Abstract
Polyomaviruses induce cell proliferation and transformation through different oncoproteins encoded within the early region (ER): large T antigen (LT), small T antigen (sT) and, in some cases, additional components. Each virus utilizes different mechanisms to achieve transformation. For instance, the LTs of Simian virus 40 (SV40), BK and/or JC virus can induce transformation; but Merkel Cell Polyomavirus (MCPyV) requires expression of sT. Lymphotropic Papovavirus (LPV) is closely related to Human Polyomavirus 9 (HuPyV9) and, under similar conditions, mice expressing LPV.ER exhibit higher rates of tumor formation than mice expressing SV40.ER. We have investigated the contributions of individual LPV.ER components to cell transformation. In contrast to SV40, LPV.ER transforms mouse embryonic fibroblasts (MEFs), but expression of LPV LT is insufficient to transform MEFs. Furthermore, LPV sT induces immortalization and transformation of MEFs. Thus, in the case of LPV, sT is the main mediator of oncogenesis.
Keywords: LPV, transformation, sT, polyomavirus, SV40
INTRODUCTION
Lymphotropic Papovavirus (LPV) is a primate polyomavirus originally isolated from the B-lymphoblast cells of African green monkey (1), and later found to grow in B-lymphoblast cells of human and primate origin (2, 3). About 30% of normal human adults contain antibodies recognizing LPV or a similar virus (3).
LPV is similar to other known polyomaviruses in terms of genome size, coding potential and genome organization (4, 5). Based on sequence homology of the LT proteins, LPV is a distant relative of both SV40 and MuPyV, but is closely related to the recently discovered human HuPy9 (6). The ER from LPV possesses a high capacity of inducing cellular transformation, both in cell culture and in vivo systems. For instance, the infection of hamster embryonic cells with LPV induces anchorage independence and allows the cells to grow in low serum, and LPV transformed cells are tumorigenic in newborn hamsters (7)(8). LPV.ER is also capable of inducing tumorigenesis in transgenic animals (9)(10), even to greater extent than SV40.ER. For instance, transgenic mice expressing either LPV.ER or SV40.ER under the same promoter show robust neoplastic growth in the choroid plexus, thymus and spleen, as well as lymphoproliferative disorders such as lymphoma and leukemia (9); but the LPV.ER transgenic mice show higher rates of neoplasic formation than those observed in mice expressing SV40.ER (9, 11).
The LPV early region has been shown to encode both LT and sT but, at present, we do not know which LPV.ER-encoded proteins contribute to tumorigenesis. However, unlike SV40 LT, some results suggest that LPV LT may not be the essential oncoprotein mediating transformation. For instance, pools of cells transformed by SV40.ER express high levels of LT, but cells transformed via LPV.ER show low LT levels, perhaps because they become genetically unstable when expressing high LT levels and are thus eliminated by negative selection (4). We have performed an extended analysis of the different T antigen products encoded by LPV.ER and examined their ability to transform mouse embryo fibroblasts and found new properties of the T antigens encoded by LPV.
MATERIALS AND METHODS
Cell Culture
Mouse Embryonic Fibroblasts (MEFs) were routinely grown in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Wild-type (wt) MEFs were generated from E13.5 old mouse embryos as previously described (12). When indicated, wt MEFs, LPV sT- and LPV LT-expressing MEFs were treated with doxorubicin at 1μM final concentration for 18 hours. After treatment, cells were collected by trypsinization and protein extracts were prepared for western blot analysis.
Cloning and Vectors
The LPV.ER, LT cDNA, LT′ cDNA and N200 cDNA were inserted into pLenti6.3-Blasticidin (Invitrogen) vectors by TOPO cloning following the manufacturer’s guidelines. LPV sT cDNA and also LPV.ER were inserted into pBABE-puromycin using Gateway cloning technology (Invitrogen), following the manufacturer’s guidelines. Total RNA from MEFs expressing LPV.ER was used to prepare cDNA, and primers specific for the 5′ and 3′ end of the ER were used to amplify the LPV LT and LPV LT′ cDNAs via PCR. The PCR products were separated through 1% agarose gel and the bands corresponding to LT and LT′ cDNAs were gel purified with the Wizard SV gel purification kit (Promega). The sequence of each isolated DNA was verified and then inserted into pLenti6.3 vector via TOPO cloning (Invitrogen). All the resulting constructs were verified by sequencing.
Production and Transduction of Retrovirus and Lentivirus
The packaging cell lines Phoenix-eco and 293FT were used to produce retroviruses and lentiviruses, respectively. Transfections were performed using lipofectamine (Invitrogen) following the manufacturer’s guidelines. MEFs were grown to 60–70% confluency and infected twice, every 24hrs, with the retrovirus- or lentivirus-containing media supplemented with 8μg/ml polybrene. Virus-containing media was replaced with regular media 24 hours after infection, and the corresponding selection with puromycin (3 μg/ml) or blasticidin (3 μg/ml) was carried out for 5 days.
Western Blots
Protein extractions and Western Blot analysis were performed with standard methods. The following primary antibodies were used: PAb416, PAb419 and PAb901 (monoclonal antibodies recognizing SV40.ER products; (13)(14)); p107 (Santa Cruz sc-318); p130 (Santa Cruz sc-317x); p53 (Santa Cruz sc-6243 HRP) and p21 (Santa Cruz sc-471). Monoclonal antibody Xt7 was raised by priming mice with recombinant HPyV10 sT fused to maltose binding protein followed by boosting with His-tagged recombinant HPyV10 J-domain. Hybridomas were generated and screened as described (15). Validation experiments revealed that Xt7 cross-reacts with the J-domain of a wide variety of polyomavirus LT proteins, including LPV. Antibody characteristics are posted at http://home.ccr.cancer.gov/Lco/BuckLabAntibodies.htm. After incubation with appropriate HRP-conjugated secondary antibodies, specific proteins were detected by chemiluminescence (Luminata Forte, Millipore).
