SUMMARY
A split or interrupted gene is defined as a gene consisting of introns and exons. Removal (splicing) of the intron (s) from a primary transcript (pre-mRNA) is an essential process to create a mRNA. Initial assignment of a potential protein coding region in KSHV genome was based on initiation codon context and predicted protein size larger than 100 amino acids, but the gene discontinuity was disregarded. Experimental investigation of the assigned ORFs has demonstrated that there are up to 25 split genes, more than one fourth of the total KSHV genes described in KSHV genome. This includes the genes involved in all phases (latent, immediate early, early and late) of KSHV infection. The complexity of a split gene expression depends upon the availability of a proximal promoter and polyadenylation (pA) signal. Sharing a single promoter or a single pA signal by two or three genes is not uncommon in the expression of KSHV split genes and the resulting transcripts are usually polycistronic. Among those of KSHV split genes, 15 genes express a bicistronic or tricistronic RNA and 10 genes express a monocistronic RNA. Alternative RNA splicing could happen in a particular pre-mRNA due to intron or exon inclusion or skipping or the presence of an alternative 5′ ss or 3′ ss. This may, respectively, result in at least 8 species of K8 and 14 species of K15 transcripts. This appears to be related to cell differentiation and stages of the virus infection, presumably involving in viral cis elements and trans splicing factors.
INTRODUCTION
Kaposi’s sarcoma (KS)-associated herpesvirus (KSHV), also called human herpesvirus 8 (HHV8), is a human gammaherpesvirus from KS tissues of AIDS patients that was recently identified using representational difference analysis [1]. Subsequent epidemiological investigations have demonstrated that infection with the newly described KSHV is a etiologically implicated in development of KS, multicentric Castleman’s disease (MCD) and primary effusion lymphoma (PEL), also known as body cavity-based B-cell lymphoma [1–4]. KS is a tumour of endothelial cell origin and MCD is a B-cell lymphoproliferative disorder. Recent reports further suggest that KSHV infection might play a role indirectly in the pathogenesis of multiple myeloma [5–7]. Like other gammaherpesvirus such as human B lymphocyte-tropic Epstein–Barr virus (EBV), monkey T lymphocyte-tropic herpesvirus saimiri and murine B lymphocyte-tropic herpesvirus 68, KSHV is another human lymphocyte-tropic virus and usually infects B lymphocytes and establishes latency in the lymphocytes following primary infection. B-cell lines derived from PEL regularly harbour KSHV DNA [3,4]. However, endothelial cells are susceptible to KSHV infection [8–10] and can be converted by KSHV infection into spindle cells seen in KS [9].
KSHV has a genome size of ~165 kb that can encode up to 90 viral proteins [11,12]. Of the 81 open reading frames (ORFs) initially assigned within KSHV’s long unique region, 66 are homologous to ORFs in herpesvirus saimiri (HVS) [11], and 63 are homologous to ORFs in murine herpesvirus 68 [13]; only 15 (K1–K15) are unique to KSHV. Recently, several other ORFs unique to KSHV but not described in the initial sequence report on the KSHV genome [11] have been designated with a decimal K number, including K4.1, K4.2, K8.1, K10.1, K10.5, K10.7, K11.1 (vIRF-2) and K14.1 [12,14–16]. Some of the KSHV-expressed gene products are homologues of cellular proteins, including Bcl-2 [17,18], cyclin D [19,20], interleukin 6 [21], MIP-1 [21], G-protein coupled receptor (GPCR) [20], interferon regulatory factor (IRF) [22] and DNA synthetic enzymes including thymidylate synthase, dihydrofolate reductase, DNA polymerase, thymidine kinase and ribonucleotide reductases [23]. Through structural and functional analysis of individual viral genes, recent studies have demonstrated that KSHV, like many other herpesviruses, expresses its genes in a sophisticated way. This includes using both the viral DNA strands for gene expression, having two or three different genes share one promoter or one single polyadenylation (pA) signal, and having the resulting polycistronic pre-mRNAs (RNAs overlapping the coding regions of other RNAs) undergo extensive alternative splicing. The present review summarizes the progress being made in KSHV split-gene studies and emphasizes the regulation of split-gene expression at the post-transcriptional level.
HOW MANY GENES IN THE KSHV GENOME ARE SPLIT GENES?
A split or interrupted gene is defined as a gene consisting of introns (intervening sequences between exons) and exons (segments of an interrupted gene that are represented in the mRNA). Thus, a simple split gene has at least two exons and one intron. Removal (splicing) of the intron(s) from a primary transcript (pre-mRNA) is an essential process for the creation of an mRNA. Thus, defining the exon–intron boundary is the first step to involve the accurate recognition of a 5′ splice site (5′ ss; the junction between the 5′ end of the intron and the 3′ end of the exon) and a 3′ splice site (3′ ss; the junction between the 3′ end of the intron and the 5′ end of the exon) by cellular splicing machinery. Most viral and eukaryotic RNA introns are GU-AG introns (see Figure 4) presumably containing consensus sequences of 5′ ss GURAGU and 3′ ss YNYURAC-Py-AG (R, purine; Y, pyrimidine; N, any nucleotide; Py, polypyrimidine tract).
FIG. 4.
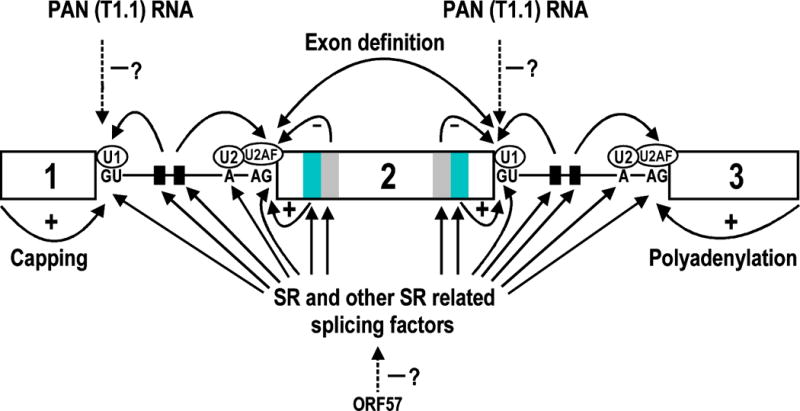
Regulation of splice site selection by cis elements and trans splicing factors and possible involvement of KSHV gene products. Diagram shows a pre-mRNA containing three exons (open boxes) and two introns (lines). Two categories of splicing regulatory elements, splicing enhancers and splicing suppressors (silencers), have been described recently in the exons (grey boxes) or introns (small black boxes) of several viral and mammalian pre-mRNA substrates (94–99). The heavy- and light-grey boxes in exon 2 are, respectively, exonic splicing enhancers (ESE) and exonic splicing suppressor (ESS) which are often juxtaposed. The small black boxes in the introns are either intronic splicing enhancers (ISE) or intronic splicing suppressors (ISS). Through interacting with SR and other SR related splicing factors, these regulatory elements positively (+) or negatively (−) control the selection of a splice site upstream or downstream. An internal exon is also usually defined as less than 500 nts in size (51). RNA 5′ capping (100, 101) and 3′ polyadenylation (102, 103) couple with RNA splicing to promote recognition of terminal splice sites. Negative effects of KSHV ORF57 and PAN RNA on RNA splicing are hypothetical.
