Abstract
The discovery of RNA interference (RNAi) gave rise to the development of new nucleic acid-based technologies as powerful investigational tools and potential therapeutics. Mechanistic key details of RNAi in humans need to be deciphered yet, before such approaches take root in biomedicine and molecular therapy.
We developed and validated an in silico-based model of siRNA-mediated RNAi in human cells in order to link in vitro-derived pre-steady state kinetic data with a quantitative and time-resolved understanding of RNAi on the cellular level. The observation that product release by Argonaute 2 is accelerated in the presence of an excess of target RNA in vitro inspired us to suggest an associative mechanism for the RNA slicer reaction where incoming target mRNAs actively promote dissociation of cleaved mRNA fragments. This novel associative model is compatible with high multiple turnover rates of RNAi-based gene silencing in living cells and accounts for target mRNA concentration-dependent enhancement of the RNAi machinery.
INTRODUCTION
In mammalians, RNA interference (RNAi) is a vital post-transcriptional gene silencing mechanism controlling development and basic cellular processes such as cell growth, heterochromatin formation and tissue differentiation (1). Its dysfunction is linked to cancer, cardiovascular disease and other degenerative disorders (2–4). Since early on, there has been great interest in RNAi (5–10), not least because of its potential application in biomedicine and molecular therapy. Novel classes of oligonucleotides, i.e. small interfering RNA (siRNA), are becoming powerful investigational tools and potential therapeutics that allow to selectively turning off genes in living organisms via the RNAi pathway (11). Especially promising is its high potential against heretofore undruggable targets. However, before RNAi-based drugs fully reach the market, many mechanistic details of the complex system have to be deciphered and quantified to yet tackle challenges like increasing the efficacy of artificial siRNA, pinpointing delivery or reducing harmful off-target effects (12).
The main effector of RNAi is a ribonucleoprotein complex known as RNA-induced silencing complex (RISC) (13). Minimally, it consists of an Argonaute-family protein and a short (19–21 nt) single-stranded (ss) RNA (14). The origin of the short ssRNA is either endogenous double-stranded (ds) miRNA (15,16) or siRNA duplex from exogenous sources (synthetic or infectious) (17). Initially, the short RNA duplex with two nucleotides (nt) overhang at each 3′-end and a phosphate group at each 5′-end (14,18) binds to RISC. For RISC activation, the so-called passenger strand needs to be removed, while the complementary guide strand is loaded onto Ago2. The Ago2-bound guide strand recognizes the target gene's mRNA with high specificity via base-pairing. In case of siRNA, cleavage of the captured mRNA is induced by the endonuclease Ago2, the key component of RISC (19–21). Eventually, multiple turnover of target recognition and degradation reduces the cellular level of the given target mRNA (22–24).
Though, overall the mechanism of RNAi is reasonably well understood, RISC remains a complex molecular machinery, embedded in an intricate intracellular network with many yet unknown molecular players (25–30). In an attempt to overcome this inherent experimental limitation, research in the field of RNAi has been complemented by theoretical biology. In the past, quantitative modelling of RNAi has been performed using worms and plants as experimental system (31,32). Other studies focused on the miRNA pathway (33–35). While siRNA-mediated RNAi was extensively modelled by Bartlett et al. (36–38) and Cuccato et al. (39), it was Larsson et al. (40), who shed light on the effect of target mRNA turnover rates on RNAi and, to our knowledge, were the first to touch upon the influence of target abundance on RNAi efficacy.
Several X-ray crystallographic studies on bacterial and eukaryotic proteins provided valuable conclusive insights concerning crucial steps during the enzymatic cycle (41–47). In combining these data with own biochemical studies, we recently presented a comprehensive minimal mechanistic model describing fundamental steps during RNAi (48). The model defines hAgo2-mediated target mRNA binding and cleavage, dissecting binary as well as ternary complex formation and target mRNA cleavage into individual substeps. Thus, it provides in-depth insights into the complicated interplay of hAgo2 with small RNAs and corresponding target molecules.
In the present study, we combined these in vitro data with quantitative data derived from cell culture experiments. We developed a computational model, which is capable to quantitatively predict important parameters of the RNAi process in living mammalian cells and to unravel mechanistic details of the underlying machinery. Cell culture experiments suggest the RNAi machinery adapts to large variations in target mRNA levels, independent of siRNA or Ago2 concentrations. These experimental findings cannot be explained by the common literature view of RNAi, which may be described by a dissociative process, where the departing ligands (here, cleaved RNA fragments) leave the complex in a slow step with positive entropy of activation, before the next incoming ligand (i.e. uncleaved target mRNA) can enter. In vitro, the release of target RNA from Ago2 is accelerated in the presence of an excess of free target RNA (48). This inspired us to suggest an associative mechanism of target strand recognition by Ago2, which involves binding of the next incoming target mRNA to hAgo2 before dissociation of the cleaved mRNA fragments. Here, the departing ligand is not any longer the rate-limiting step. This new associative model, parameterized with abovementioned pre-steady state kinetic rate constants, is compatible with high multiple turnover rates of RNAi-based gene silencing in living cells and accounts for target mRNA concentration-dependent acceleration of the RNAi machinery.
This alternative view likely might have an impact on the selection of siRNA-based therapeutics, as the associative model suggests that the efficacy of a siRNA or miRNA depends on the expression level of its target mRNA in a way that high target levels allow for better inhibitory effects.
MATERIALS AND METHODS
Cell lines, cell culture and Lipofectamine™ 2000 (LF2000)-mediated siRNA transfection
HeLa-TetOff (HTO; BD Biosciences, Heidelberg, Germany) and HeLa-TetOff Luc (HTOL; stably transfected with firefly luciferase) (49) cells were cultured in Dulbecco's Modified Eagle Medium with 4500 mg/l glucose. All media were supplemented with 10% foetal calf serum (FCS) and were purchased from Invitrogen. Cells were cultured as exponentially growing subconfluent monolayers in a humidified atmosphere containing 5% CO2.
Twenty four hours prior to transfection, cells were seeded into 12 or 96-well plates (Greiner, Frickenhausen, Germany). Cell numbers were chosen to finally reach 80–90% confluency at the time of transfection. LF2000/siRNA complexes were allowed to form in OptiMEM (Life Technologies) according to the manufacturer's protocol with a final concentration of 5–10 μg/ml LF2000. Routinely, the cell culture supernatant was discarded and replaced by medium supplemented with 10% FCS 4 h after the start of transfection. All information concerning incubation times given in the manuscript refer to the time from the beginning of the transfection procedure.
