Abstract
PCBP2, a member of the poly(C)-binding protein (PCBP) family, is involved in posttranscriptional and translational regulation by interacting with single-stranded poly(C) motifs in target mRNAs. Recent studies have shown that PCBP2 is overexpressed and plays an important role in human cancers, including glioma. However, the molecular basis for its up-regulation remains poorly understood. Here, we show that microRNA-214 (miR-214) interacts with the 3’-untranslated region of PCBP2 mRNA and induces its degradation, leading to reductions in its protein expression. As a result, overexpression of miR-214 mimics significantly inhibited, while its antisense oligos proliferation and growth of glioma cells. Restoration of PCBP2 remarkably reversed the tumor-suppressive effects of miR-214 on cell proliferation and growth. In summary, our data indicate that miR-214 may function as tumor suppressor in glioma by targeting PCBP2.
Keywords: PCBP2, miR-214, glioma, tumor suppressor
Introduction
Glioma, derived from glial cells, has become one of the most common primary malignant brain tumor in China [1]. Despite multimodal therapies, such as surgery, radiotherapy, and chemotherapy, the average life expectancy remains limited [2,3]. Therefore, a better understanding of the molecular mechanism of glioma may provide novel therapeutic strategies for its treatment.
Poly (C)-binding protein 2 (PCBP2), characterized by its high-affinity and sequence-specific interactions with polycytosine (poly (C), regulates gene expression at various levels, including transcription, mRNA processing, mRNA stabilization, and translation [4,5]. It has been reported that PCBP2 participated in the replication and translation of many RNA viruses, including poliovirus, coxsackievirus, and rhinovirus [6-8]. Recent studies demonstrate that PCBP2 may play a crucial role in the cell proliferation and tumorigenesis. For instance, depletion of PCBP2 led to an induction of cyclin-dependent kinase inhibitor (CDKN1A) and G1 arrest in the human hematopoietic cells [9]. Besides, PCBP2 promoted gastric cancer cell proliferation and colony formation [10]. More importantly, Han et al. found that PCBP2 was up-regulated in human glioma tissues and cell lines. Depletion of PCBP2 inhibited glioma growth in vitro and in vivo through suppression of cell-cycle progression and induction of caspase-3-mediated apoptosis [11]. However, the mechanism by which PCBP2 is regulated in glioma remains poorly understood.
Increasing studies have highlighted the importance of microRNAs, a class of small and non-coding RNAs, in the regulation of gene expression [12,13]. Given that multiple miRNAs are deregulated and involved in the glioma progression [14-16], whether PCBP2 is regulated by miRNAs has not been previously studied.
Materials and methods
Human samples collection
Twenty-five pairs of glioma tissues and adjacent normal tissues were collected from routine therapeutic surgery at our department. All samples were obtained with informed consent and approved by the hospital institutional review board.
Cell culture
Colon cancer cells (U251 and SHG-44 cells) were purchased from American Type Culture Collection (Rockville, MD, USA). Cells were culture in RPMI 1640 medium (GIBCO, Shanghai, China) supplemented with 10% fetal bovine serum (GIBCO). Cultures were incubated at 37°C in a humidified atmosphere containing 5% CO2.
RNA isolation and quantitative real-time PCR
Total RNA from tissue samples and cell lines was harvested using the RNA Isolation Kit (Ambion, USA). Expression of mature miRNAs was assayed using Taqman MicroRNA Assay (Applied Biosystems) specific for hsa-miR-214. Quantitative real-time PCR was performed by using an Applied Biosystems 7900 Real-time PCR System and a TaqMan Universal PCR Master Mix. All the primers were obtained from the TaqMan miRNA Assays. Small nuclear U6 snRNA (Applied Biosystems) was used as an internal control. Differences in gene expression, expressed as fold-changes, were calculated using the 2-ΔΔCt method.
miR-214 mimics, antisense and transfection
Human miR-214 mimics, antisense or negative controls were obtained from Genepharm Company (Shanghai). For transfection, cells were cultured in a 6-well plate and transiently transfected at 70-80% confluence using the LipofectamineTM 2000 reagent (Invitrogen, CA, USA) as per the manufacturer’s instructions.
BrdU and cell invasion assays
A cell proliferation enzyme-linked immunosorbent assay (BrdU kit; Beyotime) was used to analyze the incorporation of BrdU during DNA synthesis following the manufacturer’s protocols. All experiments were performed in triplicate. Absorbance was measured at 450 nm in the Spectra Max 190 ELISA reader (Molecular Devices, Sunnyvale, CA). For cell invasion assays, cells were analyzed using extracellular matrix-coated invasion chambers (Millipore, CA, USA), and quantitated with a colorimetric microplate reader at 570 nm, according to the manufacturer’s instructions.
