A unique regulator of proteasome-mediated degradation of photomorphogenesis-promoting factors affects Arabidopsis seedling development.
Abstract
Arabidopsis (Arabidopsis thaliana) Short Hypocotyl in White Light1 (SHW1) encodes a Ser-Arg-Asp-rich protein that acts as a negative regulator of photomorphogenesis. SHW1 and Constitutive Photomorphogenic1 (COP1) genetically interact in an additive manner to suppress photomorphogenesis. Elongated Hypocotyl5 (HY5) is a photomorphogenesis promoting a basic leucine zipper transcription factor that is degraded by COP1 ubiquitin ligase in the darkness. Here, we report the functional interrelation of SHW1 with COP1 and HY5 in Arabidopsis seedling development. The in vitro and in vivo molecular interaction studies show that SHW1 physically interacts with both COP1 and HY5. The genetic studies reveal that SHW1 and HY5 work in an antagonistic manner to regulate photomorphogenic growth. Additional mutation of SHW1 in hy5 mutant background is able to suppress the gravitropic root growth defect of hy5 mutants. This study further reveals that the altered abscisic acid responsiveness of hy5 mutants is modulated by additional loss of SHW1 function. Furthermore, this study shows that SHW1 promotes COP1-mediated degradation of HY5 through enhanced ubiquitylation in the darkness. Collectively, this study highlights a mechanistic view on coordinated regulation of SHW1, COP1, and HY5 in Arabidopsis seedling development.
Plants have evolved with developmental plasticity to respond to environmental changes. Light acts as one of the most influential environmental factors for plant growth and development. Arabidopsis (Arabidopsis thaliana) seedlings follow two distinct developmental patterns in the presence and absence of light. The dark-grown seedlings have long hypocotyl with apical hooks and small and closed cotyledons. The light-grown seedlings, however, have short hypocotyl with open and expanded cotyledons (Nagatani et al., 1993; Whitelam et al., 1993; Neff et al., 2000; Chen et al., 2004; Bae and Choi, 2008). Downstream to photoreceptors, several positive and negative regulators have been identified that are intimately involved in Arabidopsis seedling development (Jiao et al., 2007; Chen and Chory, 2011).
Constitutive Photomorphogenic1 (COP1) is a repressor of photomorphogenesis in the darkness (Wei and Deng, 1999; Jiao et al., 2007; Lau and Deng, 2012). COP1 acts as an E3 ubiquitin ligase and targets photomorphogenesis promoting factors, such as Elongated Hypocotyl5 (HY5), Elongated Hypocotyl5 Homolog (HYH), Long After Far-Red Light1 (LAF1), Long Hypocotyl in Far-Red1 (HFR1), Blue Insensitive Trait1, and LIGHT-REGULATED ZINC FINGER PROTEIN1, for degradation in the dark (Osterlund et al., 2000; Holm et al., 2002; Saijo et al., 2003; Seo et al., 2003; Yang et al., 2005a, 2005b; Chang et al., 2011). However, COP1 has been shown to be required for the optimum accumulation of G-box Binding Factor1 (GBF1)/Z-box Binding Factor2 (ZBF2) protein in light (Mallappa et al., 2006, 2008). A COP1 suppressor, CSU1, has recently been shown to play a major role in maintaining the COP1 homeostasis in dark (Xu et al., 2014a). A group of SPA proteins (Suppressor of PhytochromeA1 [SPA1]–SPA4), which functions redundantly to suppress photomorphogenesis, has been shown to physically interact with COP1 and enhance its function (Saijo et al., 2003; Laubinger et al., 2004; Zhu et al., 2008). It has been shown that spa quadruple-mutant seedlings with defects in all four SPA genes display constitutive photomorphogenesis in the dark. However, such morphological defects are not observed in any of the single mutants in the darkness (Laubinger et al., 2004). Also, a group of phytochrome-interacting factors (PIFs; PIF1, PIF3, PIF4, PIF5, and PIF7) that functions redundantly in the dark to suppress photomorphogenesis has been reported (de Lucas et al., 2008; Leivar et al., 2008; Leivar and Quail, 2011). Recent studies have shown that PIF1 promotes the E3 ligase activity of COP1 in the dark (Xu et al., 2014b).
Transcriptional regulatory networks play an important role in light signaling pathways through the coordinated activation and repression of downstream genes (Ma et al., 2001; Tepperman et al., 2001; Jiao et al., 2007). HY5 is a positive regulator of light signaling pathways that acts at various wavelengths of light. HY5 has been genetically defined as a positive regulator of photomorphogenesis based on its partially etiolated phenotype in light-grown mutant seedlings (Ang and Deng, 1994). HY5 encodes a basic leucine zipper (bZIP) protein that can physically interact with COP1 (Ang et al., 1998; Osterlund et al., 2000). DNA-protein interaction studies have revealed that HY5 specifically interacts with the G box and is required for the proper activation of G box-containing promoters in light (Chattopadhyay et al., 1998; Yadav et al., 2002). It has recently been reported that bZIP protein GBF1/ZBF2 physically interacts with HY5 and its other bZIP partner HYH in blue light (BL)-mediated seedling development (Mallappa et al., 2006, 2008; Singh et al., 2012; Ram et al., 2014). HY5 is shown to cross talk with multiple hormonal signaling pathways, including auxin, cytokinin, GA3, and abscisic acid (ABA; Jiao et al., 2007; Vandenbussche et al., 2007; Alabadí et al., 2008; Chen et al., 2008). Recent chromatin immunoprecipitation-on-chip studies have shown that HY5 binds to the promoters of a large number of regulatory genes in Arabidopsis (Lee et al., 2007; Zhang et al., 2011). Recent studies have shown that HY5 can directly bind to its own promoter in association with Calmodulin7 to promote its own expression (Abbas et al., 2014).
Short Hypocotyl in White Light1 (SHW1), a Ser-Arg-Asp-rich protein, is constitutively localized in the nucleus, and its expression is developmentally regulated. SHW1 acts as a negative regulator of light-mediated inhibition of hypocotyl elongation (Bhatia et al., 2008). The shw1 mutants also display shorter hypocotyl in the darkness. It has been shown that SHW1 acts nonredundantly with COP1 to control hypocotyl elongation in the darkness and thereby, is functionally interrelated to COP1 in photomorphogenesis (Bhatia et al., 2008). In this study, we have investigated the biochemical interactions of SHW1 with HY5 and COP1 through in vitro and in vivo studies. We have further analyzed the in vivo functional interactions of SHW1 with HY5 and COP1 through mutational studies. Our data strongly support that SHW1 is an associated factor of COP1 that helps in the degradation of HY5 in the darkness.