Determination of luciferase activity
Luciferase activity was quantified 24 h after transfection, subsequent to the determination of cell viability by a fluorescein diacetate (FDA, Sigma-Aldrich, Deisenhofen, Germany) assay (49). Luminescence was measured in a microplate reader (Fluoroskan Ascent FL, Thermo Labsystems, Dreieich, Germany) using 50–100 μl of a buffer containing 28 mM Tricine (pH 7.8), 500 μM adenosine triphosphate, 250 μM coenzyme A, 250 μM D-luciferin, 33 mM DTT, 200 μM EDTA, 15 mM MgSO4, 1.5% (v/v) Triton X-100 and 5% (v/v) glycerol (all reagents purchased from Sigma-Aldrich). The luminescence was normalized to cell viability to account for cell loss due to cytotoxicity or washing procedures. For the determination of IC50 values RNAi-mediated downregulation of the luciferase activity was plotted as the percentage of active siR206 (50) versus inactive siINV (49) and the experimental data were fitted with an ‘IC50 full 4 param’ equation using the GraFit5 software (Erithacus Software, Surrey, UK).
Quantification of intracellular siRNA concentration via liquid hybridisation assay
Transfections were carried out as described above in a 12-well format. Cells were detached by trypsin treatment or by incubation in cold phosphate buffered saline. A certain percentage of each sample was used to measure luciferase activity, while the rest of the sample was subjected to a liquid hybridisation protocol. Cells were pelleted, incubated in phosphate buffered saline containing 1% NP-40 for 10 min on ice followed by total RNA extraction according to standard protocols. The quantification of the siRNA was achieved by hybridisation with the corresponding 32P-labelled sense strand for 10 min at 95°C followed by 1 h incubation at 37°C before the samples were resolved by 20% polyacrylamide gel electrophoresis under nondenaturing conditions. After blotting of the gel onto a nylon membrane (Hybond N+, Amersham, Freiburg, Germany), the signals were quantified with a PhosphorImager (TyphoonTM 8600 Variable Mode Imager, GE Healthcare, München, Germany). Absolute amounts of siRNA in the samples were calculated in relation to the included standards. For this purpose, defined amounts of siRNA were added to control cell lysates before the extraction step and treated simultaneously with the other samples. The amounts of total cellular RNA were determined spectrophotometrically and used for normalisation. Supplementary Figure S1 shows the correlation between the siRNA concentration in the transfection mix and siRNA molecules taken up intracellularly. Since previous investigations (49,51–53) have shown that of those molecules only about 4% of siRNA taken up are bioavailable (i.e. available for the RNAi machinery), numbers determined with the liquid hybridisation assay were multiplied with a factor of 0.04 to account for this fact.
Quantification of intracellular luciferase mRNA concentration by quantitative polymerase chain reaction
RNA was extracted from either HTOL or HTO (in this case, transfected with different amounts (0.1, 1, 10 and 100 ng/well) of pTRE2hyg-luc plasmid by LF2000) cells (grown in 96-well plates at about 15 000 cells) with Qiazol (Qiagen), followed by DNA digestion with TURBO DNase (Ambion/Life Technologies), phenol/chloroform and chloroform extractions. After resuspension of the RNA in water, nucleic acid concentration was determined with a NanoDrop Instrument (Thermo Scientific). cDNA synthesis was performed with 1 μg of RNA and 200 ng random primers in 20 μl reaction volume using the RevertAid First Strand cDNA Kit (Fermentas). Real-time-quantitative polymerase chain reaction (qPCR) was performed in 384-well plates at 10 μl each with a 7900HT system (Applied Biosystems) using the Sequence Detection Systems Software version 2.4 and SYBR Select Master Mix (Life Technologies). The amplicon size was 104 nt using the following forward and reverse primers: 5′-GAACATCACGTACGCGGAATAC and 5′-TTTCACTGCATACGACGATTCTG, respectively. As an internal control, mRNA levels of the house keeping gene GAPDH were determined. Here, the amplicon size was 86 nucleotides using the following forward and reverse primers: 5′-GACAGTCAGCCGCATCTTCTT and 5′-TCCGTTGACTCCGACCTTC, respectively. In case of HTOL cells, we determined about 1000 copies of GAPDH mRNA molecules per cell, which nicely fit published numbers by others (54–56).
Kinetic modelling of RNAi
In this study, the RNAi process in mammalian cells is formally described by a network of elementary reactions. Their biochemical rate equations connect experimentally derived in vitro reaction rate constants with physiological concentrations of reactants and cell compartment volumes. The reaction mechanism is encoded in the reactions’ rate laws, its parameters (Supplementary Tables S1 and S2) and the way the reactions are linked to one another in the two alternative metabolic pathways (Supplementary Figure S2). The reactions for the models translate in reaction kinetics equations as given in Equations 1–16 (Supplementary Equations 1). The difference between the two models is illustrated in Figure 1. Mathematically, the kinetics of a network of biochemical reactions can be described by a set of ordinary differential equations (ODEs); Equations 17–29 and 30–45 for the dissociative and the associative model, respectively (Supplementary Equations 2 and 3). These equations are solved numerically for time course simulations. Besides time courses of molecule concentrations, two further parameters serve as indicators: t1/2 of target knockdown and IC50 of siRNA. The time of half-maximal target knockdown is described by the constant t1/2. It is used as a contracted measure of time-resolved target knockdown to be able to relate model behaviour with two free variables simultaneously, i.e. basal target mRNA level and initial siRNA concentration. IC50 is defined as the half-maximal inhibitory siRNA concentration and indicates how much of a particular siRNA is required for half-maximal reduction of basal target mRNA level within 24 h. In this study, IC50 is used to compare the potency of the RNAi machinery at different levels of target mRNA. Time course simulations, t1/2 and IC50 estimations, as well as, sensitivity analyses are performed with Copasi (57) and are described under Supplementary Information.
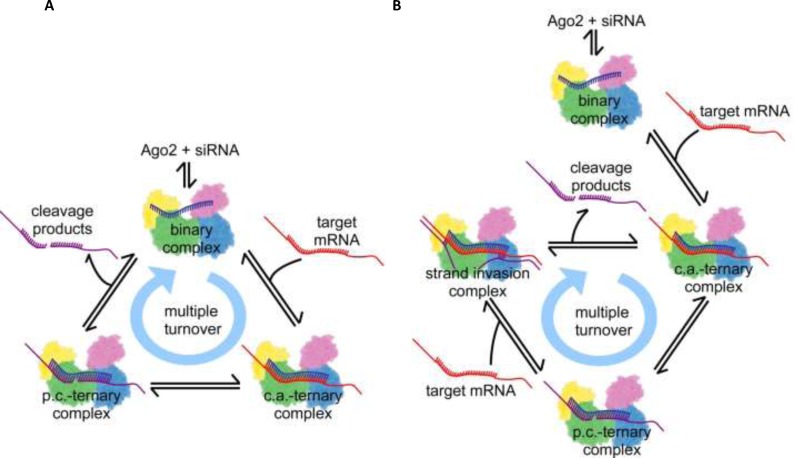
Figure 1.
