Abstract
Organismal life encounters reactive oxidants from internal metabolism and environmental toxicant exposure. Reactive oxygen and nitrogen species cause oxidative stress and are traditionally viewed as being harmful. On the other hand, controlled production of oxidants in normal cells serves useful purposes to regulate signaling pathways. Reactive oxidants are counterbalanced by complex antioxidant defense systems regulated by a web of pathways to ensure that the response to oxidants is adequate for the body’s needs. A recurrent theme in oxidant signaling and antioxidant defense is reactive cysteine thiol–based redox signaling. The nuclear factor erythroid 2–related factor 2 (Nrf2) is an emerging regulator of cellular resistance to oxidants. Nrf2 controls the basal and induced expression of an array of antioxidant response element–dependent genes to regulate the physiological and pathophysiological outcomes of oxidant exposure. This review discusses the impact of Nrf2 on oxidative stress and toxicity and how Nrf2 senses oxidants and regulates antioxidant defense.
Keywords: reactive oxygen species, redox signaling, antioxidant response element, inducer, cytoprotection
INTRODUCTION
Reactive oxygen and nitrogen species (ROS, RNS) are constantly generated in the body from internal metabolism and external exposure (1, 2). In normal cells, reactive oxidants are produced in a controlled manner and some serve useful purposes. Oxidants formed in response to physiological cues act as important signaling molecules to regulate such processes as cell division, inflammation, immune function, autophagy, and stress response (1). Uncontrolled production of oxidants results in oxidative stress that impairs cellular functions and contributes to the development of cancer, chronic disease, and toxicity (2–5). From prokaryotes to humans, reactive oxidants seemingly function as important regulators of both physiological and pathophysiological outcomes.
The nuclear factor erythroid 2 (NFE2)-related factor 2 (Nrf2) is a member of the cap ‘n’ collar (CNC) subfamily of basic region leucine zipper (bZip) transcription factors. Nrf2 was cloned by virtue of its binding to the NFE2-binding motif, a cis-regulatory sequence in the β-globin locus control region necessary for erythropoiesis and platelet development (6). Nrf2 does not appear to be essential for blood cell differentiation but was found to mediate induction of a set of drug-metabolizing enzymes (DMEs), such as glutathione S-transferase (GST) and NAD(P)H:quinone oxidoreductase 1 (NQO1), by antioxidants and electrophiles (7, 8). Induction requires a common DNA sequence called antioxidant response element (ARE) that resembles the NFE2-binding motif (9). Induction of the DMEs leads to increased detoxification and elimination of numerous exogenous and some endogenous chemicals. In this role, Nrf2 functions as a xenobiotic-activated receptor (XAR) to regulate the adaptive response to oxidants and electrophiles (10).
A major emerging function of Nrf2 from studies over the past decade is its role in resistance to oxidant stress. As follows, knockout of Nrf2 in mice (Nrf2 KO) substantially increased the susceptibility of mice to a broad range of chemical toxicity and disease conditions associated with oxidative pathology (5, 11–15). Pharmacological boosting of the Nrf2 activity with chemoprotective agents protected animals from oxidative damage (16). Genomic-scale search for Nrf2 target genes identified a number of ARE-containing genes involved in the control of oxidant homeostasis in addition to drug metabolism (17). Molecular and structural analyses of Nrf2 signaling uncovered a “dedepression” regulatory mechanism, wherein Nrf2 is suppressed under a basal condition through Keap1 (Kelch-like erythroid cell-derived protein with CNC homology-associated protein 1)-dependent ubiquitination-proteasomal degradation and is activated by oxidants and electrophiles via modification of critical cysteine thiols of Keap1 and Nrf2 (18). The protective nature of Nrf2 could also be appropriated by cancer cells to create a prosurvival microenvironment for tumor growth and drug resistance (19). This review focuses on the emerging role and molecular mechanism of action of Nrf2 in the regulation of oxidative stress and associated physiology and toxicity.
OXIDANT RESISTANCE AND REDOX SIGNALING
Reactive oxidants include ROS (i.e., O2•−, H2O2, •OH, RO2•, RO•, 1O2, and O3) and RNS (i.e., •NO, •NO2, and ONOO−). Reactive oxidants are produced from numerous sources in multiple compartments within the cell, either normally or as a result of exposure to toxic or pathologic insults (2, 20). The mitochondria are considered a primary site of ROS production from aerobic respiration under physiological and many pathophysiological conditions. Nonetheless, nearly all enzymes that utilize molecular oxygen as a substrate, including plasma membrane–bound NADPH oxidase (NOX), microsomal cytochrome P450 (CYP), and cytoplasmic xanthine oxidase, produce ROS either intentionally or as by-products. RNS are formed in cells starting with the synthesis of nitric oxide (•NO) by NO synthase. NO reacts with superoxide (O2•−) to form a stronger oxidant peroxynitrite anion (ONOO−). ONOO− reacts with other molecules to generate other RNS such as •NO2 and N2O3.
ROS and RNS are counterbalanced by intricate antioxidant systems to maintain the redox homeostasis in the cell. Major antioxidants consist of low-molecular-weight antioxidants, including reduced glutathione (GSH), vitamins C and E, bilirubin, and urate; noncatalytic antioxidant proteins, such as thioredoxin (Trx), glutaredoxin (Grx), and metallothioneins (MTs); and enzymes, such as superoxide dismutase (SOD), catalase, peroxiredoxin (Prx), and glutathione peroxidase (GPx). Ultimately, redox reactions in cells are enabled by nicotinamide pairs NADP+/NADPH and NAD+/NADH. NADPH is used to reduce oxidized Trx (Trxox) and glutathione (GSSG) through Trx reductase (TrxR) and glutathione reductase (GSR), respectively. Sulfiredoxin (Srx) reduces oxidized Prx from sulfinic (inactive) to sulfenic (active) acid in an ATP- and GSH-dependent manner. On top of antioxidant systems is a web of regulators that controls antioxidant defense at multiple levels to ensure that the response to oxidants is adequate in time and space.
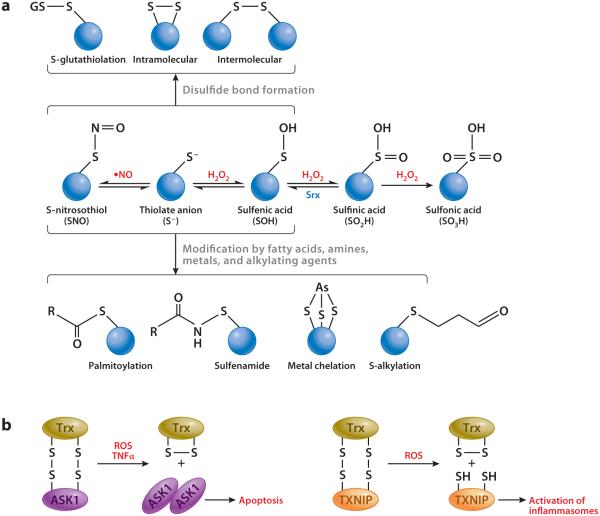
A recurrent theme emerging from oxidant signaling and antioxidant regulation is reactive cysteine thiol–based redox signaling. Reactive cysteine residues are a small set of protein cysteines with pKa values as low as 4 and 5 due to influence from the surrounding amino acid microenvironment, which is in contrast to most protein cysteine thiols that have pKa values of approximately 8.5 (21) (Figure 1a). At physiological pH, reactive cysteine thiols exist as thiolate anions (S−) and are more reactive toward ROS/RNS than are sulfhydryl groups (-SH). Reactive thiols interact with ROS and RNS to generate a range of cysteine oxidation products, including sulfenic, sulfinic, and sulfonic acids; S-nitrosothiol and thionitrates; S-glutathiolation products; and inter- and intraprotein disulfides. Reactive cysteine thiols are also modified by a variety of chemicals through alkylation, palmitoylation (thiol-ether), and metal chelation, providing a means of chemical sensing for environmental and endogenous cues such as electrophiles and metals. The versatility and reversibility of reactive cysteine thiols serve as a basis for the specificity and diversity of many redox signaling pathways.
Figure 1.
Versatile reactive cysteine thiol reactions. (a) Biochemistry of reactive cysteine thiols. Reactive cysteine thiols exist as thiolate anions and are prone to oxidation by reactive oxygen and nitrogen species (ROS, RNS). Formation of SNO, S−, and SOH is reversible and serves as an intermediate step for disulfide bond formation or modification by other agents, such as fatty acids, amines, metals, and alkylating agents. Reduction of SO2H to SOH is catalyzed by sulfiredoxin (Srx), whereas formation of SO3H is irreversible. (b) Redox sensors. In addition to being an antioxidant, thioredoxin (Trx) acts as a sensor of H2O2 to activate ASK1-dependent apoptosis or TXNIP-dependent inflammasome assembly.
The utility of reactive cysteine thiols in redox signaling and oxidant resistance is illustrated for growth factor signal transduction near the plasma membrane. Ligand binding to plasma membrane receptors, such as the platelet-derived growth factor receptor and epidermal growth factor receptor, stimulates production of O2•− and H2O2 through NOX. H2O2 inactivates protein tyrosine phosphatase (PTP) by oxidizing PTP reactive cysteine thiols to increase local protein tyrosine phosphorylation. Prx removes H2O2 to limit the influence of ROS, whereas Src activated by growth factor receptors inactivates Prx through phosphorylation and thereby boosts growth factor signaling (22, 23). Modification of reactive cysteine thiols also serves as a means of chemical sensing. Reversible oxidation of active site cysteine thiols of Trx by H2O2 enables Trx to act as a sensor of H2O2, leading to activation of the downstream signaling pathway of apoptosis or activation of inflammasome assembly (24, 25) (Figure 1b). As discussed below, modification of critical cysteine thiols of Keap1 and Nrf2 by oxidants and electrophiles is a principal mechanism by which ARE inducers activate Nrf2.
Nrf2-Keap1 SIGNALING
The importance of induction of ARE-regulated DMEs was noted from studies on chemoprevention, in which phenolic antioxidants, such as butylated hydroxyanisole and tert-butylhydroquinone (tBHQ), protected animals from chemical carcinogenesis and protection correlated with induction of GST and NQO1 (16, 26). Induction has since become a mechanistic model for analyzing antioxidant biological effects, leading to the discovery of the Nrf2-Keap1 signaling pathway.
Induction Components
Induction of DMEs by antioxidants results from the increased rate of mRNA synthesis from the DMEs’ genes and requires several regulatory components, reflecting an XAR-mediated adaptive transcriptional response to small chemicals.
ARE
Transfection experiments identified a 41-base-pair enhancer sequence upstream of the rat Gsta2 gene for induction of a reporter gene (27). The enhancer was required for the basal expression of the gene and was responsive to phenolic antioxidants and redox-active diphenols for induction, from which the name antioxidant response element was derived. Induction did not require the aryl hydrocarbon receptor that mediates induction of CYP1A1, GST, and NQO1 by planar aromatic hydrocarbons such as benzo(a)pyrene (Bap), suggesting an ARE-specific pathway for induction. ARE also controls the induction of mouse Gsta1 and rat and human NQO1 genes by antioxidants and electrophiles (28–30).
The 41-base-pair ARE contains a core sequence, 5′-TGACnnnGC-3′ (n = any base) (31) that was later expanded into a 16-base-pair consensus sequence, 5′-TMAnnRTGAYnnnGCR-3′ (M = A or C,R = A, Y = C or T,W = A or T) (32, 33). ARE consensus resembles the DNA-binding elements of several other bZip proteins (12). The tetranucleotide sequence 5′-TGAC of the ARE core is the same as the half-site of the TPA (12-O-tetradecanoylphorbol 13-acetate) response element of AP-1 proteins. ARE is similar to the NFE2-binding motif 5′-TGCTGAGTCAC-3′ and, in part, to the Maf (musculoaponeurotic fibrosarcoma) protein recognition element (MARE), 5′-TGCTGAG/C(or GC/CG)TCAGCA-3′, recognized by Maf dimers. The GC dinucleotide of MARE is critical for Maf binding, much like Nrf2/Maf heterodimer binding to ARE. bZip proteins including AP-1, NFE2, Nrf1, Nrf2, Nrf3, Bach1 and 2, small Mafs, and CREB/ATF exhibit overlapping binding activities toward the DNA elements. Cross-interaction among the bZip proteins through overlapping DNA binding and heterodimerization expands the repertoire of target genes of individual bZip proteins including Nrf2.
