Abstract
Gliomas are the most common and aggressive type of primary adult brain tumor. Although high expression and prognostic value of TMEM45A has been recently reported in various types of human tumors, the association of TMEM45A expression and glioma is still unknown. Here, we reported that TMEM45A was significantly overexpressed in glioma tissues compared to non-tumorous brain tissues. Furthermore, TMEM45A mRNA levels were gradually increased with the increasing severity of histological grade of glioma. Moreover, high TMEM45A expression level was correlated with short survival time of glioma patients. Down-regulation of TMEM45A in two glioma cell lines, U251 and U373 by transected with TMEM45A siRNA resulted in a significant reduction of cell proliferation and G1-phase arrest. Additionally, we found that suppressing of TMEM45A expression in glioma cells remarkably suppressed cell migration and cell invasion. More importantly, TMEM45A siRNA treatment significantly down-regulated the proteins promoting cell cycles transition (Cyclin D1, CDK4 and PCNA) and cell invasion (MMP-2 and MMP-9), which indicted a possible mechanism underlying its functions on glioma. In summary, our study suggests that TMEM45A may work as an oncogene and a new effective therapeutic target for glioma treatment.
Keywords: TMEM45A, glioma, proliferation, invasion
Introduction
Gliomas are the most common and aggressive type of primary adult brain tumor and persist as serious clinical and scientific problems [1]. There are three types of gliomas: astrocytoma, oligodendroglioma and ependymoma [2]. Current therapies are not effective in treating gliomas. Therefore, glioma remains as one of the leading causes of cancer deaths worldwide [3,4]. The median survival of patients with glioblastoma multiforme, the most malignant glioma, remains less than one year, even with aggressive surgery, radiation and chemotherapy [3]. New prognostic indicators and effective therapeutic targets for gliomas still needed to be identified, which urges a better understanding of the molecular mechanisms governing disease manifestation and progression.
Transmembrane protein 45A (TMEM45A), a multi-pass membrane protein, belongs to the large family of genes encoding predicted transmembrane (TMEM) proteins. TMEM45A has been reported to be involved in epidermal keratinization [5,6]. Recently, high expression of TMEM45A has been linked to poor prognostic in patients with different types of tumors [7-10]. In liver and breast cancer cells, TMEM45A expression is implicated in protecting liver and breast cancer cells from drug-induced apoptosis [7]. Lee et al. reported that TMEM45A inhibits the progression of ductal carcinoma into invasive breast cancer [11]. However, how alterations of TMEM45A expression are involved in the malignancies remains to be solved.
In the present study, we explored the role of TMEM45A in gliomas and sought to identify the involved mechanisms. TMEM45A mRNA level was significantly higher in glioma tissues than in non-tumorous brain tissues. Furthermore, TMEM45A mRNA levels were gradually increased with the increasing severity of histological grades of gliomas. Our in vitro experiments indicated that TMEM45A was involved in multiple cellular progress including cell proliferation, cell cycle progression, migration and invasion. Furthermore, the mRNA and protein levels of cell cycle related-genes and invasion related-genes were decreased in TMEM45A knockdown cells. Collectively, these data suggest that TMEM45A is a potent oncogene in glioma and it may be an effective therapeutic target for this disease.
Materials and methods
Tissue samples and gene expression data
Fresh frozen samples of 45 glioma tissues and 11 non-neoplastic brain tissues from surgical procedures for epilepsy were obtained from Shanghai Changzheng Hospital. In 45 patients with glioma enrolled in this study, 10 patients were with WHO (World Health Organization) Grade II glioma, 12 were with Grade III glioma, and 23 were with Grade IV glioma. Informed consent was obtained from all patients. This study was approved by the ethics committee of Second Military Medical University.
The Cancer Genome Atlas (TCGA) glioblastoma (GBM) dataset of 529 patients and 10 normal brain tissues (version: 2014-08-22) were downloaded from TCGA website (https://tcga-data.nci.nih.gov/tcga/).
Immunohistochemical analysis
Immunohistochemistry (IHC) was performed as previously described [12]. The results of IHC staining were evaluated independently by two trained pathologists without knowledge of clinical data.
Cell lines
The human glioblastoma cell lines, U87, SHG44, U251, T98G and U373 cells were from cell bank of Shanghai biology institute, Chinese Academy of Science (Shanghai, China) and maintained at 37°C in 5% CO2 atmosphere. All culture media were supplemented with 10% fetal bovine serum (FBS, Life Technologies, Carlsbad, CA, USA), 100 mg/ml penicillin G, and 50 g/ml streptomycin (Life Technologies). U251 and SHG44 cells were cultured in DMEM (Life Technologies), while U87, U373 and T98G were cultured in Eagle’s Minimum Essential Medium (Life Technologies).