The initial assignment of a potential protein coding region in the KSHV genome was based on the initiation codon context and the predicted protein in size larger than 100 amino acids [11,12]. Although the studies were milestones in KSHV research, their difficulties in taking account of gene discontinuity based on the inital KSHV genome sequence constituted a weak point in their ORF assignments and underestimated the potential complexity of the gene structure. Experimental investigation of the assigned ORFs has identified up to 25 genes in the KSHV genome as split genes (Table 1). The number of split genes reported so far is more than one-fourth of the total numbers of KSHV genes described, and includes the genes involved in all phases of KSHV infection (latent, immediate early, early, and late). More KSHV split genes are expected to be found as more careful studies are completed. Among the KSHV split genes, the most complex is K15, which has eight exons and seven introns [24,25].
Table 1.
Split genes in the KSHV genomea
| Name | cell homolog | Startb | Stopb | # of exons | # of introns | Messagec | Function | References |
|---|---|---|---|---|---|---|---|---|
| ORF4 | RCA | 1142 | 2794 | 2 | 1 | M | Complement regulator | 85 |
| ORF29 | 54676 | 49362 | 2 | 1 | M | Virion package | 86,87 | |
| ORF29B | 50417 | 49362 | 2 | 1 | B | 87 | ||
| ORF40 | 60368 | 61681 | 2 | 1 | B | Primase | 88,89 | |
| ORF41 | 61827 | 62444 | 2 | 1 | M | Primase | 88,89 | |
| ORF48 | 71381 | 70173 | 2 | 1 | B | 87 | ||
| ORF50 | 71596 | 74629 | 5 | 4 | T | Transactivator | 36,49 | |
| ORF57 | 82717 | 83544 | 2 | 1 | M | ICP27-like | 79,80,84 | |
| ORF72 | Cyclin D | 123567 | 122793 | 2 | 1 | B,T | 44,47 | |
| ORF73 | 127297 | 123808 | 2 | 1 | T | LANA 1 | 44,47 | |
| ORF74 | GPCR | 129372 | 130399 | 2 | 1 | B | vIL8 receptor | 47,59,90 |
| K3 | 19609 | 18608 | 3 | 2 | B | 91 | ||
| K5 | 26483 | 25713 | 2 | 1 | M | 48 | ||
| K8 | 74850 | 75791 | 4 | 3 | B | K-bZIP | 37,38,49 | |
| K8.1 | 75915 | 76695 | 2 | 1 | M | Glycoprotein | 38,52,53 | |
| K10 | IRF | 88478 | 86074 | 3 | 2 | M | 14 | |
| K10.1 | IRF | 88910 | 86074 | 2 | 1 | B | 14 | |
| K10.5 | IRF | 91394 | 89600 | 2 | 1 | M | LANA 2 | 15,92 |
| K10.7 | IRF | 91394 | 90936 | 2 | 1 | B | 14 | |
| K11 | IRF | 93367 | 91964 | 2 | 1 | B | 14 | |
| K11.1 | IRF | 94127 | 93636 | 2 | 1 | B | 14,16 | |
| K12 | 118101 | 117912 | 2 | 1 | M | Kaposin A | 93 | |
| K13 | FLIP | 122711 | 122145 | 2 | 1 | B,T | 44 | |
| K14 | OX-2 | 127884 | 128929 | 2 | 1 | B | 47,59 | |
| K15 | 136772 | 134672 | 8 | 7 | M | LMP2A | 24,25,54 |
RCA, regulator of complement activation; GPCR, G-protein-coupled receptor; IRF, interferon regulatory factor; FLIP, Fas-ligand interleukin converting enzyme (flice)-like caspase inhibitory protein; K-bZIP, K8-encoded basic-leucine zipper protein; LANA 1, latency-associated nuclear antigen 1; LANA 2, latency-associated nuclear antigen 2; LMP2A, latent membrane protein 2A; OX-2, glycoprotein CD200; vIL8 receptor, viral interleukin-8 like receptor.
Nucleotide positions of the coding region in the KSHV (BC-1) genome (GenBank accession number U75698) based on experimental data.
B, bicistronic; M, monocistronic; T, tricistronic.
It remains unclear why there are so many split genes in the KSHV genome. The virus might have been evolved to have many split genes to provide more coding capacity in a compact genome. Alternatively, a genome containing split genes might have more efficient expression [26,27]. In addition, a virus containing more split genes could be more versatile in diversifying its gene expression in response to cell activation and differentiation, which could make it a better pirate for taking over cellular machinery. It has been known for some time that lymphocyte-tropic human herpesviruses including EBV [28,29], HHV6 [30–32] and CMV [33–35], usually have more split genes than dermatotropic herpesviruses (HSV-1, HSV-2 and VZV). With more split genes in their genome, the lymphocyte-tropic viruses, which use lymphocytes as their latent sites, put their gene expression under the regulation by lymphocyte activation and differentiation. KSHV is a B lymphocyte-tropic virus and expression of its genes in B cells resembles that of other lymphocyte-tropic herpesviruses, especially EBV.
EXPRESSION OF KSHV SPLIT GENES BY ALTERNATIVE TRANSCRIPTION INITIATION AND ALTERNATIVE POLYADENYLATION
The molecular events involved in the expression of KSHV split genes are just beginning to be understood. Considerable insight has been gained from studies of similar issues in the related virus EBV. The complexity of split gene expression depends upon the availability of a proximal promoter and pA signal, and upon how each gene is organized in the genome. In general, each gene has a promoter upstream and a pA signal downstream of its ORF. In KSHV, however, not every ORF has an available promoter or pA site immediately upstream or downstream. Expression of these ORFs requires the use of another gene’s promoter or pA site far upstream or downstream. The resulting transcription initiation or RNA polyadenylation often produces a bicistronic or tricistronic RNA.
Expression of ORF 50, K8 and K8.1 in lytic infection involves alternative transcription initiation (Figure 1). The KSHV ORF50 (RTA), K8 (K-bZIP) and K8.1 locus encompasses about 5400 nts and is located at map position 0.43–0.46 of the KSHV genome. Although the three genes are positioned in the virus genome side by side, they belong to three different categories of genes, immediate early (ORF50), early (K8) and late (K8.1), and thus are expressed at different stages of the viral lytic cycle by using individual promoters for each gene: promoter P71560 for ORF 50 [36], promoter P74845 for K8 [37] and promoter P75901 for K8.1 [38]. ORF50 (110 kDa) [36] functions as a transactivator of lytic induction, K8α (38 kDa) [39] is a bZIP protein that might relate to viral DNA replication, and K8.1A (35–37 kDa) is a viral envelope glycoprotein responsible for cell attachment [40,41]. Interestingly, the ORF50, K8 and K8.1 transcripts all share a single pA signal at nt 76 714 for their RNA processing (Figure 1) [36–38,42]. Thus, expression of the ORF50 and K8 produces, respectively, a tricistronic and a bicistronic pre-mRNA.