RESULTS
SHW1 Physically Interacts with COP1
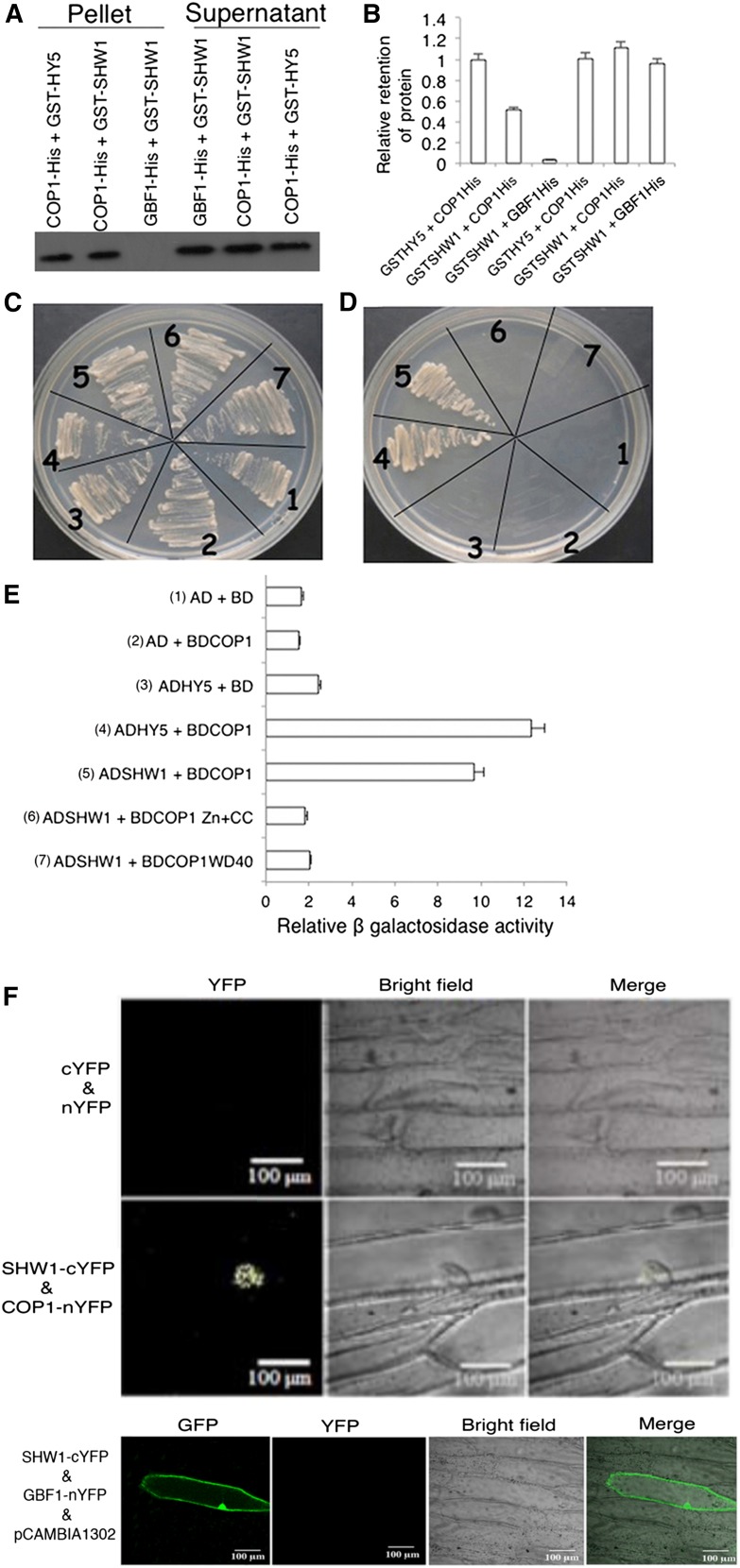
The genetic interaction studies between COP1 and SHW1 had earlier revealed that shw1 and cop1 acted in an additive manner to repress photomorphogenic growth in the dark (Bhatia et al., 2008). However, enhanced inhibition in hypocotyl elongation of shw1 mutants in white light (WL) requires functional COP1 protein (Bhatia et al., 2008). Therefore, we ask whether SHW1 physically interacts with COP1. To examine the physical interactions between SHW1 and COP1, in vitro pull-down assays were carried out using glutathione S-transferase (GST)-SHW1 and COP1-His fusion proteins. In these experiments, GST-HY5 and GBF1-His were used as positive and negative controls, respectively (Ang et al., 1998; Singh et al., 2012). GST-SHW1 and GST-HY5 proteins were separately passed through columns containing Ni-NTA Agarose (Qiagen) beads bound to GBF1-His or COP1-His protein. As shown in Figure 1A, SHW1 was retained in COP1 column, and the level of retention of SHW1 was comparable with the amount of HY5 retained in COP1 column. SHW1 was hardly detectable, if at all, in GBF1-His column (Fig. 1, A and B; Supplemental Fig. S1). Taken together, these results indicate that SHW1 physically interacts with COP1.
Figure 1.
SHW1 physically interacts with COP1. A, In vitro binding of SHW1 and COP1: 2 µg of COP1-6His or GBF1-6His (negative control) was bound to Ni-NTA beads, washed, and incubated with GST-SHW1 or GST-HY5 (positive control). Beads were washed and fractionated in 12% (w/v) SDS-PAGE. The blot was probed with anti-GST antibodies. B, Quantification of the data (by Bio-Rad multi-imager): retention of GST-SHW1 and GST-HY5 by COP1-6His or GBF1-6His is shown in the graph. Error bars indicate sem of three replicate experiments. C to E, Yeast two-hybrid interactions between SHW1, COP1, and the truncated versions of COP1. Growth of cotransformed yeast AH109 strain on double-dropout media (C) and quadruple-dropout media (D). The protein-protein interactions were examined by β-galactosidase assays. The relative β-galactosidase activities were calculated according to Clontech instructions (E). The error bars indicate se (n = 3). The numbers 1 to 7 refer to the constructs used. F, BiFC assay showing that COP1-nYFP and SHW1-cYFP interact to form a functional YFP in the nucleus, whereas SHW1-cYFP and GBF1-nYFP do not interact. GFP containing pCAMBIA-1302 vector was used as a control for transformation. The images show transiently transformed onion (Allium cepa) epidermal cells.
To further examine the observed physical interaction of SHW1 and COP1, yeast (Saccharomyces cerevisiae) two-hybrid protein-protein interaction assays were carried out. In this study, we have used full-length and truncated versions of COP1, such as WD40-repeat or zinc-finger and coiled-coil domain of COP1. The growth of cotransformed yeast cells on two-dimensional (deficient in Trp and Leu) and four-dimensional (deficient in Trp, Leu, His, and adenine) synthetically defined plates was monitored. As shown in Figure 1, C and D, the full-length SHW1 was able to interact with full-length COP1. Comparison of β-galactosidase activity in SHW1 and COP1 interaction revealed that the β-galactosidase activity was about 5-fold increased than the background and comparable with the increased activity in COP1 and HY5 interaction (approximately 6-fold) used as control (Fig. 1E). However, neither of the truncated versions of COP1 showed increased activity compared with background level (Fig. 1E). These results suggest that SHW1 physically interacts with COP1 in yeast cells. These results further indicate that full-length COP1 is required for the interaction with SHW1.
To further substantiate the physical interactions between SHW1 and COP1, bimolecular fluorescence complementation (BiFC) experiments were carried out. For these experiments, SHW1 full-length coding sequence was fused to the C terminus of YFP in pUC-SPYCE vector (SHW1-cYFP), and COP1 full-length coding sequence was fused to the N terminus of YFP in pUC-SPYNE vector (COP1-nYFP; Sethi et al., 2014). Whereas empty vectors did not produce any YFP fluorescence (Fig. 1F), interaction of SHW1 and COP1 produced strong YFP fluorescence in the nucleus with the characteristic speckles known to be formed by COP1 (Fig. 1F). Taken together, these results show that SHW1 physically interacts with COP1 in vivo.
SHW1 and HY5 Genetically Interact to Regulate Hypocotyl Growth
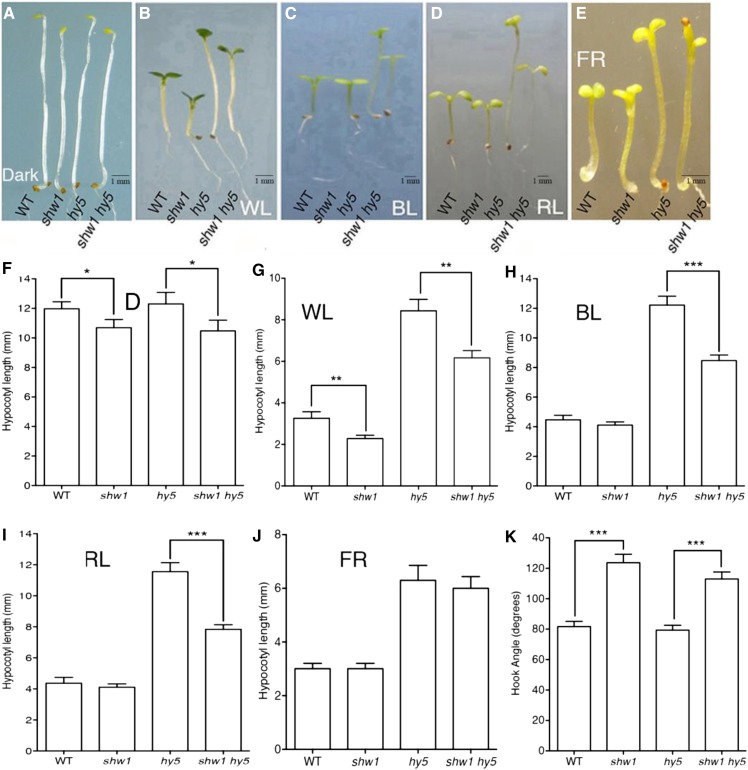
Because SHW1 physically interacts with COP1 and because earlier studies have shown that shw1 cop1 double mutants display enhanced photomorphogenic growth in the darkness (Bhatia et al., 2008), we ask whether, similar to COP1, SHW1 also genetically interacts with HY5, one of the targets of COP1 in the darkness (Osterlund et al., 2000). The hy5 mutants exhibit elongated hypocotyl, whereas shw1 mutants display shorter hypocotyl in WL (Ang et al., 1998; Bhatia et al., 2008). We generated shw1 hy5 double mutants and examined the seedling growth in the dark and at various wavelengths of light. In the darkness, the shw1 mutants displayed shorter hypocotyl with partially opened apical hooks, consistent with the previous observation (Bhatia et al., 2008). The shw1 hy5 double mutants showed hypocotyl length and hook angle similar to shw1 single mutants in the darkness (Fig. 2, A, F, and K).
