The plant hormone ethylene controls fruit ripening through a complex network of transcriptional regulations and interplay between multiple signaling pathways.
Abstract
The plant hormone ethylene plays a key role in climacteric fruit ripening. Studies on components of ethylene signaling have revealed a linear transduction pathway leading to the activation of ethylene response factors. However, the means by which ethylene selects the ripening-related genes and interacts with other signaling pathways to regulate the ripening process are still to be elucidated. Using tomato (Solanum lycopersicum) as a reference species, the present review aims to revisit the mechanisms by which ethylene regulates fruit ripening by taking advantage of new tools available to perform in silico studies at the genome-wide scale, leading to a global view on the expression pattern of ethylene biosynthesis and response genes throughout ripening. Overall, it provides new insights on the transcriptional network by which this hormone coordinates the ripening process and emphasizes the interplay between ethylene and ripening-associated developmental factors and the link between epigenetic regulation and ethylene during fruit ripening.
As a developmental process, fruit ripening is coordinated by a complex network of endogenous and exogenous cues. Indeed, the making of a fruit is a genetically regulated process unique to plants involving three distinct stages: fruit set, development, and ripening. Fruit development is characterized by a series of developmental transitions tightly coordinated by a network of interacting genes and signaling pathways. Among these, ripening has received the greatest attention from both geneticists and breeders. From the scientific point of view, fruit ripening is seen as a process in which the biochemistry and physiology of the organ are developmentally altered to influence the appearance, texture, flavor, and aroma (Giovannoni, 2004). Since most of the fruit sensory and nutritional quality traits are elaborated at the ripening stage, deciphering the key genetic and molecular factors regulating ripening becomes a major task toward improving overall fruit quality (Carrari and Fernie, 2006). In addition, the control of fruit ripening is also instrumental to maintain the quality attributes of the fruit during the postharvest shelf life.
Based on their mode of ripening, fleshy fruits are divided into two categories, climacteric and nonclimacteric, depending on the presence or absence of the climacteric rise in respiration and of autocatalytic ethylene production (Lelièvre et al., 1997). In climacteric fruit, the plant hormone ethylene is the major cue that controls most aspects of ripening. By contrast, the ripening of nonclimacteric fruit does not strictly depend on ethylene, and the nature of the triggers of ripening in this type of fruit remains yet to be elucidated. Since the upstream components of the ethylene transduction pathway are common to all ethylene responses, the apparent simplicity of the ethylene signaling pathway cannot account for the wide diversity of ethylene responses. A plausible hypothesis is that differential responses to ethylene are directed at the level of ethylene response factor (ERF) transcription factors, which are encoded by one of the largest families of plant transcription factors, and therefore, are most suited to conferring such a large diversity and specificity of ethylene responses.
A rich literature indicates that the alteration of most components of ethylene signaling and responses has an impact on the course of maturation (Grierson, 2013). Nevertheless, the understanding of the control mechanisms underlying the specificity of ethylene action requires the uncovering of the components mediating ethylene responses that are specific to each developmental process. For instance, the identification of ripening-associated transcriptional regulators acting upstream or in concert with ethylene has brought new insights into understanding the ripening control mechanisms. Functional characterization of key ripening-related transcriptional regulators, such as RIPENING-INHIBITOR (RIN; Vrebalov et al., 2002; Ito et al., 2008), COLORLESS NONRIPENING (CNR; Manning et al., 2006), NON-RIPENING (NOR; Giovannoni, 2004), TOMATO AGAMOUS-LIKE1 (TAGL1; Itkin et al., 2009; Vrebalov et al., 2009; Giménez et al., 2010), Homeodomain-leucine zipper homeobox protein (LeHB-1; Lin et al., 2008c), MADS-boxS1 (MADS1; Dong et al., 2013), APETALA2a (AP2a; Karlova et al., 2011), SlERF6 (Lee et al., 2012), and SlERF.B3 (Liu et al., 2014), indicates that transcription factors play key roles in relaying ripening-inducing signals and controlling ethylene biosynthesis and signaling. Taking advantage of the newly generated tools and resources on the tomato species, the present review aims to revisit the role of ethylene in fruit ripening by integrating the latest advances on the transcriptional network by which this hormone orchestrates the ripening process. Because most of our knowledge on the role of ethylene in fleshy fruit ripening has been achieved using tomato (Solanum lycopersicum), we will mainly focus on this reference species. In addition, several publicly accessible databases, such as the Tomato Expression Database (Fei et al., 2006) and the TomExpress online tool (http://gbf.toulouse.inra.fr/tomexpress), are used to explore the expression of relevant ripening-related genes.
ETHYLENE BIOSYNTHESIS AND PERCEPTION IN TOMATO FRUIT RIPENING
The involvement of ethylene in fruit ripening was initially reported a long time ago (Burg and Burg, 1962), and since then, direct evidences have accumulated to demonstrate that ethylene mediates fruit ripening at the physiological, biochemical, and molecular levels. Altering ethylene at the level of its biosynthesis, perception, signal transduction, or gene transcription was shown to impact fruit ripening (Hamilton et al., 1990; Oeller et al., 1991; Lanahan et al., 1994; Tieman et al., 2001; Lee et al., 2012; Liu et al., 2014). According to the currently accepted model (Fig. 1), ethylene signaling relies on a linear transduction pathway where the hormone is perceived by a specific receptor, which initiates a signaling cascade by releasing the block exerted by CTR1 on EIN2. This activates a transcriptional cascade, involving EIN3/EIL1 as the primary transcription factor and then ERFs, which in turn regulate genes underlying ripening-related traits, such as color, firmness, aroma, taste, and postharvest shelf life (Solano and Ecker, 1998; Ju et al., 2012; Chang et al., 2013).
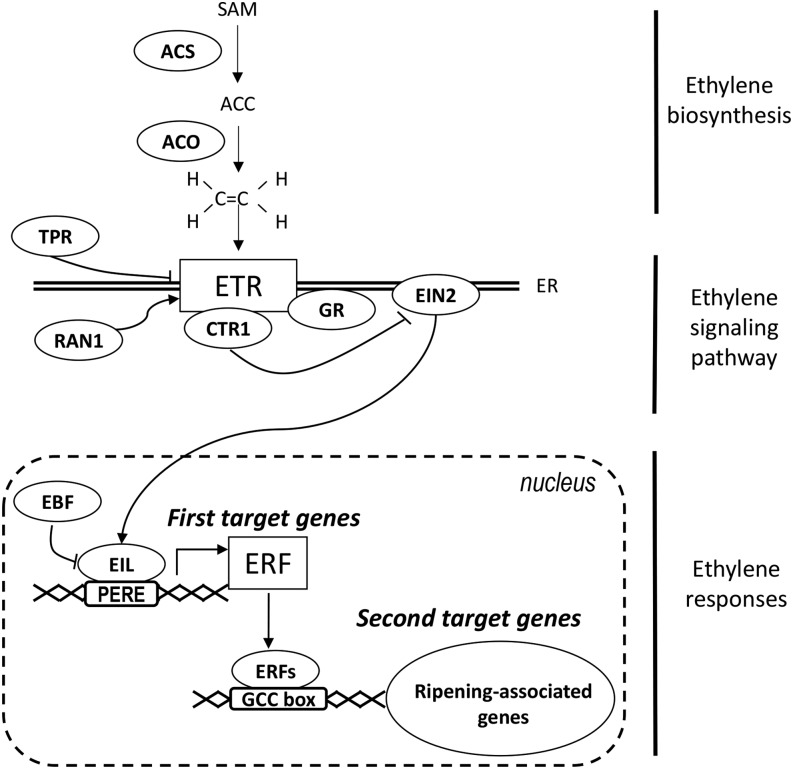
Figure 1.
Simplified scheme showing ethylene synthesis and response in tomato. Ethylene synthesis results from the activity of 1-aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS) and 1-aminocyclopropane-1-carboxylic acid oxidase (ACO), which transform S-adenosyl-l-Met (SAM) into ACC and convert ACC into ethylene, respectively. Ethylene is perceived by the receptor proteins (ETR), located in the endoplasmic reticulum (ER). RAN1 delivers the copper cofactor required for ethylene binding. GR is probably associated with the receptor and mediates the receptor signal output. It is suggested that TPR binds to ethylene receptors and leads to receptor degradation. The receptors are negative regulators of ethylene signaling, and in the absence of ethylene, the receptors activate Constitutive Triple-Response1 (CTR1), which suppresses the ethylene response via inactivation of Ethylene Insensitive2 (EIN2). The transcription factors EIN3/Ethylene Insensitive3-Like1 (EIL1) undergo a degradation process mediated by the Ethylene Insensitive3-binding F-box (EBF) proteins. In the absence of EIL, transcription of ethylene response genes is shut off. Ethylene binding to the receptors induces their inactivation, and by consequence, switches off CTR1 phosphorylation activity. Active EIN2 stabilizes EIL transcription factors, which can activate the expression of target genes, including those encoding the ERF transcription factors via binding to primary ethylene response elements (PEREs; Solano et al., 1998). ERFs, in turn, modulate the transcription of ethylene-regulated genes through binding to GCC-box type cis-elements present in their target promoters. Arrowheads represent positive regulatory interactions, and bar heads represent negative regulation.
