Summary
One of the cardinal features of neural development and adult plasticity is the contribution of activity-dependent signaling pathways. However, the interrelationships between different activity-dependent genes are not well understood. The immediate early gene, neuronal activity-regulated pentraxin (NPTX2 or Narp), encodes a protein that has been associated with excitatory synaptogenesis, AMPA receptor aggregation and the onset of critical periods. Here we show that Narp is a direct transcriptional target of Brain-derived neurotrophic factor (BDNF), another highly regulated activity-dependent gene involved in synaptic plasticity. Unexpectedly, Narp is bi-directionally regulated by BDNF. Acute BDNF withdrawal results in downregulation of Narp, whereas transcription of Narp is greatly enhanced by BDNF. Furthermore, our results show that BDNF directly regulates Narp to mediate glutamatergic transmission and mossy fiber plasticity. Hence, Narp serves as a significant epistatic target of BDNF to regulate synaptic plasticity during periods of dynamic activity.
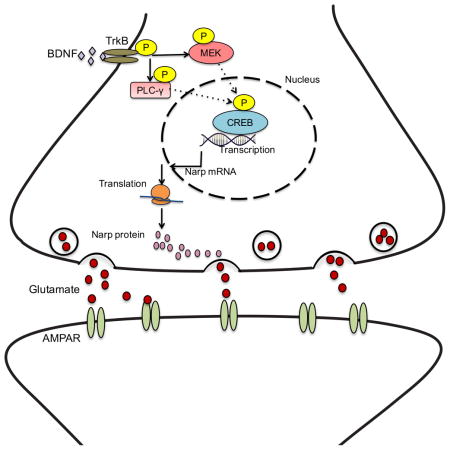
Graphical Abstract
Introduction
Activity-dependent gene expression forms the basis of many periods of heightened brain plasticity, which are influenced by the balance between excitation and inhibition, synaptogenesis and competition between different environmental signals. These events can affect the onset and closure of critical periods, neurodevelopmental disorders and behavioral flexibility (West and Greenberg, 2011; Takesian and Hensch, 2013). While transcription factors were originally identified as immediate early genes (IEGs) induced by growth factors, a growing list of diverse proteins has been discovered as activity-dependent genes. These include cytoskeletal proteins, such as Arc (Lyford et al., 1995), chromatin modification enzymes (Wijayatunge et al., 2014) and extracellular proteases, such as tissue-plasminogen activator (Qian et al., 1993). How the activities of immediate early genes are coordinated and inter-connected is an open question.
Many activity-dependent genes were identified using differential cloning techniques in the hippocampus following conditions of electro-convulsive seizure. A major IEG protein is neuronal activity-regulated pentraxin (Narp), which is involved in experience-dependent synaptic plasticity and is upregulated following long-term potentiation (LTP) induction (Tsui et al., 1996). Narp is highly expressed in the hippocampus and cortex, where it undergoes induction by synaptic activity and is present in both pre- and post-synaptic compartments (Reti et al., 2002; Chang et al., 2010). Overexpression of Narp results in co-localization and aggregation of AMPA receptor subunits in heterologous cells and spinal neurons (O’Brien et al., 1999). Deletion of Narp leads to a loss in excitatory inputs to fast-spiking parvalbumin-positive interneurons in the visual cortex and interferes with the timing and establishment of ocular dominance plasticity (Gu et al., 2013). Experience-dependent expression of Narp therefore contributes to cellular adaption to the environment.
Another gene that is a sensor of neuronal activity is Brain-derived neurotrophic factor (BDNF). Because BDNF mRNA is significantly upregulated after seizures compared to other neurotrophins NGF and NT-3 (Ernfors et al., 1991; Isackson et al., 1991), BDNF was implicated as a gene directly involved in synaptic plasticity (Thoenen, 1995). This idea has been borne out by the close association of BDNF with hippocampal plasticity (Kang and Schuman, 1995; Patterson et al., 1996). Consistent with this function, BDNF modulates local protein synthesis, cytoskeleton dynamics, synaptic neurotransmission, neuronal excitability, as well as LTP (Park and Poo, 2013; Panja and Bramham, 2014). Mice lacking the BDNF receptor, TrkB, or carrying a targeted mutation in the PLCγ site of TrkB show abnormal hippocampal LTP (Minichiello et al., 1999; 2002).
Changing the levels of BDNF has a number of consequences. For example, the human BDNF Val66Met polymorphism, results in an impairment of episodic memory and hippocampal function (Egan et al., 2003). Measurement of BDNF levels in BDNFMet/Met mice revealed a 30% reduction in activity-dependent release (Chen et al., 2006). Another prominent phenotype of the BDNF Val66Met polymorphism is anxiety, in both humans and mice (Soliman et al., 2010). The mechanism is due to intracellular trafficking of pro-BDNF and a reduction of regulated release by the pro-BDNF Met polymorphism (Egan et al., 2003; Chen et al., 2006). Accordingly, a small decrease in BDNF can have a dramatic impact.
There are hints that Narp and BDNF expression are related from microarray analyses of different environmental and developmental states (Tong et al., 2001; Wibrand et al., 2006; Spiegel et al., 2014). However, the interrelationships of these genes are not well understood. Here we report that Narp is transcriptionally up-regulated through BDNF-TrkB signaling mechanisms. Conversely, a loss of BDNF results in significant decrease in expression of Narp. More importantly, these changes manifest in an appreciable impact upon synaptic transmission and LTP in the mossy fiber pathway. Our studies reveal a heretofore unrecognized activity-dependent pathway for synaptic function.
