Abstract
The ribonuclease Dicer excises mature miRNAs from a diverse group of precursors (pre-miRNAs), most of which contain various secondary structure motifs in their hairpin stem. In this study, we analyzed Dicer cleavage in hairpin substrates deprived of such motifs. We searched for the factors other than the secondary structure, which may influence the length diversity and heterogeneity of miRNAs. We found that the nucleotide sequence at the Dicer cleavage site influences both of these miRNA characteristics. With regard to cleavage mechanism, we demonstrate that the Dicer RNase IIIA domain that cleaves within the 3′ arm of the pre-miRNA is more sensitive to the nucleotide sequence of its substrate than is the RNase IIIB domain. The RNase IIIA domain avoids releasing miRNAs with G nucleotide and prefers to generate miRNAs with a U nucleotide at the 5′ end. We also propose that the sequence restrictions at the Dicer cleavage site might be the factor that contributes to the generation of miRNA duplexes with 3′ overhangs of atypical lengths. This finding implies that the two RNase III domains forming the single processing center of Dicer may exhibit some degree of flexibility, which allows for the formation of these non-standard 3′ overhangs.
INTRODUCTION
The RNase III endonucleases Drosha and Dicer are responsible for generating mature microRNA (miRNA) ends in the canonical miRNA biogenesis pathway in mammalian cells (1,2). Drosha, together with DiGeorge syndrome critical region gene 8 protein (DGCR8) (3,4) and several auxiliary proteins form Microprocessor Complex (5–7), the role of which is to recognize and cleave the primary miRNA precursor (pri-miRNA) to the miRNA precursor (pre-miRNA) (8). The cleavages by the two RNase III domains of Drosha in each pri-miRNA hairpin arm do not occur with perfect precision, and pre-miRNA variants with differing ends are typically formed (9,10). After the pre-miRNA is exported from the cell nucleus to the cytoplasm, it becomes a substrate for Dicer, which forms a complex with TRBP (HIV-1 TAR RNA-binding protein)/PACT (protein activator of PKR) and Ago2 (11–13). Dicer-dependent cleavage is also imprecise (14,15), and two or more miRNA duplex variants are usually formed from each pre-miRNA variant. The miRNA strands of each duplex compete for Ago loading to produce a core of miRNA-induced silencing complex (miRISC) (16,17). The strand loaded into the miRISC preferentially or directly bound to the target mRNA is protected from degradation by cellular ribonucleases (18–20) and is detected at higher levels by small RNA discovery methods (21,22).
Human Dicer also contains two non-identical RNase III domains, RIIIA and RIIIB, a dsRNA-binding domain—dsRBD, a PAZ domain, a platform domain, DUF283 (Domain of unknown function) and an N-terminal putative helicase domain (23–26). Dicer is known to function as a molecular ruler that measures the distance from the pre-miRNA terminus, either the 3′ or 5′ end, which are both docked in the PAZ domain, to the RNase III domain-dependent cleavage site (24,27). The single processing center of human Dicer implies that the cleavage of each RNA strand involves both RNase III domains (26,28). The RNase IIIA domain, which is located closer to the PAZ domain in the protein sequence, cleaves the pre-miRNA 3′ arm that contains the 3′ overhangs and the RNase IIIB domain cleaves the arm that contains the 5′ phosphate (26). The typical Dicer product is an ∼22-nt long duplex that contains two nucleotides 3′ overhangs.
Nearly 2000 human pre-miRNAs are currently known (29) and they differ in their nucleotide sequence and structure: stem length, terminal loop size, type, number and localization of hairpin destabilizing motifs (30). The presence of the secondary structure elements of the stem, particularly asymmetric motifs, may influence the length of the excised miRNAs, i.e. products of different lengths may be generated from different precursors (miRNA length diversity) (14), as well as precision with which Dicer cleavage is performed (31). Imprecise cleavage of the individual pre-miRNAs by Dicer produces a population of miRNAs heterogeneous in length from each precursor's arm (miRNA length heterogeneity) (14). In contrast to the more widely investigated effects of pre-miRNA structure on miRNA biogenesis (14,31–32), fewer studies were devoted to the effects of the RNA sequence on Dicer cleavage site selection (33,34). The predominance of miRNAs having U and A nucleotides at their 5′ end and the rare presence of G nucleotide at this position revealed by deep-sequencing (35,36) was explained by Argonaute binding preferences (37). However, this raised the question of whether Ago loading is the only step implicated in strong discrimination of miRNAs that have a G nucleotide at their 5′ end. Can such discrimination also occur during pre-miRNA cleavage by Dicer? The results from recent bioinformatics studies in which some sequence biases at the Drosha and Dicer cleavage sites were observed are in favor of this possibility (32,38–39).
Here, we analyzed Dicer cleavage in natural pre-miRNAs and artificial hairpin substrates with regular, fully base-paired stem structures. Using these substrates, we aimed to identify factors other than secondary structure motifs, such as internal loops and bulges, that could be implicated in generating miRNA length diversity and heterogeneity. We found that in the absence of such motifs, Dicer generates products of non-uniform length from different substrates and that the degree of heterogeneity of the products released from individual hairpin substrates varies. These observations imply that factors such as the fixed distance measured by Dicer from pre-miRNA termini to its cleavage site (24) and the structural features of pre-miRNAs (14,32) are not the only determinants of Dicer-dependent cleavage site selection and precision. The RNA sequence could also contribute to this process.
MATERIALS AND METHODS
Oligonucleotides
Chemically synthesized RNAs were purchased from Metabion, Integrated DNA Technology and Sigma-Aldrich (Supplementary Table S1). The DNA oligonucleotides used in this study were obtained from IBB (PAS, Warsaw, Poland) (Supplementary Table S2).
Preparation of the RNA substrates
The sequences of the pre-miRNAs were reconstructed based on the rules described in (40). All RNAs were purified by 10% polyacrylamide gel electrophoresis. Prior to the Dicer cleavage assay, the synthetic RNAs were phosphorylated with Optikinase (USB Corp.) in an appropriate buffer. The RNAs were stored at −80°C until use.
Reconstitution of the Dicer–TRBP complex
Recombinant Dicer (1.875 pmole, Genlantis) was incubated with three-fold molar excess of rec. TRBP-HIS-tag (5.6 pmole) (kindly provided by Dr Kretschmer-Kazemi Far) in gel filtration buffer (100 mM KCl, 5% glycerol, 1 mM DTT, 20 mM HEPES, pH 7.5) on ice for 30 min, as described by Doudna (12).
Pull-down assay of Dicer–TRBP complex and western blotting
Prior to Dicer–TRBP complex formation, Dynabeads Protein G (Life Technologies) was used to bind 2 μg anti-TRBP antibody (Santa Cruz sc-514124) for 4 h at room temperature in 0.02% phosphate buffered saline (PBS)-Tween. After washing, 250 μl of reconstituted Dicer–TRBP complex was added to the antibody-conjugated Dynabeads and rotated overnight at 4°C.
Prior to western blotting for Dicer detection, the beads, after washing, were resuspended in a 10 μl of PBS and 10 μl 3× sodium dodecylsulphate (SDS) sample buffer, heated for 5 min at 95°C and loaded onto a 5% Tris-acetate SDS-page (acrylamide:bis = 49:1) in XT Tricine buffer (Bio-Rad) as previously described (41,42). The blots were probed with anti-Dicer antibody (Cell Signaling Technology, 1:1000) and then HRP-conjugated secondary antibody, anti-rabbit (Sigma, 1:1000). The immunoreactions were detected using Westernbright Quantum HRP substrate (Avansta).