Western blot
Cells or tissues were harvested and lysed with lysis buffer (150 mM NaCl, 50 mM Tris-HCl, 1% NP-40, pH 7.5). Proteins were quantified and separated by 8% SDS-agarose gel, transferred to NC membrane (Amersham Bioscience, Buckinghamshire, U.K.). After blocking with 10% nonfat milk in PBS, membranes were immunoblotted with antibodies as indicated, followed by HRP-linked secondary antibodies (Cell Signaling). The signals were detected by SuperSignal West Pico Chemiluminescent Substrate kit (Pierce, Rockford, IL) according to manufacturer’s instructions. Anti-PCBP2 antibody was purchased from Abcam Company (USA). Protein levels were normalized to total GAPDH, using a mouse anti-GAPDH antibody (Santa Cruz, USA).
Luciferase reporter assay
3’-UTR of human PCBP2 gene that were predicted to interact with miR-214 were synthesized and inserted into pMir-Report (Ambion, USA), yielding pMir-Report-PCBP2. Mutations within potential miR-214 binding sites were generated by nucleotide replacement of wild type sequence to inhibit miR-214 binding. Cells were transfected with the pMir-Report vectors containing the 3’-UTR variants, and miR-214 mimics for 36 hours. The pRL-SV40 vector (Promega, USA) carrying the Renilla luciferase gene was used as an internal control to normalize the transfection efficiency. Luciferase values were determined using the Dual-Luciferase Reporter Assay System (Promega).
Statistics
Data are expressed as the mean ± SEM from at least four separate experiments. Differences between groups were analyzed using Student’s t-test. A value of P < 0.05 was considered statistically significant.
Results
miR-214 down-regulates PCBP2 expression by targeting its 3’-untranslated region
Using the three bioinformatics software (miRWalk, miRanda and Targetscan) based on seed recognition, we identified several miRNAs that could potentially interact with the PCBP2 transcript. To determine whether these miRNAs could regulate PCBP2, the 3’-untranslated region (UTR) of PCBP2 gene was cloned and inserted into a luciferase reporter construct. As shown in the Figure 1A, overexpression of miR-214 mimics, but not others, significantly inhibited the reporter activity of PCBP2 3’-UTR in U251 cells. Next, the endogenous expression of PCBP2 was examined by quantitative real-time PCR and western blot analysis. As expected, forced expression of miR-214 mimics decreased PCBP2 mRNA and protein levels (Figure 1B, 1C). In agreement, inhibition of miR-214 by its antisense oligos increased PCBP2 expression (Figure 1D, 1E). Similar results were also observed in SHG-44 cells (Supplementary Figure 1A-D).
Figure 1.
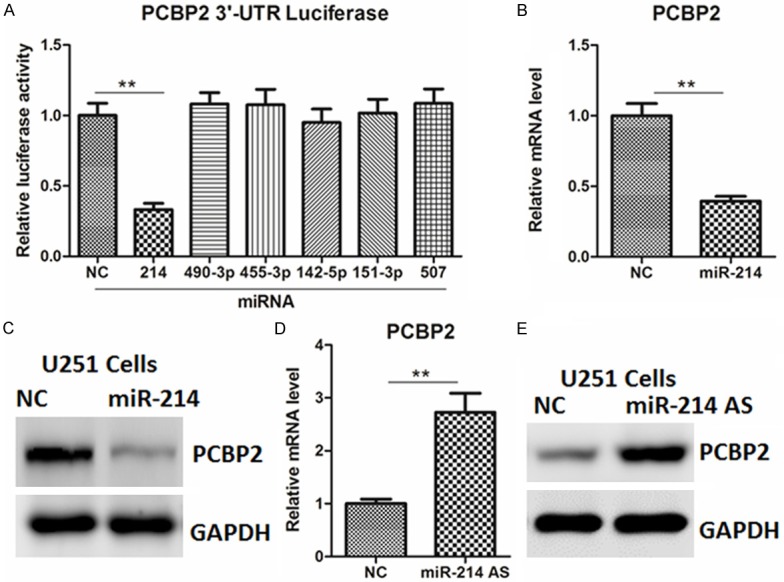
Effects of miR-214 on the regulation of PCBP2 in glioma cells. (A) List of miRNAs that could potentially interact with the PCBP2 transcript. (B, C) mRNA (B) and protein (C) levels of PCBP2 in U251 cells transfected with miR-214 mimics or negative control (NC) for 24 or 48 hr, respectively. (D, E) mRNA (D) and protein (E) levels of PCBP2 in U251 cells transfected with miR-214 antisense oligos or negative control (NC) for 24 or 48 hr, respectively.
