MR, Trognitz F, Trognitz B, Leonard G, Egan D. , et al. (2008) Components of the gene network associated with genotype-dependent response of wheat to the Fusarium mycotoxin deoxynivalenol. Funct Integr Genomics 8: 421–427 [DOI] [PubMed] [Google Scholar]
- Walter S, Kahla A, Arunachalam C, Perochon A, Khan MR, Scofield SR, Doohan FM (2015) A wheat ABC transporter contributes to both grain formation and mycotoxin tolerance. J Exp Bot 66: 2583–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter S, Nicholson P, Doohan FM (2010) Action and reaction of host and pathogen during Fusarium head blight disease. New Phytol 185: 54–66 [DOI] [PubMed] [Google Scholar]
- Williams SP, Rangarajan P, Donahue JL, Hess JE, Gillaspy GE (2014) Regulation of Sucrose non-Fermenting Related Kinase 1 genes in Arabidopsis thaliana. Front Plant Sci 5: 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ (2015) Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol 16: 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Song M, Wei Z, Tong J, Zhang L, Xiao L, Ma Z, Wang Y (2011) A jacalin-related lectin-like gene in wheat is a component of the plant defence system. J Exp Bot 62: 5471–5483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Liu H, Li Y, Li X, Xu C, Long M, Wang S (2009) A rice gene of de novo origin negatively regulates pathogen-induced defense response. PLoS One 4: e4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadeta KA, Valkenburg DJ, Hanemian M, Marco Y, Thomma BP (2014) The Brassicaceae-specific EWR1 gene provides resistance to vascular wilt pathogens. PLoS One 9: e88230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14: 415–421 [Google Scholar]
- Zhang H, Mao X, Wang C, Jing R (2010) Overexpression of a common wheat gene TaSnRK2.8 enhances tolerance to drought, salt and low temperature in Arabidopsis. PLoS One 5: e16041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.