For Dicer–TRBP cleavage assay, the pull-down was performed in the same manner as for Dicer detection, except the beads-Ab-TRBP/Dicer complex, after washing, was resuspended in a 20 μl of PBS.
Dicer in vitro cleavage products visualized by northern blotting
The phosphorylated RNAs (50 ng, (∼2.5 pmole)) were cleaved with 0.75 U (0.187 pmole) of Dicer (Genlantis) for 12 min at 37°C, as previously described (14,43) or with 2 μl of immunoprecipitated (10% of total pull-down eluate volume) by the pull-down Dicer–TRBP complex for 15 min at 37°C. The reactions were terminated after the addition of an equal volume of gel loading buffer (7.5 M urea and 20 mM ethylenediaminetetraacetic acid, with dyes).
Northern blotting was performed as previously described (44). Briefly, the RNA substrates and products (25 ng) were separated on a 12% denaturing polyacrylamide gel along with a [γ32P]ATP-labeled 17–25 nt RNA marker, a labeled 10- to 100-nt RNA Low Molecular Weight Marker (USB Corp.) and appropriate phosphorylated M5 and M3 markers (indicated in the figure legends) (Supplementary Table S2). After electrophoresis, the RNAs were transferred to a GeneScreen Plus hybridization membrane (Perkin Elmer). After UV crosslinking and baking at 80°C, the membrane was probed with 32P-labeled DNA oligonucleotides (Supplementary Table S2) that were specifically designed to detect Dicer cleavage products excised from either the 5′ or the 3′ hairpin arm. The radioactive signals were quantified by phosphoimaging (Multi Gauge; Fujifilm).
The product lengths were assayed based on the 17–25 nt end-labeled marker. For few miRNA products, the sequence composition deviated from average of a 50% GC content. Therefore, we calibrated the 17–25 nt RNA marker to the homologous 22-nt-long, labeled synthetic RNAs, whose sequence corresponded to the 22-nt-long product that was derived from the 5′ and 3′ arm of each of the analyzed natural pre-miRNAs (Supplementary Figure S2).
Statistical analysis
The WALDI parameter (Weighted Average Length of Diced RNA) was calculated as previously described (14). In all of the tests in which the P-value was calculated, P ≤ 0.05 was considered significant. Length-heterogeneity was calculated using the equation H = 1 – F max, where H represents the heterogeneity and F max is the fraction of the most abundant product (%). Student's t-test (GraphPad Prism) was used to compare length diversity of the products derived from fully base-paired pre-miRNAs and distorted pre-miRNAs. To generate a bubble-chart graph (MS Excel), we analyzed the lengths of the Dicer cleavage products, and the cleavage intensities were represented as bubble areas [as calculated from the densitometric analysis (Multi Gauge; Fujifilm)]. To calculate the efficiency of the miRNA production by Dicer, we quantified the intensity of the signal for the fraction of the miRNA and the sum of all signals derived from the miRNA, intermediate and precursor in each lane and presented the data as the% of miRNA fraction in Supplementary Figure S1.
RESULTS
miRNAs with length diversity and heterogeneity are released by Dicer from pre-miRNAs with undistorted hairpin stem structures
We first asked whether pre-miRNAs that are devoid of secondary structure motifs in their hairpin stem are similarly cleaved by Dicer to generate predominant miRNA products with the same length from different precursors and show a similar degree of miRNA heterogeneity. To answer this question, we analyzed Dicer cleavage in eight arbitrarily chosen pre-miRNAs, which contained an undistorted stem (20–25 bp long), a 2-nt 3′ overhangs, and a terminal loop of various sizes (8–11 nt). We performed the cleavage assay with recombinant Dicer under identical conditions and analyzed the cleavage products by northern blotting (Figure 1). Hybridization probes were designed to separately detect the miRNAs released by the Dicer RIIIB and RIIIA domains from the 5′ and 3′ pre-miRNA arms, respectively. The predominant product of Dicer cleavage from each pre-miRNA 5′ arm is 22 nt long, with the exception of the pre-miR-548am, which is 21 nt long (Figures 1 and 2). The lengths of major products generated from the 3′ arm of the pre-miRNAs are more diverse: 21 nt for pre-miR-4782, -549, -629 and -548am; and 22 nt for pre-miR-4679, -625, -4779 and -1258. Thus, we observed a considerable length disparity in the major miRNA variants released by the two Dicer RNase III domains.
Figure 1.
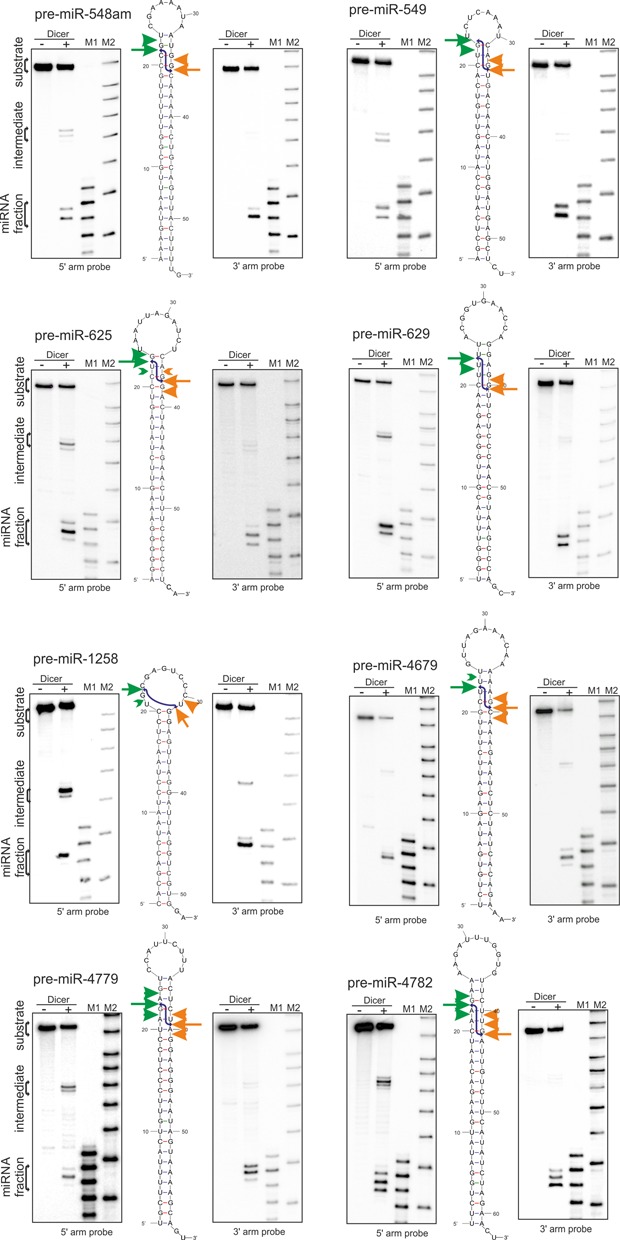
Processing of pre-miRNAs with undistorted stems. The products from the Dicer cleavage assay of the phosphorylated, unlabeled pre-miRNAs with no stem distorting elements were subjected to northern blotting with specific probes detecting the miRNAs derived from the 5′ or 3′ arm, respectively (Supplementary Table S2). In the middle, the structure of the pre-miRNA is marked with arrows representing the cleavages in the 5′ (green) or 3′ arm (orange) of precursors: the predominant cleavage site marked by an arrow, while other cleavages are marked by arrowhead. Signals that accounted for <5% of all of the cleavage products were marked in green (>) or orange (<). The blue lines indicate the duplex resulting from the predominant Dicer cleavage in both the 5′ and 3′ arms of the precursor. (−)-reaction without Dicer, (+)-reaction with Dicer. M1 denotes the end-labeled 17–25-nt RNA marker. M2 denotes the Low Molecular Weight Marker (USB Corp.). The calibration of the M1 marker to the synthetic RNA resembling the 22 nt product of each pre-miRNA arm (Supplementary Table S2) is presented in Supplementary Figure S2.
