We used whole genome sequencing with epidemiologic data to determine if USA300 methicillin-resistant Staphylococcus aureus (MRSA) transmission networks exist in an urban community. We observed potential MRSA transmission networks and identified community factors or exposures that may facilitate spread of USA300.
Keywords: MRSA, whole genome sequencing
Abstract
Background. In a community, it is unknown what factors account for transmission of methicillin-resistant Staphylococcus aureus (MRSA). We integrated whole genome sequencing (WGS) and epidemiologic data to identify factors associated with MRSA transmission networks in an urban community.
Methods. WGS was performed on colonizing USA300 MRSA isolates from 74 individuals within 72 hours of admission to a public hospital in Chicago, IL. Single nucleotide variants (SNVs) were used to reconstruct the phylogeny of sequenced isolates, and epidemiologic data was overlaid to identify factors associated with transmission networks.
Results. The maximum within-patient SNV difference for an individual with multisite colonization was 41 SNVs, with no systematic divergence among body sites. We observed a minimum of 7 SNVs and maximum of 153 SNVs between isolates from different individuals. We identified 4 pairs of individuals whose isolates were within 40 SNVs of each other. Putting our isolates in the context of previously sequenced USA300 isolates from other communities, we identified a 13-member group and two 4-member groups that represent samples from putative local transmission networks. Individuals in these groups were more likely to be African American, to be human immunodeficiency virus–infected, to reside in high detainee release areas, and to be current users of illicit drugs.
Conclusions. Using WGS, we observed potential transmission networks in an urban community and that certain epidemiologic factors were associated with inclusion in these networks. Future work with contact tracing and advanced molecular diagnostics may allow for identification of MRSA “epicenters” in the community where interventions can be targeted.
Methicillin-resistant Staphylococcus aureus (MRSA) has emerged as an important pathogen outside of healthcare settings [1, 2], with USA300 being the most common strain isolated in communities in the United States [3]. Invasive MRSA infections in the community have remained stable since 2005, in contrast to the downward trend observed in hospitals [4]. In several US cities, USA300 MRSA strains cause the majority of community-onset skin and soft tissue infections diagnosed in the emergency room [5] and are considered endemic [6]. Studies have documented uneven geographic distribution of infection due to USA300 MRSA [7–9]. We found that residence in an area of high detainee release was associated with community-associated MRSA (CA-MRSA) [10] and infection [9]. Incarceration, illicit drug use, residence in temporary housing, and human immunodeficiency virus (HIV) infection have been associated with increased risk for CA-MRSA [8–14].
It has been speculated that exposure to areas in the community with a high burden of MRSA (eg, jails) allows for more efficient person-to-person transmission of USA300 MRSA strains [7, 9, 15]. However, traditional molecular epidemiologic methods (eg, pulsed-field gel electrophoresis [PFGE]) are unable to sufficiently discriminate between USA300 MRSA strains [16], limiting our ability to identify transmission routes. While hospital prevention efforts have been highly successful at reducing the incidence of MRSA infections [4], optimal strategies for reducing MRSA outside of healthcare settings and where these interventions should be targeted are unknown.
Whole genome sequencing (WGS) is highly discriminatory for detecting strain differences among infectious agents and has been used as an epidemiologic tool in outbreak [17], hospital [18], and household settings [19, 20]. Single nucleotide variants (SNVs) identified through genomic comparisons can distinguish strains that would appear identical by conventional typing. For epidemiologic studies that use WGS, identification of genetically similar isolates suggests close proximity in a transmission network, thereby guiding the search for epidemiologic factors associated with transmission. Our objective in this study was to use WGS to evaluate the genetic diversity of USA300 MRSA strains in an urban population seeking care in a safety-net hospital and to determine if transmission clusters of genetically similar USA300 MRSA strains exist in this disadvantaged urban community where USA300 is endemic.
METHODS
Study Population
Isolates were obtained from individuals who had been enrolled in previously reported MRSA colonization surveillance [11]; enrollment was from March 2011 to April 2012 at Stroger (formerly Cook County) Hospital (CCH), a 464-bed facility and major public hospital in Chicago, Illinois. The enrollment population included 374 HIV-infected and 371 HIV-negative individuals who had surveillance swabs obtained within 72 hours of admission to CCH. Detainees who were incarcerated at the Cook County Jail and who were transferred to CCH for medical care were also enrolled. Demographic data for this study population have been previously reported; a large proportion of both HIV-infected and HIV-negative individuals were African American [11]. Surveillance swabs for MRSA colonization were collected from anterior nares, throat, axilla, groin, and perirectum; swabs were processed with broth enrichment as described elsewhere [11, 21]. One colony per anatomic site underwent PFGE to identify USA300 strains [22].
