Abstract
Here we report investigations, based on circular dichroism, nuclear magnetic resonance spectroscopy, molecular modelling, differential scanning calorimetry and prothrombin time assay, on analogues of the thrombin binding aptamer (TBA) in which individual thymidines were replaced by 5-fluoro-2′-deoxyuridine residues. The whole of the data clearly indicate that all derivatives are able to fold in a G-quadruplex structure very similar to the ‘chair-like’ conformation typical of the TBA. However, only ODNs TBA-F4 and TBA-F13 have shown a remarkable improvement both in the melting temperature (ΔTm ≈ +10) and in the anticoagulant activity in comparison with the original TBA. These findings are unusual, particularly considering previously reported studies in which modifications of T4 and T13 residues in TBA sequence have clearly proven to be always detrimental for the structural stability and biological activity of the aptamer. Our results strongly suggest the possibility to enhance TBA properties through tiny straightforward modifications.
INTRODUCTION
DNA and RNA aptamers are nucleic acid ligands characterized by an outstanding ability to bind with high affinity and specificity to various molecular targets such as small molecules, proteins, nucleic acids, and even cells, tissues and organisms. Aptamers can be discovered and engineered through repeated rounds of an in vitro selection strategy called SELEX (Systematic Evolution of Ligands by Exponential enrichment) (1,2). Among the DNA aptamers the thrombin binding aptamer (TBA, 5′-GGTTGGTGTGGTTGG-3′) has been one of the first to be discovered (3,4). However, despite this fact, TBA is still the subject of several researches, both in therapeutics, being endowed with anticoagulant properties, and in analytics, thank to its ability of binding potassium ions and, more importantly, also heavy metal ions of toxicological interest.
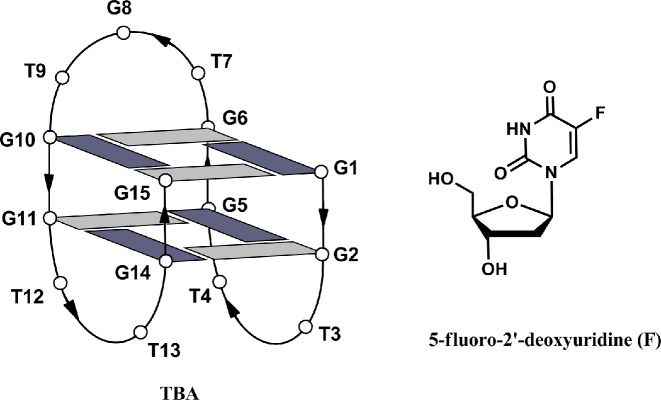
According to nuclear magnetic resonance (NMR) and X-ray structural investigations TBA adopts a monomolecular, antiparallel G-quadruplex structure with a ‘chair-like’ conformation (5,6). The central part of the G-quadruplex consists of two syn-anti-syn-anti stacked G-quartets, which are connected to each other by three edge-wise loops: a central TGT loop and two lateral TT loops (Figure 1). Quite soon after its discovery, TBA has been subjected to several chemical modifications, most of which with the targets to: (i) improve its thermal stability in physiological conditions, which is connected to the anticoagulant activity; (ii) render it resistant to the exonucleases, ubiquitous in biological environments and (iii) enhance its affinity to thrombin.
Figure 1.
Schematic representation of the TBA G-quadruplex and chemical structure of 5-fluoro-2′-deoxyuridine (F). Guanosines in syn and anti glycosidic conformations are in dark and light grey, respectively.
As far as the TBA/thrombin interaction is concerned, a compelling number of investigations have highlighted that the loops are the parts of the aptamer mostly involved in the interaction with the protein (6–8). In particular, these studies indicated that the minor loops TT interact with the thrombin anion exosite I, while the larger TGT loop is in close proximity to the heparin binding site of a further thrombin molecule. Therefore, TBA seems to interact with two thrombin molecules, inactivating only one of them. The availability of rather detailed information on which residues were involved in the interaction of TBA with its target allowed researchers to propose a number of interesting site specific modifications aimed at obtaining data concerning the quantitative structure-activity and/or structure-stability relationship. In fact, by limiting our discussion to single thymidine replacements, the physical-chemical and biological properties of several properly synthesized TBA analogues have been investigated. For example, concerning the sugar-phosphate backbone, TBA derivatives have been proposed in which thymidines have been substituted with locked nucleic acid residues (LNA-T) (9), unlocked nucleic acid residues (UNA-U) (10,11) and other types of acyclic thymidines (12–14), and L-residues (15). Recently, studies concerning TBA analogues containing D- and L-isothymides have been also described (16). Furthermore, expressly synthesized TBA derivatives have been investigated in which formacetal (17), thiophosphoryl (18) and triazole (19) internucleotide linkages were present.
The general overview, which can be inferred by the literature data, is that enhancements of the thermal stability or anticoagulant activity or, in some cases both, are more likely if the modifications concern residues T3, T7 and T12 (9-14). Conversely, residues T4, T13 and, in a less extent, T9 have resulted very critical both for thermal stability and/or anticoagulant activity (10,12,16). As a matter of fact, almost all replacements concerning these residues have caused a drop for both the physical-chemical and biological properties of the resulting modified aptamers. Surprisingly, only TBA derivatives containing a few types of thymines analogues (13,14,20) or abasic sites (21,22) have been studied, notwithstanding the not negligible availability of modified thymidines phosphoramidites from commercial sources. However, although only a few studies are available concerning the replacement of thymines with modified (13,14,20) or other natural bases (23,24), the collected data confirm the critical role of residues T4 and T13 already observed for backbone modifications.
In this frame, we have undertaken a systematic investigation of new TBA derivatives containing modified thymines in order to acquire further data concerning both the structure-stability and the structure-activity relationship in the aptamer/target interaction. Here we describe the structural and biological properties of TBA analogues in which single thymidines have been replaced with 5-fluoro-2′-deoxyuridine residues (F) (Figure 1 and Table 1). For the first time and in contrast to most of the results described in literature up to date, the collected data from circular dichroism (CD) experiments, NMR techniques (1H and 19F), molecular modelling, differential scanning calorimetry (DSC) and prothrombin time (PT) assay measurements have clearly shown significant improvements for both the physical-chemical and biological properties in TBA derivatives, where residues T4 and T13 have been replaced with F. These results are particularly noteworthy taking into account that they have been obtained with TBA analogues in which a single fluorine atom replaces a methyl group in the original structure.