FIG. 1.
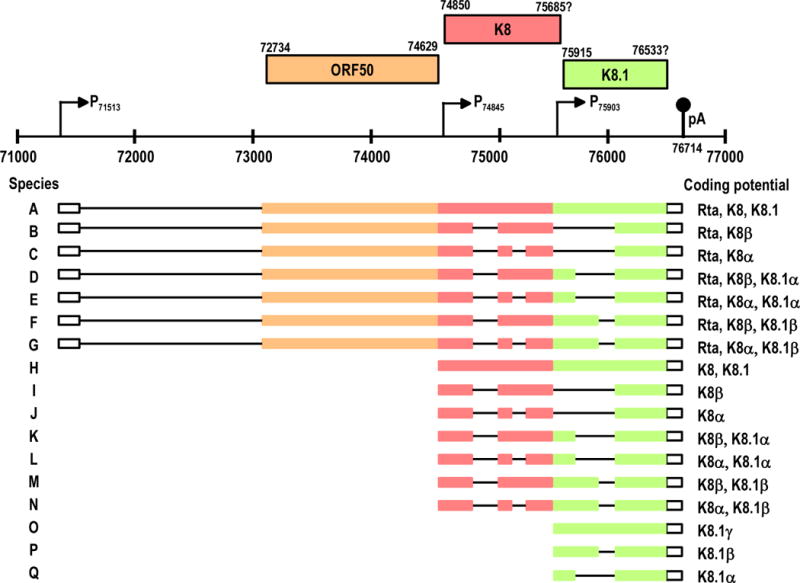
The ORF50 (immediately early), K8 (early), and K8.1 (late) transcripts are initiated from alternative promoters but share a common pA site at nt 76714, and produce an array of spliced and unspliced transcripts (36–38, 49). Because of alternative splicing, a total of approximately 19 RNA species (A to S) are predicted from three pre-mRNAs: tricistronic ORF50/K8/K8.1, bicistronic K8/K8.1, and monocistronic K8.1. Splicing of these pre-mRNAs often results in inclusion of the intron at nt 75563 to nt 75645 (RNAs C, E, G, K, M and O) due to suboptimal features of the splice sites. Among those transcripts, RNA D encodes for Rta, RNA L encodes for K-bZip and RNA R encodes for K8.1A in lytic infection.
Expression of ORF 73/72/K13 in viral latent phase involves alternative RNA splicing and alternative polyadenylation [43–47]. Three ORFs initiate their transcription from the opposite strand at the same promoter P127880 and produce three transcript isoforms of 5.4, 3.3 and 1.7 kb in size, derived from alternative RNA splicing and alternative polyadenylation. Due to the nature of sharing a single promoter for their transcription, all three RNAs have the same leader exon (Figure 2). The leader exon alternatively splices to a coding exon at nt 127313 30 ss or at nt 123773 3′ ss, producing the 5.4 (from usage of the nt 127313 3′ ss) or the 1.7 kb transcript (from usage of the nt 123773 3′ss). The mechanism by which the alternative splice sites are chosen is unknown. Both transcripts share the same pA signal at nt 122094 [43–45] and co-exist during the latent phase of KSHV infection. A minor form of the 3.3 kb transcript that uses the nt 127313 3′ ss is derived from alternative usage of a non-canonical pA signal (AGUAAA) at nt 124061 (Figure 2) (Canham M and Talbot SJ, personal communication). How this non-canonical pA signal is selected in latent KSHV infection remains to be investigated.
FIG. 2.
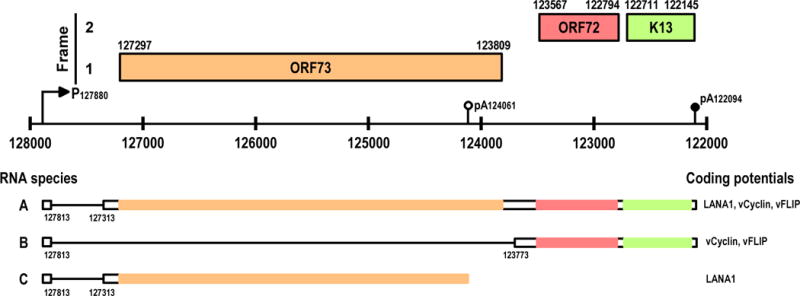
Alternative splicing and polyadenylation of ORF 73/72/K13 transcripts initiated from a single promoter, P127880 (44, 46). The coding direction of these genes is backward because they are transcribed from the opposite strand. The presence of an alternative 3′ ss in exon 2 and an alternative non-canonical pA signal (AGUAAA) at nt 124061 leads to the production of a 1.7 kb bicistronic RNA (RNA B, alternative 3′ ss selection) and a 3.3 kb monocistronic RNA (RNA C, alternative polyadenylation) (Canham, M & Talbot, SJ., personal communication). Tricistronic RNA A (ORF73/72/K13) is mainly expressed in lymph node cells and bicistronic RNA B (ORF72/K13) is most common in late-stage KS lesions (70% spindle cells) (58).
Due to the nature of such gene structures, regulation of both transcription and RNA processing are common scenarios for expression of these genes and such regulation is closely related to activation and differentiation of the host cells. As discussed above, not all exons in a split KSHV gene have coding potential. Some are leader exons preceding a coding exon, as can be found in the ORF 73/72/K13 [43–46] and K5 transcripts [48]. Others are terminal exons that make the transcripts accessible to a proximal pA signal for RNA processing. The latter can be found in the transcripts of ORF50 and K8 [38,49].