Results
BDNF regulates expression of Narp
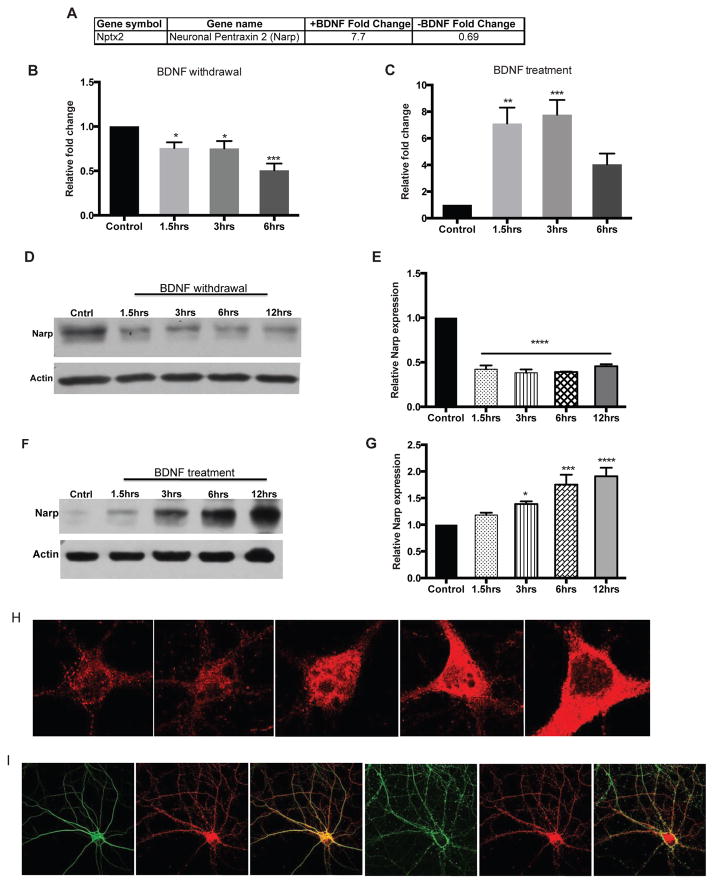
We previously investigated the transcriptional events that occur when hippocampal neurons were deprived of BDNF. A significant down-regulation of genes involved in synaptic function was observed from microarray analysis (Mariga et al., 2014). One of the target genes that was down-regulated following withdrawal of BDNF is the activity-dependent gene, Narp (Figure 1A). The down-regulation of Narp transcription was particularly striking in light of the previous transcriptional profiling of cortical neurons treated with BDNF, which showed a 8-fold induction of Narp mRNA (Figure 1A). These findings prompted us to test whether Narp is transcriptionally regulated by BDNF.
Figure 1. BDNF regulates Narp in hippocampal neurons.
(A) Microarray analysis of Narp gene expression in response to changes in BDNF. Narp mRNA levels after BDNF withdrawal, n=4 (B) BDNF treatment, n=4. (C). Narp protein levels after BDNF withdrawal, n=3 (D) and BDNF treatment, n=4 (F). (E and G) Quantitation of Narp levels. (H) Narp enrichment in the somatodendritic area after BDNF treatment, scale bar (10μm). (I) Co-localization of Narp, MAP2 and Tau, scale bar (50μm). Data is presented as mean ± SEM, p<0.05,**p<0.001,****p<0.0001.
We utilized qPCR to validate our microarray findings in hippocampal cultures deprived of BDNF using a Trk receptor ligand scavenger, TrkB-Fc. In the presence of TrkB-Fc, Narp transcription was markedly decreased in a time-dependent manner, reaching a 50% reduction by 6hrs (Figure 1B). Conversely, when hippocampal cells were treated with BDNF in a similar time frame we observed a consistent increase in Narp levels as early as 1.5hrs (Figure 1C). Narp displayed a 6-fold increase 1.5hrs following BDNF treatment, remained upregulated up to 3hrs, then started decreasing toward baseline levels by 6hrs. These findings confirmed a significant bidirectional regulation of Narp by BDNF.
The regulation of Narp mRNA by BDNF suggested that crosstalk existed between these two activity-dependent genes. To further confirm this regulation, we examined Narp protein levels in response to BDNF changes. We deprived hippocampal neurons of BDNF in a similar time-course and analyzed Narp protein expression. Consistent with the mRNA findings, Narp expression began decreasing 1.5hrs after BDNF sequestration and maintained a significant decrease (50%) up to 12hrs (Figures 1D and E). When hippocampal neurons were treated with BDNF, Narp expression increased in a time-dependent manner reaching over 200% of basal levels by 6–12hrs (Figures 1F and G). We also performed immunocytochemical analysis of the cellular distribution of Narp protein following BDNF treatment and observed a significant enrichment in the somatodendritic areas (Figure 1H). The enrichment in Narp expression was coincident with MAP2 staining and time-dependent with peak levels between 6–12 hrs (Figure 1I).
BDNF regulates Narp in vivo
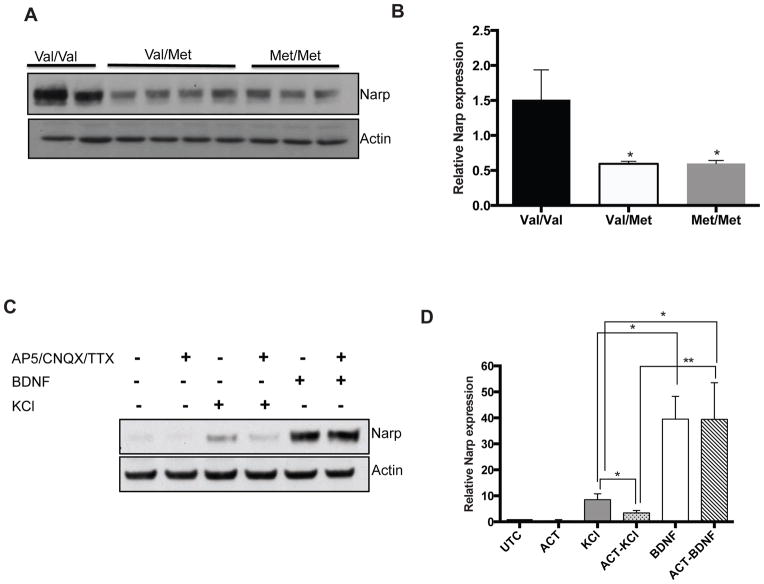
Given the profound regulation of Narp by BDNF in vitro, we asked if Narp was also regulated by BDNF in vivo. To address this question, we examined Narp levels in BDNF knock-in mice that carry the Val66Met single nucleotide polymorphism (SNP) in the prodomain of BDNF (Chen et al., 2006). The Val66Met SNP selectively affects the regulated secretion of BDNF and has been associated with learning and memory deficits (Egan et al., 2003; Chen et al., 2006). Narp expression was significantly reduced in both heterozygous (val/met) and homozygous knock-in (met/met) mice compared to wild-type (val/val) mice (Figures 2A and B). The decrease was striking; a 30% decrease in regulated secretion of BDNF resulted in a 67% decrease in Narp expression highlighting a significant contribution of BDNF signaling in Narp regulation (Chen et al., 2006).
Figure 2. Narp is downstream of BDNF signaling.
(A–B) Expression of Narp in Val66Met mice, n=2 (val/val), n=4 (val/met), n=3 (met/met). (C–D) Role of activity in BDNF-dependent Narp induction. BDNF can induce Narp when neuronal activity is blocked, (lane 6) and the induction is comparable to BDNF alone and higher than KCl. Untreated culture: UTC, AP5/CNQX/TTX: ACT. *p<0.05, **p<0.01, ****p<0.001, n=3, mean ± SEM, for each experiment.
