Abstract
Meier-Gorlin syndrome (MGS) is a genetically heterogeneous primordial dwarfism syndrome known to be caused by biallelic loss-of-function mutations in one of five genes encoding pre-replication complex proteins: ORC1, ORC4, ORC6, CDT1, and CDC6. Mutations in these genes cause disruption of the origin of DNA replication initiation. To date, only an autosomal-recessive inheritance pattern has been described in individuals with this disorder, with a molecular etiology established in about three-fourths of cases. Here, we report three subjects with MGS and de novo heterozygous mutations in the 5′ end of GMNN, encoding the DNA replication inhibitor geminin. We identified two truncating mutations in exon 2 (the 1st coding exon), c.16A>T (p.Lys6∗) and c.35_38delTCAA (p.Ile12Lysfs∗4), and one missense mutation, c.50A>G (p.Lys17Arg), affecting the second-to-last nucleotide of exon 2 and possibly RNA splicing. Geminin is present during the S, G2, and M phases of the cell cycle and is degraded during the metaphase-anaphase transition by the anaphase-promoting complex (APC), which recognizes the destruction box sequence near the 5′ end of the geminin protein. All three GMNN mutations identified alter sites 5′ to residue Met28 of the protein, which is located within the destruction box. We present data supporting a gain-of-function mechanism, in which the GMNN mutations result in proteins lacking the destruction box and hence increased protein stability and prolonged inhibition of replication leading to autosomal-dominant MGS.
Main Text
Meier-Gorlin syndrome (MGS [MIM 224690, 613800, 613803, 613804, 613805]) is recognized as an autosomal-recessive disorder characterized by dwarfism, absent or hypoplastic patellae, and microtia.1, 2 Additional phenotypic features include pulmonary emphysema, feeding difficulties, urogenital abnormalities, mammary hypoplasia, and characteristic facial features.3 The hallmark of distinct ear abnormalities and full lips, down-slanting palpebral fissures, narrow nose, high nasal bridge, microstomia, and micro/retrognathia distinguish this genetic disorder, enabling clinical diagnosis in most cases.4, 5 Intellectual disability or cognitive deficits are rare in MGS.4, 5
Human subjects manifesting a MGS clinical phenotype have provided important insights into DNA replication mechanisms and both cell and organism growth. MGS is a form of microcephalic primordial dwarfism, and thus, the syndrome is associated with proportionate growth deficits with microcephaly.4 These growth deficits are noted prenatally and persist postnatally.4, 5 Pathogenic changes in one of five genes that encode pre-replication complex proteins, namely ORC1 (MIM: 601902), ORC4 (MIM: 603056), ORC6 (MIM: 607213), CDT1 (MIM: 605525), and CDC6 (MIM: 602627), have been associated with MGS.6, 7, 8 Of these five genes, three (ORC1, ORC4, ORC6) encode proteins that are members of the origin of replication complex which consists of six proteins (ORC1–ORC6) that bind to origins of replication and initiate the process of DNA replication.9, 10 Binding of the six members of the origin of replication complex then recruits the DNA replication factors CDT1 and CDC6, which facilitates loading of the mini-chromosome maintenance (MCM) helicase.11, 12, 13 During S phase of the cell cycle, MCM helicase unwinds DNA and allows for the initiation of replication.14 It is hypothesized that the autosomal-recessive forms of MGS result from hypomorphic alleles because no homozygous or compound heterozygous loss of function mutations have been described in individuals with this syndrome.5 Defects in this process of the initiation of DNA replication are the primary mechanism underlying growth failure in this disorder. However, mutations in the five pre-replication complex genes only explain the phenotype in approximately 78% of individuals with a clinical diagnosis of MGS.4, 6, 7, 8 The molecular etiology in about 20% of cases remains unknown.
We now describe three subjects with a simplex autosomal-dominant form of MGS who carry de novo heterozygous mutations in the 5′ end of GMNN (MIM: 602842). GMNN maps to 6p22.3 and encodes the DNA replication inhibitor geminin. Geminin regulates cell-cycle progression and replication by interacting with the DNA replication factor CDT1, which is encoded by a gene with a known association to MGS, CDT1.15, 16, 17, 18 Written informed consent was obtained in accordance with protocols approved by the appropriate human subjects ethics committees at Baylor College of Medicine and Radboud university medical center. A summary of clinical findings in all three subjects is provided in Table 1.
Table 1.