Figure 2.
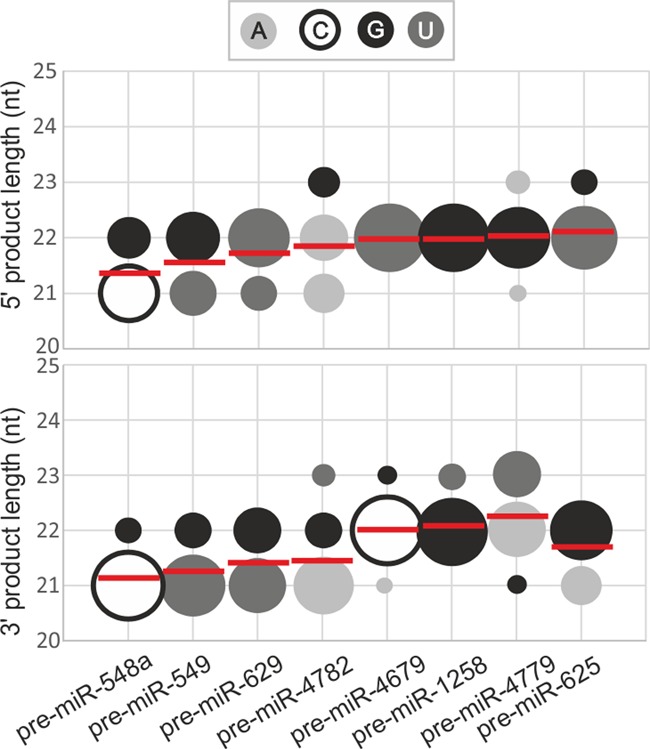
Length diversity and heterogeneity of miRNAs derived from undistorted precursors. A bubble-chart (MS Excel) depicting the fractions of the miRNA products that were generated by Dicer in the 5′ -arm (upper diagram) and 3′ -arm (lower diagram) of the precursors analyzed in Figure 1 (x-axis). The y-axis represents the lengths of the products, and the bubble size (area) represents the fraction of the product that was generated from a given arm (as calculated from the densitometric analysis of the results presented in Figure 1). The colors of the bubbles represent the terminal nucleotide of miRNA being cut by Dicer, as indicated in the legend. The WALDI parameter values for the miRNAs generated from 5′ arm and 3′ arm from each precursor are presented as horizontal red lines. Signals that accounted for <5% of all the cleavage products were not considered in the analysis.
In addition to the most prominent cleavages described above, northern analysis revealed also other cuts at neighboring positions, which resulted from imprecise pre-miRNA cleavage by Dicer. In most cases, there are either two cleavages that have a similar intensity (miR-549–5p, miR-629–3p) or one prevalent cleavage. This results in miRNA fractions that fall within the 21–23 nt length range and components of this fraction are represented by the differently sized bubbles in Figure 2. The mean degree of heterogeneity of the miRNAs is ∼25% for the products derived from each arm of the analyzed pre-miRNAs.
Our northern blotting results also demonstrate that aside from the ∼22 nt miRNA fraction, which represents the majority of Dicer cleavage products, there are ∼35 nt intermediate products (Figure 1) that result from pre-miRNA cleavage by a single RIII domain (11,14,43,45). We observe stronger signals from intermediates generated by pre-miRNA cleavage in the 3′ arm by the Dicer RIIIA domain (products detected by probes specific for the 5′ arm). The intermediates resulting from RIIIB domain activity (detected by probes specific for the 3′ arm miRNAs) are exceptions (e.g., pre-miR-1258–3p). Together, these results suggest that the semi-stable Dicer cleavage intermediates, which were previously described for typical pre-miRNAs with stem structures distorted by various secondary structure motifs (14), are also observed in atypical pre-miRNAs devoid of such motifs.
To analyze whether Dicer substrates with similar overall secondary structures are processed with a similar rate, we quantified the relative efficiency of miRNA production from these precursors (Supplementary Figure S1). Usually, the production of miRNA from one arm prevails; for pre-miR-4782, -549, -1258, -548am and -4779, the 3′ arm products dominate, whereas for pre-miR-4679, -629 and -625, the efficiency of miRNA production is greater from the 5′ arm. Of note, the latter three miRNAs that are produced most efficiently from 5′ arm are all terminated with U nucleotide (Supplementary Figure S1). This observation agrees with results of our bioinformatics data which shows that miRNAs terminate most often at a U residue, regardless of the length of the released products (38).
Nucleotide sequence at Dicer cleavage site influences site selection
When we simulated pairing of major miRNAs derived from the 5′ and 3′ arms of the pre-miRNAs, we found that products generated by major cuts in these arms form a duplex with 2-nt overhangs in five cases (miR-548am, -625, -1258, -4679 and -4779) (blue lines in Figure 1). In three cases (miR-549, -4782 and -629), these duplexes have 3-nt 3′ overhangs. From the latter pre-miRNAs, the major Dicer cleavage generates a 22-nt miRNA from the 5′ arm and a 21-nt miRNA from the 3′ arm. When we examined the nucleotide sequence of the pre-miRNAs from which Dicer cleavages generate these atypical overhangs, we noticed that a specific sequence, 5′-GpN-3′, occurs at their cleavage sites in the 3′ arm. These results suggest that Dicer tends to avoid cleaving at sites that produce miRNAs with a G nucleotide at the 5′ end. When tandem G nucleotides occur at the Dicer cleavage site, as in pre-miR-625 and pre-miR-1258, cleavage leaves G residue at the miRNA 5′ end. This conclusion is supported by the results of bioinformatics studies (32,38–39), which show that miRNAs derived from the 3′ arm of the pre-miRNA very rarely start with a G nucleotide. This result suggests that the nucleotide sequence at Dicer cleavage site is indeed involved in cleavage site selection.
Sequence modifications of pre-miRNA change length diversity and heterogeneity of miRNA
To provide further support for the effect of the nucleotide sequence on Dicer processing, we analyzed Dicer-generated cleavage in pre-miRNA mutants. We addressed the question of whether we can change the diversity and heterogeneity of miRNAs or the efficiency of pre-miRNA processing. We created sequence mutants of four pre-miRNAs, pre-miR-549, -4679, -4779 and -1258, in which we changed the nucleotide sequence at or near the Dicer cleavage sites, as shown in Figure 3A and Supplementary Figure S3. We performed a cleavage assay with Dicer followed by northern blotting.
Figure 3.