Reverse Transcription PCR (RT-PCR) analysis
cDNA was synthesized from total RNA using superscript II reverse transcriptase (Invitrogen). PCR with GoTaq polymerase (Promega) was performed using equal amounts of cDNA from different samples and gene specific primers. The PCR primers used were: Set A (5′-ACCATGGACCAAACGCTGTCTAAG-3′ and 5′-TTACATTTGTTCTTCAATTACAATTCC-3′) and Set B (5′-ACCATGGACCAAACGCTGTCTAAG-3′ and 5′-AAAGCTGGGTCTTAGAATCCCAGTT-3′).
Real-time PCR
Total RNA was isolated from approximately 1×107 cells using the Qiagen RNeasy mini kit, following the manufacturer’s instructions and including the DNase-in column treatment. Total RNA (1μg) was used to perform reverse transcription as previously described (16). Real-time semi-quantitative PCR was performed using Maxima SYBR Green (Thermo Scientific) in a 7300 Real-time PCR system (Applied Biosystems) with gene specific primers as previously described (16). Each reaction was performed in quadruplicate and the endogenous values of Rpl5 within each cDNA were used to normalize the samples.
Transformation Assays
Soft-agar assay
1.6×104 exponentially growing cells were plated in 0.33% noble agar in duplicates. About 0.5ml of MEF media was added on top of the agar layer every 3–4 days to prevent it from drying. Colonies were counted after 3–4 weeks of plating by the following method: Each plate was divided into 4 quadrants, and movies of colonies formed in each quadrant were recorded using the ZEN software package from ZEISS corporation. Each movie would capture colonies growing in the different layers of agar, and the size of colonies at 21 days was calculated by determining the area under the curve using the same software. A wilcox statistical test (wilcox.test) in R (http://www.r-project.org/) was used to calculate whether variations between populations were significant. All experiments were performed in duplicate.
Growth in low serum
5×103 cells were seeded into 12 well plates in normal serum media containing 10% FBS. After 24hrs, the cells were washed 3 times with PBS and low serum media with 1% FBS was added. Two wells per cell type were trypsinized every two days and the cells were counted using a hemocytometer.
Proliferation Assays
Growth curve
Each well within 6 well plates received 2×104 cells of different cell types, and two wells per cell type were counted every 48 hours using a hemocytometer to generate proliferation curves.
Senescence assay
Wild type MEFs were infected with retroviruses containing either either empty vector or SV40 sT cDNA. Cells were seeded into 6 well plates after 5 days of puromycin (3 μg/ml) selection. Beta-Galactosidase (β-Gal) staining was performed as previously described (17). Briefly, cells were washed with PBS and fixed with 2% formaldehyde plus 0.2% glutaraldehyde for 5 minutes at room temperature. Cells were then washed with PBS and incubated overnight, at 37°C with fresh β-Gal staining solution (Bluo-Gal, halogenated-indoyl-beta-D-galactosidase, Invitrogen). Blue staining was monitored the next day by light microscopy, and all the images were taken at same magnification.
RESULTS
LPV.ER immortalizes and transforms primary MEFs in cell culture
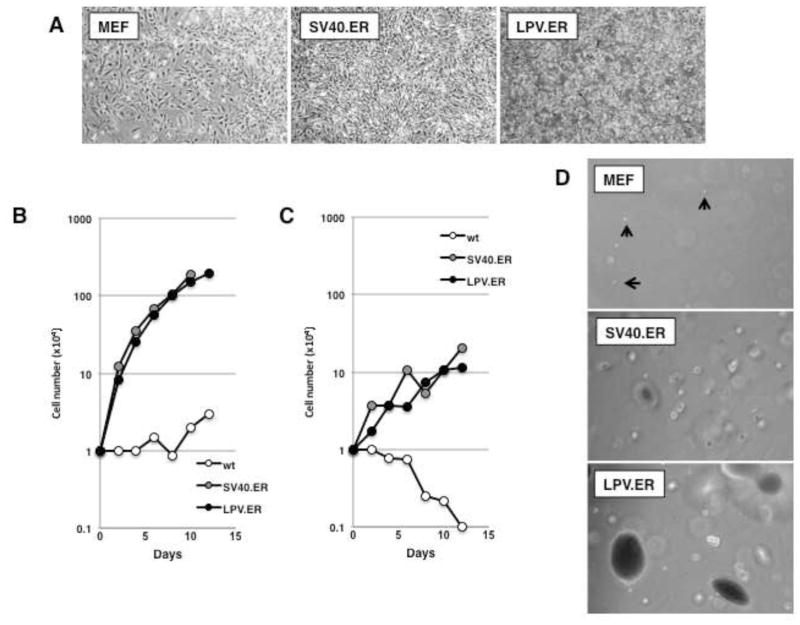
We expressed LPV.ER in primary mouse embryonic fibroblasts (MEFs) under the control of the CMV promoter. For comparison, we also expressed in parallel the SV40.ER. Two independent pools of cells expressing each ER were analyzed. Expression of either the LPV or SV40 ER resulted in morphological transformation (Fig. 1A) and enhanced proliferation in 10% serum compared to wild type MEFs (Fig. 1B).
Figure 1. LPV.ER expression leads to enhanced growth and transformation of MEFs.
A) Equal amounts of wild-type MEFs, MEFs expressing SV40.ER or MEFs expressing LPV.ER were plated in cell culture and allowed to proliferate. Images (original magnification: 50×) were taken 2–3 days after plating. B) Growth curve analysis of MEFs expressing SV40 or LPV.ER. Wild-type MEFs or MEFs expressing the SV40.ER or LPV.ER were plated in 6 cm dishes and counted a various times as described in materials and methods. C) Growth curve analysis in media containing 1% serum. Wild-type MEFs or MEFs expressing the SV40 or LPV ER were plated in 6 cm dishes and grown in media supplemented with 1% FBS as described in materials and methods. Cells were counted at various times after plating. D) Anchorage independent growth of wild type, SV40.ER and LPV.ER-expressing MEFs. Cell were suspended in media containing agar supplemented with 10% FBS (materials and methods), and monitored for 3–4 weeks until colonies developed. Images were taken at the same time for all cultures at same magnification.