Clinical Features of the Three Subjects with De Novo Heterozygous GMNN Mutations
| Phenotype | Subject 1 | Subject 2 | Subject 3 |
|---|---|---|---|
| Sex | Female | Male | Female |
| Age at diagnosis | 5 months | 4 months | 3 years |
| Age at recent examination | 34 months | 17 years | 3 years and 4 months |
| Prematurity | Yes (32 4/7 weeks) | No (38 2/7 weeks) | No (38 5/7 weeks) |
| Classic Triad | |||
| Short stature (Height Z score) | 67.5 cm (−6.8)a | 150.3 cm (−3.9)22,b | 76.5 cm (−6.0)23 |
| Microtia | Yes | Yes | Yes |
| Patella aplasia | Yes | Yes | Yes |
| Growth Parameters | |||
| Birth weight (Z score) | 980 g (−2.2)19 | 1900 g (−3.5)24 | 2750 g (−1.1)24 |
| Birth length (Z score) | NA | 40 cm (−4.9)24 | 45 cm (−2.2)24 |
| Birth head circumference | NA | 30.5 cm (−2.7)24 | NA |
| Weight on examination (Z score) | 5.6 kg (−11.98)a | 36.4 kg (−3.0)22 | 7.7 kg (−3.1 (weight for length))23 |
| Head circumference on examination (Z score) | 45.5 cm (−1.9)20,a | NA | 43.5 cm (−3.8)24 |
| Delayed bone age | Yes | Yes | NA |
| Facial Characteristics | |||
| Frontal bossing or high forehead | Yes | Yes | Yes |
| Down-slanted palpebral fissures | Yes | No | No |
| Posteriorly rotated ears | Yes | No | Yes |
| Upturned nose | Yes | No | No |
| Hypoplastic nares | Yes | Yes | Yes |
| Full lips | Yes | Yes | Yes |
| Micro-/retrognathia | Yes | Yes | Yes |
| Neurologic Features | |||
| Motor delay | No | Yes | Yes |
| Speech delay | No | Yes | Yes |
| Developmental assessment | NA | IQ = 61 (at age 14 years) | IQ = 59 (at age 10 years) |
| Respiratory Features | |||
| Pulmonary emphysema | No | Yes | No |
| Laryngomalacia | Yes | No | No |
| Tracheo-/Bronchomalacia | Yes | No | No |
| Gastrointestinal Features | |||
| Feeding problems in infancy | Yes | Yes | Yes |
| Failure to thrive | Yes | Yes | No |
| Gastresophageal reflux | Yes | No | No |
| Urogenital Anomalies | |||
| Abnormal genitalia | Yes | No | NA |
| Hypospadias | NA | No | NA |
| Cryptorchidism | NA | Yes | NA |
| Hypoplastic labia majora | Yes | NA | NA |
| Renal anomalies | NA | No | No |
| Other | Trichiasis Nanophthalmos | Increased/marked lumbar lordosis Urethral stenosis/dysplasia | Increased/marked lumbar lordosis Bilateral congenital hip subluxation Strabismus Cleft palate (palatum molle) |
Abbreviations are as follows: NA, Not available.
Not corrected for gestational age, CDC 2000 growth chart.
After use of growth hormone therapy.
Subject 1 was born at 32 4/7 weeks gestation by emergency cesarean section. Family history revealed that the mother had multiple spontaneous miscarriages. There was no known consanguinity. The pregnancy was complicated by twin gestation, placental insufficiency and severe intrauterine growth restriction in the proband. Her birth weight was 980 g (Z score = −2.2).19 The proband had a prolonged hospitalization of about 2 months due to feeding issues necessitating nasogastric tube feeding, and she was diagnosed with gastresophageal reflux. She was hospitalized again at 5 months of age for failure to thrive. She had cochlear implants placed for conductive hearing loss secondary to aural atresia/microtia and was diagnosed with grade II subglottic stenosis, tracheomalacia, and bronchomalacia by diagnostic bronchoscopy and laryngoscopy. She had delayed bone age (−3.5 SD from mean for age) with low IGF1 and normal IGFBP-3. Brain magnetic resonance imaging (MRI) study showed a mildly simplified gyral pattern with delayed myelination even after correction for prematurity. She continued to meet her developmental milestones as expected for age. When evaluated again at 2 years 9 months of age, her growth parameters remained below the expected range for her age (Table 1).19, 20 Other distinct craniofacial findings included frontal bossing, downslanting palpebral fissures, bilateral entropion, upturned nose with hypoplastic nares, severe microtia with no external auditory canal openings, midface hypoplasia, full lips, and micrognathia (Figure 1). She also had absent patellae bilaterally, midphalangeal hypoplasia of the 5th digit of the upper extremities bilaterally, single transverse palmar crease on the right hand, under-developed distal finger creases, sandal gap between first two toes bilaterally, a small umbilical hernia, and hypoplastic female genitalia. Given her features of microtia, absent patellae, and short stature/primordial dwarfism, a clinical diagnosis of MGS was made. Chromosome microarray (Baylor version 8.3) was normal, and whole-exome sequencing was performed.
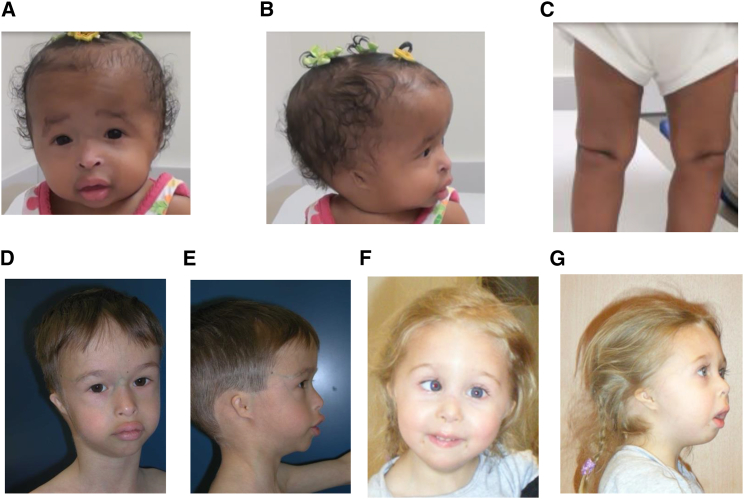
Figure 1.
Phenotypic Features of Subjects with GMNN Mutations
(A) Subject 1 with frontal bossing, fine facial features, downslanting palpebral fissures, bilateral entropion, upturned and beaked nose with hypoplastic nares, full lips, and micrognathia.
(B) Bilateral microtia with posteriorly rotated ears (subject 1).
(C) Knees with absent patellae (subject 1).
(D) Subject 2 with high forehead, short palpebral fissures with long eyelashes, small mouth with full lips, micrognathia, and prominence of the veins over both nose and forehead.
(E) Small and simple ear with small external auditory canals (subject 2).
(F) Subject 3 with high forehead, medial sparse/decreased eyebrows, upturned nose with hypoplastic nares, full lips, and micrognathia.
(G) Microtia with posteriorly rotated ears, hypoplastic nares, convex profile of the nose, everted lower lip, and micrognathia (subject 3).