Role of activity in BDNF-dependent Narp induction
Since Narp is an activity-dependent gene product, we asked if Narp induction by BDNF is downstream of neuronal activity. We incubated hippocampal neurons with AP5, TTX and CNQX to block neuronal activity prior to BDNF treatment. When neuronal activity was suppressed, BDNF induced Narp to levels comparable to BDNF alone (Figures 2C and D) suggesting that BDNF is sufficient to induce Narp, even in the absence of neuronal activity.
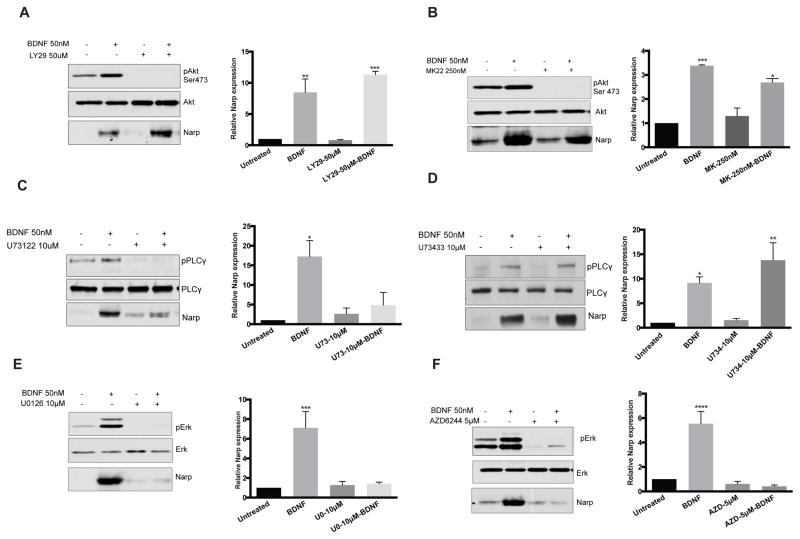
BDNF regulates Narp through the MAP kinase and PLC-γ pathways
When BDNF binds to TrkB receptor, it activates three main signaling pathways: MAPK/Erk, phosphatidylinositide 3-kinase (PI3K) and phospholipase C-gamma (PLCγ) to mediate cellular processes such as neuronal differentiation, survival and neuroplasticity (Chao 2003; Huang and Reichardt, 2003). We used selective pharmacological inhibitors of each pathway to identify the signaling mechanism responsible for induction of Narp downstream of BDNF. Blocking PI3K using LY29004, did not interfere with the induction of Narp by BDNF (Figure 3A). Similarly, inhibiting AKT with MK2206 did not block BDNF-induced Narp expression (Figure 3B), suggesting that induction of Narp downstream of BDNF signaling is independent of PI3K-Akt signaling. Blocking phosphorylation of PLC-γ with U73122 prior to BDNF treatment resulted in a partial reduction in Narp expression (Figure 3C), while its inactive analog U73433 did not interfere with Narp expression after BDNF stimulation (Figure 3D). These results suggest that the PLC-γ pathway contributes to activation of Narp expression downstream of BDNF signaling. To test the involvement of the MAP kinase pathway, we used U0126 and AZD6244, to block phosphorylation of MAPK/ERK. Unlike the lack of effect of PI3K/Akt inhibitors and the modest effect of PLC-γ inhibition, preincubation of hippocampal neurons with MAP kinase inhibitors abolished Narp activation by BDNF, supporting an important role for the MAP kinase pathway in BDNF-mediated Narp induction (Figures 3E and 3F).
Figure 3. BDNF regulates Narp through the MAP kinase and PLC-γ pathways.
Effect of PI3K-AKT inhibitors [ LY29004 (50μM) and MK2206 (250nM), n=4, A,B]. PLC-γ inhibitor [U73122 (10μM) and inactive analog U73433 (10μM), n=3, C,D] and MAPK/Erk inhibitors [U0126 (10μM) and AZD6244 (5μM),n=3, E,F] on BDNF-induced increase in Narp. Data is represented as mean ± SEM, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Transcriptional induction of Narp
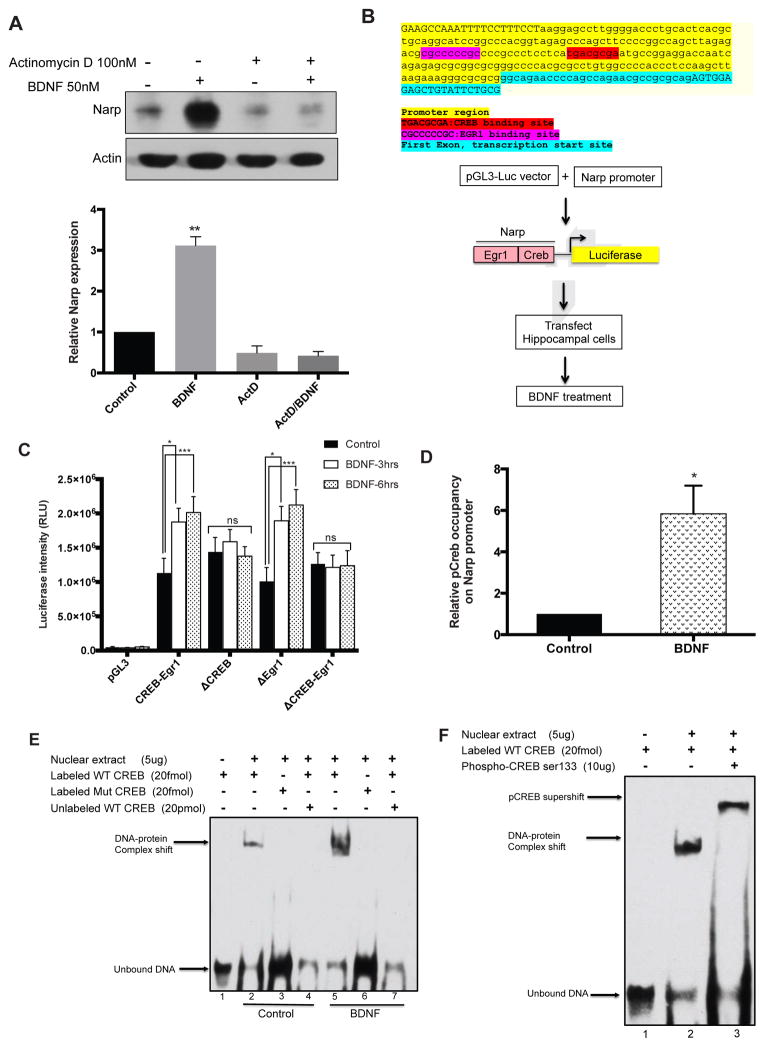
Is Narp regulated transcriptionally by BDNF? Transcription inhibition with actinomycin D prior to BDNF treatment blocked BDNF-induced Narp expression (Figure 4A). We then sought to identity the transcriptional mechanism by examining the Narp promoter for potential transcription factor binding sites. We identified putative binding sites for CREB and Early growth response1 (Egr1) within a 300 bp fragment upstream of the transcription start site (Figure 4B). We cloned the fragment into a pGL3-luciferase reporter construct and assessed luciferase expression after transfection in hippocampal neurons. Transfected neurons were treated with BDNF and luciferase responses were monitored. Luciferase intensity increased with time peaking to a 2-fold increase by 3–6hrs (Figure 4C). The profile of increase in luciferase intensity was consistent with the time-course of Narp expression in response to BDNF that we had observed by qPCR and immunoblotting (Figure 1).