Figure 2.
HY5 genetically interacts with SHW1. In A to E, wild-type (WT; Col-0), shw1, hy5, and shw1 hy5 seedlings are shown. A to E, Visible phenotypes of 6-d-old seedlings grown in constant dark (D), WL (30 µmol m−2 s−1), BL (30 µmol m−2 s−1), RL (30 µmol m−2 s−1), and FR light (30 µmol m−2 s−1) are shown as indicated. F to J, Quantification of hypocotyl length of wild-type, shw1, hy5, and shw1 hy5 seedlings grown under various wavelengths of light. K, Hook angle of 6-d-old constant dark-grown seedling of wild-type and various mutant lines. Results presented are obtained from five biological repeats, each having at least 30 seedlings. Error bars represent se (Student’s t test). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The shw1 mutants display shorter hypocotyl in WL but not in BL, red light (RL), and far-red (FR) light. When we examined the hypocotyl growth in WL, 6-d-old shw1 hy5 double mutants displayed shorter hypocotyl than hy5 single mutants in WL, suggesting that they seem to work antagonistically to regulate the hypocotyl growth in WL (Fig. 2, B and G). Furthermore, although shw1 mutants did not display any alteration in the hypocotyl length in BL, RL, and FR light (Fig. 2, C–E and H–J; Bhatia et al., 2008) conditions, the hypocotyl length of shw1 hy5 double mutants was significantly reduced compared with hy5 single mutants in BL and RL (Fig. 2, C, D, H, and I). These results suggest that functional SHW1 is required for the optimum hypocotyl phenotype of hy5 mutants in BL and RL conditions. However, the hypocotyl length of shw1 hy5 double mutant was similar to hy5 in FR light (Fig. 2, E and J), indicating that additional mutation of SHW1 in hy5 mutant does not affect the phenotype of hy5 in FR light.
SHW1 Modulates HY5-Mediated Regulation of Root Growth
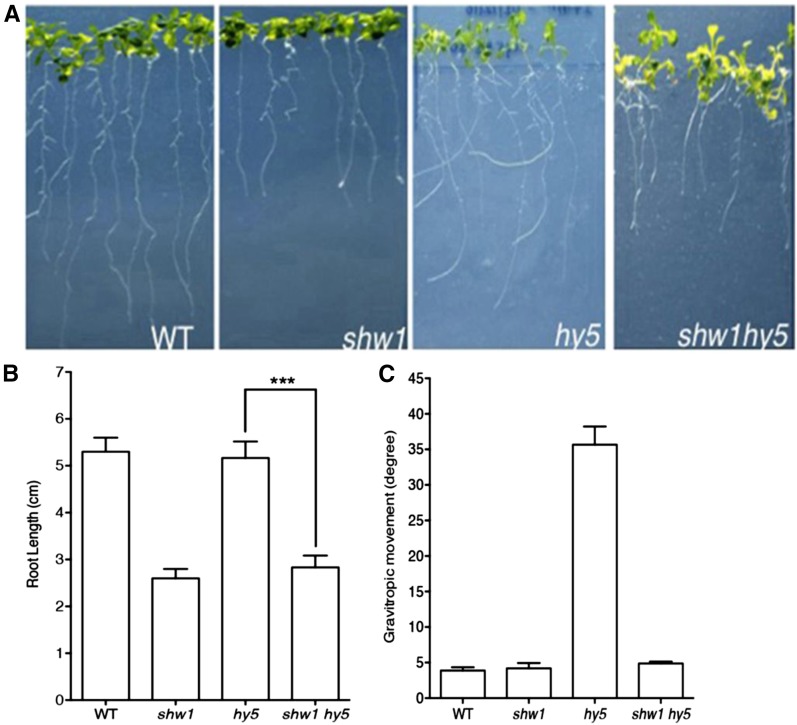
The shw1 and hy5 mutants display altered root growth (Oyama et al., 1997; Ang et al., 1998; Bhatia et al., 2008). To determine whether SHW1 and HY5 can modulate each other’s function on root growth, we examined the root growth of shw1 hy5 double mutants compared with single mutants. The shw1 mutants developed shorter roots than the wild type, and the root length of shw1 hy5 double mutants was found to be similar to shw1 (Fig. 3, A and B). Because hy5 mutants do not display any altered root length, these results indicate that SHW1 works independently of HY5 in the regulation of root length.
Figure 3.
SHW1 and HY5 act antagonistically to regulate gravitropic root growth. A, The root growth of 16-d-old WL-grown (30 µmol m−2 s−1) wild type (WT) and shw1, hy5, and shw1 hy5 mutants. B, Quantification of the root length of 16-d-old wild type and various mutants grown in WL (30 µmol m−2 s−1). C, Quantification of gravitropic root movement of the wild type and various mutants grown in WL (30 µmol m−2 s−1). Results presented are obtained from three biological repeats, each having at least 20 plants. Error bars represent se (Student’s t test). ***, P < 0.001.
Previous studies have shown that one of the obvious phenotypes of hy5 mutants is the direction of root growth. It was nearly horizontal rather than downward, indicating alteration of the gravitropic response. The angles of lateral roots of hy5 mutants were larger than the wild type (Oyama et al., 1997). As shown in Figure 3, A and C, the roots of shw1 hy5 double-mutant plants did not exhibit any defect in gravitropic responses. These results suggest that additional mutation of SHW1 in hy5 mutant background is able to suppress the gravitropic root growth defect of hy5 mutants.
HY5 and SHW1 Additively Work in ABA Responsiveness
Recent studies have revealed that light signaling pathways cross talk with multiple hormone signaling pathways. For example, HY5 acts as a point of cross talk in light and ABA signaling pathways. It has been shown that mutations in HY5 caused Arabidopsis plants to be less sensitive to ABA (Chen et al., 2008). To determine further genetic relationship between SHW1 and HY5, we ask whether mutations in SHW1 can modulate the altered ABA responsiveness of hy5 mutants. Freshly harvested seeds of wild-type and single- or double-mutant plants were plated on Murashige and Skoog medium (MS) plates without or with ABA. The rate of germination was found to be similar in the wild type and various mutants in untreated seeds (Supplemental Fig. S2A). Whereas 1 µm ABA reduced the rate of germination of wild-type seeds (34%), the effect was significantly suppressed in hy5 background (70%). The shw1 mutant seeds showed less sensitivity to ABA-mediated inhibition of seed germination compared with the wild type (48%). As shown in Supplemental Figure S2, the rate of seed germination was found to be significantly higher in shw1 hy5 double mutants than respective single mutants (82%). These results indicate that HY5 and SHW1 work in an additive manner in response to ABA-mediated inhibition of seed germination.