Illustrations of multiple turnover hAgo2 mRNA cleavage cycle according to the proposed models of RNAi in mammalian cells. Ago2 is represented as a cartoon deduced from the electron density cloud of X-ray structures with the four domains N, PAZ, Mid and PIWI coloured individually. RNAs are indicated by lines and colour blue, red or purple for guide, target or product strands. The relative spatial positions of protein and RNA substrates are indicated and formation of Watson-Crick base pairs is illustrated by short connecting lines between RNA strands. (A) Dissociative model; once a stable binary complex between ss guide siRNA and Ago2 is formed the system undergoes multiple rounds of target binding, cleavage and product release. (B) Associative model; in contrast to the dissociative model, here target cleavage is followed by the associative binding of another target, which may invade the hydrogen bonds between guide and target mRNA cleavage products, eventually replacing the product strands. The cycle is closed by dissociation of the product fragments, which leads to the formation of the next catalytically active complex. For clarity, only the most relevant steps are shown; a detailed scheme including all steps of the complex reaction is shown in Supplementary Figure S2. c.a.: catalytically active, p.c.: product complex.
Model validation
The dissociative model was designed according to the common literature view on RNAi in mammalian systems and parameterized according to in vitro determined rate constants (48) and cellular parameters for reactant concentrations (siRNA, hAgo2 and target mRNA) and cytoplasm volume (Supplementary Table S2). Due to a discrepancy between the dissociative model and time course data from cell culture experiments, a second model was developed. The associative model is based on the fact that recent cell culture experiments revealed that gene silencing rates can be increased by raising the target mRNA concentrations, while the concentrations of siRNA and Ago2 stayed constant. Likewise, this model was also parameterized according to the above given rate constants and cellular parameters for reactant concentrations and reaction compartment volume.
To test whether either of the two models agrees with RNAi activity in living mammalian cells, several cell culture experiments were performed and the validation datasets were compared with the simulation output of both models. Essentially, two different sets of validation experiments were performed: (i) time courses and t1/2 of the siRNA-mediated knockdown of target gene activity for different initial siRNA concentrations, and (ii) concentration response curves and IC50 determinations of the siRNA-mediated knockdown of target gene activity at different basal target mRNA concentrations.
RESULTS
Dissociative model of siRNA-mediated RNAi in mammalian systems
A computational model of siRNA-mediated RNAi in mammalian cells was developed in order to link experimental results with a quantitative and time-resolved understanding of RNAi. Mechanistically, it is based on the current literature view on RNAi. The model is parameterized with precise quantitative kinetic and molecular data. For each reaction step, described as elementary reaction equilibrium, either a pair of forward and backward rate constants, or in case of quasi-irreversible steps, a forward rate constant is provided (Supplementary Table S1). The parameters of the reaction compartment used for computational modelling (e.g. volume of the cytoplasm of a mammalian cell and concentrations of reactants) are provided in Supplementary Table S2. In order to take into account transcriptional bursts, which are frequently observed for mRNA in mammalian cells (58), occasional high target RNA concentrations could be assumed. Nevertheless, to not needlessly complicate the system, a setup with assumed constant mRNA concentration was chosen, i.e. constant mRNA synthesis and elimination rates (Supplementary Table S2). The name, dissociative model, is based on the biochemical ligand substitution mechanism it resembles. In the dissociative model, the departing ligand (here, cleaved RNA fragments) leaves the complex in a slow step with positive entropy of activation, before the incoming ligand (target mRNA) can enter (Figure 1A).
In a first pass, hAgo2 and possibly other cofactors, bind free ss guide siRNA (below referred to as guide) in a three-step reaction to form a binary Ago2/guide substrate complex (siRNA binding I-III). The entire reaction pathway is illustrated in Supplementary Figure S2. Corresponding rate equations and rate constants for each individual step are listed under Supplementary Information (Supplementary Equations 1) and given in Supplementary Table S1, respectively. Potential cofactors, passenger strand cleavage and release during binary complex formation are left out for clarity but essentially, the proposed pathway also holds true for ds siRNA. Next, a target RNA enters the binary Ago2/guide complex and, if complementary to the bound guide RNA, associates in a three-phase reaction, to form a catalytically active ternary Ago2/guide/target RNA substrate complex (target binding I-III). Once a catalytically active complex is formed, the system can undergo multiple rounds of target binding and cleavage, each time followed by a slow product release, without the binary Ago2/guide complex falling apart. The bound target mRNA is cleaved by Ago2 (target cleavage reaction) leading to a ternary Ago2/guide/product complex. Subsequently, the cleavage product is released from the complex in a three-step reaction (product dissociation I-III). Accordingly, the target mRNA population is reduced by one and released product strands are prone to degradation by cellular nucleases, leaving behind a binary complex comprising guide bound to Ago2, which can bind another target mRNA and thus closing the cycle of subsequent passes of target turnover.
RNAi in mammalian cells suggests an alternative model structure
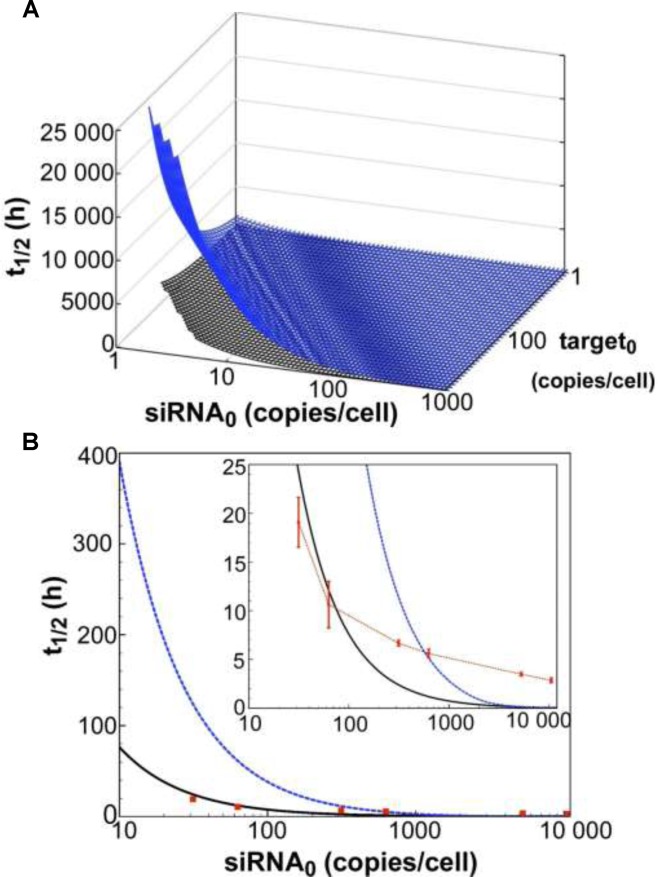
In a first attempt to test the model's relevance in mammalian cells, time-course simulations of mRNA target knockdown were compared to cell culture experiments (Figure 2). As a cellular model system, we used the HTOL cell line, stably transfected with a firefly luciferase reporter construct as previously described (49). For reporter knockdown, different amounts of an antiluciferase siRNA, namely siR206 (50), were transfected with Lipofectamine™ 2000 (LF2000). Luciferase mRNA levels were determined by qPCR as described under Materials and Methods. For quantification of the amount of siRNA internalized, a so-called liquid hybridisation assay enabling us to detect the guide strand of the siRNA with a sensitivity of >10 molecules per cell was applied as described previously (49). In Figure 2A, t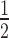 spans the surface of the parameter space of two initial concentrations, siRNA [S]0 and target [T]0. In general, t
spans the surface of the parameter space of two initial concentrations, siRNA [S]0 and target [T]0. In general, t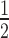 is proportional to [T]0 and antiproportional to [S]0. It is also evident that t
is proportional to [T]0 and antiproportional to [S]0. It is also evident that t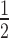 is very sensitive to changes in [S]0 while also being sensitive to changes in [T]0. Figure 2B shows a cross-section of the 3D-plot in 2A, at a basal target concentration of 35 copies per cell, which corresponds to the average basal target mRNA concentration in the control experiment (i.e. copy number of luciferase mRNA in stably transfected HTOL cells). A first discrepancy between the dissociative model and experimentally determined values (red squares) was observed. It was not possible to eliminate the discrepancy between the model and experiment by merely adjusting the model parameters within physiological meaningful boundaries. For example, a 20-fold increase of the measured in vitro constants of the two most sensitive parameters (i.e. product release III k+10 and target binding III k+6; compare Supplementary Information on sensitivity analysis, Supplementary Figure S6 and Supplementary Table S1), which may be justified by facilitating cofactors in vivo, was not sufficient to solve the discrepancy.