Next, we compared the abilities of LPV.ER and SV40.ER to induce growth in low serum and growth independent of anchorage. As expected, wt MEFs failed to proliferate in 1% serum, while expression of ERs from SV40 or LPV induced similar levels of cell proliferation (Fig. 1C). Similarly, while wt MEFs failed to proliferate in soft agar and remained as single cells, the expression of the early region of LPV or SV40 induced colony formation (Fig. 1D). The soft agar colonies formed by LPV.ER-expressing MEFs were considerably larger than those formed by MEFs expressing SV40.ER.
LPV.ER encodes four differentially spliced products
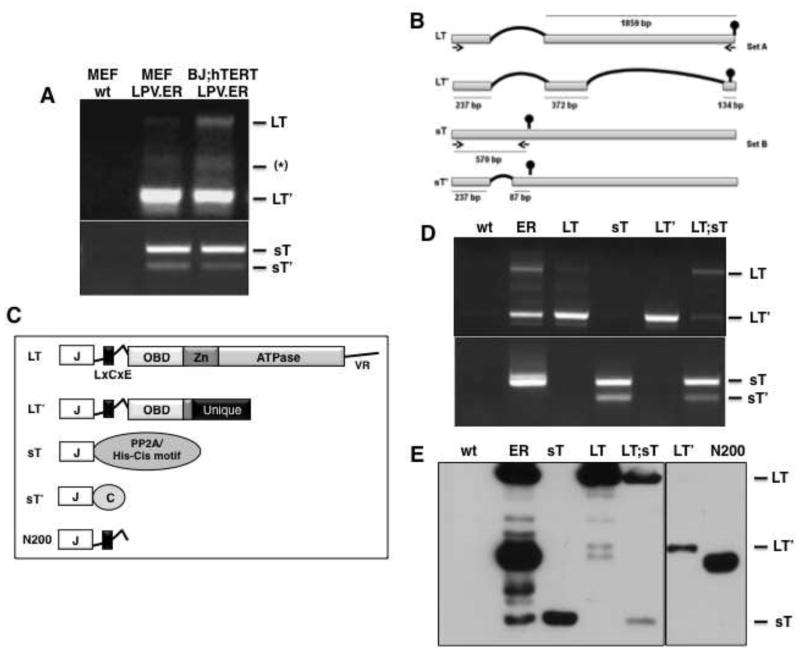
Next we examined the mRNAs expressed by the LPV.ER. Total RNA was extracted from MEFs or human fibroblasts (BJ;hTERT) expressing LPV.ER and subjected to RT-PCR. We first used primers specific for the 5′ and 3′ end of the ER (Set A, materials and methods) and detected three viral PCR products (Fig. 2A, upper panel). The identity of a PCR product showing the expected size for LPV LT (2094 bp; Fig. 2A, B) was confirmed by DNA sequencing. A second product, which we named Large T prime (LT′), migrated around 750 bp (Fig. 2A) and corresponded to a newly identified mRNA generated by removal of two introns from the ER-full length transcript (Fig. 2B). LT′ mRNA utilizes the same splice acceptor and donor sites as the LT mRNA, and thus includes the complete exon 1 of LT. In addition, a second splice results in exclusion of most of the remaining exon 2 from LT, though it retains 134 bp from the 3′ end of exon 2. The second splicing event also produces a change of reading frame and introduces a stop codon 17 bps earlier than the one observed in LT (Fig. 2B). Therefore, the putative LT′ protein encoded by this alternative transcript shares the first 203 amino acids with LT, followed by 38 unique amino acids in the carboxy-terminal region (Fig. 2B,C). We were unable to confirm the identity of the third amplified product(s) migrating around 1Kbp (*, Fig. 2B).
Figure 2. LPV.ER encodes four differentially spliced products.
A) RT-PCR analysis of MEFs and BJ;hTERT cells expressing LPV.ER. Three bands amplified with primer set A (materials and methods) are shown in the upper panel and correspond to LT (~2.1kb), LT′ (~750bs) and a PCR artifact (~1kb) confirmed by sequencing. Purified plasmids were used as controls (not shown). Two bands were amplified with primer set B (materials and methods) and are shown in the lower panel, corresponding to sT (~550b) and its spliced product sT′ (~300b). B) Schematic diagram of the splicing pattern generating LT, LT′, sT and sT′ from LPV.ER. The length of the segment in nts is indicated below each bar, and the vertical round head arrow shows the position of the stop codon for each transcript. Two sets of primers, A and B (materials and methods), were used to amplify the different components within LPV.ER and are indicated. C). Schematic representation of LPV proteins. Protein domains are inferred from sequence homology with SV40 LT. J indicates the J domain, OBD indicates the origin-binding-domain, Zn indicates the Zn binding domain, and ATPase indicates the ATPase domain. LPV LT contains similar domains and unstructured regions to SV40 LT, but lacks the C-terminal HR region and possess a longer linker region. LPV LT′ contains a J-domain, linker region and 4 amino acids of the OBD domain, and a unique C-terminus region due to a change in reading frame. LPV sT shares the J-domain with LT and LT′, but its C-terminus region contains putative PP2A interaction sites and a histidine -cysteine rich motif. sT′ contains a J domain and a unique C-terminus. N200 is a truncation mutant sharing the J domain and LxCxE motif with LT and LT′, but lacking the C-terminus unique region. D) RT-PCR analysis of MEFs expressing LPV.ER components independently or in combinations. The upper panel shows detection of LT and LT′ transcripts with primer set A; lower panel shows detection of sT and sT′ transcripts with primer set B. E). Western blot analysis of MEFs expressing LPV.ER products independently or in different combinations. Extracts were probed with Xt7 monoclonal antibody, which recognizes the J domain.
To ensure detection of LPV sT expression by RT-PCR, we then used primers specific to the 5′ and 3′ ends of the LPV sT coding sequences (Set B, materials and methods, Fig. 2B), and detected two distinct products (Fig. 2A, lower panel). An amplification product of approximately 570 bp was sequenced and confirmed to correspond to sT. An additional band of approximately 324 bp, which we called small T prime (sT′), corresponded to a spliced variant of the sT transcript. This transcript shares the first 273 bp with the sT transcript, but a unique 246 bp region is removed by splicing (Fig. 2B). The corresponding sT′ protein retains the J-domain but lacks the PP2A interacting motifs and the metal binding clusters present in sT (Fig. 2C), and instead contains a unique carboxy-terminal region.