Figure 4. Regulation of Narp by BDNF involves activation of CREB.
(A) Blocking transcription with actinomycin D (100nM) blocks Narp expression.
(B) Top: 300bp fragment upstream of the Narp transcription start site. Promoter fragment (yellow), Egr1 binding site (pink), CREB binding site (red), transcription start site (blue). Bottom: Outline of the luciferase reporter experiments.
(C) Luciferase induction after 3 and 6hrs of BDNF treatment. Increase in luciferase intensity with CREB-Egr1 and ΔEgr1 but not with ΔCREB or ΔCREB-Egr1. n=3, *p<0.05 ***p<0.001, mean ± SEM.
(D) 6-fold enrichment of pCREB on the Narp promoter after BDNF treatment, n=4, p<0.05, mean ± SEM. (E) EMSA confirmation of transcription factor binding. Gel shift observed in both untreated and BDNF treated nuclear extracts with labeled wild-type CREB oligo (lanes 2 and 5) but not with mutant oligo (lane 3 and 6) or excess unlabeled wild-type CREB oligo (lane 4 and 7). (F) Protein-DNA supershift in the presence of pCREB.
Since the Narp promoter fragment contained two promoter binding sites, Egr1 and CREB, we asked if these sites were both important for driving expression of luciferase upon BDNF treatment. We used site-directed mutagenesis to independently delete CREB or Egr1 binding sites and monitored luciferase intensity in response to BDNF treatment. When the CREB binding site (ΔCREB) was deleted, there was no increase in induction of luciferase after BDNF stimulation (Figure 4C, ΔCREB panel). Conversely, deleting the Egr1 binding site (ΔEgr1) had no effect on BDNF-induced increase in luciferase expression; there was a 2-fold increase in luciferase levels at 3 and 6hrs which was comparable to the wild-type construct at the same time-points. These results suggest that Egr1 is not required for mediating Narp induction downstream of BDNF signaling. To confirm the involvement of CREB in Narp induction, both the CREB and Egr1 binding sites (ΔCREB-Egr1) were deleted. Assaying for luciferase induction after BDNF stimulation indicated an absence of luciferase expression. Hence, CREB is necessary and sufficient to induce Narp downstream of BDNF signaling.
Luciferase reporter experiments strongly supported the importance of CREB in transcriptional activation of Narp. To further characterize the occupancy of CREB on the Narp promoter we conducted chromatin immunoprecipitation on hippocampal neurons treated with BDNF. Phospho-CREB was highly enriched on the Narp promoter in BDNF-treated neurons relative to untreated control, further supporting the role of CREB in BDNF-mediated induction of Narp expression (Figure 4D).
We further confirmed the binding of CREB to the CREB binding site on the Narp promoter, with electrophoretic mobility shift analysis. Hippocampal nuclear extracts were incubated with 3′-biotin labeled 25mer oligonucleotides containing the consensus CREB binding sequence followed by resolution of the protein-DNA complexes on a non-denaturing polyacrylamide gel. A gel shift was observed in both untreated and BDNF-treated extracts (Figure 4E, lanes 2 and 5), which competed with the unlabeled CREB sequence (Figure, 4E lanes 4 and 7). Moreover, the shift was not observed when nuclear extracts were incubated with a CREB oligomer mutated on the CREB binding motif (Figure 4E lanes 3 and 6). Supershift assays with phospho-CREB antibody confirmed the presence of a DNA-protein complex containing CREB bound to the Narp oligomer (Figure 4F). Taken together, these results confirmed CREB as the transcription factor that is responsible for inducing Narp downstream of BDNF signaling.
Narp is important for BDNF-dependent synaptic function
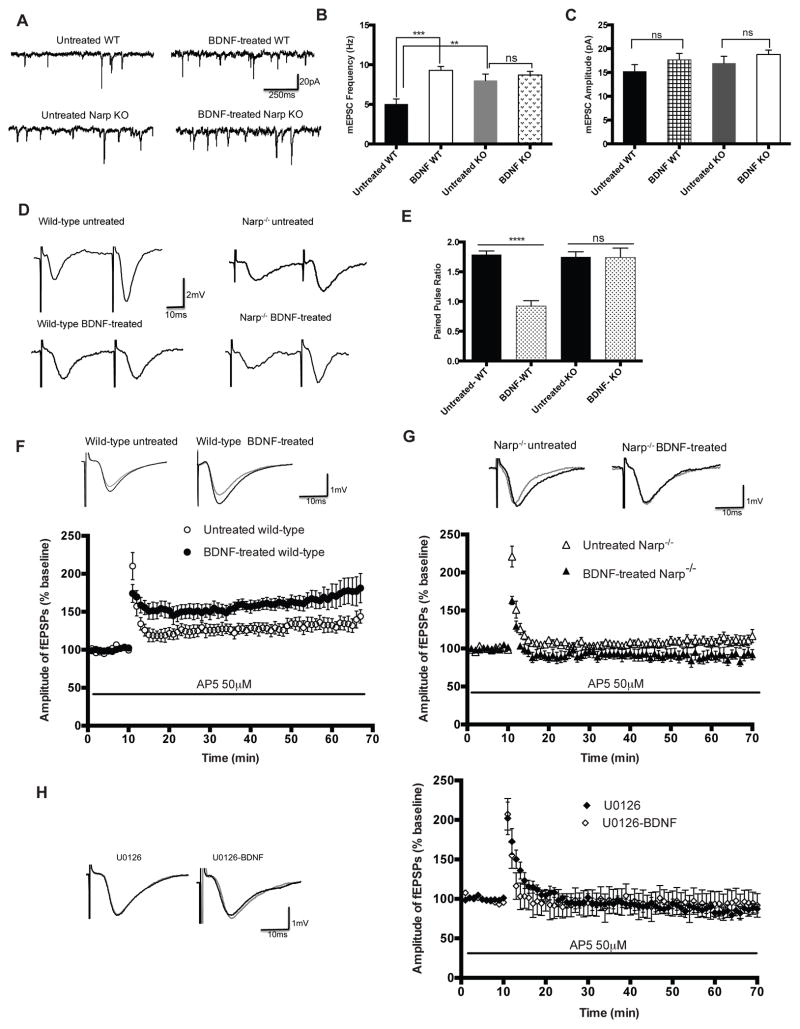
Given the bidirectional regulation of Narp by BDNF, we asked if Narp plays a role in BDNF-dependent modulation of synaptic function. To determine whether BDNF requires Narp for its effect on glutamatergic transmission, we measured miniature excitatory post-synaptic currents (mEPSCs) from BDNF-treated (50nM, 4hrs) and untreated cultured hippocampal neurons from Narp−/− and wild-type littermates. We observed an increase in frequency of mEPSCs in BDNF-treated wild-type cultures (9.3 ± 0.48 Hz, n=10; p<0.0001) relative to untreated neurons (5.04 ± 0.65 Hz, n=11; Figures 5A and B) while the effect of BDNF on mEPSC amplitude was statistically insignificant (untreated: 15.3 ± 1.39 pA, n=11; BDNF-treated: 17.7 ± 1.34 pA, n=10; p= 0.23, Figures 5A and 5C). In contrast to the effect of BDNF on wild-type hippocampal cultures, Narp−/− neuronal cultures failed to respond to BDNF treatment. Both the frequency (untreated: 8.02 ± 0.8 Hz, n=10; BDNF-treated: 8.71 ± 0.46, n=10; p=0.46) and amplitude (untreated: 16.97± 1.43 pA, n=9; BDNF treated: 18.78 ± 1.34pA, n=10; p=0.29) of mEPSCs were not significantly different (Figures 5A, B and C).