EXPRESSION OF KSHV SPLIT GENES BY ALTERNATIVE RNA SPLICING
In addition to alternative promoter and pA site usage, alternative RNA splicing is another common way to control expression of KSHV split genes. Alternative RNA splicing in the expression of KSHV split genes occurs in all phases of the virus life cycle, as was seen with ORF50, K8 and K8.1 transcripts. Alternative splicing of those transcripts may result in at least 19 species of spliced products (Figure 1). As discussed above, the KSHV ORF 50, K8 and K8.1 genes all share a common pA signal at nt 76714 downstream of the K8.1 coding region, but each gene has its own promoter (Figure 1) [36–39]. As a result, a transcribed ORF 50 pre-mRNA transverses the K8 and K8.1 coding regions and is tricistronic with five exons and four introns (RNA D), whereas a K8 pre-mRNA transcript overlaps the K8.1 coding region and thus is bicistronic with four exons and three introns (RNA L) (Figure 1). Interestingly, intron 3 of the ORF50 transcript (RNA D) or intron 2 of the K8 transcripts (RNA L) could be either spliced out or retained as part of exon 3 of ORF 50 (RNA C) or exon 2 of K8 (RNA K). Exclusion of this intron produces an α form of the mRNAs and inclusion of the intron creates a β form. However, the α forms of the K8 mRNAs are dominant in chemical-activated KSHV-positive cells and encode a large amount of the K-bZIP protein. Recent studies on splicing of the K8 pre-mRNAs showed that the β form of the message might be the precursor of the α message, since splicing of intron 2 of K8 requires removal of intron 3 [38]. In other words, selection of the nt 75838 5′ ss activates splicing of the intron immediately upstream. More importantly, exon 3 of the K8 has three 5′ ss, at nt 75838, nt 76155 and nt 76338. Selection of the nt 75838 5′ ss dictates production of K8 α mRNAs and overwhelms the K8 pre-mRNA processing. However, alternative selection of the other two 5′ ss downstream of the nt 75838 5′ ss is feasible and leads to production of two additional bicistronic mRNAs: K8/K8.1α and β. The selection of these alternative 5′ ss in combination with the inclusion or exclusion of intron 2 results in the production of eight different K8 RNA species. Since all primary ORF50 transcripts transverse the K8 region, theoretically, at least eight other species of ORF50 RNA might exist. However, exon 2 of the ORF50 pre-mRNA has a size of 2751 nts. As an internal exon, it is defined by cross-talk of a 3′ ss and a 5′ ss over the exon [50]. According to current exon definition, an oversized internal exon (>500 nts) will be difficult for the cellular splicing machinery to recognize [51].
KSHV K8.1 transcription does not initiate from its promoter, P75901, until the late stage of the virus life cycle. The transcribed pre-mRNA shares its 3′ terminal exon with ORF50 and K8, but uses a 3′ terminal intron of ORF50 and K8 as its own exon 1. Compared with the ORF50 and K8 transcripts, the K8.1 pre-mRNA is much simpler and has only two exons and one intron. However, its exon 1 has two 5′ ss positioned at nt 76115 and nt 76338 that can be alternatively selected for the pre-mRNA splicing (Figure 1). Selection of two alternative 5′ ss produces two spliced mRNAs: K8.1α and K8.1β Interestingly, data published from different laboratories [52,53] show that the nt 76338 5′ ss is preferentially selected for production of the K8.1β RNA. Why the K8.1 pre-mRNAs prefer the 3′ ss-proximal nt 76338 5′ ss over the distal nt 76155 5′ss remains unknown.
The KSHV K15 gene (either P [prototype] or M [minor] type) is a split gene containing eight exons and seven introns and is located at the right end of the KSHV genome (Figure 3). The K15 RNA structure and the predicted K15 protein sequence resemble those of EBV LMP2A [24,25,54] and LMP1 [25]. The finding that K15 inhibits B-cell receptor-mediated signalling [54] and interacts with TNF receptor-associated factors (TRAFs) [25] as well as with HS1 associated protein X-1 (HAX-1) [55] suggests that K15 plays a role in maintaining viral latency in vivo and possibly has some growth promotion potential. The full-length K15 cDNA with eight exons encodes a protein of approximately 50 kDa in an in vitro transcription and translation assay [55], but the 50 kDa K15 protein is usually cleaved as 35- and 23-kDa forms in transiently transfected cells, and predominantly as the 23 kDa form in PEL cells, including BC3 and JSC-1 cells [55]. Expression of the K15 gene in latent and activated KSHV-positive PEL cells features extensive alternative RNA splicing and thus greatly increased mRNA diversity. There are at least 14 different spliced forms of the K15 RNAs that have been discovered in PEL cells [24,25,54]. Studies in several laboratories have suggested that these RNA isoforms are created by two major splicing mechanisms: alternative 5′ ss usage and intron/exon skipping (Figure 3). However, there is not much known about how this alternative splicing is controlled, nor the coding capacity of each spliced product. The full-length K15 RNA transcript has eight exons with sizes of 7–7.5 and 10 kb, but the spliced isoforms have fewer exons (usually missing 50-half of the K15 exons) with a variable size of 3, 4–4.5 and 5.5 kb. These isoforms are usually weakly expressed in latently infected PEL cells, but are inducible by TPA [24,25,54]. However, a probe comprising the 5′-half of K15 might not detect the spliced isoforms in response to chemical treatment [24,25,54] because of alternative splicing in this region. Nevertheless, the TPA increased expression of K15 RNAs [24,25,54] does not seem to correlate with expression of a K15-specific 23 kDa protein in PEL cells [55].
FIG. 3.
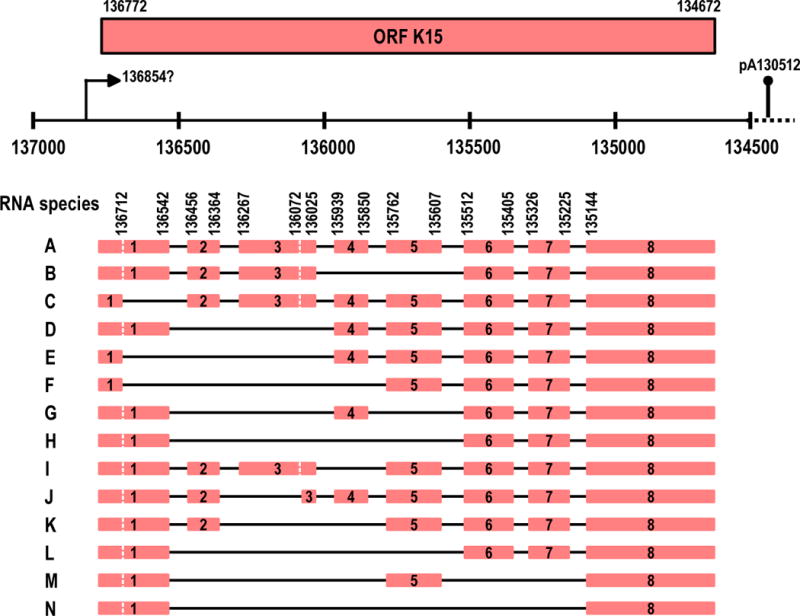
Extensive exon and intron skipping and alternative splicing of K15 pre-mRNA (24,25). A dashed vertical line on exon 1 and 3 of each RNA species represents a possible alternative 5′ ss. The depiction also shows a potential K15 promoter P136854 (TATATAA) and a putative K15 pA site at nt 130512 downstream of ORF75 (43). K15 is latently expressed in KSHV-positive PEL cell lines and plasmablasts in MCD (55).
