Summary
In response to cellular genome breaks, MRE11/RAD50/NBS1 (MRN) activates a global ATM DNA damage response (DDR) that prevents cellular replication. Here we show that MRN-ATM also has critical functions in defending the cell against DNA viruses. We reveal temporally distinct responses to adenovirus genomes: a critical MRN-ATM DDR that must be inactivated by E1B-55K/E4-ORF3 viral oncoproteins and a global MRN independent ATM DDR to viral nuclear domains that does not impact viral replication. We show that MRN binds to adenovirus genomes and activates a localized ATM response that specifically prevents viral DNA replication. In contrast to chromosomal breaks, ATM activation is not amplified by H2AX across megabases of chromatin to induce global signaling and replicative arrest. Thus, γH2AX foci discriminate ‘self’ and ‘non-self’ genomes and determine if a localized anti-viral or global ATM response is appropriate. This provides an elegant mechanism to neutralize viral genomes without jeopardizing cellular viability.
Introduction
Central to life is the faithful replication, inheritance, and maintenance of genomic DNA. The MRE11/RAD50/NBS1 (MRN) complex and ATM play a critical role in this biological mandate (Ciccia and Elledge, 2010). Cellular double strand breaks (DSBs) are sensed by MRN and trigger the assembly of DNA damage response (DDR) foci that amplify global ATM signaling to induce cell cycle arrest and DNA repair (Polo and Jackson, 2011). DNA viruses are an ancient and persistent threat to both cellular genome integrity and viability. Adenovirus has a 36 kb linear double strand DNA genome that is delivered to the cell nucleus where it is replicated concomitant with cellular DNA. Thus, the discovery of adenovirus 5 (Ad5) early proteins that target MRN excited great interest, suggesting that the cellular DDR also has an antiviral role (Figure 1A) (Stracker et al., 2002). However, despite numerous studies, the role of MRN and global cellular DDR signaling in defending against adenovirus replication has been difficult to decipher.
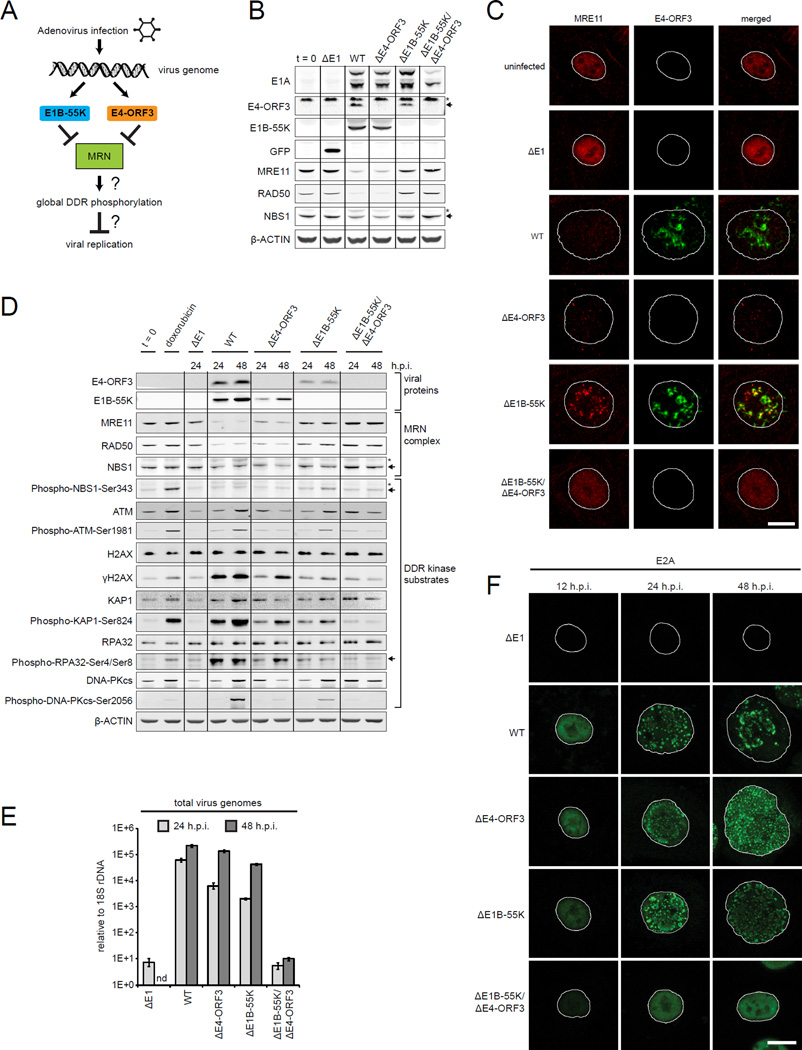
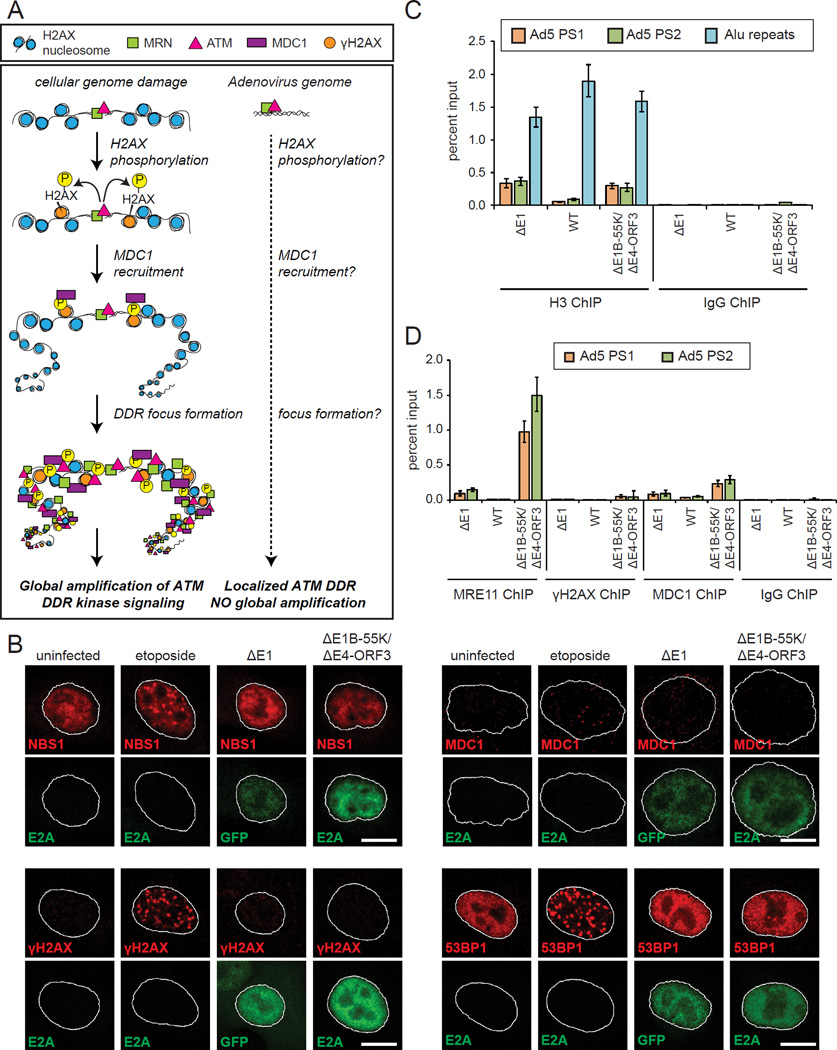
Figure 1. E1B-55K and E4-ORF3 inactivate MRN and are critical for viral genome replication but do not prevent global DDR kinase signaling.
(A) Adenovirus 5 (Ad5) early proteins target MRN through two independent mechanisms.
(B) Human small airway epithelial cells (SAECs) were infected with mock (ΔE1), wild type Ad5 (WT), ΔE4-ORF3, ΔE1B-55K or ΔE1B-55K/ΔE4-ORF3 viruses. Protein lysates were collected at 24 h.p.i. and immunoblotted as indicated. “t = 0” indicates uninfected cells. Arrows and asterisks indicate specific and non-specific bands, respectively.
(C–E) SAECs were infected as indicated. (C) SAECs were fixed at 24 h.p.i. and stained for E4-ORF3 (green) and MRE11 (red). DNA was counterstained with Hoechst. Nuclei are outlined in white. Scale bar: 10 μm. (D) Protein lysates were immunoblotted as indicated. Doxorubicin was used as a positive control for cellular DNA damage. (E) Viral genomes were quantified by Q-PCR and normalized to 18S rDNA; error bars indicate standard deviation (nd: not done).
(F) Virus genome replication domains were visualized in infected SAECs by E2A immunofluorescence (green). Scale bar: 10 µm.
(See also Figure S1.)
Cellular DSBs are first sensed by MRN that recruits and activates the apical DDR kinase, ATM, triggering ATM autophosphorylation at Ser1981 (Lee and Paull, 2004, 2005; Uziel et al., 2003). The activation of ATM at sites of cellular DSBs is globally amplified across megabases of flanking chromatin through ATM phosphorylation of H2AX at Ser139 (γH2AX) (Rogakou et al., 1999). The MDC1 scaffolding protein binds γH2AX and recruits additional MRN, ATM, DDR kinases and effectors into nuclear foci readily visualized by light microscopy (Polo and Jackson, 2011). γH2AX DDR foci play an important role in nucleating ATM and effector kinases to induce the global phosphorylation of DDR substrates, including KAP1, RPA32, 53BP1, and p53, that elicit cell cycle arrest, repair, senescence or apoptosis (Polo and Jackson, 2011; Soutoglou and Misteli, 2008). ATR and DNA-PK share some overlapping substrates with ATM, such as H2AX, but also have independent targets (Ciccia and Elledge, 2010).