Figure 5. Narp is required for BDNF-dependent synaptic function.
(A) Representative mEPSC traces from untreated and BDNF-treated wild-type and Narp−/− cultures. Average mEPSC frequency (B) and amplitude (C) in untreated and BDNF-treated wild-type and Narp−/− cultures. (D) Representative PPR traces from untreated and BDNF-treated wild-type and Narp−/− hippocampal slices. (E) Average PPR for untreated and BDNF-treated wild-type and Narp−/− slices. (F) Mossy fiber LTP in untreated (n=8) and BDNF-treated (n=6) wild-type slices. Upper panel shows representative fEPSP traces before (grey trace) and after (black trace) HFS. (G) Mossy fiber LTP is impaired in Narp−/− group (n=6) and is unaffected by BDNF (n=5). Upper panel shows representative fEPSPs traces before (grey trace) and after (black trace) HFS. (H) Mossy fiber LTP and its enhancement by BDNF are impaired in the presence of U0126. Left panels shows representative fEPSP traces fEPSPs before (grey trace) and after (black trace) HFS. Data is reported as mean ± SEM.
The lack of an effect of BDNF in Narp−/− neurons suggested that Narp is necessary for BDNF-induced enhancement of glutamatergic transmission in hippocampal neurons. Interestingly, a comparison of mEPSCs measured in untreated wild-type and Narp−/− cultures showed a higher mEPSC frequency in Narp−/− cultures (Figure 5B; p< 0.009). These results suggest that Narp may have a functional role in regulating spontaneous synaptic transmission that is developmentally regulated.
Next, we asked if Narp plays a role in activity-dependent synaptic plasticity in the mossy fiber pathway that is mediated by BDNF (Schildt et al., 2013). Based on our biochemical data, abundant presence of BDNF and Narp in the mossy fiber-CA3 pathway and the role of BDNF in mossy fiber LTP (Xu et al., 2003, Schildt et al., 2013, Huang et al., 2008), we reasoned that synaptic plasticity in the mossy fiber pathway requires Narp. First, we studied paired pulse facilitation (PPF), a form of short-term plasticity that involves a pre-synaptic mechanism (Hess et al., 1987; Zucker et al., 1989). The paired pulse ratio (PPR) was significantly reduced in BDNF-treated wild-type slices (0.92 ± 0.09, n=10; p<0.0001) compared to untreated slices (1.79 ± 0.063, n=10; Figures 5D and E), suggesting enhanced glutamate release in the BDNF-treated group. Untreated Narp−/− group showed a robust PPR similar to wild-type group (1.75 ± 0.09, n=8; Figures 5D and E). However, in contrast to BDNF-treated wild-type group, PPR remained unchanged in BDNF-treated Narp−/− group (1.74 ± 0.15, n=5; p=0.972) (Figures 5D and E). These results suggest that BDNF-mediated enhancement of glutamate release at the mossy fiber synapses depends upon Narp.
Given the aforementioned results showing the role Narp in BDNF-induced pre-synaptic plasticity in the mossy fiber pathway, we asked whether Narp is required for mossy fiber LTP, which is BDNF-dependent (Schildt et al., 2013). BDNF-treated wild-type slices showed a significantly higher LTP compared to untreated wild-type slices (F(2,20)=17.9, p<0.0001; Figure 5F). However, we observed a significant impairment of LTP in Narp−/− mice, which did not undergo any further increase following BDNF treatment (F2,10)=1.08; p=0.38; Figure 5G). These results suggested that Narp is not only required for mossy fiber LTP, a BDNF-dependent plasticity, but also BDNF-induced enhancement of mossy fiber LTP. Consistent with the role of MAPK in the regulation of Narp by BDNF, U0126 (20μM) blocked LTP and BDNF-induced enhancement of LTP in wild-type slices (F(2,11)=1.15; p=0.353; Figure 5H). Taken together, these results suggest that the mossy fiber LTP and its enhancement by BDNF is mediated by a MAP kinase-dependent transcriptional mechanism that culminates in increased Narp expression.
Discussion
In this study, we have demonstrated a fundamental relationship between BDNF and Narp that is important for activity-dependent synaptic plasticity. We have shown that Narp is transcriptionally regulated by BDNF in a time-dependent manner. This regulation occurs through phosphorylation of the MAP kinase and PLC-γ signaling pathways and consequent activation of CREB to induce Narp expression. Narp and BDNF have been separately reported to be induced by neuronal activity (Tsui et al., 1999, Enfors et al., 1991); however, the interplay between neuronal activity and BDNF or Narp regulation was unclear. Here we show that when neuronal activity is blocked, BDNF can still activate Narp, suggesting that BDNF is sufficient to induce Narp expression. This regulation was also replicated in vivo. Narp levels were dramatically reduced in mice carrying a BDNF (Val66Met) SNP that leads to a decrease in regulated secretion of BDNF. Despite a moderate decrease in regulated secretion of BDNF in Val66Met mice (Chen et al., 2006), Narp expression is profoundly reduced in these mice highlighting an important bidirectional transcriptional control of Narp by BDNF.