Nrf2 and small Mafs
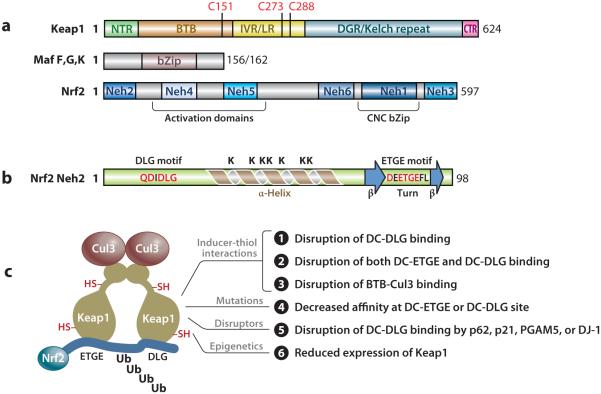
Expression cloning with tandem NFE2-binding motifs as a screening probe led to the cloning of Nrf2 cDNA (6). Nrf2 has a characteristic CNC bZip domain in the C terminal region (Figure 2a). The basic region contributes to DNA binding and the leucine zipper to heterodimerization with a small Maf. The CNC region (~36 amino acids) was named after the Drosophila segmentation protein CNC and shares a high homology among the CNC bZip proteins. Transcription activation is conferred by two regions in the N terminal half, as well as the C terminal end region. Peptides between residues 494 and 511 and between 545 and 554 host a bipartite nuclear localization and a nuclear export signal, respectively. The human and chicken Nrf2 peptide sequences are highly conserved at several locations designated Nrf2-ECH homology (Neh) domains 1 to 6 (ECH = erythroid cell-derived protein with CNC homology = chicken Nrf2) (34). The Neh2 domain at the N terminus negatively regulates Nrf2 activity through Nrf2 suppressor Keap1. Nrf2 KO mice are viable (35) but exhibit reduced fertility (36), have a tendency to develop autoimmune lesions (37–39) and leukoencephalopathy (40), and are sensitive to a variety of toxicity and disease conditions (5, 12).
Figure 2.
Domain structure and activation mechanism. (a) Domain structures of Nrf2, Keap1, and small Mafs F, G, and K. (b) Secondary structure of Neh2. (c) Model for activation of Nrf2. Abbreviations: BTB, bric-a-brac, tramtrack, broad-complex domain; bZip, basic region leucine zipper; CNC, cap ‘n’ collar; Cul3, Cullin 3; DGR, double glycine repeat; IVR, intervening region; Keap1, Kelch-like ECH (erythroid cell-derived protein with CNC homology)-associated protein 1; LR, linker region; Maf, musculoaponeurotic fibrosarcoma protein; Neh, Nrf2-ECH homology; Nrf2, nuclear factor erythroid 2–related factor 2.
The small Maf proteins (~18 kDa) consist of Maf F, G, K, and T. Small Mafs have a bZip domain for DNA binding and dimerization but lack a transcription activation domain present in large Mafs (v-Maf, c-Maf, Maf A, Maf B, and NRL) (Figure 2a). Small Mafs function as an obligatory heterodimeric partner for large CNC bZip proteins and bind to the GC dinucleotide of ARE and MARE. Homo- and heterodimers among small Mafs form transcription repressors owing to their lack of a transactivation domain.
Keap1
Keap1 was identified as a novel Nrf2-binding protein from yeast two-hybrid screening using the inhibitory Neh2 domain of Nrf2 as a bait (34). Keap1 contains two known protein-interacting domains: the BTB (bric-a-brac, tramtrack, broad-complex) domain in the N terminal region and the Kelch repeats in the C terminal region homologous to Drosophila actin-binding protein Kelch [Kelch repeat, double glycine repeat (DGR) domain] (Figure 2a). BTB mediates homodimerization and binding of Keap1 to Cullin (Cul) 3, a scaffold protein of Nrf2 ubiquitin ligase (E3). DGR mediates binding of Keap1 with Nrf2 Neh2 (41–43). Between BTB and DGR is the intervening region (IVR) or linker region (LR) rich in cysteine residues. Similar to Nrf2, Keap1 is expressed broadly in tissues and resides in the cytoplasm. Keap1 KO mice die before weaning due to malnutrition from hyperkeratinization and occlusion of the esophagus and forestomach (44). Hyperkeratinization was attributed to constitutive activation of Nrf2, a primary molecular phenotype of Keap1 knockout, in keratinocytes.
Signaling Mechanism
Nrf2 mRNA is expressed broadly and independently of inducers, suggesting a post-transcriptional mechanism(s) for Nrf2 activation. Mechanistically, activation of Nrf2 for induction is twofold: suppression of Nrf2 under a basal condition and activation of Nrf2 by inducers.
Suppression of Nrf2 by Keap1
Inhibition of protein synthesis by cycloheximide totally blocked the basal and induced expression of ARE-controlled DMEs, suggesting protein turnover as a major mechanism of regulation (45). Nrf2 is rapidly degraded by proteasomes under a basal condition with a half-life of ~20 min, resulting in a low protein level of Nrf2 in many types of cells (46–48). Degradation of Nrf2 is triggered by polyubiquitination through the Keap1/Cul3 ubiquitin ligase. Keap1 acts as a substrate adaptor to bring Nrf2 into the E3 complex, in which it binds to Nrf2 via its DGR domain and to Cul3 N terminal region via the BTB domain. RING box protein 1 recruits the catalytic function of ubiquitin-conjugating enzyme (E2) by binding to Cul3 C terminal region. E2 catalyzes polyubiquitination of Nrf2 protein on the lysine residues of Neh2 domain. Knockout of Keap1 in mice resulted in constitutive activation of Nrf2 from stabilization of Nrf2 protein (44).
The mechanism by which Keap1 interacts with Nrf2 and direct Nrf2 ubiquitination was revealed from structural resolution of Keap1 and Nrf2. The overall structure of Nrf2 is not available, but the structure of Nrf2 Neh2 was shown by nuclear magnetic resonance spectroscopy to be intrinsically disordered with certain secondary structures (49) (Figure 2b). A 33-residue α-helix (a.a. 39–71) resides in the center of rod-shaped Neh2, followed by mini antiparallel β-sheets (a.a. 74–76 and 82–85). Six of the seven lysine residues of the helix face one side. A conserved ETGE sequence (a.a. 79–82) locates in the hydrophobic hairpin loop between the two β-sheets. A second conserved sequence DLG (a.a. 29–31) follows another β-hairpin structure (a.a. 25–29).
The overall structure of mouse Keap1 resembles a cherry-bob, in which two Keap1 molecules form a dimer with two large, globular spheres attached by short linker arms to the sides of a small forked-stem structure (41) (Figure 2c). The forked stem reflects dimerization through two BTB domains. The globular structures each contain IVR and DC domains (DC = DGR + CTR) with IVR on the outer surface and DC in the interior. Crystallography of human Keap1 DGR (a.a. 321–609) at 1.85-Å resolution and mouse DC (a.a. 309–624) at 1.60-Å resolution revealed that the Kelch repeats fold into a drum-shaped, 6-bladed β-propeller structure with an inner cavity buried in the central core opening on the top and bottom of the globular spheres (42, 43).
Neh2 interacts with Keap1 DC in a ratio of 1:2 through its ETGE and DLG motifs (43, 49, 50). Each motif binds to a binding cleft of Keap1 DC near the entrance of the tunnel at the bottom of the DC sphere. ETGE has a binding affinity two orders of magnitude higher than that of DLG owing to more electrostatic interactions between DC and ETGE (~13) than between DC and DLG (~8) (51). In this two-site-binding, or hinge-and-latch, model (49) (Figure 2c), Keap1 recruits newly synthesized Nrf2 by binding to Neh2. Because of its higher binding affinity, ETGE binds to one binding cleft first, which promotes binding of DLG to the other DC sphere. The α-helix between DLG and ETGE is presented to E2 with six lysine residues facing the enzyme for polyubiquitination.
Activation of Nrf2 by ARE Inducers
A variety of chemicals, including phytochemicals and derivatives (CDDO, sulforaphane), therapeutics (oltipraz, auranofin), environmental agents (paraquat, arsenic), and endogenous chemicals [NO, 15d-PGJ2, nitro-fatty acids, and 4-hydroxynonenal (4-HNE)], induce ARE genes through Nrf2 (12, 52). Inducers are structurally diverse and have few common properties, except for their ability to modify -SH at rates closely correlated with their potency for induction of NQO1 (53). Evidence for binding of inducers to Keap1 cysteine thiols was provided using labeled inducers, stoichiometry, ultraviolet spectroscopy, mass spectrometry, and mutational studies (53–56). The IVR region contains multiple cysteine residues that were frequently labeled by inducers, in particular, C273, C288, and C297. Mutational studies confirmed that C151, C273, and C288 are critical for Nrf2 regulation. In mice, C151 was required for activation of Nrf2 by electrophiles and C272 and C288 for suppression of Nrf2 under a basal condition (57). In a zebrafish model, Keap1 was shown to recognize a number of electrophilic inducers through distinct sets of cysteine residues, coined “cysteine codes,” to activate Nrf2 (58). In a recent study, C288 was shown to be a sensor for alkenals such as 4-HNE; a combination of H225, C226, and C613, a Zn2+ sensor; and C151 together with a cluster of neighboring basic amino acids (H129, K131, R135, K150, and H154), an NO sensor (59). Compared with Keap1 that has 25 or more cysteine residues, Nrf2 has only 6 (human) or 7 (mouse and rat) cysteine residues. The cysteines are highly conserved and have multiple effects on Nrf2 function, including activation of Nrf2 by arsenic (60). The chemistry of inducer-cysteine code interaction for Keap1 and Nrf2 remains largely unaddressed.
Modification of cysteine thiols of Keap1 and Nrf2 by inducers presumably alters the structure of the Nrf2/Keap1/Cul3 complex, leading to inhibition of Nrf2 ubiquitination (Figure 2c). Inducers, such as tBHQ, bind to Keap1 IVR cysteines that are spatially close to Keap1 DC to cause disruption of DC-DLG binding. This change does not interrupt the stronger DC-ETGE binding but prevents Nrf2 Neh2 from being ubiquitinated, creating a “dead” Keap1 complex. Newly synthesized Nrf2 bypasses Keap1 and enters the nucleus to mediate induction. Some inducers, such as toxic metals As3+, Cd2+, and Cr6+, dissociate Nrf2 from Keap1 to stabilize Nrf2 (46, 61, 62). Modification of cysteine residues close to the BTB region, such as C151, disrupts Keap1-Cul3 binding, thereby inhibiting Nrf2 ubiquitination. Elucidation of the three-dimensional structure of the E3 complex and the spatial relationship of critical cysteine residues to components of the complex is key to a full understanding of the molecular interactions among inducers, specific cysteine thiols, and components of the E3 complex for Nrf2 activation.
Other Mechanisms of Nrf2 Regulation
In addition to inducer-cysteine thiol interaction, several other mechanisms have been described for the regulation of Nrf2 (Figure 2c). These mechanisms modulate the above-described Keap1-Nrf2 signaling to regulate Nrf2 in cell type–, target gene–, and inducer-dependent manners. Covalent modification of Nrf2 by phosphorylation/dephosphorylation and acetylation/deacetylation affects the nuclear translocation/export, transcription activation, and degradation of Nrf2 in response to inducing signals (63–68). Some proteins, such as autophagy substrate p62, p53-regulated p21, and Bcl-XL-interacting protein PGAM5, are called disruptors of Keap1-Nrf2 binding because they bind to Keap1 DC in a manner similar to Nrf2 DLG or ETGE through their DLG- or ETGE-like sequences and thereby disrupt Nrf2-Keap1 binding, resulting in persistent activation of Nrf2 (69–71) (Figure 2c). DJ-1 (PARK7), a multifunctional protein associated with early-onset Parkinson’s disease (PD) and elevated in cancers, stabilizes Nrf2 by interfering with Nrf2-Keap1 binding to protect against oxidative stress in dopaminergic neurons and cancer cells (72). Tumor cells hijack the Nrf2 pathway through somatic mutations and epigenetic mechanisms to cause persistent activation of Nrf2 (18). Several serine residues in Nrf2 Neh6 (a.a. 331–381) can be phosphorylated by GSK-3, creating a recognition motif of DpSGX(1–4)pS that is recognized by the WD substrate recognition domain of β-TrCP. β-TrCP is an adaptor protein for the Cul1-dependent SCF E3 and thereby promotes the ubiquitination and proteasomal degradation of Nrf2 independently of Keap1 (73). The physiological importance of this pathway remains unclear.