MECHANISMS POSSIBLY INVOLVED IN THE REGULATION OF KSHV RNA SPLICING
Pre-mRNA splicing involves five small U RNAs (U1, U2, U4, U5 and U6) and many cellular splicing factors, including 10 SR proteins (serine/arginine-rich proteins, such as ASF/SF2, SC35, SRp40) and 12 SR-related proteins (such as U1-70K, U2AF65, U2AF35). Recognition of a 5′ ss by U1 and a 3′ ss branch point by U2 requires an interconnection of many of those splicing factors [56,57]. Depending on the features of the splice sites and presence or absence of cis-acting elements in the pre-mRNAs, the recruitment of additional trans-splicing factors to the splice sites may strengthen the recognition (Figure 4). The presence of an alternative 5′ ss or 3′ ss in a pre-mRNA complicates the recognition and the switch to alternative splice sites usually correlates with cell differentiation and stages of the virus life cycle. Although the expression and phosphorylation status of SR proteins also correspond with tissue development and cell differentiation, the mechanisms that regulate RNA alternative splicing remain largely unknown.
Expression of KSHV ORF73/72/K13 tricistronic and bicistronic RNAs appears to be KS stage- and cell-specific. As discussed previously, KSHV ORFs 73, 72 and K13 are transcribed as a tricistron from the same promoter, P127880 in KSHV-infected cells. Since this tricistron has one 5′ ss, but two alternative 3′ ss (Figure 2), splicing of an intron of 499 nts from the tricistron leads to the selection of a 5′ ss proximal 3′ ss and results in production of the spliced tricistronic transcript (ORF73/72/K13) predominately responsible for LANA 1 translation [47,58,59]. However, the majority of the tricistrons splice over to the distal 3′ ss by removing a 4 kb intron, and consequently produce the bicistronic transcript (ORF72/K13) responsible for v-cyclin and v-FLIP translation. Translation of v-FLIP is carried out on this bicistronic RNA mainly via an internal ribosome entry site residing in the v-cyclin coding region [45,60,61]. Selection of the distal over the proximal 3′ ss appears to occur at a constant ratio, but can be enhanced by chemical stimulation of latently infected B cells [47,62]. Interestingly, the transcripts from the ORF 73/72/K13 locus are spliced differentially in KS lesions at different stages of development. The distal 3′ ss are preferentially selected in late-stage nodular KS lesions, but not in early lesions. Thus, the bicistronic RNAs are highly expressed and dominate in almost 70% of spindle cells. In contrast, tricistronic ORF73/72/K13 is expressed abundantly in KSHV-infected lymph node cells [58]. The observations in KS lesions of the choice of a distal over a proximal 3′ ss or vice versa strongly imply that the stage- or cell differentiation-related splicing events are the consequence of the regulation by viral cis elements or cellular splicing factors. It will be highly important to identify and characterize the viral cis elements and cellular splicing factors involved in the alternative splicing of KSHV RNAs.
Two KSHV ORFs, ORF 57 and PAN (T1.1 or nut-1), might be involved in the regulation of RNA splicing (Figure 4), including KSHV, but direct evidence is lacking. PAN RNA is a polyadenylated viral early gene transcript having no coding function [63–65]. It is abundant in nuclei of KSHV-infected B cells, has persistent expression into the late viral cycle, and bears stretches of U1 [63] and a region complementary to U12 RNAs [64,65], implying that it may modulate the recognition of splice sites in spliceosome-mediated RNA splicing (Figure 4). KSHV ORF57 is a split gene (Table 1), and during lytic replication encodes a nuclear-protein that is a homologue of HSV ICP27 [66,67], HVS ORF57 [68–70], EBV SM protein [71,72], human CMV UL69 [73] and VZV ORF4 [74]. Among these, ICP27 is a prototype protein that inhibits RNA splicing [75–77] and mediates viral intronless RNA export [78]. Similar to its homologues, KSHV ORF57 has been demonstrated to be a posttranscriptional regulator that increases the accumulation of target mRNAs, particularly in the cytoplasm, but that has no global inhibitory effects on the expression of intron-containing genes [79,80]. Although HSV ICP27 [78], EBV SM [81,82] and HVS ORF57 [69,83] all bind viral RNAs and mediate viral RNA export (possibly via the CRM-1 export pathway), no cytoplasmic translocation of KSHV ORF57 has been observed in transiently transfected cells [79,80,84]. This suggests that the interaction of KSHV ORF57 with cellular export factors differs from that of its homologues in other herpesviruses. Nevertheless, KSHV ORF57 co-localizes with the cellular splicing factor SC35 [84]. The finding that HVS ORF57, like HSV ICP27[67], is capable of redistributing the cellular SR proteins SC35 and snRNP U2 [70], active components of the spliceosome, suggests that its homologue KSHV ORF57 may also potentially function in RNA splicing.
Acknowledgments
The author thanks Simon J. Talbot at the University of Edinburgh for sharing unpublished data for the review.
References
- 1.Chang Y, Cesarman E, Pessin MS, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Boshoff C, Weiss RA. Kaposi’s sarcoma-associated herpesvirus. Adv Cancer Res. 1998;75:57–86. doi: 10.1016/s0065-230x(08)60739-3. [DOI] [PubMed] [Google Scholar]
- 3.Cesarman E, Chang Y, Moore PS, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 4.Soulier J, Grollet L, Oksenhendler E, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 5.Beksac M, Ma M, Akyerli C, et al. Frequent demonstration of human herpesvirus 8 (HHV-8) in bone marrow biopsy samples from Turkish patients with multiple myeloma (MM) Leukemia. 2001;15:1268–1273. doi: 10.1038/sj.leu.2402190. [DOI] [PubMed] [Google Scholar]
- 6.Hsu HC, Lee YM, Yang CF, et al. Detection of Kaposi sarcoma-associated herpesvirus in bone marrow biopsy samples from patients with multiple myeloma. Cancer. 2001;91:1409–1413. doi: 10.1002/1097-0142(20010415)91:8<1409::aid-cncr1146>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 7.Tedeschi R, Kvarnung M, Knekt P, et al. A prospective seroepidemiological study of human herpesvirus-8 infection and the risk of multiple myeloma. Br J Cancer. 2001;84:122–125. doi: 10.1054/bjoc.2000.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cannon JS, Ciufo D, Hawkins AL, et al. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi’s sarcoma herpesvirus-containing supernatant. J Virol. 2000;74:10187–10193. doi: 10.1128/jvi.74.21.10187-10193.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciufo DM, Cannon JS, Poole LJ, et al. Spindle cell conversion by Kaposi’s sarcoma-associated herpesvirus: Formation of colonies and plaques with mixed lytic and latent gene expression in infected primary dermal microvascular endothelial cell cultures. J Virol. 2001;75:5614–5626. doi: 10.1128/JVI.75.12.5614-5626.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lagunoff M, Bechtel J, Venetsanakos E, et al. De novo infection and serial transmission of Kaposi’s sarcoma-associated herpesvirus in cultured endothelial cells. J Virol. 2002;76:2440–2448. doi: 10.1128/jvi.76.5.2440-2448.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russo JJ, Bohenzky RA, Chien MC, et al. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neipel F, Albrecht JC, Fleckenstein B. Cell homologous genes in the Kaposi’s sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Virgin HW, Latreille P, Wamsley P, et al. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenner RG, Alba MM, Boshoff C, et al. Kaposi’s sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J Virol. 2001;75:891–902. doi: 10.1128/JVI.75.2.891-902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rivas C, Thlick AE, Parravicini C, et al. Kaposi’s sarcoma-associated herpesvirus LANA2 is a B-cells pecific latent viral protein that inhibits p53. J Virol. 2001;75:429–438. doi: 10.1128/JVI.75.1.429-438.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burysek L, Yeow WS, Pitha PM. Unique properties of a second human herpesvirus 8-encoded interferon regulatory factor (vIRF-2) J Hum Virol. 1999;2:19–32. [PubMed] [Google Scholar]