Table 1. TBA analogues investigated and their thermodynamic parameters, obtained from the analysis of the CD melting curves.
| Name | Sequence (5′-3′) | Tm (°C) (± 1) | ΔTm | ΔHv.H. (kJ/mol)a | ΔSv.H. (kJ/mol K)a |
|---|---|---|---|---|---|
| TBA | GGTTGGTGTGGTTGG | 50 | 154 | 0.48 | |
| TBA-F3 | GGFTGGTGTGGTTGG | 52 | +2 | 178 | 0.55 |
| TBA-F4 | GGTFGGTGTGGTTGG | 61 | +11 | 196 | 0.59 |
| TBA-F7 | GGTTGGFGTGGTTGG | 53 | +3 | 163 | 0.50 |
| TBA-F9 | GGTTGGTGFGGTTGG | 52 | +2 | 201 | 0.62 |
| TBA-F12 | GGTTGGGTTGGFTGG | 51 | +1 | 178 | 0.55 |
| TBA-F13 | GGTTGGGTTGGTFGG | 60 | +10 | 204 | 0.61 |
aThe errors on ΔHv.H. and ΔSv.H. are within the 10%.
F = 5-fluoro-2′-deoxyuridine.
MATERIALS AND METHODS
Oligonucleotides synthesis and purification
The oligonucleotides reported in Table 1 were synthesized on a Millipore Cyclone Plus DNA synthesizer using solid phase β-cyanoethyl phosphoramidite chemistry at 15 μmol scale. The modified monomer was introduced in the sequences using commercially available 5′-O-(4,4′-dimethoxytrityl)-5-fluoro-2′-deoxyuridine-3′-O-(2-cyanoethyl-N,N-diisopropyl)phosphoramidite (Glen Research). The oligomers were detached from the support and deprotected by treatment with concentrated aqueous ammonia at room temperature for 24 h.
The combined filtrates and washings were concentrated under reduced pressure, redissolved in H2O, analysed and purified by high-performance liquid chromatography on a Nucleogel SAX column (Macherey-Nagel, 1000–8/46), using buffer A: 20 mM NaH2PO4/Na2HPO4 aqueous solution (pH 7.0) containing 20% (v/v) CH3CN and buffer B: 1 M NaCl, 20 mM NaH2PO4/Na2HPO4 aqueous solution (pH 7.0) containing 20% (v/v) CH3CN; a linear gradient from 0 to 100% B for 45 min and flow rate 1 ml/min were used. The fractions of the oligomers were collected and successively desalted by Sep-pak cartridges (C-18). The isolated oligomers proved to be >98% pure by NMR.
CD and UV spectroscopy
CD samples of modified oligonucleotides and their natural counterpart were prepared at an oligodeoxynucleotide (ODN) concentration of 100 μM using a potassium phosphate buffer (10 mM KH2PO4/K2HPO4, 70 mM KCl, pH 7.0) and submitted to the annealing procedure (heating at 90°C and slowly cooling at room temperature). CD spectra of all quadruplexes and CD melting curves were registered on a Jasco J-715 CD spectrophotometer. For the CD spectra, the wavelength was varied from 220 to 320 nm at 100 nm min−1 scan rate, and the spectra recorded with a response of 16 s, at 2.0 nm bandwidth and normalized by subtraction of the background scan with buffer. The temperature was kept constant at 20°C with a thermoelectrically-controlled cell holder (Jasco PTC-348). CD melting curves were registered as a function of temperature from 20 to 80°C for all quadruplexes at their maximum Cotton effect wavelengths. The CD data were recorded in a 0.1 cm pathlength cuvette with a scan rate of 0.5°C/min. The CD melting curves were modelled by a two-state transition according to the van't Hoff analysis (25). The melting temperature (Tm), the enthalpy change (ΔHv.H.) and the entropy change (ΔSv.H.) values (Table 1) provide the best fit of the experimental melting data.
The UV melting experiments were carried out with a Cary 5000 UV-vis spectrophotometer equipped with a thermostated cell compartment, using quartz cuvettes with 1 cm path lengths. UV samples were prepared at a concentration of 4 μM in the same buffer used for CD experiments. The UV melting curves were obtained by following the absorbance change at 295 nm in the 20–85°C temperature range using a scan rate of 0.5°C/min.
Differential scanning calorimetry
Differential scanning measurements were performed on a last generation nano-DSC (TA Instruments). The excess molar heat capacity function ΔCP was obtained after a baseline subtraction, assuming that the baseline is given by the linear temperature dependence of the native-state heat capacity (26). A buffer versus buffer scan was subtracted from the sample scan. All systems were tested for reversibility by running the heating and cooling curves at the same scan rate of 1°C min−1. The process enthalpies, ΔH°, were obtained by integrating the area under the heat capacity versus the temperature curves. Tm is the temperature corresponding to the maximum of each DSC peak. Entropy values were obtained by integrating the curve ΔCp/T versus T (where ΔCp is the molar heat capacity and T is the temperature in Kelvin). The thermodynamic parameters in Supplementary Table S1 represent averages of heating curves from three to five experiments. The reported errors for thermodynamic parameters are the standard deviations of the mean from the multiple determinations. The measurements were performed with a DNA strand concentrations in the range 250–300 μM.
Polyacrylamide gel electrophoresis
All oligonucleotides were analysed by non-denaturing polyacrylamide gel electrophoresis (PAGE). Samples in the NMR buffer (10 mM KH2PO4, 70 mM KCl and 0.2 mM EDTA, pH = 7) were loaded on a 20% polyacrylamide gel containing Tris–Borate-EDTA (TBE) 2.5× and KCl 50 mM. The run buffer was TBE 1× containing 100 mM KCl. For all samples, a solution of glycerol/TBE 1× 100 mM KCl 2:1 was added just before loading. Electrophoresis was performed at 8 V/cm at a temperature close to 10°C. Bands were visualized by UV shadowing.
NMR spectroscopy
NMR samples were prepared at a concentration of about 3 mM, in 0.6 ml (H2O/D2O 9:1 v/v), buffer solution having 10 mM KH2PO4/K2HPO4, 70 mM KCl and 0.2 mM EDTA (pH 7.0). All the samples were heated for 5–10 min at 80°C and slowly cooled (10–12 h) to room temperature. The solutions were equilibrated for several weeks at 4°C. The annealing process was assumed to be complete when 1H-NMR spectra were super-imposable on changing time. NMR spectra were recorded with Varian Unity INOVA 500 MHz spectrometers and Varian Mercury Plus 400 MHz. One-dimensional (1D) proton spectra of the sample in H2O were recorded using pulsed-field gradient DPFGSE for H2O suppression (27). 1H-chemical shifts were referenced relative to external sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) and 19F resonances were referenced relative to external CCl3F. Pulsed-field gradient DPFGSE sequence was used for NOESY (28) (250 and 180 ms mixing times) and TOCSY (29) (120 ms mixing time) experiments in H2O. All experiments were recorded using STATES-TPPI procedure for quadrature detection (30). In all two-dimensional (2D) experiments, the time domain data consisted of 2048 complex points in t2 and 400–512 fids in t1 dimension. A relaxation delay of 1.2 s was used for all experiments.