To rule out possible promoter-specific effects, we also expressed the LPV.ER under the control of MoMuLV (Mouse Moloney leukemia virus) promoter. The same splicing pattern was observed upon expression from CMV or MoMuLV promoter (Fig. 2A and data not shown).
Next, we cloned the cDNAs of LPV LT, LT′ and sT in retroviral vectors, and obtained at least two independent MEF pools expressing each cDNA individually or in different combinations. Transcript and protein analysis of the MEF pools was subsequently done by RT-PCR and western blot. Using either primer set A to amplify LT and LT′ transcripts, or primer set B to detect sT and sT′ transcripts, we observed only the expected products corresponding to each retroviral construct. As the LT′ splicing sites are present within the LT cDNA, MEFs transduced with this construct show both LT and LT′ transcripts (Fig 2D). Similarly, MEFs transduced with LPV sT contain LPV sT and sT′ transcripts (Fig 2D). We estimated the sizes of the four LPV early transcripts to be 2449 bp (sT), 2203 bp (ST′), 2094 bp (LT), and 743 bp (LT′). In agreement with our results, previous studies of human B-lymphoblastoid cells infected with the LPV virus also suggested the presence of two or more transcripts encoded by the LPV.ER (18).
Our transcript analysis indicated that all the putative ER products share the N-terminus J-domain. Therefore, we used a monoclonal antibody (Xt7) specific for epitopes present within the J-domain to monitor protein expression in different pools of MEF cells (materials and methods). As expected, strong expression of LT was observed in MEFs after transduction with either LPV.ER, LT or LT plus sT; LT′ product was present in cells transduced with LPV.ER or LT′ alone; and sT expression was observed either in MEFs expressing LPV.ER, sT alone or sT in combination with LT (Fig. 2E). However, despite containing sT′ transcripts (Fig. 2D), cell pools expressing neither sT nor LT plus sT showed sT′ protein expression (Fig. 2E and data not shown). Similar transcript and protein expression patterns were observed in human fibroblasts (BJ;hTERT and IMR90) expressing LPV.ER (data not shown).
LPV LT and sT independently induce immortalization and increase cell proliferation in MEFs
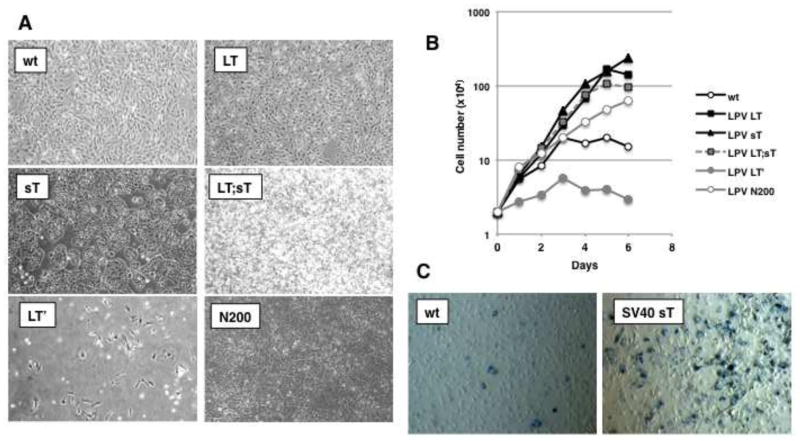
To determine the contributions of different LPV.ER proteins to immortalization and transformation of MEFs, we analyzed the properties of each product individually or in different combinations. As expected, MEFs expressing LPV LT alone exhibited very robust proliferation (Fig. 3A, B). Surprisingly, sT-expressing MEFs also showed enhanced proliferation (Fig. 3A, B) at rates comparable to those of cells expressing LT (Fig. 3B). These results are in clear contrast to observations in MEFs expressing SV40 sT, whose presence is growth inhibitory and results in cellular senescence (Fig 3C).
Figure 3. Different effects of LPV.ER components on cell proliferation.
A) Morphology of MEFs expressing different components of LPV.ER. Plating and manipulation of the cells was as described previously (Fig. 1A, materials and methods). B) Growth curve analysis of MEFs stably expressing LPV proteins. Pools of cells expressing different LPV components were allowed to proliferate in medium supplemented with 10% FBS, and the number of cells was monitored every two days (materials and methods). Each data point is an average of 2 separate experiments, performed using 2 biological replicates of each cell type. C) Beta-galactosidase staining to determine senescence in MEFs transduced with retroviruses containing empty vector (MEF) or SV40 sT cDNA (SV40 sT). The experiment was performed in triplicates and a representative image is shown. Blue-violet positive staining and enlarged morphology were observed only in SV40 sT expressing MEFs and not in control MEFs.
In contrast to wt MEFs, which are primary cells and normally growth arrest after 6–8 passages in culture, MEFs expressing either LT or sT continued proliferation for over (>20) passages. Finally, the proliferation rates observed in MEFs upon combined LT and sT expression were similar to those produced by either LT or sT alone (Fig. 3B). We conclude that both the LPV LT and sT proteins are capable of inducing immortalization and enhancing proliferation in MEFs.
LPV sT per se induces transformation, and cooperates with LPV LT to enhance transformation
Cells expressing LPV sT displayed morphological signs of transformation and formed tight groups of cells (Fig. 3A). Furthermore, the morphology of MEFs expressing both sT and LT closely resembled the morphology observed in LPV.ER expressing cells (Fig. 3A, compare to Fig. 1A). Thus, we next assessed the capacity of LPV sT and LT to induce cell transformation. We performed low serum and soft agar transformation assays with MEFs expressing sT or LT, using wt MEFs as controls. While wt MEFs ceased to proliferate in low serum conditions, sT-expressing MEFs showed continuous cell proliferation (Fig. 4A). Surprisingly, although LT induced proliferation of MEFs in the presence of 10% serum (Fig. 3B), it failed to do so in media containing 1% serum (Fig. 4A). Thus, LPV sT, but not LPV LT, allows MEFs to grow in low serum conditions.
Figure 4. LPV sT is necessary and sufficient to transform MEFs.