Ethylene Biosynthesis Is Instrumental to Climacteric Fruit Ripening
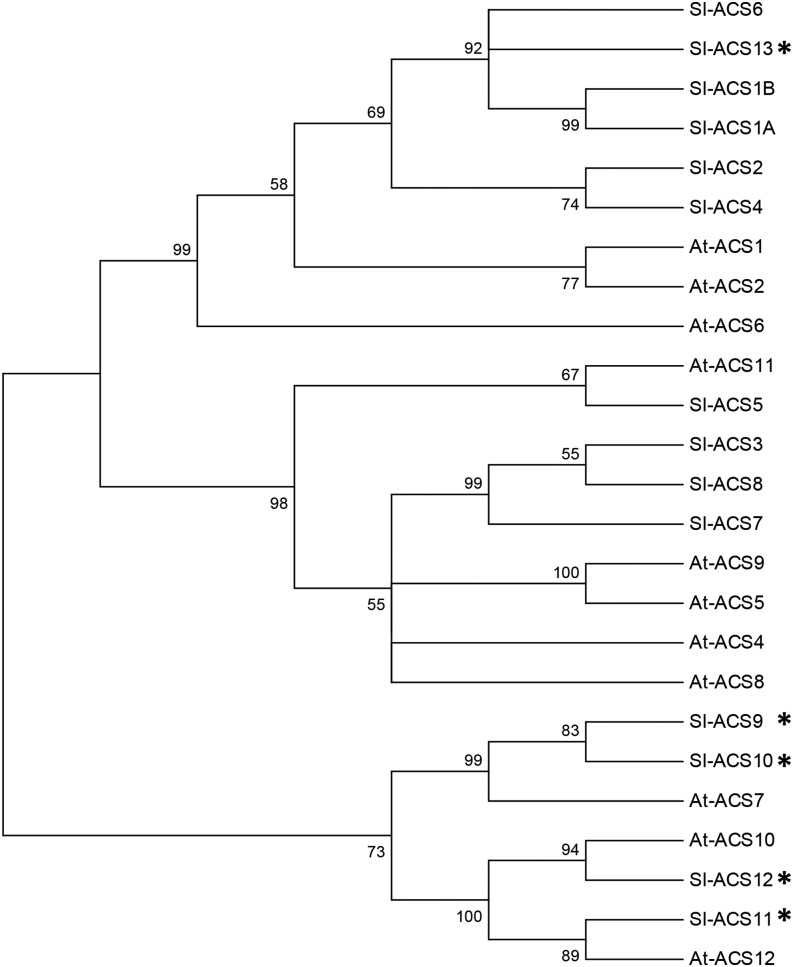
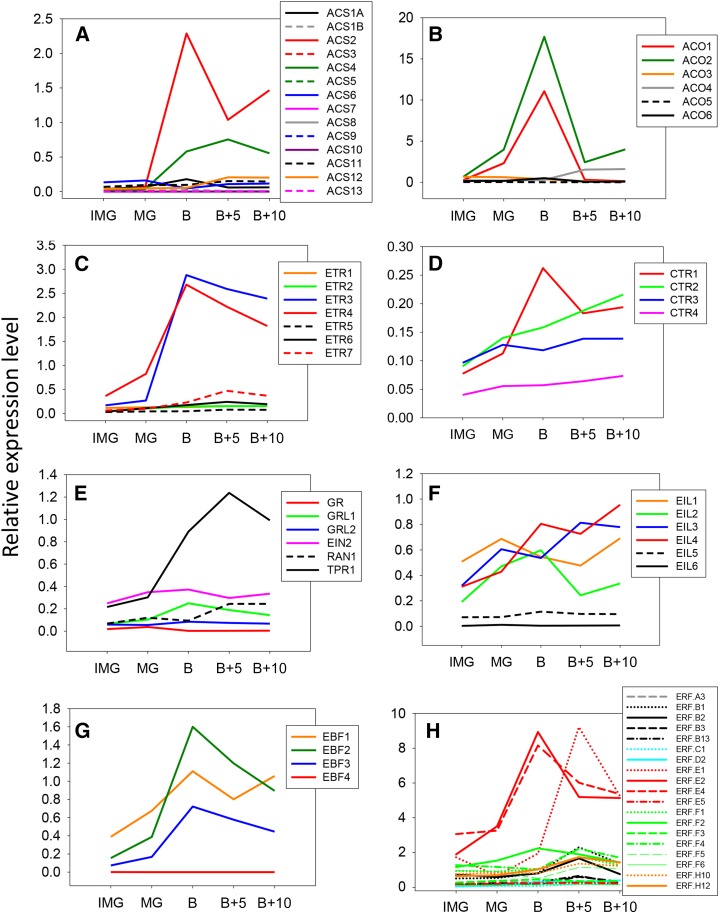
In higher plants, ethylene biosynthesis (Fig. 1) originates from S-adenosyl-Met and comprises two steps catalyzed by ACS and ACO, the latter converting ACC into ethylene (Yang and Hoffman, 1984). The genome-wide search for ACS and ACO genes performed using tBLASTn as the program and SlACS1A and SlACO1 as query identified 14 sequences corresponding to putative ACS and 6 to ACO in the most recent tomato genome sequence (Tomato Genome Consortium, 2012). For the ACS and ACO described here, InterProScan analysis confirmed the presence of specific domains characteristic of these proteins, and the Kyoto Encyclopedia of Genes and Genomes orthology analysis validated the presence of the enzymatic domains, EC:4.4.1.14 and EC:1.14.17.4, characteristic of ACS and ACO, respectively. Even though the ACS proteins have not been biochemically characterized, phylogenetic analysis clustered the 14 putative ACS proteins in the same branch as Arabidopsis (Arabidopsis thaliana) ACSs (Fig. 2). Moreover, the putative tomato ACS genes were further checked for the presence of the pyridoxal 5′-phosphate-binding site, a necessary feature of ACS enzymes (Jakubowicz, 2002), showing that this domain is well conserved in the five new ACS genes identified (ACS9–13), allowing them to be assigned to the ACS group. All genes related to ethylene biosynthesis, perception, or signaling are listed in Supplemental Table S1, providing the correspondence between gene names, Solyc numbers, and, when relevant, other names cited in the literature. The implementation of the newly available TomExpress pipeline (http://gbf.toulouse.inra.fr/tomexpress) allowed in silico mining of the expression pattern of all ACS genes in the tomato based on the publicly available RNA-seq data sets (Fig. 3A). While confirming that ACS2 and ACS4 are the main family members expressed during ripening (Oeller et al., 1991; Theologis et al., 1992), the new expression study also confirmed that ACS1A transcript accumulation peaks at the breaker stage (Barry et al., 2000), suggesting its potential contribution to the climacteric ethylene production, although its expression level is quantitatively lower than ACS2 and ACS4. Moreover, among the new ACS genes, ACS11 and 12 also display a significant up-regulation during fruit ripening, whereas ACS1B, 5, 7, 8, 9, 10, and 13 transcripts are almost undetectable in tomato fruit (Fig. 3A).
Figure 2.
Phylogenetic tree of tomato and Arabidopsis ACS. The phylogenetic tree was inferred using the neighbor-joining method. The optimal tree with the sum of branch length = 3.82205137 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. The evolutionary distances were computed using the Poisson correction method and are the number of amino acid substitutions per site. The analysis involved 25 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 138 positions in the final data set. Phylogenetic trees were conducted in MEGA7. *, New tomato ACS genes identified in the current study.
Figure 3.
Expression data of ethylene biosynthesis and signaling genes during fruit ripening extracted from public databases and processed using the TomExpress platform. Five fruit developmental stages have been studied: immature green (IMG), mature green (MG), breaker (B), 5 d after breaker (B+5), and 10 d after breaker (B+10). Expression patterns for the following gene families are presented: ACS (A), ACO (B), ETR (C), CTR (D), ETR partners (E), EIL (F), EBF (G), and ERF (H). For each gene, the plot represents normalized counts per base for RNA-seq data released from transcriptome analyses in multiple tomato cultivars.