Silencing of TMEM45A by small interfering RNA
Three siRNAs targeting human TMEM45A mRNA were synthesized (TMEM45A-siRNA1: 5’-GGCCUUUAUCUUCUACAACUU-3’; TMEM45A-siRNA2: 5’-GUUCCUUGUUCGGAACAAUUU-3’; TMEM45A-siRNA2: 5’-AGUGUACUGUUUGCAUUUCUU-3’). A non-specific scramble siRNA sequence was used as negative control (NC: 5’-UUGUACUACACAAAAGUACUG-3’). The siRNAs were transiently transfected into U251 and U373 cells using Lipofectamine 2000 (Invitrogen) according to the manufacture’s instruction. Assays were performed 48 h after transfection.
Reverse transcription and Real-Time PCR
Total RNA was extracted from cultured cells or tissue samples using TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. Total RNA (1 μg) was reverse-transcribed using M-MuLV Reverse Transcriptase (Thermo Fisher Scientific, Rockford, IL, USA). The resulting cDNA was used as template in real-time quantitative PCR by using a standard SYBR Green PCR kit (Thermo) on an ABI 7300 Thermocycler Real-Time PCR machine (Applied Biosystems, Foster City, CA, USA). GAPDH was used as control of the input RNA level. Primer sequences were listed in Table 1. All reactions were conducted using the following cycling parameters, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C for 45 s. GAPDH was served as an internal control. The gene expression was calculated using the ΔΔ Ct method. All data represent the average of three replicates.
Table 1.
Primers sequences for quantitative PCR
| Primer | Primer sequence | Size (bp) |
|---|---|---|
| TMEM45A (NM_018004.1) | F: 5’-CTCCTGGGCTGGCATCATTTC-3’ | 167 |
| R: 5’-CGGCCATGAGTGTGGTTGTAG-3’ | ||
| Cyclin D1 (NM_053056) | F: 5’-GCTGCGAAGTGGAAACCATC-3’ | 135 |
| R: 5’-CCTCCTTCTGCACACATTTGAA-3’ | ||
| CDK4 (NM_000075) | F: 5’-ATGGCTACCTCTCGATATGAGC-3’ | 124 |
| R: 5’-CATTGGGGACTCTCACACTCT-3’ | ||
| CDK2 (NM_001798) | F: 5’-CCAGGAGTTACTTCTATGCCTGA-3’ | 90 |
| R: 5’-TTCATCCAGGGGAGGTACAAC-3’ | ||
| PCNA (NM_002592.2) | F: 5’-GCCTGACAAATGCTTGCTGAC-3’ | 223 |
| R: 5’-GCTGTTCTTGGCCTCAGTC-3’ | ||
| MMP-2 (NM_004530) | F: 5’-TACAGGATCATTGGCTACACACC-3’ | 90 |
| R: 5’-GGTCACATCGCTCCAGACT-3’ | ||
| MMP-9 (NM_004994) | F: 5’-TGTACCGCTATGGTTACACTCG-3’ | 97 |
| R: 5’-GGCAGGGACAGTTGCTTCT-3’ | ||
| GADPH (NM_001256799.1) | F: 5’-CACCCACTCCTCCACCTTTG-3’ | 110 |
| R: 5’-CCACCACCCTGTTGCTGTAG-3’ |
Immunoblotting
Treated and untreated cells were lysed in radioimmunoprecipitation assay buffer (JRDUN Biotehnology, Shanghai, China) and protein concentration was measured by BCA protein assay kit (Thermo Fisher Scientific) [13]. Protein isolates were then subjected to SDS gel electrophoresis and analyzed by western blot using enhanced chemiluminescence system (ECL, Millipore). Antibodies against TMEM45A, PCNA and MMP-9 were purchased from Abcam (Cambridge, MA, USA). Anti-Cyclin D1 was from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibodies against CDK4 and GAPDH were purchased from CST Biotech. (Danvers, MA, USA). Anti-MMP-2 was from Epitmics (Burlingame, CA, USA).
Cell proliferation assay
Treated and untreated cells were seeded at a density of 5×103 cells per well in a 96-well plate. Cell proliferation was detected by using the Cell Count Kit-8 (CCK-8, Dojindo Laboratories) according to the manufacturer’s instructions. At indicated time point, CCK8 solution was added to each well and incubated for 1 h. Absorbance was detected at a wavelength of 450 nm by a microplate reader (Bio-Rad Laboratories Inc., Hercules, CA, USA). Three wells were measured for cell viability in each treatment group, and all independent treatments were performed in triplicate.