Most importantly, our findings demonstrate that Narp plays a critical role in BDNF-dependent synaptic modulation. Unlike the effect of BDNF on excitatory synaptic transmission in wild-type hippocampal neurons, BDNF did not affect glutamatergic transmission in Narp−/− neurons suggesting that Narp is important for BDNF-mediated modulation of glutamatergic synapses. In the dentate gyrus, both Narp and BDNF are highly expressed and Narp is enriched in fiber terminals in the CA3 stratum lucidum (Xu et al., 2003). Furthermore, mossy fiber LTP depends on BDNF (Huang et al., 2008; Schildt et al., 2013); however, the mechanism by which BDNF modulates mossy fiber plasticity had not been described. We observed impaired mossy fiber LTP in Narp−/− mice, a finding that was not observed before. Moreover, BDNF treatment failed to rescue the lack of plasticity in Narp−/− mice underscoring the importance of Narp in modulating mossy fiber plasticity downstream of BDNF. Despite compelling evidence for the role of BDNF in experience dependent-plasticity both in vivo and in vitro and its impact upon behavior, the downstream signaling mechanisms by which BDNF is mediating these functions have not been described. Our study demonstrates a critical transcriptional regulation of Narp by BDNF and identifies Narp as a downstream target of BDNF at the synapse.
How does Narp modulate BDNF-dependent plasticity? Both cell culture and brain slice experiments revealed that Narp−/− neurons exhibit an impairment in BDNF-dependent increase in glutamate release suggesting that Narp plays a critical role in the effect of BDNF on pre-synaptic function. These results are further supported by an impairment of activity-dependent LTP in the mossy fiber pathway, a synaptic plasticity event mediated by a pre-synaptic mechanism (Zalutsky and Nicoll, 1990; Castillo et al., 1994). Furthermore, our findings are consistent with earlier studies demonstrating the role of the PLC-γ pathway and pre-synaptic proteins downstream of BDNF-TrkB signaling in mediating enhanced neurotransmitter release (Jovanovic et al., 2000; Pan et al., 2011). Previous reports have also shown that MAPK-dependent phosphorylation of CREB results in transcriptional regulation involved in LTP (Davis et al., 2000). Also, the release of BDNF and consequent nuclear translocation of activated MAPK is involved in hippocampal plasticity (Patterson et al., 2001). Similarly, long-term plasticity and memory in Aplysia involves BDNF-induced MAPK signaling (Purcell et al., 2003). The effect of exogenous BDNF upon the enhancement of LTP in the wild-type mice may also be mediated by a post-synaptic mechanism as Narp is known to facilitate the synaptic targeting and stabilization of AMPA receptors on excitatory synapses (Chang et al., 2010; Pelkey et al., 2015).
In summary, we have unraveled the relationship between BDNF and Narp; two genes that have been widely thought to be independently regulated by activity. BDNF signaling primarily activates the MAPK pathway leading to increase in Narp, which orchestrates the plasticity process at the synapse. These findings provide insight on the interrelationship between activity-dependent genes and their functional relevance in regulating network stability during synaptic plasticity.
Experimental Procedures
Animals
Timed-pregnant Sprague-Dawley rats (Taconic), C57BL6 mice (Charles River), and C57BL6 Narp−/− transgenic mice (Worley laboratory, Johns Hopkins University) were allowed ad libitum access to food and water and maintained on a 12-hr light- dark cycle. All protocols were in compliance with the NYULMC guidelines for the care and use of laboratory animals.
Cell culture
Primary hippocampal neurons were prepared from E18 timed-pregnant Sprague–Dawley rats. Neurons were cultured in neurobasal medium (Life Technologies) containing NeuroBrew-21 supplement (Miltenyl Biotech), 0.5 mM L-glutamine (Life Technologies), 5-fluoro- uridine/uridine (10 μM each). The mouse primary culture protocol was adapted from Chang et al., 2010, supplemental methods.
Biochemistry
Day in vitro 7 (DIV7) neurons were treated with BDNF (50nM) or (TrkB-Fc (100ng/mL) for indicated time-points. For inhibitor experiments, neurons were starved in supplement-free media (4hrs) then pre-incubated with the inhibitor for 1hr prior to a 4hr BDNF treatment. For neuronal activity experiments, neurons were incubated with supplement-free neurobasal medium with TTX(1μM) /CNQX(10μM)/AP5 (50μM) for 4hrs before adding BDNF (50nM) or KCl (25mM) for 4hrs. After treatments, cells were lysed in NP-40 lysis buffer. Hippocampal tissues from 2-months old Val66Met knock-in mice were lysed in RIPA buffer. Changes in protein expression was assessed by western blotting and quantified by densitometry in Image J.
Luciferase Reporter Assays
Hippocampal neurons were transfected with a PGL3 basic luciferase vector (Promega) containing fire-fly luciferase reporter gene under the control of a 300bp fragment of the NPTX2 (Narp) promoter (NCBI Reference Sequence: NM_001034199.1) and a renilla luciferase internal control. Two days after transfection, neurons were treated with BDNF for 3 and 6hrs and luciferase activity was assayed using the Dual Glo luciferase kit (Promega) per manufacturer’s instructions. Luciferase intensity was normalized to renilla. Mutagenesis of the Narp promoter fragment was performed using the site directed mutagenesis kit (Agilent Biotechnologies).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation of BDNF-treated hippocampal neurons was performed using the Magna ChIP A Kit (Millipore). Syber Green qPCR was performed with primers flanking a putative CREB binding site on the Narp promoter; forward:5′-CCTCCTCATGACGCGAATGC3′, reverse:5′CCGCGCGCCCTTTCTTA-3′. Fold enrichment of pCREB enrichment on Narp promoter was determined by normalizing gene expression to input DNA and IgG control.
Electrophysiology
Cultured hippocampal neurons
mEPSCs were recorded from DIV 12–14 hippocampal neurons at −60mV in the presence of tetrodotoxin and bicuculline. Recordings were done using Clampex 10 software (Molecular Devices) and analyzed with Mini Analysis program (Synaptosoft).
Hippocampal slices
Field excitatory post-synaptic potentials (fEPSPs) were recorded from mossy fiber pathway in hippocampal slices at 32°C (Castillo et al., 1994). PPR was measured at 40ms interstimulus interval. Mossy fiber LTP was induced at 30% maximal stimulation strength by a 1s train stimulation protocol (3x 100Hz at 20s interval). Data analysis was performed with Clampex 10 software (Molecular Devices)
Statistics
Statistical analysis was conducted using Graph Pad Prism version 6.0 and IBM SPSS Statistical programs. Statistical significance was defined at P<0.05.
Supplementary Material
Acknowledgments
We thank Pablo Castillo (Albert Einstein); Stephen Ginsberg (Nathan Kline Institute); Andrew Sproul and Scott Noggle (New York Stem Cell Foundation); Freddy Jeanneteau and Francis Lee (Cornell Medical School) for technical advice and protein samples. The work was supported by NIH grants (AG025870; NS21072) to M.V. Chao and NIH grant (HD076914) to I. Ninan.