REGULATION OF ANTIOXIDANT DEFENSE BY Nrf2
Evolutionary Conservation
The function of Nrf2 in regulating antioxidant defense has an evolutionary basis. Nrf2 is highly conserved across vertebrates. Compared with human Nrf2, mouse Nrf2 has DNA and protein homology scores of 83.4% and 82.5%, respectively; rat Nrf2, 84% and 83%; cow Nrf2, 91% and 89%; dog Nrf2, 89.4% and 88.9%; chicken Nrf2, 72% and 67%; and zebrafish Nrf2, 55% and 49% (74). The CNC bZip proteins possess considerable similarities in their structure, DNA recognition sequence, and target genes, reflecting an evolutionary link to erythropoiesis essential for oxygen utilization in vertebrates. Among the proteins, NFE2 is critical for erythrocyte and platelet development (6); Bach1 and Bach2 act as suppressors of transcription by NFE2 (75); Nrf1 is required for embryonic hepatic hematopoiesis but has also evolved to aid in resistance to ER stress, oxidant stress, and metal toxicity (76); Nrf2 deviates from most CNC bZip proteins to become specialized in oxidant resistance; Nrf3 is less well understood but may contribute to smooth muscle cell differentiation from stem cells toward vascular lineage and to in vivo protection against hematopoietic malignancy (77–79). In invertebrates, Caenorhabditis elegans protein skinhead-1 (SKN-1) has significant homology with Nrf2 and binds to an ARE-like element to regulate oxidative stress–related genes (80). Unlike Nrf2, however, SKN-1 binds DNA as a homodimer and has a function in the regulation of mesodermal differentiation and intestinal development. Drosophila proteins CncA, CncB, and CncC have marked sequence similarity to mammalian CNC bZip proteins. Like Nrf2, CncC dimerizes with a small Maf to activate oxidative defense genes and to resist paraquat and arsenic toxicity (81).
Signaling through Keap1 is evolutionarily conserved. Drosophila Keap1 (dKeap1) is expressed in tissues where CncC is expressed, and dKeap1 represses induction of GST D1 in the fly (81). Zebrafish expresses two forms of Keap1, zKeap1a and zKeap1b. Both proteins contain cysteine residues equivalent to mouse Keap1 C273 and C288, and both cysteines are critical for suppression of zebrafish Nrf2. Like mammalian Keap1, zKeap1a and zKeap1b function through dimerization (82). The alkenal sensor C288 of mammalian Keap1 has an ancient origin common for bilaterians, but the NO sensor C151 (and surrounding basic residues) evolved more recently, emerging coincidently with the NOS gene family in vertebrates (59). The amino acid residues composing the pocket of Keap1 for Nrf2 binding are highly conserved in vertebrates and certain invertebrates, including flies and mosquitoes. The Nrf2 ETGE motif is present in Drosophila CncC (82). Therefore, the Nrf2-Keap1 signaling pathway appears to be evolutionarily conserved with respect to cis- and trans-acting factors, target genes, cysteine codes, and Nrf2-Keap1 binding motifs for resistance to oxidants.
Target Genes and Antioxidant Defense
Genome-wide search for Nrf2 target genes has led to the identification of an array of ARE-regulated genes, providing a rational explanation for the multiple functions of Nrf2 (5, 12, 17). A central theme emerging from the identification of these target genes and their functions is resistance to oxidants and electrophiles. Notably, three major groups of Nrf2 target genes regulate drug metabolism and disposition, antioxidant defense, and oxidant signaling, respectively, to impact the response to oxidants and electrophiles (see Supplemental Table 1 and accompanying references; follow the Supplemental Materials link from the Annual Reviews home page at http://www.annualreviews.org). In addition, Nrf2 regulates proteasomal protein degradation (83), cell proliferation (84), and metabolic reprogramming (85), which are not discussed in this review.
Induction of drug metabolism and drug transport through ARE may have originated as a strategy of detoxification of endogenous oxidants and electrophiles but evolved as an adaptation to environmental toxicants. By controlling the basal and induced expression of the DMEs and transporters, Nrf2 regulates the metabolic fate of numerous pro-oxidants and electrophiles in the body. ARE-regulated DMEs catalyze a collection of heterogeneous reactions, including oxidation by CYP2A5, ALDH3A1, and ADH7; reduction by NQO1 and AKRs; conjugation by UGTs and SULT3A1; and nucleophilic trapping reactions by GSTs, mEH, and ES-10 (Supplemental Table 1). Drug transporters, such as MRP2 and MRP3, transport drugs and metabolites out of cells.
Nrf2 directly affects the homeostasis of ROS and RNS by regulating the antioxidant defense systems through several mechanisms (Supplemental Table 1). These include (a) induction of catabolism of superoxide and peroxides through SOD, Prx, and GPx; (b) regeneration of oxidized cofactors and proteins, where GSSG is reduced by GSR, Trxox by TrxR, and Prx-SO2H by Srx; (c) synthesis of reducing factors, i.e., GSH by GCLC and GCLM, and NADPH by G6PDH and 6PGD; (d ) expression of antioxidant protein Trx and inhibition of expression of Trx inhibitor TXNIP; (e) the increase of redox transport, such as cystine/glutamate transport through xCT; (f) metal-chelation by MT1, MT2, and ferritin; and ( g) induction of stress response proteins, such as HO-1. Many of the antioxidant enzymes/proteins regulated by Nrf2 localize in specific compartments within the cell to regulate redox signaling in the local environment.
Nrf2 also regulates the expression of several oxidant signaling proteins to impact a number of programmed cellular functions (see Supplemental Table 1 and discussion below). Some regulators, such as p62 and DJ-1, activate Nrf2 and are induced by oxidants through Nrf2, creating a positive feedback loop with Nrf2.
Nrf2 AND OXIDANT-STIMULATED PHYSIOLOGIC FUNCTIONS
Oxidants stimulate several programmatic functions (20). Recent findings reveal that Nrf2 plays a role in the regulation of these functions.
Autophagy
Macroautophagy is a conserved bulk protein degradation pathway responsible for the turnover of long-lived proteins, disposal of excess or damaged organelles, and clearance of aggregation-prone proteins (86). Autophagy is essential for housekeeping functions and is induced in a number of stress responses. Autophagy is accompanied by accumulation of ROS from damaged mitochondria (87). Mitochondria-derived H2O2 may stimulate autophagy by oxidizing reactive cysteine residues of proteins encoded by essential autophagy genes, such as ATG4, to increase the overall autophagosome formation.
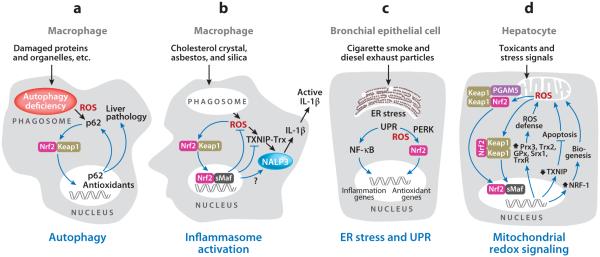
In autophagy-defective mice, Nrf2 is overactivated and associated with the liver pathology observed in the mice, because loss of Nrf2 alleviated but loss of Keap1 exacerbated autophagy deficiency–induced liver injury (69). A key link between autophagy and Nrf2 is p62, an autophagy substrate and autophagic cargo receptor (69, 88, 89) (Figure 3a). Loss of autophagy causes marked accumulation of p62 that in turn suppresses Keap1 to activate Nrf2 and increase ARE gene expression. p62 binds to Keap1 through its KIR motif (a.a. 345–359); KIF contains a STGE sequence (a.a. 351–355) that has a binding affinity to Keap1 DC similar to that of Nrf2 DLG. p62 is also induced by oxidative stress through Nrf2 and ARE, creating a positive feedback loop between p62 accumulation and Nrf2 activation in autophagy deficiency. The Toll-like receptors (TLRs) influence several innate immune responses by regulating autophagy. p62 is upregulated in TLR4-mediated, selective autophagy of aggresome-like induced structures (ALIS). Both accumulation of p62 and ALIS required activation of Nrf2 through ROS-p38 axis–dependent TLR4/MyD88 signaling (90).
Figure 3.
Nrf2 and oxidant-stimulated programmatic functions. (a) Autophagy. (b) Inflammasome activation. (c) Endoplasmic reticulum (ER) stress and unfolded protein response (UPR). (d ) Mitochondrial redox signaling. Abbreviations: GPx, glutathione peroxidase; Keap1, Kelch-like ECH-associated protein 1; Maf, musculoaponeurotic fibrosarcoma protein; Nrf2, nuclear factor erythroid 2–related factor 2; PERK, protein kinase RNA-like endoplasmic reticulum kinase; Prx, peroxiredoxin; ROS, reactive oxygen species; Srx, sulfiredoxin; Trx, thioredoxin; TrxR, thioredoxin reductase.
Inflammation and Inflammasome Signaling
Nrf2 is anti-inflammatory, as evidenced by several observations. Nrf2 KO mice have a tendency to develop age-dependent autoimmune and inflammatory lesions in multiple tissues (37, 38). Inflammation is commonly observed in chemically induced pathology with Nrf2 deficiency (91– 96). Many ARE inducers are potent anti-inflammatory agents, and their potency for induction of ARE genes correlates well with their potency for inhibition of inflammation (97–99). Inhibition of inflammation by Nrf2 is associated with inhibition of the NF-κB pathway and inhibition of proinflammatory cytokine production (100, 101). The molecular events underlying the interaction between Nrf2 and inflammatory regulators remain largely unclear.
The inflammasomes are a molecular platform required for fully functional innate immune systems (102). Inflammasomes consist of a pattern recognition receptor, such as NALP3, the adaptor protein ASC, and procaspase-1. NALP3 senses a wide range of damage signals, including microbes (pathogen-associated patterns), endogenous danger signals (damage-associated patterns), and environmental irritants such as silica and asbestos. Activated NLRP3 oligomerizes and recruits ASC and procaspase 1, triggering the activation of caspase 1 and the maturation of proinflammatory cytokines, such as IL-1β and IL-18. ROS derived from mitochondria (103) or “frustrated phagocytosis” from the engulfing of silica and asbestos particles by macrophages activate NLRP3 (104); in the latter case, activation of NOX is involved. ROS oxidize Trx active site cysteine thiols, causing dissociation of TXNIP from Trx; freed TXNIP binds to NLRP3 to induce robust activation of the inflammasome (25) (Figures 1b and 3b). Efficient inflammasome activation depends on the redox state of macrophages that is regulated by cystine/cysteine cycling and the cystine transporter, xCT (105).
Cholesterol crystals are engulfed by macrophages to stimulate inflammation during atherogenesis (106). Cholesterol crystals activated Nrf2 and induced Nrf2-regulated genes Nqo1, Hmox1, and Prdx1; moreover, Nrf2 was required for cholesterol crystal–induced inflammasome response (107). In this scenario, Nrf2 was activated as an adaptive defense against oxidative stress during inflammation. At the same time, Nrf2 enhanced atherogenesis by supporting IL-1β-mediated vascular inflammation induced by cholesterol crystals. Therefore, the consequence of Nrf2 activation by activators of the NLRP3 inflammasomes may vary depending on the activators, cell type, and pathology. Nrf2 may affect inflammasome signaling by several means: induction of antioxidative genes to counteract ROS increase; induction of xCT and Trx to regulate the redox state of monocytes; suppression of TXNIP transcription to reduce the amount of TXNIP; and promotion of IL-1β production for atherogenesis (Figure 3b and Supplemental Table 1).
ER Stress and Unfolded Protein Response
Accumulation of unfolded and misfolded proteins or excessive protein trafficking in the lumen of the ER causes ER stress, which triggers the unfolded protein response (UPR). UPR uses evolutionarily conserved signaling pathways to restore the normal function of the cell in the presence of ER stress and, if homeostasis is not achieved, initiates apoptosis (108). Among the signaling molecules of UPR, PERK (protein kinase RNA-like endoplasmic reticulum kinase) phosphorylates the α subunit of eIF2 causing attenuation of general protein synthesis and promotion of cell survival in ER stress. UPR is regulated by ROS and the redox systems in ER (109). ER-resident peroxidases Prx4, GPx7, and GPx8 catalyze the transfer of electrons from protein disulfide isomerase (PDI)-like oxidoreductases to H2O2, leading to detoxification of H2O2 in the ER lumen.