The assembly of DDR foci is conserved from yeast to humans and is considered one of the most sensitive hallmarks of cellular genotoxic stress (Lisby et al., 2004). However, in contrast to MRN components (Luo et al., 1999; Xiao and Weaver, 1997; Zhu et al., 2001), H2AX- and MDC1-null mice are viable and only partially defective in DSB repair (Bassing et al., 2002; Celeste et al., 2002; Lou et al., 2006). Thus, the functional logic of modifying megabases of flanking chromatin to protect the genome against tiny breaks remains one of the most cryptic aspects of the cellular DNA damage response (Cleaver, 2011; Fernandez-Capetillo et al., 2003).
An outstanding question is if MRN activates a similar signaling response to protect the cell against viral DNA genomes and cellular DSBs. A unifying feature of viruses is that they have smaller genomes compared to their hosts. In principle, small viral DNA genomes would not support the contiguous spreading of DDR proteins across megabases of nucleosome-bound DNA to induce global DNA damage signaling and cell cycle arrest. Given the daily onslaught we face from viruses, the induction of cell cycle arrest or apoptosis in response to viral DNA may be untenable in terms of tissue homeostasis.
Ad5 encodes early viral proteins that target MRN through two independent mechanisms (Figure 1A). E1B-55K binds to MRE11 and forms a complex with E4-ORF6 that targets MRE11 for degradation in the proteasome (Querido et al., 2001; Stracker et al., 2002). E4-ORF3 assembles a multivalent polymer network in the nucleus that mislocalizes and sequesters MRN (Ou et al., 2012; Stracker et al., 2002). The prevailing model is that MRN inactivation is necessary to prevent the activation of a global cellular DDR to adenovirus genomes (Carson et al., 2003; Weitzman et al., 2010). However, other studies have shown that a global DDR is activated in Ad5 infected cells (Blackford et al., 2008; Forrester et al., 2011; Nichols et al., 2009). Thus, the role of MRN and DDR signaling in virus infection remains contentious and confounding.
Here we show that MRN binds to viral genomes and activates a localized ATM anti-viral response that prevents viral replication at the earliest stages. In contrast to cellular genome breaks, MRN-ATM activation at viral genomes is not amplified by γH2AX to induce a global DDR. This provides an elegant mechanism to selectively arrest viral DNA replication without jeopardizing cellular viability and proliferation. E1B-55K/E4-ORF3 inactivate MRN and enable logarithmic virus genome replication. The subsequent assembly of newly replicated virus genomes in nuclear domains triggers global ATM signaling through an MRN independent mechanism but does not impact viral replication.
Results
E1B-55K and E4-ORF3 inactivate MRN and are critical for viral genome replication
To determine the role of viral proteins that target MRN we infected quiescent primary human small airway epithelial cells (SAECs) with wild type Ad5 (WT) or viruses that have deletions of E1B-55K and/or E4-ORF3 (Figures 1B, 1C and S1A). ΔE1 is a non-replicating vector control for virus infection that expresses GFP instead of viral genes. Previous studies have shown that E1B-55K targets MRE11 for degradation in the proteasome (Querido et al., 2001; Stracker et al., 2002). Consistent with this, MRE11 and RAD50 protein levels are decreased in WT and ΔE4-ORF3 virus infected cells that express E1B-55K (Figure 1B). E4-ORF3 assembles a multivalent nuclear matrix that binds and mislocalizes MRN (Ou et al., 2012). Thus, even in the absence of E1B-55K induced MRN degradation, E4-ORF3 sequesters MRE11 and NBS1 in ΔE1B-55K infected cells (Figures 1C and S1A). However, ΔE1B-55K/ΔE4-ORF3 viruses do not degrade or mislocalize MRN and enable its role in viral replication to be determined (Figures 1B, 1C and S1A).
In response to cellular DSBs, MRN activates global ATM phosphorylation of DDR substrates (Ciccia and Elledge, 2010). To determine if MRN activates a global DDR to viral genomes, we analyzed canonical ATM and DDR kinase substrates in lysates from infected SAECs. As a positive control, we used doxorubicin to induce cellular DNA breaks. As expected, cellular DNA damage triggers the global phosphorylation of DDR kinase substrates, including NBS1-Ser343, ATM-Ser1981, H2AX-Ser139, KAP1-Ser824, RPA32-Ser4/Ser8, and DNA-PKcs-Ser2056 (Figure 1D).
However, contrary to expectations, MRN does not activate the global phosphorylation of DDR kinase substrates in response to virus genomes in ΔE1B-55K/ΔE4-ORF3 infected cells (Figures 1D and S1B). Furthermore, in WT, ΔE4-ORF3 and ΔE1B-55K infected cells, where MRN is inactivated by E1B-55K and/or E4-ORF3, global DDR phosphorylated substrates are induced (Figure 1D). These data suggest that global DDR signaling is activated independently of MRN in WT, ΔE1B-55K and ΔE4-ORF3 virus infected cells.
To determine if there is a correlation between global DDR signaling and virus genome replication we harvested total viral and cellular DNA. Virus genomes were quantified using Q-PCR and normalized relative to cellular 18S rDNA. The levels of virus genomes in WT, ΔE1B-55K and ΔE4-ORF3 infected cells are 104 times higher than in ΔE1B-55K/ΔE4-ORF3 virus infected cells (Figure 1E). We conclude that E1B-55K or E4-ORF3 is required for viral genome replication.
The replication of viral genomes is coincident with the induction of global DDR phosphorylated substrates. In WT, ΔE4-ORF3, and ΔE1B-55K infected cells virus genomes are concentrated in specialized domains in the nucleus demarcated by the viral protein E2A (Figure 1F). The size and morphology of E2A viral replication domains change over the course of the viral life cycle (Boyer et al., 1957). In ΔE1B-55K/ΔE4-ORF3 virus infected cells, E2A is nuclear diffuse, indicating that viral genome replication is blocked at the earliest stages (Figure 1F).
In vitro, adenovirus DNA replication does not require E1B-55K and E4-ORF3 (Challberg and Kelly, 1979). This suggests that E1B-55K/E4-ORF3 inactivate a cellular target that prevents early virus DNA replication in vivo. A compelling overlapping candidate is MRN. Therefore, we determined if MRN knockdown rescues genome replication and activates global DDR phosphorylation in ΔE1B-55K/ΔE4-ORF3 virus infected cells.
The inactivation of the MRN complex is required for virus genome replication
The stable knockdown of MRN is lethal and primary SAECs are refractory to transient siRNA transfection. However, A549 cells are amenable to siRNA transfection. Analogous to SAECs, in WT, ΔE4-ORF3, ΔE1B-55K but not ΔE1B-55K/ΔE4-ORF3 infected A549 cells, global DDR kinase phosphorylated substrates are induced (Figure 2A). Adenovirus replication is accelerated by about 12 hours in A549 cells. However, similar to SAECs, the levels of virus genomes are 104 times higher in WT versus ΔE1B-55K/ΔE4-ORF3 infected A549 cells (Figure 2B). Therefore, for experiments that required siRNA transfection or large numbers of cells, we used A549 cells.
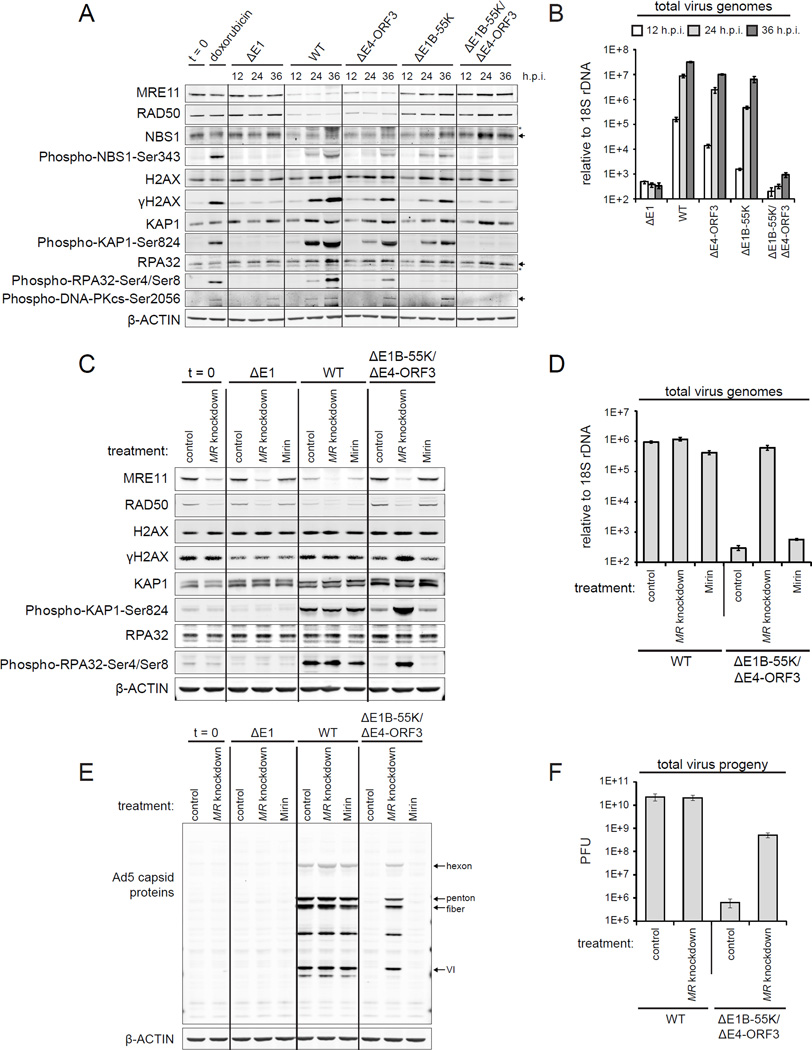
Figure 2. The inactivation of the MRN complex is critical for viral genome replication.