Footnotes
Author Contributions
A.M., I.N. and M.V.C. conceived and designed the study. A.M. performed all the experiments, with assistance from J.G. and L.M. D.X., M.X. and P.W. provided essential reagents for the experiments. I.N. arranged and oversaw the electrophysiology experiments. M.V.C., A.M. and I.N. wrote the manuscript and P.W. contributed to manuscript editing.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Castillo PE, Weisskopf MG, Nicoll RA. The role of Ca2+ channels in hippocampal mossy fiber synaptic transmission and long-term potentiation. Neuron. 1994;12:261–269. doi: 10.1016/0896-6273(94)90269-0. [DOI] [PubMed] [Google Scholar]
- Chang MC, Park JM, Pelkey KA, Grabenstatter HL, Xu D, Linden DJ, Sutula TP, McBain CJ, Worley PF. Narp regulates homeostatic scaling of excitatory synapses on parvalbumin-expressing interneurons. Nature Neurosci. 2010;13:1090–1097. doi: 10.1038/nn.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MV. Neurotrophins and their receptors: A convergence point for many signaling pathways. Nature Rev Neurosci. 2003;4:299–309. doi: 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Jing DQ, Bath KG, Ieraci A, Khan T, Siao CJ, Herrera DG, Toth M, Yang C, McEwen BS, Hempstead BL, Lee FS. Variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science. 2006;314:140–143. doi: 10.1126/science.1129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer H, Chazai G, Carleton A, Goridis C, Vincent JD, Lledo PM. Long-term but short-term plasticity at mossy fiber synapses is important in neural cell adhesion molecule-deficient mice. Proc Natl Acad Sci USA. 1998;95:13242–13247. doi: 10.1073/pnas.95.22.13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S, Vanhoutte P, Pages C, Caboche J, Laroche S. The MAPK/ERK cascade targets both Elk-1 and cAMP response element-binding protein to control long-term potentiation-dpependent gene expression in the dentate gyrus in vivo. J Neuroscience. 2000;20:4563–4572. doi: 10.1523/JNEUROSCI.20-12-04563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, Lu B, Weinberger DR. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal formation. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Ernfors P, Bengzon J, Kokaia Z, Persson H, Lindvall O. Increased levels of messenger RNAs for neurotrophic factors in the rbain during kindling epileptogenesis. Neuron. 1991;7:165–176. doi: 10.1016/0896-6273(91)90084-d. [DOI] [PubMed] [Google Scholar]
- Gu Y, Huang S, Chang MC, Worley P, Kirkwood A, Quinlan EM. Obligatory role for the immediate early gene NARP in critical period plasticity. Neuron. 2013;79:335–346. doi: 10.1016/j.neuron.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XP, Minichiello L, Klein R, McNamara JO. Immunochemical evidence of seizure-induced activation of trkB receptor in the mossy fiber pathway of mouse hippocampus. J Neurosci. 2002;22:7502–7508. doi: 10.1523/JNEUROSCI.22-17-07502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess G, Kuhnt U, Voronin LL. Quantal analysis of pair-pulse facilitation in guinea pig hippocampal slices. Neurosci Letters. 1987;77:187–192. doi: 10.1016/0304-3940(87)90584-2. [DOI] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. Trk receptors: roles in neuronal signal transduction. Ann Rev Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- Huang YZ, Pan E, Xiong ZQ, McNamara JO. Zinc-mediated trasactivation of TrkB potentiates the hippocampal mossy fiber-CA3 pyramid synapse. Neuron. 2008;57:546–558. doi: 10.1016/j.neuron.2007.11.026. [DOI] [PubMed] [Google Scholar]
- Isackson PJ, Huntsman MM, Murray KD, Gall CM. BDNF mRNA expression is increased in adult rat forebrain after limbic seizures: temporal patterns of induction distinct from NGF. Neuron. 1991;6:97–948. doi: 10.1016/0896-6273(91)90234-q. [DOI] [PubMed] [Google Scholar]
- Jovanovic JN, Czernik AJ, Fienberg AA, Greengard P, Sihra TS. Synapsins as mediators of BDNF-enhanced neurotransmitter release. Nature Neurosci. 2000;3:323–329. doi: 10.1038/73888. [DOI] [PubMed] [Google Scholar]
- Kang H, Schuman EM. Long-lasting neurotrophin-induced enhancement of synaptic transmission in the adult hippocapaus. Science. 1995;267:1658–1662. doi: 10.1126/science.7886457. [DOI] [PubMed] [Google Scholar]
- Li YX, Zhang Y, Lester HA, Schuman EM, Davidson N. Enhancement of neurotransmitter release induced by Brain-derived neurotrophic factor in cultured hippocampal neurons. J Neuroscience. 1998;18:10231–10240. doi: 10.1523/JNEUROSCI.18-24-10231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene encodes a novel cytoskelton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- Mariga A, Zavadil J, Ginsberg SD, Chao MV. Withdrawal of BDNF from hippocampal cultures leads to changes in genes involved in synaptic function. Dev Neurobiology. 2014;75:173–192. doi: 10.1002/dneu.22216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minichiello L, Korte M, Wolfer D, Kuhn R, Unsicker K, Cestari V, Rossi-Arnaud C, Lipp HP, Bonhoeffer T. Essential role for TrkB receptors in hippocampus-mediated learning. Neuron. 1999;24:401–414. doi: 10.1016/s0896-6273(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Minichiello L, Calella AM, Medina DL, Bonhoeffer T, Klein R, Korte M. Mechanism of TrkB-mediated hippocampal long-term potentiation. Neuron. 2002;36:121–137. doi: 10.1016/s0896-6273(02)00942-x. [DOI] [PubMed] [Google Scholar]
- O’Brien RJ, Xu D, Petralia RS, Steward O, Huganir RL, Worley PF. Synaptic clustering of AMPA receptors by the extracellular immediate-early gene product Narp. Neuron. 1999;23:309–323. doi: 10.1016/s0896-6273(00)80782-5. [DOI] [PubMed] [Google Scholar]