Whole Genome Sequencing
WGS was performed on isolates from 81 individuals with USA300 MRSA colonization [11]. For 7 isolates (all from African-American males; 6 were from HIV-infected individuals), the number of SNVs relative to the reference was >1000; these isolates were unlikely to be USA300 and were therefore excluded from analysis. As previously reported, the mean number of body sites colonized with USA300 MRSA was 2.8 (standard deviation, 1.5), with 53/74 (72%) individuals having colonization at more than 1 body site [11, 23]. Thirty-six individuals with multisite colonization had isolates sequenced from different body sites to determine the maximum within-host diversity. For within-host analysis, isolates were preferentially chosen from individuals who had colonization in the nares, throat, or perirectal area. The latter 2 sites are common sites of extranasal colonization [11], and we hypothesized that due to various extrinsic factors, isolates from these sites would be the most divergent. For between-host comparisons, a single isolate from each individual was chosen for sequencing.
Genomic DNA extracted from isolates was prepared for sequencing on an Illumina HiSeq2000 instrument using standard library preparation approaches and sample-specific bar coding as described previously [24]. Libraries from each strain were pooled together before sequencing to an average depth of 200–300× coverage per genome. Comparative genomic analyses were performed using SPANDx version 2.4 [25]. SPANDx is a fully automated pipeline designed to identify genetic variants for medium to large haploid next-generation resequencing datasets. The software was upgraded using the annotation tool snpEff in SPANDx to version 4.1. The reference genome of S. aureus, NC_010079, was downloaded from the National Center for Biotechnology Information and normalized with Picard NormalizeFasta module. The SPANDx data output consisted of an SNV matrix of 3417 positions per genome. SNVs were used as a measure of genetic distance between pairs of USA300 strains.
Phylogenetic Analysis and Linkage of Epidemiologic Data
For the within- and between-host comparative genome evaluations, the SNV matrix was imported into the software package MEGA6 [26] for phylogenetic analyses. Boot-strapped maximum likelihood and neighbor-joining trees were generated within MEGA6 [26]. The SNV matrix was also used for Bayesian phylogenetic analysis using the software package MrBayes [27]. Clusters of USA300 strains were identified when all 3 phylogenetic analyses indicated support for nodes, including bootstrap values of >70% for maximum likelihood and neighbor-joining analysis, and posterior probability values of >95% for Bayesian analysis.
We then used phylogenetic analysis to test whether there was evidence of long-term transmission networks in this urban population seeking care at the major safety-net hospital in Chicago. To do this, we used previously sequenced USA300 MRSA isolates from New York, Chicago, and Los Angeles [19, 20]. SNVs were identified for these isolates relative to the aforementioned USA300 reference genome, and a maximum likelihood tree that included isolates from current and previous studies was created with RAxML (GTRCAT substitution model and 100 bootstrap replicates). Clusters were defined as subtrees with >90% bootstrap support that contained ≥3 isolates from the current study and did not contain any isolates from the other city studies.
Demographic, comorbidity, and exposure data were collected on all enrolled individuals by questionnaire and medical record review [11]. Exposures of interest, based on prior studies, included illicit drug use, incarceration, zip code of residence, and residence in temporary housing (homeless, shelter, substance abuse center) [9–11, 28, 29]. Areas of high detainee release were defined as described previously [9]. Data elements were compared for individuals in any identified genomic clusters to individuals whose isolates were not in a cluster.
Statistical Analyses
For statistical analyses, SAS software version 9.2 (SAS Institute, Cary, North Carolina) was used. In addition, χ2 analysis was used for comparison of epidemiologic data between individuals whose MRSA strains were and were not in identified clusters; the Fisher exact test was used for small samples. The institutional review boards at Rush University Medical Center and CCH approved the study.
Data Access
The genome sequence data from this study have been submitted to the NCBI Sequence Read Archive under BioProject PRJNA275322.
RESULTS
Study Population
Of the 74 individuals confirmed with WGS to have USA300 MRSA colonization, 53 were HIV infected and 21 were HIV negative. Seventy-seven percent of individuals in the study were male and 70% were African American; only 12% were of Hispanic ethnicity. Thirty-six (49%) individuals were current (17) or former (19) illicit drug users. Twelve percent of individuals resided in temporary housing currently or in the past year, and 11% were currently incarcerated or had been released from the Cook County Jail in the prior 3 months.