miR-214 regulates glioma cell growth in vitro
Next, to clarify the roles of miR-214 in glioma, we introduced miR-214 mimics into U251 and SHG-44 cells. As a result, miR-214 mimics significantly inhibited the abilities of cell viability, proliferation and invasion, compared with negative control (NC) (Figure 2A-C, Supplementary Figure 2A-C). Moreover, anti-miR-214 promoted the growth and invasion of glioma cells, compared to NC-transfected cells (Figure 3A-C, Supplementary Figure 3A-C). These results provide strong evidence that miR-213 could regulate glioma cell growth in vitro.
Figure 2.
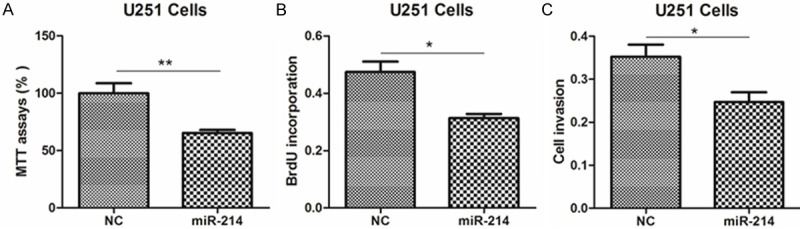
miR-214 mimics inhibits glioma cell proliferation. (A-C) The cell viability (A), proliferative potential (B) and invasion abilities were determined in U251 cells transfected with miR-214 mimics or negative control (NC).
Figure 3.
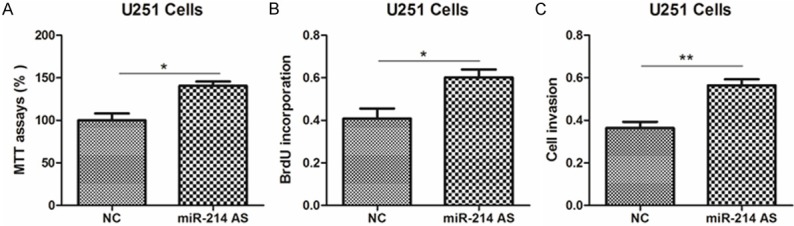
Inhibition of miR-214 promotes glioma cell proliferation. (A-C) The cell viability (A), proliferative potential (B) and invasion abilities were determined in U251 cells transfected with miR-214 antisense or negative control (NC).
PCBP2 re-introduction reversed the anti-proliferative role of miR-214
To further verify the functional connection between miR-214 and PCBP2, U251 cells were transfected with PCBP2 plasmids or empty vector (EV) (Figure 4A). As shown in Figure 4B-D, PCBP2 overexpression reversed the tumor suppressive roles of miR-214, underlining the specific importance of the PCBP2 for miR-214 action in the cell proliferation and invasion.
Figure 4.
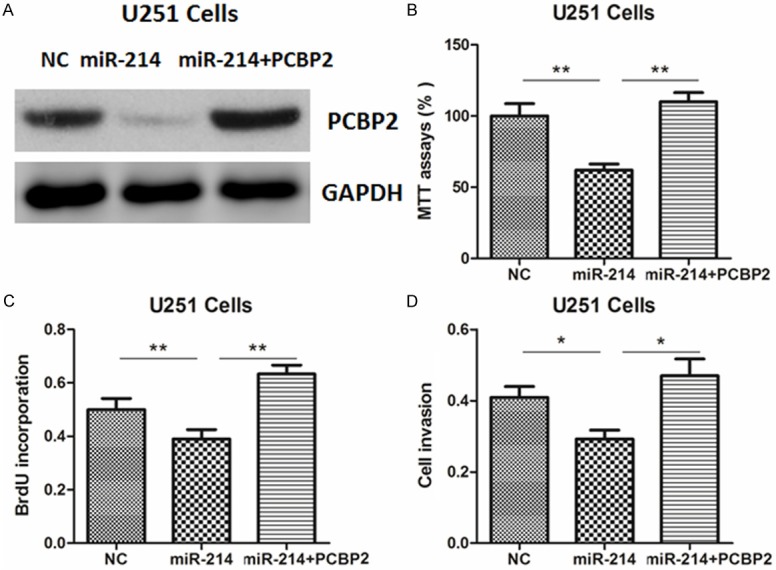
PCBP2 re-introduction reverses the anti-proliferative roles of miR-214. (A) PCBP2 protein expression was determined by western blot in U251 cells. Cells were pre-transfected with miR-214 mimics or negative control (NC) for 24 hr, and then transfected with expression plasmids for PCBP2 or empty vector (EV) for another 24 hr. (B-D) The cell viability (B), proliferation (C) and invasion abilities (D) were determined in U251 cells.