is very sensitive to changes in [S]0 while also being sensitive to changes in [T]0. Figure 2B shows a cross-section of the 3D-plot in 2A, at a basal target concentration of 35 copies per cell, which corresponds to the average basal target mRNA concentration in the control experiment (i.e. copy number of luciferase mRNA in stably transfected HTOL cells). A first discrepancy between the dissociative model and experimentally determined values (red squares) was observed. It was not possible to eliminate the discrepancy between the model and experiment by merely adjusting the model parameters within physiological meaningful boundaries. For example, a 20-fold increase of the measured in vitro constants of the two most sensitive parameters (i.e. product release III k+10 and target binding III k+6; compare Supplementary Information on sensitivity analysis, Supplementary Figure S6 and Supplementary Table S1), which may be justified by facilitating cofactors in vivo, was not sufficient to solve the discrepancy.
Figure 2.
Dependency of observed rate of target knockdown on siRNA and target mRNA concentration. (A) t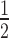 , the time of half-maximal target knockdown, is plotted against logarithmically-scaled initial siRNA concentration and logarithmically-scaled basal target concentration. t
, the time of half-maximal target knockdown, is plotted against logarithmically-scaled initial siRNA concentration and logarithmically-scaled basal target concentration. t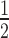 of dissociative and associative model span the blue and the black surface over the parameter space, respectively. (B) It shows a cross-section of the 3D-plot at a basal target concentration of 35 copies/cell (experimentally determined luciferase mRNA copy number in stably transfected HTOL cells). For decreasing siRNA concentrations, the difference in t
of dissociative and associative model span the blue and the black surface over the parameter space, respectively. (B) It shows a cross-section of the 3D-plot at a basal target concentration of 35 copies/cell (experimentally determined luciferase mRNA copy number in stably transfected HTOL cells). For decreasing siRNA concentrations, the difference in t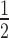 between the dissociative (blue line) and the associative (black line) model increases progressively. Experimentally determined t
between the dissociative (blue line) and the associative (black line) model increases progressively. Experimentally determined t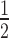 values of knockdown of luciferase reporter gene activity after LF2000-mediated transfection of different amounts of siR206 into HTOL cells are depicted by red squares. Transfection of 0.05, 0.1, 0.5, 1, 50 and 100 nM siRNA in the transfection mix translates into 31, 63, 314, 628, 5261 and 10 523 copies of bioavailable siR206 per cell (Supplementary Figure S1). For details see Materials and Methods and references (49,53). The corresponding t1/2 values are: 19 (± 2.5), 10.6 (± 2.4), 6.7 (± 0.3), 5.6 (± 0.4), 3.5 (± 0.2) and 2.9 (± 0.25) h (for experimental details see Supplementary Figure S7). The data are an average of at least two independent experiments. The inset shows the graph on a shorter y-axis scale including corresponding error bars.
values of knockdown of luciferase reporter gene activity after LF2000-mediated transfection of different amounts of siR206 into HTOL cells are depicted by red squares. Transfection of 0.05, 0.1, 0.5, 1, 50 and 100 nM siRNA in the transfection mix translates into 31, 63, 314, 628, 5261 and 10 523 copies of bioavailable siR206 per cell (Supplementary Figure S1). For details see Materials and Methods and references (49,53). The corresponding t1/2 values are: 19 (± 2.5), 10.6 (± 2.4), 6.7 (± 0.3), 5.6 (± 0.4), 3.5 (± 0.2) and 2.9 (± 0.25) h (for experimental details see Supplementary Figure S7). The data are an average of at least two independent experiments. The inset shows the graph on a shorter y-axis scale including corresponding error bars.
Associative model: accelerated product release possibly via strand exchange between Ago2-bound cleaved target RNA and next incoming target mRNA
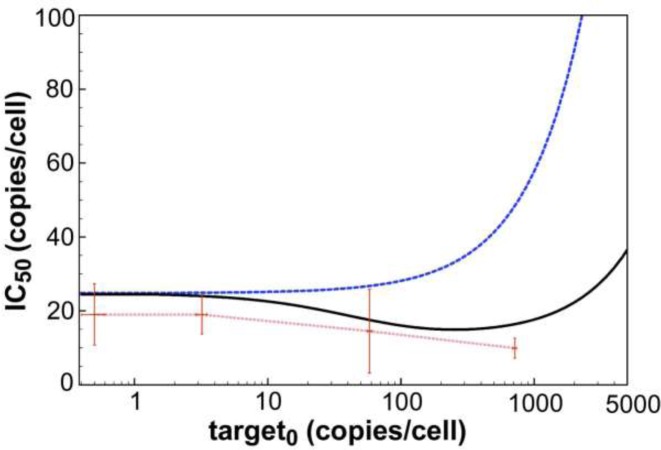
Cell culture experiments revealed that despite rising target mRNA concentrations over several orders of magnitude, gene knock-down efficiency, given as half-maximal inhibitory siRNA concentration (IC50) 24 h post transfection, stayed constant (compare Figure 4). Thus, in order to compensate for the increase in target mRNA cleavage, the RNAi machinery must become more effective. Along these lines, transient in vitro binding experiments showed that the release of a target or product RNA strand from a ternary hAgo2/guide/target or product complex is accelerated in the presence of an excess of the next incoming target RNA (48) (Supplementary Figure S3). Taken together, these two experimental findings imply that RNAi-mediated gene knockdown is capable to adapt to large changes in target mRNA level without affecting efficacy by target-induced acceleration of the overall rate-limiting step, namely release of cleavage products. The lack of the above described dissociative model to account for such a scenario motivates the development of an alternative so-called associative model.
Figure 4.