Within-Host Comparative Genome Analysis
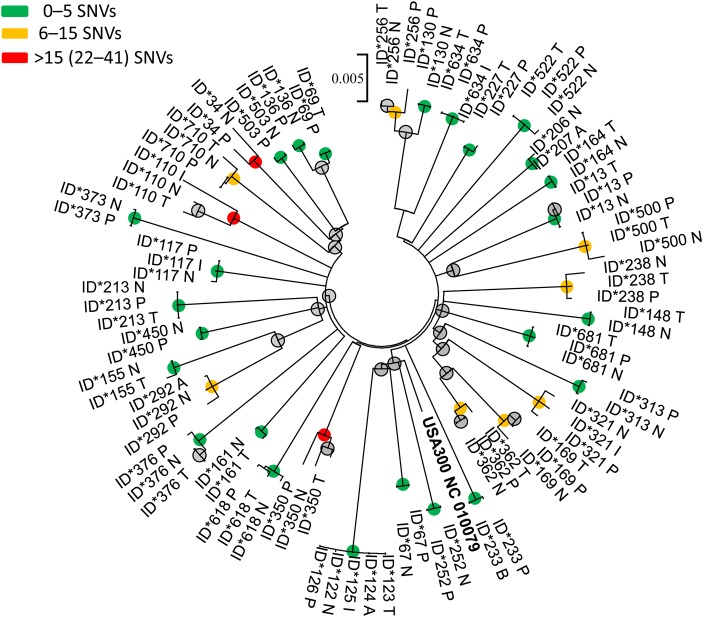
Thirty-six individuals with multisite USA300 MRSA colonization were included; 39% had nares, throat, and perirectal colonization; 21 had colonization at 2 of these sites. Within-host isolates were similar across body sites (Figure 1), and within-host pairwise comparisons between nares–throat, throat–perirectal, and nares–perirectal were similar (Table 1).
Figure 1.
Maximum-likelihood phylogenetic tree of USA300 methicillin-resistant Staphylococcus aureus isolates from 36 individuals with multisite body colonization. Note: USA300 NC 010079 represents the reference genome. Nodes supported by neighbor-joining and maximum-likelihood bootstrap analysis and Bayesian inference are indicated with a colored circle. The node color indicates the maximum number of single nucleotide variant (SNV) differences between isolates from the same individual within the clade. Abbreviations: A, axilla; I, inguinal area; N, nares; P, perirectal area; T, throat.
Table 1.
Within-Host Comparative Genome Analysis: Pairwise Analysis by Body Site for 35 Individuals With Multisite USA300 Methicillin-Resistant Staphylococcus aureus Colonization in the Nares, Throat, or Perirectal Area
| Body Sites | Mean SNV Difference (Standard Deviation) | Median SNV Difference (Range) |
|---|---|---|
| Nares–throat (n = 19) | 4.2 (3.8) | 3 (0–11) |
| Nares–perirectal (n = 27) | 4.7 (6.4) | 2 (0–25) |
| Throat–perirectal (n = 17) | 4 (5.5) | 2 (0–20) |
Thirty-two individuals had nares colonization, 22 had throat colonization, and 30 had perirectal colonization.
Abbreviation: SNV, single nucleotide variant.
Forty-three percent (15/35) of individuals had no SNV differences for nares–throat, nares–perirectum, or throat–perirectum isolate comparisons. Eleven (31%) individuals had more than 5 SNV differences and 4 (11%) had more than 10 SNV differences for nares–throat, throat–perirectum, or nares–perirectum isolate comparisons. One individual who had USA300 isolates sequenced from 5 body sites (nares, throat, axilla, groin, and perirectum) had a maximum of 1 SNV difference across sites. The maximum SNV difference observed was in an individual who had nares, throat, and inguinal colonization; there were 41 SNV differences between the throat and inguinal isolate and 39 SNV differences between the nares and inguinal isolate.
Between-Host Comparative Genome Analysis
Isolates from 74 individuals were included in between-host analysis; 1 isolate per person was chosen for comparison. There were 3471 informational positions for which at least a single sequenced genome contained an SNV relative to the USA300 reference. We observed that there was a wide range of genetic diversity, with a maximum distance of 153 SNVs and a minimum of 7 SNVs between strains isolated from different individuals. The mean and median SNV differences between strains were both 105 SNVs.