A) Growth in low serum of MEFs expressing LPV LT and sT, on their own or in combination. Cells were plated and counted as described (Fig. 3B, materials and methods). While wt MEFs and LPV LT MEFs ceased to proliferate, MEFs expressing LPV sT -by itself on with LT- proliferated continuously. B) Soft agar formation induced by LPV products. LPV ER and sT were able to induce colonies in soft agar, but not LT. C) Box plots showing the size distribution of colonies for each cell type. Size was determined from images taken from each colony, as described in materials and methods, also Fig. 1D. Box plots showing the size distribution of colonies for each cell type were prepared using R programming. Each box plot is divided into four quartiles, the thick line in the middle of the box represents the median for the sample. Significance testing was performed by wilcox test using R programming. The size of colonies present in LPV LT expressing MEFs was not significantly different than that observed in wt MEFs (p value 0.36). MEFs expressing either LPV.ER, sT, or LT plus sT formed colonies in soft agar at a significantly larger rate (p value≪0.05). LPV sT expressing MEFs induced soft agar colonies of larger size than those formed by wt MEFs or LPV LT MEFs (p value= 0.0002), but the colonies were significantly smaller than those formed upon LPV.ER expression (p value= 0.02). Interestingly, co-expression of LPV LT and sT in trans resulted into significant increase in soft agar colonies with similar size to those found upon LPV.ER expression (p value= 0.622).
Next we performed soft agar assays to determine the ability of sT and LT to allow MEFs to grow in anchorage independent conditions. As expected, wt MEFs failed to proliferate in soft agar and remained as single cells, while LPV.ER expressing MEFs readily formed large soft agar colonies (Fig. 4B, C, see also Fig. 1D). The performance of LPV LT and sT promoting growth in soft agar was similar to their behavior in low serum conditions: While LT was unable to support colony formation, MEFs expressing sT formed colonies in soft agar (Fig. 4B, C). However, the size of colonies induced by sT was significantly smaller than the size of colonies formed after LPV.ER expression (Fig. 4B, C). Indeed, the combined expression of both LT and sT produced colonies of significantly larger size than those observed in cells expressing either oncogene independently (Fig. 4B, C). These results indicate that LPV sT is sufficient to induce transformation of MEFs, as determined by low serum and soft-agar assays. However, a complete recapitulation of the phenotype induced by LPV.ER requires additional contribution/s by LPV LT.
LPV LT′ induces growth arrest in MEFs
As MEFs expressing LPV.ER also expressed LT′, we tested the contributions of LPV LT′ to cell proliferation and immortalization. In contrast to the cell expressing LT or sT, MEFs expressing LPV LT′ alone underwent growth arrest, displayed an enlarged morphology (Fig 3A), and failed to reach confluency (Fig. 3A, B). LT′-expressing cells failed to produce signs of senescence when tested by β-galactosidase assay staining (data not shown). The results, indicating that expression of LT′ produces growth arrest, were surprising, as LT′ contains the LXCXE motif and thus should be able to interact with and disrupt the RB family of proteins, an action that normally results in cell cycle progression and cell proliferation. Therefore, we generated a LT truncation mutant (N200), which shares the first 200 amino acids with LT and LT′ but lacks the unique LT′ C-terminus region (Fig. 2C). N200 contains the J domain and LXCXE motif, and is thus expected to interact with the RB proteins. The expression of N200 was confirmed by western blot analysis (Fig. 2E). In contrast to LT′-containing cells, MEFs expressing N200 continue proliferating at similar rates to those observed in LT-expressing MEFs (Fig. 3A, B). These results indicate that LT′ possesses additional function/s within its C-terminus unique region, linked to either prevent proliferation or to inhibit cell growth.
Differential effects of the LPV.ER products on the RB pathway
The transformation abilities of SV40.ER are mostly attributed to the LT-mediated inhibition of two major growth suppressive pathways, RB/E2F and p53. SV40 LT interacts with all three members of the RB family and induces disruption of the pRB/E2F repressor complexes. While LT expression does not change pRb levels significantly, it induces proteosomal degradation of p130 and results in increased p107 protein levels (19, 20). A significant increase in E2F target gene expression results from the inhibition of pRBs by LT (21), inducing cell proliferation.
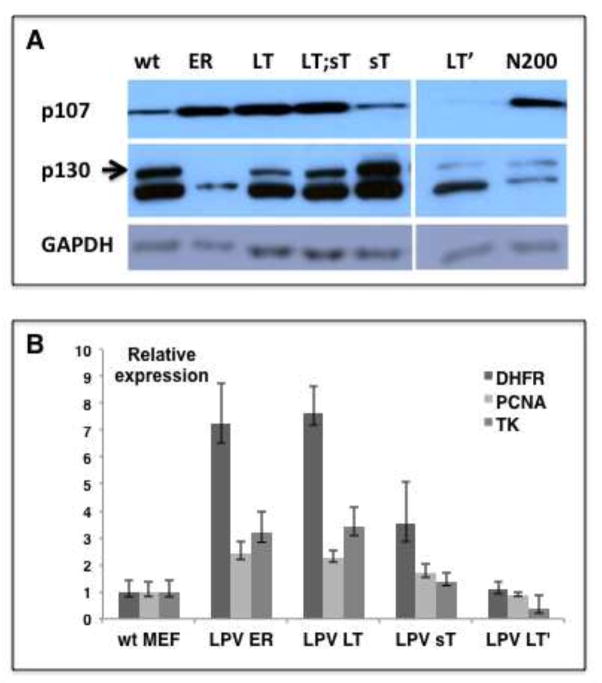
We first assessed the status of the retinoblastoma proteins p107 and p130 in MEFs expressing LPV.ER or its different components. In a similar way to SV40 LT, we found that the LPV.ER induced a decrease in p130 and an increase in p107 protein levels, as determined by western blot analysis (Fig. 5A). In addition, real-time PCR analysis of MEFs expressing either LPV.ER or LPV LT showed at least a 2-fold upregulation of E2F target genes (Fig. 5B) in comparison to wt controls. We conclude that LPV LT blocks RB tumor suppressors and elevates E2F-dependent transcription. Despite our findings that LPV sT induces MEF immortalization and robust proliferation, we found that sT does not affect the RB/E2F pathway significantly. On one hand, sT neither decreased p130 levels nor raised p107 levels (Fig. 5A) and, on the other, sT only induced a modest upregulation of E2F genes (Fig. 5B). These results suggest that LPV sT might trigger pathways other than RB/E2F to elicit cell immortalization and proliferation.