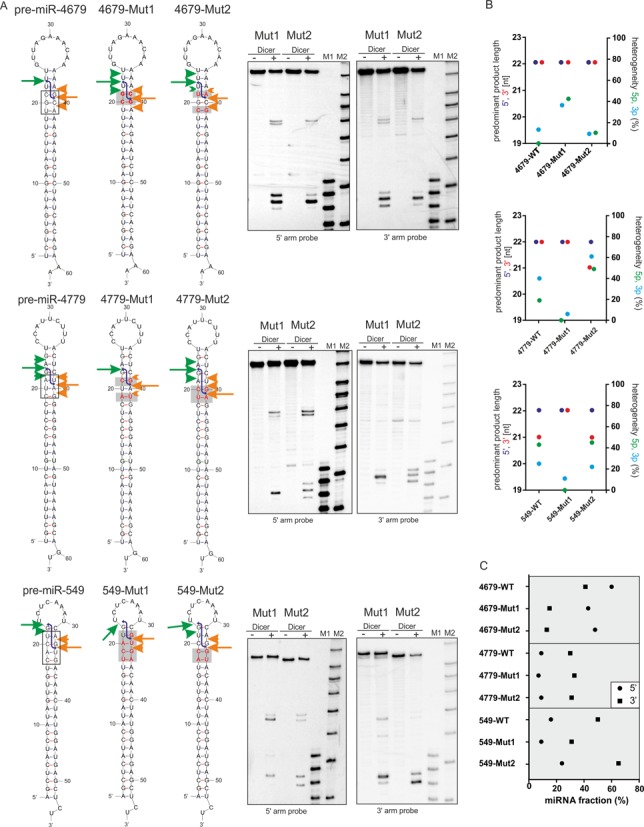
Dicer cleavages in the pre-miRNAs mutants. (A) Recombinant Dicer-mediated cleavage of the mutant variants of pre-miR-4679, -4779 and -549 were visualized by northern blotting (on the right). The cleavage sites are marked by arrows and arrowheads as indicated in Figure 1. The black frame in the WT precursor models marks the RNA fragment that were mutated. The red letters in the sequence of mutants and the gray rectangle mark the positions mutated compared to the WT precursors. Other designations are as described in the legend for Figure 1. (B) Sequence mutations in the Dicer cleavage sites of the pre-miRNA influence the diversity and heterogeneity of the miRNAs. Left Y- axis—the length of the predominant miRNA product generated by Dicer from the 5′ (violet) or 3′ (red) arm of the pre-miRNAs and their mutant variants, based on the data presented in Figures 1 and 3A. Right Y-axis—the heterogeneity (%) of the products that were generated by Dicer from the 5′ arm (green) and 3′ arm (blue) of pre-miRNAs and their mutant variants was calculated based on the data presented in Figures 1 and 3A. Signals that accounted for <5% of all the cleavage products were not considered in these analyses. (C) The processing efficiency of the pre-miRNAs was calculated separately for the miRNAs derived from 5′ arm and 3′ arm (see inset) of the pre-miRNA and their mutant variants, based on the results of northern blotting. The signal for a miRNA fraction was compared to the signals derived from the entire lane and expressed as the % of miRNA fraction.
pre-miR-4679 is one of the pre-miRNAs from which RNase Dicer generated miRNAs most effectively (Supplementary Figure S1). The mutations introduced in 4679-Mut1 and 4679-Mut2 (Figure 3A), substantially decreased the miRNA production from 3′ arm of the mutants (Figure 3C) without changing the length of the released products (Figure 3B). However, only mutations in Mut1 increased the heterogeneity of released miRNAs, up to ∼40% from both pre-miRNA arms (Figure 3B). This set of RNAs shows that the introduction of G nucleotides at positions 21 or 22 in the 3′ arm of pre-miRNA decreases the Dicer processing efficiency, as measured by the amount of miRNA generated from that arm.
The dinucleotide change introduced in 1258-Mut1, and the single-nucleotide changes in 1258-Mut2 and 1258-Mut3 did not affect much the length of the released miRNAs (Supplementary Figure S3A and B). However, the precision of Dicer cleavage in both pre-miRNA arms in 1258-Mut3 was slightly improved, as measured by the degree of miRNA heterogeneity (Supplementary Figure S3B). As for the efficiency, each mutant was cleaved only slightly less effectively compared with 1258-WT (Supplementary Figure S3C).
pre-miR-4779 (hereafter named 4779-WT) was one of the worst substrates for Dicer, as measured by the amount of the miRNA fraction released from the pre-miRNA (Supplementary Figure S1). To determine whether we can change its processing efficiency and specificity, we created 4779-Mut1 and 4779-Mut2 (Figure 3A). These mutations did not improve the processing efficiency (Figure 3C) but drastically changed the heterogeneity of the miRNAs derived from these precursors (Figure 3B). The mutations in 4779-Mut1 generated nearly homogeneous miRNAs from both arms, whereas the mutations in 4779-Mut2 increased the miRNA heterogeneity to ∼50% (Figure 3B). Additionally, the major miRNA variant excised by Dicer from the 3′ pre-miRNA arm of 4779-Mut2 was 1 nt shorter (21 nt) than from 4779-WT (Figure 3B), which resulted in a duplex with a 3-nt 3′ overhangs. This effect can be explained by the G nucleotide introduced at position 22 from the 3′ end of 4779-Mut2. This position is occupied by an A nucleotide in 4779-WT which is preferentially cleaved.
pre-miR-549 (hereafter named 549-WT) was one of the precursors that produced a duplex with 3-nt 3′ overhangs from the major cleavage events in both arms (Figure 1). To determine whether the nucleotide sequence changes at the Dicer cleavage site can shift the major cleavage site, we generated 549-Mut1 and 549-Mut2 (Figure 3A). The heterogeneity and diversity of products released from Mut2 was comparable to those derived from 549-WT (Figure 3B), with slightly improved efficiency of miRNA production (Figure 3C). Analyses of miRNA heterogeneity demonstrated that Dicer generates products of lower heterogeneity from both arms of 549-Mut1 than from 549-WT, with homogeneous miRNAs being produced from the 5′ pre-miRNA arm in 549-Mut1 (Figure 3B). More importantly, we also observed a change in the length of the products released from the 3′ arm of 549-Mut1. For 549-Mut1, the predominant product is 1 nt longer (22 nt) than for 549-WT (21 nt), which produces now a duplex with a 2-nt 3′ overhangs. This effect may be explained by the fact that Dicer cleaves at GpU in the 3′ pre-miRNA arm of 549-WT to have U nucleotide at the miRNA 5′ end. When G nucleotide was shifted by 1 nt in the 3′ arm of 549-WT from position 22 into position 21 in 549-Mut1, Dicer still preferred to cleave at GpU and thereby generate a miRNA with a U nucleotide at its 5′ end.
These results support the notion that the nucleotide sequence at the Dicer cleavage site contributes to cleavage site selection, as shown for 549-Mut1 and 4779-Mut2. Additionally, we observed that changing a few nucleotides at Dicer cleavage site influenced the heterogeneity of the products released by Dicer, by either increasing (4779-Mut2) or decreasing the heterogeneity of the miRNAs (4779-Mut1, 549-Mut1 and 1258-Mut3).