Subject 2 (Figure 1) was previously described by Bongers et al. (patient 2) and de Munnik et al. (patient 43).3, 5 His short stature has persisted, and he has been diagnosed with growth hormone deficiency. Growth hormone was initiated at age 3 years (height −6.8 SD) and ended at 14 years (height −3 SD). In addition, he was diagnosed with delayed puberty. Puberty was induced with testosterone at the age of 13.5 years, but his progression through puberty has remained slow. His growth parameters at most recent physical examination are presented in Table 1. His pulmonary complications ultimately led to a diagnosis of congenital emphysema. In addition, his developmental delays persisted, and a full developmental assessment revealed an IQ of 61. Karyotype analysis and molecular testing of the five known MGS genes were normal.
Subject 3 was the first child of healthy, non-consanguineous Dutch parents. The infant was delivered in the occiput posterior position at 38 5/7 weeks. Her birth weight was 2,750 g (Z score −1.1),21 and her birth length was 45 cm (Z score −2.2).22 She had a cleft palate, feeding difficulties and required nasogastric tube feeds. Moreover, she was diagnosed with congenital hip dysplasia, strabismus convergens, and recurrent respiratory infections. Psychomotor development, especially speech, was mildly delayed. Growth parameters assessed during the most recent visit are presented in Table 1.23, 24 Other findings on physical examination included posteriorly rotated ears with a simple shape and narrow auditory canals, small nose with flat nasal bridge, aplastic patellae, feet in a planovalgus position, and marked lumbar lordosis (Figure 1). Echocardiogram and renal ultrasound were normal. The combination of short stature, absent patellae, and microtia led to a clinical diagnosis of MGS. Karyotype analysis and Sanger sequencing for the five genes with known associations to MGS, CHD7 (MIM: 608892) for CHARGE syndrome (MIM: 214800), and TCOF1 (MIM: 606847) for Treacher Collins syndrome (TCS1 [MIM: 154500]) revealed no abnormalities.
Whole-exome sequencing (WES) was completed in the Exome Laboratory at Baylor Miraca Genetics Laboratories (subject 1) and at Baylor College of Medicine Human Genome Sequencing Center (BCM-HGSC) through the Baylor Hopkins Center for Mendelian Genomics (subject 2) (Table S1). Sequencing and data analyses were conducted as previously described, targeting approximately 20,000 genes, including the coding and UTR exons.25, 26, 27 Sequencing data from Illumina HiSeq 2000 were mapped to the reference haploid human genome sequence (GRCh37/hg19) using the Burrows-Wheeler Alignment (BWA) algorithm.28 Variant calling from the aligned BAM files was performed using the in-house developed ATLAS2 suite.29 Annotation was performed using the in-house developed annotation pipelines CASSANDRA and SACBE that make use of ANNOVAR and custom scripts.29, 30 Variants were classified into exonic, intronic, or intergenic as well as their potential functional effects, and their frequencies in different populations and databases by the mentioned annotation pipelines. Variants were filtered by their observed frequencies in databases including dbSNP, the 1000 Genomes Project, and the NHLBI Exome Sequencing Project Exome Variant Server (EVS) to filter out common polymorphisms of high frequency in healthy control populations that are likely to be benign variants.25, 27
Whole-exome sequencing identified a heterozygous c.16A>T (p.Lys6∗) nonsense (stopgain) mutation in GMNN (GenBank: NM_015895.4) of subject 1 and a heterozygous c.35_38delTCAA (p.Ile12Lysfs∗4) frameshift deletion in the same gene of subject 2, which is predicted to result in a premature stop four positions downstream (Table 2, Figure 2). To determine whether mutations in GMNN are present in other individuals with MGS, we performed subsequent Sanger sequencing of all coding regions and splice sites of GMNN in seven additional subjects at the Radboud university medical center with suspected MGS but without a molecular diagnosis (Table S2). In subject 3, we identified a missense mutation in the second-to-last nucleotide of exon 2, c.50A > G (p.Lys17Arg), which was predicted to eliminate an exonic splice enhancer (ESE) target site of serine/arginine-rich splicing factor 5 (SRSF5) by ESE Finder (Table 2, Figure 2).31, 32 The downstream splice site at the exon 2 and intron 2 boundary was also predicted to be diminished by another in silico prediction program, Mutation Taster (Table 2, Figure 2).33 All three changes were located in exon 2, which is the first coding exon of GMNN (GenBank: NM_015895.4).
Table 2.
Summary of GMNN Mutations in Three Unrelated Subjects with Meier-Gorlin Syndrome
| Subject | Coordinate (hg19) | Mutation Type | Nucleotide Change | Predicted Amino Acid Change | Exon | Inheritance | ExAC Frequency | Mutation Taster | ESE Finder |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Chr6: 24777490 |
Stop gain | c.16A>T | p.Lys6∗ | 2 a | de novo | NRb | deleterious | NAd |
| 2 | Chr6: 24777509-24777512 |
Frameshift deletion | c.35_38delTCAA | p.Ile12Lysfs∗4 | 2 a | de novo | NRb | deleterious | NAd |
| 3 | Chr6: 24777524 |
Missense | c.50A>G | p.Lys17Arg | 2 a | de novo | NRb | deleteriousc | Elimination of SRSF5 |
Exon 2 is the first coding exon for GenBank NM_015895.4.
NR, not reported.
Predicted to affect downstream splicing at the exon 2 and intron 2 boundary.
NA, not applicable.