Figure 5. Interaction of LPV products with the RB/E2F pathway.
A) Western blot analysis of p130 and p107 proteins in MEFs expressing different LPV products. SV40.ER-expressing MEFs were used as positive control. GAPDH was used as loading control. B) Real-time PCR analysis for canonical E2F target genes using gene specific primers. The gene expression levels are represented relative to the corresponding expression levels in wt MEFs, and Rpl5 expression levels were used for cDNA normalization. The error bars represent standard deviation; within each sample, each gene was analyzed in quadruplicate.
Finally, we found that LT′ was unable to efficiently disrupt the RB/E2F pathway. While expression of LPV LT′ decreased p130 protein levels in a similar way to LPV LT, it did not increase p107 levels (Fig. 5A). Furthermore, E2F gene expression in LPV LT′-expressing MEFs did not increase and was similar to the basal levels observed in wt MEFs. In contrast, expression of the truncation mutant N200 decreased p130 and increased p107 protein levels in a similar way to LPV LT (Fig. 5A). These results suggest that disruption of the RB pathway is absent or incomplete in LT′ and might contribute to explain why LT′ fails to elicit immortalization and cell proliferation in MEFs. However, the incomplete inhibition of the RB/E2F pathway observed in LT′-expressing MEFs might be explained, at least in part, by the relatively low expression levels of LT′ protein observed in comparison to levels of N200 protein found in MEFs (Fig 2E).
Interaction between LPV T antigens and p53
SV40 LT binds to p53 and blocks its transcriptional activity, and through this process LT enhances the protein stability of p53 (22). However, previous studies regarding LPV LT interaction with p53 proved controversial (7, 10), thus we analyzed the effects of LPV.ER products on p53 status.
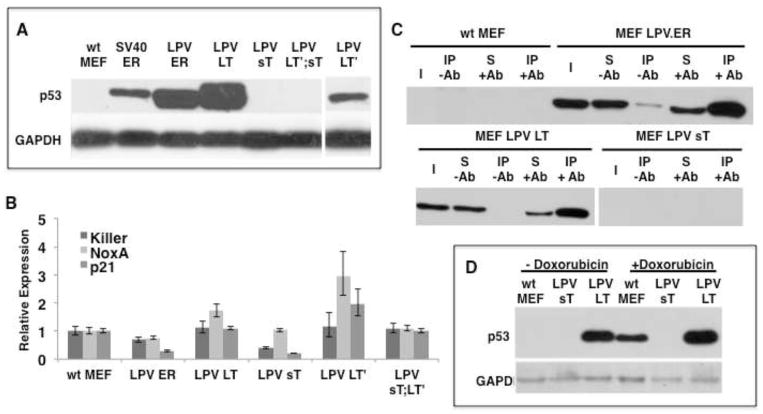
We found increased levels of p53 in MEFs expressing LPV.ER or LPV LT (Fig. 6A). Furthermore, the transcriptional levels of several p53-target genes were not upregulated in cells expressing LPV LT (Fig. 6B), suggesting that the p53 protein present in LPV.ER or LPV LT-expressing MEFs is inactive. We then performed co-immunoprecipitation analysis to determine if LPV LT binds p53 in MEFs expressing LPV.ER or LT. Protein extracts from different MEF cells were immunoprecipitated using a monoclonal antibody specific for the J-domain of LPV T antigens, and the precipitates were analyzed by western blot using a p53-specific antibody.
Figure 6. Differential effects of LPV products on the tumor suppressor p53.
A) Evaluation of p53 protein levels in various cell types as determined by western blot. B). Real time PCR analysis of p53 target genes in MEFs expressing different LPV products. The target genes were normalized against the Rpl5 gene and the expression levels in wt MEFs were used as baseline control. C) Differential interactions between LPV products and p53 as revealed by Immunoprecipitation with Xt7 (anti J-domain antibody recognizing the different LPV products) and subsequent western blot analysis with anti-p53. Immunoprecipitations were carried out in the absence (−Ab) or presence (+Ab) of anti-LPV J-domain antibody. Input (I), supernantant (S), immunoprecipitated complex (IP). D) LPV sT prevents p53 up-regulation induced by a DNA damaging agent. Cells were treated with or without doxorubicin and the levels of p53 evaluated by Western Blot. GAPDH was used as loading control.
Both LPV.ER or LT expressing MEFs contained high levels of p53, which in both cases co-precipitated by the J-domain specific antibody, suggesting the existence of a complex between LPV T antigens and p53 (Fig. 6C). The LPV LT′ product was also able to raise p53 levels (Fig. 6A), but not to the extent observed with LPV.ER or LPV LT. However, p53 seems to be activated and not neutralized by the expression of LT′, as p53 target levels increased significantly in MEFs containing LT′ (Fig. 6B). This suggests that LT′ elicits an active p53 response and is unable to block p53, and is consistent with the growth arrest observed in MEFs expressing LT′. In a similar fashion, the carboxy terminus of MCV LT has been shown to posses a growth inhibitory function and its expression leads to upregulation of p53 target genes and cell cycle arrest (23) (24).
As expected, we observed neither increased p53 protein levels in MEFs expressing LPV sT (Fig. 6A), nor upregulation of p53 target genes (Fig. 6B). Surprisingly, we found that not only sT does not trigger a p53 response, but that, in addition, it seems to prevent p53 activation. First, the presence of sT in LT′-expressing cells eliminates the surge in p53 levels observed upon LT′ expression alone (Fig. 6A). Furthermore, the upregulation of p53 target genes observed after LT′ expression is eliminated when sT is co-expressed (Fig. 6B). We then decided to test the ability of LPV sT to prevent p53 activation by the cytotoxic agent doxorubicin (adriamycin). Control MEFs and those expressing LPV LT or LPV sT were treated with doxorubicin, and the presence of p53 was monitored by western blot. Untreated MEFs contained no detectable p53 unless expressing LPV LT. The doxorubicin treatment increased p53 levels in wt MEFs, but the increase in p53 was obliterated by LPV sT expression and not LT expression (Fig 6D). These results suggest that LPV sT is able to prevent p53 activation.