(A and B) A549 cells were infected as indicated and harvested at 12, 24, and 36 h.p.i. (A) Protein lysates were immunoblotted as indicated. (B) Viral genomes were quantified by Q-PCR; error bars indicate standard deviation.
(C–F) A549 cells were transfected with either control siRNA (control) or MRE11 and RAD50 siRNAs (MR knockdown). Cells were infected 48 hours post transfection as indicated. Mirin was added at 2 h.p.i. (Mirin) to control siRNA treated cells. Protein lysates and DNA were harvested at 24 h.p.i. (C and E) Protein lysates were immunoblotted as indicated. (D) Viral genomes were quantified by Q-PCR. (F) Total virus plaque forming units (PFU) were quantified at 48 h.p.i. Error bars indicate standard deviation of triplicates.
(See also Figure S2.)
We optimized two independent sets of siRNAs against MRE11 and RAD50 (MR knockdown) to knockdown the MRN complex (Figures S2A–C). We also used Mirin to inhibit MRE11 exonuclease activity (Dupre et al., 2008). In WT virus infected cells, neither MR knockdown nor Mirin prevents the global phosphorylation of DDR substrates (Figure 2C). Thus, the activation of global DDR signaling in WT virus infected cells is MRN independent and is not due to residual MRN expression or activity. MR knockdown and Mirin have no effect on DDR phosphorylated substrates in uninfected and AE1 infected cells (Figure 2C).
Strikingly, in ΔE1B-55K/ΔE4-ORF3 infected cells MR knockdown, but not Mirin, induces the global phosphorylation of DDR substrates (Figure 2C). To determine if the phosphorylation is a downstream consequence of rescuing viral genome replication, we quantified viral genomes. MR knockdown rescues ΔE1B-55K/ΔE4-ORF3 virus genome replication by over 1000-fold to WT virus levels (Figure 2D). The knockdown of individual MRN complex components is similar to the knockdown of MRE11 and RAD50 together (Figures S2D and S2E). In comparison, Mirin results in a nominal 1.9-fold rescue of ΔE1B-55K/ΔE4-ORF3 virus genome replication (Figure 2D). These data indicate that the MRN complex as opposed to MRE11 exonuclease activity prevents viral genome replication.
Adenovirus genome replication is required for the expression of late viral proteins that form the capsid (Thomas and Mathews, 1980). In WT virus infected cells, MR knockdown and Mirin do not further increase viral genome replication, late protein expression or virus titres (Figures 2D–F). However, MR knockdown results in a significant rescue of both capsid protein expression and virus titres in ΔE1B-55K/ΔE4-ORF3 infected cells (Figures 2E and 2F). A complete rescue is not expected as E1B-55K and E4-ORF3 have additional functions in viral replication, such as, p53 inactivation and late viral RNA export (O’Shea et al., 2004; Soria et al., 2010).
We conclude that the MRN complex prevents virus genome replication through a mechanism that does not activate global ATM signaling or require MRE11 exonuclease activity. We show that the inactivation of MRN enables virus genome replication and triggers downstream global DDR signaling.
The assembly of virus replication domains activates global ATM phosphorylation independently of MRN
Adenovirus E1A induces both cellular and viral DNA replication. E1A and oncogene induced replicative stress has been linked to DNA damage signaling (Halazonetis et al., 2008; Singhal et al., 2013). To determine if viral or cellular DNA replication activates global DDR signaling we exploited hydroxyurea (HU) and aphidicolin. At low concentrations, aphidicolin inhibits cellular but not virus DNA replication (Figures S3A–C). HU prevents both cellular and virus DNA replication (Figures S3A-C). We show that the inhibition of viral but not cellular DNA replication prevents ATM, H2AX, KAP1, RPA32 and DNA-PKcs phosphorylation in WT virus infected cells (Figure 3A). We conclude that global DDR signaling is activated downstream of viral DNA replication.
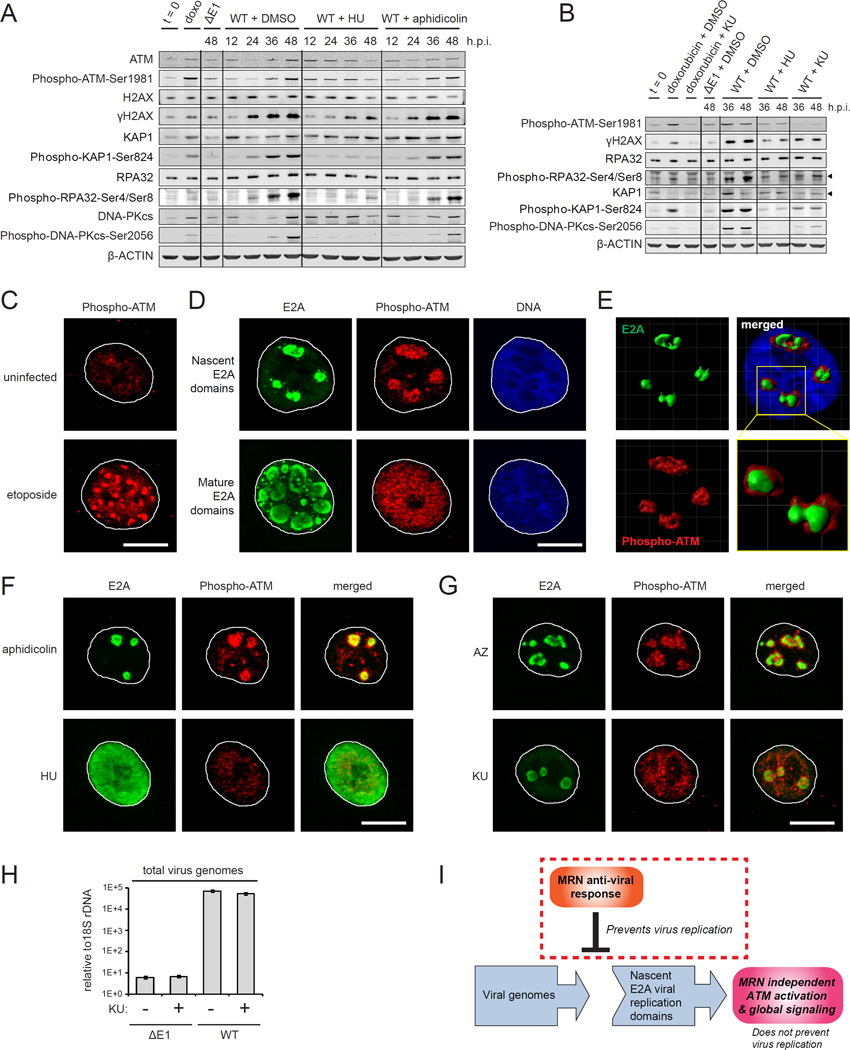
Figure 3. The assembly of viral genome domains activates global ATM phosphorylation independently of MRN.
(A) SAECs were infected as indicated and treated with DMSO, hydroxyurea (HU) or aphidicolin at 2 h.p.i. Protein lysates were immunoblotted as indicated.
(B) SAECs were infected as indicated and treated with DMSO, HU or the ATM kinase inhibitor KU-55933 (KU) at 2 h.p.i. Doxorubicin treatment was used as a positive control. Protein lysates were harvested and immunoblotted as indicated.
(C) SAECs were treated with DMSO or etoposide for 12 hours and stained for Phospho-ATM-Ser1981 (red). Scale bar: 10 μm.
(D–G) WT virus infected cells were treated with DMSO, HU, aphidicolin, KU or AZ20 (AZ) at 2 h.p.i. Cells were fixed at 18 h.p.i. and stained for E2A (green) and Phospho-ATM-Ser1981 (red). DNA was counterstained with Hoechst (blue). (D) Representative images of WT virus infected cells with nascent (upper) and more mature E2A domains (lower). (E) 3D rendering, merge and zoom of nascent E2A domains. (F) Aphidicolin and HU treated WT virus infected cells. (G) KU and AZ treated WT virus infected cells. Scale bar: 10 μm.
(H) SAECs were infected as indicated and treated with DMSO or KU at 2 h.p.i. Virus genomes were quantified by Q-PCR at 48 h.p.i. Error bars indicate standard deviation of triplicates.
(I) Model: In WT Ad5 infection E1B-55K/E4-ORF3 inactivate MRN and enable logarithmic viral genome replication. The assembly of viral genomes in nuclear domains activates global ATM phosphorylation but does not impact viral replication.
To determine if viral DNA replication activates ATM phosphorylation of global DDR substrates, we treated infected cells with the ATM kinase inhibitor KU-55933 (Hickson et al., 2004). Similar to HU, KU-55933 prevents the phosphorylation of ATM, H2AX, RPA32 and KAP1 in WT virus infected cells (Figure 3B). HU and KU-55933 also prevent the phosphorylation of DNA-PKcs (Figure 3B), indicating that DNA-PK is activated downstream of ATM in response to virus DNA replication. The ATR kinase inhibitor, AZ20 (Foote et al., 2013), prevents the induction of ATR and RAD17 phosphorylation, but not global ATM phosphorylated substrates, such as ATM, H2AX, KAP1, RPA32 and DNA-PKcs (Figure S3D).