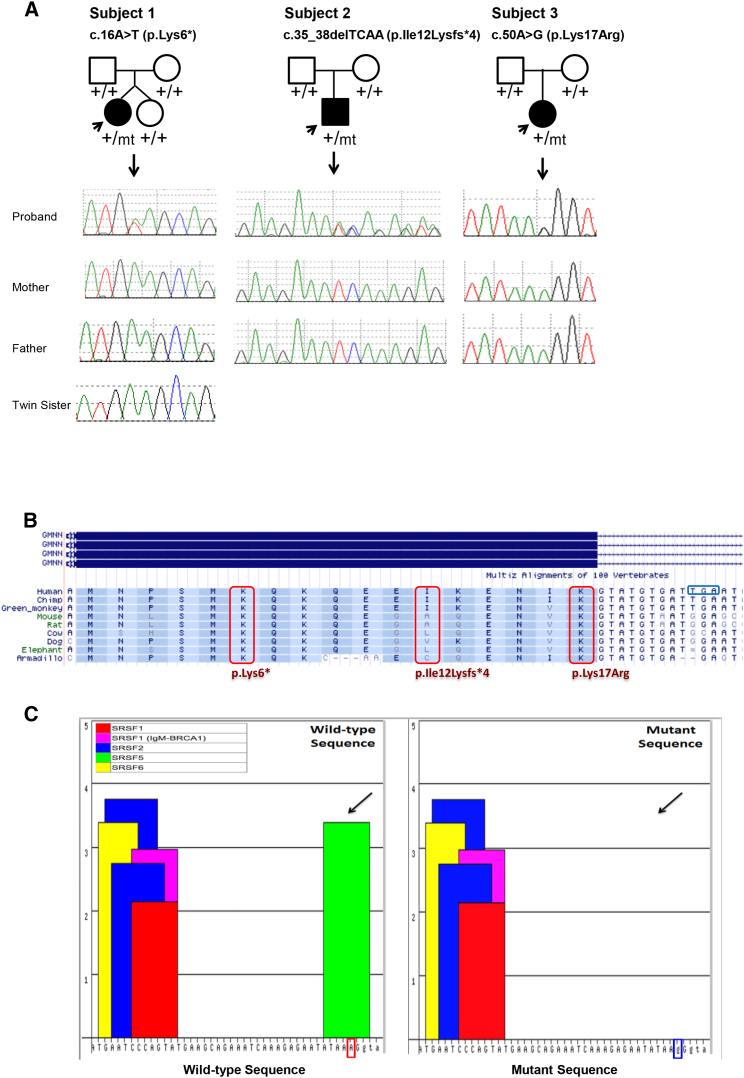
Figure 2.
GMNN Mutations in the Three Families with Meier-Gorlin Syndrome and the Predicted Effects
(A) Sanger sequencing of genomic DNA from peripheral blood of subjects 1, 2, 3, and parents suggesting the three mutations occurred de novo in the probands. The genotypes are shown below each individual with “+” representing the wild-type allele and “mt” representing the mutant allele.
(B) Annotation of the three amino acid residues in exon 2 (Lys6, Ile12, Lys17) affected in the subjects shows conservation across different species. Ile12 is the least conserved; however, it is a frameshift mutation that is predicted to affect not only Ile12 but also result in a premature stop four positions downstream. The p.Lys17Arg substitution is predicted to disrupt splicing possibly leading to either skipping of exon 2 or retention of intron 2 and introduction of a premature stop codon (TGA) in intron 2 that is highlighted with a blue box.
(C) ESE Finder predicted the downstream effects of the c.50A>G (p.Lys17Arg) mutation in subject 3. Different exonic splicing enhancer (ESE) motifs represented in different colors are listed in the lower boxes with SFRS5 represented in green. The mutant sequence (ATAAGGg, right panel) was predicted to result in loss of SFRS5, which presented in the wild-type sequence (ATAAAGg, left panel) with a score of 3.39 (threshold 2.67). The thresholds are values above which a score for a given sequence is considered to be significant (high-score motif).
Sanger sequencing confirmed the heterozygous mutations in the subjects and showed that the mutations were absent in DNA samples from all biological parents in the three families, indicating that the three mutations arose de novo in the subjects (Figure 2). Moreover, the mutation was not identified in a sample from the unaffected twin of subject 1. These mutations were not found in control databases including the 1000 Genomes Project and Exome Aggregation Consortium (ExAC) databases. Moreover, we were unable to detect any likely contributing mutations by exome sequencing in the five genes previously reported to harbor mutations that cause MGS or in other candidate genes such as ORC2 (MIM: 601182), ORC3 (MIM: 604972), and ORC5 (MIM: 602331) encoding other members of the origin of replication complex in subjects 1 and 2. In subject 3, we previously excluded the five known MGS genes by Sanger sequencing. In summary, three distinct, rare and likely pathogenic de novo alleles were identified in the same gene in three unrelated subjects with the same rare clinically recognizable disease trait. Our data suggested that GMNN dysfunction is the most likely molecular explanation for the subjects’ phenotypes.
GMNN encodes geminin, a nuclear regulator protein that is a known inhibitor of replication.34 Geminin is present during S, G2, and M phases of the cell cycle but is degraded as cells exit mitosis, and it is absent in G1 when origins of replication are licensed.34 During S phase, geminin binds DNA replication factor CDT1 and prevents MCM helicase from loading onto origins of replication and thus prevents re-licensing of origins of replication.15, 16 In addition, geminin inhibits histone acetyltransferase HB01, and this inhibition, which requires CDT1, is believed to be another mechanism by which geminin and CDT1 regulate cell-cycle progression, in particular, re-replication licensing.17, 18 The degradation of geminin at the end of mitosis is mediated by a nine-amino-acid destruction box at the N terminus of the protein from Arg23 to Pro31 (RRTLKMIQP), which is encoded by DNA sequences in exon 3 of GMNN and located downstream of the three exon 2 mutations identified in this study (Figure 3).34 The destruction box sequence is ubiquitinated by the anaphase-promoting complex (APC), resulting in protein degradation at the end of mitosis.34 Previous studies showed that when this destruction box is absent, the half-life of geminin is increased, and cell proliferation is inhibited in vitro.34, 35, 36
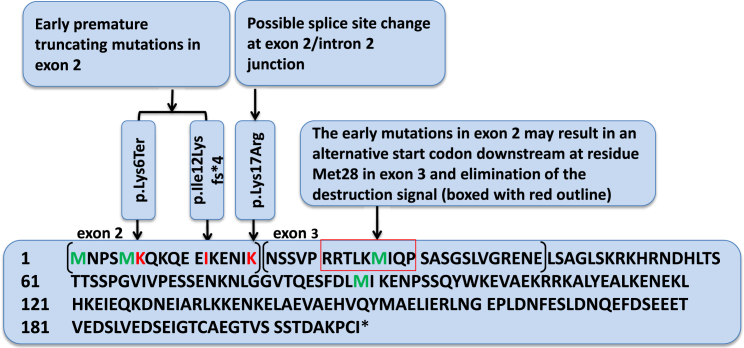
Figure 3.