DISCUSSION
Previous studies have shown that the early region of LPV is able to cause transformation of hamster cell lines, as well as to induce robust tumorigenesis in transgenic mice (7, 9, 11). However, no study has evaluated the different oncoproteins encoded by the LPV.ER, or their role and contribution to cell transformation. In this study, we have uncovered novel splice products encoded by LPV.ER, and have extended the current knowledge of their individual contributions to cell transformation. At least four different splice products are encoded by LPV.ER: large T antigen (LT), small T antigen (sT), a large T antigen splice form (LT′, large T prime) and a small T antigen splice form (sT′, small T prime). While we found evidence for the presence of all four transcripts in LPV.ER-expressing MEFs, we detected LT, sT and LT′ protein expression but failed to detect sT′. This could indicate that either sT′ protein levels were very low or that the protein product is highly unstable.
LPV.ER expression induced transformation in MEFs to similar levels as that observed in MEFs expressing SV40.ER. Our results also indicate that, like SV40 LT, LPV LT inhibits the RB/E2F and p53 tumor suppressor pathways, thus suggesting similarities between the mechanisms of action used by both LT oncoproteins to control cellular proliferation. However, further analysis of the contributions provided by other LPV oncoproteins showed major differences between the two systems. While SV40 LT was sufficient to induce transformation of MEFs in cell culture, LPV LT was able to immortalize MEFs but failed to transform them. In addition, we found that the roles of LPV sT and SV40 sT in cell immortalization and transformation are remarkably different. In SV40, sT cooperates with LT to transform human fibroblasts and enhances the LT-transformation phenotype in cell culture, but its sole expression in primary or immortalized cells fails to induce proliferation and results in growth arrest (25). In agreement, we found that expression of SV40 sT in primary MEFs results in senescence but, in contrast, the expression of LPV sT is sufficient to immortalize and transform MEFs, and cells expressing LPV sT managed to proliferate in low serum conditions. Furthermore, expression of LPV sT induced the formation of colonies in soft agar, but the presence of both sT and LT increased transformation significantly. These results suggest that LPV sT is sufficient to induce immortalization and transformation in MEFs, but cooperation with LPV LT is required to fully recapitulate the transformation observed in LPV.ER expressing cells.
On one hand, we found that LPV sT is unable to degrade p130 or increase the transcriptional activity of E2F genes. On the other, albeit lacking p53-binding domains, sT is able to prevent the activation of p53 triggered by LT′ expression and/or by a DNA damaging agent. Murine polyomavirus sT also seems to posses a p53-supressing activity (26), although induction of cell death by MuPyV sT is independent of the status of p53 (27). These results suggests that LPV sT uses pathways other than RB/E2F and p53 to elicit cell proliferation and transformation.
To date, the only other polyomavirus sT shown to posses transforming properties is encoded by MCPyV (28, 29). However, in this case the transformation was observed in immortalized cell types, thus it is not clear if other factors are needed to contribute to cell immortalization prior to MCPyV sT action to elicit transformation. Other than LPV sT, no other polyomavirus sT has been shown to immortalize or transform primary cells in culture.
Many of the biological activities of sT are attributed to its interactions with the protein phosphatase 2A (PP2A) protein complex. An amino acid sequence comparison between homologous sT proteins from different polyomaviruses shows high conservation of the Hsc70 interacting J-domain HPD motif and PP2A interacting motifs. However, major differences have been observed regarding the mode of interaction. For instance, the sT proteins from SV40, MuPyV and MCPyV all bind to PP2A, but while SV40 sT oncogenic activity depends upon its inhibition of PP2A mediated dephosphorylation of Akt, transformation mediated by MCPyV sT is independent of PP2A interaction (29). Instead MCPyV sT bypasses the Akt-mTOR pathway and increases phosphorylation of 4E-BP1, a cap-dependent translation regulator (29). In contrast, SV40 sT induces dephosphorylation of 4E-BP1 via Akt-mTOR signaling. Therefore, while sT products from different polyomaviruses might disrupt the same cellular pathways, their effects might vary considerably. Investigating the transcriptional profiles of cells expressing LPV sT should yield novel ways to approach cell transformation.
CONCLUSIONS
The comparative study of homologous oncoproteins from different polyomaviruses has helped us to identify their molecular targets and cell/tissue type specific roles. To date, only one polyomavirus sT (MCPyV sT) has been shown to transform cells in culture by itself. We now report that LPV induction of tumorigenesis proceeds in different ways than previously suspected, and that the small T antigen of LPV is necessary and sufficient to immortalize and transform primary MEFs in cell culture.