Molecular modelling
The main conformational features of quadruplex adopted by TBA-F4 and TBA-F13 were explored by means of a molecular modelling study. The CFF91 force field using CFF91 atom types set was used (31,32). The initial coordinates for the starting model of TBA-F4 and TBA-F13 were taken from the NMR solution structure of the quadruplex d(GGTTGGTGTGGTTGG) (Protein Data Bank entry number 148D), with the first and best representative conformer of the twelve available ones submitted by Feigon et al. (33) The initial d(GGTFGGTGTGGTTGG) (TBA-F4) and d(GGTTGGTGTGGTFGG) (TBA-F13) G-quadruplex models were built by replacing the 5-methyl group of the thymine in the fourth and thirteenth position with a fluorine, using the Biopolymer building tool of Discover. The calculations were performed using a distance-dependent macroscopic dielectric constant of 4r and an infinite cut-off for non-bonded interactions to partially compensate for the lack of solvent used (34). Using steepest descent and conjugate gradient methods, the conformational energy of the complexes was minimized until convergence to an Root Mean Square (RMS) gradient of 0.1 kcal/mol Å was reached. Illustrations of the structure were generated using the INSIGHT II program, version 2005 (Accelrys, San Diego, CA, USA). All the calculations were performed on a PC running Linux ES 2.6.9.
PT assay
PT assay on human plasma samples was measured by using Koagulab MJ Coagulation System with a specific kit HemosIL RecombinPlasTin 2G (Instrumentation Laboratory, Milan, Italy). Briefly, this method relies on the high sensitivity of thromboplastin reagent based on recombinant human tissue factors. The addition of recombiplastin to the plasma, in presence of calcium ions, initiates the activation of extrinsic pathway converting the fibrinogen into fibrin, with a formation of solid gel. The procedure was according to the manufacturer's instructions. In our experimental conditions, each ODN or vehicle was incubated with 100 μl of plasma at 37°C for 3 and 15 min, after that 200 μl of the kit solution containing recombiplastin was added with consequent activation of extrinsic pathway. For the evaluation of PT at 20 μM, in the apposite microtube, 2 μl of the corresponding ODN solution (1 mM in phosphate buffered saline (PBS) buffer) or vehicle was added. The PT at final ODN concentration of 2 μM was determined using 2 μl of a diluted solution (0.1 mM ODN solution in PBS buffer). The PT measurement, for each incubation time, was produced in triplicate and the average and its standard error values were calculated and expressed as seconds. The basal clotting time was determined by measuring the clotting time in absence of any ODN.
Nucleases stability assay
Given the molecular complexity and the large biological variability among foetal bovine sera - making of each serum batch a unique biological sample - nuclease biostability experiments of anti-thrombin aptamers were conducted by using a range of concentrations (from 10 to 30%) of foetal bovine serum (FBS, Gibco), diluted with Dulbecco's Modified Eagle's Medium (DMEM, Lonza) at 37°C. Approximately 19 nmol of stock solution of each ODN (3 O.D.U) were evaporated to dryness under reduced pressure and then incubated with 200 or 250 μl of FBS solutions at 37°C. At established times, ranging from 0 to 6 h, 50 μl of samples were collected and stored at −20°C for at least 20 min. The samples were evaporated to dryness and then resuspended in 10 μl of gel loading buffer consisting of 30% glycerol, 30% formamide, 40% TE buffer 1× (10 mM Tris–HCl, 1 mM EDTA, pH 8.0) and 10 μl of sterile water. A total of 10 μl of the mixture were used for denaturing Urea PAGE, carried out at room temperature using 20% polyacrylamide gel in 0.5 × TBE buffer (Tris-borate-EDTA) and 7 M urea. The degradation pattern on the gel was visualized by UV shadowing.
RESULTS AND DISCUSSION
CD spectroscopy
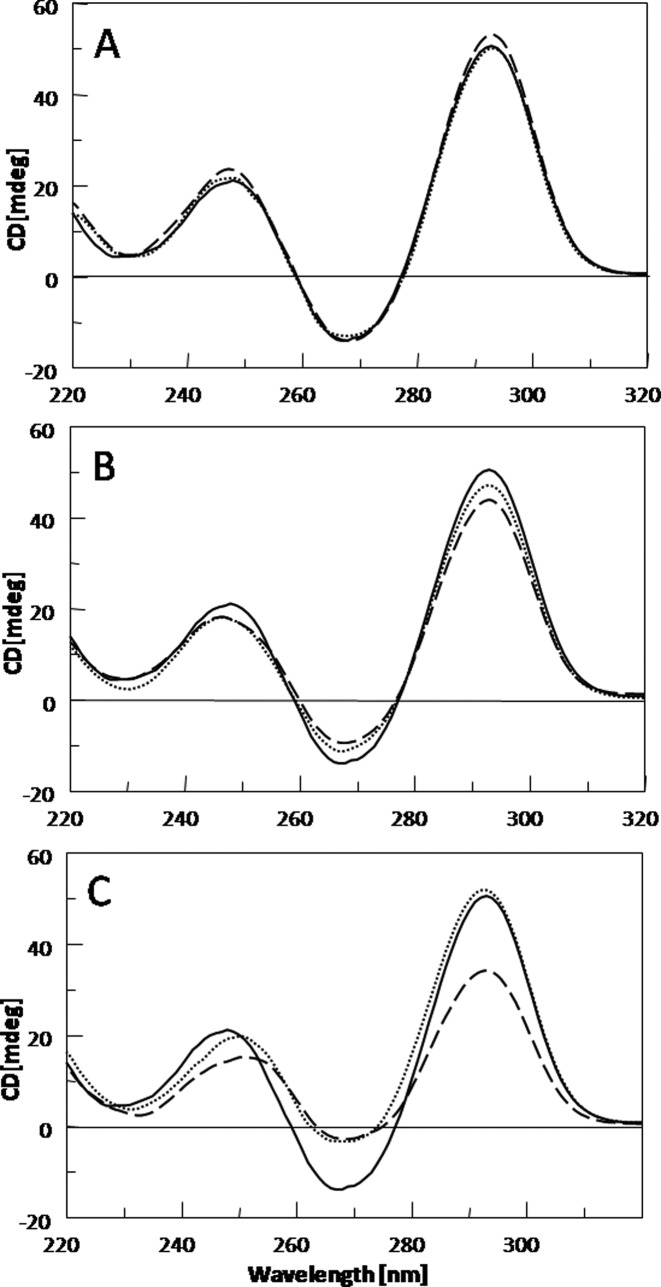
The CD spectrum of the TBA is characterized by two positive bands at 247 and 295 nm, and a negative band at 266 nm. This profile is typical of an antiparallel G-quadruplex in which anti and syn guanosines alternate along the strands. Considering the distinctiveness of this profile, a close comparison between CD spectra of the modified TBAs and that of their natural counterpart can be considered a useful method to straightforwardly evaluate the effects of the F residues on the G-quadruplex structure. In Figure 2 the CD profiles of the TBA analogues and their natural counterpart are shown. In general, all TBA analogues show CD spectra quite similar to that of the original TBA. However subtle differences suggest that CD profiles of the TBA derivatives containing F can be conveniently divided in three groups. In the first one, derivatives TBA-F4 and TBA-F13 are included, showing CD profiles practically super-imposable to that of the original TBA (Figure 2A). Modified aptamers TBA-F3 and TBA-F12 belong to the second group, that is characterized by CD profiles in which only differences in the band intensities are evident in comparison with the CD spectrum of TBA (Figure 2B). Finally, derivatives TBA-F7 and TBA-F9 show CD profiles characterized by differences in both the band intensities and wavelengths (Figure 2C). The collected CD data clearly suggest different effects of the F residues on the G-quadruplex structure, according to the position in the sequence.