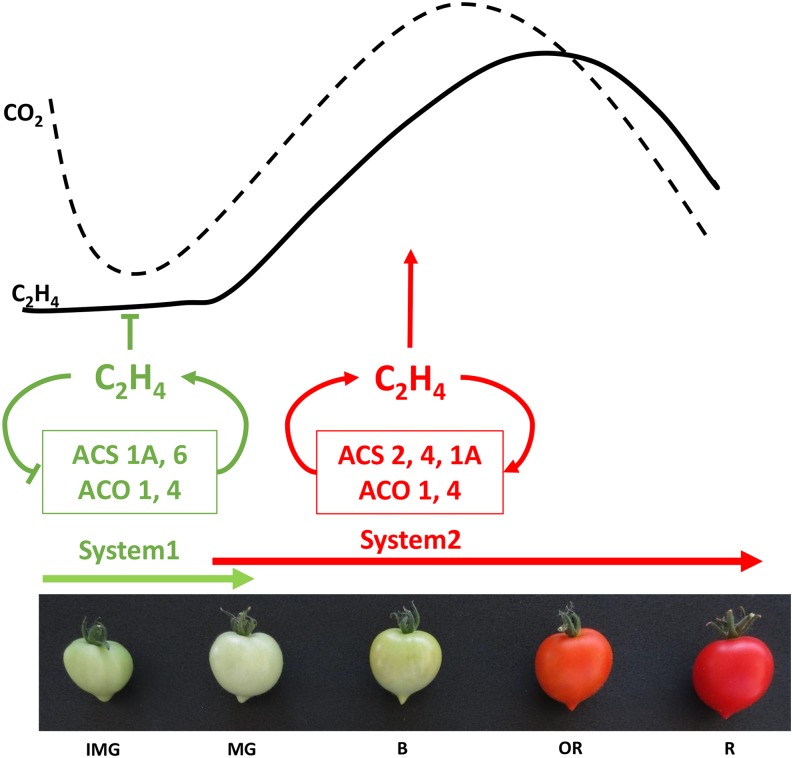
Genome-wide analysis confirmed the presence of six ACO genes in the tomato genome (Seymour et al., 2013), and mining their expression with TomExpress pipeline (Fig. 3B) indicated that ACO1 and ACO2 (Supplemental Table S1) display the most striking ripening-regulated pattern of expression peaking at the breaker stage, whereas ACO4 expression undergoes a steady but slight increase throughout ripening (Barry et al., 1996; Nakatsuka et al., 1998; Van de Poel et al., 2012). The present expression analysis confirms previous studies pointing to ACO1 and ACO4 as the main ACO genes supporting ripening-associated ethylene production (Nakatsuka et al., 1998). The transcript level of ACO3, ACO5, and ACO6 remains very low, suggesting that their contribution to climacteric ethylene production is negligible. Two systems of ethylene biosynthesis have been proposed in climacteric fruits (McMurchie et al., 1972). System 1 is responsible for producing basal ethylene levels that are detected in all tissues, including those of nonclimacteric fruit (Fig. 4). System 1 is known to be ethylene autoinhibitory and is reported to function during fruit growth, whereas system 2 operates during the climacteric ripening and is autocatalytic (Fig. 4). System 1 relies on ACS1A and ACS6, both being negatively regulated by ethylene, whereas the up-regulation of ACS2 and ACS4 through a positive feedback by ethylene is responsible for the activation of system 2 (Nakatsuka et al., 1998; Barry et al., 2000). ACO1 and ACO4 are both expressed at low levels in immature green fruit where system 1 is operating, but their transcripts accumulate with the climacteric rise of ethylene production and are therefore responsible for the transition to system 2 (Fig. 4). Moreover, ACO4 maintains a sustained expression during fruit ripening (Nakatsuka et al., 1998). The expression of ethylene biosynthesis genes was shown to be regulated by developmental regulators, such as RIN and LeHB-1, which modulate the expression of ACS2 and ACO1 through direct binding to their promoter (Lin et al., 2008c; Fujisawa et al., 2013). It is therefore possible that system 2 ethylene production is not the only mechanism contributing to the autocatalytic regulation of climacteric ethylene. Recent data showed that ethylene biosynthesis displays a tissue-specific and developmental differentiation throughout tomato fruit growth, indicating that it is organized and regulated in a well-defined tissue-specific way (Van de Poel et al., 2014).
Figure 4.
Two different systems of ethylene production operate during fruit development and ripening. At immature stages, ethylene biosynthesis is mediated by system 1, whereas system 2 takes over during ripening and is characterized by autocatalytic ethylene production. The main genes involved in system 1 are ACS6 and ACS1A, both genes being down-regulated by ethylene. From mature green stage onward, system 2 ethylene production is driven mainly by ACS2 and ACS4, the expression of which is stimulated by ethylene. ACS1A transcripts show a transient increase at the onset of ripening, suggesting that this gene may be important in regulating the transition from system 1 to system 2. ACO1 and ACO4 transcript levels are low in immature green stages, but undergo sharp increase at the climacteric peak when system 2 ethylene production is operating. IMG, Immature green; MG, mature green; B, breaker; OR, orange; R, red. Arrowheads represent positive regulatory interactions, and bar heads represent negative regulation.
Altered Ethylene Perception Impairs Fruit Ripening
The ethylene receptors have been studied in detail in tomato, where six genes have been initially described, named LeETR1 to LeETR6 (Wilkinson et al., 1995; Lashbrook et al., 1998; Tieman and Klee, 1999; Klee and Tieman, 2002; Gapper et al., 2013). The genome-wide search identified LeETR7 as a new member of the tomato ethylene receptor family. Phylogenetic analysis validated its similarity with other receptors, and its expression pattern was established using the TomExpress pipeline (Fig. 3C; Supplemental Figs. S1 and S2). Like in other plant species, two subfamilies of ethylene receptors are present in tomato. LeETR1, LeETR2, and LeETR3 (also named NR for never ripe) belong to subfamily I, and harbor three transmembrane domains (Supplemental Fig. S1) and His kinase and Histidine kinase-like ATPase (HATPase_c) domains predicted by the SMART online tool (http://smart.embl-heidelberg.de/). LeETR1 and LeETR2, but not LeETR3, have a receiver domain at the C-terminal position containing a phosphoacceptor described as important in eukaryotic two-component systems (Schaller et al., 2011). Subfamily II gathers four receptors, LeETR4 to LeETR7, containing four transmembrane domains as confirmed by the TMpred online tool (http://www.ch.embnet.org/software/TMPRED_form.html). In the Nr mutant, a point mutation, leading to a substitution of Pro to Leu in the N-terminal ethylene binding pocket, results in impaired fruit ripening (Lanahan et al., 1994; Wilkinson et al., 1995). While confirming LeETR3 and LeETR4 as the main receptor genes expressed at the inception of tomato fruit ripening (Kevany et al., 2007; Klee and Giovannoni, 2011), the TomExpress tool revealed that LeETR7 also displays a ripening-regulated expression, being the third most highly abundant receptor transcript during ripening (Fig. 3C).
It has been reported that receptor proteins are regulated by GREEN-RIPE (GR), a small protein made of around 240 amino acids, and the Gr mutant was described to display impaired fruit ripening (Barry and Giovannoni, 2006). A genome-wide search identified three GR genes in the tomato (GR, Green-Ripe Like1 [GRL1], and GRL2). GRL1 is the closest ortholog of the Arabidopsis REVERSION TO ENTHYLENE SENSITIVITY1 (RTE1) gene (Resnick et al., 2006). The rte1 mutants were able to restore ethylene sensitivity in the etr1-2 mutant, suggesting that RTE1 and GR homologs may act at the receptor levels (Resnick et al., 2006). GRL1 is the most expressed homolog during fruit development, and its transcript levels peak at the breaker stage (Fig. 3E). This is consistent with the work by Ma et al. (2012) who were the first to report the expression of GR/GRL1/GRL2 in fruit development, showing that GRL1 displays ripening-related expression. It was also suggested that GR and GRL1 may confer a subfunctionalization of the receptors by mediating different responses to ethylene (Ma et al., 2012). Nevertheless, overexpression of GRL1 or GRL2 does not seem to impact fruit ripening (Klee and Giovannoni, 2011).
Two other proteins, Response to Antagonist1 (RAN1) and tetratricopeptide repeat1 (TRP1), play important roles at the receptor levels. SlRAN1 is the ortholog to the Arabidopsis AtRAN1 that delivers the copper ion, essential for ethylene binding activity (Binder et al., 2010). SlRAN1 shows continuous low expression levels with a slight rise at late ripening stages (Fig. 3E). SlTPR1, known to bind the ethylene receptors, has been suggested to lead to receptor degradation (Lin et al., 2008b). Interestingly, SlTPR1 expression is high in the late ripening stages (Fig. 3E) when LeETR3 and LeETR4, its potential targets, are also highly expressed (Fig. 3C).