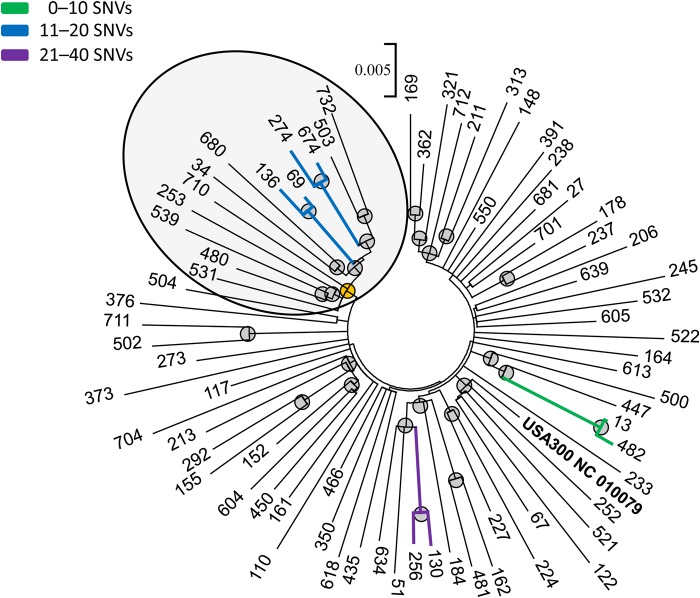
Consistent with prior work [18, 30], we observed that approximately 40 SNVs was the maximum within-host genetic variation and therefore hypothesized that cases where isolates from different hosts were within 40 SNVs were indicative of close proximity in a transmission network. We found four 2-member pairs who had isolates that were within 40 SNVs of each other and that were statistically supported by maximum likelihood, neighbor joining, and Bayesian analysis (Table 2, Figure 2). One pair included isolates within 10 SNVs of each other, isolates from 2 other pairs were within 20 SNVs of each other, and isolates for 1 pair were within 40 SNVs of each other.
Table 2.
Between-Host Comparative Genome Analysis: Two-Member Pairs Identified in Phylogenetic Analysis That Were Supported by Maximum Likelihood, Neighbor-Joining, and Bayesian Analysis at Defined Single Nucleotide Variant Limits
| SNV Limit | Number of Pairs at SNV Limit | Number of Total Pairs | Number of Individuals in Any Pair | Percent of the Population in Any Pair, % |
|---|---|---|---|---|
| <10 | 1 | 1 | 2 | 3 |
| <20 | 2 | 3 | 6 | 5 |
| <40 | 1 | 4 | 8 | 11 |
The table is based on isolates from 74 individuals.
Abbreviation: SNV, single nucleotide variant.
Figure 2.
Maximum-likelihood phylogenetic tree of USA300 methicillin-resistant Staphylococcus aureus isolates among 74 human immunodeficiency virus (HIV)-infected and HIV-negative patients seeking care at the major public hospital in Chicago, Illinois. Note: USA300 NC 010079 represents the reference genome. Nodes supported by neighbor-joining and maximum-likelihood bootstrap analysis and Bayesian inference are indicated with a gray circle. Clusters containing genomes with fewer than 40 single nucleotide variant (SNV) differences are highlighted by branch colors. A “super cluster,” described in the text, is highlighted with a yellow node and circled.
Linkage of Genomic and Epidemiologic Data
For the 8 individuals who were included in the four 2-member pairs of genetically similar isolates, 7 (88%) were African American, 6 (75%) were HIV infected, 4 (50%) were current or former illicit drug users, and 3 (38%) were homeless or residing in a shelter (Table 3). The 2 members of the <10 SNV cluster were both HIV-infected with illicit drug use; 1 individual was homeless and the other had been released from jail in the prior 3 months. Members of the clusters at the <20 SNV limit were all HIV infected. The cluster at the <40 SNV limit did not have any apparent epidemiologic linkages or demographic similarities; both were HIV negative. Individuals in these pairs were more likely to be homeless or in a shelter than others (odds ratio [OR], 6; 95% confidence interval [CI], 1.1, 31.5; P = .05).
Table 3.
Association of Epidemiologic Factors Between Pairs of Individuals Whose Colonizing USA300 Methicillin-Resistant Staphylococcus aureus Isolates had <40 Single Nucleotide Variant Differences Between Them
| SNVs Between Strains | Epidemiologic Factors |
|
|---|---|---|
| Person 1 | Person 2 | |
| <10 | Illicit drugs, HIV infection, incarceration in jail within the prior 3 months | Illicit drugs, HIV infection, currently homeless |
| <20 | HIV infection, MSM | HIV infection, illicit drugs, incarceration within the prior 3 months, currently resides in a shelter |
| <20 | HIV infection | HIV infection, illicit drugs, currently resides in a shelter |
| <40 | History of nursing home residence | None |
Abbreviations: HIV, human immunodeficiency virus; MSM, men who have sex with men; SNV, single nucleotide variant.