SHW1 Physically Interacts with HY5
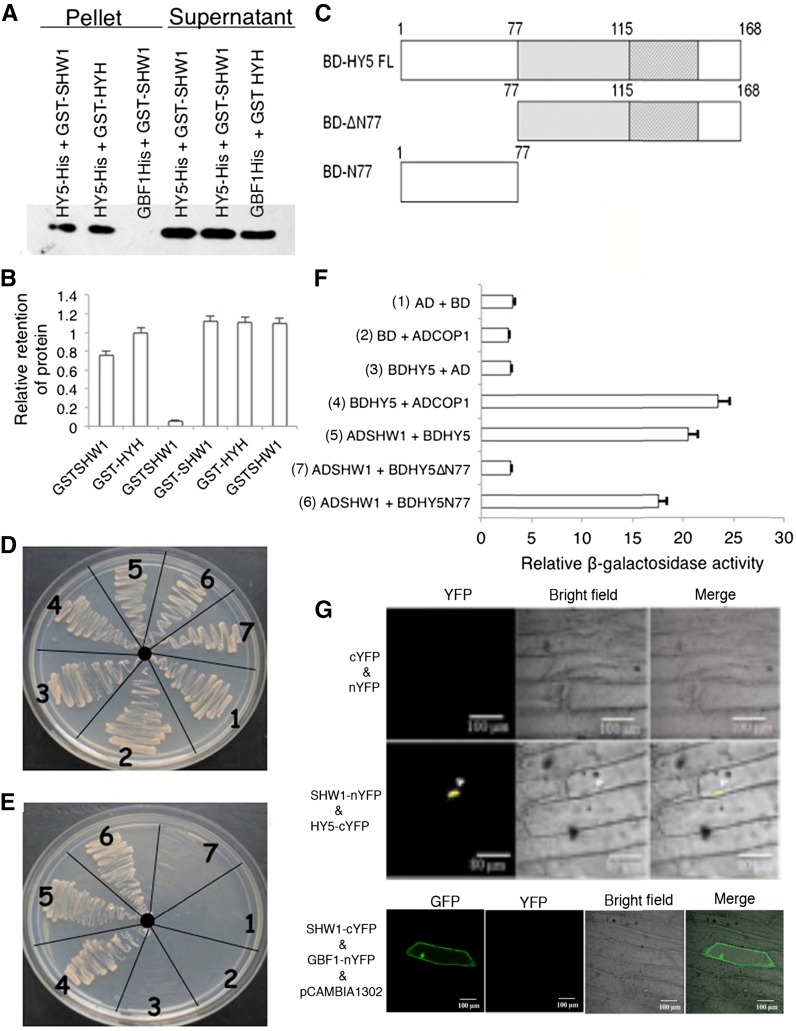
The genetic interaction between SHW1 and HY5 prompted us to investigate the possible physical interactions between SHW1 and HY5. To examine that, in vitro pull-down assays were performed using GST-SHW1 and HY5-His fusion proteins. In these experiments, we used GST-HYH as positive control and GBF1-His as negative control (Holm et al., 2002; Singh et al., 2012). GST-SHW1 and GST-HYH proteins were separately passed through columns containing Ni-NTA beads bound to GBF1-His or HY5-His protein. The level of SHW1 retained by HY5 was similar to the level of retention of HYH by HY5, and practically no retention of SHW1 protein was detected in GBF1 column (Fig. 4, A and B; Supplemental Fig. S3).
Figure 4.
SHW1 physically interacts with HY5. A, In vitro binding of SHW1 and HY5: 2 µg of HY5-6His or GBF1-6His (negative control) was bound to the Ni-NTA beads, washed, and incubated with GST-SHW1 or GST-HYH (positive control; 2 µg). Beads were washed and fractionated in 12% (w/v) SDS-PAGE. The blot was probed with anti-GST antibodies. B, Quantification of the data (by Bio-Rad multi-imager): retention of GST-SHW1 and GST-HYH by COP1-6His or GBF1-6His is shown in the graph. Error bars indicate sem of three replicate experiments. C, Diagram of the domain structure of HY5 and truncated versions of HY5. D and E, Yeast two-hybrid interactions between SHW1, HY5, and the truncated versions of HY5. Growth of cotransformed yeast AH109 strain on double-dropout media (D) and quadruple-dropout media (E). F, The protein-protein interactions were examined by β-galactosidase assays. The relative β-galactosidase activities were calculated according to Clontech instructions. The error bars indicate se (n = 3). The numbers 1 to 7 refer to the constructs used. G, BiFC assay showing that SHW1-nYFP and HY5-cYFP interact to form a functional YFP in the nucleus, whereas SHW1-cYFP and GBF1-nYFP do not interact. GFP containing pCAMBIA-1302 vector was used as a control for transformation. The images show transiently transformed onion epidermal cells.
To further substantiate the observed physical interaction of SHW1 and HY5, yeast two-hybrid assays were performed. In these experiments, we used full-length and truncated versions of HY5 proteins (Fig. 4C). The growth of cotransformed yeast cells on two-dimensional (deficient in Trp and Leu) and four-dimensional (deficient in Trp, Leu, His, and adenine) synthetically defined media plates was monitored. As shown in Figure 4, D and E, the full-length HY5 was able to interact with full-length SHW1. Furthermore, the truncated version of HY5, HY5N77, was also able to interact with SHW1. The comparison of β-galactosidase activity in SHW1 with HY5 and HY5N77 interactions revealed that the β-galactosidase activity was about 6- to 7-fold increased than the background and similar to the increased activity in COP1 and HY5 interaction, used as control (Fig. 4F). These results suggest that SHW1 is able to interact with the full length and the N-terminal domain of HY5 protein.
To further substantiate the physical interactions between SHW1 and HY5, BiFC experiments were carried out. For these experiments, SHW1 full-length coding sequence was fused to the N terminus of YFP in pUC-SPYNE vector (SHW1-nYFP), and HY5 full-length coding sequence was fused to the C terminus of YFP in pUC-SPYCE vector (HY5-cYFP; Singh et al., 2012). The empty vectors or a combination of SHW1-cYFP and GBF1-nYFP did not produce any YFP fluorescence; however, interaction of SHW1 and HY5 produced strong YFP fluorescence in the nucleus (Fig. 4G). Taken together, these results show that SHW1 physically interacts with HY5.
SHW1 Promotes COP1-Mediated Degradation of HY5 in the Darkness
It has earlier been shown that the extent of abundance of HY5 protein directly correlates with the level of photomorphogenesis (Osterlund et al., 2000). Subsequently, it was shown that COP1-SPA complexes interact with various photomorphogenesis-promoting factors, such as HYH, HFR1, and LAF1, and degrade these photomorphogenesis-promoting factors through the ubiquitin/26S proteasome pathway in the dark (Saijo et al., 2003; Seo et al., 2003; Jang et al., 2005; Yang et al., 2005a, 2005b; Lau and Deng, 2012). Earlier studies have shown that additional mutation of SHW1 in cop1 mutants enhances the photomorphogenic phenotype of cop1 mutants in the darkness (Bhatia et al., 2008). This study further shows that SHW1 physically interacts with both COP1 and HY5 (Figs. 1 and 4). Therefore, we ask whether enhanced photomorphogenesis in shw1 cop1 background in the dark is caused by an increased abundance of HY5.
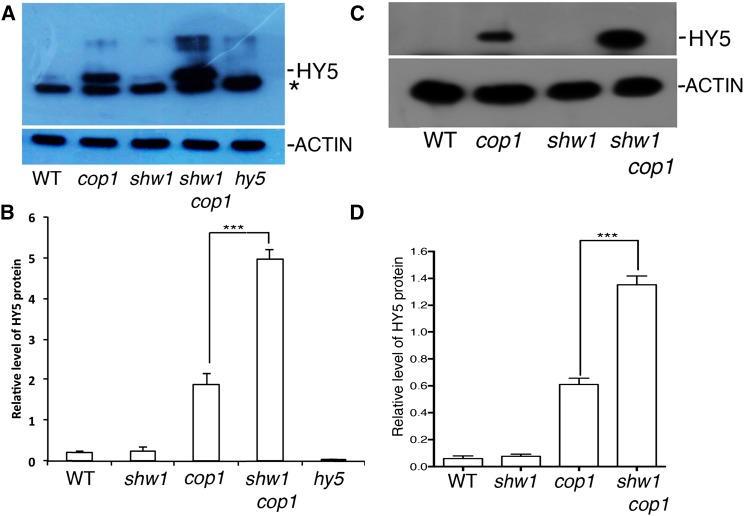
To determine that, we first examined the level of HY5 protein in shw1 and shw1 cop1 double-mutant backgrounds in constant dark-grown seedlings. Whereas HY5 protein was barely detectable in wild-type background, it was accumulated in cop1 mutant background as expected (Fig. 5A; Osterlund et al., 2000). Similar to the wild type, HY5 protein was not detectable in shw1 mutants (Fig. 5A). However, although the transcript level of HY5 remains the same in shw1 and shw1 cop1 backgrounds (Supplemental Fig. S4), the level of accumulation of HY5 protein was drastically enhanced in shw1 cop1 double mutants compared with cop1 single mutants (Fig. 5A). Whereas the level of HY5 protein was elevated to approximately 3-fold in cop1 mutants, the level of accumulation in shw1 cop1 double mutant was increased to approximately 7-fold compared with wild-type background (Fig. 5B).