CTR1 and Fruit Ripening
The Mitogene-activated protein kinase kinase kinase, known as CTR1, acts directly downstream of the ethylene receptors. The ctr1 loss-of-function mutations result in the constitutive activation of ethylene response in seedlings and adult plants, indicating that the encoded protein acts as a negative regulator of ethylene signaling (Lin et al., 2008a; Klee and Giovannoni, 2011). So far, four CTR1 homologs (SlCTR1, SlCTR2, SlCTR3, and SlCTR4) have been identified in the tomato, three of which can completely (SlCTR3) or partially (SlCTR1 and SlCTR4) complement the Arabidopsis ctr1-8 mutation (Leclercq et al., 2002; Adams-Phillips et al., 2004; Lin et al., 2008a), suggesting a conserved function for tomato CTR proteins. All tomato CTRs display ability to interact with one or more ethylene receptors in yeast two-hybrid systems (Zhong et al., 2008). Tomato and CTR1, 2, 3, and 4 show differential expressions in various plant tissues (Adams-Phillips et al., 2004; Lin et al., 2008a), and the ethylene-responsive CTR1 (Zegzouti et al., 1999; Leclercq et al., 2002) displays a ripening-related expression pattern. Our present study indicates that SlCTR1 displays a typical ripening-regulated expression, whereas SlCTR2 shows a steady increase in its expression during ripening (Fig. 3D) and was up-regulated in ripening-impaired mutants Nr and rin (Lin et al., 2008a), suggesting its putative role in the ripening process. The suppression of SlCTR1 via Virus-induced gene silencing (VIGS) strategy was reported to promote tomato fruit ripening, consistent with CTR being a negative regulator of climacteric ripening (Fu et al., 2005).
EIN2, Another Component of Ethylene Signaling Influencing Ripening
In Arabidopsis, EIN2 is required for all ethylene responses, and based on genetic analyses, EIN2 acts downstream of the receptor/CTR1 complex to positively regulate ethylene responses. It constitutes a critical step in the signal transduction pathway and acts between CTR1 and the EIN3/EIL transcription factors (Alonso et al., 1999; Guo and Ecker, 2003). Although the expression of EIN2 in tomato is ethylene independent and does not exhibit substantial changes during fruit growth and ripening (Fig. 3E), its down-regulation by a cosuppression mechanism or via VIGS strategy resulted in ethylene insensitivity and ripening inhibition associated with reduced expression of ethylene- and ripening-related genes (Fu et al., 2005; Hu et al., 2010), suggesting that LeEIN2 is a positive regulator of ethylene-mediated responses during fruit ripening.
Posttranslational Regulation of Ethylene Perception Proteins and Fruit Ripening
Ethylene receptors are negative regulators of ethylene signaling, and it is therefore rather intriguing that the corresponding genes undergo dramatic up-regulation during fruit ripening (Fig. 3C). Pioneering studies addressing the evolution of ethylene receptor proteins during tomato fruit ripening showed that the levels of receptor transcripts are not correlated with the amount of receptor proteins, thus suggesting that the posttranslational regulation of ethylene perception is an essential mechanism (Kevany et al., 2007). Indeed, exogenous ethylene treatment of immature fruits results in enhanced accumulation of ETR transcripts concomitant with a decrease in the corresponding encoded proteins, and the use of the MG132, an inhibitor of proteasome, suggested that ETR protein degradation was mediated by the proteasome (Kevany et al., 2007). Moreover, these authors developed the hypothesis of a relationship between the phosphorylation status of the receptor proteins and their degradation. In support of this hypothesis, it was reported that the amount of LeETR3 and LeETR4 receptor proteins increases at the onset of ripening, and that the phosphorylation level of some N-terminal residues plays a critical role in switching on or off the downstream ethylene signal transduction (Kamiyoshihara et al., 2012). The phosphorylation status of LeETR4 was shown to decrease over the transition from immature green to breaker stage, and exogenous ethylene induces dephosphorylation of the receptor protein. Taken together, these studies (Kevany et al., 2007; Kamiyoshihara et al., 2012) suggest that, during fruit ripening, ethylene signaling is modulated at the level of the receptor proteins either quantitatively by tuning their amount or by adjusting their phosphorylation status.
TRANSCRIPTIONAL CASCADE LEADING TO THE ACTIVATION OF ETHYLENE-RESPONSIVE GENES
EIL Proteins in Fruit Ripening
Ethylene regulates ripening-related genes through a transcriptional cascade that comprises primary (EIL) and secondary response factors (ERFs). Four tomato EIL genes (SlEIL1, SlEIL2, SlEIL3, and SlEIL4) were initially described (Tieman et al., 2001; Yokotani et al., 2003), and mining the most updated tomato genome sequence identified two additional genes (named here as SlEIL5 and SlEIL6) based on the presence of the typical domains characteristic of Arabidopsis EIL proteins, including the acidic and basic domains as well as the Pro-rich domain. Tomato SlEIL1, SlEIL2, SlEIL3, and SlEIL4 genes exhibit a ripening-associated pattern of expression, with SlEIL1 and SlEIL2 transcripts accumulating at the onset of ripening and declining at later stages, whereas those corresponding to SlEIL3 and SlEIL4 show a steady increase throughout ripening (Fig. 3F). Notably, the expression of SlEIL5 and SlEIL6 is not regulated during fruit ripening, which may suggest distinct roles among EILs. Down-regulation of SlEIL genes in transgenic tomato plants altered fruit ripening (Tieman et al., 2001), and overexpression of SlEIL1 in the tomato Nr mutant partially restored normal fruit ripening and stimulated the expression of some ethylene-responsive genes, supporting the role of EILs in ethylene-mediated fruit ripening (Chen et al., 2004). Moreover, down-regulation of SlEIL genes resulted in limited increase in SlACS2 and SlACS4 expression (Yokotani et al., 2009), suggesting that EILs might be essential for the activation of genes involved in autocatalytic ethylene production. A new phosphorylation region, named EIN3/EIL phosphorylation region1, has been shown to be essential for the transcriptional activity of tomato SlEIL1 and dimerization of SlEIL1 proteins (Li et al., 2012). In Arabidopsis, EIL proteins are known to be regulated by EBFs at the posttranslational level (Guo and Ecker, 2003). Two tomato homologs of these F-box proteins, EBF1 and EBF2, have been shown to regulate ethylene signaling and fruit ripening through mediating the degradation of EIN3/EIL proteins (Yang et al., 2010). Mining the annotated tomato genome sequence identified two new EBF proteins (SlEBF3 and SlEBF4) based on the presence of conserved F-box domains and Leu-rich repeats. SlEBF1, SlEBF2, and SlEBF3 exhibit a typical ripening-associated expression pattern with a peak of transcript accumulation at the onset of ripening (Fig. 3G), suggesting that EBFs may actively contribute to the control of ripening-associated ethylene signaling.
ERFs and the Regulation of Fruit Ripening
The ethylene signaling cascade ends with transcriptional activation of the transcription factors termed ERFs. ERFs belong to the AP2/ERF superfamily shown to regulate the expression of ethylene-responsive genes through direct binding to their promoter regions (Ohme-Takagi and Shinshi, 1995; Pirrello et al., 2012). ERFs represent one of the largest plant multigene families of transcription factors, which makes these components suited to channel the ethylene signaling toward specific responses through recruiting the appropriate ethylene-responsive genes. Taking advantage of the recently released annotated tomato genome sequence (Tomato Genome Consortium, 2012), 146 genes were postulated to encode proteins containing the AP2/ERF domain, of which 77 belong to the ERF subfamily (Pirrello et al., 2012). Although our knowledge of the specific functions assigned to tomato ERFs is still scarce, in recent years, an increasing number of studies showed that ERF proteins play an important role in fruit ripening. Most of the tomato ERF genes identified so far are ethylene inducible and show ripening-related expression (Pirrello et al., 2012; Liu et al., 2014). Comprehensive expression analysis using the TomExpress online tool revealed that 55 out of 77 ERF family genes exhibit a ripening-associated pattern of expression, with 27 being up-regulated during ripening, whereas the remaining 28 are down-regulated, which suggests that different ERFs may have contrasting roles during fruit ripening (M. Liu, B. Lima Gomes, E. Purgatto, L.E.P. Peres, E. Maza, M. Zouine, J.P. Roustan, M. Bouzayen, and J. Pirrello, unpublished data). SlERF.E1, SlERF.E2, and SlERF.E4 exhibit the highest level of expression during ripening (Fig. 3H) and show dramatic down-regulation in rin, nor, and Nr tomato ripening mutants (M. Liu, B. Lima Gomes, E. Purgatto, L.E.P. Peres, E. Maza, M. Zouine, J.P. Roustan, M. Bouzayen, and J. Pirrello, unpublished data), suggesting that members of subclass E may have the most prominent role in regulating the ripening process. Interestingly, these three ERFs are among the 23 ERFs identified by chromatin immunoprecipitation on chip (ChIP-chip) and chromatin immunoprecipitation coupled to sequencing (ChIP-seq) approaches as potential direct targets of the RIN key ripening regulator (Fujisawa et al., 2013; Zhong et al., 2013). Altogether, these data are consistent with the assumption that ERF genes are important components of ethylene- and RIN/NOR-dependent ripening and suggest that ERFs may represent the link between ethylene signaling and developmental regulation of fruit ripening. Further supporting the active role of ERFs in fruit ripening, overexpressing SlERF.H1 (Supplemental Table S1) resulted in constitutive ethylene response and accelerated tomato fruit ripening (Li et al., 2007). A ripening-related pattern of expression has also been shown for SlERF.E1 (LeERF2) and SlERF.A3 in tomato fruit (Tournier et al., 2003; Chen et al., 2008; Supplemental Table S1). Moreover, a systems biology approach identified SlERF.E4 as a negative regulator of ethylene and carotenoid biosynthesis in fruit ripening (Lee et al., 2012). More recently, the use of a dominant repression strategy revealed that SlERF.B3 is involved in the control of fruit ripening by regulation of climacteric ethylene production and carotenoid accumulation (Liu et al., 2013, 2014). In other climacteric fruits, such as apple (Malus domestica), banana (Musa spp.), plum (Prunus salicina), and papaya (Carica papaya), although direct evidence showing the involvement of ERF family genes in fruit ripening is lacking, some ERFs were reported to exhibit a ripening-associated expression pattern (Wang et al., 2007; El-Sharkawy et al., 2009; Li et al., 2013). In concert with the master regulator RIN, ERFs regulate autocatalytic ethylene biosynthesis in climacteric fruit ripening, and can directly modulate the expression of ripening-related genes involved in various metabolic pathways activated during fruit ripening.