Evaluation for MRSA Transmission Networks
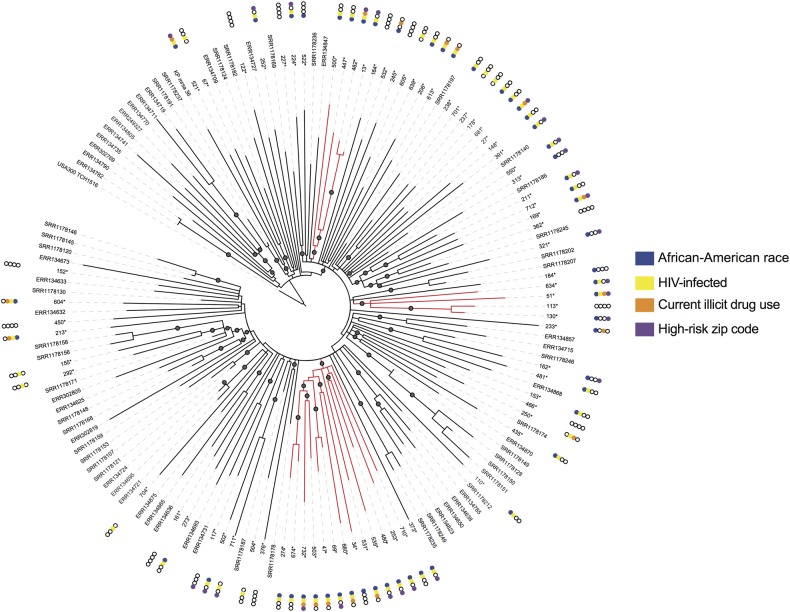
To expand our search beyond recent transmissions and to try to uncover larger transmission networks, we incorporated previously sequenced USA300 isolates from individuals in New York City, Los Angeles, and Chicago in our analysis (Figure 3). As seen in prior studies, we observed intermixing of our isolates with those from disparate geographies, suggesting that a migration of strains between communities is not uncommon. However, we did identify 3 phylogenetic clusters that included only isolates from the current study. These clusters included a 13-member “super cluster” and 2 additional 4-member clusters. Isolates from the 13-member super cluster shared 5 SNVs unique to these strains (Supplementary Table 1).
Figure 3.
Maximum-likelihood phylogenetic analysis of USA300 methicillin-resistant Staphylococcus aureus transmission networks in our urban study population relative to other US cities. Note: Asterisk indicates isolates from current study. Gray circles indicate >90% confidence by bootstrap analysis. Isolates highlighted in red indicate those belonging to identified transmission networks. Abbreviation: HIV, human immunodeficiency virus.
We examined whether there were epidemiologic associations with cluster membership (Table 4) and found that USA300 isolates from African-American individuals were significantly more likely to be in a cluster (OR, 13; 95% CI, 1.6, 105.3; P = .004). In contrast, no isolates from individuals of Hispanic ethnicity were included in a cluster. Forty-seven percent of individuals in these transmission networks lived in areas (based on zip code) with high detainee release. Isolates from individuals who were HIV infected, current users of illicit drugs, or residents of areas with high detainee release or temporary housing were more likely to be included in genomic clusters, although associations did not attain statistical significance (Figure 3, Table 4).
Table 4.
Epidemiologic Predictors for Inclusion in a Genomic Cluster Supported by Maximum Likelihood and Bootstrap Analysis
| Epidemiologic Factor | Number (%) of Individuals in a Cluster (n = 21)a | Number (%) of Individuals not in Any Cluster (n = 53) | P Value |
|---|---|---|---|
| Human immunodeficiency virus infected | 18 (86) | 35 (66) | .15 |
| Gender | |||
| Male | 16 (76) | 41 (77) | .91 |
| Female | 5 (24) | 12 (23) | |
| African-American race | 20 (95) | 32 (60) | .004 |
| Hispanic ethnicity | 0 (0) | 9 (17) | .05 |
| Men who have sex with men | 6 (29) | 15 (28) | .99 |
| Current residence in a high detainee-release zip code | 10 (47) | 15 (28) | .11 |
| Current drug use | 7 (33) | 10 (19) | .18 |
| Temporary housing residence currently or in the past yearb | 4 (19) | 5 (9) | .26 |
| Current incarceration | 0 | 5 (9) | .31 |
| Healthcare exposure in the past yearc | 12 (57) | 30 (57) | .82 |
a Cluster defined as a collection of 3 or more USA300 methicillin-resistant Staphylococcus aureus isolates with genetic similarity supported by bootstrap (>90%) and maximum likelihood analysis.
b Temporary housing residence includes being homeless or residing in a homeless shelter or substance abuse center.
c Healthcare exposure in the past year included visits to the emergency room or hospitalization at Stroger Hospital of Cook County.