Dependency of RNAi efficacy on target mRNA concentration. For both models (dissociative: blue line; associative: black line), simulated IC50 values (specified as copies/cell; conversion given under Materials and Methods) are plotted against logarithmically-scaled basal target mRNA concentration. Experimentally determined IC50 values of siR206 are shown by red dots. Here, siR206 (0.0005, 0.005, 0.05, 0.5, 5 and 50 nM) and pTRE2hyg-luc plasmid (0.1, 1, 10 and 100 ng/well) were cotransfected into HTO cells. In the absence of siRNA 0.5 (±0.1), 3.2 (±0.3), 58 (±2), and 715 (±23) luciferase mRNA copies/cell were determined by qPCR for the different amounts of plasmid transfected. For each concentration of pTRE2hyg-luc plasmid, IC50 values were determined by plotting qPCR-derived mRNA copy numbers versus concentrations of transfected siR206 yielding the following values: 0.1 ng plasmid/well = IC50 19 (±8), 1 ng plasmid/well = IC50 19 (±5), 10 ng plasmid/well = IC50 15 (±11) and 100 ng plasmid/well = IC50 10 (±2) copies/cell. Standard deviations are given by horizontal and vertical error bars.
Different from the dissociative mechanism, the associative model involves the association of an incoming ligand with the enzyme/substrate complex before departure of the leaving ligand. In this model, hAgo2 supposedly facilitates the interaction of an incoming target strand with a ternary hAgo2/guide/product complex, most likely by a kind of strand invasion mechanism as proposed for in vitro RNA/RNA strand exchange (59,60). As shown in Supplementary Figures S9 and S10, such interactions only take place in case of fully complementary targets. Figure 1B pictures the target-product-exchange mechanism, which is fundamental to the multiple turnover cycle of the associative model.
The first passes of siRNA binding, followed by target binding, as well as, target cleavage are equivalent to the reaction steps of the dissociative mode: ‘siRNA binding I-III’, ‘target binding I-III’ and ‘target cleavage’, respectively. However, after target mRNA cleavage and formation of the hAgo2/guide/product complex, the two models differ. In contrast to the dissociative model, target cleavage is followed by an associative binding of another target (associative target binding I-III). A newly incoming target interacts with the ternary hAgo2/guide/product complex, forming a transient quaternary complex eventually replacing the product. The cycle is closed by the complete release of the product in three reaction steps (product release I-III), leading to the formation of the next catalytically active ternary complex. Like in the dissociative model, released product fragments are subsequently degraded by cellular nucleases. The remaining ternary complex can pass through the next cycle of target cleavage, association of a new target and product release. The full reaction scheme for the associative model is shown in Supplementary Figure S2B. Analogous to the dissociative model, the rate constants for complex formation and dissociation are based on presteady state binding data (Supplementary Table S1) by Deerberg et al. (48).
The associative model closely resembles siRNA-mediated target knockdown in cell culture
As described above, time-course simulations were repeated with the associative model (Figure 2A) and t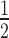 of target knockdown was likewise compared to experimental data. For a combination of high siRNA0 and low target0 concentrations, surfaces of both models superimpose well. However, they increasingly diverge with higher target0 concentration and to a lesser extend with lower siRNA0 concentration. Experimentally determined t
of target knockdown was likewise compared to experimental data. For a combination of high siRNA0 and low target0 concentrations, surfaces of both models superimpose well. However, they increasingly diverge with higher target0 concentration and to a lesser extend with lower siRNA0 concentration. Experimentally determined t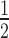 values (red squares in Figure 2B), more closely match the behaviour of the associative model.
values (red squares in Figure 2B), more closely match the behaviour of the associative model.
Time-resolved target knockdown with respect to initial siRNA concentration
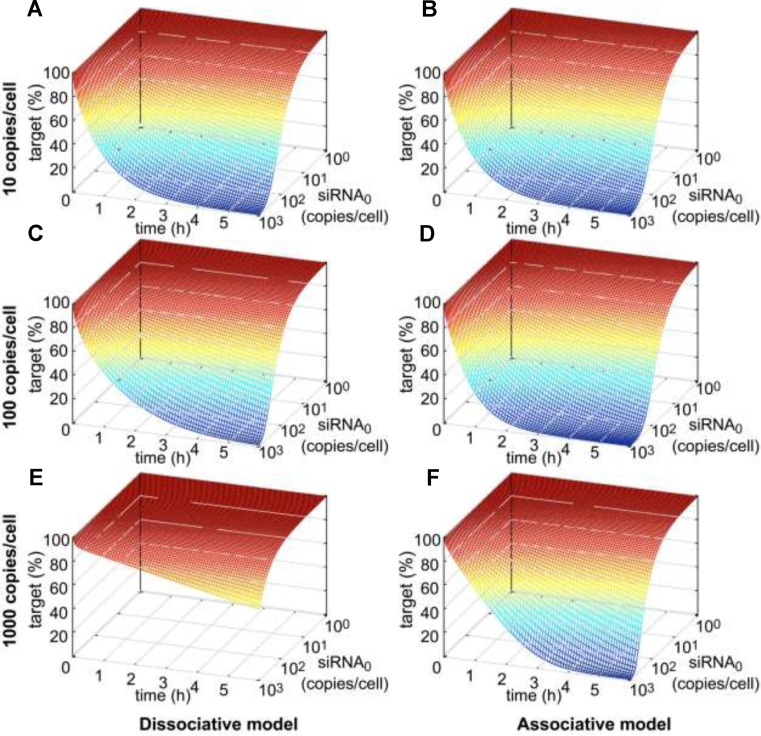
After administration of siRNA, target mRNA concentration and as a consequence gene activity decrease within a defined time span until reaching a minimum. In cell culture, recovery of mRNA concentration back to basal levels takes 2–5 days and is likely to be controlled by dilution of siRNA or loaded RISC due to cell division (36). In this work, the time-resolved knockdown of target mRNA concentration as a function of administered siRNA dose as well as of basal mRNA level was analysed on a short-term time scale of 6 h (Figure 3). hAgo2 concentration was set to 200 000 copies/cell (61) and did not limit model performance down to 10 000 or 50 000 copies/cell for neither the associative or dissociative model (Supplementary Figure S4). Thus, initial hAgo2 concentration was not of concern during the simulations performed, where a number of time course simulations with different combinations of administered siRNA dose and basal mRNA level were calculated for both models. Figure 3 shows the time-resolved onset of target knockdown within the first 6 h after siRNA administration as a function of initial siRNA concentration, where relative target concentration is given as function of reaction time and initial siRNA concentration. At lower target levels (i.e. 10 copies/cell), the RNAi machinery according to both models, respond likewise to changes in siRNA concentration of several orders of magnitudes (Figure 3A/B). However, in case of the dissociative model, the RNAi machinery requires increased siRNA concentrations to maintain the rate of target knockdown at higher target levels, i.e. 100 copies/cell (Figure 3C/D). In contrast, for the associative model, the relative time courses of target knockdown are far more similar for different levels of basal target concentrations. This difference, in time-dependent model behaviour between dissociative and associative model, becomes more recognizable at even higher target levels of ≥1000 copies/cell (Figure 3E/F). Supplementary Figure S8 shows a direct comparison between experimentally determined and simulated data at concentrations where the difference among the two models is most significant; i.e. high target and moderate siRNA concentrations.