We hypothesized that the assembly of virus genome domains within the nucleus (Figure 1F) triggers the MRN independent activation of ATM. ATM activation triggers ATM autophosphorylation at Ser1981 (Bakkenist and Kastan, 2003). Etoposide induces cellular DNA breaks and the phosphorylation of ATM at discrete DDR foci (Figure 3C). Strikingly, ATM phosphorylation is also induced at nascent E2A viral genome replication domains (Figure 3D). Phospho-ATM is concentrated towards the center of nascent viral genome domains as well as immediately surrounding cellular chromatin (Figure 3E and Movie S1). However, in contrast to cellular DSBs, phospho-ATM is not retained at mature E2A domains and diffuses throughout the nucleus to induce global DDR phosphorylated substrates (Figure 3D). A similar pattern is observed in WT virus infected A549 cells (data not shown).
HU but not aphidicolin prevents viral genome replication and the activation of ATM at E2A viral replication domains (Figure 3F). Furthermore, treatment with KU-55933, but not AZ20 or Mirin, prevents ATM phosphorylation at nascent E2A viral genome domains (Figures 3G and S3E). Taken together, we conclude that the assembly of nascent virus genome replication domains activates ATM through an MRN independent mechanism.
To determine if global ATM activation impacts virus replication we analyzed WT virus infected cells treated with KU-55933 or DMSO. KU-55933 has no impact on WT virus genome replication levels (Figure 3H). Furthermore, capsid protein expression and total virus titres are similar in WT virus infected cells treated with DMSO, KU-55933 or AZ20 (Figures S3F and S3G). ATM inhibition also does not impact the cytopathic effect associated with WT virus replication (data not shown). We conclude that the MRN independent activation of global ATM phosphorylation does not impact viral replication (Figure 3I) in cell culture. Therefore, we focused on the critical early MRN-dependent checkpoint that prevents viral genome replication.
MRN senses replicating virus genomes and activates a local ATM DDR that prevents viral DNA replication
In the cellular DDR, MRN is the initial sensor that binds to DSBs where it recruits and activates ATM (Figure 4A) (Ciccia and Elledge, 2010). To determine if MRN binds to adenovirus genomes, we performed MRE11 chromatin immunoprecipitations (ChIP) on A549 cells that had been infected with ΔE1, WT, ΔE4-ORF3, ΔE1B-55K, or ΔE1B-55K/ΔE4-ORF3 viruses. Viral genomes were quantified using primer sets that amplify the right (PS1) and left (PS2) ends of virus genomes.
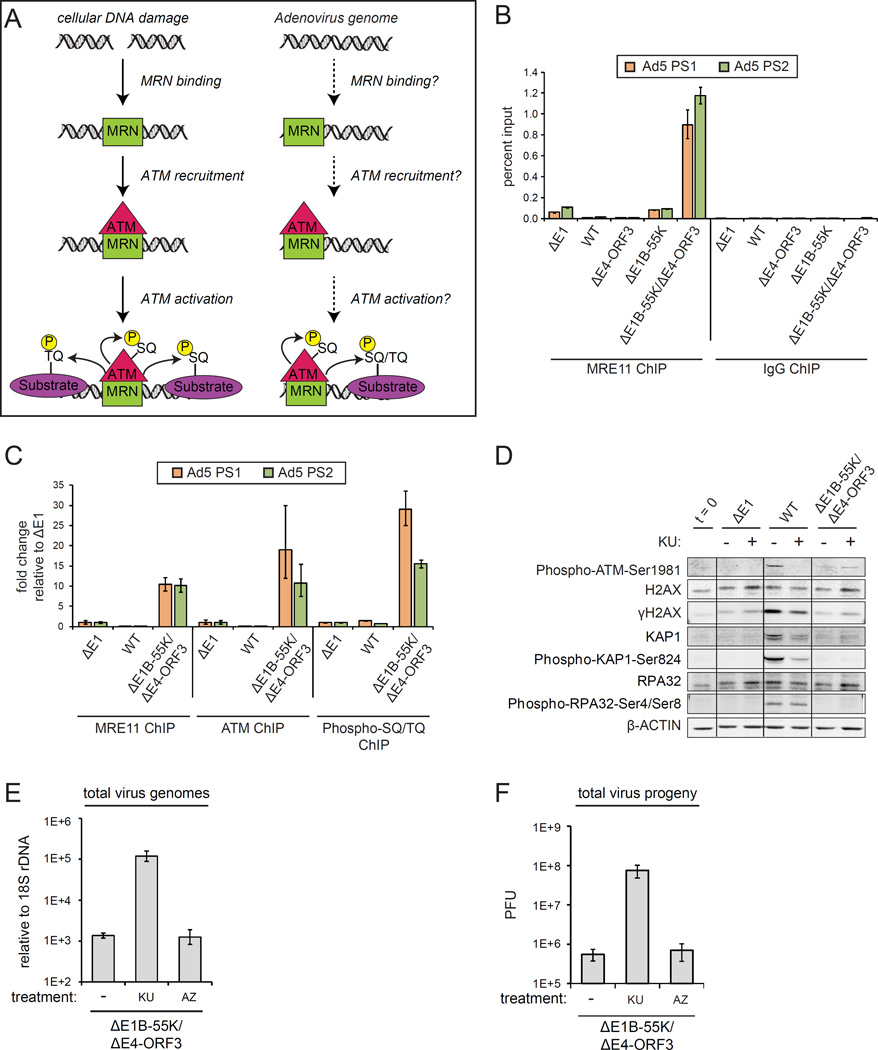
Figure 4. MRN senses replicating virus genomes and recruits ATM that activates a local DDR to prevent viral DNA replication.
(A) Cellular DSBs are sensed by MRN that activates ATM phosphorylation of DDR substrates at SQ/TQ motifs. (B) A549 cells were infected as indicated and harvested at 12 h.p.i. ChIP was performed using MRE11 antibodies and an IgG control. The left and right ends of the virus genome were quantified by Q-PCR (Ad5 PS1 and PS2). Error bars indicate the standard deviation of quadruplicates. (C) A549 cells were infected as indicated and harvested at 12 h.p.i. ChIP was performed using MRE11, ATM and Phospho-SQ/TQ substrate motif antibodies and plotted as fold enrichment relative to ΔE1 samples.
(D–F) A549 cells were infected as indicated and treated with DMSO (−), KU or AZ20 (AZ) at 2 h.p.i. (D) Protein lysates at 24 h.p.i. were immunoblotted as indicated. (E) Virus genomes at 24 h.p.i. were quantified by Q-PCR. (F) Total virus plaque forming units (PFU) at 48 h.p.i. Error bars indicate standard deviation of triplicates.
(See also Figure S4.)
MRE11 binds to viral genomes in ΔE1B-55K/ΔE4-ORF3 infected cells but not WT, ΔE4-ORF3 or ΔE1B-55K infected cells (Figure 4B). Thus, either MRN degradation by E1B-55K or mislocalization by E4-ORF3 is sufficient to prevent MRN binding to virus genomes (Figure 4B). Interestingly, MRE11 does not bind to ΔE1 virus genomes (Figure 4B). ΔE1 is a replication-incompetent vector (Figures 1E and S4A). In contrast to ΔE1, ΔE1B-55K/ΔE4-ORF3 viruses initiate limited replication as evidenced by a 2–3 fold increase in viral genomes over the timecourse of infection (Figures 1E and S4A). These data indicate that MRN senses early replicating viral genomes.
To determine if MRN recruits ATM to viral genomes we performed ChIP for MRE11 and ATM. We also performed ChIP for epitopes with the preferred ATM and ATR SQ/TQ phosphorylated substrate motif (Kim et al., 1999). ATM and SQ/TQ phosphorylated epitopes are enriched at viral genomes in ΔE1B-55K/ΔE4-ORF3 infected cells but not WT or ΔE1 infected cells (Figure 4C). We conclude that MRN recruits and activates ATM phosphorylated substrates at viral genomes.
Despite the phosphorylation of ATM substrates at viral genomes (Figure 4C), global ATM phosphorylated substrates are not induced in total lysates from ΔE1B-55K/ΔE4-ORF3 infected cells (Figures 2A, 4D, S4B and S4C). To determine if ATM activation at ΔE1B-55K/ΔE4-ORF3 viral genomes prevents replication, we inhibited ATM with KU-55933. In parallel, we also tested the AZ20 ATR kinase inhibitor since ATR and ATM phosphorylate substrates with similar SQ/TQ motifs. KU-55933, but not AZ20, rescues ΔE1B-55K/ΔE4-ORF3 viral genome replication and titres in A549 cells (Figures 4E and 4F). Similarly, in SAECs, KU-55933 and ATM shRNA rescue ΔE1B-55K/ΔE4-ORF3 viral replication (Figures S4C–E). We conclude that MRN activates a localized ATM DDR that restricts viral genome replication.
MRN-ATM activation at viral genomes is not amplified through γH2AX foci and global signaling
At cellular DSBs, ATM signaling is amplified by the phosphorylation of H2AX across megabases of chromatin (Polo and Jackson, 2011). MDC1 binds to γH2AX in a feed-forward loop that recruits additional MRN, DDR kinases and effectors into foci that facilitate global phosphorylation (Figure 5A). We hypothesized that ATM activation at adenovirus genomes is not amplified through H2AX phosphorylation and the assembly of DDR foci. As a positive control, we used etoposide, which induces cellular DNA damage and the assembly of NBS1, γH2AX, MDC1, and 53BP1 DDR foci, as expected (Figure 5B). However, in ΔE1B-55K/ΔE4-ORF3 infected cells, the activation of MRN-ATM at virus genomes does not induce the assembly of NBS1, γH2AX, MDC1, and 53BP1 in DDR foci (Figures 5B and S5). These data are consistent with the absence of global DDR signaling in ΔE1B-55K/ΔE4-ORF3 infected cells (Figure 1).
Figure 5. MRN-ATM activation at viral genomes is not amplified by H2AX to induce DDR foci and global signaling.
(A) At cellular genomes ATM activation is amplified by H2AX phosphorylation that recruits MDC1 and DDR proteins into nuclear foci. A key question is if small viral genomes meet the threshold for amplifying ATM activation through H2AX.