Cigarette smoke (CS) and smoke from biomass fuels, such as diesel exhaust, trigger inflammation in the respiratory tract that potentially leads to chronic obstructive pulmonary disease (COPD). CS and diesel exhaust contain small-sized particles that do not appear to activate the NALP3 inflammasome in human macrophage-like cells as asbestos and silica particles do (104). On the other hand, smokers’ lung tissues exhibited increased expression of proteins characteristic of ER stress and UPR, including calreticulin, PDIs, and glucose-regulated protein 78; moreover, CS induced UPR and activated Nrf2 to induce several antioxidant genes in transformed human airway epithelial cells (110). The mechanism by which UPR activates Nrf2 may involve PERK. PERK phosphorylates Nrf2 on Thr-80 to activate Nrf2 and induce ARE genes, which increase GSH level, reduce ROS in ER, and promote survival (67, 111) (Figure 3c). Nrf2 upregulates ER-resident antioxidant enzymes, such as GPx8 (see Supplemental Table 1; follow the Supplemental Materials link from the Annual Reviews home page at http://www.annualreviews.org). UPR triggered by CS and diesel exhaust also activates the NF-κB signaling pathway to stimulate proinflammatory cytokine production, which is inhibited by Nrf2.
Apoptosis and Mitochondrial Biogenesis
Mitochondria play multiple functions in addition to being the power house for ATP synthesis. Mitochondria are a major site of ROS production and a target of many toxicants. Mitochondria serve as a central site where most apoptotic pathways intersect (112). Mitochondria contain nonchromosomal DNA and undergo biogenesis to replenish, which is coordinated by several transcription factors, including nuclear respiratory factor 1 (NRF-1) (113).
Nrf2 affects mitochondrial physiology and pathology in several ways (Figure 3d and Supplemental Table 1). Nrf2 KO cells exhibit increased spontaneous apoptosis and are highly sensitive to chemically induced mitochondrial damage, whereas activation of Nrf2 by chemoprotective agents protects against mitochondrial damage (96, 114–116). Nrf2 promotes mitochondrial biogenesis in mouse cardiomyocytes (117). Nrf2 directly regulates mitochondrial ROS homeostasis by (a) promoting detoxification of peroxides through Prx3 and GPx; (b) regenerating Trx2, GSH, and Prx3-SO2H through TrxR, GSR, and Srx1, respectively; (c) increasing the synthesis of GSH and NADPH; and (d ) inhibiting TXNIP expression. Nrf2-Keap1 forms a ternary complex with PGAM5, which localizes at the mitochondrial outer membrane through the PGAM5 N-terminal mitochondrial-localization sequence; this mitochondrial localization may allow Nrf2 to directly sense and respond to mitochondrial ROS (71). Inhibition of TXNIP expression by Nrf2 reduces the concentration of TXNIP in the mitochondria to free Trx from TXNIP; reduced Trx binds to ASK1 and inhibits ASK1-dependent mitochondrial apoptosis. For mitochondrial biogenesis, endogenous carbon monoxide generated by HO-1 stimulates MnSOD upregulation and mitochondrial H2O2 production to activate Akt/PKB; Akt deactivates GSK-3β, which activates Nrf2; Nrf2 binds to the four AREs in the enhancer of the gene encoding NRF-1 to induce NRF-1; NRF-1 coordinates gene activation for mitochondrial biogenesis to increase resistance to oxidative toxicants (117). DJ-1 translocates into the mitochondria upon oxidative stress to protect the mitochondria (118). Loss of DJ-1 increases ROS production, mitochondrial damage, autophagy, and mitophagy, accompanied by downregulation of Nrf2 and ARE genes (72, 119). DJ-1 activates Nrf2 by interfering with Nrf2-Keap1 binding, and Nrf2 may upregulate DJ-1 expression through an ARE-like enhancer sequence of DJ-1 (72).
Stem Cell Function
Several types of stem and progenitor cells exhibit low intracellular concentrations of ROS that may serve as a critical condition for stemness and pluripotency; on the other hand, a rise in ROS appears to be required for proper differentiation of myeloid progenitor cells (120). Thus, the intracellular redox balance is carefully regulated within stem cells to modulate regenerative processes (121). A connection between Nrf2 and stem cell functions in high-turnover tissues was suggested in recent studies. The Drosophila intestinal stem cells (ISCs) respond to oxidative stress and inflammation by increasing proliferation, which is a regenerative response but may lead to hyperproliferation and epithelial degeneration in aged fly (122). Nrf2 is constitutively active in ISCs, and repression of Nrf2 by Keap1 is required for ISC proliferation. Loss of Nrf2 in ISCs caused accumulation of ROS and accelerated age-dependent degeneration of intestinal epithelium. Thus, Nrf2 promotes intestinal homeostasis of fly by regulating the intracellular redox balance of ISCs. Nrf2 is also required for hematopoietic stem progenitor cell (HSPC) survival and myeloid development in mice; Nrf2 KO bone marrow showed defective stem cell function, as evidenced by reduced chimerism after transplantation (123). The basal level of ROS in Nrf2 KO bone marrow was not elevated compared with wild type; however, Nrf2 KO HSPCs had increased rates of spontaneous apoptosis and decreased survival when exposed to oxidative stress, indicating a critical role of Nrf2 in hematopoiesis and stem cell survival in mice.
Nrf2 IN TOXICITY AND DISEASE
Nrf2 KO mice show increased sensitivity to a variety of chemically induced toxicities and disease pathologies. A few selected examples are discussed below to illustrate the importance and versatility of Nrf2 in toxicity and disease.
Acetaminophen Hepatotoxicity
Acetaminophen is a widely used analgesic and antipyretic drug and a prototypical hepatotoxicant for drug-induced liver injury (DILI). DILI is the leading cause of liver failure in the United States, the United Kingdom, and Australia (124). Most DILI-induced liver failure is due to acetaminophen overdose. Acetaminophen causes characteristic centrilobular hepatic necrosis and sinusoidal congestion, which to a large extent reflects the expression pattern of CYP2E1. Acetaminophen undergoes glucuronidation and sulfation, but a small portion is converted to N-acetyl-p-benzoquinone imine (NAPQI) via CYP2E1-mediated metabolic activation. At a high dose, the glucuronidation and sulfation pathways are saturated, resulting in increased production of NAPQI. NAPQI is highly reactive and binds covalently to protein and nonprotein thiols, causing oxidative stress and cell death. At doses relatively nontoxic to wild-type mice, Nrf2 KO mice showed increased serum ALT enzyme activity, severe hepatocellular injury, and even death from liver failure (13, 125). Boosting the Nrf2 activity by pharmacological agents or by genetic manipulation, i.e., hepatocyte-specific Keap1 knockout, effectively inhibited acetaminophen toxicity (126, 127). Several mechanisms may explain the protective effect of Nrf2. Nrf2 increases glutathione synthesis by inducing GCLC and GCLM to replenish the liver GSH pool, which is depleted by acetaminophen. Nrf2 mediates the induction of UGTA6, which increases glucuronidation of acetaminophen. Nrf2 controls the induction of MRP3 to increase excretion of acetaminophen glucuronide through the drug transporter. Although acetaminophen-induced injury is morphologically similar to necrosis, acetaminophen induces BAX-translocation to mitochondria to increase apoptosis, which is inhibited by Nrf2 (13, 124, 128) (Supplemental Table 1).
Ethanol and Alcohol-Induced Liver Disease
Chronic alcohol consumption increases the production of reactive oxidants, a major factor in the development of alcohol-induced liver disease (AILD) (129). Ethanol-induced oxidative damage in the liver involves depletion of antioxidants (especially GSH in the mitochondria), lipid peroxidation, and protein modifications (carbonylation, addition of 4-HNE, and nitration of tyrosine residues). Ethanol consumption induces the accumulation of CYP2E1 (130), which is a major contributor to ethanol-induced ROS production in the liver. CYP2E1 metabolizes low-molecular-weight molecules, including ethanol, acetone, benzene, and acetaminophen, but the reaction is leaky, resulting in the formation of O2•− and H2O2. The antioxidant systems are critical for defense against alcohol hepatotoxicity. A pivotal role of Prx1 and Srx1 in protection against AILD was demonstrated in mice with Prx1- or Srx1-deficiency (131). When fed with ethanol for a prolonged time, the mice showed more severe oxidative damage in the liver as compared with wild type. Among Prxs 1–4, Prx1 is localized at the cytoplasmic side of ER where CYP2E1 is embedded and is the most active Prx for elimination of ROS in the livers of ethanol-fed mice. Prx1 is hyperoxidized to an inactive form (Prx1-SO2H) that is markedly elevated in Srx1 KO mice, indicating that Srx1 is critical for reducing Prx1-SO2H to Prx1-SOH. Srx1 is translocated into the mitochondria upon ethanol-induced oxidative stress to reduce Prx3-SO2H in the mitochondria. Mitochondrial Trx2 is induced by ethanol, which together with Prx3 and Srx1 forms the primary line of defense against mitochondrial H2O2 induced by ethanol. Nrf2 is activated by chronic alcohol consumption and by overexpression of CYP2E1 (131, 132). Chemoprotective agents activate Nrf2 and protect animals from ethanol-induced lesions, including fetal alcohol spectrum disorders (133, 134). Nrf2 controlled the basal and ethanol-induced expression of Srx1, HO-1, and NQO1 in mouse liver (131, 135). TXNIP is present in the mitochondria and inhibits mitochondrial Trx2. Nrf2 suppresses the expression and induction of TXNIP and thereby enhances the Trx-Prx function (see Supplemental Table 1; follow the Supplemental Materials link from the Annual Reviews home page at http://www.annualreviews.org). In this manner, Nrf2 coordinates a concerted action of Srx1, Prx1, Prx3, and Trx, which are essential for protection against AILD.
Cigarette Smoke and COPD
Chronic exposure to cigarette smoke in the lungs is typically associated with the development of COPD (in particular, emphysema) and lung cancer. Nrf2 is clearly protective against CS-induced COPD (92, 136, 137), but its role in CS-associated lung cancer is less certain (discussed below) (19). A fully functional Nrf2 genotype is required to protect smokers against acquiring lung emphysema. Nrf2 KO mice chronically exposed to CS developed emphysema with earlier onset and more extensive pathology compared with wild type (92, 136). Besides compromised alveolar structures, the mice exhibited increased oxidative genotoxic stress (high levels of 8-OH-dG), increased apoptosis, pronounced inflammation, and reduced function in Nrf2 KO lungs. Selective deletion of Keap1 in Clara cells in mouse lungs elevated the expression of Nrf2-dependent genes (Nqo1 and Gclm), increased the total level of glutathione in the lungs, protected Clara cells against oxidative stress ex vivo, and protected the lungs against oxidative stress and CS-induced inflammation in intact animals (138). Pharmacological activation of Nrf2 protected mice against CS-induced emphysema (139). When included in the diet, CDDO-Im, a potent triterpenoid ARE inducer, significantly reduced lung oxidative stress, alveolar cell apoptosis, alveolar destruction, and pulmonary hypertension in Nrf2 wild-type mice exposed to CS for 6 months (139). Protection was dependent on Nrf2, because Nrf2 KO mice failed to show significant reduction in alveolar cell apoptosis and alveolar destruction after treatment with CDDO-Im.
The effect of Nrf2 on CS lung toxicity involves several mechanisms. The Nrf2 pathway is activated by CS. CS is a complex aerosol with >60 carcinogens and >5,000 chemicals, many of which have strong reactivity toward -SH (140). Among the chemicals, diphenols, polyenes (1,3-butadiene), polycyclic aromatic hydrocarbons and reactive metabolites, metals (As3+, Cd2+), and endogenous chemicals generated upon exposure to CS (NO, 4-HNE) activate the Nrf2-Keap1 pathway by modifying critical cysteine residues of Keap1 and Nrf2. Alternatively, CS particles induce ER stress and activate UPR to activate the Nrf2 pathway via PERK (Figure 3c). The adaptive activation of Nrf2 leads to increased expression of Nrf2-controlled genes that encode detoxification enzymes (NQO1), antioxidant enzymes (GPx2, Srx1), and enzymes for the synthesis of low-molecular-weight antioxidants (GSH, bilirubin), all of which suppress CS-induced ROS production and oxidative damage (see Supplemental Table 1 and accompanying references). Induction of DMEs and transporters through Nrf2 inhibits the metabolic activation of CS carcinogens, such as Bap, to reduce DNA-adduct formation, carcinogenesis, and toxicity, as demonstrated for Bap-induced tumorigenesis (141). Nrf2 also controls the induction of secretory leukoprotease inhibitor (SLPI) and α-antitrypsin in the lungs that are important for maintaining the protease/antiprotease balance and elasticity of the lungs (92, 136). A normal Nrf2 function is required for cell cycle progression (137, 142) and for inhibition of apoptosis and inflammation induced by CS in the lungs (92, 136), both of which contribute to protection against CS-induced COPD by Nrf2. Chronic exposure to CS may also compromise the adaptive response of Nrf2, leading to exacerbation of CS-induced oxidative stress and pathology in smokers’ lungs. Nrf2 functions were found to be downregulated and Keap1 and Bach1 to be upregulated in the lung tissues and alveolar macrophages from smokers with emphysema, which promotes a proemphy-sematous microenvironment in the lungs (143). Defective Nrf2 in CS-induced emphysema was attributed to a decreased level of DJ-1 that supports the stability of Nrf2 (72, 144), and to decreased histone deacetylase 2 (HDAC2) activity that increases the acetylation and inhibition of Nrf2 in CS-exposed lungs (145, 146).