Figure 3.
Time resolved siRNA-mediated target knockdown with respect to initial siRNA concentration at three different levels of basal target mRNA. In all six plots, target concentration (in percent of basal target level) is plotted against reaction time (in h) and logarithmically-scaled initial siRNA concentration (in copies/cell). Simulation results of the dissociative model in the first column are presented in parallel with the ones of the associative model in the second column. Row-wise, absolute basal target levels increase by 10-fold from 10 to 100 to 1000 copies/cell.
Messenger RNA synthesis may be triggered by transcriptional bursts (58) and mRNA levels in mammalian cells can vary by magnitudes between 1–10 000 copies/cell (62). Thus, RNAi-mediated knockdown should be able to adapt to such massive changes in target mRNA levels, as reflected by the proposed associative, but not the dissociative model.
Adaptation of the RNAi machinery to variations in target mRNA level indicated by IC50 values
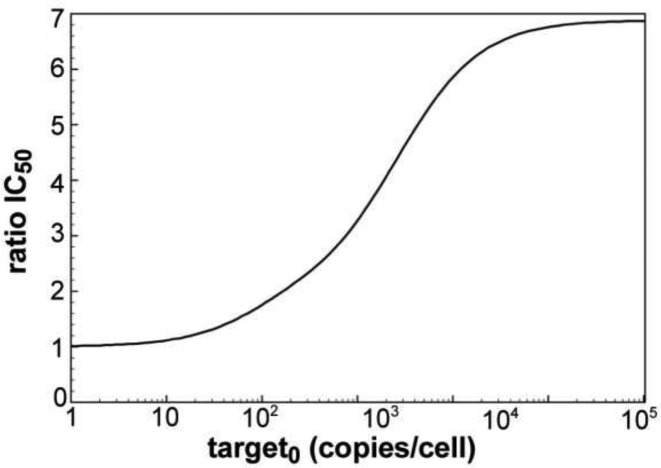
To further investigate the adaptiveness of the RNAi machinery to changes in target mRNA levels, calculated IC50 values (given as siRNA copies/cell) were plotted against basal target mRNA levels (Figure 4). The simulated values were then compared to experimentally determined numbers. The experimentally derived IC50 values are essentially constant over four magnitudes of LF2000 transfected target pTREhyg-luc plasmid into HTO cells (0.1–100 ng/well), which translates into 0.5–715 luciferase mRNA copies/cell, as determined by qPCR. This indicates a robustness of the RNAi machinery against a certain extend of variation in target mRNA levels. For the dissociative model, the IC50 stays relatively constant for small target concentrations of <50 copies/cell, and then increases rapidly. In the associative model, the IC50 slowly decreases with increasing target concentration until it reaches its optimum between 200 and 300 target copies/cell from where the IC50 starts increasing with the target copy number. All in all, the associative model describes the observed adaption to variations in target mRNA level much more convincing than the dissociative model. Figure 5 shows the difference between the two models by plotting IC50 ratios (quotient IC50_diss/IC50_ass) versus target copies per cell. Here, the effect is most obvious for the physiologically significant range of 50–10 000 mRNA copies/cell.
Figure 5.
Quotient IC50_diss/IC50_ass as function of initial target mRNA. The simulated IC50 values for the dissociative model divided by IC50 values for the associative model are plotted against logarithmically-scaled basal target mRNA concentration.
DISCUSSION
Precise kinetic data made it possible to model siRNA-mediated RNAi with never observed precision. Our experimental observations suggest high multiple turnover rates of RNAi-based gene silencing in cell tissue culture and stability of gene silencing efficacy over several magnitudes of target mRNA concentration. That is, gene silencing rates increase when target mRNA concentration rises, while siRNA concentration stays constant.
First modelling approaches (later dubbed dissociative model) were not consistent with experimental data derived from tissue culture. Sensitivity analyses (Supplementary Figure S6) revealed product release step III (k+10; see Supplementary Figure S2A) as the limiting step for mRNA target knockdown in this model explaining the discrepancy to the experimental findings. In contrast, the cell culture data can be sufficiently described by an associative model using measured model parameters. Here, product release (k+16; see Supplementary Figure S2B) is not limiting (Supplementary Figure S6). The associative model accounts for target concentration-dependent acceleration of product release from Ago2 and is compatible with a high multiple turnover rate of RNAi-based gene silencing in tissue culture. Thus, we propose an associative mechanism of mRNA target binding and recognition by the postcleavage guide/product duplex, facilitated by hAgo2 during the catalytic cycle. Such a scenario is supported by data from Wünsche et al. (60), which quantified increased RNA/RNA association kinetics during strand exchange between siRNA duplexes and corresponding target RNA strands in vitro.
Comparison between simulated and experimentally determined IC50 values suggests that the associative mechanism interlinks target mRNA loading with acceleration of unloading of cleaved product fragments from Ago2 under multiple turnover conditions in a target concentration-dependent fashion. Our findings are supported by observations of other groups. Already in 2003, Harborth et al. (63) detected differences in siRNA-dependent silencing efficiencies targeting sequence segments identical among human and mouse lamin A/C. They assumed that among others the different levels of target mRNA in human and mouse cells might account for different silencing efficiencies. Another study showed that transcripts with low abundancy are less susceptible to siRNA-dependent Ago-mediated cleavage suggesting the necessity of a certain target transcript threshold for efficient gene silencing (64). In contrast to these conceptions, Krueger et al. (65) excluded a correlation between transcript levels and gene silencing efficiencies in their studies using a random set of validated siRNAs. This discrepancy could possibly be explained by findings demonstrating that the effect of target transcript abundance is relativized if the turnover rates of the target are very high or the siRNA used is very efficient (40). But since it could be shown that transcripts with the highest abundance are usually correlated with low turnover numbers (40), the importance of target transcript levels for efficient siRNA-mediated RNAi is underscored. Interestingly, the correlation between target RNA abundance and gene silencing efficiency seems to be restricted to siRNA-dependent RNAi. Analyses of a possible connection between transcript abundance and miRNA-dependent gene silencing revealed similar gene silencing efficiencies for lowly and highly expressed target transcripts (66). These results might be correlated with the fact that in case of miRNAs, which are not fully paired to their targets, dissociation rate constants of ternary Ago/guide/target complexes are faster than in case of their nearly fully paired siRNA counterparts (67), and therefore, may not limit Ago-mediated target turnover.