TRANSCRIPTION FACTORS REGULATING FRUIT RIPENING IN CONCERT WITH ETHYLENE
It is widely accepted that climacteric fruit ripening involves a complex interplay between ethylene and ripening-associated developmental regulators (Fig. 5). Indeed, the cloning of genes responsible for impaired-ripening mutations in the tomato, including RIN, NOR, and CNR, represents a major breakthrough in deciphering the transcriptional control underlying fruit ripening. Fruits produced by rin, nor, and Cnr mutants exhibit inhibited ripening that cannot be rescued by exogenous ethylene treatment (Klee and Giovannoni, 2011; Karlova et al., 2014). The RIN gene encodes a MADS-box transcription factor, and molecular studies showed that RIN protein can directly bind to the promoters of ACS2, ACS4, and ACO1 ethylene biosynthesis genes, NR and ETR4 ethylene receptor genes, and ERF genes (Fujisawa et al., 2013; Zhong et al., 2013; M. Liu, B. Lima Gomes, E. Purgatto, L.E.P. Peres, E. Maza, M. Zouine, J.P. Roustan, M. Bouzayen, and J. Pirrello, unpublished data). These data provide convincing evidence for a link between the RIN-mediated transcriptional regulation and ethylene during fruit ripening. On the other hand, ethylene was shown to regulate the expression of RIN, suggesting an active interplay between RIN and ethylene signaling (Fujisawa et al., 2013). The Cnr mutant is due to an epigenetic change that alters the methylation of a gene encoding a putative SQUAMOSA promoter-binding (SBP) protein, which results in pleiotropic ripening inhibition and inhibited expression of ethylene-associated genes, including ACO1, E8, and NR (Manning et al., 2006; Osorio et al., 2011). Ethylene biosynthesis is impaired in the tomato nor mutant, and it was recently shown that nor has a more global effect on ethylene-related gene expression than rin (Osorio et al., 2011). LeHB-1, another transcription factor, can bind the LeACO1 promoter, and silencing of LeHB-1 via VIGS strategy results in down-regulation of LeACO1 expression associated with delayed fruit ripening (Lin et al., 2008c). The TAGL1 gene, which is highly expressed during fruit ripening, was reported to act as a positive regulator of fruit ripening, and TAGL1 knock-down fruits produce lower amounts of ethylene with a reduced expression of LeACS2, suggesting that TAGL1 controls fruit ripening by regulating ethylene biosynthesis (Itkin et al., 2009; Vrebalov et al., 2009). The putative transcription factor SlAP2a, a member of the AP2/ERF superfamily gene, was described as a negative regulator of fruit ripening and ethylene production and signaling since its down-regulation leads to higher levels of ethylene and fast ripening (Chung et al., 2010; Karlova et al., 2011). Likewise, SlMADS1 is a negative regulator of fruit ripening, and its down-regulation via RNA interference strategy results in early ripening and increased ethylene production (Dong et al., 2013). More recently, SlNAC1 (for tomato NAM, ATAF1/2, CUC2), a new tomato NAC domain protein whose expression increases in ripening fruit, was described as a negative regulator of ripening. Its overexpression resulted in altered carotenoid pathway and decreased ethylene synthesis mainly due to the reduced expression of system 2 ethylene biosynthetic genes (Ma et al., 2014). These data indicate that both positive and negative ripening regulators are involved in the control of fruit ripening, at least partially in an ethylene-dependent pathway. Interestingly, although the transcription factors Fruitfull1 (FUL1) and FUL2 were initially reported to impact fruit ripening in an ethylene-independent manner (Bemer et al., 2012), recent evidences support the involvement of FUL1/FUL2 in the regulation of ethylene biosynthesis during fruit ripening (Fujisawa et al., 2014; Shima et al., 2014; Wang et al., 2014).
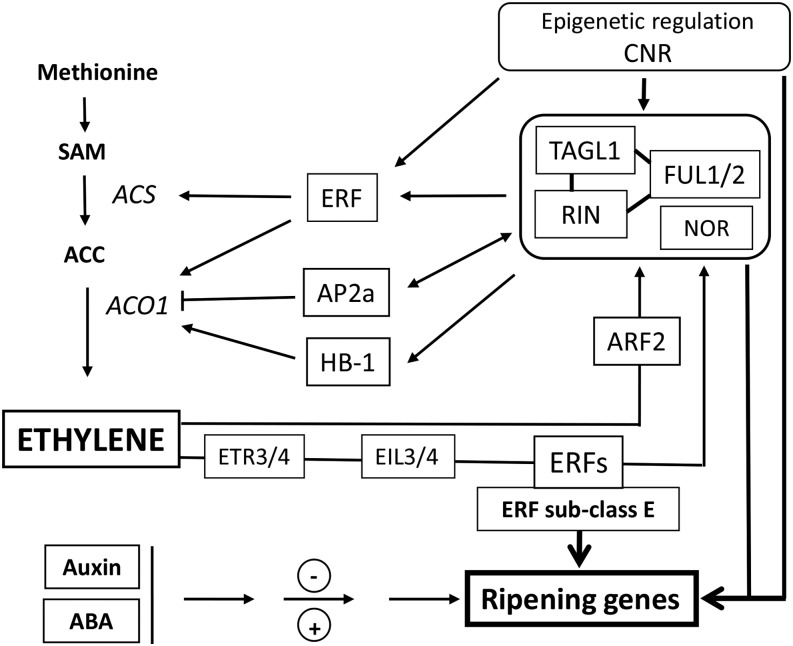
Figure 5.
Schematic overview of the multifactor regulatory network involved in ethylene biosynthesis and signaling during fruit development and ripening. RIN, TAGL1, and FUL1/2 are linked since they probably function as complexes of varying composition. The ripening master regulator NOR is placed in the same box. Along with CNR, these factors are master regulators of climacteric ripening. CNR affects the expression of RIN, LeHB1, SlAP2a, and SlTAGL1. FUL1 and FUL2 can potentially regulate ethylene biosynthesis, perception, and signaling genes. RIN promotes ripening via direct regulation of some transcription factors, such as ERFs. SlERF.B1 and SlERF.E1 are hypermethylated in cnr and rin mutants. ERFs regulate ethylene production in tomato by interaction with the promoters of ACO. Another transcription factor, LeHB-1, can bind in vivo to the promoter of ACO. The putative transcription factor Sl-AP2a was described as a negative regulator of fruit ripening and ethylene production. In addition, the control of ethylene biosynthesis can be regulated by RIN through direct interaction with the promoters of ACS2, ACS4, and ACO1. The ethylene biosynthesis pathway is controlled by a feedback mechanism, where ethylene regulates the expression of RIN. Moreover, there is evidence that ARFs also contribute to this complex feedback mechanism. ERF-type transcription factors are involved in fruit ripening through the control of ethylene and carotenoid biosynthesis pathways in tomato. Other hormones, such as Auxin and abscisic acid (ABA), also play a role in tuning fruit ripening. In particular, ARF2 was reported to be an essential component of the regulatory network controlling fruit ripening in tomato. Arrowheads represent positive regulatory interactions, and bar heads represent negative regulation. SAM, S-adenosyl-l-Met.
EPIGENETIC REGULATION OF ETHYLENE-REGULATED FRUIT RIPENING
Deciphering the basis of the tomato Cnr epimutation provided the initial clue on the epigenetic control of fruit ripening by demonstrating that the impaired ripening phenotype is due to hypermethylated cytosines in the promoter of SQUAMOSA Promoter Binding Protein-like (LeSPL)-CNR, a gene encoding the SBP-box transcription factor (Manning et al., 2006). Subsequently, it was shown that demethylation is essential for climacteric ethylene production, and that treatment of immature fruit with an inhibitor of methyltransferases results in early ripening, indicating that DNA methylation impacts the transition from system 1 to system 2 of ethylene production (Zhong et al., 2013). Demethylation is critical to the binding of RIN protein to the promoter of ripening genes (Zhong et al., 2013), and repression of a DEMETER-like DNA demethylase in tomato results in DNA hypermethylation, ripening inhibition, and a dramatic decrease in climacteric ethylene production (Liu et al., 2015). Furthermore, the hypermethylated cnr mutant can be rescued by down-regulating the tomato Chromomethylase3 gene, a plant-specific CHROMOMETHYLASE (Chen et al., 2015).