Figure 5.
HY5 accumulates at a higher level in shw1 cop1 compared with cop1. A, Immunoblot shows HY5 protein levels in wild-type (WT) and cop1, shw1, shw1 cop1, and hy5 mutant seedlings (6-d-old constant dark-grown seedlings). ACTIN bands show the loading control. Total protein was separated on an 8% (w/v) SDS-PAGE gel, blotted onto a polyvinylidene difluoride membrane, and probed with anti-HY5 antibody. *, Cross reacting bands. B, Quantitation of the accumulated HY5 level in the wild type and different mutant lines as in A. Results presented are obtained from three biological repeats. Error bars indicate sd. The Student’s t tests show the significant differences. ***, P < 0.001. C, Immunoblot shows HY5 protein levels in wild-type and cop1, shw1, and shw1 cop1 mutant seedlings (4-d-old WL-grown seedlings were transferred to dark for 2 d). For experimental details, see above. D, Quantitation of the accumulated HY5 level in the wild type and different mutant lines as in C. Results presented are obtained from three biological repeats. Error bars indicate sd. The Student’s t test shows the significant differences. ***, P < 0.001.
To further examine the dark-induced degradation of HY5 in shw1 cop1 background, we grew the seedlings for 4 d in WL and then, transferred them to dark for 2 d and determined the level of HY5 proteins in various single- and double-mutant backgrounds. As shown in Figure 5, C and D, the level of accumulation of HY5 was increased in cop1 mutant (approximately 6-fold), and the level of HY5 was further increased (approximately 14-fold) in shw1 cop1 double mutant. These results indicate that SHW1 promotes COP1-mediated degradation of HY5 in the darkness.
SHW1 Enhances COP1-Mediated Ubiquitination of HY5
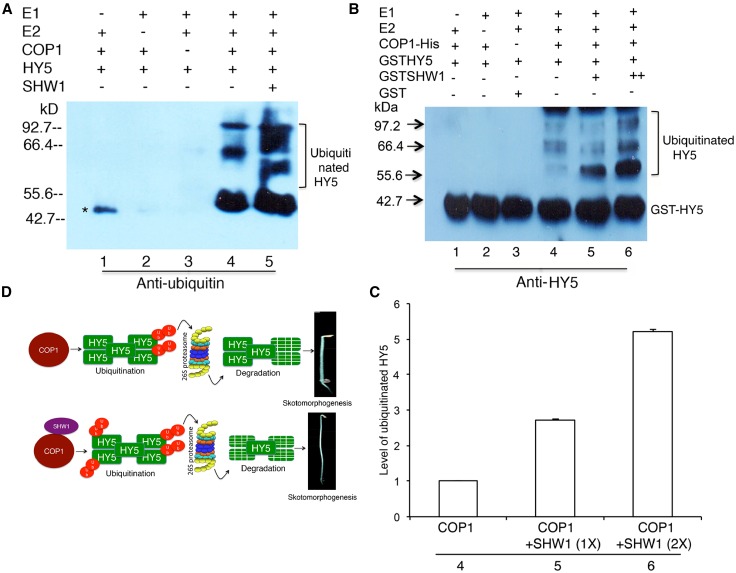
COP1 is an E3 ubiquitin ligase involved in the degradation of multiple positive regulators of photomorphogenesis, including HY5, in the darkness (Osterlund et al., 2000; Holm et al., 2002; Saijo et al., 2003; Seo et al., 2003; Jang et al., 2005; Yang et al., 2005a, 2005b; Hong et al., 2008). To examine the level of ubiquitination of HY5 in the presence or absence of SHW1, we performed in vitro transubiquitylation assays (Saijo et al., 2003; Seo et al., 2003). The western-blot analysis with antiubiquitin antibody revealed that SHW1 enhanced the ubiquitylation of HY5 by COP1 (Fig. 6A). Furthermore, the western-blot results with anti-HY5 antibody also showed the ubiquitylated HY5 (Fig. 6, B and C). No ubiquitylation of SHW1 was detected (Supplemental Fig. S5). The ubiquitylated bands of HY5 in the presence or absence of SHW1 showed that SHW1 was able to increase approximately 2- to 3-fold transubiquitylation of HY5 mediated by COP1. Taken together, these results show that SHW1 promotes COP1-mediated degradation of HY5 by the enhancement of the level of ubiquitination of HY5.
Figure 6.
SHW1 enhances the ability of COP1 to degrade HY5. Recombinant COP1-His, GST-SHW1, and GST-HY5 fusion proteins were purified from Escherichia coli (BL21DE3), and in vitro ubiquitination assays were performed. A, Ubiquitination assays were performed using UBE1 (E1), UbcH5b (E2), and His-tagged ubiquitin (His-6-Ub). Recombinant COP1-His was preincubated with 20 mm zinc chloride. Ubiquitinated HY5 was detected using antiubiquitin antibody. *, Nonspecific band. B, Immunoblot against HY5 antibody shows that increasing concentration of SHW1 results in more ubiquitinated HY5 in vitro. GST was used as a negative control. C, Quantification of the ubiquitinated HY5 level in the presence of COP1 with increasing concentrations SHW1. For experimental detail, see Figure 5B. D, A working model illustrates the regulatory role of SHW1 on HY5 stability. SHW1 interacts with COP1 and promotes COP1-mediated degradation of HY5.
DISCUSSION
This study provides evidence that SHW1, a unique negative regulator of photomorphogenesis, works in concert with COP1 ubiquitin ligase and promotes the degradation of HY5 in the darkness to suppress photomorphogenesis. Several lines of experimental evidences, including protein-protein interactions by two-hybrid and in vitro pull-down assays, show the physical interactions of SHW1 with both HY5 and COP1 proteins. The shw1 cop1 double mutant has earlier been shown to display increased photomorphogenic growth compared with cop1 single mutants in the dark (Bhatia et al., 2008). One plausible mechanism of such genetic and physical interaction of SHW1 with COP1 may be that it controls the COP1-mediated degradation of HY5. Indeed, the level of accumulation of HY5 protein in shw1 cop1 double mutant is increased compared with cop1 mutants in dark-grown seedlings. Furthermore, COP1-mediated ubiquitination of HY5 protein has been enhanced in the presence of SHW1. The higher level of accumulation of HY5 protein in shw1 cop1 double mutant than cop1 mutant alone thereby explains the enhanced photomorphogenic growth of shw1 cop1 double mutants in the darkness.
Negative regulators play important roles in light signaling pathways (Kim et al., 2003; Jiao et al., 2007; Lau and Deng, 2012; Li et al., 2015). COP1 is an essential suppresser of photomorphogenesis in dark-grown Arabidopsis seedlings. COP1 and its interacting partners, SPA proteins, have been shown to establish the skotomorphogenesis in the darkness and prevent hyperphotomorphogenic growth in light-grown seedlings (Hoecker et al., 1998; Laubinger and Hoecker, 2003; Saijo et al., 2003; Seo et al., 2003). However, whereas mutation in any single SPA gene family member does not display any morphological defects in the darkness, shw1 mutants show partial photomorphogenic growth in the darkness. After all four SPA genes are mutated, the quadruple mutant shows strong phenotype similar to cop1 (Laubinger et al., 2004). Recent studies have shown that PIF1 promotes COP1-mediated degradation of HY5 (Xu et al., 2014a, 2014b).