Cell cycle distribution analysis
The cell cycle was evaluated by flow cytometry using propidium iodide (PI) staining on a flow cytometer (BD Biosciences, San Jose, CA, USA). Briefly, treated and untreated cells were harvested, re-suspended in PBS and fixed ice-cold 70% ethanol at -20°C for at least 2 h. The fixed cells were washed with PBS, incubated with ribonuclease A (Sigma) and PI (0.05 mg/ml, Sigma, St. Louis, MO, USA) at room temperature in the dark for 30 min. DNA content was then analyzed using a FACScan flow cytometry. The percentage of cells in the G0/G1, S, and G2/M phases was determined by the FlowJo software (Tree Star). Experiments were performed in triplicate and 3×104 cells were analyzed per sample.
Boyden chamber assay for migration and invasion
Quantitative cell migration and invasion assays were performed using Boyden chambers containing polycarbonate filters with a pore size of 8 μm (Coring Incorporated, NY, USA).
For transwell migration assay, treated and untreated cells were serum starved for 24 h, 5×104 cells were plated into the upper well of the Boyden chambers with serum-free medium in the top chamber and medium containing 10% FBS in the lower chamber. After 24 h of incubation, the cells on the upper surface of the filter were completely removed by wiping with a cotton swab. The remaining migrated cells were washed with PBS, fixed in 4% paraformaldehyde and stained with 0.2% crystal violet. The migrated cells were observed under Leica inverted microscope (Deerfield, IL, USA). Cell number was counted in 10 random fields for each condition. The experiments were performed in triplicate.
For in vitro invasion assay, the upper wells of the Boyden chambers were coated with Matrigel (BD Biosciences) at 37°C in a 5% CO2 incubator for 1 h. The rest of the assay was performed as described above.
Statistical analysis
The two-tailed Student’s t-test was used to evaluate statistical differences between two groups. Statistically significant differences were defined as having a P<0.05. Where appropriate, data are expressed as mean ± S.D. Overall survival time of patients was analyzed by the Kaplan-Meier survival curves and log-rank nonparametric test.
Results
Elevated expression of TMEM45A was correlated with the histological grade of primary human glioma
We first compared TMEM45A mRNA level in glioma tissues (n=45) and normal brain tissues (n=11) by using real-time PCR. Statistical analysis with student’s t-test suggested that TMEM45A expression was significantly elevated in glioma tissues compared with that in normal tissues (Figure 1A, P<0.0001). Then we re-analyzed high throughput RNA-sequencing data of the glioblastomas cohort of The Cancer Genome Atlas (TCGA) and found that TMEM45A expression was significantly increased in glioma tissues compared with normal brain tissues (Figure 1B, P<0.0001). We also assessed the expression level of TMEM45A by immunohistochemistry and found that TMEM45A protein level was also elevated in glioma tissues (Figure 1D).
Figure 1.
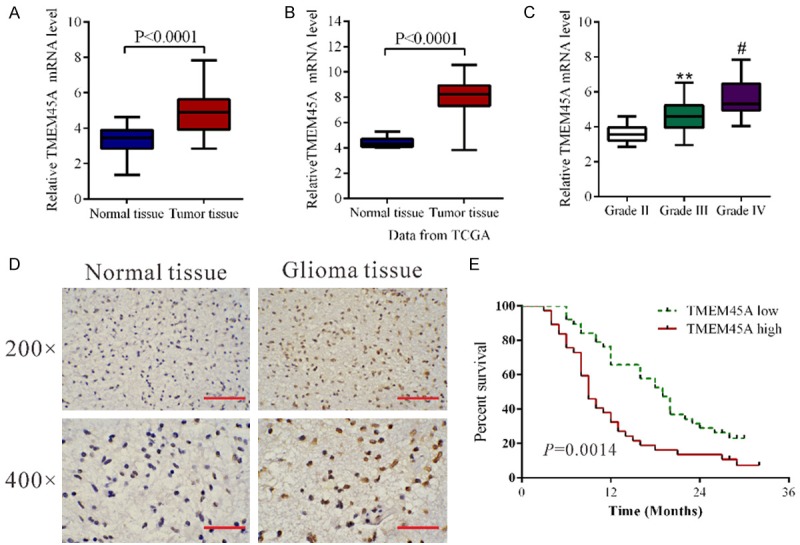
TMEM45A was overexpressed in glioma tissues. A. TMEM45A mRNA level was significantly higher in glioma tissues (n=45) than that in non-tumorous brain tissues (n=11) from patients admitted to Shanghai Changzheng Hospital between 2010 and 2013 (P<0.0001). B. TMEM45A expression was significantly increased in glioma tissues (n=529) when compared with normal tissues of patients (n=10) from TCGA dataset (P<0.0001). C. TMEM45A mRNA expression levels in 10 WHO Grade II glioma, 12 WHO Grade III glioma and 23 WHO Grade IV glioma (**P<0.01 as compared with Grade II glioma, #P<0.05 as compared with Grade III glioma). D. The expression of TMEM45A detected by immunohistochemisty in glioma and non-tumorous brain tissues. Low power (200×) scale bars: 100 μm, high power (400×) scale bars: 50 μm. E. Overall survival time of patients with gliomas.