Global methylation level at the 5′ end of genes gradually declines during fruit development while remaining high in the tomato ripening-deficient Cnr and rin mutants. The RIN binding sites in ACS4 and ACO1 genes undergo decreased methylation during tomato fruit ripening; by contrast, these sites remain hypermethylated in cnr and rin mutants (Fig. 5). Likewise, the ethylene response components, SlERF.B1 and SlERF.E1, are hypermethylated in cnr and rin mutants compared with the wild type (Zhong et al., 2013). These data suggest that regulation of the ethylene pathway through RIN is strongly controlled by the methylation status of target genes.
ETHYLENE AND OTHER PHYTOHORMONES IN FRUIT RIPENING
It has long been considered that other plant hormones besides ethylene are likely required for climacteric fruit ripening (Dostal and Leopold, 1967; Frenkel and Dyck, 1973; Mizrahi et al., 1975; Fan et al., 1998). ABA is known to promote ripening, whereas auxin seems to have an antagonistic effect (Frenkel and Dyck, 1973; Mizrahi et al., 1975; Zhang et al., 2009; Su et al., 2015). The expression of ACS2, ACS4, and ACO1 genes is induced by exogenous ABA, revealing an ABA/ethylene interplay operating at the level of ethylene biosynthesis (Chernys and Zeevaart, 2000; Jiang et al., 2000; Zhang et al., 2009). Down-regulation of the key ABA biosynthesis enzyme 9-cis-epoxycarotenoid dioxygenase1 in tomato fruit resulted in altered firmness and color but surprisingly higher ethylene production, indicating the complexity of the ABA/ethylene interplay during ripening (Sun et al., 2012). In tomato, ABA might also be perceived through an ethylene-independent pathway that is mediated by tomato Zinc Finger Transcription Factor (Weng et al., 2015).
The expression of ethylene biosynthesis and signaling genes is regulated by auxin in tomato and other fleshy fruits, such as peach (Prunus persica; Gillaspy et al., 1993; Jones et al., 2002; Trainotti et al., 2007; Pirrello et al., 2012). The auxin inhibitor p-Chlorophenoxyisobutyric acid mimics ACC treatment, confirming the antagonistic action of the two hormones during fruit ripening, and auxin delays tomato ripening by affecting a set of key factors, such as RIN, ethylene, and ABA (Su et al., 2015). Consistent with the role of auxin in fruit ripening, tomato fruit firmness was shown to be partly regulated by tomato Auxin Response Factor4 (SlARF4), a transcription factor known to mediate auxin responses (Jones et al., 2002; Guillon et al., 2008; Sagar et al., 2013). More recently, SlARF2, a tomato auxin response factor, was described as an essential component of the regulatory network controlling fruit ripening. Indeed, tomato fruits underexpressing SlARF2 exhibited dramatic ripening defects associated with reduced climacteric ethylene production and dramatic down-regulation of the key ripening regulators RIN, CNR, and NOR (Hao et al., 2015). These data highlight the complex interplay between ethylene and other hormone-signaling components during fruit ripening. Further sustaining the idea of an interplay between ethylene and auxin during fruit ripening is the ethylene-induced expression of PIN-FORMED1 auxin transporter and the requirement of high auxin levels to produce large amounts of system 2 ethylene in peaches (Trainotti et al., 2007; Tatsuki et al., 2013).
CONCLUSION
During the last decade, the implementation of advanced high-throughput technologies in genomics, metabolomics, and proteomics threw new light on the mechanisms by which ethylene regulates the ripening process. Although these studies confirmed ethylene as the main hormone regulating climacteric ripening, they provided evidence supporting the intervention of a complex network of interacting signaling pathways (Fig. 5). Indeed, it is now clear that hormonal and developmental factors act in concert to tune the whole set of ripening-associated pathways. The emerging idea is that fruit development and ripening are complex multilevel processes depending on the coordinated action of master regulators, including multiple hormone signaling, microRNAs, epigenetic maintenance, and epigenetic modifying genes. Future challenges will consist of unraveling the molecular mechanisms underlying the specificity of ethylene responses during plant development and fruit ripening. It is particularly important to uncover how the ethylene perception system evolves at the protein level and to address the functional significance of individual ERF genes. Deciphering the function of ERF genes in both ethylene-dependent and ethylene-independent processes during ripening and identifying the target genes of individual ERFs will be instrumental to better clarify their specific contribution to fruit ripening. Moreover, deciphering the ethylene receptor subfunctionalization and assigning specific roles to ERF members will open new avenues toward engineering fruit development and ripening via targeted approaches, especially when aiming to enhance some desirable traits and metabolic pathways and to reduce unwanted ones.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic tree of tomato ETRs.
Supplemental Figure S2. Expression pattern of ETR genes during fruit ripening.
Supplemental Table S1. Correspondence between common names for the genes and their Solyc numbers.
Supplementary Material
Acknowledgments
The authors thank Mohamed Zouine, Elie Maza, and Pierre Frasse for assistance in the setup of the TomExpress platform.
Glossary
- ACC
1-aminocyclopropane-1-carboxylic acid
- ABA
abscisic acid
Footnotes
This work was supported by the “Laboratoire d'Excellence” entitled TULIP (grant no. ANR–10–LABX–41 to M.B.) and the networking activities within the European Cooperation In Science and Technology Action FA1106.
References
- Adams-Phillips L, Barry C, Kannan P, Leclercq J, Bouzayen M, Giovannoni J (2004) Evidence that CTR1-mediated ethylene signal transduction in tomato is encoded by a multigene family whose members display distinct regulatory features. Plant Mol Biol 54: 387–404 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Barry CS, Blume B, Bouzayen M, Cooper W, Hamilton AJ, Grierson D (1996) Differential expression of the 1-aminocyclopropane-1-carboxylate oxidase gene family of tomato. Plant J 9: 525–535 [DOI] [PubMed] [Google Scholar]
- Barry CS, Giovannoni JJ (2006) Ripening in the tomato Green-ripe mutant is inhibited by ectopic expression of a protein that disrupts ethylene signaling. Proc Natl Acad Sci USA 103: 7923–7928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry CS, Llop-Tous MI, Grierson D (2000) The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol 123: 979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemer M, Karlova R, Ballester AR, Tikunov YM, Bovy AG, Wolters-Arts M, Rossetto P de B, Angenent GC, de Maagd RA (2012) The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 24: 4437–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder BM, Rodríguez FI, Bleecker AB (2010) The copper transporter RAN1 is essential for biogenesis of ethylene receptors in Arabidopsis. J Biol Chem 285: 37263–37270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burg SP, Burg EA (1962) Role of ethylene in fruit ripening. Plant Physiol 37: 179–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Fernie AR (2006) Metabolic regulation underlying tomato fruit development. J Exp Bot 57: 1883–1897 [DOI] [PubMed] [Google Scholar]
- Chang KN, Zhong S, Weirauch MT, Hon G, Pelizzola M, Li H, Huang SS, Schmitz RJ, Urich MA, Kuo D, et al. (2013) Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2: e00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Alexander L, Grierson D (2004) Constitutive expression of EIL-like transcription factor partially restores ripening in the ethylene-insensitive Nr tomato mutant. J Exp Bot 55: 1491–1497 [DOI] [PubMed] [Google Scholar]
- Chen G, Hu Z, Grierson D (2008) Differential regulation of tomato ethylene responsive factor LeERF3b, a putative repressor, and the activator Pti4 in ripening mutants and in response to environmental stresses. J Plant Physiol 165: 662–670 [DOI] [PubMed] [Google Scholar]
- Chen W, Kong J, Qin C, Yu S, Tan J, Chen YR, Wu C, Wang H, Shi Y, Li C, et al. (2015) Requirement of CHROMOMETHYLASE3 for somatic inheritance of the spontaneous tomato epimutation Colourless non-ripening. Sci Rep 5: 9192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernys JT, Zeevaart JA (2000) Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiol 124: 343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, Klein P, Giovannoni J (2010) A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J 64: 936–947 [DOI] [PubMed] [Google Scholar]
- Dong T, Hu Z, Deng L, Wang Y, Zhu M, Zhang J, Chen G (2013) A tomato MADS-box transcription factor, SlMADS1, acts as a negative regulator of fruit ripening. Plant Physiol 163: 1026–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostal HC, Leopold AC (1967) Gibberellin delays ripening of tomatoes. Science 158: 1579–1580 [DOI] [PubMed] [Google Scholar]