- 17.Sarid R, Sato T, Bohenzky RA, et al. Kaposi’s sarcoma-associated herpesvirus encodes a functional bcl-2 homologue. Nat Med. 1997;3:293–298. doi: 10.1038/nm0397-293. [DOI] [PubMed] [Google Scholar]
- 18.Cheng EH, Nicholas J, Bellows DS, et al. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc Natl Acad Sci USA. 1997;94:690–694. doi: 10.1073/pnas.94.2.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang Y, Moore PS, Talbot SJ, et al. Cyclin encoded by KS herpesvirus. Nature. 1996;382:410. doi: 10.1038/382410a0. [DOI] [PubMed] [Google Scholar]
- 20.Cesarman E, Nador RG, Bai F, et al. Kaposi’s sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi’s sarcoma and malignant lymphoma. J Virol. 1996;70:8218–8223. doi: 10.1128/jvi.70.11.8218-8223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholas J, Ruvolo VR, Burns WH, et al. Kaposi’s sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat Med. 1997;3:287–292. doi: 10.1038/nm0397-287. [DOI] [PubMed] [Google Scholar]
- 22.Zimring JC, Goodbourn S, Offermann MK. Human herpesvirus 8 encodes an interferon regulatory factor (IRF) homolog that represses IRF-1-mediated transcription. J Virol. 1998;72:701–707. doi: 10.1128/jvi.72.1.701-707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore PS, Chang Y. Molecular virology of Kaposi’s sarcoma-associated herpesvirus. Philos Trans R Soc Lond B Biol Sci. 2001;356:499–516. doi: 10.1098/rstb.2000.0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poole LJ, Zong JC, Ciufo DM, et al. Comparison of genetic variability at multiple loci across the genomes of the major subtypes of Kaposi’s sarcoma- associated herpesvirus reveals evidence for recombination and for two distinct types of open reading frame K15 alleles at the right-hand end. J Virol. 1999;73:6646–6660. doi: 10.1128/jvi.73.8.6646-6660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glenn M, Rainbow L, Aurad F, et al. Identification of a spliced gene from Kaposi’s sarcoma-associated herpesvirus encoding a protein with similarities to latent membrane proteins 1 and 2A of Epstein-Barr virus. J Virol. 1999;73:6953–6963. doi: 10.1128/jvi.73.8.6953-6963.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brinster RL, Allen JM, Behringer RR, et al. Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci USA. 1988;85:836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitelaw CB, Archibald AL, Harris S, et al. Targeting expression to the mammary gland: intronic sequences can enhance the efficiency of gene expression in transgenic mice. Transgenic Res. 1991;1:3–13. doi: 10.1007/BF02512991. [DOI] [PubMed] [Google Scholar]
- 28.Rogers RP, Woisetschlaeger M, Speck SH. Alternative splicing dictates translational start in Epstein-Barr virus transcripts. EMBO J. 1990;9:2273–2277. doi: 10.1002/j.1460-2075.1990.tb07398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lau R, Packham G, Farrell PJ. Differential splicing of Epstein-Barr virus immediate-early RNA. J Virol. 1992;66:6233–6236. doi: 10.1128/jvi.66.10.6233-6236.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfeiffer B, Thomson B, Chandran B. Identification and characterization of a cDNA derived from multiple splicing that encodes envelope glycoprotein gp105 of human herpesvirus 6. J Virol. 1995;69:3490–3500. doi: 10.1128/jvi.69.6.3490-3500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirandola P, Menegazzi P, Merighi S, et al. Temporal mapping of transcripts in herpesvirus 6 variants. J Virol. 1998;72:3837–3844. doi: 10.1128/jvi.72.5.3837-3844.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flebbe-Rehwaldt LM, Wood C, Chandran B. Characterization of transcripts expressed from human herpesvirus 6A strain GS immediate-early region B U16-U17 open reading frames. J Virol. 2000;74:11040–11054. doi: 10.1128/jvi.74.23.11040-11054.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kerry JA, Sehgal A, Barlow SW, et al. Isolation and characterization of a low-abundance splice variant from the human cytomegalovirus major immediate-early gene region. J Virol. 1995;69:3868–3872. doi: 10.1128/jvi.69.6.3868-3872.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stenberg RM, Witte PR, Stinski MF. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J Virol. 1985;56:665–675. doi: 10.1128/jvi.56.3.665-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rawlinson WD, Barrell BG. Spliced transcripts of human cytomegalovirus. J Virol. 1993;67:5502–5513. doi: 10.1128/jvi.67.9.5502-5513.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukac DM, Kirshner JR, Ganem D. Transcriptional activation by the product of open reading frame 50 of Kaposi’s sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J Virol. 1999;73:9348–9361. doi: 10.1128/jvi.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin SF, Robinson DR, Miller G, et al. Kaposi’s sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J Virol. 1999;73:1909–1917. doi: 10.1128/jvi.73.3.1909-1917.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang S, Zheng ZM. Kaposi’s sarcoma-associated herpesvirus K8 exon 3 contains three 5′-splice sites and harbors a K8.1 transcription start site. J Biol Chem. 2002;277:14547–14556. doi: 10.1074/jbc.M111308200. [DOI] [PubMed] [Google Scholar]