To analyze the physiological significance of the observed protein-protein interactions between SHW1 and HY5, the genetic interactions between these two respective genes were analyzed. These studies have revealed that SHW1 and HY5 act in an antagonistic manner to regulate photomorphogenic growth. The etiolated phenotype of hy5 mutants is partly suppressed in shw1 hy5 double mutants in WL. Although shw1 mutants do not display any significant difference in hypocotyl length in RL, FR light, or BL compared with wild-type background, the additional mutation in SHW1 in hy5 mutant was able to partly suppress the hy5 mutant phenotype in RL and BL. These results establish that SHW1 and HY5 act antagonistically with each other to regulate photomorphogenic growth in RL and BL conditions. The gravitropic root growth defect of hy5 mutants is also suppressed by additional loss of function of SHW1. Although Bhatia et al. (2008) did not find any significant change in root length in shw1 mutants, careful examination in this study revealed that, although not drastic, shw1 mutants indeed display significantly shorter root length than wild-type plants.
Cross talk of light signaling pathways with other signaling cascades has been shown (Boter et al., 2004; Lorenzo et al., 2004; Yadav et al., 2005; Henriques et al., 2009; Huang et al., 2014). Both shw1 and hy5 mutants display less sensitivity to ABA-mediated inhibition of seed germination. This individual effect on ABA responsiveness on seed germination has been enhanced in shw1 hy5 double mutants. Therefore, although SHW1 and HY5 work in an antagonistic manner in photomorphogenic growth, these two proteins function nonredundantly to regulate ABA-mediated inhibition of seed germination. Several reports have earlier shown the differential regulation of a gene in two different signaling pathways (Anderson et al., 2004; Kazan and Manners, 2008). MYC2 works as a negative regulator of light signaling pathways (Yadav et al., 2005; Gangappa et al., 2010; Sethi et al., 2014). However, it works as both positive and negative regulators of jasmonic acid signaling pathways (Dombrecht et al., 2007; Kazan and Manners 2008, 2013). Furthermore, differential genetic interaction of HY5 and GBF1 has been shown to work in a light intensity-dependent manner (Singh et al., 2012). Recent studies have revealed the cross talk between light and temperature signaling pathways (Delker et al., 2014; Johansson et al., 2014; Toledo-Ortiz et al., 2014). It would be interesting to investigate the possible involvement of SHW1 and HY5 in temperature signaling pathways. Furthermore, it would also be interesting to determine the possible involvement of SHW1, as an associated factor of COP1, in COP1-mediated proteasomal degradation of other photomorphogenesis-promoting factors, such as HYH, HFR1, and LAF1, in future studies.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Generation of Double Mutants
The growth conditions and light sources are described in Singh et al. (2012). For the generation of the hy5 shw1 double mutant, homozygous hy5 mutant plants (hy5-215) in Columbia-0 (Col-0) background were crossed with shw1 (shw1-1) in Col-0 background. In the F2 generation, seedlings were grown in WL (30 μmol m−2 s−1) for the identification of hy5 mutant phenotype, and seedlings were selected and transferred to soil. To determine the genotype at the SHW1 locus, approximately 40 seedlings from each line were tested by genomic PCR (Bhatia et al., 2008). For this, individual plants were examined by PCR using the left border-specific primer. F3 progeny that are homozygous for shw1 and hy5 mutant plants were further examined by reverse transcription (RT)-PCR and considered as hy5 shw1 double mutants. The cop1 mutant used in this study is cop1-6 allele (Bhatia et al., 2008).
Root Growth
Seeds were plated on one-half-strength MS (1% [w/v] Suc and 1% [w/v] agar) on vertical square plates and stratified at 4°C in dark conditions for 4 d to induce uniform germination. The plates were placed vertically in racks, and the seedlings were grown under constant WL conditions (40 μmol m−2 s−1). The numbers of lateral roots of the wild type and single and double mutants were counted from 10 to 15 d after germination and tabulated. The root angle was measured by using ImageJ 1.x software (NIH).
Effect of ABA on Seed Germination
The seeds of wild-type, shw1, hy5, and shw1 hy5 mutant plants were plated onto MS plates with or without ABA (Sigma-Aldrich). The plates were kept in cold and dark for 4 d for stratification and then transferred to constant WL, and the seeds were monitored for germination.
Cloning of Constructs
For making the recombinant constructs in each case, the desired fragment was amplified by PCR using a specific pair of forward and reverse primers with restriction sites at the 5′ end of the primers (as indicated in the primers list). Then, the desired fragment and the vectors were digested with the same pair of enzymes. The digested sample was purified from the agarose gel using the gel extraction kit (Qiagen). The different DNA fragments used in this study were ligated into respective vectors with T4 DNA ligase using different molar ratios (insert:vector, 3:1 to 5:1) by incubating overnight at 16°C or 22°C (as per the company’s recommendation for T4 DNA ligase enzyme). Ligated DNA fragments were transformed into Escherichia coli DH5α-competent cells. Colony PCR was performed to confirm the transformed cells with recombinant plasmid. The clones were confirmed by restriction digestion of plasmids and additional sequencing.
For in vitro protein-protein interaction studies, DNA fragments encoding full-length SHW1 or HY5 were cloned into pGEX-4T2 vector using the respective primers at BamHI and EcoRI restriction sites to get GST fusion protein. The DNA fragments encoding full-length HY5 and COP1 were cloned into pET-20b (+) vector using the respective primers at NdeI-ClaI and EcoRI-PstI restriction sites, respectively, to obtain 6× His tag at the C terminus of the protein. For yeast (Saccharomyces cerevisiae) two-hybrid protein-protein interaction assay, full-length SHW1 was cloned into the pGADT7 vector using the respective primers at EcoRI and BamHI restriction sites to produce translational fusion proteins with the activation domain. To generate full-length COP1 constructs, COP1-FL (for full length), COP1-Zn+CC (for coiled coil), and COP1-WD40 were cloned into pGBKT7 vector using the respective primers (Supplemental Table S1) at EcoRI, PstI, and BamHI restriction sites to produce translational fusion with the binding domain. The HY5-FL, HY5-N77 (for N-terminal 77 amino acids), and HY5ΔN77 have been cloned into the pGBKT7 vector using the respective primers (Supplemental Table S1) at EcoRI and BamHI restriction sites to produce translational fusion proteins with the binding domain. For BiFC experiments, full-length CDS (coding sequence) of SHW1 was cloned in the vectors pUC-SPYNE and pUC-SPYCE (Walter et al., 2004) using the respective primers at BamHI and XhoI sites to obtain SHW1-YFPN-ter and SHW1-YFPC-ter fusion proteins, respectively. To obtain COP1-YFPC-ter and COP1-YFPN-ter fusion proteins, full-length CDS of COP1 was cloned in pUC-SPYCE and pUC-SPYNE vectors, respectively, using the respective primers at AscI and XhoI restriction sites.
Pull-Down Assay
In vitro protein-protein interaction studies were carried out as described in Abbas et al. (2014) with slight modifications. DNA fragments encoding full-length SHW1 or HY5 were cloned into pGEX-4T2 vector, obtaining translational fusion constructs with the GST protein. GST-SHW1 and GST-HY5 proteins were overexpressed and purified from E. coli by Glutathione Sepharose 4B Beads (GE). The DNA fragments encoding full-length COP1 or HY5 were cloned into pET-20b (+) vector with 6× His tag at the C terminus of the protein. COP1-His protein was overexpressed and purified from E. coli cells by Ni-NTA Agarose Beads (Qiagen). For in vitro binding experiments, HY5-His and COP1-His (2.0 μg each) proteins were bound to His column by incubating with in vitro pull-down buffer for 2 h at 4°C. Excess unbound protein was washed off, and GST-HYH, GST-SHW1, and GST proteins were added in equimolar ratio and incubated in 500 μL of in vitro pull-down buffer (50 mm Tris-Cl, pH 7.5, 100 mm NaCl, 0.2% [v/v] glycerol, 1 mm EDTA, 0.1% [v/v] Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, and full-strength protease inhibitors cocktail; Sigma) at 4°C. The Ni-NTA beads were collected by brief centrifugation (supernatant was collected separately and saved for further analysis) and washed three times with 1 mL of in vitro pull-down buffer. Pellet was resuspended in full-strength SDS loading buffer, boiled for 5 min, and analyzed by SDS-PAGE for protein binding. Both pellet and supernatant (2%) were probed with anti-GST antibodies.