Positions of the Early GMNN Mutations and Destruction Signal in Geminin
The early de novo mutations could lead to elimination of the destruction signal (boxed) with use of Met28 as an alternative initiation codon. Affected amino acids are represented in red; start codon (Met1), the proposed alternative start codon (Met28), and residues Met4 and Met89 are represented in green. The amino acids encoded by exon 2 and exon 3 of GMNN are in separate brackets with exon numbers labeled above.
The heterozygous de novo mutations identified in exon 2 of GMNN in subjects 1 and 2 are predicted to result in premature stops that occur before the destruction box encoded by exon 3.34 In subject 3, the de novo substitution, which is also located before the destruction box sequence, affects the second-to-last nucleotide of exon 2 and predicted a moderately conservative missense change of lysine to arginine, because both are di-basic amino acids. However, this mutation is also predicted to result in aberrant splicing possibly leading to either skipping of exon 2 or retention of intron 2 and introduction of a premature stop codon (TGA) in intron 2 (Figure 2). Further studies for subject 3 using peripheral blood samples or cultured cells to assess the effects of the mutation at the cDNA level were not conducted as they were not consented by the family.
For subjects 1 and 2, we conducted transcriptional studies on cells from each subject to monitor the effect of the mutations at the mRNA level. PCR amplification and Sanger sequencing of cDNA generated from lymphoblast-derived cell lines (subjects 1 and 2) as well as cultured fibroblasts (subject 2) demonstrated the presence of the mutant transcript with approximate 1:1 ratios of the wild-type and mutant alleles (Figure S1) suggesting that the mutant transcript is stable and escapes nonsense mediated decay (NMD). To corroborate this observation, we cloned GMNN RT-PCR product of subject 1 using TOPO TA-cloning (Invitrogen) and Sanger sequenced eight individual clones. Of the sequenced clones, 50% (4/8 clones) harbored the mutant allele c.16A>T (p.Lys6∗) and the other 50% (4/8 clones) harbored the wild-type allele (Figure S1). Because RNA studies demonstrated that the mutant alleles did not appear to be subject to NMD, in the setting of early truncation, we hypothesize that an alternative start codon at residue Met28, which is located in exon 3 within the destruction box and downstream of the exon 2 mutations (Figure 3), might be used for translation initiation in the subjects resulting in a protein lacking the 27 amino acids at the N terminus including most of the destruction box. Western blot analysis of protein lysates from subject 1 lymphoblast-derived cell lines offered preliminary confirmation of this posit; we detected both full length and truncated GMNN isoforms using a previously described antibody raised against the full-length human protein (Figure S2).35
To investigate further whether the AUG codon corresponding to Met28 in subject 1 and 2 can be used as an alternative start codon, we tagged mutant GMNN cDNA as well as the wild-type GMNN cDNA with a HA epitope at the N terminus and a V5 epitope at the C terminus. The mutation in subject 3 was not studied because its effect on cDNA cannot be determined due to lack of consent for further research studies. Both in vitro and ectopic expressions of the GMNN mutants produced a truncated protein 4–5 kDa shorter than the wild-type protein, consistent with what would be predicted if the translation starts from the Met28 residue (Figures 4A and 4B). Additionally, the wild-type GMNN construct produced a full-length protein with both the HA and V5 epitopes, whereas mutant GMNN constructs produced truncated proteins tagged only with V5 at the C terminus but lacking the HA epitope at the N terminus, consistent with the translation initiation at the downstream Met28 (Figures 4A and 4B). Although we also detected additional products predicted to be translated from Met89 further downstream in the in vitro assays (Figure 4A), the resulting protein is predicted to lack the interaction domain with CDT1 and is thus unlikely to be functional.
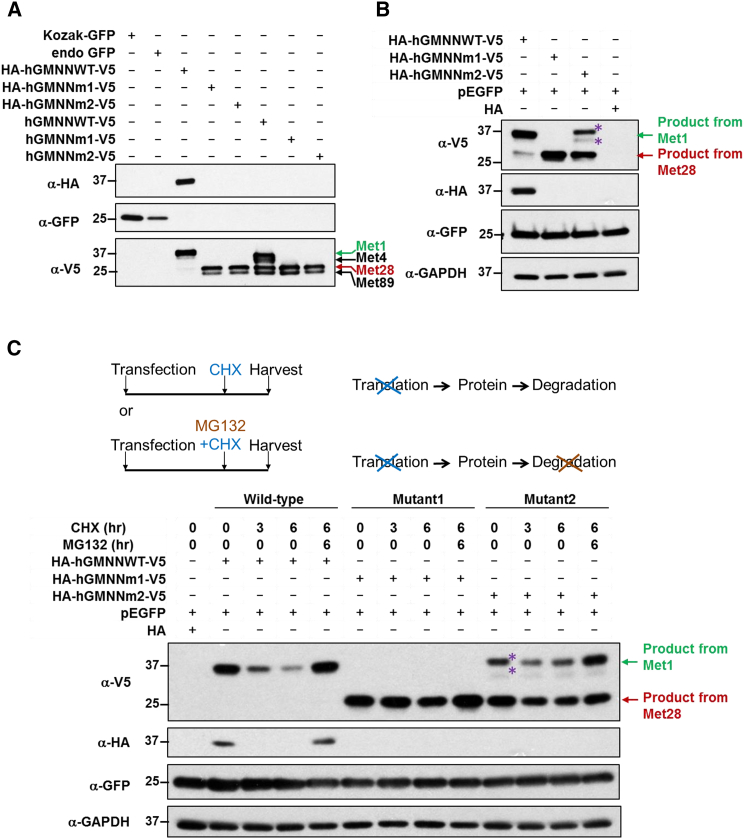
Figure 4.