(B) A549 cells were treated as indicated, fixed at 12 h.p.i. and stained for E2A (green) and NBS1 (red), γH2AX (red), MDC1 (red) or 53BP1 (red). ΔE1 infected cells were identified by GFP expression. Scale bar: 10 μm.
(C) A549 cells were infected as indicated and harvested at 12 h.p.i. ChIP was performed using histone H3 antibodies or an IgG control. Virus genomes and cellular Alu sequences were quantified by Q-PCR.
(D) A549 cells were infected as indicated and harvested at 12 h.p.i. ChIP was performed using MRE11, γH2AX, MDC1 and IgG control antibodies. Virus genomes were quantified by Q-PCR.
(See also Figure S5.)
The adenovirus genome is only 36 kb, which by definition precludes H2AX phosphorylation across megabases of chromatin. Adenovirus DNA is compacted by protein VII in capsids but upon early gene transcription may associate with cellular histones (Komatsu and Nagata, 2012). To compare the levels of nucleosomes at cellular and viral genomes in infected cells, we performed ChIP for histone H3 at multi-copy cellular Alu sequences and adenovirus genome sequences. There is almost five times more H3 associated with cellular versus adenovirus genomes in ΔE1 and ΔE1B-55K/ΔE4-ORF3 infected cells (Figure 5C).
H2AX comprises approximately 10% of the H2A nucleosome complement in cellular chromatin (Fernandez-Capetillo et al., 2004). We hypothesized that the smaller genome size and lower nucleosome occupancy of adenovirus genomes is below the threshold for amplifying MRN-ATM activation through H2AX phosphorylation and MDC1 recruitment. To test this, we performed MRE11, γH2AX and MDC1 ChIPs. γH2AX is below the limits of detection at ΔE1B-55K/ΔE4-ORF3 viral genomes and at background IgG levels (0.053% input) (Figure 5D). MDC1 is enriched above background IgG levels (0.24% input); however, normalizing relative to ΔE1 samples, there is four fold more MRE11 than MDC1 at ΔE1B-55K/ΔE4-ORF3 virus genomes (Figure 5D). We conclude that MRN-ATM activation at viral genomes is not amplified through γH2AX to induce DDR foci and a global cellular DDR.
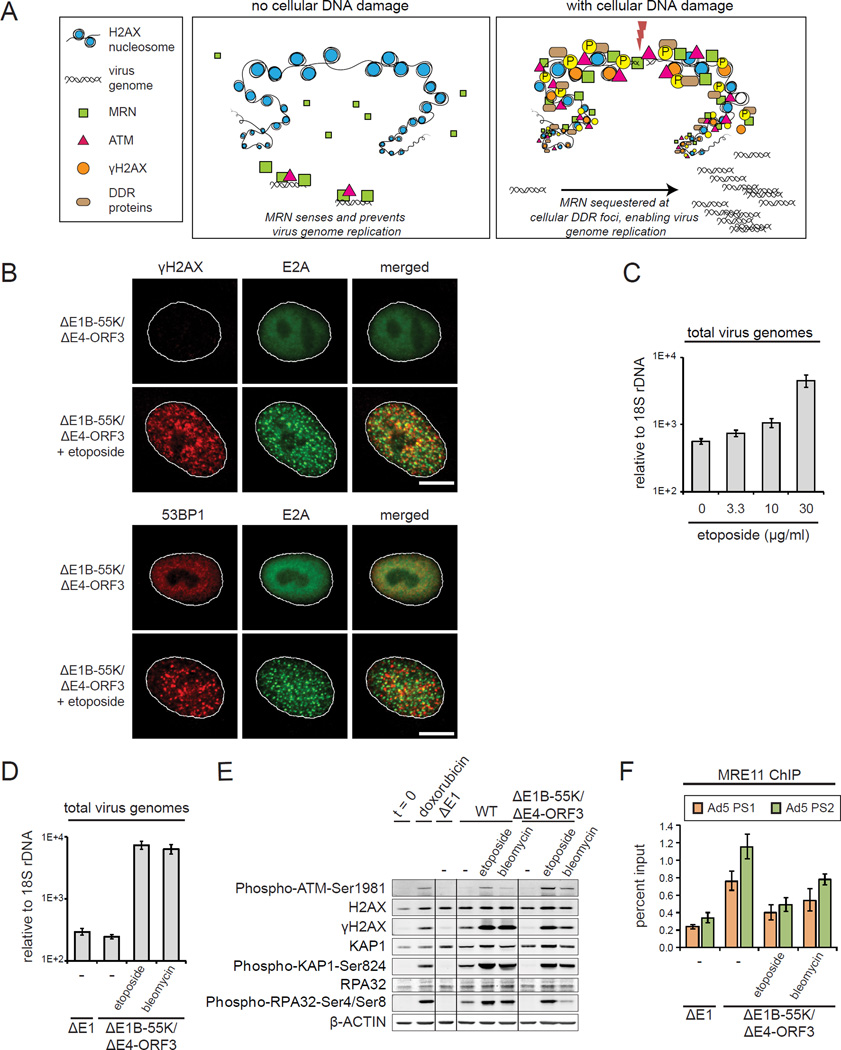
Cellular DNA damage prevents MRN binding and restriction of virus genome replication
The role of MRN in responding to both cellular DSBs and virus genomes has profound implications. We reasoned that the recruitment of MRN to cellular DDR foci could sequester MRN and prevent the cell sensing and restricting the replication of virus genomes (Figure 6A). To test this, we used etoposide to induce cellular DSBs in ΔE1B-55K/ΔE4-ORF3 infected cells. In contrast to viral genomes, cellular genome breaks trigger γH2AX and 53BP1 DDR foci in ΔE1B-55K/ΔE4-ORF3 infected cells (Figure 6B). Importantly, these data demonstrate that H2AX phosphorylation and the assembly of DDR foci are not suppressed by viral proteins or infection. Instead these data indicate that viral genomes do not support the amplification of MRN-ATM activation through γH2AX DDR foci. Thus, DDR foci discriminate viral and cellular genomes and determine if a local or global ATM response is more appropriate.
Figure 6. Cellular DNA damage prevents MRN sensing and restriction of viral genome replication.
(A) Model: Cellular DSBs compete for MRN binding and prevent MRN restriction of viral genome replication.
(B) A549 cells were infected with ΔE1B-55K/ΔE4-ORF3 and treated with DMSO or 10 μg/ml etoposide at 2 h.p.i. Cells were fixed at 12 h.p.i. and co-stained for E2A (green) and γH2AX or 53BP1 (red). Scale bar: 10 μm.
(C) A549 cells were infected with ΔE1B-55K/ΔE4-ORF3 and treated with different concentrations of etoposide at 6 h.p.i. and harvested at 24 h.p.i. Virus genomes were quantified by Q-PCR.
(D–E) A549 cells were infected as indicated and treated with DMSO (−), 30 μg/ml etoposide or 10 μM bleomycin at 6 h.p.i. Protein lysates and DNA were harvested at 24 h.p.i, (D) Virus genomes were quantified by Q-PCR. (E) Protein lysates were immunoblotted as indicated.
(F) A549 cells were infected and treated as indicated. Cells were harvested for MRE11 ChIP analysis at 12 h.p.i. Virus genomes were quantified by Q-PCR. Error bars indicate standard deviation of quadruplicates.
(See also Figure S6.)
Strikingly, etoposide induced cellular DNA damage rescues the assembly of E2A domains in ΔE1B-55K/ΔE4-ORF3 infected cells (Figure 6B). Consistent with this, using Q-PCR, we show that virus genome replication is rescued by etoposide in a dose dependent manner (Figures 6C and S6A). A similar rescue of virus genome replication is observed in ΔE1B-55K/ΔE4-ORF3 infected SAECs that tolerate higher concentrations of etoposide and doxorubicin (Figures S6B and S6C). In contrast, the induction of cellular DNA damage has no effect on the levels of WT virus genome replication (Figure S6D).
In addition to etoposide, we also used bleomycin, which induces cellular DNA damage through a distinct mechanism (Povirk, 1996). Both etoposide and bleomycin rescue ΔE1B-55K/ΔE4-ORF3 viral genome replication, despite the activation of global DDR kinase signaling (Figures 6D and 6E). Thus, localized but not global ATM activation prevents virus genome replication.
To determine if cellular DSBs compete for MRN binding, we performed MRE11 ChIP. We show that MRE11 recruitment to viral genomes is inhibited by the concomitant induction of cellular DNA damage in ΔE1B-55K/ΔE4-ORF3 infected cells treated with etoposide or bleomycin (Figure 6F). Similar conclusions were obtained with doxorubicin treatment (Figure S6E). These data demonstrate that the binding of MRN and activation of a localized ATM response prevents viral genome replication. Furthermore, the induction of cellular DNA breaks sequesters MRN and prevents the restriction of viral replication.