The expression level of miR-214 is down-regulated and inversely correlated with PCBP2 mRNA in glioma tissues
Given that miR-214 could regulate PCBP2 expression and cell growth in vitro, its expression level was determined in glioma tissues. As shown in the Figure 5A and 5B, the expression levels of miR-214 were significantly higher, while PCBP2 was up-regulated in glioma tissues, compared with adjacent normal tissues. Furthermore, across all specimens tested, we found an inverse correlation between the expression of miR-214 and the level of PCBP2 mRNA (Figure 5C), further suggesting that miR-214 may be involved in the regulation of PCBP2 in glioma.
Figure 5.
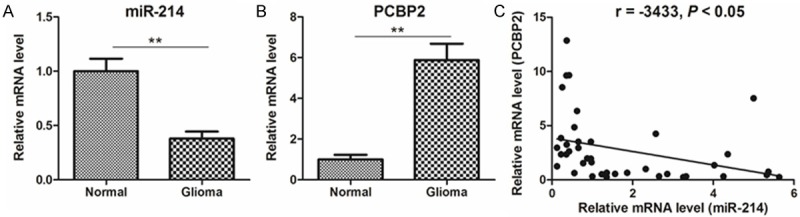
Expression of miR-214 and PCBP2 in glioma tissue specimens. A, B. Relative expression levels of miR-214 and PCBP2 in glioma and adjacent normal tissues. C. An inverse correlation between the expression of miR-214 and the level of PCBP2 mRNA in two groups of tissues.
Discussion
The results from our study indicate that mRNA and protein levels of PCBP2 are negatively regulated by miR-214. Although the functional significance of PCBP2 in glioma has been studied in vitro and in vivo [11], the molecular mechanisms for its up-regulation remain poorly understood. Therefore, our data highlight an important role of miR-214 in the regulation of PCBP2. However, whether this regulatory pathway occurs in other human cancers remain to be determined.
Moreover, we found that miR-214 was down-regulated in glioma tissues and could inhibit cell proliferation and growth. It has been shown that miR-214 is down-regulated in human cervical cancer tissue compared with normal tissue [17]. miR-214 represses HeLa cell proliferation by targeting the noncoding regions of MEK3 and JNK1 mRNAs [17]. Besides, miR-214 reduces cell survival and enhances cisplatin-induced cytotoxicity via down-regulation of Bcl2l2 in cervical cancer cells [18]. On the other hand, miR-214 was up-regulated in pancreatic cancer tissues and the elevation of miR-214 expression in pancreatic cancer specimens might be related to the poor response of pancreatic cancer cells to chemotherapy [19]. Moreover, miR-214 was noted to be highly overexpressed in gastric cancer tissues and cell lines [20]. As a result, knockdown of miR-214 could significantly inhibit proliferation, migration and invasion of gastric cancer cells [20]. Therefore, miR-214 could act as an oncogenic miRNA or a tumor suppressor. Although the inconsistence for these observations remains largely unknown, we speculate that the precise role of miR-214 might be tissue or cell-specific.