Questions like ‘what determines the cellular response to a siRNA or miRNA stimulus?’’ and ‘how to quantitatively predict it?’ are sought after for improved and secure application of RNAi in medicine and life science. It is commonly accepted that factors like local RNA sequence context (68) or structure (69–72) are important for RNAi efficiency. However, inconsistencies in state-of-the-art siRNA design and prediction of RNAi efficacy hint that limitations may not be solely in the design of the siRNA molecule itself. In fact, it was shown that system-level properties such as RNA elimination rates have important, but still inconclusive effects on RNAi efficacy (40). Likewise, our study strongly suggests considering the target as a whole when choosing appropriate targets or when predicting gene silencing efficacy. This implies the need of a new generation of bioinformatics tools for siRNA (and miRNA) target prediction, which consider system-level properties such as target transcript abundance and mRNA decay rates. For instance, an mRNA in one cell type may not be targetable to the same extent in another cell type, depending on the abundance of the mRNA.
Transcriptional bursts and the resulting variability in mRNA abundance (i.e. transcriptional noise) among isogenic cell populations impact upon the effectiveness of clinical treatment and the resistance of bacteria to antibiotics (73,74). Variability in gene expression may also contribute to resistance of subpopulations of cancer cells to chemotherapy (75). Robustness of RNAi drugs to transcriptional bursts as shown in our IC50 experiments and the here proposed associative model, may be an important property of RNAi-based drugs to overcome microbial and cancer cell resistance to conventional drugs.
Supplementary Material
Acknowledgments
We thank W. Wünsche for experimental help. S.D. cordially thanks Prof. Pedro Mendes and Dr. Juergen Pahle for support with COPASI and for many fruitful discussions.
Footnotes
Present addresses:
Sarah Willkomm, Department of Microbiology, Faculty of Biology and Preclinical Medicine, University of Regensburg, 93053 Regensburg, Germany.
Sandra D. Laufer, Hamburg Centre for Experimental Therapy Research (HEXT), Universitätsklinikum Hamburg-Eppendorf, Martinistr. 52, 20246 Hamburg, Germany.
Jens Christian Claussen, Systems Biology Lab, Jacobs University Bremen, Campus Ring 1, 28759 Bremen, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Graduate School for Computing in Medicine and Life Sciences, University of Lübeck funded by Germany's Excellence Initiative [DFG GSC 235/1 to S.D. and S.W.]. Funding for open access charge: institutes' budget.
Conflict of interest statement. None declared.
REFERENCES
- 1.Stefani G., Slack F.J. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 2.Esquela-Kerscher A., Slack F.J. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 3.Tili E., Michaille J.J., Calin G.A. Expression and function of micro-RNAs in immune cells during normal or disease state. Int. J. Med. Sci. 2008;5:73–79. doi: 10.7150/ijms.5.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu M., Zhang Q., Deng M., Miao J., Guo Y., Gao W., Cui Q. An analysis of human microRNA and disease associations. PloS one. 2008;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Napoli C., Lemieux C., Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Krol A.R. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell Online. 1990;2:291–299. doi: 10.1105/tpc.2.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romano N., Macino G. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol. 1992;6:3343–3353. doi: 10.1111/j.1365-2958.1992.tb02202.x. [DOI] [PubMed] [Google Scholar]
- 8.Lindbo J.A., Silva-Rosales L., Proebsting W.M., Dougherty W.G. Induction of a highly specific antiviral state in transgenic plants: implications for regulation of gene expression and virus resistance. Plant Cell. 1993;5:1749–1759. doi: 10.1105/tpc.5.12.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 10.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 11.Wilson R.C., Doudna J.A. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiemann K., Rossi J.J. RNAi-based therapeutics-current status, challenges and prospects. EMBO Mol. Med. 2009;1:142–151. doi: 10.1002/emmm.200900023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond S.M., Bernstein E., Beach D., Hannon G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 14.Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21-and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jinek M., Doudna J.A. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 16.Macfarlane L.A., Murphy P.R. MicroRNA: biogenesis, function and role in cancer. Curr. Genomics. 2010;11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghildiyal M., Zamore P.D. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nykanen A., Haley B., Zamore P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 19.Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 21.Tolia N.H., Joshua-Tor L. Slicer and the argonautes. Nat. Chem. Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 22.Haley B., Zamore P.D. Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 23.Rivas F.V., Tolia N.H., Song J.J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 24.Hutvagner G., Zamore P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 25.Sen G.L., Blau H.M. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 26.Jakymiw A., Lian S., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell. Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- 27.MacRae I.J., Ma E., Zhou M., Robinson C.V., Doudna J.A. In vitro reconstitution of the human RISC-loading complex. Proc. Natl. Acad. Sci. U. S. A. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eulalio A., Huntzinger E., Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 29.Grosshans H., Chatterjee S. MicroRNAses and the regulated degradation of mature animal miRNAs. Adv. Exp. Med. Biol. 2010;700:140–155. [PubMed] [Google Scholar]
- 30.Fabian M.R., Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 31.Bergstrom C.T., McKittrick E., Antia R. Mathematical models of RNA silencing: unidirectional amplification limits accidental self-directed reactions. Proc. Natl. Acad. Sci. U. S. A. 2003;100:11511–11516. doi: 10.1073/pnas.1931639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groenenboom M.A., Maree A.F., Hogeweg P. The RNA silencing pathway: the bits and pieces that matter. PLoS Comput. Biol. 2005;1:155–165. doi: 10.1371/journal.pcbi.0010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raab R.M., Stephanopoulos G. Dynamics of gene silencing by RNA interference. Biotechnol. Bioeng. 2004;88:121–132. doi: 10.1002/bit.20216. [DOI] [PubMed] [Google Scholar]
- 34.Vohradsky J., Panek J., Vomastek T. Numerical modelling of microRNA-mediated mRNA decay identifies novel mechanism of microRNA controlled mRNA downregulation. Nucleic Acids Res. 2010;38:4579–4585. doi: 10.1093/nar/gkq220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gokhale S.A., Gadgil C.J. Analysis of miRNA regulation suggests an explanation for ‘unexpected’ increase in target protein levels. Mol. Biosyst. 2012;8:760–765. doi: 10.1039/c1mb05368j. [DOI] [PubMed] [Google Scholar]
- 36.Bartlett D.W., Davis M.E. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;34:322–333. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartlett D.W., Davis M.E. Effect of siRNA nuclease stability on the in vitro and in vivo kinetics of siRNA-mediated gene silencing. Biotechnol. Bioeng. 2007;97:909–921. doi: 10.1002/bit.21285. [DOI] [PubMed] [Google Scholar]