Cancer
The impact of Nrf2 on cancer is complex, precluding a simple conclusion pertaining to its role in cancer. An emerging concept is that Nrf2 acts as a double-edged sword: On one hand, Nrf2 is required for protecting the body against cancer by endogenous and pharmacological anticancer agents; on the other, it is persistently activated in some tumors, resulting in a prosurvival phenotype to promote tumor growth and resistance to oxidants and anticancer drugs (12, 18, 19). Nrf2 KO mice are significantly more susceptible to the carcinogenic effects of several known carcinogens in animal models, and Nrf2 is needed for protection against chemical carcinogenesis by chemopreventive agents. Increased tumor formation in Nrf2 KO mice correlated with reduced, or lack of, expression of ARE genes, whereas inhibition of tumorigenesis via chemoprevention in wild-type mice is associated with elevated expression of the cytoprotective genes. Examples include protection against Bap-induced forestomach cancer by oltipraz (a dithiolethione antischistosomiasis drug) (141, 147) and by sulforaphane (an isothiocyanate inducer derived from glucoraphanin of cruciferous vegetables) (148); inhibition of N-nitrosobutyl(4-hydroxybutyl)amine (BNN)-induced urinary bladder cancer by oltipraz (149); inhibition of 7,12-dimethylbenz(a)anthracene (DBA)/TPA-induced skin tumor by sulforaphane (150); azoxymethane/dextran sulfate sodium-induced colorectal cancer (151); pulmonary metastasis of implanted mouse lung carcinoma cells (152); and progression of DBA/medroxyprogesterone acetate–induced mammary carcinoma (153). The chemopreventive effects of oltipraz and sulforaphane were examined in humans in Qidong, China, where high incidences of hepatocellular carcinoma were observed, possibly owing to consumption of aflatoxin-contaminated foods. Oltipraz increased urinary excretion of aflatoxin-mercapturic acid, when taken at 125 mg by mouth daily, and reduced the excretion of aflatoxin M1, a primary oxidative metabolite of aflatoxin B1, when taken at 500 mg once a week (154). Consumption of broccoli sprout glucosinolate (precursor of sulforaphane) reduced the excretion of aflatoxin-DNA adducts, though large individual variability in the bioavailability of the agents was noted (155).
The most potent inducers of ARE genes were found among the newly synthesized derivatives of oleanolic triterpenoids. The semisynthetic inducers contain one or two cyano enone motifs that are highly electrophilic Michael acceptors that react with Keap1 cysteine thiols, thereby activating Nrf2 with high potency (i.e., at low nanomolar concentrations). For example, TP-225, the most potent inducer tested to date, has a Dm value of 0.27 nM for induction of Nqo1, which is 1,074 times more potent than that of sulforaphane (290 nM) (Dm = the concentration of an inducer at which 50% effect is produced) (156). The inducers potently inhibit cancer formation in several animal models, such as vinyl carbamate–induced lung cancer and aflatoxin-induced liver cancer (157). In addition to being potent inducers of ARE genes and anticancer agents, cyano enone inducers exhibit strong anti-inflammatory, hypoglycemic, antihyperlipidemic, antiproliferative, and proapoptotic activities (97, 157). Therefore, triterpenoids and derivatives also become promising drug candidates for prevention and therapy against a range of chronic diseases and toxicities (12, 14, 97, 126, 157). As an example, CDDO-Me (bardoxolone methyl) has completed a Phase II clinical trial for treatment of chronic kidney disease associated with type 2 diabetes; the treatment was associated with persistent improvement in the estimated glomerular filtration rate in patients and a relatively mild safety profile (158).
The potentially more-harm-than-benefit effect of Nrf2 on cancer was recognized from the observations that some tumors have persistently elevated expression of ARE genes, indicating that cancer cells hijack the protective power of Nrf2 to increase resistance to oxidants and cancer-killing agents. The mechanism responsible for persistent activation of Nrf2 in cancer is severalfold (Figure 2c). Somatic mutations of NRF2 were often found in squamous cell carcinoma and were mostly located in ETGE (57%) and DLG (43%) motifs, both of which disrupt Keap1 DC–Nrf2 Neh2 binding (159, 160). Mutations in KEAP1 were frequently associated with adenocarcinomas and were found in the DC (65%), IVR (29%), BTB (3%), and NTR (3%) domains, causing inhibition of Keap1 and stabilization of Nrf2 protein (161–164). Lung and prostate tumors exhibit increased methylation of the KEAP1 promoter that downregulates KEAP1 expression (165). DJ-1 is a putative oncoprotein, is expressed at high levels in primary lung and prostate cancer biopsies, and correlates inversely with clinical outcomes. As in neuronal cells, DJ-1 stabilizes Nrf2 by inhibiting Nrf2-Keap1 binding to upregulate cytoprotective genes in cancer cells (72). Expression of endogenous oncogenic alleles of Kras, Braf, and Myc stimulates the transcription of Nrf2, resulting in activation of Nrf2 and suppression of ROS; the findings suggest a role of Nrf2 in oncogene tumorigenesis (166). Nrf2 promotes aggressive proliferation of cancer cells by reinforcing the metabolic programming triggered by proliferative signals; in this scenario, Nrf2 redirects glucose and glutamine into anabolic pathways to support aggressive proliferation (85). Activation of Nrf2 in tumors is unlikely to be a cancer-initiating event but rather a result of selection during tumor development, because Keap1 KO mice with elevated Nrf2 functions did not develop spontaneous cancer over a 2-year period (167). These findings suggest that specific inhibitors of Nrf2 could be developed for therapy against cancers that have elevated Nrf2 activity (12).
Neurodegeneration
A role of oxidative stress in neurodegenerative pathology is well recognized (2, 3, 168). Mutations in Cu/ZnSOD (SOD1) cause familial amyotrophic lateral sclerosis (ALS). Modification and mutations of at least five separate gene products are associated with Parkinson’s disease, including α-synuclein, parkin, ubiquitin C-terminal hydrolase-1, DJ-1, and PINK1, all of which trigger ROS formation in neurons. Formation of extracellular amyloid plaques in the brain is a hallmark of Alzheimer’s disease, where aggregation of amyloid-β (Aβ) and ROS production promote each other for the development of brain atrophy.
Several studies support a protective role of Nrf2 against neurodegenerative diseases. Crossing mice that overexpress Nrf2 with two ALS mouse models showed significant delay in the onset of ALS and extended survival of the mice with ALS (169). Nrf2 KO mice were significantly more sensitive to MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine)-induced PD-like lesions in mice, which are alleviated by overexpression of Nrf2 in astrocytes (170). Loss of Nrf2 also increased the sensitivity of mice to myelin oligodendrocyte glycoprotein (MOG 35–55)-stimulated autoimmune attack on myelin and oligodendrocytes in the central nervous system that causes autoimmune encephalomyelitis, an experimental model of multiple sclerosis (91). ARE inducers were shown to protect against chemically induced neuronal lesions in several models (115, 116). A molecular link between Nrf2 and neurodegeneration was provided by a study on DJ-1. Loss of DJ-1 leads to early-onset PD in humans; however, DJ-1 KO mice did not show widespread loss of neurons but were more susceptible to neurotoxicant MPTP and oxidative stress (171). Loss of DJ-1 leads to reduced activity of Nrf2 and, consequently, decreased resistance to neurotoxicants (72). This relationship between DJ-1 and Nrf2 may also be important in other neurodegenerative pathology, as both proteins were found consistently upregulated in inflammatory multiple sclerosis lesions (172).
CONCLUSIONS
Since it was discovered as the transcription factor to mediate induction of ARE-dependent DMEs over a decade ago, Nrf2 has quickly emerged as a major regulator of oxidant resistance and has been implicated in a range of toxicities and chronic diseases that are characteristically associated with oxidative stress. Nrf2 is activated through a Keap1-dependent, evolutionarily conserved, dedepression signaling mechanism, wherein Nrf2 is suppressed under a basal condition through Keap1-controlled ubiquitination-proteasomal degradation and is activated by oxidants and electrophiles via modification of critical cysteine thiols of Keap1 and Nrf2. Activated Nrf2 mediates induced expression of an array of enzymes and signaling proteins to regulate drug metabolism, antioxidant defense, and oxidant signaling, thereby influencing oxidant physiology and pathology. By regulating oxidant levels and oxidant signaling, Nrf2 participates in the control of several programmatic functions, such as autophagy, inflammasome signaling, UPR, apoptosis, mitochondrial biogenesis, and stem cell regulation. Nrf2 exhibits multiple protective effects against toxicity and chronic disease naturally or through pharmacological means, opening new avenues for drug development. The protective capacity of Nrf2 could be hijacked by cancer cells to promote cancer growth and drug resistance, triggering potential side effects of ARE inducers but, at the same time, raising a possibility of developing Nrf2 inhibitors to treat, through personalized medicine, cancers that have elevated Nrf2 activities.
Supplementary Material
SUMMARY POINTS.
Intracellular oxidant signaling and antioxidant defense appear to be governed by reactive cysteine thiol–based redox signaling.
Nrf2 controls the cellular oxidant level and oxidant signaling by regulating the expression of three groups of ARE-dependent genes: drug metabolizing enzymes/transporters, antioxidant enzymes/proteins, and oxidant signaling proteins.
Nrf2-Keap1 signaling is evolutionarily conserved and involves suppression of Nrf2 through Keap1/Cul3-dependent ubiquitination/proteasomal degradation and activation of Nrf2 by inducers via modification of Keap1/Nrf2 cysteine codes.
Nrf2 participates in the regulation of oxidant-stimulated programmatic functions, including autophagy, inflammasome assembly, ER stress/UPR, mitochondrial biogenesis, and stem cell regulation.
Nrf2 broadly protects against toxicity and chronic diseases in normal cells or through pharmacological interventions.
The dedepression mechanism is hijacked in a number of pathological conditions by interfering with Nrf2-Keap1 or Keap1-Cul3 binding through somatic mutations, accumulation of disruptor proteins, and downregulation of Keap1 expression, resulting in persistent activation of Nrf2 in cancer and autophagy deficiency.
FUTURE ISSUES.
Although several genes important for ROS metabolism have been identified as target genes of Nrf2, detailed mechanisms by which Nrf2 regulates oxidant levels in cellular compartments remain to a large extent unclear.
Understanding of the role and mechanism of action of Nrf2 in oxidant-stimulated cellular programs, such as autophagy, inflammasome assembly, and ER stress/UPR, is only beginning. The impact of these functions of Nrf2 on toxicity and disease remains to be defined.
The chemistry of inducer-cysteine code interaction remains largely unaddressed. Resolution of the three-dimensional structure of the Nrf2-Keap1-Cul3 E3 complex is needed for a full understanding of inducer-cysteine code interaction for Nrf2 activation.
Pharmacological application of the Nrf2-Keap1 pathway in disease prevention and therapy is at an early stage. New and potent ARE inducers open new possibilities for clinical trials and animal models, but drug safety for ARE inducers demands increasing attention.
Developing specific Nrf2 inhibitors is desirable for the treatment of cancers with persistently activated Nrf2.
Footnotes
DISCLOSURE STATEMENT
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review. The findings and conclusions presented in this review are those of the author and do not necessarily represent the views of the National Institute for Occupational Safety and Health.