- 39.Polson AG, Huang L, Lukac DM, et al. Kaposi’s sarcoma-associated herpesvirus K-bZIP protein is phosphorylated by cyclin-dependent kinases. J Virol. 2001;75:3175–3184. doi: 10.1128/JVI.75.7.3175-3184.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang FZ, Akula SM, Pramod NP, et al. Human herpesvirus 8 envelope glycoprotein K8.1A interaction with the target cells involves heparan sulfate. J Virol. 2001;75:7517–7527. doi: 10.1128/JVI.75.16.7517-7527.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Birkmann A, Mahr K, Ensser A, et al. Cell surface heparan sulfate is a receptor for human herpesvirus 8 and interacts with envelope glycoprotein K8.1. J Virol. 2001;75:11583–11593. doi: 10.1128/JVI.75.23.11583-11593.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun R, Lin SF, Gradoville L, et al. A viral gene that activates lytic cycle expression of Kaposi’s sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rainbow L, Platt GM, Simpson GR, et al. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarid R, Wiezorek JS, Moore PS, et al. Characterization and cell cycle regulation of the major Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J Virol. 1999;73:1438–1446. doi: 10.1128/jvi.73.2.1438-1446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grundhoff A, Ganem D. Mechanisms governing expression of the v-FLIP gene of Kaposi’s sarcoma associated herpesvirus. J Virol. 2001;75:1857–1863. doi: 10.1128/JVI.75.4.1857-1863.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeong J, Papin J, Dittmer D. Differential regulation of the overlapping Kaposi’s sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J Virol. 2001;75:1798–1807. doi: 10.1128/JVI.75.4.1798-1807.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Talbot SJ, Weiss RA, Kellam P, et al. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology. 1999;257:84–94. doi: 10.1006/viro.1999.9672. [DOI] [PubMed] [Google Scholar]
- 48.Haque M, Chen J, Ueda K, et al. Identification and analysis of the K5 gene of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2000;74:2867–2875. doi: 10.1128/jvi.74.6.2867-2875.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu FX, Cusano T, Yuan Y. Identification of the immediate-early transcripts of Kaposi’s sarcoma-associated herpesvirus. J Virol. 1999;73:5556–5567. doi: 10.1128/jvi.73.7.5556-5567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berget SM. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 51.Sterner DA, Carlo T, Berget SM. Architectural limits on split genes. Proc Natl Acad Sci USA. 1996;93:15081–15085. doi: 10.1073/pnas.93.26.15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raab MS, Albrecht JC, Birkmann A, et al. The immunogenic glycoprotein gp35-37 of human herpesvirus 8 is encoded by open reading frame K8.1. J Virol. 1998;72:6725–6731. doi: 10.1128/jvi.72.8.6725-6731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chandran B, Bloomer C, Chan SR, et al. Human herpesvirus-8 ORF K8.1 gene encodes immunogenic glycoproteins generated by spliced transcripts. Virology. 1998;249:140–149. doi: 10.1006/viro.1998.9316. [DOI] [PubMed] [Google Scholar]
- 54.Choi JK, Lee BS, Shim SN, et al. Identification of the novel K15 gene at the rightmost end of the Kaposi’s sarcoma-associated herpesvirus genome. J Virol. 2000;74:436–446. [PMC free article] [PubMed] [Google Scholar]
- 55.Sharp TV, Wang HW, Koumi A, et al. K15 protein of Kaposi’s sarcoma-associated herpesvirus is latently expressed and binds to HAX-1, a protein with antiapoptotic function. J Virol. 2002;76:802–816. doi: 10.1128/JVI.76.2.802-816.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharp PA. Split genes and RNA splicing. Cell. 1994;77:805–816. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 57.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sturzl M, Hohenadl C, Zietz C, et al. Expression of K13/v-FLIP gene of human herpesvirus 8 and apoptosis in Kaposi’s sarcoma spindle cells. J Natl Cancer Inst. 1999;91:1725–1733. doi: 10.1093/jnci/91.20.1725. [DOI] [PubMed] [Google Scholar]
- 59.Kirshner JR, Staskus K, Haase A, et al. Expression of the open reading frame 74 (G-protein-coupled receptor) gene of Kaposi’s sarcoma (KS)-associated herpesvirus: implications for KS pathogenesis. J Virol. 1999;73:6006–6014. doi: 10.1128/jvi.73.7.6006-6014.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bieleski L, Talbot SJ. Kaposi’s sarcoma-associated herpesvirus vCyclin open reading frame contains an internal ribosome entry site. J Virol. 2001;75:1864–1869. doi: 10.1128/JVI.75.4.1864-1869.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Low W, Harries M, Ye H, et al. Internal ribosome entry site regulates translation of Kaposi’s sarcoma-associated herpesvirus FLICE inhibitory protein. J Virol. 2001;75:2938–2945. doi: 10.1128/JVI.75.6.2938-2945.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dittmer D, Lagunoff M, Renne R, et al. A cluster of latently expressed genes in Kaposi’s sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun R, Lin SF, Gradoville L, et al. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1996;93:11883–11888. doi: 10.1073/pnas.93.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhong W, Ganem D. Characterization of ribonucleoprotein complexes containing an abundant polyadenylated nuclear RNA encoded by Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) J Virol. 1997;71:1207–1212. doi: 10.1128/jvi.71.2.1207-1212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhong W, Wang H, Herndier B, et al. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sandri-Goldin RM, Hibbard MK. The herpes simplex virus type 1 regulatory protein ICP27 coimmunoprecipitates with anti-Sm antiserum, and the C terminus appears to be required for this interaction. J Virol. 1996;70:108–118. doi: 10.1128/jvi.70.1.108-118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sandri-Goldin RM, Hibbard MK, Hardwicke MA. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J Virol. 1995;69:6063–6076. doi: 10.1128/jvi.69.10.6063-6076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whitehouse A, Cooper M, Meredith DM. The immediate-early gene product encoded by open reading frame 57 of herpesvirus saimiri modulates gene expression at a posttranscriptional level. J Virol. 1998;72:857–861. doi: 10.1128/jvi.72.1.857-861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goodwin DJ, Hall KT, Stevenson AJ, et al. The open reading frame 57 gene product of herpesvirus saimiri shuttles between the nucleus and cytoplasm and is involved in viral RNA nuclear export. J Virol. 1999;73:10519–10524. doi: 10.1128/jvi.73.12.10519-10524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cooper M, Goodwin DJ, Hall KT, et al. The gene product encoded by ORF 57 of herpesvirus saimiri regulates the redistribution of the splicing factor SC-35. J Gen Virol. 1999;80:1311–1316. doi: 10.1099/0022-1317-80-5-1311. [DOI] [PubMed] [Google Scholar]
- 71.Cook ID, Shanahan F, Farrell PJ. Epstein-Barr virus SM protein. Virology. 1994;205:217–227. doi: 10.1006/viro.1994.1637. [DOI] [PubMed] [Google Scholar]
- 72.Ruvolo V, Wang E, Boyle S, et al. The Epstein-Barr virus nuclear protein SM is both a post-transcriptional inhibitor and activator of gene expression. Proc Natl Acad Sci USA. 1998;95:8852–8857. doi: 10.1073/pnas.95.15.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Winkler M, Rice SA, Stamminger T. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J Virol. 1994;68:3943–3954. doi: 10.1128/jvi.68.6.3943-3954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perera LP, Kaushal S, Kinchington PR, et al. Varicella-zoster virus open reading frame 4 encodes a transcriptional activator that is functionally distinct from that of herpes simplex virus homology ICP27. J Virol. 1994;68:2468–2477. doi: 10.1128/jvi.68.4.2468-2477.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hardy WR, Sandri-Goldin RM. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J Virol. 1994;68:7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bryant HE, Wadd SE, Lamond AI, et al. Herpes simplex virus IE63 (ICP27) protein interacts with spliceosome-associated protein 145 and inhibits splicing prior to the first catalytic step. J Virol. 2001;75:4376–4385. doi: 10.1128/JVI.75.9.4376-4385.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lindberg A, Kreivi JP. Splicing inhibition at the level of spliceosome assembly in the presence of herpes simplex virus protein ICP27. Virology. 2002;294:189–198. doi: 10.1006/viro.2001.1301. [DOI] [PubMed] [Google Scholar]