- 38.Bartlett D.W., Su H., Hildebrandt I.J., Weber W.A., Davis M.E. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15549–15554. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cuccato G., Polynikis A., Siciliano V., Graziano M., di Bernardo M., di Bernardo D. Modeling RNA interference in mammalian cells. BMC Syst. Biol. 2011;5:19. doi: 10.1186/1752-0509-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larsson E., Sander C., Marks D. mRNA turnover rate limits siRNA and microRNA efficacy. Mol. Syst. Biol. 2010;6:433. doi: 10.1038/msb.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y., Sheng G., Juranek S., Tuschl T., Patel D.J. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y., Juranek S., Li H., Sheng G., Tuschl T., Patel D.J. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y., Juranek S., Li H., Sheng G., Wardle G.S., Tuschl T., Patel D.J. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schirle N.T., MacRae I.J. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elkayam E., Kuhn C.D., Tocilj A., Haase A.D., Greene E.M., Hannon G.J., Joshua-Tor L. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012;150:100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakanishi K., Weinberg D.E., Bartel D.P., Patel D.J. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368–374. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schirle N.T., Sheu-Gruttadauria J., MacRae I.J. Structural basis for microRNA targeting. Science. 2014;346:608–613. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deerberg A., Willkomm S., Restle T. Minimal mechanistic model of siRNA-dependent target RNA slicing by recombinant human Argonaute 2 protein. Proc. Natl. Acad. Sci. U. S. A. 2013;110:17850–17855. doi: 10.1073/pnas.1217838110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Veldhoen S., Laufer S.D., Trampe A., Restle T. Cellular delivery of small interfering RNA by a non-covalently attached cell-penetrating peptide: quantitative analysis of uptake and biological effect. Nucleic Acids Res. 2006;34:6561–6573. doi: 10.1093/nar/gkl941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W.S., Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 51.Laufer S.D., Restle T. Peptide-mediated cellular delivery of oligonucleotide-based therapeutics in vitro: quantitative evaluation of overall efficacy employing easy to handle reporter systems. Curr. Pharm. Des. 2008;14:3637–3655. doi: 10.2174/138161208786898806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laufer S.D., Recke A.L., Veldhoen S., Trampe A., Restle T. Noncovalent peptide-mediated delivery of chemically modified steric block oligonucleotides promotes splice correction: quantitative analysis of uptake and biological effect. Oligonucleotides. 2009;19:63–80. doi: 10.1089/oli.2008.0160. [DOI] [PubMed] [Google Scholar]
- 53.Laufer S.D., Detzer A., Sczakiel G., Restle T. Selected strategies for the delivery of silencing RNA in vitro and in vivo. In: Erdmann VA, Barciszewski J, editors. RNA Technologies and Their Applications. Springer; 2010. pp. 29–58. [Google Scholar]
- 54.Kihara T., Yoshida N., Kitagawa T., Nakamura C., Nakamura N., Miyake J. Development of a novel method to detect intrinsic mRNA in a living cell by using a molecular beacon-immobilized nanoneedle. Biosens. Bioelectron. 2010;26:1449–1454. doi: 10.1016/j.bios.2010.07.079. [DOI] [PubMed] [Google Scholar]
- 55.White A.K., VanInsberghe M., Petriv O.I., Hamidi M., Sikorski D., Marra M.A., Piret J., Aparicio S., Hansen C.L. High-throughput microfluidic single-cell RT-qPCR. Proc. Natl. Acad. Sci. U. S. A. 2011;108:13999–14004. doi: 10.1073/pnas.1019446108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fox B.C., Devonshire A.S., Baradez M.O., Marshall D., Foy C.A. Comparison of reverse transcription-quantitative polymerase chain reaction methods and platforms for single cell gene expression analysis. Anal. Biochem. 2012;427:178–186. doi: 10.1016/j.ab.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 57.Hoops S., Sahle S., Gauges R., Lee C., Pahle J., Simus N., Singhal M., Xu L., Mendes P., Kummer U. COPASI–a COmplex PAthway SImulator. Bioinformatics. 2006;22:3067–3074. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- 58.Raj A., Peskin C.S., Tranchina D., Vargas D.Y., Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Homann M., Nedbal W., Sczakiel G. Dissociation of long-chain duplex RNA can occur via strand displacement in vitro: biological implications. Nucleic Acids Res. 1996;24:4395–4400. doi: 10.1093/nar/24.22.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wünsche W., Sczakiel G. The activity of siRNA in mammalian cells is related to the kinetics of siRNA-target recognition in vitro: mechanistic implications. J. Mol. Biol. 2005;345:203–209. doi: 10.1016/j.jmb.2004.10.041. [DOI] [PubMed] [Google Scholar]
- 61.Wang D., Zhang Z., O'Loughlin E., Lee T., Houel S., O'Carroll D., Tarakhovsky A., Ahn N.G., Yi R. Quantitative functions of Argonaute proteins in mammalian development. Genes Dev. 2012;26:693–704. doi: 10.1101/gad.182758.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Velculescu V.E., Madden S.L., Zhang L., Lash A.E., Yu J., Rago C., Lal A., Wang C.J., Beaudry G.A., Ciriello K.M., et al. Analysis of human transcriptomes. Nat. Genet. 1999;23:387–388. doi: 10.1038/70487. [DOI] [PubMed] [Google Scholar]
- 63.Harborth J., Elbashir S.M., Vandenburgh K., Manninga H., Scaringe S.A., Weber K., Tuschl T. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 2003;13:83–105. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- 64.Hu X., Hipolito S., Lynn R., Abraham V., Ramos S., Wong-Staal F. Relative gene-silencing efficiencies of small interfering RNAs targeting sense and antisense transcripts from the same genetic locus. Nucleic Acids Res. 2004;32:4609–4617. doi: 10.1093/nar/gkh790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krueger U., Bergauer T., Kaufmann B., Wolter I., Pilk S., Heider-Fabian M., Kirch S., Artz-Oppitz C., Isselhorst M., Konrad J. Insights into effective RNAi gained from large-scale siRNA validation screening. Oligonucleotides. 2007;17:237–250. doi: 10.1089/oli.2006.0065. [DOI] [PubMed] [Google Scholar]
- 66.Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wee L.M., Flores-Jasso C.F., Salomon W.E., Zamore P.D. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012;151:1055–1067. doi: 10.1016/j.cell.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kretschmer-Kazemi Far R., Sczakiel G. The activity of siRNA in mammalian cells is related to structural target accessibility: a comparison with antisense oligonucleotides. Nucleic Acids Res. 2003;31:4417–4424. doi: 10.1093/nar/gkg649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Overhoff M., Alken M., Far R.K., Lemaitre M., Lebleu B., Sczakiel G., Robbins I. Local RNA target structure influences siRNA efficacy: a systematic global analysis. J. Mol. Biol. 2005;348:871–881. doi: 10.1016/j.jmb.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 71.Brown K.M., Chu C.Y., Rana T.M. Target accessibility dictates the potency of human RISC. Nat. Struct. Mol. Biol. 2005;12:469–470. doi: 10.1038/nsmb931. [DOI] [PubMed] [Google Scholar]
- 72.Dornseifer S., Sczakiel G. Computational identification of biologically functional non-hairpin GC-helices in human Argonaute mRNA. BMC Bioinformatics. 2013;14:122. doi: 10.1186/1471-2105-14-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moyed H.S., Bertrand K.P. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 1983;155:768–775. doi: 10.1128/jb.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lewis K. Persister cells. Annu. Rev. Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 75.Sharma S.V., Lee D.Y., Li B., Quinlan M.P., Takahashi F., Maheswaran S., McDermott U., Azizian N., Zou L., Fischbach M.A., et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.