Dicer cleavage of specifically designed hairpin substrates supports the role of the nucleotide sequence in cleavage site selection
To gain further insight into the role of nucleotide sequence in Dicer cleavage site selection, we designed a series of four model hairpin substrates, with stems composed of the repeated tetranucleotide sequence motif AGUC. All of the analyzed model hairpins had the same terminal loop and 2-nt overhang sequence derived from natural pre-miR-496, but the stem composed of the same tetranucleotide motifs was shifted in frame in each of the four RNAs (Figure 4A). Each hairpin started with a different nucleotide; consequently, position 22 (counting from pre-miRNA termini), which is most frequently cleaved by Dicer, would be occupied by G in A-UC, by A in C-UC, by U in G-UC and by C in U-UC in both arms. This approach to Dicer substrate design allowed us to more systematically analyze the nucleotide sequence preference at Dicer cleavage sites. We examined this set of artificial Dicer substrates with recombinant Dicer followed by northern blotting with specific probes to the 5′ and 3′ arms (Figure 4B). Two probes were designed, one for each hairpin arm, that were universal for all analyzed RNAs (Supplementary Figure S4).
Figure 4.

Northern blotting analysis to trace Dicer cleavage sites in hairpin RNAs with shifted, repeated sequences. (A) The hairpin RNAs subjected to Dicer cleavage assay were shown with the positions of the Dicer cleavage sites marked by arrows. The positions of the G residues at the Dicer cleavage site are marked in red. Other designations are as in Figure 1. (B) RNAs with 2-nt 3′ overhangs were subjected to the in vitro Dicer cleavage assay (lane +) followed by northern blotting analysis with probes detecting the products from the 5′ arm (left) or 3′ arm (right). As a control, RNA incubations were also performed in the absence of Dicer (lane −). Appropriate markers were included; M5 and M3 represent the unlabeled 22-nt homologous markers for RNA products derived from the 5′ arm or 3′ arm, respectively. M1 denotes the end-labeled 17–25 nt RNA marker; M2 denotes a Low Molecular Weight Marker (USB Corp.). The calculated WALDI parameter is shown below each autoradiogram, as well as a quantitative representation of the RNA length variants using the peaks obtained from the phosphoimaging analysis, with dotted lines indicating specific lengths. Other designations are as in Figure 1. (C) The hairpin models show sequence mutations introduced in the U-UC RNA. The sequences that were mutated in the U-UC RNA are marked with gray rectangles. The Dicer cleavage sites are shown on the secondary structures of the RNA, based on the quantification of the northern blotting results which are presented next to the structures. Other designations are as in Figure 4A and B.
The cleavages generated by the Dicer RNase IIIA domain did not show identical patterns in the series of RNAs with a 2-nt overhangs (Figure 4B, right). The predominant cleavage site in the 3′ arm occurred after A22 in C-UC, after U22 in G-UC, after C22 in U-UC and, exceptionally, after A21 in A-UC (counting from the 3′ end of the hairpin). The last RNA hairpin contained a G nucleotide at position 22, which was cleaved considerably less efficiently than other nucleotides occupying this position. Similarly, different cleavage patterns were generated by the Dicer RNase IIIB domain in the 5′ arm of these hairpin substrates (Figure 4B, left). Again, the most prominent cuts occurred after A22 in C-UC, after U22 in G-UC, after C22 in U-UC and after A21 in A-UC (counting from the 5′ end of the hairpin). In this series, the main cut after residue 21 also occurred only when position 22 was occupied by a G residue. These results demonstrate that the nucleotide sequence of the stem portion of an RNA substrate has a significant effect on the selection of the scissile phosphodiester bond and that G residues located in the cleavage region may be considered as antideterminants for Dicer cleavage. It appears that in RNAs containing a G residue at nucleotide position 21 or 22, the RNase IIIA and RNase IIIB domains tend to skip these residues and cleave at the neighboring residues.
To verify the hypothesis that Dicer selects the cleavage site based on the nucleotide sequence, we designed six mutants of a U-UC model (Figure 4C). We changed the base-pairs around the cleavage site to observe the sequence-dependent effects on Dicer cleavage. Mutants Mut1, Mut2 and Mut3 (Figure 4C) exhibited changes in the WT U-UC sequence by introducing a G nucleotide at positions 21, 22 and 23 in the 5′ arm and a corresponding mutation on the opposite strand to maintain base-pairing. Other mutants (Figure 4C) had G residues introduced in the 3′ arm of RNA hairpin at position 22 (Mut4), a G nucleotide removed from position 23 (Mut5) and a G nucleotide introduced in position 21 (Mut6). This design disrupted staggered appearance of the same nucleotides on opposite sides of the strand, which were shifted by a 2-nt distance in the RNA hairpins presented in Figure 4A.
Mut1, Mut2 and Mut3 (Figure 4C) have a G nucleotide introduced in the 5′ arm of the RNA hairpin and show only minor changes in the diversity of the 5′ arm products released by the RIIIB domain compared with WT U-UC, as measured by the Weighted Average Length of Diced RNA (WALDI parameter) (14,43). The predominant Dicer cleavage produces a 22-nt-long product. In the 3′ arm of these RNAs, a Dicer cleavage site is shifted, which results in 1 nt shorter products from Mut1 and Mut3 (21 nt) compared with WT U-UC (22 nt), which has its consequence in decreased WALDI values from 21.9 nt in WT U-UC to 21.34 nt in Mut1 and 21.7 nt in Mut3 (Figure 4C). When there is no G residue restriction at the Dicer cleavage site in the 3′ arm of the pre-miRNA, Dicer often prefers to cleave RNAs at positions that leave an miRNA starting with a U residue. This shows that sequence restrictions at the Dicer cleavage site are stronger in the 3′ arm of the RNA hairpin, which corroborates the results obtained for pre-miRNAs (Figure 1). The heterogeneity of Dicer products was changed most significantly for products derived from Mut3, where the introduction of a G at position 23 in the 5′ arm increased heterogeneity up to 47%. For Mut4, Mut5, Mut6 (Figure 4C), the RIIIB domain also produces the same length (22 nt) of miRNA products, regardless of the sequence at the Dicer cleavage sites, with slightly changed heterogeneity. In the 3′ pre-miRNA arm of Mut4, Mut5 and Mut6, the RIIIA domain generates products with more diverse lengths: 22 nt for Mut4 and Mut6 and 21 nt for Mut5. The results presented for this set of mutants show that cleavage in the 3′ arm of RNA occurs preferentially at a U residue and omits a G residue.
To provide further support for the contribution of the nucleotide sequence at the Dicer cleavage site to the selection of the cleaved bond, we investigated an additional two sets of RNAs, which were designed as those described above (Figure 4A) but had either 1-nt or 3-nt overhangs (Figure 5A,B). In a set of RNAs with a 3-nt overhangs (Figure 5A), the Dicer RIIIA domain produces a major 22-nt-long product for A-UCC and C-UCC. A 23-nt-long product is generated by RIIIA for G-UCC to avoid the production of miRNA starting with a G nucleotide as a consequence of cleavage in the NpG sequence. In a set of RNAs with a 1-nt overhang (Figure 5B), RIIIA also escapes cleavage in the NpG sequence in a C-U model, where a 21-nt-long product is generated compared with the longer (22 and 23 nt) predominant products in the A-U, G-U, U-U RNA hairpins. These analyses demonstrate that regardless of the length of the 3′ overhangs (1, 2 and 3 nt), cleavage by the Dicer RNase domains generates a population of products with heterogeneous ends and that the major cleavage products do not contain G residues at the 5′ end.
Figure 5.