To assess the clinical relevance of TMEM45A, we compared its mRNA expression among a series of gliomas of different grades (WHO (World Health Organization) grades II, III and IV) using real-time PCR data. As shown in Figure 1C, TMEM45A mRNA levels were gradually increased with the increasing grade of disease severity, which indicates the role of TMEM45A in the progression of glioma. Moreover, Kaplan-Meier analysis showed that the overall survival time of lower-TMEM45A-expressing was notably higher than that of higher-TMEM45A-expressing patients (Figure 1E).
Silencing of TMEM45A expression by RNA interference (RNAi)
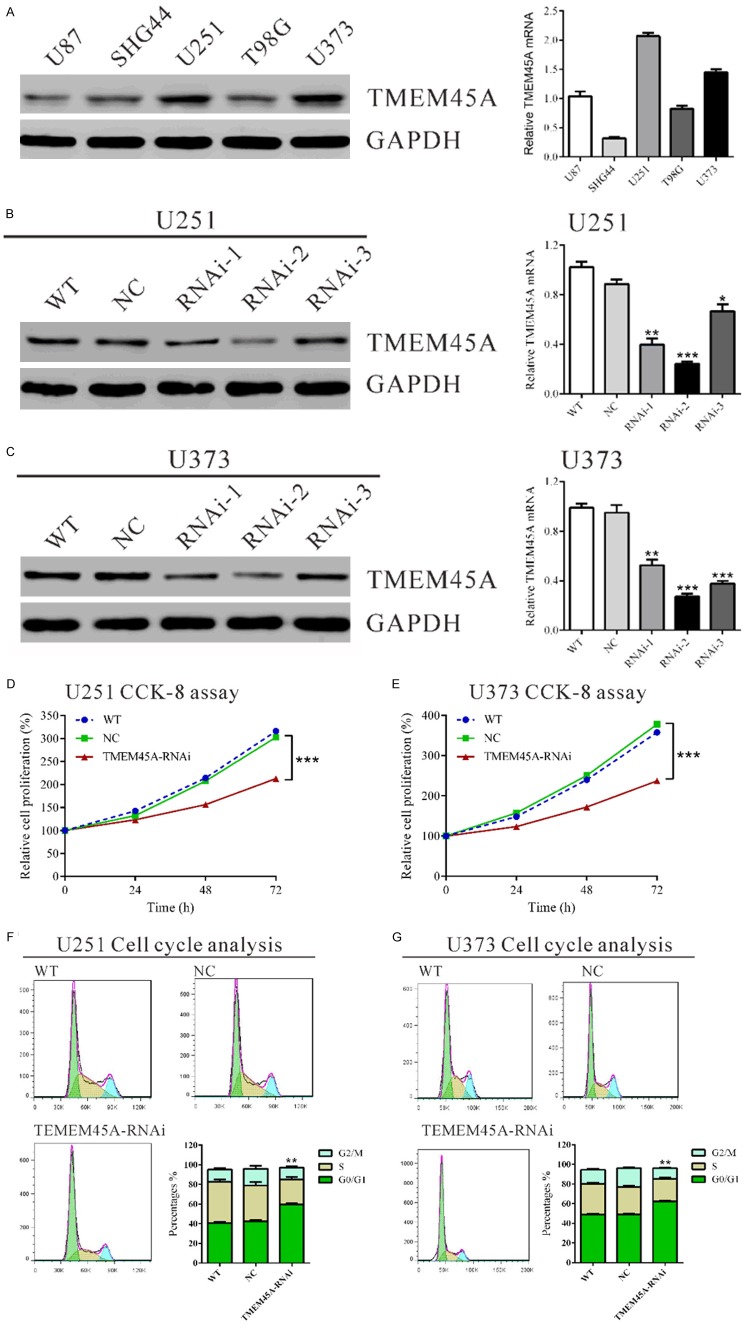
We evaluated the protein and mRNA levels of TMEM45A in five glioma cell lines, U87, SHG44, U251, T98G and U373 by western blot and real-time PCR, respectively. Two cell lines, U251 and U373 cells, showed higher TMEM45A mRNA and protein expression than the other three cell lines, U87, SHG44 and T98G cells (Figure 2A).