GMNN Mutations Produce a More Stable Protein Starting at Residue Met28 with the Destruction Box Eliminated
(A) Western blot of geminin proteins from in vitro transcription and translation: GFP is used as a positive control for the reaction. Kozak-GFP (lane 1): GFP control with optimal Kozak sequence (for translation initiation); endogenous GFP (lane 2): GFP control with human GMNN endogenous Kozak sequence; HA-hGMNNWT-V5 (lane 3), HA-hGMNNm1-V5 (lane 4), and HA-hGMNNm2-V5 (lane 5): human GMNN from wild-type, subject 1, and subject 2, respectively, carrying endogenous Kozak sequence and tagged with N-terminal HA epitope and C-terminal V5 epitope; hGMNNWT-V5 (lane 6), hGMNNm1-V5 (lane 7), and hGMNNm2-V5 (lane 8): human GMNN from wild-type, subject 1, and subject 2 respectively carrying endogenous Kozak sequence and tagged only with C-terminal V5 epitope (this is designed for testing the translation preference for the start codon staring from HA). Same amount of templates (69 ng) are used in each reaction. Our data showed the mutant constructs produced a truncated protein of 4–5 kDa shorter than the wild-type protein, consistent with what would be predicted if the translation starts from Met28 residue (lanes 3–7). Although there is also product translated from the 4th start codon (p.Met89), it lacks CDT1 interaction domain and is thus likely to be non-functional. Wild-type GMNN construct can be translated into a full-length protein with both HA and V5 epitopes while mutant GMNN constructs can only produce truncated protein with V5 tag, but not HA tag. The HA-tagged products from both mutant GMNN constructs would be too small and thus are not presented on the Western blot.
(B) Western blot of ectopically expressed geminin in HEK293T cells. GMNN cDNA of wild-type, subject 1, and subject 2, are cloned into the pCMV-HA expressing vector and are also tagged with V5 epitope at C terminus. GFP is co-transfected in every condition as a transfection internal control and GAPDH is used as loading control. We again observed that the mutant constructs produced a truncated protein that would be expected if the translation starts from Met28 residue. Wild-type GMNN construct can be translated into a full-length protein with both HA and V5 epitopes while mutant GMNN constructs can only produce truncated protein with V5 tag but not HA tag.
(C) Western blot analysis of protein stability of the ectopically expressed proteins in HEK293T cells. The transfected cells were treated with cycloheximide (CHX, 100 μg/ml, blocker of protein synthesis) or CHX plus MG132 (10 μM, inhibitor of proteasome) and lysed at 0, 3, and 6 hr. The Western blot data showed that CHX treatment revealed degradation of the wild-type protein, while no obvious degradation was observed in mutant proteins under the same conditions indicating that the mutant proteins, which were translated using Met28 as the start and without the destruction box, were more stable. The addition of MG132 conferred stability for both wild-type and mutant proteins as no obvious degradation was observed for protein with the CHX plus MG132 treatment. In (B) and (C), asterisks denote that the product is likely to be a posttranslational modification or dimer since this is not recognized by HA but only V5 antibody and the molecular weight of the top band is slightly different from the wild-type protein.
To compare the stability of the wild-type and mutant proteins, the transfected HEK293T cells were treated with cycloheximide (CHX, which blocks protein synthesis) or CHX plus MG132 (a proteasome inhibitor), and lysed at 0, 3, and 6 hr. Western blot data showed that CHX treatment led to degradation of the wild-type protein, while no obvious degradation was observed in mutant proteins under the same conditions, indicating that the mutant proteins, which were translated using Met28 as the start and without the intact destruction box, were more stable (Figure 4C). For both wild-type and mutant proteins, no obvious degradation was observed when treated with the CHX plus MG132 (Figure 4C), consistent with the reported role of the proteasome in geminin degradation and suggesting that lack of the destruction box signal confers stability for the protein.
We also performed flow cytometry to investigate the distribution of cells in different cell-cycle phases in asynchronous lymphoblasts of subject 1 and her father (as a control) (Figure S3). The percentages of cells (average from the triplicate studies) in G1 phase were 60% and 48% in cells from subject 1 and the control respectively (p = 0.0005) while the percentages in S phase were 23% and 32% in subject 1 and the control, respectively (p = 0.0024). Although we observed a small increase in G1 content in subject 1, it is unknown whether the mutant geminin causes delayed S-phase entry, as previously observed in ORC1-deficient cells.6 Additional investigations on the mutant geminin protein’s effects on the cell cycle and its expression in vivo are warranted in order to further understand its role in MGS pathogenicity.
Interestingly, the usage of the same alternative start codon (Met28) and increased geminin stability was also observed in a previous study, which showed that an engineered mutation abolishing the original translation start site of geminin in vitro results in a truncated protein that likely resulted from use of the same Met28 alternative start codon in a human colon cancer cell line.35 When Met28, which is located within the destruction box, is used as the alternative start codon, the destruction box is eliminated, resulting in increased protein stability and decreased replication.35 Deletions involving the destruction box sequence in Xenopus produce similar results in vitro.34 A partial rescue of these phenotypes occurs with overexpression of CDT1.35
As the most parsimonious explanation of the aggregate data, we propose that the de novo GMNN mutations identified in the subjects result in truncated proteins (initiated at residue Met28) lacking the destruction box, leading to increased geminin protein stability and prolonged inhibition of replication. The mutations cause gain-of-function effects mimicking the molecular defects caused by bi-allelic mutations in ORC1, ORC4, ORC6, CDT1, or CDC6, leading to autosomal-dominant MGS.