Increasing reports have demonstrated that some miRNAs are dys-regulated and play a key role in the development of human glioma [14-16]. Together with these studies, our results suggest that identification of specific miRNAs and their targets may provide novel insight for the diagnosis and treatment of glioma.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Chang L, Su J, Jia X, Ren H. Treating malignant glioma in Chinese patients: update on temozolomide. Onco Targets Ther. 2014;7:235–44. doi: 10.2147/OTT.S41336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y, Jiang T. Understanding high grade glioma: molecular mechanism, therapy and comprehensive management. Cancer Lett. 2013;331:139–146. doi: 10.1016/j.canlet.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 3.Kumthekar PU, Macrie BD, Singh SK, Kaur G, Chandler JP, Sejpal SV. A review of management strategies of malignant gliomas in the elderly population. Am J Cancer Res. 2014;4:436–44. [PMC free article] [PubMed] [Google Scholar]
- 4.Tommerup N, Leffers H. Assignment of human KH-box-containing genes by in situ hybridization: HNRNPK maps to 9q21.32-q21.33, PCBP1 to 2p12-p13 and PCBP2 to 12q13.12-q13.13, distal to FRA12A. Genomics. 1996;32:297–298. doi: 10.1006/geno.1996.0121. [DOI] [PubMed] [Google Scholar]
- 5.Choi HS, Hwang CK, Song KY, Law PY, Wei LN, Loh HH. Poly (C)-binding proteins as transcriptional regulators of gene expression. Biochem Biophys Res Commun. 2009;380:431–436. doi: 10.1016/j.bbrc.2009.01.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans JR, Mitchell SA, Spriggs KA, Ostrowski J, Bomsztyk K, Ostarek D, Willis AE. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene. 2003;22:8012–2020. doi: 10.1038/sj.onc.1206645. [DOI] [PubMed] [Google Scholar]
- 7.Sean P, Nguyen JH, Semler BL. The linker domain of poly (rC) binding protein 2 is a major determinant in poliovirus cap-independent translation. Virology. 2008;378:243–253. doi: 10.1016/j.virol.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sean P, Nguyen JH, Semler BL. Altered interactions between stem-loop IV within the 5’ noncoding region of coxsackievirus RNA and poly (rC) binding protein 2: effects on IRES-mediated translation and viral infectivity. Virology. 2009;389:45–58. doi: 10.1016/j.virol.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waggoner SA, Johannes GJ, Liebhaber SA. Depletion of the poly (C)-binding proteins alphaCP1 and alphaCP2 from K562 cells leads to p53-independent induction of cyclin-dependent kinase inhibitor (CDKN1A) and G1 arrest. J Biol Chem. 2009;284:9039–49. doi: 10.1074/jbc.M806986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu CE, Liu YC, Zhang HD, Huang GJ. The RNA-binding protein PCBP2 facilitates gastric carcinoma growth by targeting miR-34a. Biochem Biophys Res Commun. 2014;448:437–42. doi: 10.1016/j.bbrc.2014.04.124. [DOI] [PubMed] [Google Scholar]
- 11.Han W, Xin Z, Zhao Z, Bao W, Lin X, Yin B, Zhao J, Yuan J, Qiang B, Peng X. RNA-binding protein PCBP2 modulates glioma growth by regulating FHL3. J Clin Invest. 2013;123:2103–18. doi: 10.1172/JCI61820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–64. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs--the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 14.Low SY, Ho YK, Too HP, Yap CT, Ng WH. MicroRNA as potential modulators in chemoresistant high-grade gliomas. J Clin Neurosci. 2014;21:395–400. doi: 10.1016/j.jocn.2013.07.033. [DOI] [PubMed] [Google Scholar]
- 15.Zhao B, Bian EB, Li J, Li J. New advances of microRNAs in glioma stem cells, with special emphasis on aberrant methylation of microRNAs. J Cell Physiol. 2014;229:1141–7. doi: 10.1002/jcp.24540. [DOI] [PubMed] [Google Scholar]
- 16.Besse A, Sana J, Fadrus P, Slaby O. MicroRNAs involved in chemo- and radioresistance of high-grade gliomas. Tumour Biol. 2013;34:1969–78. doi: 10.1007/s13277-013-0772-5. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z, Chen S, Luan X, Li Y, Liu M, Li X, Liu T, Tang H. MicroRNA-214 is aberrantly expressed in cervical cancers and inhibits the growth of HeLa cells. IUBMB Life. 2009;61:1075–82. doi: 10.1002/iub.252. [DOI] [PubMed] [Google Scholar]
- 18.Wang F, Liu M, Li X, Tang H. MiR-214 reduces cell survival and enhances cisplatin-induced cytotoxicity via down-regulation of Bcl2l2 in cervical cancer cells. FEBS Lett. 2013;587:488–95. doi: 10.1016/j.febslet.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 19.Zhang XJ, Ye H, Zeng CW, He B, Zhang H, Chen YQ. Dysregulation of miR-15a and miR-214 in human pancreatic cancer. J Hematol Oncol. 2010;3:46. doi: 10.1186/1756-8722-3-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang TS, Yang XH, Wang XD, Wang YL, Zhou B, Song ZS. MiR-214 regulate gastric cancer cell proliferation, migration and invasion by targeting PTEN. Cancer Cell Int. 2013;13:68. doi: 10.1186/1475-2867-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