Figure 2.
Suppressing TMEM45A expression inhibited cell growth and G1/S phase transition in glioma cells. A. TMEM45A expression level in five glioma cell lines was analyzed by Western blot (left panel) and real-time PCR (right panel). B, C. Expression of TMEM45A in U251 and U373 cells was analyzed by Western blot (left panel) and real-time PCR (right panel). WT: wild type cells; NC: scrambled siRNA transfected cells; RNAi-1, RNAi-2 and RNAi-3: TMEM45A-siRNA-1, -2 and -3 transfected cells. D, E. Cell proliferation was detected in siRNA treated and untreated U251 and U373 cells. Results of cell growth were normalized to the initial cell numbers (100%). F, G. U251 and U373 cells were transfected with indicated siRNA and 48 hours later cells were collected. Cell cycle profile was analyzed using flow cytometry. Data were based on at least 3 independent experiments, and shown as mean ± S.D. (*P<0.05, **P<0.01, ***P<0.001 as compared with NC).
As high levels of TMEM45A were related to severity of glioma, we wonder whether TMEM45A might be an oncogene in glioma. We thus knockdown the expression of TMEM45A in U251 and U373 cells, which expressed high levels of TMEM45A by siRNA transfection. Three siRNAs targeting human TMEM45A (TMEM45A-RNAi) and negative control (NC, a non-specific scramble siRNA) were synthesized. As shown in Figure 2B and 2C, all siRNAs were able to efficiently suppress endogenous TMEM45A expression in both glioma cells, whereas TMEM45A expression remained unaffected in NC cells. TMEM45A-RNAi-2 was the most effective one and used for the following assays.
Depletion of TMEM45A suppressed the proliferation and induced G1-phase arrest of glioma cells
To examine the effects of TMEM45A silencing on the proliferation of glioma cells, we assessed the proliferation of TMEM45A knockdown glioma cells by using CCK-8 assay. In U251 (Figure 2D) and U373 cells (Figure 2E) with TMEM45A silenced, cell growth was significantly inhibited at 24 h, 48 h and 72 h (***P<0.001). These results indicated the proliferation-promoting role of TMEM45A in glioma cells.
To determine whether TMEM45A influences the cell cycle of glioma cells, cell cycle distribution was assessed in TMEM45A knockdown cells. Flow cytometry analysis revealed that the population of G0/G1 phase cells in U251 (Figure 2F) transfected with TMEM45A siRNA was significantly increased by 40.6% (**P<0.01), and S phase cells was decreased by 30.9%, compared with NC and WT cells. Similar results were obtained in U373 cells (Figure 2G).
Silencing of TMEM45A inhibited the metastasis of glioma cells
To investigate the involvement of TMEM45A in cell motility, transwell assays were carried out to quantitatively determine the effect of TMEM45A knockdown on cell migration. Suppressing of TMEM45A expression brought about a significant reduction in the migration of U251 and U373 cells. As shown in Figure 3A, similar numbers of WT and NC-transfected cells migrated to the lower face of Boyden chambers (U251: WT, 76 ± 1; NC, 75 ± 1; U373: WT, 62 ± 2; NC, 61 ± 2), whereas a strongly inhibited motility was observed in TMEM45A knockdown cells (U251: 46 ± 1; U373: 27 ± 1).
Figure 3.
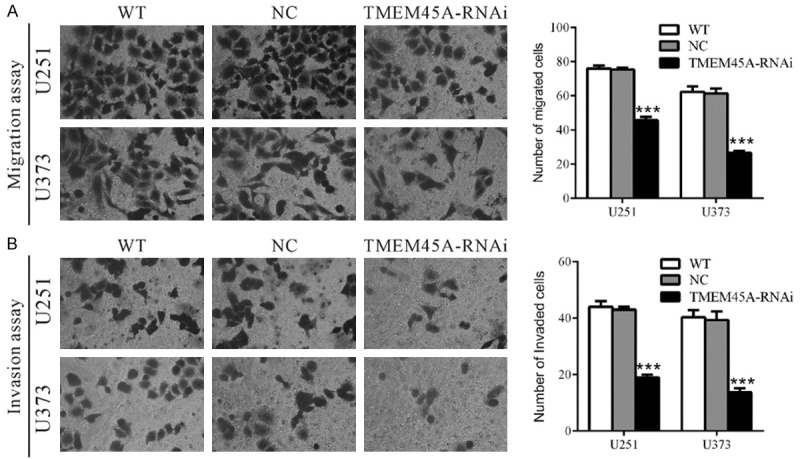
Silencing of TMEM45A inhibited cell migration and invasion in glioma cells. U251 and U373 cells were transfected with indicated siRNA and transwell migration assay and invasion assay were performed 48 h later. A. For transwell migration assay, cells were serum starved for 24 h, and then plated into the upper well of the Boyden chambers with serum-free medium in the top chamber and medium containing 10% FBS in the lower chamber. After 24 h of incubation, cells that migrated into the lower well were stained, photographed and counted. B. For invasion assay, the upper wells of Boyden chambers were pre-coated with Matrigel. The rest of the assay was performed as described above. Data were based on at least 3 independent experiments, and shown as mean ± S.D. (***P<0.001 as compared with NC).