Total Protein Extraction
The seedlings (100 mg) were frozen in liquid nitrogen and ground in 300 µL of grinding buffer (400 mm Suc, 50 mm Tris-Cl, pH 7.5, 10% [v/v] glycerol, and 2.5 mm EDTA), and phenylmethylsulfonyl fluoride (1 mm stock) was added (0.5 µL for every 100 µL of grinding buffer). The protein extract was transferred to fresh microcentrifuge tube and centrifuged at 5,000 rpm for 5 min to pellet down the debris. The supernatant was transferred to a fresh tube, and an aliquot of 5 µL was taken out in a separate tube for the estimation of protein by Bradford assay. To the rest of the protein extract, appropriate volume of 4× sample buffer (200 mm Tris-Cl, pH 6.8, 400 mm dithiothreitol, 4% [w/v] SDS, 0.025% [w/v] Bromophenol Blue, and 20% [v/v] glycerol) was added and boiled for 5 min before loading on SDS-PAGE.
Yeast Two-Hybrid Assay
Yeast two-hybrid assays were performed using the Matchmaker GAL4-Based Two-Hybrid System as recommended (Clontech Laboratories, Inc.). To investigate the protein-protein interaction, full-length SHW1 was cloned into the pGADT7 vector (Clontech Laboratories, Inc.) with EcoRI-BamHI restriction sites to produce translational fusion proteins with the activation domain. To generate full-length COP1 constructs, COP1-FL, COP1-Zn+CC, and COP1-WD40 were cloned into pGBKT7 vector (Clontech Laboratories, Inc.) with EcoRI-PstI restriction site to produce translational fusion with the binding domain. The HY5-FL, HY5-N77, and HY5∆N77 have been individually cloned into the pGBKT7 vector (Clontech Laboratories, Inc.) with EcoRI-BamHI restriction sites to produce translational fusion proteins with the binding domain. To assess protein-protein interactions, the corresponding plasmids were cotransformed into yeast strain AH109 according to the protocol given by Clontech Laboratories, Inc. Successfully transformed colonies were identified on quadruple-dropout media lacking Trp, Leu, His, and adenine. Expression of GAL4 Activation Domain fused-SHW1 fusion protein was examined by Hemagglutinin antibodies, and GAL4 Binding Domain fused (BD)-COP1, BD-HY5, and their truncated versions were examined by c-Myc antibodies (Supplemental Fig. S6). The protein-protein interactions were also examined by β-galactosidase assays using chlorophenol red-β-d-galactopyranoside as a substrate. The relative β-galactosidase activities were calculated according to Clontech Laboratories, Inc. instructions.
BiFC Assay
Coding sequences corresponding to full-length SHW1, COP1, and HY5 were amplified using respective primers (Supplemental Table S1) cloned under the control of the 35S promoter and fused to the N- and/or C-terminal part of YFP (pUC-SPYNE/pUC-SPYCE; Walter et al., 2004). The desired constructs were mixed in equal proportions (5 μg each) and cobombarded into onion (Allium cepa) epidermal cells as described in Abbas et al., 2014. DNA particle bombardment was performed using the helium-driven particle accelerator (PDS-1000) following the manufacturer’s instructions (Bio-Rad). The bombarded onion peels were kept in the dark for approximately 20 h at 22°C to allow the expression of the transfected DNA and reconstruction of the functional YFP, and then, they were mounted onto glass slide and observed under a confocal laser-scanning microscope (Leica-TCS-SP-2) with a visible aquistic optical tunable filter standard filter set.
RT-PCR Analysis
For RT-PCR, RNA from dark-adapted 6-d-old seedlings was extracted using the RNeasy Plant Mini Kit (Qiagen). One microgram of total RNA was converted into complementary DNA by using Thermo Scientific RevertAid H Minus First-Strand cDNA Synthesis Kit followed by PCR by using gene-specific and ACTIN primers. Primer sequences are given in Supplemental Table S1. The intensity of each band was quantified by the Gel Doc EZ Imager (BIO-RAD), and the ratio of the HY5 versus ACTIN2 band was determined and plotted.
In Vitro Ubiquitylation Assays
In vitro ubiquitination assays were performed as described previously (Seo et al., 2003) with minor modifications. Ubiquitination reaction mixtures (30 μL) contained 30 ng of UBE1 (E1; Boston Biochem), 30 ng of UbcH5b (E2; Boston Biochem), 10 μg of His-tagged ubiquitin (Ub; Boston Biochem), 500 ng of COP1-His (previously incubated with 20 mm zinc chloride), 400 ng of GST-HY5, and 200 ng of GST-SHW1 in a reaction buffer containing 50 mm Tris, pH 7.5, 5 mm MgCl2, 2 mm ATP, and 0.5 mm dithiothreitol. GST was used as a negative control. After 2 h of incubation at 30°C, the reactions were stopped by adding 5× sample buffer. The reaction mixtures (30 μL) were then separated onto 8% (w/v) SDS-PAGE gels. Ubiquitinated HY5 was detected using anti-HY5 (antibody of HY5 was raised against the peptide [IKEGIESDEEIRRVPC] of HY5 by Bangalore GeneI) and anti-GST antibody (Sigma) and anti-ubiquitin antibody. The intensity of the GST-HY5 bands detected by anti-HY5 antibody from three independent blots was quantified using ImageJ software. Briefly, protein levels in western-blot analyses were quantified by Gel Doc EZ Imager (BIO-RAD) using Image Lab software (version 4.0). Bands present in all of the lanes are selected automatically. The band that corresponds to ACTIN in the wild type is taken as relative. By taking this band as standard, the machine automatically quantifies the value of relative intensities of other bands. Conventionally, the machine takes the relative intensity of ACTIN band of the wild type as 1.
Western-Blot Analysis
Western blot was performed using the Super Signal West Pico Chemiluminescent Substrate Kit (Pierce) following the instructions as described in the user’s manual provided by the manufacturer. The samples were then run on SDS-PAGE gel and transferred to a polyvinylidene difluoride membrane (GE) at 130 mA for 1 h in transfer buffer (5.82 g of Tris, 2.93 g of Gly, and 20% [v/v] methanol in 1 L) in a Genei Dual Transfer Unit Apparatus (Banglore Genei). The membrane was stained with Ponceau-S to confirm the protein transfer and washed with sterile milli-Q water. The membrane was then incubated for 1 h in 25 mL of blocking buffer (5% [w/v] nonfat dry milk [Himedia] in phosphate-buffered saline [PBS] and 0.05% [v/v] Tween 20) at room temperature on a dancing rocker. The blocking reagent was removed, and the affinity-purified primary antibody was diluted (1:500 to 1:10,000) in 10 mL of PBS with 0.05% (v/v) Tween 20 and incubated for 2 h with shaking at room temperature. The membrane was then washed with 15 mL of wash buffer (PBS and 0.05% [v/v] Tween 20) three times for 5 min each. The secondary antibody conjugated with horseradish peroxidise diluted 1:10,000 in 10 mL of blocking buffer with 0.05% (v/v) Tween 20 was added and incubated for 1 h with shaking at room temperature. The membrane was washed with 15 mL of wash buffer three times at room temperature. The working solution of substrate was prepared by mixing peroxide solution:luminol/enhancer solution (1:1), and the blot was incubated in that working solution for 5 min in dark. The blot was then removed from the working solution, covered with plastic wrap in cassette, and exposed to x-ray film for different times.
Primers Used in Various Experiments
The primers used in this study are summarized in Supplemental Table S1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Physical interaction between SHW1 and COP1 as shown by in vitro binding assay.
Supplemental Figure S2. ABA-mediated responsiveness of shw1 hy5 double mutants.
Supplemental Figure S3. Physical interaction between SHW1 and HY5 as shown by in vitro binding assay.
Supplemental Figure S4. Transcript levels of HY5.
Supplemental Figure S5. SHW1 enhances the ubiquitylation activity of COP1 but is not itself ubiquitinated by COP1.
Supplemental Figure S6. The expressed proteins in yeast cells.
Supplemental Table S1. Primers used in this study.
Acknowledgments
We thank Xing-Wang Deng for providing the cop1 mutant seeds and Jay Prakash Maurya for critically reading and commenting on this article.