Figure 2.
CD spectra at 20°C of modified TBAs and their natural counterpart (solid line) at 100 μM ODN strand concentration in a buffer solution 10 mM KH2PO4/K2HPO4, 70 mM KCl (pH 7.0). (A) TBA-F4 (dotted line) and TBA-F13 (dashed line); (B) TBA-F3 (dotted line) and TBA-F12 (dashed line); (C) TBA-F9 (dotted line) and TBA-F7 (dashed line).
Thermal denaturation measurements
The differences in the behaviour of the TBA analogues are clearly more evident by comparing their CD melting profiles (Supplementary Figure S1), useful to evaluate the structural stability in comparison with the original TBA. The thermodynamic parameters obtained by the van't Hoff analysis of the CD melting curves are shown in Table 1. Inspection of Table 1 reveals that none or only modest increases of the melting temperature (ΔTm ∼ +3) have been observed for TBA-F3, TBA-F12, TBA-F7 and TBA-F9 whereas, in the cases of TBA-F4 and TBA-F13, a remarkable enhancement (ΔTm ∼ +10) of the thermal stability has been obtained. Furthermore, TBA-F9, TBA-F4 and TBA-F13 show the higher enthalpy and entropy change upon unfolding whereas TBA-F3, TBA-F12 and TBA-F7 have ΔHv.H. and ΔSv.H values comparable to the unmodified TBA, within the experimental error (Table 1). Analysis of the thermodynamic parameters has revealed that the extra stability of TBA-F4 and TBA-F13 is mainly due to the enthalpic term, thus suggesting the formation of additional interactions in the folded TBA-F4 and TBA-F13 in comparison with natural TBA. Interestingly, TBA-F9 shows a ΔHv.H value similar to TBA-F4 and TBA-F13 but a Tm value comparable to the unmodified TBA. The unfolding thermodynamics of the most stable TBA analogues, namely TBA-F4 and TBA-F13, was further investigated by using DSC. Supplementary Figure S2 in Supplementary Data shows the DSC melting profiles of TBA-F4 and TBA-F13 whereas the thermodynamic parameters are collected in Supplementary Table S1. The Tm values are in good agreement with those obtained by CD measurements. The comparison of the observed enthalpy changes with the one reported from calorimetric study of TBA (35), confirms that the modified sequences have a larger favourable enthalpy change being the ΔΔH ∼ 50–60 kJ mol−1. In order to confirm the monomolecular nature of TBA-F4 and TBA-F13 suggested by PAGE (vide infra), UV melting experiments at lower concentration (4 μM) were also performed (Supplementary Figure S3). As expected for monomolecular structures, the Tm values are independent of the concentration.
Polyacrylamide gel electrophoresis
In order to confirm that all TBA analogues preserve the monomolecular G-quadruplex structure characteristic of the natural TBA, the TBA modified aptamers containing F residues were further investigated by PAGE and compared to their natural counterpart. The electrophoretic profile (Supplementary Figure S4) clearly shows that all the TBA analogues form G-quadruplex structures with electrophoretic motilities very similar to that of the natural TBA, thus strongly suggesting the occurring of G-quadruplex conformations comparable to that of the original TBA and with the same molecularity, in agreement with the CD data.
Nuclear magnetic resonance
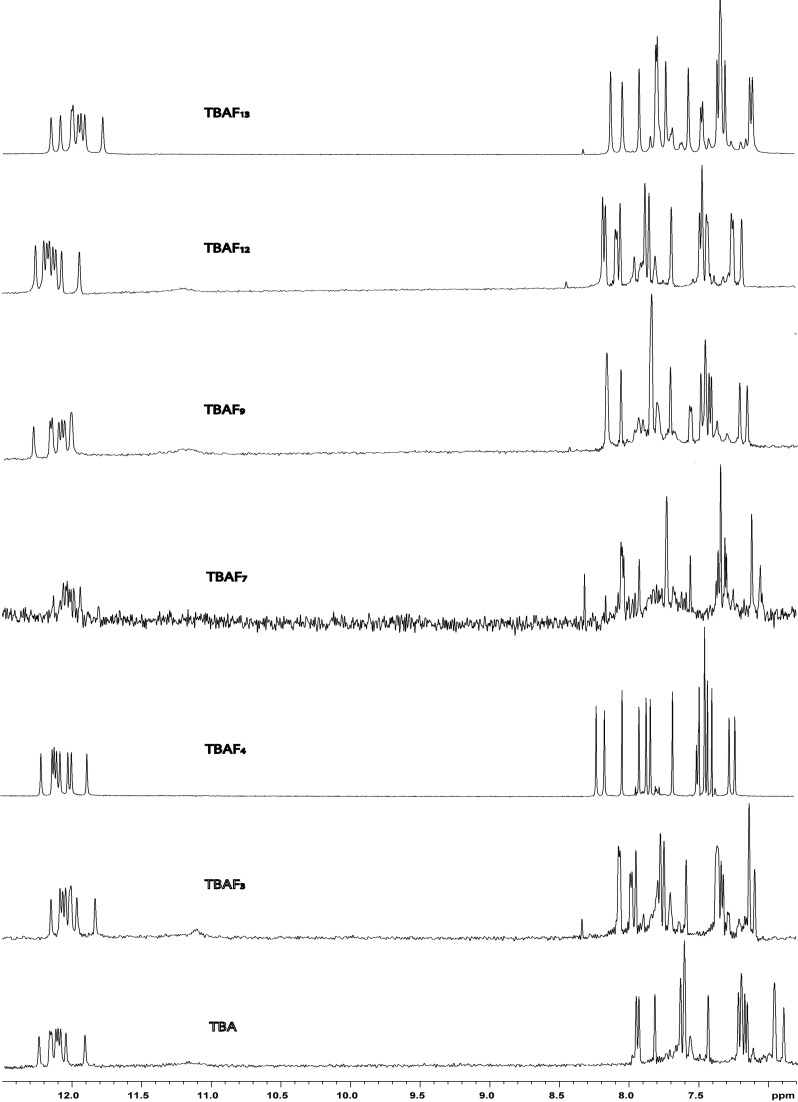
The ability of all the TBA analogues incorporating F to fold into a TBA-like antiparallel G-quadruplex conformation was also assessed by NMR spectroscopy. The NMR samples were prepared at a concentration of about 3.0 mM in strands (0.6 ml, 90% H2O/10% D2O), having a 10 mM potassium phosphate, 70 mM KCl and 0.2 mM EDTA (pH 7.0) buffer. The samples were heated for 5–10 min at 80°C and slowly cooled down (10–12 h) to room temperature. The solutions were equilibrated at least for 1 week at 4°C and then their 1H-NMR spectra were recorded by using pulsed-field gradient DPFGSE for H2O suppression (27). A completed annealing process was guaranteed by the achievement of super-imposable 1H-NMR spectra on changing time. With the exclusion of some weak resonances, whose relative intensities turned out to be sensitive to temperature changes and attributable to unfolded species also present in solution, the simple appearance of all 1H-NMR spectra indicates that, in the conditions used here, the modified oligomers form mainly a single well-defined hydrogen-bonded conformation. In fact their 1D 1H-NMR spectra (500 MHz, T = 25°C) (Figure 3) in the buffer conditions utilized show the presence of eight well-defined signals in the region 11.8–12.5 p.p.m., attributable to imino protons involved in Hoogsteen hydrogen bonds of at least two G-quartets and, moreover, 15 main signals in the aromatic region, due to the presence