- El-Sharkawy I, Sherif S, Mila I, Bouzayen M, Jayasankar S (2009) Molecular characterization of seven genes encoding ethylene-responsive transcriptional factors during plum fruit development and ripening. J Exp Bot 60: 907–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Mattheis JP, Fellman JK (1998) A role for jasmonates in climacteric fruit ripening. Planta 204: 444–449 [Google Scholar]
- Fei Z, Tang X, Alba R, Giovannoni J (2006) Tomato Expression Database (TED): a suite of data presentation and analysis tools. Nucleic Acids Res 34: D766–D770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel C, Dyck R (1973) Auxin inhibition of ripening in Bartlett pears. Plant Physiol 51: 6–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu DQ, Zhu BZ, Zhu HL, Jiang WB, Luo YB (2005) Virus-induced gene silencing in tomato fruit. Plant J 43: 299–308 [DOI] [PubMed] [Google Scholar]
- Fujisawa M, Nakano T, Shima Y, Ito Y (2013) A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25: 371–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M, Shima Y, Nakagawa H, Kitagawa M, Kimbara J, Nakano T, Kasumi T, Ito Y (2014) Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell 26: 89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapper NE, McQuinn RP, Giovannoni JJ (2013) Molecular and genetic regulation of fruit ripening. Plant Mol Biol 82: 575–591 [DOI] [PubMed] [Google Scholar]
- Gillaspy G, Ben-David H, Gruissem W (1993) Fruits: A Developmental Perspective. Plant Cell 5: 1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giménez E, Pineda B, Capel J, Antón MT, Atarés A, Pérez-Martín F, García-Sogo B, Angosto T, Moreno V, Lozano R (2010) Functional analysis of the Arlequin mutant corroborates the essential role of the Arlequin/TAGL1 gene during reproductive development of tomato. PLoS One 5: e14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ. (2004) Genetic regulation of fruit development and ripening. Plant Cell 16(Suppl): S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson D (2013) Ethylene and the control of fruit ripening. In GB Seymour, M Poole, JJ Giovannoni, GA Tucker, eds, The Molecular Biology and Biochemistry of Fruit Ripening. Blackwell Publishing Ltd., Ames, IA, pp 43–73 [Google Scholar]
- Guillon F, Philippe S, Bouchet B, Devaux M-F, Frasse P, Jones B, Bouzayen M, Lahaye M (2008) Down-regulation of an Auxin Response Factor in the tomato induces modification of fine pectin structure and tissue architecture. J Exp Bot 59: 273–288 [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Lycett GW, Grierson D (1990) Antisense gene that inhibits synthesis of the hormone ethylene in transgenic plants. Nature 346: 284–287 [Google Scholar]
- Hao Y, Hu G, Breitel D, Liu M, Mila I, Frasse P, Fu Y, Aharoni A, Bouzayen M, Zouine M (2015) Auxin Response Factor SlARF2 Is an Essential Component of the Regulatory Mechanism Controlling Fruit Ripening in Tomato. PLoS Genet http://dx.doi.org/10.1371/journal.pgen.10.05649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu ZL, Deng L, Chen XQ, Wang PQ, Chen GP (2010) Co-suppression of the EIN2-homology gene LeEIN2 inhibits fruit ripening and reduces ethylene sensitivity in tomato. Russ J Plant Physiol 57: 554–559 [Google Scholar]
- Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A (2009) TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J 60: 1081–1095 [DOI] [PubMed] [Google Scholar]
- Ito Y, Kitagawa M, Ihashi N, Yabe K, Kimbara J, Yasuda J, Ito H, Inakuma T, Hiroi S, Kasumi T (2008) DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J 55: 212–223 [DOI] [PubMed] [Google Scholar]
- Jakubowicz M. (2002) Structure, catalytic activity and evolutionary relationships of 1-aminocyclopropane-1-carboxylate synthase, the key enzyme of ethylene synthesis in higher plants. Acta Biochim Pol 49: 757–774 [PubMed] [Google Scholar]
- Jiang Y, Joyce DC, Macnish AJ (2000) Effect of Abscisic Acid on Banana Fruit Ripening in Relation to the Role of Ethylene. J Plant Growth Regul 19: 106–111 [DOI] [PubMed] [Google Scholar]
- Jones B, Frasse P, Olmos E, Zegzouti H, Li ZG, Latché A, Pech JC, Bouzayen M (2002) Down-regulation of DR12, an auxin-response-factor homolog, in the tomato results in a pleiotropic phenotype including dark green and blotchy ripening fruit. Plant J 32: 603–613 [DOI] [PubMed] [Google Scholar]
- Ju C, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang J, Garrett WM, Kessenbrock M, Groth G, Tucker ML, et al. (2012) CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci USA 109: 19486–19491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiyoshihara Y, Tieman DM, Huber DJ, Klee HJ (2012) Ligand-induced alterations in the phosphorylation state of ethylene receptors in tomato fruit. Plant Physiol 160: 488–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R, Chapman N, David K, Angenent GC, Seymour GB, de Maagd RA (2014) Transcriptional control of fleshy fruit development and ripening. J Exp Bot 65: 4527–4541 [DOI] [PubMed] [Google Scholar]
- Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, Fraser PD, Baxter C, Angenent GC, de Maagd RA (2011) Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23: 923–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevany BM, Tieman DM, Taylor MG, Cin VD, Klee HJ (2007) Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J 51: 458–467 [DOI] [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ (2011) Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet 45: 41–59 [DOI] [PubMed] [Google Scholar]
- Klee H, Tieman D (2002) The tomato ethylene receptor gene family: Form and function. Physiol Plant 115: 336–341 [DOI] [PubMed] [Google Scholar]
- Lanahan MB, Yen HC, Giovannoni JJ, Klee HJ (1994) The Never ripe mutation blocks ethylene perception in tomato. Plant Cell 6: 521–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashbrook CC, Tieman DM, Klee HJ (1998) Differential regulation of the tomato ETR gene family throughout plant development. Plant J 15: 243–252 [DOI] [PubMed] [Google Scholar]
- Leclercq J, Adams-Phillips LC, Zegzouti H, Jones B, Latché A, Giovannoni JJ, Pech JC, Bouzayen M (2002) LeCTR1, a tomato CTR1-like gene, demonstrates ethylene signaling ability in Arabidopsis and novel expression patterns in tomato. Plant Physiol 130: 1132–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Joung JG, McQuinn R, Chung MY, Fei Z, Tieman D, Klee H, Giovannoni J (2012) Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J 70: 191–204 [DOI] [PubMed] [Google Scholar]
- Lelièvre JM, Latchè A, Jones B, Bouzayen M, Pech JC (1997) Ethylene and fruit ripening. Physiol Plant 101: 727–739 [Google Scholar]
- Li J, Li Z, Tang L, Yang Y, Zouine M, Bouzayen M (2012) A conserved phosphorylation site regulates the transcriptional function of ETHYLENE-INSENSITIVE3-like1 in tomato. J Exp Bot 63: 427–439 [DOI] [PubMed] [Google Scholar]
- Li X, Zhu X, Mao J, Zou Y, Fu D, Chen W, Lu W (2013) Isolation and characterization of ethylene response factor family genes during development, ethylene regulation and stress treatments in papaya fruit. Plant Physiol Biochem 70: 81–92 [DOI] [PubMed] [Google Scholar]
- Li Y, Zhu B, Xu W, Zhu H, Chen A, Xie Y, Shao Y, Luo Y (2007) LeERF1 positively modulated ethylene triple response on etiolated seedling, plant development and fruit ripening and softening in tomato. Plant Cell Rep 26: 1999–2008 [DOI] [PubMed] [Google Scholar]
- Lin Z, Alexander L, Hackett R, Grierson D (2008a) LeCTR2, a CTR1-like protein kinase from tomato, plays a role in ethylene signalling, development and defence. Plant J 54: 1083–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Arciga-Reyes L, Zhong S, Alexander L, Hackett R, Wilson I, Grierson D (2008b) SlTPR1, a tomato tetratricopeptide repeat protein, interacts with the ethylene receptors NR and LeETR1, modulating ethylene and auxin responses and development. J Exp Bot 59: 4271–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Hong Y, Yin M, Li C, Zhang K, Grierson D (2008c) A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J 55: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Diretto G, Pirrello J, Roustan JP, Li Z, Giuliano G, Regad F, Bouzayen M (2014) The chimeric repressor version of an Ethylene Response Factor (ERF) family member, Sl-ERF.B3, shows contrasting effects on tomato fruit ripening. New Phytol 203: 206–218 [DOI] [PubMed] [Google Scholar]
- Liu M, Pirrello J, Kesari R, Mila I, Roustan JP, Li Z, Latché A, Pech JC, Bouzayen M, Regad F (2013) A dominant repressor version of the tomato Sl-ERF.B3 gene confers ethylene hypersensitivity via feedback regulation of ethylene signaling and response components. Plant J 76: 406–419 [DOI] [PubMed] [Google Scholar]