- O’Brien R, Xu D, Mi R, Tang X, Hopf C, Worley P. Synaptically targeted narp plays an essential role in the aggregation of AMPA receptors at excitatory synapses in cultured spinal neurons. J Neuroscience. 2002;22:4487–4498. doi: 10.1523/JNEUROSCI.22-11-04487.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan E, Zhang X, Huang Z, Krezel A, Zhao M, Tinberg CE, Lippard SJ, McNamara JO. Vesicular zince promotes presynaptic and inhibits postsynaptic long-term potentiation of mossy fiber-CA3 synapse. Neuron. 2011;71:1116–1126. doi: 10.1016/j.neuron.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panja D, Bramham CR. BDNF mechanisms in late LTP formation: A synthesis and breakdown. Neuropharmacology. 2014;76:664–676. doi: 10.1016/j.neuropharm.2013.06.024. [DOI] [PubMed] [Google Scholar]
- Park H, Poo MM. Neurotrophin regulation of neural circuit development and function. Nature Rev Neurosci. 2013;14:7–23. doi: 10.1038/nrn3379. [DOI] [PubMed] [Google Scholar]
- Patterson SL, Abel T, Deuel TA, Martin KC, Rose JC, Kandel ER. Recombinant BDNF rescues deficits in basal synaptic transmission and hippocampal LTP in BDNF knockout mice. Neuron. 1996;16:123–140. doi: 10.1016/s0896-6273(00)80140-3. [DOI] [PubMed] [Google Scholar]
- Patterson SL, Pittenger C, Morozov A, Martin KC, Scanlin H, Drake C, Kandel ER. Some forms of cAMP-mediated long-lasting potentiation are associated with release of BDNF and nuclear translocation of phospho-MAP kinase. Neuron. 2001;32:123–140. doi: 10.1016/s0896-6273(01)00443-3. [DOI] [PubMed] [Google Scholar]
- Pelkey KA, Barksdale E, Craig MT, Yuan X, Sukumaran M, Vargish GA, Mitchell RM, Wyeth MS, Petralia RS, Chittajallu R, Karlsson RM, Cameron HA, Murata Y, Colonnese MT, Worley PF, McBain CJ. Pentraxins coordinate excitatory synapse maturationa nd circuit integration of parvalbumin interneurons. Neuron. 2015;85:1257–1272. doi: 10.1016/j.neuron.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell AJ, Sharma SK, Bagnall MW, Sutton MA, Carew TJ. Activation of tyrosine kinase-MAPS cascade enhances the induction of long-term synaptic facilitation and long-term memory in Aplysia. Neuron. 2003;37:473–484. doi: 10.1016/s0896-6273(03)00030-8. [DOI] [PubMed] [Google Scholar]
- Qian Z, Gilbert ME, Colicos MA, Kandel ER, Kuhl D. Tissue-plasminogen activator is induced as an immediate-early gene during seizure, kindling and long-term potentiation. Nature. 1993;361:453–457. doi: 10.1038/361453a0. [DOI] [PubMed] [Google Scholar]
- Reti IM, Reddy R, Worley PF, Baraban JM. Prominent Narp expession in projection pathways and terminal fields. J Neurochem. 2002;82:935–944. doi: 10.1046/j.1471-4159.2002.01051.x. [DOI] [PubMed] [Google Scholar]
- Schildt S, Endres T, Lessmann V, Edelmann E. Acute and chronic interference with BDNF/TrkB-signaling impair LTP selectively at mossy fiber synapses in the CA3 region of mouse hippocampus. Neuropharmacology. 2013;71:247–254. doi: 10.1016/j.neuropharm.2013.03.041. [DOI] [PubMed] [Google Scholar]
- Soliman F, Glatt CE, Bath KG, Levita L, Jones RM, Pattwell SS, Jing D, Tottenham N, Amso D, Somerville LH, Voss HU, Glover G, Ballon DJ, Liston C, Teslovich T, Van Kempen T, Lee FS, Casey BJ. A genetic variant BDNF polymorphism alters extinction learning in both mouse and human. Science. 2010;327:863–866. doi: 10.1126/science.1181886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel I, Mardinly AR, Gabel HW, Bazinet JE, Couch CHG, Tzeng CP, Harmin DA, Greenberg ME. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell. 2014;157:1216–1229. doi: 10.1016/j.cell.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takesian AE, Hensch TK. Balancing plasticity/stability across brain development. Prog Brain Res. 2013;207:3–34. doi: 10.1016/B978-0-444-63327-9.00001-1. [DOI] [PubMed] [Google Scholar]
- Thoenen H. Neurotrophins and neuronal plasticity. Science. 1995;270:593–598. doi: 10.1126/science.270.5236.593. [DOI] [PubMed] [Google Scholar]
- Tong L, Shen H, Perreau VM, Balazs R, Cotman CW. Effects of exercise on gene-expession profile in the rat hippocampus. Neurobiol of Disease. 2001;8:1046–1056. doi: 10.1006/nbdi.2001.0427. [DOI] [PubMed] [Google Scholar]
- Tsui CC, Copeland NG, Gilbert DJ, Jenkins NA, Barnes C, Worley PF. Narp, a novel member of the pentraxin family, promotes neurite outgrowth and is dynamically regulated by neuronal activity. J Neuroscience. 1996;16:2463–2478. doi: 10.1523/JNEUROSCI.16-08-02463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011;3:1–21. doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayatunge R, Chen LF, Cha YM, Zannas AS, Frank CL, West AE. The histone lysiine demethylase Kdm6b is required for activity-dependent preconditioning of hippocampal neuronal survival. Mol Cell Neuroscience. 2014;61:187–200. doi: 10.1016/j.mcn.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbrand K, Messaoudi E, Havik B, Steenslid V, Lovlie R, Steen VM, Bramham CR. Identification of genes co-upregulated with Arc during BDNF-induced long-term potentiation in adult rat dentate gyrus in vivo. Eur J Neuroscience. 2006;23:1501–1511. doi: 10.1111/j.1460-9568.2006.04687.x. [DOI] [PubMed] [Google Scholar]
- Xu D, Hopf C, Reddy R, Cho RW, Guo L, Lanahan A, Petralia RS, Wenthold RJ, Worley P. Narp and NP1 form heterocomplexes that function in developmental and activity-dependent synaptic plasticity. Neuron. 2003;39:513–528. doi: 10.1016/s0896-6273(03)00463-x. [DOI] [PubMed] [Google Scholar]
- Ying S-W, Futter M, Rosenblum K, Webber MJ, Hunt SP, Bliss TVP, Bramham CR. Brain-derived neurotrophic factor induces long-term potentiation in intact adult hippocampus: Requirement for ERK activation coupled to CREB and upregulation of Arc synthesis. J Neuroscience. 2002;22:1532–1540. doi: 10.1523/JNEUROSCI.22-05-01532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalutsky RA, Nicoll RA. Comparison of two forms of long-term potentiation in single hippocampal neurons. Science. 1990;248:1619–1624. doi: 10.1126/science.2114039. [DOI] [PubMed] [Google Scholar]
- Zucker RS. Short-term synaptic plasticity. Ann Rev Neuroscience. 1989;12:13–31. doi: 10.1146/annurev.ne.12.030189.000305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.