The localized MRN-ATM anti-viral DDR prevents viral but not cellular replication
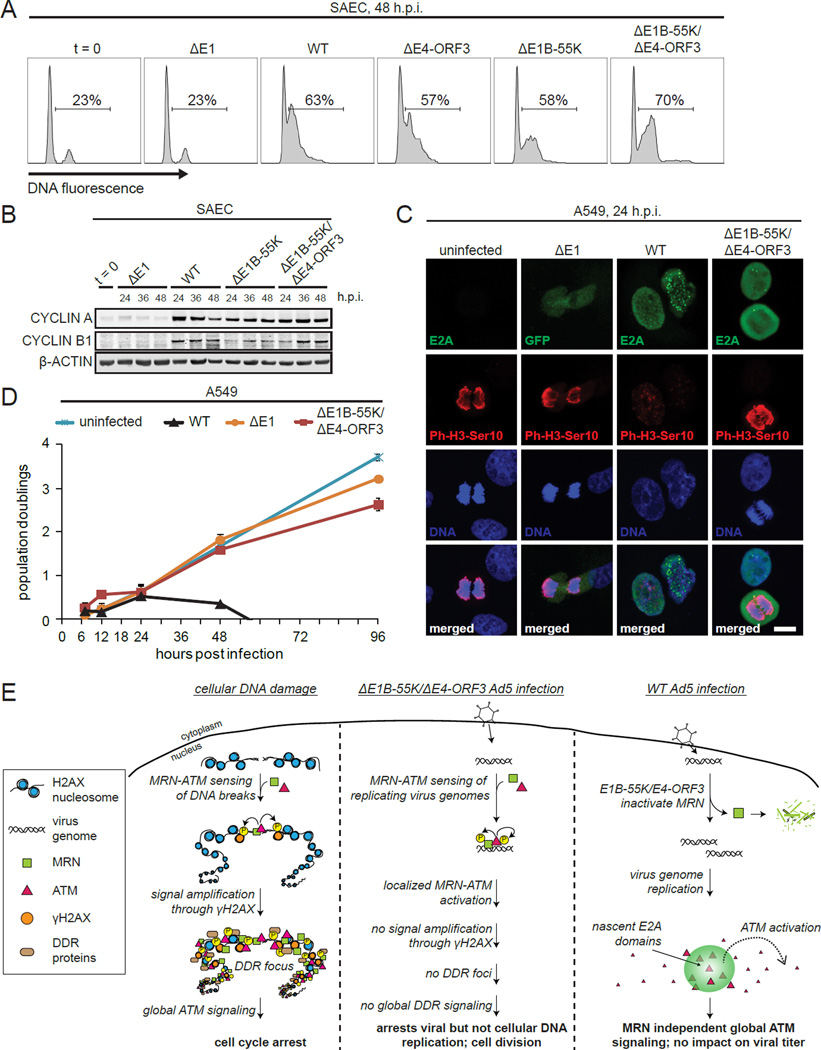
DDR foci play an important role in amplifying global DDR signaling, so that even a single DSB is sufficient to elicit cell cycle arrest (Polo and Jackson, 2011). To determine if the localized MRN-ATM anti-viral DDR specifically prevents viral but not cellular DNA replication, we analyzed S phase entry in quiescent SAECs that had been infected with ΔE1, WT, ΔE4-ORF3, ΔE1B-55K, and ΔE1B-55K/ΔE4-ORF3 viruses. In contrast to ΔE1 virus infected cells, WT, ΔE4-ORF3, ΔE1B-55K, and ΔE1B-55K/ΔE4-ORF3 virus infections all induce S phase entry to equivalent levels (Figure 7A). Consistent with this, CYCLIN A and CYCLIN B are also induced (Figure 7B).
Figure 7. The localized MRN-ATM anti-viral DDR specifically prevents viral but not cellular replication.
(A) SAECs were infected as indicated, fixed at 48 h.p.i., stained with propidium iodide and analyzed by flow cytometry. The % of cells with DNA content > 2N is indicated.
(B) SAECs were infected and immunoblotted as indicated.
(C) A549 cells were treated as indicated, fixed at 24 h.p.i. and stained for the mitotic marker, Phospho-H3-Ser10 (P-H3-Ser10) (red). Infected cells were identified by E2A staining or GFP (green). DNA was counterstained with Hoechst (blue). Scale bar: 10 μm.
(D) A549 cells were untreated or infected as indicated. Population doublings are plotted against h.p.i. Error bars indicate standard deviation of triplicates.
(E) Model: At cellular DSBs MRN-ATM activation is amplified by H2AX to induce global DDR signaling and arrest (left). At small viral genomes MRN activates a local ATM DDR that prevents viral but not cellular genome replication (middle). In WT Ad5 infection, MRN is inactivated by E1B-55K and E4-ORF3. The assembly of virus genomes in nuclear domains activates ATM independently of MRN. ATM phosphorylates global DDR substrates throughout the nucleus but does not impact virus replication.
We also examined if the localized MRN-ATM anti-viral response is uncoupled from mitotic arrest. We seeded A549 cells infected with ΔE1 or ΔE1B-55K/ΔE4-ORF3 viruses at subconfluent densities and stained for Phospho-H3-Ser10 and mitotic bodies. Despite the activation of MRN-ATM at virus genomes (Figures 2D, 4B and 4C) ΔE1B-55K/ΔE4-ORF3 infected cells induce Phospho-H3-Ser10 and form mitotic bodies (Figure 7C).
To determine if activation of the MRN-ATM anti-viral DDR induces a subsequent cell cycle arrest, we performed a population doubling analysis of infected cells over four days. WT virus undergoes productive lytic replication that kills infected cells after two days (Figure 7D). However, ΔE1B-55K/ΔE4-ORF3 infected cells double at similar rates to uninfected cells (Figure 7D). Thus, the localized MRN-ATM anti-viral DDR selectively prevents viral replication and maintains cellular proliferative potential.
Discussion
Our studies identify a critical localized MRN-ATM response that prevents viral replication and is inactivated by viral oncoproteins. In contrast to chromosomal DSBs, MRN-ATM activation at virus genomes is not amplified across megabases of chromatin by γH2AX to trigger the assembly of DDR foci, global signaling and cell cycle arrest. The localized MRN-ATM response provides an elegant mechanism to selectively neutralize viral replication without jeopardizing cellular replication and viability.
Our data provide the following model (Figure 7E). MRN senses and binds to early replicating virus genomes where it recruits ATM and activates a localized signaling response that selectively prevents virus genome replication. In WT Ad5 infected cells, E1B-55K and E4-ORF3 inactivate MRN and enable virus genome replication. The assembly of virus genomes in nascent domains within the nucleus triggers the MRN independent activation of ATM. ATM diffuses throughout the nucleus and induces global DDR phosphorylated substrates. However, in contrast to the localized MRN-ATM anti-viral DDR at viral genomes, global ATM phosphorylation does not impact viral replication.
The existence of two temporally distinct ATM DDRs to adenovirus genome replication reconciles the confounding observations of numerous studies. The majority of previous studies used E4 deleted viruses in cancer cell lines (Carson et al., 2009; Carson et al., 2003; Gautam and Bridge, 2013; Mathew and Bridge, 2007, 2008; Stracker et al., 2002). In contrast to ΔE1B-55K/ΔE4-ORF3 viruses, E4 deleted viruses express E1B-55K and induce the assembly of mature E2A virus genome replication domains. E1B-55K can bind to MRE11 in the absence of E4-ORF6 (Carson et al., 2003), which may be sufficient to inactivate the early MRN checkpoint to viral genome replication.
Our studies reveal adenovirus infection as a powerful system to identify MRN independent mechanisms that activate ATM. MRN is generally thought to be critical for ATM activation (Ciccia and Elledge, 2010). However, in WT virus infected cells, ATM is activated at viral replication domains despite MRN degradation by E1B-55K/E4-ORF6 and sequestration by E4-ORF3 (Figures 1A–C). Furthermore, neither MR siRNA knockdown nor Mirin prevents ATM activation in WT virus infected cells (Figure 2C). We show that ATM is activated and phosphorylated at nascent E2A viral genome replication domains (Figure 3D). Phospho-ATM is induced at the center of nascent viral domains as well as immediately surrounding cellular chromatin (Figure 3E and Movie S1). Previous studies have shown that the disruption of cellular chromatin can activate ATM (Bakkenist and Kastan, 2003). This raises the intriguing possibility that the assembly of virus genomes in nuclear domains is sensed through the disruption of surrounding cellular chromatin. In contrast to cellular DNA breaks (Figure 3C) (So et al., 2009), phospho-ATM is not retained at E2A domains (Figure 3D) and diffuses throughout the nucleus where it phosphorylates global cellular DDR substrates. In contrast to the localized MRN dependent activation of ATM at virus genomes, global ATM phosphorylation has no impact on viral genome replication, capsid protein expression or total virus titres (Figures 3H, S3F and S3G). However, global ATM signaling could play an important role in vivo in modulating the host immune response to virus infection (Brzostek-Racine et al., 2011; Gasser and Raulet, 2006; Mboko et al., 2012).
We show that either E1B-55K or E4-ORF3 is sufficient to prevent MRN binding to virus genomes (Figure 4B). The prevailing model is that the linear ends of adenovirus genomes resemble cellular DSBs and are targets for MRN binding (Stracker et al., 2002; Weitzman et al., 2010). However, TP/pTP is covalently attached to the 5’ ends of adenovirus genomes (Rekosh et al., 1977). We show that MRE11 binds to viral genomes in ΔE1B-55K/ΔE4-ORF3 but not ΔE1 virus infected cells (Figures 4B, 4C, 5D, 6F and S6E). In contrast to ΔE1 vectors, ΔE1B-55K/ΔE4-ORF3 viruses undergo limited genome replication (Figures 1E, 2B and S4A). The initial replication of adenovirus genomes is semi-conservative, similar to cellular DNA. These data suggest that MRN specifically senses early replicating as opposed to incoming viral genomes.
MRE11 nuclease activity plays a critical role in the response to cellular DSBs and DNA repair (Stracker and Petrini, 2011) and is inhibited by Mirin (Dupre et al., 2008). In contrast to the siRNA-mediated knockdown of MRN, Mirin has a nominal impact in rescuing ΔE1B-55K/ΔE4-ORF3 virus genome replication (Figures 2D and S2E). Thus, the binding of the MRN complex as opposed to MRE11 exonuclease activity protects the cell against viral replication. MRN recruits and activates ATM phosphorylated substrates at viral genomes (Figure 4C). ATM kinase inhibitors rescue ΔE1B-55K/ΔE4-ORF3 viral replication (Figures 4E and S4D) but to a lesser extent than MRN knockdown (Figures 2D and S2E). These data indicate that ATM has an important albeit subsidiary role to MRN binding in preventing viral genome replication. We favor the model that MRN binding near viral replication origins (located at both ends of the adenovirus genome) physically prevents the progression of viral DNA replication.