- Liu R, How-Kit A, Stammitti L, Teyssier E, Rolin D, Mortain-Bertrand A, Halle S, Liu M, Kong J, Wu C, et al. (2015) A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc Natl Acad Sci USA 112: 10804–10809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Feng H, Meng X, Li D, Yang D, Wu C, Meng Q (2014) Overexpression of tomato SlNAC1 transcription factor alters fruit pigmentation and softening. BMC Plant Biol 14: 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Du W, Brandizzi F, Giovannoni JJ, Barry CS (2012) Differential control of ethylene responses by GREEN-RIPE and GREEN-RIPE LIKE1 provides evidence for distinct ethylene signaling modules in tomato. Plant Physiol 160: 1968–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38: 948–952 [DOI] [PubMed] [Google Scholar]
- McMurchie EJ, McGlasson WB, Eaks IL (1972) Treatment of fruit with propylene gives information about the biogenesis of ethylene. Nature 237: 235–236 [DOI] [PubMed] [Google Scholar]
- Mizrahi Y, Dostal HC, McGlasson WB, Cherry JH (1975) Effects of abscisic acid and benzyladenine on fruits of normal and rin M=mutant tomatoes. Plant Physiol 56: 544–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuka A, Murachi S, Okunishi H, Shiomi S, Nakano R, Kubo Y, Inaba A (1998) Differential expression and internal feedback regulation of 1-aminocyclopropane-1-carboxylate synthase, 1-aminocyclopropane-1-carboxylate oxidase, and ethylene receptor genes in tomato fruit during development and ripening. Plant Physiol 118: 1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeller PW, Lu MW, Taylor LP, Pike DA, Theologis A (1991) Reversible inhibition of tomato fruit senescence by antisense RNA. Science 254: 437–439 [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7: 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S, Alba R, Damasceno CMB, Lopez-Casado G, Lohse M, Zanor MI, Tohge T, Usadel B, Rose JKC, Fei Z. , et al. (2011) Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol 157: 405–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrello J, Prasad BCN, Zhang W, Chen K, Mila I, Zouine M, Latché A, Pech JC, Ohme-Takagi M, Regad F. , et al. (2012) Functional analysis and binding affinity of tomato ethylene response factors provide insight on the molecular bases of plant differential responses to ethylene. BMC Plant Biol 12: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick JS, Wen C-K, Shockey JA, Chang C (2006) REVERSION-TO-ETHYLENE SENSITIVITY1, a conserved gene that regulates ethylene receptor function in Arabidopsis. Proc Natl Acad Sci USA 103: 7917–7922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M, Chervin C, Mila I, Hao Y, Roustan JP, Benichou M, Gibon Y, Biais B, Maury P, Latché A, et al. (2013) SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol 161: 1362–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Shiu SH, Armitage JP (2011) Two-component systems and their co-option for eukaryotic signal transduction. Curr Biol 21: R320–R330 [DOI] [PubMed] [Google Scholar]
- Seymour GB, Chapman NH, Chew BL, Rose JKC (2013) Regulation of ripening and opportunities for control in tomato and other fruits. Plant Biotechnol J 11: 269–278 [DOI] [PubMed] [Google Scholar]
- Shima Y, Fujisawa M, Kitagawa M, Nakano T, Kimbara J, Nakamura N, Shiina T, Sugiyama J, Nakamura T, Kasumi T. , et al. (2014) Tomato FRUITFULL homologs regulate fruit ripening via ethylene biosynthesis. Biosci Biotechnol Biochem 78: 231–237 [DOI] [PubMed] [Google Scholar]
- Solano R, Ecker JR (1998) Ethylene gas: perception, signaling and response. Curr Opin Plant Biol 1: 393–398 [DOI] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Diretto G, Purgatto E, Danoun S, Zouine M, Li Z, Roustan JP, Bouzayen M, Giuliano G, Chervin C (2015) Carotenoid accumulation during tomato fruit ripening is modulated by the auxin-ethylene balance. BMC Plant Biol 15: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Yuan B, Zhang M, Wang L, Cui M, Wang Q, Leng P (2012) Fruit-specific RNAi-mediated suppression of SlNCED1 increases both lycopene and β-carotene contents in tomato fruit. J Exp Bot 63: 3097–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuki M, Nakajima N, Fujii H, Shimada T, Nakano M, Hayashi K, Hayama H, Yoshioka H, Nakamura Y (2013) Increased levels of IAA are required for system 2 ethylene synthesis causing fruit softening in peach (Prunus persica L. Batsch). J Exp Bot 64: 1049–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theologis A, Zarembinski TI, Oeller PW, Liang X, Abel S (1992) Modification of fruit ripening by suppressing gene expression. Plant Physiol 100: 549–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman DM, Ciardi JA, Taylor MG, Klee HJ (2001) Members of the tomato LeEIL (EIN3-like) gene family are functionally redundant and regulate ethylene responses throughout plant development. Plant J 26: 47–58 [DOI] [PubMed] [Google Scholar]
- Tieman DM, Klee HJ (1999) Differential expression of two novel members of the tomato ethylene-receptor family. Plant Physiol 120: 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier B, Sanchez-Ballesta MT, Jones B, Pesquet E, Regad F, Latché A, Pech JC, Bouzayen M (2003) New members of the tomato ERF family show specific expression pattern and diverse DNA-binding capacity to the GCC box element. FEBS Lett 550: 149–154 [DOI] [PubMed] [Google Scholar]
- Trainotti L, Tadiello A, Casadoro G (2007) The involvement of auxin in the ripening of climacteric fruits comes of age: the hormone plays a role of its own and has an intense interplay with ethylene in ripening peaches. J Exp Bot 58: 3299–3308 [DOI] [PubMed] [Google Scholar]
- Van de Poel B, Bulens I, Markoula A, Hertog MLATM, Dreesen R, Wirtz M, Vandoninck S, Oppermann Y, Keulemans J, Hell R, et al. (2012) Targeted systems biology profiling of tomato fruit reveals coordination of the Yang cycle and a distinct regulation of ethylene biosynthesis during postclimacteric ripening. Plant Physiol 160: 1498–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Poel B, Vandenzavel N, Smet C, Nicolay T, Bulens I, Mellidou I, Vandoninck S, Hertog ML, Derua R, Spaepen S, et al. (2014) Tissue specific analysis reveals a differential organization and regulation of both ethylene biosynthesis and E8 during climacteric ripening of tomato. BMC Plant Biol 14: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Pan IL, Arroyo AJM, McQuinn R, Chung M, Poole M, Rose J, Seymour G, Grandillo S, Giovannoni J. , et al. (2009) Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21: 3041–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Wang A, Tan D, Takahashi A, Li TZ, Harada T (2007) MdERFs, two ethylene-response factors involved in apple fruit ripening. J Exp Bot 58: 3743–3748 [DOI] [PubMed] [Google Scholar]
- Wang S, Lu G, Hou Z, Luo Z, Wang T, Li H, Zhang J, Ye Z (2014) Members of the tomato FRUITFULL MADS-box family regulate style abscission and fruit ripening. J Exp Bot 65: 3005–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng L, Zhao F, Li R, Xu C, Chen K, Xiao H (2015) The zinc finger transcription factor SlZFP2 negatively regulates abscisic acid biosynthesis and fruit ripening in tomato. Plant Physiol 167: 931–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Yen HC, Giovannoni JJ, Klee HJ (1995) An ethylene-inducible component of signal transduction encoded by never-ripe. Science 270: 1807–1809 [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol 35: 155–189 [Google Scholar]
- Yang Y, Wu Y, Pirrello J, Regad F, Bouzayen M, Deng W, Li Z (2010) Silencing Sl-EBF1 and Sl-EBF2 expression causes constitutive ethylene response phenotype, accelerated plant senescence, and fruit ripening in tomato. J Exp Bot 61: 697–708 [DOI] [PubMed] [Google Scholar]
- Yokotani N, Nakano R, Imanishi S, Nagata M, Inaba A, Kubo Y (2009) Ripening-associated ethylene biosynthesis in tomato fruit is autocatalytically and developmentally regulated. J Exp Bot 60: 3433–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani N, Tamura S, Nakano R, Inaba A, Kubo Y (2003) Characterization of a novel tomato EIN3-like gene (LeEIL4). J Exp Bot 54: 2775–2776 [DOI] [PubMed] [Google Scholar]
- Zegzouti H, Jones B, Frasse P, Marty C, Maitre B, Latch A, Pech JC, Bouzayen M (1999) Ethylene-regulated gene expression in tomato fruit: characterization of novel ethylene-responsive and ripening-related genes isolated by differential display. Plant J 18: 589–600 [DOI] [PubMed] [Google Scholar]
- Zhang M, Yuan B, Leng P (2009) The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J Exp Bot 60: 1579–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Fei Z, Chen YR, Zheng Y, Huang M, Vrebalov J, McQuinn R, Gapper N, Liu B, Xiang J. , et al. (2013) Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol 31: 154–159 [DOI] [PubMed] [Google Scholar]
- Zhong S, Lin Z, Grierson D (2008) Tomato ethylene receptor-CTR interactions: visualization of NEVER-RIPE interactions with multiple CTRs at the endoplasmic reticulum. J Exp Bot 59: 965–972 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.