of nine guanine H8 and five thymine H6 singlets, and one FH6 doublet, splitted for the spin-spin coupling between proton and fluorine. Since the whole of data collected from CD melting experiments and PT assays (vide infra) clearly indicate that TBA-F4 and TBA-F13 are the most stable and active modified aptamers of the series, we have focused our attention on these two samples, performing further NMR investigations. A combination of the analysis of 2D NOESY (Supplementary Figures S5 and S6) and TOCSY spectra (data not shown) allowed us to get the almost complete assignment (Supplementary Table S2) of the non-exchangeable protons. The intensities of NOESY crosspeaks between the aromatic base proton and sugar H1' resonances of the modified thrombin aptamers indicate that four Gs (G1, G5, G10 and G14) adopt syn glycosidic conformations, while five Gs (G2, G6, G8, G11 and G15) adopt anti conformations, where the H8 resonances of the syn G are upfield shifted with respect to those of the anti ones (4,36,37). All the thymidines and the modified residue F show anti glycosidic conformations. Then, four anti-Gs (G2, G6, G11 and G15) have classical H8/H2′-H2″ sequential connectivities to 5′ neighbouring syn-Gs (G1, G5, G10 and G14, respectively) indicating that the subunits G1-G2, G5-G6, G10-G11 and G14-G15 are involved in the formation of a four-stranded helical structure (underlined residues adopt a syn glycosidic conformation). In summary, as observed for the parent TBA, there are 5′-GsynGanti-3′ steps along each strand of the two quartets and moreover, the entire pattern of Nuclear Overhauser Effects (NOEs) observed for all cited Gs indicates that the backbone conformations of these tracts resemble those of the unmodified TBA possessing a right-handed helix structure. The alternation of syn and anti G residues within each strand suggests that, as for TBA, the two modified oligomers fold into a monomolecular foldback G-quadruplex, characterized by two G tetrads. Two stretches of 5′-TT-3′ and 5′-TF-3′ residues and one of 5′-TGT-3′ could also be identified on the basis of anti–anti connectivities. All the stretches of sequential connectivities were arranged using the information in long-range NOE connectivities. Similarly to the case of the parent TBA, for both samples, there is a number of NOE connectivities observed between residues not adjacent in sequence. In particular, there are NOE connectivities between H8 of G8 and Hl′, H2′ and H2″ of G6 and between H1′ of T9 and H8 of G15, thus indicating that G8 and T9 residues in the TGT loop are near to the G1-G6-G10-G15 tetrad both in the TBA-F4 that in the TBA-F13. NOEs are also present between H8 of G2 and H2′ and H2″ of F4 and between H8 of G11 and methyl, H2′ and H2″ of T13 in the case of TBA-F4. Correspondingly, as far as TBA-F13 is concerned, there are NOE connectivities between H8 of G2 and methyl, H2′ and H2″ of T4 and between H8 of G11 and H2′ and H2″ of F13. Complementary information is provided by the NOEs from the methyl of T13 with H1′, H2′ and H2″ of G2, in the case of TBA-F4, and from the methyl of T4 with H1′ H2′ and H2″ of G11, in the case of TBA-F13. For both the samples, this collection of NOEs places the residues in position 4 and 13 close to the G2-G5-G11-G14 tetrad. The whole of NMR data showed that, as strongly suggested by the CD experiments, these two modified aptamers adopt G-quadruplex structures strictly resembling the parent one.
Figure 3.
Aromatic and imino protons regions of the 1H-NMR spectra (500 MHz, T = 25°C) of the modified TBAs and their natural counterpart in 10 mM KH2PO4/K2HPO4, 70 mM KCl and 0.2 mM EDTA (pH 7.0).
The presence of a fluorine atom in each sequence has given us the opportunity to further investigate the modified TBAs by 19F-NMR spectroscopy. Since it is generally accepted that TBA binds and inhibits thrombin through the TT loops, only G-quadruplex structures formed by TBA-F3, TBA-F4, TBA-F12 and TBA-F13 have been analysed by this technique. Proton-decoupled fluorine 1D NMR spectra (400 MHz, T = 25°C) of all the samples exhibit the occurrence of one uppermost signal in the region −62.0 to −59.0 p.p.m. (Supplementary Figure S7), attributable to the fluorine nucleus of the modified residue F, thus confirming the presence in solution of a main conformation. Furthermore, taking into account that the 19F chemical shifts are particularly sensitive to the chemical environment (38,39), the very similar values for TBA-F3 and TBA-F12, on one hand, and TBA-F4 and TBA-F13, on the other hand, has allowed us to confirm the almost identical conformation and behaviour of the residues in each couple.