LITERATURE CITED
- 1.Finkel T. Signal transduction by reactive oxygen species. J. Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma Q. Transcriptional responses to oxidative stress: pathological and toxicological implications. Pharmacol. Ther. 2010;125:376–93. doi: 10.1016/j.pharmthera.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Bossy-Wetzel E, Schwarzenbacher R, Lipton SA. Molecular pathways to neurodegeneration. Nat. Med. 2004;10(Suppl.):S2–9. doi: 10.1038/nm1067. [DOI] [PubMed] [Google Scholar]
- 4.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–95. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 6.Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the β-globin locus control region. Proc. Natl. Acad. Sci. USA. 1994;91:9926–30. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–22. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 8.Venugopal R, Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc. Natl. Acad. Sci. USA. 1996;93:14960–65. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu. Rev. Pharmacol. Toxicol. 2003;43:233–60. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 10.Ma Q. Xenobiotic-activated receptors: from transcription to drug metabolism to disease. Chem. Res. Toxicol. 2008;21:1651–71. doi: 10.1021/tx800156s. [DOI] [PubMed] [Google Scholar]
- 11.Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med. 2004;10:549–57. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Ma Q, He X. Molecular basis of electrophilic and oxidative defense: promises and perils of Nrf2. Pharmacol. Rev. 2012;64:1055–81. doi: 10.1124/pr.110.004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan K, Han XD, Kan YW. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc. Natl. Acad. Sci. USA. 2001;98:4611–16. doi: 10.1073/pnas.081082098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klaassen CD, Reisman SA. Nrf2 the rescue: effects of the antioxidative/electrophilic response on the liver. Toxicol. Appl. Pharmacol. 2010;244:57–65. doi: 10.1016/j.taap.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walters DM, Cho HY, Kleeberger SR. Oxidative stress and antioxidants in the pathogenesis of pulmonary fibrosis: a potential role for Nrf2. Antioxid. Redox Signal. 2008;10:321–32. doi: 10.1089/ars.2007.1901. [DOI] [PubMed] [Google Scholar]
- 16.Talalay P, Dinkova-Kostova AT, Holtzclaw WD. Importance of phase 2 gene regulation in protection against electrophile and reactive oxygen toxicity and carcinogenesis. Adv. Enzyme Regul. 2003;43:121–34. doi: 10.1016/s0065-2571(02)00038-9. [DOI] [PubMed] [Google Scholar]
- 17.Hayes JD, McMahon M, Chowdhry S, Dinkova-Kostova AT. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid. Redox Signal. 2010;13:1713–48. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- 18.Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells. 2011;16:123–40. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- 19.Kensler TW, Wakabayashi N. Nrf2: Friend or foe for chemoprevention? Carcinogenesis. 2010;31:90–99. doi: 10.1093/carcin/bgp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finkel T. Signal transduction by mitochondrial oxidants. J. Biol. Chem. 2012;287:4434–40. doi: 10.1074/jbc.R111.271999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janssen-Heininger YM, Mossman BT, Heintz NH, Forman HJ, Kalyanaraman B, et al. Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic. Biol. Med. 2008;45:1–17. doi: 10.1016/j.freeradbiomed.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sundaresan M, Yu ZX, Ferrans VJ, Irani K, Finkel T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science. 1995;270:296–99. doi: 10.1126/science.270.5234.296. [DOI] [PubMed] [Google Scholar]
- 23.Lee SR, Kwon KS, Kim SR, Rhee SG. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J. Biol. Chem. 1998;273:15366–72. doi: 10.1074/jbc.273.25.15366. [DOI] [PubMed] [Google Scholar]
- 24.Saitoh M, Nishitoh H, Fujii M, Takeda K, Tobiume K, et al. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 1998;17:2596–606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat. Immunol. 2010;11:136–40. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- 26.Watternberg LW. Inhibition of carcinogenic and toxic effects of polycyclic hydrocarbons by phenolic antioxidants and ethoxyquin. J. Natl. Cancer Inst. 1972;48:1425–30. [PubMed] [Google Scholar]
- 27.Rushmore TH, Pickett CB. Transcriptional regulation of the rat glutathione S-transferase Ya subunit gene. Characterization of a xenobiotic-responsive element controlling inducible expression by phenolic antioxidants. J. Biol. Chem. 1990;265:14648–53. [PubMed] [Google Scholar]
- 28.Friling RS, Bensimon A, Tichauer Y, Daniel V. Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proc. Natl. Acad. Sci. USA. 1990;87:6258–62. doi: 10.1073/pnas.87.16.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Favreau LV, Pickett CB. Transcriptional regulation of the rat NAD(P)H:quinone reductase gene. Identification of regulatory elements controlling basal level expression and inducible expression by planar aromatic compounds and phenolic antioxidants. J. Biol. Chem. 1991;266:4556–61. [PubMed] [Google Scholar]
- 30.Jaiswal AK. Human NAD(P)H:quinone oxidoreductase (NQO1) gene structure and induction by dioxin. Biochemistry. 1991;30:10647–53. doi: 10.1021/bi00108a007. [DOI] [PubMed] [Google Scholar]
- 31.Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 1991;266:11632–39. [PubMed] [Google Scholar]
- 32.Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem. J. 2003;374:337–48. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wasserman WW, Fahl WE. Functional antioxidant responsive elements. Proc. Natl. Acad. Sci. USA. 1997;94:5361–66. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan K, Lu R, Chang JC, Kan YW. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc. Natl. Acad. Sci. USA. 1996;93:13943–48. doi: 10.1073/pnas.93.24.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu X, Roberts JR, Apopa PL, Kan YW, Ma Q. Accelerated ovarian failure induced by 4-vinyl cyclohexene diepoxide in Nrf2 null mice. Mol. Cell. Biol. 2006;26:940–54. doi: 10.1128/MCB.26.3.940-954.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoh K, Itoh K, Enomoto A, Hirayama A, Yamaguchi N, et al. Nrf2-deficient female mice develop lupus-like autoimmune nephritis. Kidney Int. 2001;60:1343–53. doi: 10.1046/j.1523-1755.2001.00939.x. [DOI] [PubMed] [Google Scholar]
- 38.Ma Q, Battelli L, Hubbs AF. Multiorgan autoimmune inflammation, enhanced lymphoproliferation, and impaired homeostasis of reactive oxygen species in mice lacking the antioxidant-activated transcription factor Nrf2. Am. J. Pathol. 2006;168:1960–74. doi: 10.2353/ajpath.2006.051113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee JM, Chan K, Kan YW, Johnson JA. Targeted disruption of Nrf2 causes regenerative immune-mediated hemolytic anemia. Proc. Natl. Acad. Sci. USA. 2004;101:9751–56. doi: 10.1073/pnas.0403620101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hubbs AF, Benkovic SA, Miller DB, O’Callaghan JP, Battelli L, et al. Vacuolar leukoencephalopathy with widespread astrogliosis in mice lacking transcription factor Nrf2. Am. J. Pathol. 2007;170:2068–76. doi: 10.2353/ajpath.2007.060898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogura T, Tong KI, Mio K, Maruyama Y, Kurokawa H, et al. Keap1 is a forked-stem dimer structure with two large spheres enclosing the intervening, double glycine repeat, and C-terminal domains. Proc. Natl. Acad. Sci. USA. 2010;107:2842–47. doi: 10.1073/pnas.0914036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, Zhang D, Hannink M, Beamer LJ. Crystal structure of the Kelch domain of human Keap1. J. Biol. Chem. 2004;279:54750–58. doi: 10.1074/jbc.M410073200. [DOI] [PubMed] [Google Scholar]
- 43.Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, et al. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 44.Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat. Genet. 2003;35:238–45. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 45.Ma Q, De-Fede KK. A labile factor regulates induction of NQOR by TCDD and phenolic antioxidants. Toxicologist. 2001;60:364. [Google Scholar]
- 46.He X, Chen MG, Lin GX, Ma Q. Arsenic induces NAD(P)H-quinone oxidoreductase I by disrupting the Nrf2•Keap1•Cul3 complex and recruiting Nrf2•Maf to the antioxidant response element enhancer. J. Biol. Chem. 2006;281:23620–31. doi: 10.1074/jbc.M604120200. [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 2004;24:7130–39. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 2004;24:10941–53. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol. Cell. Biol. 2006;26:2887–900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 2006;25:3605–17. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tong KI, Padmanabhan B, Kobayashi A, Shang C, Hirotsu Y, et al. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol. Cell. Biol. 2007;27:7511–21. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dinkova-Kostova AT, Fahey JW, Talalay P. Chemical structures of inducers of nicotinamide quinone oxidoreductase 1 (NQO1) Methods Enzymol. 2004;382:423–48. doi: 10.1016/S0076-6879(04)82023-8. [DOI] [PubMed] [Google Scholar]
- 53.Dinkova-Kostova AT, Massiah MA, Bozak RE, Hicks RJ, Talalay P. Potency of Michael reaction acceptors as inducers of enzymes that protect against carcinogenesis depends on their reactivity with sulfhydryl groups. Proc. Natl. Acad. Sci. USA. 2001;98:3404–9. doi: 10.1073/pnas.051632198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eggler AL, Liu G, Pezzuto JM, van Breemen RB, Mesecar AD. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc. Natl. Acad. Sci. USA. 2005;102:10070–75. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hong F, Sekhar KR, Freeman ML, Liebler DC. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. J. Biol. Chem. 2005;280:31768–75. doi: 10.1074/jbc.M503346200. [DOI] [PubMed] [Google Scholar]
- 56.He X, Ma Q. Critical cysteine residues of Kelch-like ECH-associated protein 1 in arsenic sensing and suppression of nuclear factor erythroid 2-related factor 2. J. Pharmacol. Exp. Ther. 2010;332:66–75. doi: 10.1124/jpet.109.160465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamamoto T, Suzuki T, Kobayashi A, Wakabayashi J, Maher J, et al. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol. Cell. Biol. 2008;28:2758–70. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kobayashi M, Li L, Iwamoto N, Nakajima-Takagi Y, Kaneko H, et al. The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol. Cell. Biol. 2009;29:493–502. doi: 10.1128/MCB.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McMahon M, Lamont DJ, Beattie KA, Hayes JD. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc. Natl. Acad. Sci. USA. 2010;107:18838–43. doi: 10.1073/pnas.1007387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He X, Ma Q. NRF2 cysteine residues are critical for oxidant/electrophile-sensing, Kelch-like ECH-associated protein-1–dependent ubiquitination-proteasomal degradation, and transcription activation. Mol. Pharmacol. 2009;76:1265–78. doi: 10.1124/mol.109.058453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He X, Chen MG, Ma Q. Activation of Nrf2 in defense against cadmium-induced oxidative stress. Chem. Res. Toxicol. 2008;21:1375–83. doi: 10.1021/tx800019a. [DOI] [PubMed] [Google Scholar]
- 62.He X, Lin GX, Chen MG, Zhang JX, Ma Q. Protection against chromium (VI)-induced oxidative stress and apoptosis by Nrf2. Recruiting Nrf2 into the nucleus and disrupting the nuclear Nrf2/Keap1 association. Toxicol. Sci. 2007;98:298–309. doi: 10.1093/toxsci/kfm081. [DOI] [PubMed] [Google Scholar]
- 63.Apopa PL, He X, Ma Q. Phosphorylation of Nrf2 in the transcription activation domain by casein kinase 2 (CK2) is critical for the nuclear translocation and transcription activation function of Nrf2 in IMR-32 neuroblastoma cells. J. Biochem. Mol. Toxicol. 2008;22:63–76. doi: 10.1002/jbt.20212. [DOI] [PubMed] [Google Scholar]
- 64.Huang HC, Nguyen T, Pickett CB. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J. Biol. Chem. 2002;277:42769–74. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 65.Jain AK, Jaiswal AK. Phosphorylation of tyrosine 568 controls nuclear export of Nrf2. J. Biol. Chem. 2006;281:12132–42. doi: 10.1074/jbc.M511198200. [DOI] [PubMed] [Google Scholar]