Alternative potential mechanisms of action include that the disease is caused by an autosomal-recessive paradigm like observed for the other pre-replication complex defects and that the second mutation lies within deep intronic regions or regulatory regions not covered by exome or Sanger analysis. However, the function of geminin as an inhibitor of replication is not consistent with such a mechanism and thus an undetected mutation seems the least likely mechanism to account for the phenotype of these individuals. Additionally, all three mutations that we identified in GMNN are de novo, which would be extremely rare for autosomal-recessive disorders, suggesting further that an undetected second mutation is unlikely.
The three subjects in this study with de novo mutations in GMNN have classic features of primordial dwarfism, microtia, and absent patellae, fulfilling the clinical diagnostic criteria of MGS. Prior studies of individuals with mutations in ORC1, ORC4, ORC6, CDT1, or CDC6 have noted that individuals with ORC1 mutations were shorter in stature and had a smaller head circumference compared to individuals with mutations in the other four genes.4 Relative to published cohorts, individuals with de novo mutations in GMNN described herein are also on the extreme end of the height spectrum with a height Z score ranging from −3.9 to −6.8. In addition, subjects 2 and 3 have marked lumbar lordosis and developmental delay and/or cognitive impairment, a rare finding in MGS. Moreover, subjects 1 and 2 share fullness of the peri-orbital region, not previously recognized in other individuals with MGS who have mutations in the five known genes. Based on the severity of the phenotype, we speculate that ORC1 and GMNN mutations might perturb DNA replication to a greater extent than the other MGS associated genes, and perhaps GMNN mutations have a more profound effect on the replicative burst and cell growth required for brain development, head size expansion, and height.
In conclusion, we describe an autosomal-dominant form of MGS resulting from de novo heterozygous mutations in GMNN, likely caused by a gain-of-function mechanism in replication inhibitor geminin. These de novo GMNN mutations lead to more stable geminin proteins that lack the intact destruction box and interfere with the cell cycle. We speculate that other types of mutations such as small in-frame deletions or missense changes that affect the normal function of the destruction box might also cause MGS. Mutations in GMNN should be considered in individuals with MGS who do not have contributing mutations in one of the genes associated with the autosomal-recessive form of MGS. The identification of autosomal-dominant MGS caused by defects in GMNN in this study expands the genetic heterogeneity of this disorder, helps identify the molecular etiology in additional individuals with MGS, and provides further insights into the relationship between DNA replication, cell growth, and organismal development.
Acknowledgments
We thank the patients and their families for participating in this study. We also thank S.D. van der Velde-Visser for technical support. This work was funded in part by the US National Human Genome Research Institute (NHGRI)/National Heart Lung and Blood Institute (NHLBI) grant number U54HG006542 to the Baylor-Hopkins Center for Mendelian Genomics (BH-CMG). L.C.B. was supported by the Genzyme/ACMG Foundation for Genetic and Genomic Medicine Medical Genetics Training Award in Clinical Biochemical Genetics, the National Urea Cycle Disorders Foundation Fellowship, a fellowship from the Urea Cycle Disorders Consortium (UCDC; U54HD061221), which is a part of the National Institutes of Health (NIH) Rare Disease Clinical Research Network (RDCRN), supported through collaboration between the Office of Rare Diseases Research (ORDR), the National Center for Advancing Translational Science (NCATS and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), and the National Institutes of Health (T32 GM07526). W.-L.C. is supported by CPRIT training Program RP140102. N.K. is supported by P50 DK096415 and is a distinguished Jean and George Brumley Professor. J.R.L. holds stock ownership in 23andMe and Lasergen, is a paid consultant for Regeneron Pharmaceuticals, and is a co-inventor of multiple United States and European patents related to molecular diagnostics. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from molecular genetic testing offered in the Baylor Miraca Genetics Laboratories.
Published: December 3, 2015
Footnotes
Supplemental Data include three figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.11.006.
Contributor Information
Ernie M.H.F. Bongers, Email: ernie.bongers@radboudumc.nl.
Yaping Yang, Email: yapingy@bcm.edu.
Accession Numbers
The ClinVar accession numbers for the DNA variant data reported in this paper are SCV000224003, SCV000224004, and SCV000224005.