Focusing on the 13 individuals in the super cluster (Supplementary Table 2), we found that all 13 were African American and 92% were HIV infected. Two individuals had resided in the same homeless shelter in the prior year. Nearly 40% of individuals in this cluster were current drug users; strains from individuals who were current drug users were more likely to be included in this super cluster, although this association did not reach significance (OR, 2.6; 95% CI, .7, 9.2; P = .14). Healthcare exposures were not more frequent among members of the super cluster.
DISCUSSION
We performed within- and between-host comparative WGS analyses on USA300 MRSA isolates collected from HIV-infected and HIV-negative individuals at admission to the major public hospital in Chicago, Illinois. Overall, there was low within-host genetic diversity, with a maximum of 41 SNVs between isolates for an individual. We observed a wide range in the genetic diversity of USA300 strains from different individuals, with up to 153 SNV differences detected. However, even in this diverse urban community, we were able to identify 4 pairs of individuals whose isolates were within 40 SNVs of each other, smaller than the maximum within-host value we observed. In addition, we identified potential USA300 MRSA transmission networks unique to our population and found that individuals in these networks were largely African American and HIV infected. Nearly half of individuals in these genomic networks resided in geographic areas of high detainee release.
Our results suggest that individuals with multisite USA300 MRSA colonization carry strains that are highly genetically similar, consistent with prior studies in which multiple isolates were sequenced [18, 20]. Comparing isolates from different anatomical sites, we did not detect systematic divergence between nares, throat, and perirectal colonization, that is, we could not detect a conserved order by which MRSA disseminates from one body site to another. Despite the overall similarity of isolates within individuals, we identified instances of large genetic distances among an individual's isolates. While it has been hypothesized that this within-host diversity is indicative of extended colonization [31], it is also consistent with multiple independent acquisitions or acquisition of a genetically diverse population whereby genetic diversity is propagated from one host to the next. Distinguishing these possibilities will be critical in extracting maximal insights from future genomic epidemiology investigations.
We found individuals with highly genetically similar USA300 MRSA strains in our urban community. Work by Uhlemann et al [20] in New York City and Alam et al in Chicago and Los Angeles [19] documented clustering of MRSA strains in households, suggesting that households enable spread of MRSA outside the hospital. Our work extends these studies by exploring how transmission may occur outside a household in an urban community. We found that a large proportion of individuals in transmission networks resided in areas (based on zip code) of high detainee release; these areas have previously been associated with increased risk for MRSA colonization [10] and infection [9]. It is unclear if these associations represent downstream effects of a high burden of MRSA in correctional facilities [12, 15, 32] or a complex network of individuals in the community at increased risk for MRSA acquisition.
We found that illicit drug use and being homeless or residing in a shelter were common among individuals who had highly genetically similar isolates and were more common in our identified MRSA transmission networks. Prior work has suggested that individuals who use illicit drugs may represent a reservoir for MRSA strains [11, 28, 29, 33], which facilitates their dissemination among at-risk individuals. Temporary housing, for example, homeless shelters, substance abuse centers, and public housing, has been associated with CA-MRSA colonization or infection [8–10, 34] and may facilitate spread of MRSA (eg, close person-to-person contact and high MRSA prevalence). These settings may represent “hot spots” for MRSA spread where infection prevention efforts could be targeted.
We found that African-American race was significantly associated with membership in an MRSA transmission network. No individuals of Hispanic ethnicity were in our detected clusters. Previously we described differential risk for MRSA colonization and infection based on race and ethnicity, with African Americans at increased risk and individuals of Hispanic ethnicity at lower risk [8, 10, 11]. It has been speculated that different social networks, levels of incarceration, and geography of residence contribute to this observed disparity. By combing genomic and epidemiologic data, we were able to explore these associations further. Nearly all individuals in our detected MRSA transmission clusters were African American and most were HIV infected. We and others have documented the uneven geographic distribution of MRSA infection in US cities [7–9], with areas of higher MRSA burden perhaps facilitating greater spread of strains. Prior work has shown that African Americans and individuals of Hispanic ethnicity usually reside in geographically distinct neighborhoods in Chicago [35]. These geographic factors likely contribute to different daily contacts and thus different levels of opportunity for MRSA acquisition. For other pathogens, contact tracing and social network approaches have been integrated with genomic and phylogenetic analysis to identify “super spreaders” [36]. This type of analysis may be of value when characterizing how MRSA is being disseminated in community populations with the highest burden of disease.