Molecular modelling
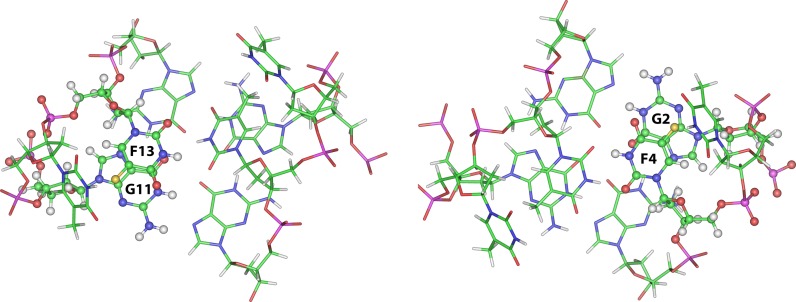
Molecular models for both TBA-F4 and TBA-F13 have been built. As expected, the replacement of a methyl group with a fluorine atom doesn't significantly affect the general folding of the modified G-quadruplex structures, compared to the original TBA, as already suggested by NMR data. However, the close inspection of the TT loop regions in the TBA-F13 model reveals a good stacking between F13 and G11 involved in the formation of the adjacent G-tetrad, in which the fluorine atom with a partially negative charge is in close proximity to C4 of the guanosine with a partially positive charge (Figure 4, left). Similarly, in the TBA-F4 model, the fluorine atom of F4 is in close proximity to the C4 of the G2 (Figure 4, right). Although more accurate investigations concerning the origin of the thermodynamic parameter differences observed for these modified aptamers are beyond the aim of this paper, the increase in the enthalpy change observed for TBA-F4 and TBA-F13 could be tentatively explained by the more effective dipole–dipole interactions between fluorine and carbon atoms of the adjacent G-tetrad, compared to the parent TBA.
Figure 4.
Bottom view representations of the molecular models of the quadruplexes formed by TBA-F13 (left) and TBA-F4 (right). The structures are oriented with the 5′ and 3′-ends upward. The TGT loop and the G1-G6-G10-G15 tetrad have been omitted for clarity. Backbones and bases are depicted in coloured ‘stick’ (carbons, green; nitrogens, blue; oxygens, red; hydrogens, white; fluorine, yellow). The F13 (left) and F4 (right) residues and the respectively overlying G11 and G2 are reported in ball and sticks, in order to point out the effectiveness of the stacking interaction between them.
PT assay
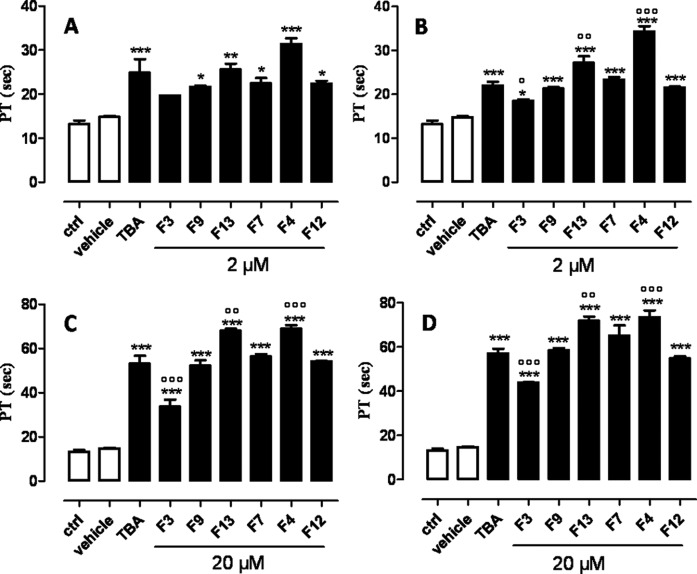
In order to evaluate the anticoagulant activity of the TBA analogues containing a F residue, the modified sequences have been subjected to PT assay. This analysis has shown that all TBA derivatives preserve their anticoagulant activity, although in different extents, depending on the position of the F residues in the sequence (Figure 5). TBA-F4 and TBA-F13 have shown a significant enhancement of the anticoagulant activity compared to the original TBA. In particular these compounds prolonged the PT values more than TBA when they are incubated for 15 min at the concentration of 2 μM (Figure 5B) and for 3 and 15 min at 20 μM (Figure 5C and 5D). Conversely, the anticoagulant activity of TBA-F3 has turned out lower than the natural TBA. As far as TBA-F7, TBA-F9 and TBA-F12 are concerned, no noteworthy differences with the original TBA have been observed. These results indicate that TBA-F4 possesses the best anticoagulant activity among all the TBA analogues tested and it is more efficient than natural TBA at both concentrations tested. The data obtained have clearly highlighted that it is possible to significantly improve the anticoagulant activity of TBA in vitro also through tiny chemical modifications.
Figure 5.
PT values of the modified TBAs and their natural counterpart at different ODN concentrations and incubation times (A and C: 3 min.; B and D: 15 min.). See experimental section for details. ***P < 0.001 versus vehicle, °P < 0.05, °°P < 0.01, °°°P < 0.001 versus TBA (n = 3).
Nucleases stability assay
In order to test the resistance in biological environments, the most active modified aptamers, namely TBA-F4 and TBA-F13, were undergone to a degradation assay in foetal bovine serum (Supplementary Figure S8) and analysed by gel electrophoresis. As expected considering the very small chemical modification compared to the parent TBA, results clearly show that TBA analogues behave as their natural counterpart and, then, the replacement of a methyl group by a fluorine atom is not enough to increase the resistance in biological environments. On the other hand, several investigations have been reported indicating that a suitable resistance to nucleases of modified TBA can be obtained by simple specific modifications of the sugar-phosphate backbone without affecting the loops involved in the interaction with the target protein (for example see 18,19,40).
CONCLUSIONS
The sequence of TBA is the final product of a selection process and of the subsequent truncation studies (3). Therefore, the peculiar ‘chair-like’ G-quadruplex structure adopted by this sequence has been selected to bind with high affinity and selectivity the thrombin. In view of a possible therapeutic use of this aptamer several modifications have been proposed in order to improve both its physical-chemical and biological properties (41). However, each loop modification, by altering the conformation of the aptamer which has been optimized by the SELEX process to bind thrombin, can revert the favourable properties, particularly if this modification concerns residues T4, T9 and T13 (10,12,16). As a matter of fact, improvements of the TBA properties have been obtained only by modifying the sugar-phosphate backbone concerning residues T7, T3 and T12 (9-14). In an attempt to introduce modifications and, at the same time, to preserve the conformation of the negatively charged sugar-phosphate backbone of the TT loop interacting with the positive exosite I of the thrombin (8), we have focused our attention on the aromatic bases of the loop residues. In a previous paper (20), we improved thermal stability and/or anticoagulant activity by substituting individual thymines (position T3, T7 and T9) with 5-hydroxymethyluracil bases. In this paper we show that the replacement of thymine T4 or T13 with 5-fluorouracil results in a significant improvement of both stability and biological activity, particularly considering the minor change performed on TBA (a fluorine atom replacing a methyl group). It should be noted that examples of enhancements of TBA properties by site-specific substitutions in positions T4 and T13 are unprecedented in literature. Taking into consideration the availability of modified thymidine phosphoramidites from commercial sources and the possibility to prepare further modified thymidines from synthetic approaches described in the literature, our results open up new perspectives of improving TBA properties through tiny and straightforward chemical modifications, which would be worth testing also for other aptamers. Recently several ‘second generation’ anticoagulant aptamers have been described in literature. Among these, the duplex-quadruplex modular aptamer (42,43,44) dimeric DNA aptamers (45) and the bivalent aptamer (46) are particularly noteworthy. It should be noted that the biological activity of all these aptamers could benefit from the site specific replacement here described, since they preserve the peculiar sequence of TBA (Table 1) folding in the antiparallel G-quadruplex, as the structural component interacting with the thrombin. Furthermore, it would be interesting to explore the properties of TBA analogues in which two or more different types of modified residues have been introduced in specific positions, with the aim to test the possibility of additive or synergic effects between the modifications. At the moment, investigations concerning TBA analogues containing both 5-hydroxymethyl-2′-deoxyuridine and 5-fluoro-2′-deoxyuridine have been planned in our laboratory.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Regione Campania under POR Campania [FESR 2007–2013-O.O. 2.1 (FarmaBioNet)]. Funding for open access charge: Dipartimento di Farmacia Università degli Studi di Napoli.