Glossary
- ABA
abscisic acid
- BiFC
bimolecular fluorescence complementation
- BL
blue light
- Col-0
Columbia-0
- FR
far-red
- MS
Murashige and Skoog medium
- PBS
phosphate-buffered saline
- RL
red light
- RT
reverse transcription
- WL
white light
Footnotes
This work was supported by the Department of Science and Technology (Fast Track Young Scientist Grant to A.K.S.), the National Institute of Technology-Durgapur (fellowship to D.S.), the University Grants Commission, Government of India (fellowships to A.S. and M.C.), and J.C. Bose (National Fellowship Grant SR/S2/JCB–85/2010 to S.C.).
Articles can be viewed without a subscription.
References
- Abbas N, Maurya JP, Senapati D, Gangappa SN, Chattopadhyay S (2014) Arabidopsis CAM7 and HY5 physically interact and directly bind to the HY5 promoter to regulate its expression and thereby promote photomorphogenesis. Plant Cell 26: 1036–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadí D, Gallego-Bartolomé J, Orlando L, García-Cárcel L, Rubio V, Martínez C, Frigerio M, Iglesias-Pedraz JM, Espinosa A, Deng XW, et al. (2008) Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J 53: 324–335 [DOI] [PubMed] [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K (2004) Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell 1: 213–222 [DOI] [PubMed] [Google Scholar]
- Ang LH, Deng XW (1994) Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6: 613–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae G, Choi G (2008) Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol 59: 281–311 [DOI] [PubMed] [Google Scholar]
- Bhatia S, Gangappa SN, Kushwaha R, Kundu S, Chattopadhyay S (2008) SHORT HYPOCOTYL IN WHITE LIGHT1, a serine-arginine-aspartate-rich protein in Arabidopsis, acts as a negative regulator of photomorphogenic growth. Plant Physiol 147: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M, Ruíz-Rivero O, Abdeen A, Prat S (2004) Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev 18: 1577–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CS, Maloof JN, Wu SH (2011) COP1-mediated degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis. Plant Physiol 156: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhang J, Neff MM, Hong SW, Zhang H, Deng XW, Xiong L (2008) Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc Natl Acad Sci USA 105: 4495–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J (2011) Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol 21: 664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher plants. Annu Rev Genet 38: 87–117 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Delker C, Sonntag L, James GV, Janitza P, Ibañez C, Ziermann H, Peterson T, Denk K, Mull S, Ziegler J, et al. (2014) The DET1-COP1-HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Reports 9: 1983–1989 [DOI] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Prasad VB, Chattopadhyay S (2010) Functional interconnection of MYC2 and SPA1 in the photomorphogenic seedling development of Arabidopsis. Plant Physiol 154: 1210–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R, Jang IC, Chua NH (2009) Regulated proteolysis in light-related signaling pathways. Curr Opin Plant Biol 12: 49–56 [DOI] [PubMed] [Google Scholar]
- Hoecker U, Xu Y, Quail PH (1998) SPA1: a new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell 10: 19–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, Kim HJ, Ryu JS, Choi H, Jeong S, Shin J, Choi G, Nam HG (2008) CRY1 inhibits COP1-mediated degradation of BIT1, a MYB transcription factor, to activate blue light-dependent gene expression in Arabidopsis. Plant J 55: 361–371 [DOI] [PubMed] [Google Scholar]
- Huang X, Ouyang X, Deng XW (2014) Beyond repression of photomorphogenesis: role switching of COP/DET/FUS in light signaling. Curr Opin Plant Biol 21: 96–103 [DOI] [PubMed] [Google Scholar]
- Jang IC, Yang JY, Seo HS, Chua NH (2005) HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev 19: 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Johansson H, Jones HJ, Foreman J, Hemsted JR, Stewart K, Grima R, Halliday KJ (2014) Arabidopsis cell expansion is controlled by a photothermal switch. Nat Commun 5: 4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM (2008) Jasmonate signaling: toward an integrated view. Plant Physiol 146: 1459–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM (2013) MYC2: the master in action. Mol Plant 6: 686–703 [DOI] [PubMed] [Google Scholar]
- Kim J, Yi H, Choi G, Shin B, Song PS, Choi G (2003) Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17: 584–593 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Fittinghoff K, Hoecker U (2004) The SPA quartet: a family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Hoecker U (2003) The SPA1-like proteins SPA3 and SPA4 repress photomorphogenesis in the light. Plant J 35: 373–385 [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Gao Z, He H, Terzaghi W, Fan LM, Deng XW, Chen H (2015) Arabidopsis DET1 represses photomorphogenesis in part by negatively regulating DELLA protein abundance in darkness. Mol Plant 8: 622–630 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallappa C, Singh A, Ram H, Chattopadhyay S (2008) GBF1, a transcription factor of blue light signaling in Arabidopsis, is degraded in the dark by a proteasome-mediated pathway independent of COP1 and SPA1. J Biol Chem 283: 35772–35782 [DOI] [PubMed] [Google Scholar]
- Mallappa C, Yadav V, Negi P, Chattopadhyay S (2006) A basic leucine zipper transcription factor, G-box-binding factor 1, regulates blue light-mediated photomorphogenic growth in Arabidopsis. J Biol Chem 281: 22190–22199 [DOI] [PubMed] [Google Scholar]
- Nagatani A, Reed JW, Chory J (1993) Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol 102: 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J (2000) Light: an indicator of time and place. Genes Dev 14: 257–271 [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram H, Priya P, Jain M, Chattopadhyay S (2014) Genome-wide DNA binding of GBF1 is modulated by its heterodimerizing protein partners, HY5 and HYH. Mol Plant 7: 448–451 [DOI] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Sethi V, Raghuram B, Sinha AK, Chattopadhyay S (2014) A mitogen-activated protein kinase cascade module, MKK3-MPK6 and MYC2, is involved in blue light-mediated seedling development in Arabidopsis. Plant Cell 26: 3343–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Ram H, Abbas N, Chattopadhyay S (2012) Molecular interactions of GBF1 with HY5 and HYH proteins during light-mediated seedling development in Arabiopsis thaliana. J Biol Chem 287: 25996–26009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA 98: 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Johansson H, Lee KP, Bou-Torrent J, Stewart K, Steel G, Rodríguez-Concepción M, Halliday KJ (2014) The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet 10: e1004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Habricot Y, Condiff AS, Maldiney R, Straeten DVD, Ahmad M (2007) HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J 49: 428–441 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wei N, Deng XW (1999) Making sense of the COP9 signalosome. A regulatory protein complex conserved from Arabidopsis to human. Trends Genet 15: 98–103 [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP (1993) Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5: 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Lin F, Jiang Y, Huang X, Li J, Ling J, Hettiarachchi C, Tellgren-Roth C, Holm M, Deng XW (2014a) The RING-finger E3 ubiquitin ligase COP1 SUPPRESSOR1 negatively regulates COP1 abundance in maintaining COP1 homeostasis in dark-grown Arabidopsis seedlings. Plant Cell 26: 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Paik I, Zhu L, Bu Q, Huang X, Deng XW, Huq E (2014b) PHYTOCHROME INTERACTING FACTOR1 enhances the E3 ligase activity of CONSTITUTIVE PHOTOMORPHOGENIC1 to synergistically repress photomorphogenesis in Arabidopsis. Plant Cell 26: 1992–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V, Kundu S, Chattopadhyay D, Negi P, Wei N, Deng XW, Chattopadhyay S (2002) Light regulated modulation of Z-box containing promoters by photoreceptors and downstream regulatory components, COP1 and HY5, in Arabidopsis. Plant J 31: 741–753 [DOI] [PubMed] [Google Scholar]
- Yadav V, Mallappa C, Gangappa SN, Bhatia S, Chattopadhyay S (2005) A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17: 1953–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lin R, Hoecker U, Liu B, Xu L, Wang H (2005a) Repression of light signaling by Arabidopsis SPA1 involves post-translational regulation of HFR1 protein accumulation. Plant J 43: 131–141 [DOI] [PubMed] [Google Scholar]
- Yang J, Lin R, Sullivan J, Hoecker U, Liu B, Xu L, Deng XW, Wang H (2005b) Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, He H, Wang X, Wang X, Yang X, Li L, Deng XW (2011) Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J 65: 346–358 [DOI] [PubMed] [Google Scholar]
- Zhu D, Maier A, Lee JH, Laubinger S, Saijo Y, Wang H, Qu LJ, Hoecker U, Deng XW (2008) Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20: 2307–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]