- 66.Salazar M, Rojo AI, Velasco D, de Sagarra RM, Cuadrado A. Glycogen synthase kinase-3β inhibits the xenobiotic and antioxidant cell response by direct phosphorylation and nuclear exclusion of the transcription factor Nrf2. J. Biol. Chem. 2006;281:14841–51. doi: 10.1074/jbc.M513737200. [DOI] [PubMed] [Google Scholar]
- 67.Cullinan SB, Diehl JA. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J. Biol. Chem. 2004;279:20108–17. doi: 10.1074/jbc.M314219200. [DOI] [PubMed] [Google Scholar]
- 68.Kawai Y, Garduno L, Theodore M, Yang J, Arinze IJ. Acetylation-deacetylation of the transcription factor Nrf2 (nuclear factor erythroid 2–related factor 2) regulates its transcriptional activity and nucleocytoplasmic localization. J. Biol. Chem. 2011;286:7629–40. doi: 10.1074/jbc.M110.208173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12:213–23. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 70.Chen W, Sun Z, Wang XJ, Jiang T, Huang Z, et al. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol. Cell. 2009;34:663–73. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lo SC, Hannink M. PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp. Cell Res. 2008;314:1789–803. doi: 10.1016/j.yexcr.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Clements CM, McNally RS, Conti BJ, Mak TW, Ting JP. DJ-1, a cancer- and Parkinson’s disease–associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc. Natl. Acad. Sci. USA. 2006;103:15091–96. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, Cuadrado A. SCF/β-TrCP promotes glycogen synthase kinase 3–dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell. Biol. 2011;31:1121–33. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maher J, Yamamoto M. The rise of antioxidant signaling–the evolution and hormetic actions of Nrf2. Toxicol. Appl. Pharmacol. 2010;244:4–15. doi: 10.1016/j.taap.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 75.Sun J, Hoshino H, Takaku K, Nakajima O, Muto A, et al. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 2002;21:5216–24. doi: 10.1093/emboj/cdf516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Biswas M, Chan JY. Role of Nrf1 in antioxidant response element–mediated gene expression and beyond. Toxicol. Appl. Pharmacol. 2010;244:16–20. doi: 10.1016/j.taap.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kobayashi A, Ito E, Toki T, Kogame K, Takahashi S, et al. Molecular cloning and functional characterization of a new cap ‘n’ collar family transcription factor Nrf3. J. Biol. Chem. 1999;274:6443–52. doi: 10.1074/jbc.274.10.6443. [DOI] [PubMed] [Google Scholar]
- 78.Pepe AE, Xiao Q, Zampetaki A, Zhang Z, Kobayashi A, et al. Crucial role of Nrf3 in smooth muscle cell differentiation from stem cells. Circ. Res. 2010;106:870–79. doi: 10.1161/CIRCRESAHA.109.211417. [DOI] [PubMed] [Google Scholar]
- 79.Chevillard G, Paquet M, Blank V. Nfe2l3 (Nrf3) deficiency predisposes mice to T-cell lymphoblastic lymphoma. Blood. 2011;117:2005–8. doi: 10.1182/blood-2010-02-271460. [DOI] [PubMed] [Google Scholar]
- 80.An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–93. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sykiotis GP, Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell. 2008;14:76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li L, Kobayashi M, Kaneko H, Nakajima-Takagi Y, Nakayama Y, Yamamoto M. Molecular evolution of Keap1. Two Keap1 molecules with distinctive intervening region structures are conserved among fish. J. Biol. Chem. 2008;283:3248–55. doi: 10.1074/jbc.M708702200. [DOI] [PubMed] [Google Scholar]
- 83.Kwak M-K, Wakabayashi N, Greenlaw JL, Yamamoto M, Kensler TW. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol. Cell. Biol. 2003;23:8786–94. doi: 10.1128/MCB.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Malhotra D, Portales-Casamar E, Singh A, Srivastava S, Arenillas D, et al. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718–34. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, et al. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 86.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scherz-Shouval R, Elazar Z. Regulation of autophagy by ROS: physiology and pathology. Trends Biochem. Sci. 2011;36:30–38. doi: 10.1016/j.tibs.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 88.Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element–driven gene transcription. J. Biol. Chem. 2010;285:22576–91. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol. Cell. Biol. 2010;30:3275–85. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fujita K, Maeda D, Xiao Q, Srinivasula SM. Nrf2-mediated induction of p62 controls Toll-like receptor-4–driven aggresome-like induced structure formation and autophagic degradation. Proc. Natl. Acad. Sci. USA. 2011;108:1427–32. doi: 10.1073/pnas.1014156108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Johnson DA, Amirahmadi S, Ward C, Fabry Z, Johnson JA. The absence of the pro-antioxidant transcription factor Nrf2 exacerbates experimental autoimmune encephalomyelitis. Toxicol. Sci. 2010;114:237–46. doi: 10.1093/toxsci/kfp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rangasamy T, Cho CY, Thimmulappa RK, Zhen L, Srisuma SS, et al. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke–induced emphysema in mice. J. Clin. Investig. 2004;114:1248–59. doi: 10.1172/JCI21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rangasamy T, Guo J, Mitzner WA, Roman J, Singh A, et al. Disruption of Nrf2 enhances susceptibility to severe airway inflammation and asthma in mice. J. Exp. Med. 2005;202:47–59. doi: 10.1084/jem.20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Garbin U, Fratta Pasini A, Stranieri C, Cominacini M, Pasini A, et al. Cigarette smoking blocks the protective expression of Nrf2/ARE pathway in peripheral mononuclear cells of young heavy smokers favouring inflammation. PLoS ONE. 2009;4:e8225. doi: 10.1371/journal.pone.0008225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cho HY, Reddy SP, Yamamoto M, Kleeberger SR. The transcription factor NRF2 protects against pulmonary fibrosis. FASEB J. 2004;18:1258–60. doi: 10.1096/fj.03-1127fje. [DOI] [PubMed] [Google Scholar]
- 96.He X, Ma Q. Disruption of Nrf2 synergizes with high glucose to cause heightened myocardial oxidative stress and severe cardiomyopathy in diabetic mice. J. Diabetes Metab. 2012 doi: 10.4172/2155-6156.S7-002. In press, doi: 10:4172/21556156.S7-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dinkova-Kostova AT, Liby KT, Stephenson KK, Holtzclaw WD, Gao X, et al. Extremely potent triterpenoid inducers of the phase 2 response: correlations of protection against oxidant and inflammatory stress. Proc. Natl. Acad. Sci. USA. 2005;102:4584–89. doi: 10.1073/pnas.0500815102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Talalay P, Fahey JW, Healy ZR, Wehage SL, Benedict AL, et al. Sulforaphane mobilizes cellular defenses that protect skin against damage by UV radiation. Proc. Natl. Acad. Sci. USA. 2007;104:17500–5. doi: 10.1073/pnas.0708710104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ma Q, Kinneer K. Chemoprotection by phenolic antioxidants. Inhibition of tumor necrosis factor α induction in macrophages. J. Biol. Chem. 2002;277:2477–84. doi: 10.1074/jbc.M106685200. [DOI] [PubMed] [Google Scholar]
- 100.Li W, Khor TO, Xu C, Shen G, Jeong WS, et al. Activation of Nrf2-antioxidant signaling attenuates NFκB-inflammatory response and elicits apoptosis. Biochem. Pharmacol. 2008;76:1485–89. doi: 10.1016/j.bcp.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ma Q, Kinneer K, Ye J, Chen BJ. Inhibition of nuclear factor κB by phenolic antioxidants: interplay between antioxidant signaling and inflammatory cytokine expression. Mol. Pharmacol. 2003;64:211–19. doi: 10.1124/mol.64.2.211. [DOI] [PubMed] [Google Scholar]
- 102.Tschopp J, Schroder K. NLRP3 inflammasome activation: The convergence of multiple signalling pathways on ROS production? Nat. Rev. Immunol. 2010;10:210–15. doi: 10.1038/nri2725. [DOI] [PubMed] [Google Scholar]
- 103.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–25. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 104.Dostert C, Petrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–77. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rubartelli A, Gattorno M, Netea MG, Dinarello CA. Interplay between redox status and inflammasome activation. Trends Immunol. 2011;32:559–66. doi: 10.1016/j.it.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 106.Duewell P, Kono H, Rayner KJ, Sirois CM, Vladimer G, et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 2010;464:1357–61. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Freigang S, Ampenberger F, Spohn G, Heer S, Shamshiev AT, et al. Nrf2 is essential for cholesterol crystal-induced inflammasome activation and exacerbation of atherosclerosis. Eur. J. Immunol. 2011;41:2040–51. doi: 10.1002/eji.201041316. [DOI] [PubMed] [Google Scholar]
- 108.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 109.Appenzeller-Herzog C. Updates on “endoplasmic reticulum redox.”. Antioxid. Redox Signal. 2012;16:760–62. doi: 10.1089/ars.2011.4463. [DOI] [PubMed] [Google Scholar]
- 110.Kelsen SG, Duan X, Ji R, Perez O, Liu C, Merali S. Cigarette smoke induces an unfolded protein response in the human lung: a proteomic approach. Am. J. Respir. Cell Mol. Biol. 2008;38:541–50. doi: 10.1165/rcmb.2007-0221OC. [DOI] [PubMed] [Google Scholar]
- 111.Cullinan SB, Zhang D, Hannink M, Arvisais E, Kaufman RJ, Diehl JA. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol. Cell. Biol. 2003;23:7198–209. doi: 10.1128/MCB.23.20.7198-7209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–33. [PubMed] [Google Scholar]
- 113.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008;88:611–38. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 114.He X, Kan H, Cai L, Ma Q. Nrf2 is critical in defense against high glucose–induced oxidative damage in cardiomyocytes. J. Mol. Cell Cardiol. 2009;46:47–58. doi: 10.1016/j.yjmcc.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 115.Calkins MJ, Jakel RJ, Johnson DA, Chan K, Kan YW, Johnson JA. Protection from mitochondrial complex II inhibition in vitro and in vivo by Nrf2-mediated transcription. Proc. Natl. Acad. Sci. USA. 2005;102:244–49. doi: 10.1073/pnas.0408487101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Calkins MJ, Townsend JA, Johnson DA, Johnson JA. Cystamine protects from 3-nitropropionic acid lesioning via induction of Nf-E2 related factor 2 mediated transcription. Exp. Neurol. 2010;224:307–17. doi: 10.1016/j.expneurol.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Piantadosi CA, Carraway MS, Babiker A, Suliman HB. Heme oxygenase-1 regulates cardiac mitochondrial biogenesis via Nrf2-mediated transcriptional control of nuclear respiratory factor-1. Circ. Res. 2008;103:1232–40. doi: 10.1161/01.RES.0000338597.71702.ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Junn E, Jang WH, Zhao X, Jeong BS, Mouradian MM. Mitochondrial localization of DJ-1 leads to enhanced neuroprotection. J. Neurosci. Res. 2009;87:123–29. doi: 10.1002/jnr.21831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McCoy MK, Cookson MR. DJ-1 regulation of mitochondrial function and autophagy through oxidative stress. Autophagy. 2011;7:531–32. doi: 10.4161/auto.7.5.14684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Owusu-Ansah E, Banerjee U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature. 2009;461:537–41. doi: 10.1038/nature08313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yanes O, Clark J, Wong DM, Patti GJ, Sánchez-Ruiz A, et al. Metabolic oxidation regulates embryonic stem cell differentiation. Nat. Chem. Biol. 2010;6:411–17. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hochmuth CE, Biteau B, Bohmann D, Jasper H. Redox regulation by Keap1 and Nrf2 controls intestinal stem cell proliferation in Drosophila. Cell Stem Cell. 2011;8:188–99. doi: 10.1016/j.stem.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Merchant AA, Singh A, Matsui W, Biswal S. The redox-sensitive transcription factor Nrf2 regulates murine hematopoietic stem cell survival independently of ROS levels. Blood. 2011;118:6572–79. doi: 10.1182/blood-2011-05-355362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nelson S, Bruschi SA. Mechanisms of acetaminophen-induced liver disease. In: Kaplowitz N, DeLeve LD, editors. Drug-Induced Liver Disease. Informa Healthcare; New York: 2007. pp. 353–88. [Google Scholar]