Conflict of interest statement. None declared.
REFERENCES
- 1.Szeitner Z., András J., Gyurcsányi R.E., Mészáros T. Is less more? Lessons from aptamer selection strategies. J. Pharm. Biomed. Anal. 2014;101:58–65. doi: 10.1016/j.jpba.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 2.Kong H.Y., Byun J. Nucleic Acid aptamers: new methods for selection, stabilization, and application in biomedical science. Biomol. Ther. 2013;21:423–434. doi: 10.4062/biomolther.2013.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bock L.C., Griffin L.C., Latham J.A., Vermaas E.H., Toole J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 4.Wang K.Y., McCurdy S., Shea R.G., Swaminathan S., Bolton P.H. A DNA Aptamer Which Binds to and Inhibits Thrombin Exhibits a New Structural Motif for DNA. Biochemistry. 1993;32:1899–1904. doi: 10.1021/bi00059a003. [DOI] [PubMed] [Google Scholar]
- 5.Macaya R.F., Schultze P., Smith F.W., Roe J.A., Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. U.S.A. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly J.A., Feigon J., Yeates T.O. Reconciliation of the X-ray and NMR Structures of the Thrombin-Binding Aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1996;256:417–422. doi: 10.1006/jmbi.1996.0097. [DOI] [PubMed] [Google Scholar]
- 7.Padmanabhan K., Tulinsky A. An ambiguous structure of a DNA 15-mer thrombin complex. Acta Crystallogr. Sect. D Biol. Crystallogr. 1996;D52:272–282. doi: 10.1107/S0907444995013977. [DOI] [PubMed] [Google Scholar]
- 8.Russo Krauss I., Merlino A., Giancola C., Randazzo A., Mazzarella L., Sica F. Thrombin-aptamer recognition: a revealed ambiguity. Nucleic Acids Res. 2011;39:7858–7867. doi: 10.1093/nar/gkr522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonifacio L., Church F., Jarstfer M. Effect of locked-nucleic acid on a biologically active G-quadruplex. A structure-activity relationship of the thrombin aptamer. Int. J. Mol. Sci. 2008;9:422–433. doi: 10.3390/ijms9030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasternak A., Hernandez F.J., Rasmussen L.M., Veste B., Wengel J. Improved thrombin binding aptamer by incorporation of a single unlocked nucleic acid monomer. Nucleic Acids Res. 2011;39:1155–1164. doi: 10.1093/nar/gkq823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen T.N., Henriksen J.R., Rasmussen B.E., Rasmussen L.M., Andresen T.L., Wengel J., Pasternak A. Thermodynamic and biological evaluation of a thrombin binding aptamer modified with several unlocked nucleic acid (UNA) monomers and a 2′-C-piperazino-UNA monomer. Bioorg. Med. Chem. 2011;19:4739–4745. doi: 10.1016/j.bmc.2011.06.087. [DOI] [PubMed] [Google Scholar]
- 12.Coppola T., Varra M., Oliviero G., Galeone A., D'Isa G., Mayol L., Morelli E., Bucci M.R., Vellecco V., Cirino G., et al. Synthesis, structural studies and biological properties of new TBA analogues containing an acyclic nucleotide. Bioorg. Med. Chem. 2008;16:8244–8253. doi: 10.1016/j.bmc.2008.07.040. [DOI] [PubMed] [Google Scholar]
- 13.Borbone N., Bucci M., Oliviero G., Morelli E., Amato J., D'Atri V., D'Errico S., Vellecco V., Cirino G., Piccialli G., et al. Investigating the role of T7 and T12 residues on the biological properties of thrombin-binding aptamer: enhancement of anticoagulant activity by a single nucleobase modification. J. Med. Chem. 2012;55:10716–10728. doi: 10.1021/jm301414f. [DOI] [PubMed] [Google Scholar]
- 14.Scuotto M., Persico M., Bucci M., Vellecco V., Borbone N., Morelli E., Oliviero G., Novellino E., Piccialli G., Cirino G., et al. Outstanding effects on antithrombin activity of modified TBA diastereomers containing an optically pure acyclic nucleotide analogue. Org. Biomol. Chem. 2014;12:5235–5242. doi: 10.1039/c4ob00149d. [DOI] [PubMed] [Google Scholar]
- 15.Virgilio A., Varra M., Scuotto M., Capuozzo A., Irace C., Mayol L., Esposito V., Galeone A. Expanding the potential of G-quadruplex structures: formation of a heterochiral TBA analogue. Chembiochem. 2014;15:652–655. doi: 10.1002/cbic.201300775. [DOI] [PubMed] [Google Scholar]
- 16.Cai B., Yang X., Sun L., Fan X., Li L., Jin H., Wu Y., Guan Z., Zhang L., Zhang L., et al. Stability and bioactivity of thrombin binding aptamers modified with D-/L-isothymidine in the loop regions. Org. Biomol. Chem. 2014;12:8866–8876. doi: 10.1039/c4ob01525h. [DOI] [PubMed] [Google Scholar]
- 17.He G.X., Williams J.P., Postich M.J., Swaminathan S., Shea R.G., Terhorst T., Law V.S., Mao C.T., Sueoka C., Coutré S., et al. In vitro and in vivo activities of oligodeoxynucleotide-based thrombin inhibitors containing neutral formacetal linkages. J. Med. Chem. 1998;41:4224–4231. doi: 10.1021/jm970766i. [DOI] [PubMed] [Google Scholar]
- 18.Zaitseva M., Kaluzhny D., Shchyolkina A., Borisova O., Smirnov I., Pozmogova G. Conformation and thermostability of oligonucleotide d(GGTTGGTGTGGTTGG) containing thiophosphoryl internucleotide bonds at different positions. Biophys. Chem. 2010;146:1–6. doi: 10.1016/j.bpc.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Varizhuk A.M., Tsvetkov V.B., Tatarinova O.N., Kaluzhny D.N., Florentiev V.L., Timofeev E.N., Shchyolkina A.K., Borisova O.F., Smirnov I.P., Grokhovsky S.L., et al. Synthesis, characterization and in vitro activity of thrombin-binding DNA aptamers with triazole internucleotide linkages. Eur. J. Med. Chem. 2013;67:90–97. doi: 10.1016/j.ejmech.2013.06.034. [DOI] [PubMed] [Google Scholar]
- 20.Virgilio A., Petraccone L., Scuotto M., Vellecco V., Bucci M., Mayol L., Varra M., Esposito V., Galeone A. 5-hydroxymethyl-2′-deoxyuridine residues in the thrombin binding aptamer: investigating anticoagulant activity by making a tiny chemical modification. Chembiochem. 2014;15:2427–2434. doi: 10.1002/cbic.201402355. [DOI] [PubMed] [Google Scholar]