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://browser.1000genomes.org
ESE Finder, http://rulai.cshl.edu/tools/ESE
ExAC Browser, http://exac.broadinstitute.org/
MutationTaster, http://www.mutationtaster.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
Supplemental Data
References
- 1.Gorlin R.J., Cervenka J., Moller K., Horrobin M., Witkop C.J., Jr. Malformation syndromes. A selected miscellany. Birth Defects Orig. Artic. Ser. 1975;11:39–50. [PubMed] [Google Scholar]
- 2.Meier Z., Poschiavo, Rothschild M. [Case of arthrogryposis multiplex congenita with mandibulofacial dysostosis (Franceschetti syndrome)] Helv. Paediatr. Acta. 1959;14:213–216. [PubMed] [Google Scholar]
- 3.Bongers E.M., Opitz J.M., Fryer A., Sarda P., Hennekam R.C., Hall B.D., Superneau D.W., Harbison M., Poss A., van Bokhoven H. Meier-Gorlin syndrome: report of eight additional cases and review. Am. J. Med. Genet. 2001;102:115–124. doi: 10.1002/ajmg.1452. [DOI] [PubMed] [Google Scholar]
- 4.de Munnik S.A., Otten B.J., Schoots J., Bicknell L.S., Aftimos S., Al-Aama J.Y., van Bever Y., Bober M.B., Borm G.F., Clayton-Smith J. Meier-Gorlin syndrome: growth and secondary sexual development of a microcephalic primordial dwarfism disorder. Am. J. Med. Genet. A. 2012;158A:2733–2742. doi: 10.1002/ajmg.a.35681. [DOI] [PubMed] [Google Scholar]
- 5.de Munnik S.A., Bicknell L.S., Aftimos S., Al-Aama J.Y., van Bever Y., Bober M.B., Clayton-Smith J., Edrees A.Y., Feingold M., Fryer A. Meier-Gorlin syndrome genotype-phenotype studies: 35 individuals with pre-replication complex gene mutations and 10 without molecular diagnosis. Eur. J. Hum. Genet. 2012;20:598–606. doi: 10.1038/ejhg.2011.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bicknell L.S., Walker S., Klingseisen A., Stiff T., Leitch A., Kerzendorfer C., Martin C.A., Yeyati P., Al Sanna N., Bober M. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat. Genet. 2011;43:350–355. doi: 10.1038/ng.776. [DOI] [PubMed] [Google Scholar]
- 7.Guernsey D.L., Matsuoka M., Jiang H., Evans S., Macgillivray C., Nightingale M., Perry S., Ferguson M., LeBlanc M., Paquette J. Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nat. Genet. 2011;43:360–364. doi: 10.1038/ng.777. [DOI] [PubMed] [Google Scholar]
- 8.Bicknell L.S., Bongers E.M., Leitch A., Brown S., Schoots J., Harley M.E., Aftimos S., Al-Aama J.Y., Bober M., Brown P.A. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat. Genet. 2011;43:356–359. doi: 10.1038/ng.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell S.P., Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 10.Micklem G., Rowley A., Harwood J., Nasmyth K., Diffley J.F. Yeast origin recognition complex is involved in DNA replication and transcriptional silencing. Nature. 1993;366:87–89. doi: 10.1038/366087a0. [DOI] [PubMed] [Google Scholar]
- 11.Nishitani H., Lygerou Z., Nishimoto T., Nurse P. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature. 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka T., Knapp D., Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 13.Ticau S., Friedman L.J., Ivica N.A., Gelles J., Bell S.P. Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading. Cell. 2015;161:513–525. doi: 10.1016/j.cell.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 15.Wohlschlegel J.A., Dwyer B.T., Dhar S.K., Cvetic C., Walter J.C., Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- 16.Tada S., Li A., Maiorano D., Méchali M., Blow J.J. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol. 2001;3:107–113. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miotto B., Struhl K. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol. Cell. 2010;37:57–66. doi: 10.1016/j.molcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suchyta M., Miotto B., McGarry T.J. An inactive geminin mutant that binds cdt1. Genes (Basel) 2015;6:252–266. doi: 10.3390/genes6020252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fenton T.R., Kim J.H. A systematic review and meta-analysis to revise the Fenton growth chart for preterm infants. BMC Pediatr. 2013;13:59. doi: 10.1186/1471-2431-13-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Department of Health and Human Services Centers for Disease Control and Prevention. National Center for Health Statistics. 2000 CDC Growth Charts for the United States: Methods and Development. http://www.cdc.gov/growthcharts/2000growthchart-us.pdf. Series 11, Number 246. Published: May 2002. [PubMed]
- 21.Visser G.H., Eilers P.H., Elferink-Stinkens P.M., Merkus H.M., Wit J.M. New Dutch reference curves for birthweight by gestational age. Early Hum. Dev. 2009;85:737–744. doi: 10.1016/j.earlhumdev.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 22.Ozer B.K. Growth reference centiles and secular changes in Turkish children and adolescents. Econ. Hum. Biol. 2007;5:280–301. doi: 10.1016/j.ehb.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Schönbeck Y., Talma H., van Dommelen P., Bakker B., Buitendijk S.E., HiraSing R.A., van Buuren S. The world’s tallest nation has stopped growing taller: the height of Dutch children from 1955 to 2009. Pediatr. Res. 2013;73:371–377. doi: 10.1038/pr.2012.189. [DOI] [PubMed] [Google Scholar]
- 24.Niklasson A., Albertsson-Wikland K. Continuous growth reference from 24th week of gestation to 24 months by gender. BMC Pediatr. 2008;8:8. doi: 10.1186/1471-2431-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y., Ward P., Braxton A., Wang M., Buhay C. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lupski J.R., Gonzaga-Jauregui C., Yang Y., Bainbridge M.N., Jhangiani S., Buhay C.J., Kovar C.L., Wang M., Hawes A.C., Reid J.G. Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome Med. 2013;5:57. doi: 10.1186/gm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen Y., Wan Z., Coarfa C., Drabek R., Chen L., Ostrowski E.A., Liu Y., Weinstock G.M., Wheeler D.A., Gibbs R.A., Yu F. A SNP discovery method to assess variant allele probability from next-generation resequencing data. Genome Res. 2010;20:273–280. doi: 10.1101/gr.096388.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cartegni L., Wang J., Zhu Z., Zhang M.Q., Krainer A.R. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith P.J., Zhang C., Wang J., Chew S.L., Zhang M.Q., Krainer A.R. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- 33.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 34.McGarry T.J., Kirschner M.W. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida K., Oyaizu N., Dutta A., Inoue I. The destruction box of human Geminin is critical for proliferation and tumor growth in human colon cancer cells. Oncogene. 2004;23:58–70. doi: 10.1038/sj.onc.1206987. [DOI] [PubMed] [Google Scholar]
- 36.Shreeram S., Sparks A., Lane D.P., Blow J.J. Cell type-specific responses of human cells to inhibition of replication licensing. Oncogene. 2002;21:6624–6632. doi: 10.1038/sj.onc.1205910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.