- 125.Enomoto A, Itoh K, Nagayoshi E, Haruta J, Kimura T, et al. High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol. Sci. 2001;59:169–77. doi: 10.1093/toxsci/59.1.169. [DOI] [PubMed] [Google Scholar]
- 126.Reisman SA, Buckley DB, Tanaka Y, Klaassen CD. CDDO-Im protects from acetaminophen hepatotoxicity through induction of Nrf2-dependent genes. Toxicol. Appl. Pharmacol. 2009;236:109–14. doi: 10.1016/j.taap.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Okawa H, Motohashi H, Kobayashi A, Aburatani H, Kensler TW, Yamamoto M. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem. Biophys. Res. Commun. 2006;339:79–88. doi: 10.1016/j.bbrc.2005.10.185. [DOI] [PubMed] [Google Scholar]
- 128.Reisman SA, Csanaky IL, Aleksunes LM, Klaassen CD. Altered disposition of acetaminophen in Nrf2-null and Keap1-knockdown mice. Toxicol. Sci. 2009;109:31–40. doi: 10.1093/toxsci/kfp047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wu D, Cederbaum AI. Oxidative stress and alcoholic liver disease. Semin. Liver Dis. 2009;29:141–54. doi: 10.1055/s-0029-1214370. [DOI] [PubMed] [Google Scholar]
- 130.Yang CS, Brady JF, Hong JY. Dietary effects on cytochromes P450, xenobiotic metabolism, and toxicity. FASEB J. 1992;6:737–44. doi: 10.1096/fasebj.6.2.1537464. [DOI] [PubMed] [Google Scholar]
- 131.Bae SH, Sung SH, Cho EJ, Lee SK, Lee HE, et al. Concerted action of sulfiredoxin and peroxiredoxin I protects against alcohol-induced oxidative injury in mouse liver. Hepatology. 2011;53:945–53. doi: 10.1002/hep.24104. [DOI] [PubMed] [Google Scholar]
- 132.Gong P, Cederbaum AI. Nrf2 is increased by CYP2E1 in rodent liver and HepG2 cells and protects against oxidative stress caused by CYP2E1. Hepatology. 2006;43:144–53. doi: 10.1002/hep.21004. [DOI] [PubMed] [Google Scholar]
- 133.Dong J, Sulik KK, Chen SY. Nrf2-mediated transcriptional induction of antioxidant response in mouse embryos exposed to ethanol in vivo: implications for the prevention of fetal alcohol spectrum disorders. Antioxid. Redox Signal. 2008;10:2023–33. doi: 10.1089/ars.2007.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kumar A, Singh CK, Lavoie HA, Dipette DJ, Singh US. Resveratrol restores Nrf2 level and prevents ethanol-induced toxic effects in the cerebellum of a rodent model of fetal alcohol spectrum disorders. Mol. Pharmacol. 2011;80:446–57. doi: 10.1124/mol.111.071126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yeligar SM, Machida K, Kalra VK. Ethanol-induced HO-1 and NQO1 are differentially regulated by HIF-1α and Nrf2 to attenuate inflammatory cytokine expression. J. Biol. Chem. 2010;285:35359–73. doi: 10.1074/jbc.M110.138636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Iizuka T, Ishii Y, Itoh K, Kiwamoto T, Kimura T, et al. Nrf2-deficient mice are highly susceptible to cigarette smoke–induced emphysema. Genes Cells. 2005;10:1113–25. doi: 10.1111/j.1365-2443.2005.00905.x. [DOI] [PubMed] [Google Scholar]
- 137.Gebel S, Diehl S, Pype J, Friedrichs B, Weiler H, et al. The transcriptome of Nrf2−/− mice provides evidence for impaired cell cycle progression in the development of cigarette smoke–induced emphysematous changes. Toxicol. Sci. 2010;115:238–52. doi: 10.1093/toxsci/kfq039. [DOI] [PubMed] [Google Scholar]
- 138.Blake DJ, Singh A, Kombairaju P, Malhotra D, Mariani TJ, et al. Deletion of Keap1 in the lung attenuates acute cigarette smoke–induced oxidative stress and inflammation. Am. J. Respir. Cell Mol. Biol. 2010;42:524–36. doi: 10.1165/rcmb.2009-0054OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sussan TE, Rangasamy T, Blake DJ, Malhotra D, El-Haddad H, et al. Targeting Nrf2 with the triterpenoid CDDO-imidazolide attenuates cigarette smoke-induced emphysema and cardiac dysfunction in mice. Proc. Natl. Acad. Sci. USA. 2009;106:250–55. doi: 10.1073/pnas.0804333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rodgman A, Perfetti TA. The Chemical Compounds of Tobacco and Tobacco Smoke. CRC Press; New York: 2009. [Google Scholar]
- 141.Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, et al. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in Nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. USA. 2001;98:3410–15. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Reddy NM, Kleeberger SR, Bream JH, Fallon PG, Kensler TW, et al. Genetic disruption of the Nrf2 compromises cell-cycle progression by impairing GSH-induced redox signaling. Oncogene. 2008;27:5821–32. doi: 10.1038/onc.2008.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Goven D, Boutten A, Lecon-Malas V, Marchal-Somme J, Amara N, et al. Altered Nrf2/Keap1-Bach1 equilibrium in pulmonary emphysema. Thorax. 2008;63:916–24. doi: 10.1136/thx.2007.091181. [DOI] [PubMed] [Google Scholar]
- 144.Malhotra D, Thimmulappa R, Navas-Acien A, Sandford A, Elliott M, et al. Decline in NRF2-regulated antioxidants in chronic obstructive pulmonary disease lungs due to loss of its positive regulator, DJ-1. Am. J. Respir. Crit. Care Med. 2008;178:592–604. doi: 10.1164/rccm.200803-380OC. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 145.Malhotra D, Thimmulappa RK, Mercado N, Ito K, Kombairaju P, et al. Denitrosylation of HDAC2 by targeting Nrf2 restores glucocorticosteroid sensitivity in macrophages from COPD patients. J. Clin. Investig. 2011;121:4289–302. doi: 10.1172/JCI45144. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 146.Mercado N, Thimmulappa R, Thomas CM, Fenwick PS, Chana KK, et al. Decreased histone deacetylase 2 impairs Nrf2 activation by oxidative stress. Biochem. Biophys. Res. Commun. 2011;406:292–98. doi: 10.1016/j.bbrc.2011.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ramos-Gomez M, Dolan PM, Itoh K, Yamamoto M, Kensler TW. Interactive effects of Nrf2 genotype and oltipraz on benzo[a]pyrene-DNA adducts and tumor yield in mice. Carcinogenesis. 2003;24:461–67. doi: 10.1093/carcin/24.3.461. [DOI] [PubMed] [Google Scholar]
- 148.Fahey JW, Haristoy X, Dolan PM, Kensler TW, Scholtus I, et al. Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[a]pyrene-induced stomach tumors. Proc. Natl. Acad. Sci. USA. 2002;99:7610–15. doi: 10.1073/pnas.112203099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Iida K, Itoh K, Kumagai Y, Oyasu R, Hattori K, et al. Nrf2 is essential for the chemopreventive efficacy of oltipraz against urinary bladder carcinogenesis. Cancer Res. 2004;64:6424–31. doi: 10.1158/0008-5472.CAN-04-1906. [DOI] [PubMed] [Google Scholar]
- 150.Xu C, Huang MT, Shen G, Yuan X, Lin W, et al. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2–related factor 2. Cancer Res. 2006;66:8293–96. doi: 10.1158/0008-5472.CAN-06-0300. [DOI] [PubMed] [Google Scholar]
- 151.Khor TO, Huang MT, Prawan A, Liu Y, Hao X, et al. Increased susceptibility of Nrf2 knockout mice to colitis-associated colorectal cancer. Cancer Prev. Res. 2008;1:187–91. doi: 10.1158/1940-6207.CAPR-08-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Satoh H, Moriguchi T, Taguchi K, Takai J, Maher JM, et al. Nrf2-deficiency creates a responsive microenvironment for metastasis to the lung. Carcinogenesis. 2010;31:1833–43. doi: 10.1093/carcin/bgq105. [DOI] [PubMed] [Google Scholar]
- 153.Becks L, Prince M, Burson H, Christophe C, Broadway M, et al. Aggressive mammary carcinoma progression in Nrf2 knockout mice treated with 7,12-dimethylbenz[a]anthracene. BMC Cancer. 2010;10:540. doi: 10.1186/1471-2407-10-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kensler TW, Curphey TJ, Maxiutenko Y, Roebuck BD. Chemoprotection by organosulfur inducers of phase 2 enzymes: dithiolethiones and dithiins. Drug Metabol. Drug Interact. 2000;17:3–22. doi: 10.1515/dmdi.2000.17.1-4.3. [DOI] [PubMed] [Google Scholar]
- 155.Kensler TW, Chen JG, Egner PA, Fahey JW, Jacobson LP, et al. Effects of glucosinolate-rich broccoli sprouts on urinary levels of aflatoxin-DNA adducts and phenanthrene tetraols in a randomized clinical trial in He Zuo township, Qidong, People’s Republic of China. Cancer Epidemiol. Biomark. Prev. 2005;14:2605–13. doi: 10.1158/1055-9965.EPI-05-0368. [DOI] [PubMed] [Google Scholar]
- 156.Dinkova-Kostova AT, Talalay P, Sharkey J, Zhang Y, Holtzclaw WD, et al. An exceptionally potent inducer of cytoprotective enzymes: elucidation of the structural features that determine inducer potency and reactivity with Keap1. J. Biol. Chem. 2010;285:33747–55. doi: 10.1074/jbc.M110.163485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liby KT, Yore MM, Sporn MB. Triterpenoids and rexinoids as multifunctional agents for the prevention and treatment of cancer. Nat. Rev. Cancer. 2007;7:357–69. doi: 10.1038/nrc2129. [DOI] [PubMed] [Google Scholar]
- 158.Pergola PE, Raskin P, Toto RD, Meyer CJ, Huff JW, et al. Bardoxolone methyl and kidney function in CKD with type 2 diabetes. N. Engl. J. Med. 2011;365:327–36. doi: 10.1056/NEJMoa1105351. [DOI] [PubMed] [Google Scholar]
- 159.Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, et al. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc. Natl. Acad. Sci. USA. 2008;105:13568–73. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kim YR, Oh JE, Kim MS, Kang MR, Park SW, et al. Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J. Pathol. 2010;220:446–51. doi: 10.1002/path.2653. [DOI] [PubMed] [Google Scholar]
- 161.Nioi P, Nguyen T. A mutation of Keap1 found in breast cancer impairs its ability to repress Nrf2 activity. Biochem. Biophys. Res. Commun. 2007;362:816–21. doi: 10.1016/j.bbrc.2007.08.051. [DOI] [PubMed] [Google Scholar]
- 162.Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, et al. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 2008;135:1358–68. doi: 10.1053/j.gastro.2008.06.082. 68.e1–4. [DOI] [PubMed] [Google Scholar]
- 163.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Takahashi T, Sonobe M, Menju T, Nakayama E, Mino N, et al. Mutations in Keap1 are a potential prognostic factor in resected non-small cell lung cancer. J. Surg. Oncol. 2010;101:500–6. doi: 10.1002/jso.21520. [DOI] [PubMed] [Google Scholar]
- 165.Wang R, An J, Ji F, Jiao H, Sun H, Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem. Biophys. Res. Commun. 2008;373:151–54. doi: 10.1016/j.bbrc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 166.DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–9. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Taguchi K, Maher JM, Suzuki T, Kawatani Y, Motohashi H, Yamamoto M. Genetic analysis of cytoprotective functions supported by graded expression of Keap1. Mol. Cell. Biol. 2010;30:3016–26. doi: 10.1128/MCB.01591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Finkel T. Radical medicine: treating ageing to cure disease. Nat. Rev. Mol. Cell Biol. 2005;6:971–76. doi: 10.1038/nrm1763. [DOI] [PubMed] [Google Scholar]
- 169.Vargas MR, Johnson DA, Sirkis DW, Messing A, Johnson JA. Nrf2 activation in astrocytes protects against neurodegeneration in mouse models of familial amyotrophic lateral sclerosis. J. Neurosci. 2008;28:13574–81. doi: 10.1523/JNEUROSCI.4099-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen PC, Vargas MR, Pani AK, Smeyne RJ, Johnson DA, et al. Nrf2-mediated neuroprotection in the MPTP mouse model of Parkinson’s disease: critical role for the astrocyte. Proc. Natl. Acad. Sci. USA. 2009;106:2933–38. doi: 10.1073/pnas.0813361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kim RH, Smith PD, Aleyasin H, Hayley S, Mount MP, et al. Hypersensitivity of DJ-1-deficient mice to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyrindine (MPTP) and oxidative stress. Proc. Natl. Acad. Sci. USA. 2005;102:5215–20. doi: 10.1073/pnas.0501282102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.van Horssen J, Drexhage JA, Flor T, Gerritsen W, van der Valk P, de Vries HE. Nrf2 and DJ1 are consistently upregulated in inflammatory multiple sclerosis lesions. Free Radic. Biol. Med. 2010;49:1283–89. doi: 10.1016/j.freeradbiomed.2010.07.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.