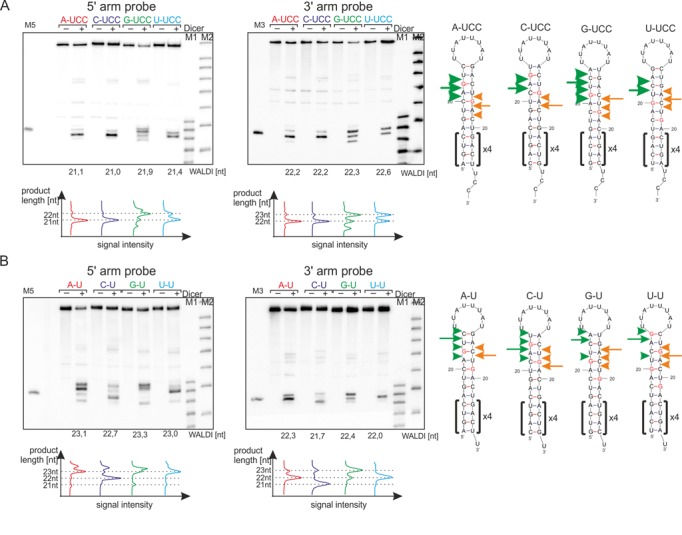
The effect of the sequence at Dicer cleavage site and overhangs length on the length of the released products. RNAs with 3-nt 3′ overhangs (A) and 1-nt 3′ overhangs (B) were subjected to the in vitro Dicer cleavage assay (lane +) followed by northern blotting analysis, with probes detecting the products from the 5′ arm (left) or 3′ arm (right). As a control, RNA incubations were also performed in the absence of Dicer (lane −). Appropriate markers were included; M5 and M3 represent the unlabeled 22-nt homologous markers for the RNA products derived from the 5′ arm or 3′ arm, respectively. M1 denotes the end-labeled 17–25 nt RNA marker; M2 denotes a Low Molecular Weight Marker (USB Corp.). The calculated WALDI parameter is shown below each autoradiogram, as well as a quantitative representation of the RNA length variants using the peaks obtained from the phosphoimaging analysis, with dotted lines indicating specific lengths. The Dicer cleavage sites were marked on the hairpin RNAs presented aside. Other designations are as in Figure 4.
Effect of TRBP on specificity of Dicer cleavages
Having characterized Dicer-induced cleavages in fully base-paired pre-miRNAs and artificial hairpin substrates we set out to determine to what extent TRBP complexed with Dicer affects miRNA length diversity and heterogeneity. We reconstituted Dicer–TRBP complex from recombinant proteins as described by Doudna (12), and this was followed by pull-down with TRBP antibody (Figure 6A). The presence of Dicer in a pull-down fraction was demonstrated by western blotting (Figure 6B). Cleavages induced in these substrates by Dicer alone and Dicer–TRBP complex were analyzed by northern blotting (Figure 6C, Supplementary Figure S5).
Figure 6.
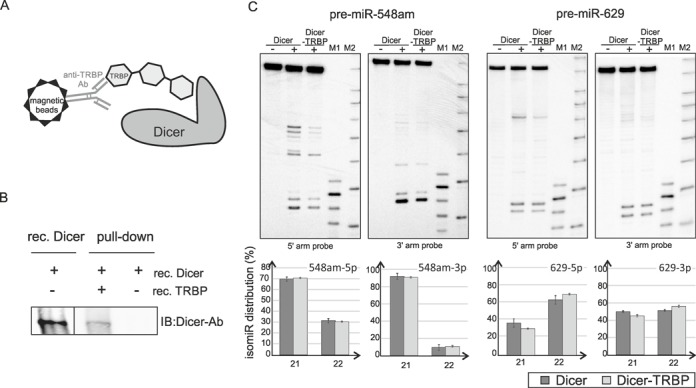
Dicer–TRBP induces cleavage in fully base-paired pre-miRNAs. (A) Schematic overview of the pull-down experiment for Dicer–TRBP complex isolation. (B) Western blotting analysis of the Dicer pull-down experiment. The indicated recombinant proteins were mixed and incubated for 30 min on ice, then incubated with beads bound with anti-TRBP specific antibody at 4°C overnight and eluted with SDS sample buffer. Eluted proteins (a half of the pull-down eluate volume) and recombinant Dicer (0.187 pmole), were analyzed by western blotting with an anti-Dicer specific antibody. (C) Dicer/TRBP cleavage assay of the fully base-paired pre-miRNAs followed by northern blotting. Recombinant Dicer (0.2U, 0.05 pmole) and reconstituted Dicer/TRBP complex (10% of the pull-down eluate volume) were used for pre-miRNA (∼2.5 pmole) cleavage for 15 min at 37°C. Below, the distribution of isomiRs generated by Dicer or Dicer in complex with TRBP in the 5′ and 3′ arms of the precursors. Data are mean ± SD for two independent experiments. Other designations are as described in the legend for Figure 1.
We found that the presence of TRBP does not considerably alter the length diversity of miRNAs released by Dicer from analyzed pre-miRNAs, as shown by densitometric quantification of signal intensity of miRNAs (Figure 6C, Supplementary Figure S5). The main cleavage site of Dicer in analyzed pre-miRNAs remains unchanged. With regard to length heterogeneity of miRNAs released from individual precursors, only small changes (up to 6%) were observed; the TRBP has the strongest effect on pre-miR-629 cleavage (Figure 6C). The sensitivity to nucleotide sequence at the Dicer cleavage site is retained in the presence of TRBP.
DISCUSSION
Numerous human miRNA hairpin precursors differ in their nucleotide sequences and structural features, and this variability creates a highly diverse group of Dicer substrates. Nevertheless, a number of pre-miRNAs is completely devoid of stem structure distorting elements. One might anticipate that in the absence of structure destabilizing motifs within the pre-miRNA stem, Dicer will produce miRNAs of the same length from the different precursors, which is determined solely by the distance between the PAZ domain and the RIII domains (24). The results of this study demonstrate that miRNAs of diverse length are consistently generated from different fully base-paired precursors, and their length diversity is greater for products derived from the 3′ arm of pre-miRNA compared to those derived from the 5′ arm. When we compared the miRNA diversity parameter for products derived from fully base-paired pre-miRNAs with those derived from pre-miRNAs distorted by secondary structure motifs analyzed in our previous study (14), we noted that the range of miRNA diversity is greater for the latter group (Supplementary Figure S6A). This could be explained the fact that asymmetric structural motifs within the pre-miRNA stem greatly increase miRNA length diversity (14).
Our current study also revealed that Dicer excises a population of heterogeneous miRNAs from fully base-paired pre-miRNAs. Moreover, the level of heterogeneity varies between miRNA products from pre-miRNAs with similar secondary structure. When we compared the heterogeneity of all of the products generated from the fully base-paired pre-miRNAs (Figures 1 and 2) and the pre-miRNAs distorted with secondary structure motifs (14), we observed heterogeneous products in both groups, but the mean heterogeneity was lower for products derived from fully base-paired pre-miRNAs (Supplementary Figure S6B). These results indicated other features of pre-miRNAs, which are different than the pre-miRNA structure that can influence the selection of the Dicer cleavage site and cleavage precision. As the analyzed in this study pre-miRNAs differed in their nucleotide sequence, we hypothesized that the pre-miRNA sequence also affects Dicer cleavage site selection, precision and efficiency (Figure 7). This hypothesis is supported by the results from other authors who studied the sequence specificity of other types of RNase III enzymes (46–48).
Figure 7.