Our study has limitations. We did not perform a longitudinal assessment of within-host diversity and could not determine if duration of colonization was associated with more genetic diversity. Future analysis of diversity in relation to host factors and duration of colonization may better characterize how persistent colonization impacts diversity of MRSA. Second, we did not perform contact tracing and could not comment on direct transmission events (as done in intensive care units). However, we selected epidemiologic factors that have been associated consistently with increased USA300 MRSA risk and highlight potential community transmission networks of MRSA. Larger sample sizes in at-risk community settings may detect MRSA epicenters outside of the hospital. Third, our study population of safety-net patients, HIV-infected inner city patients, and jail detainees may limit the generalizability of our findings. However, the framework used, that is, integrating community exposure and epidemiologic data with genomic data, could be applied to other high-risk settings for MRSA acquisition, for example, military barracks and correctional facilities, in order to improve our understanding of MRSA transmission dynamics. Also, our study population represents a large segment of urban individuals.
In conclusion, in community and healthcare settings where USA300 MRSA strains are endemic, WGS phylogenetic analysis may be a useful tool for improving our understating of MRSA transmission networks. Similar to others, our work supports the hypothesis that USA300 has been introduced repeatedly into Chicago. Strains of USA300 were then likely able to spread among particular networks of individuals characterized by certain factors or behaviors, such as illicit drug use and location or type of residence. Future work with a larger sample size and integration of contact tracing may identify MRSA “epicenters” in the community where interventions can be targeted.
Supplementary Data
Supplementary materials are available at http://cid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. We kindly acknowledge the assistance of Zhengdeng Lei at the Center for Research Informatics at the University of Illinois–Chicago for processing sequence data through the SPANDx pipeline.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases (NIAID) or the National Institutes of Health.
Financial support. This project was supported by a grant from the NIAID (grant number K23AI085029 to K. J. P.). Support was also provided by the Centers for Disease Control and Prevention (cooperative agreement number 1U54CK000161 to R. A. W.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Fridkin SK, Hageman JC, Morrison M et al. . Methicillin-resistant Staphylococcus aureus disease in three communities. N Engl J Med 2005; 352:1436–44. [DOI] [PubMed] [Google Scholar]
- 2.Herold BC, Immergluck LC, Maranan MC et al. . Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 1998; 279:593–8. [DOI] [PubMed] [Google Scholar]
- 3.Tenover FC, McDougal LK, Goering RV et al. . Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J Clin Microbiol 2006; 44:108–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dantes R, Mu Y, Belflower R et al. . National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern Med 2013; 173:1970–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moran GJ, Krishnadasan A, Gorwitz RJ et al. . Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med 2006; 355:666–74. [DOI] [PubMed] [Google Scholar]
- 6.Dukic VM, Lauderdale DS, Wilder J, Daum RS, David MZ. Epidemics of community-associated methicillin-resistant Staphylococcus aureus in the United States: a meta-analysis. PLoS One 2013; 8:e52722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diep BA, Chambers HF, Graber CJ et al. . Emergence of multidrug-resistant, community-associated, methicillin-resistant Staphylococcus aureus clone USA300 in men who have sex with men. Ann Intern Med 2008; 148:249–57. [DOI] [PubMed] [Google Scholar]
- 8.Hota B, Ellenbogen C, Hayden MK, Aroutcheva A, Rice TW, Weinstein RA. Community-associated methicillin-resistant Staphylococcus aureus skin and soft tissue infections at a public hospital: do public housing and incarceration amplify transmission? Arch Intern Med 2007; 167:1026–33. [DOI] [PubMed] [Google Scholar]
- 9.Popovich KJ, Weinstein RA, Aroutcheva A, Rice T, Hota B. Community-associated methicillin-resistant Staphylococcus aureus and HIV: intersecting epidemics. Clin Infect Dis 2010; 50:979–87. [DOI] [PubMed] [Google Scholar]
- 10.Popovich KJ, Smith KY, Khawcharoenporn T et al. . Community-associated methicillin-resistant Staphylococcus aureus colonization in high-risk groups of HIV-infected patients. Clin Infect Dis 2012; 54:1296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Popovich KJ, Hota B, Aroutcheva A et al. . Community-associated methicillin-resistant Staphylococcus aureus colonization burden in HIV-infected patients. Clin Infect Dis 2013; 56:1067–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowy FD, Aiello AE, Bhat M et al. . Staphylococcus aureus colonization and infection in New York state prisons. J Infect Dis 2007; 196:911–8. [DOI] [PubMed] [Google Scholar]