Acknowledgments
This work was supported by NIH grant 1R21AI109339-01A1 to JMP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.zur Hausen H, Gissmann L. Lymphotropic papovaviruses isolated from African green monkey and human cells. Medical microbiology and immunology. 1979;167:137–153. doi: 10.1007/BF02121180. [DOI] [PubMed] [Google Scholar]
- 2.Takemoto KK, Furuno A, Kato K, Yoshiike K. Biological and biochemical studies of African green monkey lymphotropic papovavirus. Journal of virology. 1982;42:502–509. doi: 10.1128/jvi.42.2.502-509.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takemoto KK, Segawa K. A new monkey lymphotropic papovavirus: characterization of the virus and evidence of a related virus in humans. Prog Clin Biol Res. 1983;105:87–96. [PubMed] [Google Scholar]
- 4.von Hoyningen-Huene V, Kurth M, Deppert W. Selection against large T-antigen expression in cells transformed by lymphotropic papova virus. Virology. 1992;190:155–167. doi: 10.1016/0042-6822(92)91201-5. [DOI] [PubMed] [Google Scholar]
- 5.Pawlita M, Clad A, zur Hausen H. Complete DNA sequence of lymphotropic papovavirus: prototype of a new species of the polyomavirus genus. Virology. 1985;143:196–211. doi: 10.1016/0042-6822(85)90108-4. [DOI] [PubMed] [Google Scholar]
- 6.Scuda N, Hofmann J, Calvignac-Spencer S, Ruprecht K, Liman P, Kuhn J, Hengel H, Ehlers B. A novel human polyomavirus closely related to the african green monkey-derived lymphotropic polyomavirus. J Virol. 2011;85:4586–4590. doi: 10.1128/JVI.02602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang S, Folk WR. Lymphotropic papovavirus transforms hamster cells without altering the amount or stability of p53. Virology. 1992;191:754–764. doi: 10.1016/0042-6822(92)90251-j. [DOI] [PubMed] [Google Scholar]
- 8.Takemoto KK, Kanda T. Lymphotropic papovavirus transformation of hamster embryo cells. J Virol. 1984;50:100–105. doi: 10.1128/jvi.50.1.100-105.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen JD, Neilson K, Van Dyke T. Lymphotropic papovavirus early region is specifically regulated transgenic mice and efficiently induces neoplasia. J Virol. 1989;63:2204–2214. doi: 10.1128/jvi.63.5.2204-2214.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Symonds H, Chen JD, Van Dyke T. Complex formation between the lymphotropic papovavirus large tumor antigen and the tumor suppressor protein p53. J Virol. 1991;65:5417–5424. doi: 10.1128/jvi.65.10.5417-5424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen JD, Van Dyke T. Uniform cell-autonomous tumorigenesis of the choroid plexus by papovavirus large T antigens. Mol Cell Biol. 1991;11:5968–5976. doi: 10.1128/mcb.11.12.5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Markovics JA, Carroll PA, Robles MT, Pope H, Coopersmith CM, Pipas JM. Intestinal dysplasia induced by simian virus 40 T antigen is independent of p53. J Virol. 2005;79:7492–7502. doi: 10.1128/JVI.79.12.7492-7502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tevethia MJ, Bonneau RH, Griffith JW, Mylin L. A simian virus 40 large T-antigen segment containing amino acids 1 to 127 and expressed under the control of the rat elastase-1 promoter produces pancreatic acinar carcinomas in transgenic mice. J Virol. 1997;71:8157–8166. doi: 10.1128/jvi.71.11.8157-8166.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harlow E, Crawford LV, Pim DC, Williamson NM. Monoclonal antibodies specific for simian virus 40 tumor antigens. J Virol. 1981;39:861–869. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pastrana DV, Pumphrey KA, Cuburu N, Schowalter RM, Buck CB. Characterization of monoclonal antibodies specific for the Merkel cell polyomavirus capsid. Virology. 2010;405:20–25. doi: 10.1016/j.virol.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chong JL, Wenzel PL, Saenz-Robles MT, Nair V, Ferrey A, Hagan JP, Gomez YM, Sharma N, Chen HZ, Ouseph M, Wang SH, Trikha P, Culp B, Mezache L, Winton DJ, Sansom OJ, Chen D, Bremner R, Cantalupo PG, Robinson ML, Pipas JM, Leone G. E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature. 2009;462:930–934. doi: 10.1038/nature08677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta T, Saenz Robles MT, Pipas JM. Cellular transformation of mouse embryo fibroblasts in the absence of activator E2Fs. J Virol. 2015;89:5124–5133. doi: 10.1128/JVI.03578-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abraham G, Yarom R, Manor H. Mapping of polyadenylated transcripts of a monkey lymphotropic papova virus. Virology. 1984;136:442–447. doi: 10.1016/0042-6822(84)90181-8. [DOI] [PubMed] [Google Scholar]
- 19.Stubdal H, Zalvide J, DeCaprio JA. Simian virus 40 large T antigen alters the phosphorylation state of the RB-related proteins p130 and p107. J Virol. 1996;70:2781–2788. doi: 10.1128/jvi.70.5.2781-2788.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zalvide J, DeCaprio JA. Role of pRb-related proteins in simian virus 40 large-T-antigen-mediated transformation. Mol Cell Biol. 1995;15:5800–5810. doi: 10.1128/mcb.15.10.5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rathi AV, Saenz Robles MT, Cantalupo PG, Whitehead RH, Pipas JM. Simian virus 40 T-antigen-mediated gene regulation in enterocytes is controlled primarily by the Rb-E2F pathway. J Virol. 2009;83:9521–9531. doi: 10.1128/JVI.00583-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.An P, Saenz Robles MT, Pipas JM. Large T antigens of polyomaviruses: amazing molecular machines. Annu Rev Microbiol. 2012;66:213–236. doi: 10.1146/annurev-micro-092611-150154. [DOI] [PubMed] [Google Scholar]
- 23.Cheng J, Rozenblatt-Rosen O, Paulson KG, Nghiem P, DeCaprio JA. Merkel cell polyomavirus large T antigen has growth-promoting and inhibitory activities. J Virol. 2013;87:6118–6126. doi: 10.1128/JVI.00385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Wang X, Diaz J, Tsang SH, Buck CB, You J. Merkel cell polyomavirus large T antigen disrupts host genomic integrity and inhibits cellular proliferation. J Virol. 2013;87:9173–9188. doi: 10.1128/JVI.01216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrabi S, Hwang JH, Choe JK, Roberts TM, Schaffhausen BS. Comparisons between murine polyomavirus and Simian virus 40 show significant differences in small T antigen function. Journal of virology. 2011;85:10649–10658. doi: 10.1128/JVI.05034-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moule MG, Collins CH, McCormick F, Fried M. Role for PP2A in ARF signaling to p53. Proc Natl Acad Sci U S A. 2004;101:14063–14066. doi: 10.1073/pnas.0405533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pores Fernando AT, Andrabi S, Cizmecioglu O, Zhu C, Livingston DM, Higgins JM, Schaffhausen BS, Roberts TM. Polyoma small T antigen triggers cell death via mitotic catastrophe. Oncogene. 2015;34:2483–2492. doi: 10.1038/onc.2014.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeCaprio JA, Garcea RL. A cornucopia of human polyomaviruses. Nature reviews Microbiology. 2013;11:264–276. doi: 10.1038/nrmicro2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J Clin Invest. 2011;121:3623–3634. doi: 10.1172/JCI46323. [DOI] [PMC free article] [PubMed] [Google Scholar]