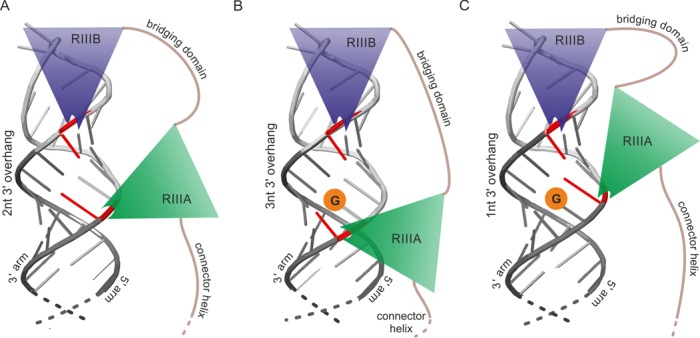
Schematic representation of the Dicer RNase III domains cleaving the pre-miRNA which fragment consisting of Dicer cleavage site and a terminal loop is shown. (A) RIIIA and RIIIB domains activity produces a duplex having standard 2 nt 3′ overhang. In red are marked terminal miRNA nucleotides resulting from Dicer cleavage. (B and C) Sequence-sensitive cleavage by RIIIA domain avoids leaving G residue at 5′ end of miRNA what results in the formation of duplexes having non-standard 3′ overhangs: 3 nt (B) or 1 nt (C).
Our analyses of natural pre-miRNAs devoid of secondary structure motifs within the stem and their mutants revealed that we could modulate the diversity and heterogeneity of the miRNAs and the cleavage efficiency of the pre-miRNAs by changing the sequence at Dicer cleavage site (Figure 3). Surprisingly, the response of each RNase III domain to sequence restrictions at Dicer cleavage site is different and the RIIIA domain exhibits greater sequence restrictions compared to the RIIIB domain. Although both domains possess consensus clusters of acidic residues that are responsible for cleavage of the phosphodiester bonds (26), the sequence identity between these domains is low (∼17%), which suggests that they may have different cleavage properties. Our data are in accord with the results from other authors (49), who demonstrated the existence of several patches of positively charged residues on the surface of Dicer, particularly positioning loop residing within the RNase IIIA domain, which is essential for Dicer activity. Together, these data imply that the RNase IIIA domain may recognize a nucleotide sequence at the Dicer cleavage site. Specifically, the Dicer RIIIA domain is reluctant to cleave an NpG sequence in both natural pre-miRNAs and artificial hairpin structures, which will produce miRNA starting with G nucleotide and it prefers to cleave at positions that result in miRNAs that start with a U nucleotide (Figure 7).
The predominant RNase III domain-mediated cleavage products form a duplex with 2-nt overhangs. However, sequence restrictions at the Dicer cleavage site force Dicer to shift the cleavage position in the 3′ arm of the pre-miRNA, which results in the formation of a duplex with 3-nt overhangs. This sheds new light on the cooperation between the two RNase III domains that form the single catalytic center. It implies that the intramolecular pseudodimer formed by the two RIII domains (26) is not a rigid structure which forms only 2-nt overhangs in the cleavage products. These domains may have some ability to move relative to each other, thus resulting in different lengths of the 3′ overhangs in the released duplex (Figure 7).
Previous reports have shown that sequence bias exists at the miRNA termini (32,35–36,38,39) and at the sequence neighboring the Drosha and Dicer cleavage sites (32,38). These observations are derived from miRNA deep-sequencing data. It is known that processes other than imprecise cleavages by Drosha and Dicer may affect the sequence at the miRNA termini in cells. These include Ago2 loading bias and 3′ end modification events (15,37). The advantage of using an in vitro Dicer cleavage assay is that the cellular processes that occur downstream of Dicer cleavage can be disregarded and that the potential of Dicer alone to generate heterogeneous miRNAs of diverse length can thus be evaluated. Using this experimental system, we demonstrated that sole Dicer recognizes the sequence at the cleavage site and, consequently, responds to the changes in cleavage site sequence by producing heterogeneous miRNAs diverse in lengths. Using a specific group of substrates with no stem distorting motifs, we could evaluate the effect of the pre-miRNA sequence on Dicer processing, without considerable influence of the pre-miRNA structure. The lack of a considerable effect of TRBP on Dicer cleavage site selection in the analyzed fully-base paired pre-miRNAs might reflect the fact that the TRBP effect has been shown for only a small fraction of sensitive pre-miRNAs (50,51). We speculate that atypical pre-miRNAs with fully base-paired stems might not be sensitive to TRBP, which senses structural imperfections in miRNA precursors, thereby helping to establish the cut site position for cleavage by Dicer (52).
From a functional perspective, it appears from our study that miRNA selection by Argonaute (37) is not the only step that decreases the functionality of miRNAs having a G residue at the 5′ end: Dicer cleavage also contributes to this sequence-specific selection. Thus, the auxiliary miRNA-sorting role of mammalian Argonautes allows these proteins to act as the final gate-keepers for functional miRNAs, which are then delivered to the target mRNAs. The low frequency of Dicer cleavage at G residues places fewer demands on Argonautes for their miRNA sorting activity. We hypothesize that the driving force for these discriminative processes could be the efficient loading of Argonautes with miRNA guide strands to increase the efficiency of miRISC driven regulatory processes in cells.
Supplementary Material
Acknowledgments
We thank Dr Edyta Koscianska for her invaluable help and discussions. We also thank Dr Kretschmer-Kazemi Far for providing TRBP protein.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Center [2011/03/B/NZ1/03259 to W.J.K; 2013/11/N/NZ1/02429 to P.G.-M.]. Ministry of Science and Higher Education of the Republic of Poland, from the quality-promoting subsidy, under the Leading National Research Centre (KNOW) programme for the years 2014–2019. Funding for open access charge: National Science Center [ 2014/15/B/NZ1/01880 to W.J.K].
Conflict of interest statement. None declared.