- 78.Sandri-Goldin RM. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 1998;12:868–879. doi: 10.1101/gad.12.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gupta AK, Ruvolo V, Patterson C, et al. The human herpesvirus 8 homolog of Epstein-Barr virus SM protein (KS-SM) is a posttranscriptional activator of gene expression. J Virol. 2000;74:1038–1044. doi: 10.1128/jvi.74.2.1038-1044.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kirshner JR, Lukac DM, Chang J, et al. Kaposi’s sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J Virol. 2000;74:3586–3597. doi: 10.1128/jvi.74.8.3586-3597.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Semmes OJ, Chen L, Sarisky RT, et al. Mta has properties of an RNA export protein and increases cytoplasmic accumulation of Epstein-Barr virus replication gene mRNA. J Virol. 1998;72:9526–9534. doi: 10.1128/jvi.72.12.9526-9534.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boyle SM, Ruvolo V, Gupta AK, et al. Association with the cellular export receptor CRM 1 mediates function and intracellular localization of Epstein-Barr virus SM protein, a regulator of gene expression. J Virol. 1999;73:6872–6881. doi: 10.1128/jvi.73.8.6872-6881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goodwin DJ, Whitehouse A. A gamma-2 herpesvirus nucleocytoplasmic shuttle protein interacts with importin alpha 1 and alpha 5. J Biol Chem. 2001;276:19905–19912. doi: 10.1074/jbc.M009513200. [DOI] [PubMed] [Google Scholar]
- 84.Bello LJ, Davison AJ, Glenn MA, et al. The human herpesvirus-8 ORF 57 gene and its properties. J Gen Virol. 1999;80:3207–3215. doi: 10.1099/0022-1317-80-12-3207. [DOI] [PubMed] [Google Scholar]
- 85.Spiller OB, Robison M, O’Donnell E, et al. Complement regulation by Kaposi’s sarcoma-associated herpesvirus ORF4 protein. J Virol. 2003;77:592–599. doi: 10.1128/JVI.77.1.592-599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Renne R, Blackbourn D, Whitby D, et al. Limited transmission of Kaposi’s sarcoma-associated herpesvirus in cultured cells. J Virol. 1998;72:5182–5188. doi: 10.1128/jvi.72.6.5182-5188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Saveliev AK, Zhu FX, Yuan Y. Transcription mapping and expression patterns of genes in the major immediate-early region of Kaposi’s sarcoma associated herpesvirus. Virology. 2002;299:301–314. doi: 10.1006/viro.2002.1561. [DOI] [PubMed] [Google Scholar]
- 88.Wu FY, Ahn JH, Alcendor DJ, et al. Origin-independent assembly of Kaposi’s sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J Virol. 2001;75:1487–1506. doi: 10.1128/JVI.75.3.1487-1506.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.AuCoin DP, Pari GS. The human herpesvirus-8 (Kaposi’s sarcoma-associated herpesvirus) ORF40/41 region encodes two distinct transcripts. J Gen Virol. 2002;83:189–193. doi: 10.1099/0022-1317-83-1-189. [DOI] [PubMed] [Google Scholar]
- 90.Chiou CJ, Poole LJ, Kim PS, et al. Patterns of gene expression and a transactivation function exhibited by the vGCR (ORF74) chemokine receptor protein of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2002;76:3421–3439. doi: 10.1128/JVI.76.7.3421-3439.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rimessi P, Bonaccorsi A, Sturzl M, et al. Transcription pattern of human herpesvirus 8 open reading frame K3 in primary effusion lymphoma and Kaposi’s sarcoma. J Virol. 2001;75:7161–7174. doi: 10.1128/JVI.75.15.7161-7174.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lubyova B, Pitha PM. Characterization of a novel human herpesvirus 8-encoded protein, vIRF-3, that shows homology to viral and cellular interferon regulatory factors. J Virol. 2000;74:8194–8201. doi: 10.1128/jvi.74.17.8194-8201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li H, Komatsu T, Dezube BJ, et al. The Kaposi’s sarcoma-associated herpesvirus K12 transcript from a primary effusion lymphoma contains complex repeat elements, is spliced, and initiates from a novel promoter. J Virol. 2002;76:11880–11888. doi: 10.1128/JVI.76.23.11880-11888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zheng ZM, He P, Baker CC. Selection of the bovine papillomavirus type 1 nucleotide 3225 3′ splice site is regulated through an exonic splicing enhancer and its juxtaposed exonic splicing suppressor. J Virol. 1996;70:4691–4699. doi: 10.1128/jvi.70.7.4691-4699.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zheng ZM, Reid ES, Baker CC. Utilization of the bovine papillomavirus type 1 late-stage-specific nucleotide 3605 3′ splice site is modulated by a novel exonic bipartite regulator but not by an intronic purine-rich element. J Virol. 2000;74:10612–10622. doi: 10.1128/jvi.74.22.10612-10622.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Amendt BA, Si ZH, Stoltzfus CM. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: evidence for inhibition mediated by cellular factors. Mol Cell Biol. 1995;15:4606–4615. doi: 10.1128/mcb.15.8.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Staffa A, Cochrane A. Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol Cell Biol. 1995;15:4597–4605. doi: 10.1128/mcb.15.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Staffa A, Acheson NH, Cochrane A. Novel exonic elements that modulate splicing of the human fibronectin EDA exon. J Biol Chem. 1997;272:33394–33401. doi: 10.1074/jbc.272.52.33394. [DOI] [PubMed] [Google Scholar]
- 99.Del Gatto F, Breathnach R. Exon and intron sequences, respectively, repress and activate splicing of a fibroblast growth factor receptor 2 alternative exon. Mol Cell Biol. 1995;15:4825–4834. doi: 10.1128/mcb.15.9.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Izaurralde E, Lewis J, McGuigan C, et al. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 101.O’Mullane L, Eperon IC. The pre-mRNA 5′ cap determines whether U6 small nuclear RNA succeeds U1 small nuclear ribonucleoprotein particle at 5′ splice sites. Mol Cell Biol. 1998;18:7510–7520. doi: 10.1128/mcb.18.12.7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cooke C, Alwine JC. Characterization of specific protein-RNA complexes associated with the coupling of polyadenylation and last-intron removal. Mol Cell Biol. 2002;22:4579–4586. doi: 10.1128/MCB.22.13.4579-4586.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fong N, Bentley DL. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev. 2001;15:1783–1795. doi: 10.1101/gad.889101. [DOI] [PMC free article] [PubMed] [Google Scholar]