We also investigated whether TMEM45A affected the invasive ability of glioma cells. In vitro invasion assay was performed in Boyden chambers with the upper wells coated with Matrigel to mimic the extracellular matrix. In sharp contrast to control cells, TMEM45A knockdown cells showed dramatically reduced invasive ability (Figure 3B). The number of invaded cells was 44.2% and 35.9% of that of the control cells in U251 and U373 cells, respectively (U251: WT, 44 ± 1; NC, 43 ± 1; RNAi, 19 ± 1; U373: WT, 40 ± 1; NC, 39 ± 2; RNAi, 14 ± 1). These data suggested a role of TMEM45A in the promotion of glioma metastasis.
TMEM45A siRNA down-regulated the expression of cell cycle related and invasion related genes in glioma cells
After 48 h of TMEM45A siRNA treatment, the mRNA and protein levels of cell cycle-related genes, Cyclin B1, CDK2 and PCNA, and invasion related genes, MMP-2 and MMP-9 were analyzed. As revealed in Figure 4A and 4B, the protein levels of Cyclin D1, CDK4, PCNA, MMP-2 and MMP-9 in U251 cells with TMEM45A silenced were 71.9%, 50.5%, 21.6%, 53.2% and 35.3% of that in the control cells, respectively. The mRNA levels of detected genes were also decreased in TMEM45A treated U251 cells (Figure 4C). Similar results were obtained in U373 cells (Figure 4D-F).
Figure 4.
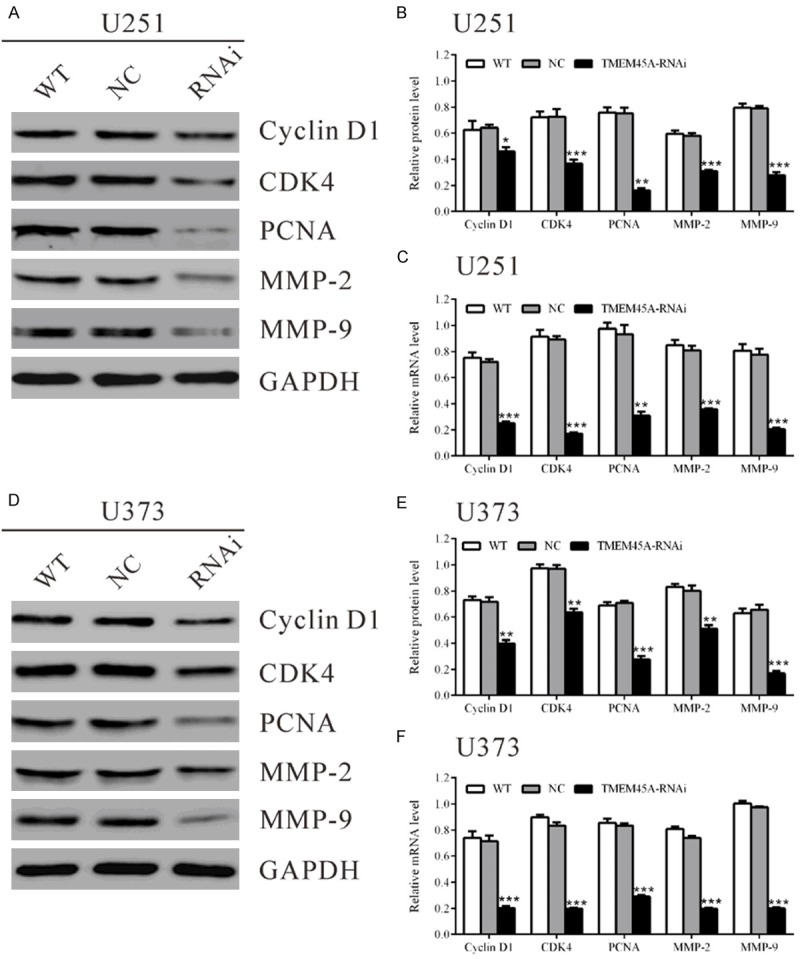
TMEM45A siRNA down-regulated the expression of cell cycle related and invasion related genes in glioma cells. Protein and mRNA levels of cell cycle related-genes (CyclinD1, CDK4 and PCNA) and invasion related-genes (MMP-2 and MMP-9) in U251 and U373 cells were detected. GAPDH was also detected as the control of sample loading. Representative western blots were shown in (A) and (D). Quantitative results were shown in (B) and (E). (C, F) mRNA levels were detected by real-time PCR. Data were based on at least three independent experiments, and shown as mean ± SD (*P<0.05, **P<0.01, ***P<0.001 as compared with NC). WT: wild type cells; NC: scrambled siRNA transfected cells; RNAi: TMEM45A-siRNA transfected cells.