Our studies suggest a role for DDR foci and the logic of modifying vast tracts of chromatin flanking a cellular DSB (Polo and Jackson, 2011). There are universal and absolute differences between the sizes, chromatin composition and diffusion of viral and cellular genomes. The entire adenovirus genome is only 36 kb and associated with protein VII in viral capsids (Knipe and Howley, 2013). We show that there are less nucleosomes associated with viral versus cellular genomes in infected cells (Figure 5C). The latter could prevent chromatin compaction, which plays an important role in the assembly of DDR foci (Ayrapetov et al., 2014; Burgess et al., 2014; Khurana et al., 2014). H2AX is prepositioned not recruited to chromatin flanking cellular DSBs. γH2AX is also below the limits of detection at ΔE1B-55K/ΔE4-ORF3 viral genomes (Figure 5D). Furthermore, in contrast to cellular chromosomal breaks (Soutoglou et al., 2007), adenovirus genomes diffuse throughout the nucleus (Pombo et al., 1994), which may impair the assembly of stable DDR domains. Thus, viral genomes may not meet several threshold criteria for the assembly of DDR foci. An alternative explanation is that viral infection or proteins prevent the assembly of DDR foci and signaling. However, in contrast to MRN-ATM activation at viral genomes, cellular genome breaks trigger the assembly of γH2AX DDR foci and global signaling in ΔE1B-55K/ΔE4-ORF3 infected cells (Figures 5 and 6B). Thus, γH2AX DDR foci function as a diagnostic device to discriminate MRN-ATM activation at ‘self and ‘non-self genomes to determine if a localized ATM anti-viral DDR or global cellular DDR is more appropriate.
DDR foci amplify global DNA damage signaling such that even a single cellular DSB is sufficient to induce cell cycle arrest and repair (Bennett et al., 1993). However, the localized anti-viral DDR selectively prevents viral genome replication but not cellular replication and division (Figures 1 and 7). Adenovirus genomes do not integrate into cellular DNA and are lost upon nuclear membrane breakdown. The localized anti-viral DDR arrests viral genome replication while enabling cellular replication to potentially purge viral genomes from the nucleus.
The reliance of both the anti-viral and cellular DDR on MRN sensing has profound consequences and is susceptible to saturation. We show that the induction of cellular DNA damage by genotoxic drugs rescues ΔE1B-55K/ΔE4-ORF3 viral genome replication in a dose-dependent manner (Figure 6). Cellular DNA breaks compete with virus genomes for MRN binding and saturate MRN’s capacity to restrict viral genome replication (Figure 6). This has important implications for virus infections in pathological conditions where genotoxic stress is common, such as, cancer and aging (Jackson and Bartek, 2009). Telomere shortening is a hallmark of aging that could sequester MRN and render ‘old’ cells more permissive for viral replication (Suram and Herbig, 2014). The prevalence of virus infections, including adenovirus, is a serious and often fatal complication in cancer patients treated with chemotherapy, especially in the case of children (Hough et al., 2005; Lee et al., 2014; Steiner et al., 2008). DNA damage is one of the earliest hallmarks of cancer (Halazonetis et al., 2008) and could explain the permissiveness of many tumor cell-lines for ΔE1B-55K/ΔE4-ORF3 viral genome replication (G.S. and C.O., unpublished data). Thus, our studies provide key mechanistic insights that could enable the development of E1B-55K/E4-ORF3 mutant viruses as novel cancer therapies (O’Shea, 2005) that selectively replicate in precancerous lesions and tumor cells that have high levels of DNA damage. These viral agents could also be exciting and rational combination therapies with drugs that selectively agonize or antagonize DDR pathways dysregulated in cancer (Curtin, 2012).
Viruses are one of the most ancient and persistent threats to cellular genome integrity from single-cell to long-lived multicellular organisms. To date, studies of an anti-viral role for MRN and DDR signaling have been predicated on the well established response to cellular DSBs and hostage to its assays, namely, the induction of DDR foci and global signaling. Here we show that the critical MRN-ATM anti-viral DDR exhibits neither hallmark and is uncoupled from the cellular DDR. It will be interesting to determine if this is peculiar to adenovirus or extends to other DNA viruses and extrachromosomal DNA. Adenovirus is thought to be a descendant of phage and some relation of it has been with us for a long time (Hendrix, 1999). There are 68 human adenoviruses. However, the evolution of independent sets of viral proteins that inactivate MRN appears to be a recent evolutionary innovation of subgroup C viruses, such as Ad5 and 2 (Carson et al., 2009; Cheng et al., 2013; Forrester et al., 2011), which are among the most prevalent in the population. The ratio of virus particles with partial or defective genomes to infectious virus particles is approximately 50:1, even with highly purified laboratory preparations. If every incoming and defective viral genome activated global DDR signaling and cell cycle arrest or death, it would present a severe threat to cell growth and tissue homeostasis, especially during early development and wound healing. Our study suggests that the assembly of DDR foci distinguishes MRN-ATM activation at ‘self’ and ‘non-self’ genomes, which may be a critical adaptation in ensuring an appropriate and proportional response.
Experimental Procedures
Cell culture and viruses
Human small airway epithelial cells (SAECs) and A549 cells were cultured as previously described (Soria et al., 2010) and infected at a multiplicity of infection (MOI) of 10 plaque forming units (PFU). Titres of virus stocks and total virus production in infected cells were quantified in 293/E4/pIX cells (O’Shea et al., 2004). For a description of viruses used in these studies, see Extended Experimental Procedures.
Drugs
Unless otherwise stated, doxorubicin (Sigma) was used at 0.5 μg/ml, etoposide (Sigma) at 30 μg/ml, hydroxyurea (Sigma) at 2 mM, aphidicolin (Sigma) at 1 μM, KU-55933 (Calbiochem) at 10 μM, AZ20 (ApexBio) at 3 µM, bleomycin (Sigma) at 10 μM, and Mirin (Santa Cruz Biotechnology) at 20 µM.
siRNA
Stealth siRNAs (Life Technologies) were transfected using PepMute Plus (Signagen). Virus infection was performed 48 hours after transfection of siRNAs. See Extended Experimental Procedures.
Immunofluorescence
Cells were stained as described previously (O’Shea et al., 2004). Images were acquired with a Zeiss LSM780 imaging system. See Extended Experimental Procedures.
Immunoblot
Lysates from an equal number of cells were analyzed by SDS-PAGE and immunoblotting (Ou et al., 2012). Protein levels were quantified using a LI-COR-Odyssey scanner.
Quantification of viral genome replication
Viral genomes were quantified using Taqman probes (Johnson et al., 2002) and normalized relative to cellular 18S rDNA. See Extended Experimental Procedures.
Chromatin immunoprecipitation
ChIP was performed as described previously (Soria et al., 2010). Virus and cellular genome sequences were quantified by Q-PCR. See Extended Experimental Procedures.
Cell cycle analysis
SAECs were stained with propidium iodide/RnaseA and cellular DNA content was analyzed using flow cytometry (O’Shea et al., 2004).
Population doubling analysis
A549 cells were seeded at subconfluent densities and counted at the indicated time to calculate population doublings. See Extended Experimental Procedures.
Antibodies
See table in Extended Experimental Procedures.