- 21.Wang K.Y., Krawczyk S.H., Bischofberger N., Swaminathan S., Bolton P.H. The tertiary structure of a DNA aptamer which binds to and inhibits thrombin determines activity. Biochemistry. 1993;32:11285–11292. doi: 10.1021/bi00093a004. [DOI] [PubMed] [Google Scholar]
- 22.Nagatoishi S., Sugimoto N. Interaction of water with the G-quadruplex loop contributes to the binding energy of G-quadruplex to protein. Mol. Biosyst. 2012;8:2766–2770. doi: 10.1039/c2mb25234a. [DOI] [PubMed] [Google Scholar]
- 23.Smirnov I., Shafer R.H. Effect of loop sequence and size on DNA aptamer stability. Biochemistry. 2000;39:1462–1468. doi: 10.1021/bi9919044. [DOI] [PubMed] [Google Scholar]
- 24.Nagatoishi S., Isono N., Tsumoto K., Sugimoto N. Loop residues of thrombin-binding DNA aptamer impact G-quadruplex stability and thrombin binding. Biochimie. 2011;93:1231–1238. doi: 10.1016/j.biochi.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 25.Marky L.A., Breslauer K.J. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 26.Pagano B., Randazzo A., Fotticchia I., Novellino E., Petraccone L., Giancola C. Differential scanning calorimetry to investigate G-quadruplexes structural stability. Methods. 2013;64:43–51. doi: 10.1016/j.ymeth.2013.02.018. [DOI] [PubMed] [Google Scholar]
- 27.Dalvit C. Efficient multiple-solvent suppression for the study of the interactions of organic solvents with biomolecules. J. Biomol. NMR. 1998;11:437–444. [Google Scholar]
- 28.Jeener J., Meier B., Bachmann H.P., Ernst R.R. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 1979;71:4546–4553. [Google Scholar]
- 29.Braunschweiler L., Ernst R.R. Coherence transfer by isotropic mixing: Application to proton correlation spectroscopy. J. Magn. Reson. 1983;53:521–528. [Google Scholar]
- 30.Marion D., Ikura M., Tschudin R., Bax A. Rapid recording of 2D NMR spectra without phase cycling. Application to the study of hydrogen exchange in proteins. J. Magn. Reson. 1989;85:393–399. [Google Scholar]
- 31.Maple J.R., Hwang M.-J., Stockfisch T.P., Dinur U., Waldman M., Ewing C.S., Hagler A.T. Derivation of class II force fields. I. Methodology and quantum force field for the alkyl functional group and alkane molecules. J. Comput. Chem. 1994;15:162–182. [Google Scholar]
- 32.Rudnicki W.R., Lesyng B. Applicability of commonly used atom-atom type potential energy functions in structural analysis of nucleic acids. The role of electrostatic interactions. Comput. Chem. (Oxford) 1995;19:253–258. [Google Scholar]
- 33.Schultze P., Macaya R.F., Feigon J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1994;235:1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- 34.Weiner S.J., Kollman P.A., Case D.A., Singh U.C., Ghio C., Alagona G., Profeta S., Weiner P.J. A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 1984;106:765–784. [Google Scholar]
- 35.Pagano B., Martino L., Randazzo A., Giancola C. Stability and binding properties of a modified thrombin binding aptamer. Biophys. J. 2008;94:562–569. doi: 10.1529/biophysj.107.117382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith F.W., Feigon J. Strand orientation in the DNA quadruplex formed from the Oxytricha telomere repeat oligonucleotide d(G4T4G4) in solution. Biochemistry. 1993;32:8682–8692. doi: 10.1021/bi00084a040. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y., Patel D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 38.Cobb S.L., Murphy C.D. 19F NMR applications in chemical biology. J. Fluorine Chem. 2009;130:132–143. [Google Scholar]
- 39.Puffer B., Kreutz C., Rieder U., Ebert M.-O., Konrat R., Micura R. 5-Fluoro pyrimidines: labels to probe DNA and RNA secondary structures by 1D 19F NMR spectroscopy. Nucleic Acids Res. 2009;37:7728–7740. doi: 10.1093/nar/gkp862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esposito V., Scuotto M., Capuozzo A., Santamaria R., Varra M., Mayol L., Virgilio A., Galeone A. A straightforward modification in the thrombin binding aptamer improving the stability, affinity to thrombin and nuclease resistance. Org. Biomol. Chem. 2014;12:8840–8843. doi: 10.1039/c4ob01475h. [DOI] [PubMed] [Google Scholar]
- 41.Avino A., Fabrega C., Tintore M., Eritja R. Thrombin binding aptamer, more than a simple aptamer: chemically modified derivatives and biomedical applications. Curr. Pharm. Des. 2012;18:2036–2047. doi: 10.2174/138161212799958387. [DOI] [PubMed] [Google Scholar]
- 42.Zavyalova E., Golovin A., Timoshenko T., Babiy A., Pavlova G., Kopylov A. DNA aptamers for human thrombin with high anticoagulant activity demonstrate target- and species-specificity. Curr. Med. Chem. 2012;19:5232–5237. doi: 10.2174/092986712803530575. [DOI] [PubMed] [Google Scholar]
- 43.Zavyalova E., Golovin A., Pavlova G., Kopylov A. Module-activity relationship of G-quadruplex based DNA aptamers for human thrombin. Curr. Med. Chem. 2013;20:4836–4843. doi: 10.2174/09298673113206660283. [DOI] [PubMed] [Google Scholar]
- 44.Zavyalova E., Samoylenkova N., Revishchin A., Golovin A., Pavlova G., Kopylov A. Evaluation of antithrombotic activity of thrombin DNA aptamers by a murine thrombosis model. PLoS One. 2014;9:e107113. doi: 10.1371/journal.pone.0107113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poniková S., Tlučková K., Antalík M., Víglaský V., Hianik T. The circular dichroism and differential scanning calorimetry study of the properties of DNA aptamer dimers. Biophys. Chem. 2011;155:29–35. doi: 10.1016/j.bpc.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 46.Müller J., Wulffen B., Pötzsch B., Mayer G. Multidomain targeting generates a high-affinity thrombin-inhibiting bivalent aptamer. Chembiochem. 2007;8:2223–2226. doi: 10.1002/cbic.200700535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.