Discussion
High expression of TMEM45A was reported in breast cancer [7], bladder cancer [9] and ovarian cancer [10,14] and may serve as poor prognostic factor for these malignancies. Here, we found that TMEM45A mRNA levels were elevated in glioma tissues, which suggested a role of TMEM45A in glioma (Figure 1A and 1B). Further analysis revealed that its expression was progressively increased with the increasing severity of histological grade (Figure 1C). Survival of patients with glioma depends heavily on the histological grade of the tumor, thus TMEM45A expression may be a useful prognosis factor for glioma.
Then we investigated the functions of TMEM45A in glioma cells by suppressing its expression. Deregulated proliferation has profound effects on the malignant phenotype of cancer. Here, we found that knockdown of TMEM45A in glioma cells significantly reduced cell growth by inhibiting cell cycle progression (Figure 2). Cyclin D1-CDK4 complex are key players for G1/S transition [15,16]. Therefore, we then evaluated the expression of the above genes. As shown in Figure 4, the expression of Cyclin D1, CDK4 and PCNA was notably decreased in TMEM45A knockdown cells, which further demonstrated the role of TMEM45A in the G1/S transition.
Previous studies indicate that gliomas tend to recur soon after resection of the initial tumor mass because of the highly invasive ability of glioma cells to invade adjacent areas by degrading extracellular matrix (ECM) components and diffusing into normal brain tissue [17]. MMP-2 and MMP-9 are well known to play a critical role in tumor invasion, angiogenesis, metastasis and recurrence [18,19]. Here, our data indicated that TMEM45A siRNA notably inhibited the migration and invasion of glioma cells (Figure 3), and decrease the expression of MMP-2 and MMP-9 (Figure 4). Taken together, we speculated that TMEM45A may perform its invasion-promoting function through regulating MMP-2 and MMP-9.
A recent study reported that inhibition of TMEM45A impaired cell proliferation, cell cycle transition and reduced cell invasion of ovarian cancer cells [14]. These data matched well with our results of in vitro experiments, suggesting that TMEM45A is a potential oncogene regardless of location and might be a therapy target for multiple cancers.
In summary, our study demonstrated for the first time that TMEM45A was overexpressed in glioma tissues and TMEM45A expression was associated with the histological tumor grade. In addition, our data indicated that TMEM45A played a key role in the proliferation and metastasis of glioma cells. Whether TMEM45A can be used as a therapeutic target for glioma remains to be further investigated.
Acknowledgements
This study was supported by grant no. 81172398 from the National Natural Science Foundation of China.
Disclosure of conflict of interest
None.