Supplementary Material
Acknowledgements
We thank Tony Hunter, James Fitzpatrick, Mike Nunn, Nicola Allen and members of the O’Shea laboratory for helpful comments. This work was supported by R01CA137094 and P30CA014195 from NCI. C.C.O. is supported by the Marshall Legacy Foundation, The Leona M. and Harry B. Helmsley Charitable Trust grant #2012-PG-MED002, the William Scandling Trust and the Price Family Foundation. G.S. was supported by T32 GM007240 and the Chapman Charitable Trust. We thank Jamie Kasuboski and Carolyn O’Connor for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, Price BD. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:9169–9174. doi: 10.1073/pnas.1403565111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K, et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:8173–8178. doi: 10.1073/pnas.122228699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CB, Lewis AL, Baldwin KK, Resnick MA. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:5613–5617. doi: 10.1073/pnas.90.12.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackford AN, Bruton RK, Dirlik O, Stewart GS, Taylor AM, Dobner T, Grand RJ, Turnell AS. A role for E1B–AP5 in ATR signaling pathways during adenovirus infection. Journal of virology. 2008;82:7640–7652. doi: 10.1128/JVI.00170-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer GS, Leuchtenberger C, Ginsberg HS. Cytological and cytochemical studies of HeLa cells infected with adeno-viruses. The Journal of experimental medicine. 1957;105:195–216. doi: 10.1084/jem.105.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzostek-Racine S, Gordon C, Van Scoy S, Reich NC. The DNA damage response induces IFN. Journal of immunology. 2011;187:5336–5345. doi: 10.4049/jimmunol.1100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess RC, Burman B, Kruhlak MJ, Misteli T. Activation of DNA damage response signaling by condensed chromatin. Cell reports. 2014;9:1703–1717. doi: 10.1016/j.celrep.2014.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson CT, Orazio NI, Lee DV, Suh J, Bekker-Jensen S, Araujo FD, Lakdawala SS, Lilley CE, Bartek J, Lukas J, et al. Mislocalization of the MRN complex prevents ATR signaling during adenovirus infection. The EMBO journal. 2009;28:652–662. doi: 10.1038/emboj.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson CT, Schwartz RA, Stracker TH, Lilley CE, Lee DV, Weitzman MD. The Mre11 complex is required for ATM activation and the G2/M checkpoint. The EMBO journal. 2003;22:6610–6620. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challberg MD, Kelly TJ., Jr Adenovirus DNA replication in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:655–659. doi: 10.1073/pnas.76.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Gilson T, Wimmer P, Schreiner S, Ketner G, Dobner T, Branton PE, Blanchette P. Role of E1B55K in E4orf6/E1B55K E3 ligase complexes formed by different human adenovirus serotypes. Journal of virology. 2013;87:6232–6245. doi: 10.1128/JVI.00384-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver JE. gammaH2Ax: biomarker of damage or functional participant in DNA repair “all that glitters is not gold!”. Photochemistry and photobiology. 2011;87:1230–1239. doi: 10.1111/j.1751-1097.2011.00995.x. [DOI] [PubMed] [Google Scholar]
- Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nature reviews Cancer. 2012;12:801–817. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- Dupre A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee JH, Nicolette ML, Kopelovich L, Jasin M, Baer R, Paull TT, et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nature chemical biology. 2008;4:119–125. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Celeste A, Nussenzweig A. Focusing on foci: H2AX and the recruitment of DNA-damage response factors. Cell cycle. 2003;2:426–427. [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA repair. 2004;3:959–967. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- Foote KM, Blades K, Cronin A, Fillery S, Guichard SS, Hassall L, Hickson I, Jacq X, Jewsbury PJ, McGuire TM, et al. Discovery of 4-{4-[(3R)-3-Methylmorpholin-4-yl]-6-[1-(methylsulfonyl)cyclopropyl]pyrimidin-2-y l}-1H–indole (AZ20): a potent and selective inhibitor of ATR protein kinase with monotherapy in vivo antitumor activity. Journal of medicinal chemistry. 2013;56:2125–2138. doi: 10.1021/jm301859s. [DOI] [PubMed] [Google Scholar]
- Forrester NA, Sedgwick GG, Thomas A, Blackford AN, Speiseder T, Dobner T, Byrd PJ, Stewart GS, Turnell AS, Grand RJ. Serotype-specific inactivation of the cellular DNA damage response during adenovirus infection. Journal of virology. 2011;85:2201–2211. doi: 10.1128/JVI.01748-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser S, Raulet DH. The DNA damage response arouses the immune system. Cancer research. 2006;66:3959–3962. doi: 10.1158/0008-5472.CAN-05-4603. [DOI] [PubMed] [Google Scholar]
- Gautam D, Bridge E. The kinase activity of ataxia-telangiectasia mutated interferes with adenovirus E4 mutant DNA replication. Journal of virology. 2013;87:8687–8696. doi: 10.1128/JVI.00376-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Hendrix RW. Evolution: the long evolutionary reach of viruses. Current biology : CB. 1999;9:914–917. doi: 10.1016/s0960-9822(00)80103-7. [DOI] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer research. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- Hough R, Chetwood A, Sinfield R, Welch J, Vora A. Fatal adenovirus hepatitis during standard chemotherapy for childhood acute lymphoblastic leukemia. Journal of pediatric hematology/oncology. 2005;27:67–72. doi: 10.1097/01.mph.0000153958.95486.6f. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Shen A, Boyle L, Kunich J, Pandey K, Lemmon M, Hermiston T, Giedlin M, McCormick F, Fattaey A. Selectively replicating adenoviruses targeting deregulated E2F activity are potent, systemic antitumor agents. Cancer cell. 2002;1:325–337. doi: 10.1016/s1535-6108(02)00060-0. [DOI] [PubMed] [Google Scholar]
- Khurana S, Kruhlak MJ, Kim J, Tran AD, Liu J, Nyswaner K, Shi L, Jailwala P, Sung MH, Hakim O, et al. A macrohistone variant links dynamic chromatin compaction to BRCA1-dependent genome maintenance. Cell reports. 2014;8:1049–1062. doi: 10.1016/j.celrep.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. The Journal of biological chemistry. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- Knipe DM, Howley PM. Fields virology. 6th edn. Philadelphia, PA: Wolters Kluwer/Lippincott Williams & Wilkins Health; 2013. [Google Scholar]
- Komatsu T, Nagata K. Replication-uncoupled histone deposition during adenovirus DNA replication. Journal of virology. 2012;86:6701–6711. doi: 10.1128/JVI.00380-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Palomino-Guilen P, Babady NE, Lamson DM, St George K, Tang YW, Papanicolaou GA. Disseminated adenovirus infection in cancer patients presenting with focal pulmonary consolidation. Journal of clinical microbiology. 2014;52:350–353. doi: 10.1128/JCM.01893-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Molecular cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Luo G, Yao MS, Bender CF, Mills M, Bladl AR, Bradley A, Petrini JH. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:7376–7381. doi: 10.1073/pnas.96.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew SS, Bridge E. The cellular Mre11 protein interferes with adenovirus E4 mutant DNA replication. Virology. 2007;365:346–355. doi: 10.1016/j.virol.2007.03.049. [DOI] [PubMed] [Google Scholar]
- Mathew SS, Bridge E. Nbs1-dependent binding of Mre11 to adenovirus E4 mutant viral DNA is important for inhibiting DNA replication. Virology. 2008;374:11–22. doi: 10.1016/j.virol.2007.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mboko WP, Mounce BC, Wood BM, Kulinski JM, Corbett JA, Tarakanova VL. Coordinate regulation of DNA damage and type I interferon responses imposes an antiviral state that attenuates mouse gammaherpesvirus type 68 replication in primary macrophages. Journal of virology. 2012;86:6899–6912. doi: 10.1128/JVI.07119-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols GJ, Schaack J, Ornelles DA. Widespread phosphorylation of histone H2AX by species C adenovirus infection requires viral DNA replication. Journal of virology. 2009;83:5987–5998. doi: 10.1128/JVI.00091-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea CC. Viruses - seeking and destroying the tumor program. Oncogene. 2005;24:7640–7655. doi: 10.1038/sj.onc.1209047. [DOI] [PubMed] [Google Scholar]
- O’Shea CC, Johnson L, Bagus B, Choi S, Nicholas C, Shen A, Boyle L, Pandey K, Soria C, Kunich J, et al. Late viral RNA export, rather than p53 inactivation, determines ONYX-015 tumor selectivity. Cancer cell. 2004;6:611–623. doi: 10.1016/j.ccr.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Ou HD, Kwiatkowski W, Deerinck TJ, Noske A, Blain KY, Land HS, Soria C, Powers CJ, May AP, Shu X, et al. A structural basis for the assembly and functions of a viral polymer that inactivates multiple tumor suppressors. Cell. 2012;151:304–319. doi: 10.1016/j.cell.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes & development. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo A, Ferreira J, Bridge E, Carmo-Fonseca M. Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. The EMBO journal. 1994;13:5075–5085. doi: 10.1002/j.1460-2075.1994.tb06837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povirk LF. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin, neocarzinostatin and other enediynes. Mutation research. 1996;355:71–89. doi: 10.1016/0027-5107(96)00023-1. [DOI] [PubMed] [Google Scholar]
- Querido E, Blanchette P, Yan Q, Kamura T, Morrison M, Boivin D, Kaelin WG, Conaway RC, Conaway JW, Branton PE. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes & development. 2001;15:3104–3117. doi: 10.1101/gad.926401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekosh DM, Russell WC, Bellet AJ, Robinson AJ. Identification of a protein linked to the ends of adenovirus DNA. Cell. 1977;11:283–295. doi: 10.1016/0092-8674(77)90045-9. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. The Journal of cell biology. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal G, Leo E, Setty SK, Pommier Y, Thimmapaya B. Adenovirus E1A oncogene induces rereplication of cellular DNA and alters DNA replication dynamics. Journal of virology. 2013;87:8767–8778. doi: 10.1128/JVI.00879-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So S, Davis AJ, Chen DJ. Autophosphorylation at serine 1981 stabilizes ATM at DNA damage sites. The Journal of cell biology. 2009;187:977–990. doi: 10.1083/jcb.200906064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria C, Estermann FE, Espantman KC, O’Shea CC. Heterochromatin silencing of p53 target genes by a small viral protein. Nature. 2010;466:1076–1081. doi: 10.1038/nature09307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nature cell biology. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320:1507–1510. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner I, Aebi C, Ridolfi Luthy A, Wagner B, Leibundgut K. Fatal adenovirus hepatitis during maintenance therapy for childhood acute lymphoblastic leukemia. Pediatric blood & cancer. 2008;50:647–649. doi: 10.1002/pbc.21120. [DOI] [PubMed] [Google Scholar]
- Stracker TH, Carson CT, Weitzman MD. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature. 2002;418:348–352. doi: 10.1038/nature00863. [DOI] [PubMed] [Google Scholar]
- Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nature reviews Molecular cell biology. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suram A, Herbig U. The replicometer is broken: telomeres activate cellular senescence in response to genotoxic stresses. Aging cell. 2014;13:780–786. doi: 10.1111/acel.12246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GP, Mathews MB. DNA replication and the early to late transition in adenovirus infection. Cell. 1980;22:523–533. doi: 10.1016/0092-8674(80)90362-1. [DOI] [PubMed] [Google Scholar]
- Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. The EMBO journal. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzman MD, Lilley CE, Chaurushiya MS. Genomes in conflict: maintaining genome integrity during virus infection. Annual review of microbiology. 2010;64:61–81. doi: 10.1146/annurev.micro.112408.134016. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Weaver DT. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic acids research. 1997;25:2985–2991. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Petersen S, Tessarollo L, Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Current biology : CB. 2001;11:105–109. doi: 10.1016/s0960-9822(01)00019-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.