- 13.Maree CL, Eells SJ, Tan J et al. . Risk factors for infection and colonization with community-associated methicillin-resistant Staphylococcus aureus in the Los Angeles County jail: a case-control study. Clin Infect Dis 2010; 51:1248–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee NE, Taylor MM, Bancroft E et al. . Risk factors for community-associated methicillin-resistant Staphylococcus aureus skin infections among HIV-positive men who have sex with men. Clin Infect Dis 2005; 40:1529–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okano JT, Blower S. Are correctional facilities amplifying the epidemic of community-acquired methicillin-resistant Staphylococcus aureus? Nat Rev 2010; 8:83. [DOI] [PubMed] [Google Scholar]
- 16.David MZ, Taylor A, Lynfield R et al. . Comparing pulsed-field gel electrophoresis with multilocus sequence typing, spa typing, staphylococcal cassette chromosome mec (SCCmec) typing, and PCR for panton-valentine leukocidin, arcA, and opp3 in methicillin-resistant Staphylococcus aureus isolates at a U.S. medical center. J Clin Microbiol 2013; 51:814–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koser CU, Holden MT, Ellington MJ et al. . Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med 2013; 366:2267–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price JR, Golubchik T, Cole K et al. . Whole-genome sequencing shows that patient-to-patient transmission rarely accounts for acquisition of Staphylococcus aureus in an intensive care unit. Clin Infect Dis 2014; 58:609–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alam MT, Read TD, Petit RA III et al. . Transmission and microevolution of USA300 MRSA in U.S. households: evidence from whole-genome sequencing. MBio 2015; 6:e00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhlemann AC, Dordel J, Knox JR et al. . Molecular tracing of the emergence, diversification, and transmission of S. aureus sequence type 8 in a New York community. Proc Natl Acad Sci U S A 2014; 111:6738–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.http://www.srga.org/Workshop/RAF-M%202007/Talks/Nordic%20protocol%20for%20MRSA%20screening.pdf Broth Enrichment. Accessed April 2012.
- 22.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol 2003; 41:5113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popovich KJ, Aroutcheva A, Hota B, Beavis KG, Hayden MK, Weinstein RA. Anatomic sites of colonization with community-associated methicillin-resistant Staphylococcus aureus. Infect Control Hosp Epidemiol 2014; 35:1192–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Himsworth CG, Miller RR, Montoya V et al. . Carriage of methicillin-resistant Staphylococcus aureus by wild urban Norway rats (Rattus norvegicus). PLoS One 2014; 9:e87983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarovich DS, Price EP. SPANDx: a genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets. BMC Res Notes 2014; 7:618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013; 30:2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003; 19:1572–4. [DOI] [PubMed] [Google Scholar]
- 28.Charlebois ED, Bangsberg DR, Moss NJ et al. . Population-based community prevalence of methicillin-resistant Staphylococcus aureus in the urban poor of San Francisco. Clin Infect Dis 2002; 34:425–33. [DOI] [PubMed] [Google Scholar]
- 29.Rhee Y, Aroutcheva A, Hota B, Weinstein RA, Popovich KJ. Evolving Epidemiology of Staphylococcus aureus Bacteremia. ID Week 2014 Philadelphia, PA October 8–12, 2014. [DOI] [PubMed] [Google Scholar]
- 30.Golubchik T, Batty EM, Miller RR et al. . Within-host evolution of Staphylococcus aureus during asymptomatic carriage. Mol Biol Evol 2013; 8:e61319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong SY, Holden MT, Nickerson EK et al. . Genome sequencing defines phylogeny and spread of methicillin-resistant Staphylococcus aureus in a high transmission setting. Genome Res 2015; 25:111–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartley DM, Furuno JP, Wright MO, Smith DL, Perencevich EN. The role of institutional epidemiologic weight in guiding infection surveillance and control in community and hospital populations. Infect Control Hosp Epidemiol 2006; 27:170–4. [DOI] [PubMed] [Google Scholar]
- 33.Charlebois ED, Perdreau-Remington F, Kreiswirth B et al. . Origins of community strains of methicillin-resistant Staphylococcus aureus. Clin Infect Dis 2004; 39:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landers TF, Harris RE, Wittum TE, Stevenson KB. Colonization with Staphylococcus aureus and methicillin-resistant S. aureus among a sample of homeless individuals, Ohio. Infect Control Hosp Epidemiol 2009; 30:801–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee BA, Reardon SF, Firebaugh G, Farrell CR, Matthews SA, O'Sullivan D. Beyond the census tract: patterns and determinants of racial segregation at multiple geographic scales. Am Sociol Rev 2008; 73:766–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gardy JL, Johnston JC, Ho Sui SJ et al. . Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med 2011; 364:730–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.