References
- 1.Sathornsumetee S, Rich JN. New treatment strategies for malignant gliomas. Expert Rev Anticancer Ther. 2006;6:1087–1104. doi: 10.1586/14737140.6.7.1087. [DOI] [PubMed] [Google Scholar]
- 2.Kleihues P, Cavenee WK. Pathology and genetics of tumours of the nervous system. International Agency for Research on Cancer. 2000 [Google Scholar]
- 3.Davis FG, Freels S, Grutsch J, Barlas S, Brem S. Survival rates in patients with primary malignant brain tumors stratified by patient age and tumor histological type: an analysis based on Surveillance, Epidemiology, and End Results (SEER) data, 1973-1991. J Neurosurg. 1998;88:1–10. doi: 10.3171/jns.1998.88.1.0001. [DOI] [PubMed] [Google Scholar]
- 4.Penas-Prado M, Gilbert MR. Molecularly targeted therapies for malignant gliomas: advances and challenges. Expert Rev Anticancer Ther. 2007;7:641–661. doi: 10.1586/14737140.7.5.641. [DOI] [PubMed] [Google Scholar]
- 5.Hayez A, Malaisse J, Roegiers E, Reynier M, Renard C, Haftek M, Geenen V, Serre G, Simon M, de Rouvroit CL, Michiels C, Poumay Y. High TMEM45A expression is correlated to epidermal keratinization. Exp Dermatol. 2014;23:339–344. doi: 10.1111/exd.12403. [DOI] [PubMed] [Google Scholar]
- 6.Mattiuzzo NR, Toulza E, Jonca N, Serre G, Guerrin M. A large-scale multi-technique approach identifies forty-nine new players of keratinocyte terminal differentiation in human epidermis. Exp Dermatol. 2011;20:113–118. doi: 10.1111/j.1600-0625.2010.01188.x. [DOI] [PubMed] [Google Scholar]
- 7.Flamant L, Roegiers E, Pierre M, Hayez A, Sterpin C, De Backer O, Arnould T, Poumay Y, Michiels C. TMEM45A is essential for hypoxia-induced chemoresistance in breast and liver cancer cells. BMC Cancer. 2012;12:391. doi: 10.1186/1471-2407-12-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jinawath N, Chamgramol Y, Furukawa Y, Obama K, Tsunoda T, Sripa B, Pairojkul C, Nakamura Y. Comparison of gene expression profiles between Opisthorchis viverrini and non-Opisthorchis viverrini associated human intrahepatic cholangiocarcinoma. Hepatology. 2006;44:1025–1038. doi: 10.1002/hep.21330. [DOI] [PubMed] [Google Scholar]
- 9.Urquidi V, Goodison S, Cai Y, Sun Y, Rosser CJ. A candidate molecular biomarker panel for the detection of bladder cancer. Cancer Epidemiol Biomarkers Prev. 2012;21:2149–2158. doi: 10.1158/1055-9965.EPI-12-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crijns AP, Fehrmann RS, de Jong S, Gerbens F, Meersma GJ, Klip HG, Hollema H, Hofstra RM, te Meerman GJ, de Vries EG, van der Zee AG. Survival-related profile, pathways, and transcription factors in ovarian cancer. PLoS Med. 2009;6:e24. doi: 10.1371/journal.pmed.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S, Stewart S, Nagtegaal I, Luo J, Wu Y, Colditz G, Medina D, Allred DC. Differentially expressed genes regulating the progression of ductal carcinoma in situ to invasive breast cancer. Cancer Res. 2012;72:4574–4586. doi: 10.1158/0008-5472.CAN-12-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayez A, Malaisse J, Roegiers E, Reynier M, Renard C, Haftek M, Geenen V, Serre G, Simon M, Rouvroit CL. High TMEM45A expression is correlated to epidermal keratinization. Exp Dermatol. 2014;23:339–344. doi: 10.1111/exd.12403. [DOI] [PubMed] [Google Scholar]
- 13.Ding DF, Li XF, Xu H, Wang Z, Liang QQ, Li CG, Wang YJ. Mechanism of resveratrol on the promotion of induced pluripotent stem cells. J Integr Med. 2013;11:389–396. doi: 10.3736/jintegrmed2013039. [DOI] [PubMed] [Google Scholar]
- 14.Guo J, Chen L, Luo N, Yang W, Qu X, Cheng Z. Inhibition of TMEM45A suppresses proliferation, induces cell cycle arrest and reduces cell invasion in human ovarian cancer cells. Oncol Rep. 2015;33:3124–3130. doi: 10.3892/or.2015.3902. [DOI] [PubMed] [Google Scholar]
- 15.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nature Reviews Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 16.Dietrich DR. Toxicological and pathological applications of proliferating cell nuclear antigen (PCNA), a novel endogenous marker for cell proliferation. Crit Rev Toxicol. 1993;23:77–109. doi: 10.3109/10408449309104075. [DOI] [PubMed] [Google Scholar]
- 17.Giese A, Westphal M. Glioma invasion in the central nervous system. Neurosurgery. 1996;39:235–252. doi: 10.1097/00006123-199608000-00001. [DOI] [PubMed] [Google Scholar]
- 18.Forsyth PA, Wong H, Laing TD, Rewcastle NB, Morris DG, Muzik H, Leco KJ, Johnston RN, Brasher PM, Sutherland G, Edwards DR. Gelatinase-A (MMP-2), gelatinase-B (MMP-9) and membrane type matrix metalloproteinase-1 (MT1-MMP) are involved in different aspects of the pathophysiology of malignant gliomas. Br J Cancer. 1999;79:1828–35. doi: 10.1038/sj.bjc.6990291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turpeenniemi-Hujanen T. Gelatinases (MMP-2 and-9) and their natural inhibitors as prognostic indicators in solid cancers. Biochimie. 2005;87:287–297. doi: 10.1016/j.biochi.2005.01.014. [DOI] [PubMed] [Google Scholar]