REFERENCES
- 1.Wilson R.C., Doudna J.A. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammond S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 4.Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 6.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 7.Davis B.N., Hilyard A.C., Lagna G., Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 9.Ma H., Wu Y., Choi J.G., Wu H. Lower and upper stem-single-stranded RNA junctions together determine the Drosha cleavage site. Proc. Natl. Acad. Sci. U.S.A. 2013;110:20687–20692. doi: 10.1073/pnas.1311639110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu H., Ye C., Ramirez D., Manjunath N. Alternative processing of primary microRNA transcripts by Drosha generates 5′ end variation of mature microRNA. PloS One. 2009;4:e7566. doi: 10.1371/journal.pone.0007566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koscianska E., Starega-Roslan J., Krzyzosiak W.J. The role of Dicer protein partners in the processing of microRNA precursors. PloS One. 2011;6:e28548. doi: 10.1371/journal.pone.0028548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacRae I.J., Ma E., Zhou M., Robinson C.V., Doudna J.A. In vitro reconstitution of the human RISC-loading complex. Proc. Natl. Acad. Sci. U.S.A. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregory R.I., Chendrimada T.P., Cooch N., Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 14.Starega-Roslan J., Krol J., Koscianska E., Kozlowski P., Szlachcic W.J., Sobczak K., Krzyzosiak W.J. Structural basis of microRNA length variety. Nucleic Acids Res. 2011;39:257–268. doi: 10.1093/nar/gkq727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neilsen C.T., Goodall G.J., Bracken C.P. IsomiRs–the overlooked repertoire in the dynamic microRNAome. Trends Genet. 2012;28:544–549. doi: 10.1016/j.tig.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Kawamata T., Yoda M., Tomari Y. Multilayer checkpoints for microRNA authenticity during RISC assembly. EMBO Rep. 2011;12:944–949. doi: 10.1038/embor.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noland C.L., Ma E., Doudna J.A. siRNA repositioning for guide strand selection by human dicer complexes. Mol. Cell. 2011;43:110–121. doi: 10.1016/j.molcel.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kai Z.S., Pasquinelli A.E. MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat. Struct. Mol. Biol. 2010;17:5–10. doi: 10.1038/nsmb.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janas M.M., Wang B., Harris A.S., Aguiar M., Shaffer J.M., Subrahmanyam Y.V., Behlke M.A., Wucherpfennig K.W., Gygi S.P., Gagnon E., et al. Alternative RISC assembly: binding and repression of microRNA-mRNA duplexes by human Ago proteins. RNA. 2012;18:2041–2055. doi: 10.1261/rna.035675.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flores O., Kennedy E.M., Skalsky R.L., Cullen B.R. Differential RISC association of endogenous human microRNAs predicts their inhibitory potential. Nucleic Acids Res. 2014;42:4629–4639. doi: 10.1093/nar/gkt1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burroughs A.M., Ando Y., de Hoon M.J., Tomaru Y., Suzuki H., Hayashizaki Y., Daub C.O. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011;8:158–177. doi: 10.4161/rna.8.1.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 24.Macrae I.J., Zhou K., Li F., Repic A., Brooks A.N., Cande W.Z., Adams P.D., Doudna J.A. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H., Kolb F.A., Brondani V., Billy E., Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H., Kolb F.A., Jaskiewicz L., Westhof E., Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 27.Park J.E., Heo I., Tian Y., Simanshu D.K., Chang H., Jee D., Patel D.J., Kim V.N. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gan J., Tropea J.E., Austin B.P., Court D.L., Waugh D.S., Ji X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. doi: 10.1016/j.cell.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 29.Kozomara A., Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozlowski P., Starega-Roslan J., Legacz M., Magnus M., Krzyzosiak W.J. Structures of microRNA precursors. In: Ying S-Y, editor. Current Perspectives in microRNAs (miRNA) Houten: Springer; 2008. pp. 1–16. [Google Scholar]
- 31.Gu S., Jin L., Zhang Y., Huang Y., Zhang F., Valdmanis P.N., Kay M.A. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warf M.B., Johnson W.E., Bass B.L. Improved annotation of C. elegans microRNAs by deep sequencing reveals structures associated with processing by Drosha and Dicer. RNA. 2011;17:563–577. doi: 10.1261/rna.2432311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vermeulen A., Behlen L., Reynolds A., Wolfson A., Marshall W.S., Karpilow J., Khvorova A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11:674–682. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiNitto J., Wang L., Wu J. Continuous fluorescence-based method for assessing dicer cleavage efficiency reveals 3′ overhang nucleotide preference. Biotechniques. 2010;48:303–311. doi: 10.2144/000113402. [DOI] [PubMed] [Google Scholar]
- 35.Seitz H., Tushir J.S., Zamore P.D. A 5′-uridine amplifies miRNA/miRNA* asymmetry in Drosophila by promoting RNA-induced silencing complex formation. Silence. 2011;2:4. doi: 10.1186/1758-907X-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu H.Y., Yan Z., Xu Y., Hu H., Menzel C., Zhou Y.H., Chen W., Khaitovich P. Sequence features associated with microRNA strand selection in humans and flies. BMC Genomics. 2009;10:413. doi: 10.1186/1471-2164-10-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frank F., Sonenberg N., Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 38.Starega-Roslan J., Witkos T.M., Galka-Marciniak P., Krzyzosiak W.J. Sequence features of Drosha and Dicer cleavage sites affect the complexity of isomiRs. Int. J. Mol. Sci. 2015;16:8110–8127. doi: 10.3390/ijms16048110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Humphreys D.T., Hynes C.J., Patel H.R., Wei G.H., Cannon L., Fatkin D., Suter C.M., Clancy J.L., Preiss T. Complexity of murine cardiomyocyte miRNA biogenesis, sequence variant expression and function. PLoS One. 2012;7:e30933. doi: 10.1371/journal.pone.0030933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krol J., Starega-Roslan J., Milanowska K., Nowak D., Kubiaczyk E., Nowak M., Majorek K., Kaminska K., Krzyzosiak W.J. Structural features of microRNAs and their precursors. In: Clarke N, Sanseau P, editors. microRNA: Biology, Function & Expression. Eagleville: DNA Press; 2006. pp. 95–110. [Google Scholar]
- 41.Mosca N., Starega-Roslan J., Castiello F., Russo A., Krzyzosiak W.J., Potenza N. Characterization of a naturally occurring truncated Dicer. Mol. Biol. Rep. 2015;42:1333–1340. doi: 10.1007/s11033-015-3878-6. [DOI] [PubMed] [Google Scholar]
- 42.Fiszer A., Mykowska A., Krzyzosiak W.J. Inhibition of mutant huntingtin expression by RNA duplex targeting expanded CAG repeats. Nucleic Acids Res. 2011;39:5578–5585. doi: 10.1093/nar/gkr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Starega-Roslan J., Krzyzosiak W.J. Analysis of microRNA length variety generated by recombinant human Dicer. Methods Mol. Biol. 2013;936:21–34. doi: 10.1007/978-1-62703-083-0_2. [DOI] [PubMed] [Google Scholar]
- 44.Koscianska E., Starega-Roslan J., Czubala K., Krzyzosiak W.J. High-resolution northern blot for a reliable analysis of microRNAs and their precursors. ScientificWorldJournal. 2011;11:102–117. doi: 10.1100/tsw.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flores-Jasso C.F., Arenas-Huertero C., Reyes J.L., Contreras-Cubas C., Covarrubias A., Vaca L. First step in pre-miRNAs processing by human Dicer. Acta Pharmacol. Sin. 2009;30:1177–1185. doi: 10.1038/aps.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamontagne B., Ghazal G., Lebars I., Yoshizawa S., Fourmy D., Elela S.A. Sequence dependence of substrate recognition and cleavage by yeast RNase III. J. Mol. Biol. 2003;327:985–1000. doi: 10.1016/s0022-2836(03)00231-6. [DOI] [PubMed] [Google Scholar]
- 47.Glow D., Pianka D., Sulej A.A., Kozlowski L.P., Czarnecka J., Chojnowski G., Skowronek K.J., Bujnicki J.M. Sequence-specific cleavage of dsRNA by Mini-III RNase. Nucleic Acids Res. 2015;43:2864–2873. doi: 10.1093/nar/gkv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang K., Nicholson A.W. Regulation of ribonuclease III processing by double-helical sequence antideterminants. Proc. Natl. Acad. Sci. U.S.A. 1997;94:13437–13441. doi: 10.1073/pnas.94.25.13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacRae I.J., Zhou K., Doudna J.A. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- 50.Wilson R.C., Tambe A., Kidwell M.A., Noland C.L., Schneider C.P., Doudna J.A. Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol. Cell. 2015;57:397–407. doi: 10.1016/j.molcel.2014.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fukunaga R., Han B.W., Hung J.H., Xu J., Weng Z., Zamore P.D. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell. 2012;151:533–546. doi: 10.1016/j.cell.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Acevedo R., Orench-Rivera N., Quarles K.A., Showalter S.A. Helical defects in microRNA influence protein binding by TAR RNA binding protein. PLoS One. 2015;10:e0116749. doi: 10.1371/journal.pone.0116749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


